Abstract
Biomarker-driven cancer therapy has revolutionized precision oncology. With a better understanding of tumor biology, tissue-agnostic targets have been characterized and explored, which ultimately led to therapeutics with pan-cancer efficacy. To date, five molecular biomarkers have obtained FDA tissue-agnostic approval for targeted therapies and immunotherapies. Those include BRAFV600E mutations, RET fusions, NTRK fusions, high tumor mutation burden (TMB), and deficient mismatch repair/high microsatellite instability (dMMR/MSI-High). Herein, we have used data from AACR project GENIE to explore the clinico-genomic landscape of these alterations. AACR GENIE is a publicly accessible registry of genomic data from multiple collaborating cancer centers. Current database (version 13.0) includes sequencing data of 168,423 samples collected from patients with different cancers. We were able to identify BRAFV600E, RET fusions, NTRK fusions, and high TMB in 2.9%, 1.6%, 1.5%, and 15.2% of pan-cancer samples, respectively. In this article, we describe the distribution of those tissue-agnostic targets among different cancer types. In addition, we summarize the current prospect on the biology of these alterations and evidence on approved drugs, including pembrolizumab, dostarilmab, larotrectinib, entrectinib, selpercatinib, and dabrafenib/trametinib combination.
Introduction
For decades, cancer management has relied primarily on microscopic examination of tumor tissue with focus on understanding its exact pathology, including the extent of infiltration and the tissue of origin (1). Tumors have been, therefore, managed according to their location in the human body and management guidelines have made this distinction clear by being developed in a site-specific approach. Moreover, cancer research, cancer clinical trials, and anticancer drugs were all developed on the basis of histology and site. However, with the rapid evolution that happened in clinical genomic testing and contemporaneous advances in the development of precision cancer therapies, genomically targeted therapies and agents that arm the immune system have been introduced.
The list of targeted therapies, that act on specific molecular alterations, has exponentially grown in the past decade (2). Moreover, the introduction of immunotherapy has brought cancer therapeutics to the next level by enabling the patients’ body itself to fight cancer (3). That being said, beyond the era of site-specific cancer management, tissue-agnostic biomarkers across different cancer histologies have emerged. This was mainly driven by actionable alterations identified by next-generation sequencing (NGS) and the availability of medicines that can target these alterations. Although this appeared hypothetical, the past few years have brought some evidence on the efficacy of such biomarker-driven tissue-agnostic therapeutics; and regulatory approvals have driven the paradigm shift for these tissue-agnostic therapies. So far, six therapies have been approved by the FDA for tissue-agnostic indications based on molecular alterations (2, 4). More such tissue-agnostic therapies albeit in rare subsets of cancers are on the horizon with similar potential and the list is only expected to grow more in the future.
Herein, we review the current landscape of tissue-agnostic targets, distribution among different tumor types, and therapeutics that are being used to target. Moreover, we review data from 148,268 patients (168,423 samples) from the AACR project GENIE (version 13; ref. 5) to elaborate on the length and breadth of the clinical and molecular landscapes of those alterations.
Molecular Biomarkers and Targets in Tissue-Agnostic Approvals
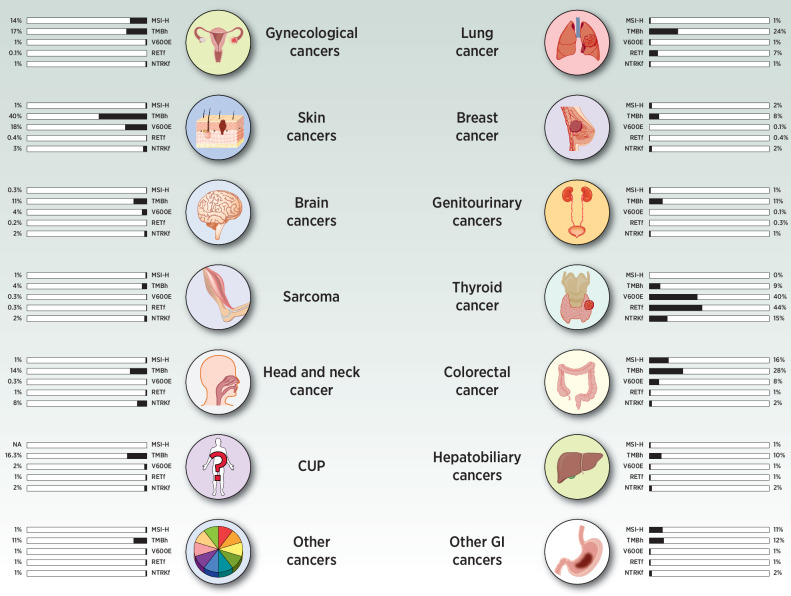
To date, five biomarkers have received regulatory approval for tissue-agnostic therapies (Table 1: (i) mismatch repair deficient (dMMR) or high microsatellite instability (MSI) cancers, (ii) tumor mutational burden-high (TMB-H) cancers (≥10 mutations per megabase; mut/Mb), (iii) NTRK fusion-positive cancers, (iv) RET fusion-positive cancers, and (v) BRAFV600E-mutant cancers other than colorectal cancers. In the AACR project GENIE, 4,912 samples harbored BRAFV600E mutations (2.9%), 410 harbored NTRK fusions (0.24% of all samples and 1.6% of samples profiled for structural variants), and 396 harbored RET fusions (0.26% of all samples and 1.5% of samples profiled for structural variants). In addition, 25,527 samples (15.2%) were classified as having high TMB. There are no data on dMMR or MSI in AACR GENIE although other studies have suggested a pan-cancer prevalence of around 3% (refs. 6, 7; Fig. 1).
Table 1.
Biomarkers currently with drugs that have FDA approval for a tissue agnostic indication.
| Biomarker/target | Approved drug(s) | Year of FDA approval |
|---|---|---|
| dMMR | Pembrolizumab | 2017 |
| Dostarlimab | 2022 | |
| MSI-H | Pembrolizumab | 2017 |
| TMB-High | Pembrolizumab | 2020 |
| BRAF V600E | Dabrafenib + Trametinib | 2022 |
| RET fusions | Selpercatinib | 2022 |
| NTRK fusions | Larotrectinib | 2018 |
| Entrectinib | 2019 |
Figure 1.
Frequency of tissue-agnostic targets across common malignancies. Data on BRAF V600E, RET fusions, NTRK fusions, and TMB are obtained from AACR GENIE database. Data on dMMR/MSI-H are obtained through literature search as they are not available in GENIE.
BRAF V600E Mutations
BRAF V600E is a mutation that commonly occurs in the MAPK pathway leading to unregulated cell growth and differentiation (8, 9). The V600E alteration is part of the BRAF class I mutations that are RAS-independent. Therefore, such mutation would lead to maintained active cell signaling via the MAPK pathway without upstream activation via RAS (10–12).
In AACR Project GENIE (version 13), we identified BRAFV600E mutations in 4,912 out of 167,387 profiled samples for BRAF mutations accounting for nearly 3% of the pan-cancer cohort. The most common cancers with BRAFV600E mutations were melanoma (n = 1,379; 28.1%), colorectal cancer (n = 1,228; 25%), thyroid cancer (n = 922; 18.8%), glioma (n = 392; 8%), non–small cell lung cancer (NSCLC; n = 329; 6.7%); among multiple other tumor types (13.5%). The most frequent cooccurring alterations were TERT and TP53 (Supplementary Table S1). Cancers with the highest frequency of BRAFV600E alterations were thyroid cancer (n = 922; 40.2%), melanoma (n = 1,379; 20.3%), histiocytosis (n = 91; 17.3%), colorectal cancer (n = 1,228; 7.9%), miscellaneous brain tumor (n = 16; 5.2%), glioma (n = 392; 3.9%), non-melanoma skin cancer (n = 46; 3.8%), gastrointestinal neuroendocrine tumors (n = 25; 3.7%), small bowel cancer (n = 12; 2.6%), and Wilms tumor (n = 4; 2.1%; Table 2). Cancers with a high frequency of BRAFV600E (≥1%) but a low number of profiled samples (<100) included primary central nervous system (CNS) melanocytic tumors (n = 3/7; 42.9%); melanocytoma (n = 4/13; 30.8%); head and neck cancer, not otherwise specified (NOS; n = 1/8; 12.5%); gastrointestinal neuroendocrine tumors of the esophagus/stomach (n = 1/11; 9.1%); angiomatoid fibrous histiocytoma (n = 1/14; 7.1%); parathyroid cancer (n = 2/29; 6.9%); tubular adenoma of the colon (n = 2/44; 4.5%); non-Hodgkin lymphoma (n = 1/62; 1.6%); and peritoneal cancer, NOS (n = 1/86; 1.2%).
Table 2.
Frequency of tissue-agnostic targets in different cancer types (alphabetically arranged) in AACR GENIE v13.
| Alteration frequency | |||||
|---|---|---|---|---|---|
| Cancer type | BRAF V600E | RET fusion | NTRK fusion | TMB-high (> 16) | TMB-high (≥ 10) |
| Ampullary cancer | 1.1% (n = 4) | 0% (n = 0) | 5.1% (n = 2) | 13% (n = 46) | 19.3% (n = 68) |
| Anal cancer | 0% (n = 0) | 2% (n = 1) | 2% (n = 1) | 12% (n = 44) | 19.9% (n = 73) |
| Appendiceal cancer | 1.2% (n = 9) | 0% (n = 0) | 2.1% (n = 1) | 14.6% (n = 108) | 18.5% (n = 137) |
| Bladder cancer | 0.1% (n = 6) | 0.3% (n = 3) | 1% (n = 9) | 20.3% (n = 947) | 38.8% (n = 1,813) |
| Breast cancer | 0.1% (n = 17) | 0.4% (n = 13) | 1.4% (n = 41) | 7.7% (n = 1,235) | 11.7% (n = 1,874) |
| Cancer of unknown primary | 1.6% (n = 86) | 1% (n = 8) | 1.5% (n = 12) | 15.2% (n = 815) | 23.4% (n = 1,249) |
| Cervical cancer | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) | 11.5% (n = 100) | 18.2% (n = 158) |
| Colorectal cancer | 7.9% (n = 1,228) | 0.9% (n = 15) | 1.7% (n = 29) | 26.8% (n = 4,145) | 31.9% (n = 4,937) |
| Endometrial cancer | 0.1% (n = 4) | 0.1% (n = 1) | 0.5% (n = 4) | 24.7% (n = 1,257) | 30.4% (n = 1,549) |
| Esophagogastric cancer | 0.1% (n = 6) | 0.7% (n = 8) | 1.8% (n = 20) | 11.4% (n = 541) | 17.2% (n = 817) |
| Gastrointestinal neuroendocrine tumor | 3.7% (n = 25) | 0% (n = 0) | 5.4% (n = 3) | 4% (n = 27) | 6.6% (n = 45) |
| Gastrointestinal stromal tumor | 0.4% (n = 6) | 0% (n = 0) | 0.8% (n = 1) | 14.4% (n = 222) | 14.6% (n = 226) |
| Germ cell tumor | 0.1% (n = 1) | 0% (n = 0) | 1.1% (n = 1) | 2.2% (n = 23) | 2.9% (n = 31) |
| Glioma | 3.9% (n = 392) | 0.3% (n = 6) | 1.8% (n = 43) | 9.8% (n = 988) | 11.1% (n = 1,121) |
| Head and neck cancer | 0.05% (n = 1) | 0.8% (n = 2) | 0.8% (n = 2) | 15.6% (n = 344) | 24.9% (n = 548) |
| Hepatobiliary cancer | 1.1% (n = 39) | 0.5% (n = 3) | 1.6% (n = 10) | 8.4% (n = 291) | 12% (n = 413) |
| Histiocytosis | 17.3% (n = 91) | 8% (n = 2) | 0% (n = 0) | 2.7% (n = 14) | 2.7% (n = 14) |
| Hodgkin lymphoma | 0% (n = 0)a | 0% (n = 0) | 0% (n = 0) | 4.8% (n = 6) | 7.9% (n = 10) |
| Leukemia | 0.2% (n = 9) | 0% (n = 0) | 0% (n = 0) | 22.8% (n = 1,351) | 28% (n = 1,660) |
| Melanoma | 20.3% (n = 1,379) | 0.1% (n = 1) | 2.6% (n = 18) | 39.8% (n = 2,709) | 49.1% (n = 3,338) |
| Mesothelioma | 0.1% (n = 1) | 0% (n = 0) | 1% (n = 2) | 2% (n = 19) | 2.8% (n = 27) |
| Non-Hodgkin lymphoma | 1.6% (n = 1) | 0% (n = 0) | 0% (n = 0) | 8.1% (n = 5) | 9.7% (n = 6) |
| NSCLC | 1.4% (n = 329) | 5.7% (n = 215) | 0.9% (n = 33) | 22.6% (n = 5,447) | 33.8% (n = 8,142) |
| Ovarian cancer | 0.9% (n = 56) | 0% (n = 0) | 0.6% (n = 7) | 10.3% (n = 630) | 12.8% (n = 783) |
| Pancreatic cancer | 0.4% (n = 26) | 0.1% (n = 1) | 1.8% (n = 15) | 10.3% (n = 706) | 11.9% (n = 820) |
| Parathyroid cancer | 6.9% (n = 2) | 0% (n = 0)a | 0% (n = 0)a | 44.8% (n = 13) | 44.8% (n = 13) |
| Penile cancer | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) | 17.5% (n = 11) | 25.4% (n = 16) |
| Prostate cancer | 0.02% (n = 1) | 0.2% (n = 4) | 0.4% (n = 8) | 4.2% (n = 239) | 5.4% (n = 312) |
| Renal cell carcinoma | 0% (n = 0) | 0.5% (n = 1) | 0% (n = 0) | 4.4% (n = 115) | 6.2% (n = 160) |
| Salivary gland cancer | 0.7% (n = 7) | 0.5% (n = 1) | 15.3% (n = 29) | 6.3% (n = 64) | 9.2% (n = 93) |
| Sex cord stromal tumor | 0% (n = 0)a | 0% (n = 0) | 0% (n = 0) | 3.8% (n = 9) | 4.6% (n = 11) |
| Skin cancer, nonmelanoma | 3.8% (n = 46) | 1.6% (n = 3) | 6.4% (n = 12) | 32.6% (n = 396) | 36.9% (n = 448) |
| Small bowel cancer | 2.6% (n = 12) | 0% (n = 0) | 0% (n = 0) | 26.7% (n = 124) | 35.3% (n = 164) |
| Small cell lung cancer | 0% (n = 0) | 0.7% (n = 1) | 0.7% (n = 1) | 16.3% (n = 151) | 35.8% (n = 332) |
| Soft tissue sarcoma | 0.3% (n = 16) | 0.3% (n = 4) | 2.3% (n = 34) | 4.1% (n = 202) | 5.8% (n = 287) |
| Thyroid cancer | 40.2% (n = 922) | 36% (n = 96) | 17.2% (n = 46) | 8.7% (n = 199) | 10.1% (n = 231) |
| Uterine sarcoma | 0.1% (n = 1) | 0.6% (n = 1) | 2.3% (n = 4) | 5.1% (n = 36) | 6% (n = 42) |
| Vaginal cancer | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) | 11.4% (n = 19) | 21.6% (n = 36) |
| Vulvar carcinoma | 0% (n = 0)a | 0% (n = 0)a | 0% (n = 0)a | 0% (n = 0) | 33.3% (n = 1) |
| Wilms tumor | 2.1% (n = 4) | 0% (n = 0) | 0% (n = 0) | 4.7% (n = 9) | 4.7% (n = 9) |
Note: Additional cancer types are presented in Supplementary Table S2.
aIt is possible that gene might have not been profiled for this alteration.
BRAF V600E can be targeted by using inhibitors of BRAF and MEK (12). On the basis of multiple studies showing dramatic efficacy in BRAFV600E-positive melanoma, three different BRAF/MEK inhibitor combinations have received FDA approval, including dabrafenib and trametinib. Further studies in NSCLC and anaplastic thyroid cancer led to the approval of dabrafenib + trametinib combination in these tumor types (2, 13–49). Dabrafenib plus trametinib is a BRAF inhibitor that when combined with the MEK inhibitor trametinib leads to synergistic blockade of the BRAF/MEK pathway and control of cellular-signaling that triggers aberrant cell proliferation (50–52). The exception for this activity is colorectal cancer due to an EGFR-dependent feedback mechanism that causes a paradoxical response to BRAF inhibitors and can be controlled by using EGFR inhibitors like cetuximab (53–55). Recently, dabrafenib and trametinib received United States FDA approval for BRAFV600E-positive solid tumors. This was based on pooled data from the ROAR study (BRF117019; NCT02034110), NCI-MATCH (NCT02465060), and CTMT212×2101 (NCT02124772; refs. 35–40), and was supported with prior analysis in melanoma and NSCLC studied as part of COMBI-d (NCT01584648), COMBI-v (NCT01597908), and BRF113928 (NCT01336634) trials (26–28, 31–34). In 131 adult and 36 pediatric patients, the objective response rate (ORR) was 41% and 25% in adult and pediatric patients; respectively (ref. 55; Fig. 2).
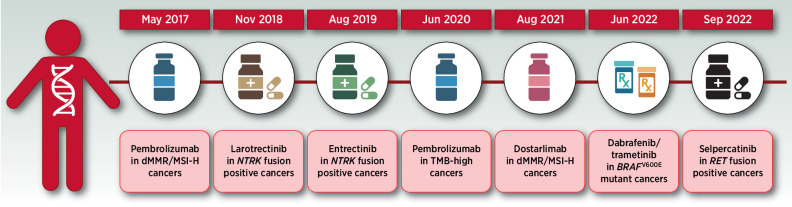
Figure 2.
Timeline of FDA tissue-agnostic approvals. (Adapted from an image created with BioRender.com.)
RET Fusions
Chromosomal rearrangements can lead to functional changes, which occur as part of oncogenesis (56–70). Fusions that occur in the RET gene, coding for a transmembrane tyrosine kinase with downstream activating action on multiple cellular pathways, were first reported in 1990 (56–70) in papillary thyroid carcinoma and demonstrated to associate with unregulated cellular-signaling via ligand-independent activation (56–70). Later, other RET fusions were detected, characterized, and shown to drive cancer development in multiple other tumor types (56–70).
In AACR Project GENIE (version 13), 396 RET fusion-positive samples were identified among 25,792 profiled samples for RET structural variants (1.5%). RET fusions were identified in patients with NSCLC (n = 215; 54.3%), thyroid cancer (n = 96; 24.2%), colorectal cancer (n = 15; 3.8%), breast cancer (n = 13; 3.3%), cancer of unknown primary (n = 8; 2%), esophagogastric cancer (n = 8; 2%), and multiple other tumor types (10.4%). Fusion partners that were most frequently observed were KIF5B, CCDC6, and NCOA4. The most frequent cooccurring alterations included TP53 and SETD2 mutations (Supplementary Table S1). In disease-specific analysis, cancers with the highest prevalence of RET fusions were thyroid cancer (n = 96; 36%), NSCLC (n = 215; 5.7%), anal cancer (n = 1; 2%), miscellaneous brain tumors (n = 1; 1.7%), non-melanoma skin cancer (n = 3; 1.6%), nerve sheath tumor (n = 1; 1.1%), cancer of unknown primary (n = 8; 1%), mature T and natural killer (NK) neoplasms (n = 1; 1%), colorectal cancer (n = 15; 0.9%), and head and neck cancer (n = 2; 0.8%; Table 2). Other tumors with a high frequency of RET fusions (≥1%) but a low number of profiled samples (<50) included pheochromocytoma (n = 1/2; 50%), histiocytosis (n = 2/25; 8%), and peripheral nervous system tumors (n = 1/49; 2%).
The ATP-dependent selective RET inhibitor, selpercatinib, is approved by FDA for solid tumors with RET fusions (71). Selpercatinib leads to the cessation of upstream RET-dependent activation which when aberrant leads to unbridled activation of multiple cellular-signaling pathways, including PI3K, RAS, and JAK (65–69, 72, 73). The FDA approval was based on data from the LIBRETTO-001 (NCT03157128) basket trial that evaluated the use of selpercatinib in multiple solid tumors. Responses were initially observed in NSCLC and thyroid cancer (ORR of 85% and 64%, 73% and 69%, for untreated and pretreated NSCLC and patients with thyroid cancer; respectively; refs. 74, 75). Further analysis in other tumor types revealed similar activity in different non-thyroid and non-lung cancers with an ORR of 43.9% which was the basis for FDA approval (ref. 4; Fig. 2).
NTRK Fusions
The first discovery of fusions involving NTRK genes was in 1998 in congenital fibrosarcoma (76). Multiple other tumors were shown to be driven by other fusions involving the NTRK family of genes; NTRK1, NTRK2, and NTRK3. Such structural abnormalities can lead to activation of downstream pathways independent on ligand-binding, and therefore can lead to uncontrolled cancerous proliferation (77).
In AACR Project GENIE (version 13), we were able to identify NTRK fusions (affecting either NTRK1, NTRK2, or NTRK3 genes) in 410 out of 25,792 profiled samples for NTRK structural variants (1.6%). NTRK fusions were primarily observed in thyroid cancer (n = 46; 11.2%), glioma (n = 43; 10.5%), breast cancer (n = 41; 10%), soft tissue sarcomas (n = 34; 8.3%), and NSCLC (n = 33; 8%), among other tumor types (52%). The most common fusion partners were ETV6, intragenic fusions, and TPM3. Again, the most frequent cooccurring mutations were detected in TP53 and TERT (Supplementary Table S1). The highest frequency of NTRK fusions was observed in thyroid cancer (n = 46; 17.2%), salivary gland cancer (n = 29; 15.3%), miscellaneous brain tumor (n = 6; 10.3%), nonmelanoma skin cancer (n = 12; 6.4%), gastrointestinal neuroendocrine tumor (n = 3; 5.4%), melanoma (n = 18; 2.6%), soft tissue sarcoma (n = 34; 2.3%), uterine sarcoma (n = 4; 2.3%), anal cancer (n = 1; 2%; Table 2), and B-lymphoblastic leukemia/lymphoma (n = 2; 2%). Other cancers with high frequency of NTRK fusions (≥1%) but low number of profiled samples (<50) included infantile fibrosarcoma (n = 3/3; 100%), ampullary cancer (n = 2/39; 5.1%), peripheral nervous system tumors (n = 2/49; 4.1%), miscellaneous neuroepithelial tumors (n = 1/41; 2.4%), and appendiceal cancer (n = 1/48; 2.1%)
NTRK fusions can be targeted using larotrectinib and entrectinib (78, 79). Both agents have received FDA approval for a tissue-agnostic indication in patients with solid tumors who harbor NTRK fusions (78, 79). The efficacy of larotrectinib in patients with NTRK gene fusions was evaluated in the context of three clinical trials: LOXO-TRK-14001 (NCT02122913), SCOUT (NCT02637687), and NAVIGATE (NCT02576431). In a pooled analysis of 55 patients, the ORR was 75%, and the median duration of response was 32.9 months (80). Entrectinib was approved on the basis of a pooled analysis of ALKA-372–001 (NCT02097810), STARTRK-1 (NCT02097810), and STARTRK-2 (NCT02568267) trials. The ORR was 59% in 54 evaluable patients with NTRK gene fusions (ref. 81; Fig. 2).
TMB-High
TMB reflects the number of genetic alterations in the genome of cancer cells. Its importance in cancer management comes from the hypothetical notion of higher odds of immune system recognition of cancerous cells that have a higher number of mutations (82). Practically, TMB can be calculated using data from NGS of either tissue or plasma and is reported as the number of mutations per megabase (mut/MB). Cutoffs used for defining TMB high and low cancers have been variable, but at least there is a consensus that cancers with 10 or more mutations per megabase would behave differently in terms of triggering immune response (82–84).
In AACR Project GENIE (version 13), nearly 15.2% (n = 25,527) of pan-cancer samples (n = 167,423) were classified as TMB-high tumors using the classification algorithm developed by GENIE. Data on calculated TMB were provided to investigators by AACR GENIE team upon request. Those included TMB calculated raw data as well as groups based on a classifier that was used in GENIE (low: <2, intermediate: 2–16, and high: >16 TMB subgroups). The most frequent diagnoses in TMB-high tumors were NSCLC (n = 5,447; 21.3%), colorectal cancer (n = 4,145; 16.2%), melanoma (n = 2,709; 10.6%), leukemia (n = 1,351; 5.3%), and endometrial cancer (n = 1,257; 4.9%), among others (41.6%). Tumors with highest frequency of TMB-high samples included melanoma (n = 2,709; 39.8%), non-melanoma skin cancer (n = 396; 32.6%), myelodysplastic/myeloproliferative neoplasms (n = 174; 28.9%), colorectal cancer (n = 4,145; 26.8%), small bowel cancer (n = 124; 26.7%), endometrial cancer (n = 1,257; 24.7%), leukemia (n = 1,351; 22.8%), NSCLC (n = 5,447; 22.6%), bladder cancer (n = 947; 20.3%), and myeloproliferative syndromes (n = 419; 19.5%). Cancers with a high frequency of TMB high (≥15%) but a low number of profiled samples (<100) included bowel cancer, NOS (n = 2/3; 66.7%), lung cancer, NOS (n = 15/27; 55.6%), parathyroid cancer (n = 13/29; 44.8%), thyroid cancer, NOS (n = 3/9; 33.3%), vulvar/vaginal cancer, NOS (n = 1/3; 33.3%), uterine cancer, NOS (n = 1/3; 33.3%), adenocarcinoma in situ (n = 1/4; 25%), penile cancer (n = 11/63; 17.5%), and bladder/urinary tract cancer, NOS (n = 1/6; 16.7%; Table 2). RET fusions, NTRK fusions, and BRAFV600E mutations were simultaneously identified in 15 (0.06%), 37 (0.1%), and 1440 (5.6%) samples with high TMB.
Because TMB data from AACR GENIE bins only tumors with TMB > 16 as high TMB cancers, we have also performed an exploratory analysis to reclassify tumors using 10 mut/mb as cutoff, which is considered the cutoff used for defining the tissue-agnostic indication of pembrolizumab by FDA. Using that classification, 34,874 samples (20.3% of pan-cancer samples) had ≥ 10 mut/mb and were classified as having high TMB. Cancers with highest frequency of TMB high samples using that classification included melanoma (n = 3,338; 49.1%), bladder cancer (n = 1,813; 38.8%), non-melanoma skin cancer (n = 448; 36.9%), small cell lung cancer (n = 332; 35.8%), small bowel cancer (n = 164; 35.3%), NSCLC (n = 8,142; 33.8%), myelodysplastic/myeloproliferative neoplasms (n = 203; 33.7%), colorectal cancer (n = 4,937; 31.9%), endometrial cancer (n = 1,549; 30.4%), and leukemia (n = 1,660; 28%; Table 2).
The FDA approval of pembrolizumab, an antibody blocker of PD-1, for solid tumors with high TMB was based on retrospective analysis of 102 patients with high TMB (≥10 mut/mb) advanced solid tumors in the KEYNOTE-158 trial (NCT02628067). The ORR was 29% with a median duration of response that was not reached at the time of data cutoff. In a subgroup analysis of patients with ≥ 13 mut/Mb (n = 70), the ORR increased to 37% with a median duration of response that is not reached (ref. 85; Fig. 2).
dMMR/MSI-H
Mismatch repair genes are responsible for fixing DNA damage that results during DNA replication. Therefore, deficiency leads to loss of genomic integrity and accumulation of mutations, including regions of repetitive DNA sequences also known as microsatellites. Cancers with dMMR/MSI-H generally confer a poor prognosis due to the hypermutable status (7).
The AACR project GENIE does not currently contain data on dMMR/MSI-H tumors. We have performed a review of literature to expand on landscape of MSI-H in different tumor types, which varied substantially between different studies. Bonneville and colleagues (7) analyzed whole exome data from 11,139 tumor-normal paired samples from 11,080 patients with 39 cancer types. They were able to detect MSI in 27 of 39 types of cancer with a variable disease-specific distribution. The highest frequency was observed in endometrial carcinoma (n = 170; 31.37%), colon adenocarcinoma (n = 85; 19.72%), stomach adenocarcinoma (n = 84; 19.09%), rectal adenocarcinoma (n = 9; 5.73%), adrenocortical carcinoma (n = 4; 4.35%), uterine carcinosarcoma (n = 2; 3.51%), cervical cancer (n = 8; 2.62%), Wilms tumor (n = 1; 2.44%), mesothelioma (n = 2; 2.41%), and esophageal carcinoma (n = 3; 1.63%; ref. 7). Another meta-analysis of published studies included data from 28,213 patients, where MSI-H was estimated to be prevalent in nearly 2.7%–2.9% of samples in the pooled analysis. Highest prevalence was reported in endometrial cancer (21.9%), small bowel cancer (14.3%), colon cancer (13%), colorectal cancer (10.2%), and gastric cancer (8.5%; ref. 6).
Immunotherapies are potential options for patients with dMMR/MSI-H tumors. Currently, the FDA approved the PD-1 blockers pembrolizumab and dostarlimab for dMMR/MSI-H tumors regardless of tissue of origin (86, 87). Data leading to FDA tissue-agnostic indication of pembrolizumab in 2017 came from five clinical trials: KEYNOTE-016 (NCT01876511), KEYNOTE-164 (NCT02460198), KEYNOTE-012 (NCT01848834), KEYNOTE-028 (NCT02054806), and KEYNOTE-158 (NCT02628067), which assessed the use of pembrolizumab in dMMR or MSI-H tumors. In a pooled analysis of 149 patients, the ORR was 39.6%, and the median duration of response was not reached at data cutoff (88). In 2021, dostarlimab was also approved for solid tumors with dMMR on the basis of data from the GARNET trial (NCT02715284) that evaluated the drug in 209 patients and showed an ORR of 41.6% and a median duration of response of 34.7 months (ref. 89; Fig. 2). More recently, there became interest in the use of PD-1 blockade in the neoadjuvant setting. In a phase II study where dostarlimab was used in patients with dMMR/MSI-H locally advanced rectal cancer, all patients treated (n = 12) had clinical complete response and no evidence of disease on PET/CT, MRI, digital rectal examination, or biopsy (90). While the study was including only patients with a rectal cancer, such outstanding result aligning with prior data that led to tissue-agnostic approvals can open the door for lots of future possibilities; including the possibility of moving tissue-agnostic treatments to neoadjuvant setting.
Special Situations: Carcinoma of Unknown Primary
One interesting finding we observed was the presence of tissue-agnostic biomarkers in a substantial portion of patients with carcinoma of unknown primary (1.6%, 1%, 1.5%, 15.2% for BRAFV600E mutations, RET fusions, NTRK fusions, and TMB high samples, respectively). With targetable alterations in those patients, the chances of improved outcomes or even cure are rising in a category of patients that long suffered from a historically poor prognosis due to the inability to determine the tissue of origin (91). In an era of changing landscape of tumor identity from microscopic tissue–based diagnosis to genomic alteration–based diagnosis, carcinoma of unknown primary emerges as the perfect prototype to prove the hypothesis given its historical resistance to conventional treatment options. In fact, newer technologies, including liquid biopsy, have made genetic testing a practical and feasible option in those patients that can drive the paradigm change led by molecularly-targeted therapeutics (92).
Special Situations: Rare Tumors
The beauty of tissue-agnostic precision oncology is its ability to treat cancers with low incidence rates that have been less frequently studied or profiled (93–95). Interestingly, our analysis of AACR GENIE revealed the presence of tissue-agnostic targets in a wide variety of cancers with low incidence and prevalence (Table 2; Supplementary Table S2). This was evident through the analysis that we performed for tumors with a low number of profiled samples that showed the strong presence of those biomarkers as aforementioned. Although data will remain statistically inconclusive given the limitation of a small sample size that is inappropriate for generalization, this can at least provide a proof of concept on the importance of molecular profiling in those patients and that it can lead to expanded treatment options beyond current standards of care.
Ongoing Promising Targets and Biomarkers
There are many emerging targets that have the potential in the future as tissue-agnostic biomarkers given promising data from early clinical studies in different tumor types and ongoing basket trials. For example, there is compelling clinical evidence supporting the use of zenocutuzumab in patients with NRG1 fusion–positive solid tumors, repotrectinib in patients with NTRK1/2/3 fusion–positive solid tumors, and PC14586 in patients with TP53 Y220C mutations (2). Other targets of potential interest that are being tested in clinical trials with promising results include HER2/neu and FGFR2 fusions (4, 96). Moreover, there is at least biological evidence to support hypothesis in favor of multiple other targets which are currently being explored in early clinical contexts (2). Whether any of these agents will succeed in proving a tissue-agnostic efficacy, either alone or in combination, will remain an area of controversy. Generally speaking, the histology-agnostic arsenal has suffered in the past from many negative results that limited expansion beyond single-site indications (97). One important challenge is the inherent molecular resistance mechanisms that can be present locally in some tumor types. However, with a better understanding of tumor biology, combination with resistance-targeting agents or the development of second-generation therapeutics that can overcome those resistance mechanisms can hopefully lead to more tissue-agnostic approvals. Therefore, cancer drug developers should balance biological, statistical, and regulatory issues that characterize the era of molecularly driven indications, which can fast-track such change in the years to come.
Limitations
In this article, we have reported data on the clinical landscape of tissue-agnostic targets using the AACR GENIE database. GENIE contains sequencing data from multiple contributing sites that allowed a wide scale analysis of large number of samples even win tumors with low frequency in individual sites. However, clinical data in AACR GENIE are limited especially in regard to prior treatments. Therefore, we were not able to explore possible differences between targeted therapy-naïve and pretreated populations. It is possible that some alterations could have emerged as a resistance mechanism to targeted therapeutics.
Conclusions
Current tissue-agnostic biomarkers are prevalent across multiple tumors. Drugs that target those alterations offer a broad spectrum of anticancer drug activity potentially changing the paradigm of cancer management from site-focused to genetic-focused care. Further analysis of cooccurring alterations and their impact on response to targeted therapies is warranted, in order to tailor precision treatments for patients harboring these alterations.
Supplementary Material
Genes most frequently mutated in cases with tissue-agnostic targets in the overall altered population and top 5 cancers with highest number of profiled samples in each alteration
Frequency of tissue-agnostic targets in other additional different tumors (alphabetically arranged) in AACR GENIE v.13
Acknowledgments
V. Subbiah is an Andrew Sabin Family Foundation Fellow at The University of Texas MD Anderson Cancer Center. V. Subbiah acknowledges support of The Jacquelyn A. Brady Fund and is supported by NIH grants (R01CA242845 and R01CA273168). MD Anderson Cancer Center Department of Investigational Cancer Therapeutics is supported by the Cancer Prevention and Research Institute of Texas (RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (1U01 CA180964), NCATS (Center for Clinical and Translational Sciences) grant (no. UL1 TR000371), and the MD Anderson Cancer Center Support Grant (P30 CA016672).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
V. Subbiah reports research funding from Novartis, Eli Lilly/Loxo Oncology, and Blueprint Medicines to conduct clinical trials; other grant support for clinical trials from AbbVie, Agensys, Inc, Alfasigma, Altum, Amgen, Bayer, BERG Health, Blueprint Medicines Corporation, Boston Biomedical, Inc, Boston Pharmaceuticals, Celgene Corporation, D3 Bio, Inc, Dragonfly Therapeutics, Inc, Exelixis, Fujifilm, GlaxoSmithKline, Idera Pharmaceuticals, Inc, Incyte Corporation, Inhibrx, Loxo Oncology/Eli Lilly, MedImmune, MultiVir, Inc, NanoCarrier Co, National Comprehensive Cancer Network, NCI-CTEP, Novartis, PharmaMar, Pfizer, Relay Therapeutics, Roche/Genentech, Takeda, Turning Point Therapeutics, UT MD Anderson Cancer Center, and Vegenics Pty Ltd; travel support from ASCO, ESMO, Helsinn Healthcare, Incyte Corporation, Novartis, and PharmaMar; consultancy or advisory board participation for Helsinn Healthcare, Incyte Corporation, Loxo Oncology/Eli Lilly, MedImmune, Novartis, QED Therapeutics, Relay Therapeutics, Daiichi-Sankyo, and R-Pharm U.S.; and other relationship with Medscape. No disclosures were reported by the other authors.
References
- 1. Russell WO. The pathologic diagnosis of cancer–a crescendo of importance in current and future therapies. Ward burdick award lecture. Am J Clin Pathol 1980;73:3–11. [DOI] [PubMed] [Google Scholar]
- 2. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Decker WK, da Silva RF, Sanabria MH, Angelo LS, Guimaraes F, Burt BM, et al. Cancer immunotherapy: historical perspective of a clinical revolution and emerging preclinical animal models. Front Immunol 2017;8:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Subbiah V, Wolf PJ, Konda B, Spira HKA, Weiss J, Takeda M, et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol 2022;23:1261–73. [DOI] [PubMed] [Google Scholar]
- 5. AACR project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang YJ, O'Haire S, Franchini F, IJzerman M, Zalcberg J, Macrae F, et al. A scoping review and meta-analysis on the prevalence of pan-tumour biomarkers (dMMR, MSI, high TMB) in different solid tumours. Sci Rep 2022;12:20495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 2017;2017:PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol 2004;5:875–85. [DOI] [PubMed] [Google Scholar]
- 9. Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 2001;22:153–83. [DOI] [PubMed] [Google Scholar]
- 10. Dankner M, Rose AAN, Rajkumar S, Siegel PM, Watson IR. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183–99. [DOI] [PubMed] [Google Scholar]
- 11. Yaeger R, Corcoran RB. Targeting alterations in the RAF-MEK pathway. Cancer Discov 2019;9:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gouda M, Subbiah V. Precision oncology for BRAF mutant cancers with BRAF and MEK inhibitors: from melanoma to tissue-agnostic therapy. ESMO Open 2023;8:100788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. New Engl J Med 2011;364:2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chapman PB, Robert C, Larkin J, Haanen JB, Ribas A, Hogg D, et al. Vemurafenib in patients with BRAFV600 mutation-positive metastatic melanoma: final overall survival results of the randomized BRIM-3 study. Ann Oncol 2017;28:2581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol 2014;15:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. New Engl J Med 2012;366:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McArthur GA, Maio M, Arance A, Nathan P, Blank C, Avril MF, et al. Vemurafenib in metastatic melanoma patients with brain metastases: an open-label, single-arm, phase 2, multicentre study. Ann Oncol 2017;28:634–41. [DOI] [PubMed] [Google Scholar]
- 18. Subbiah V, Puzanov I, Blay JY, Chau I, Lockhart AC, Raje NS, et al. Pan-cancer efficacy of vemurafenib in BRAF (V600)-mutant non-melanoma cancers. Cancer Discov 2020;10:657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diamond EL, Subbiah V, Lockhart AC, Blay JY, Puzanov I, Chau I, et al. Vemurafenib for BRAF V600-mutant erdheim-chester disease and langerhans cell histiocytosis: analysis of data from the histology-independent, Phase 2, open-label VE-BASKET study. JAMA Oncol 2018;4:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. New Engl J Med 2015;373:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012;380:358–65. [DOI] [PubMed] [Google Scholar]
- 22. Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. An update on BREAK-3, a Phase III, randomized trial: dabrafenib (DAB) versus dacarbazine (DTIC) in patients with BRAF V600E-positive mutation metastatic melanoma (MM). J Clin Oncol 31:15s, 2013. (suppl; abstr 9013). [Google Scholar]
- 23. Long GV, Trefzer U, Davies MA, Kefford RF, Ascierto PA, Chapman PB, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain (BREAK-MB): a multicentre, open-label, Phase 2 trial. Lancet Oncol 2012;13:1087–95. [DOI] [PubMed] [Google Scholar]
- 24. Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. New Engl J Med 2012;367:107–14. [DOI] [PubMed] [Google Scholar]
- 25. Robert C, Flaherty K, Nathan P, Hersey P, Garbe C, Milhem M, et al. Five-year outcomes from a phase 3 METRIC study in patients with BRAF V600 E/K-mutant advanced or metastatic melanoma. Eur J Cancer 2019;109:61–9. [DOI] [PubMed] [Google Scholar]
- 26. Long GV, Flaherty KT, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, et al. Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: long-term survival and safety analysis of a Phase 3 study. Ann Oncol 2017;28:1631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. New Engl J Med 2014;371:1877–88. [DOI] [PubMed] [Google Scholar]
- 28. Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. New Engl J Med 2015;372:30–9. [DOI] [PubMed] [Google Scholar]
- 29. Davies MA, Saiag P, Robert C, Grob JJ, Flaherty KT, Arance A, et al. Dabrafenib plus trametinib in patients with BRAF(V600)-mutant melanoma brain metastases (COMBI-MB): a multicentre, multicohort, open-label, phase 2 trial. Lancet Oncol 2017;18:863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Long GV, Hauschild A, Santinami M, Atkinson V, Mandala M, Chiarion-Sileni V, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. New Engl J Med 2017;377:1813–23. [DOI] [PubMed] [Google Scholar]
- 31. Planchard D, Besse B, Groen HJM, Hashemi SMS, Mazieres J, Kim TM, et al. Phase 2 study of dabrafenib plus trametinib in patients with BRAF V600E-mutant metastatic NSCLC: updated 5-year survival rates and genomic analysis. J Thorac Oncol 2022;17:103–15. [DOI] [PubMed] [Google Scholar]
- 32. Planchard D, Besse B, Groen HJM, Souquet PJ, Quoix E, Baik CS, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Planchard D, Kim TM, Mazieres J, Quoix E, Riely G, Barlesi F, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, Phase 2 trial. Lancet Oncol 2016;17:642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Planchard D, Smit EF, Groen HJM, Mazieres J, Besse B, Helland A, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, Phase 2 trial. Lancet Oncol 2017;18:1307–16. [DOI] [PubMed] [Google Scholar]
- 35. Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J Clin Oncol 2018;36:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, et al. Dabrafenib plus trametinib in patients with BRAF V600E-mutant anaplastic thyroid cancer: updated analysis from the Phase II ROAR basket study. Ann Oncol 2022;33:406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen PY, Stein A, van den Bent M, De Greve J, Wick A, de Vos F, et al. Dabrafenib plus trametinib in patients with BRAF(V600E)-mutant low-grade and high-grade glioma (ROAR): a multicentre, open-label, single-arm, Phase 2, basket trial. Lancet Oncol 2022;23:53–64. [DOI] [PubMed] [Google Scholar]
- 38. Subbiah V, Lassen U, Elez E, Italiano A, Curigliano G, Javle M, et al. Dabrafenib plus trametinib in patients with BRAF(V600E)-mutated biliary tract cancer (ROAR): a Phase 2, open-label, single-arm, multicentre basket trial. Lancet Oncol 2020;21:1234–43. [DOI] [PubMed] [Google Scholar]
- 39. Salama AKS, Li S, Macrae ER, Park JI, Mitchell EP, Zwiebel JA, et al. Dabrafenib and trametinib in patients with tumors with BRAF(V600E) mutations: results of the NCI-MATCH trial subprotocol H. J Clin Oncol 2020;38:3895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Geoerger B, Bouffet E, Whitlock JA, Moertel CL, Hargrave DR, Aerts I, et al. Dabrafenib plus trametinib combination therapy in pediatric patients with BRAF V600-mutant low-grade glioma: safety and efficacy results. J Clin Oncol 38:15s, 2020. (suppl; abstr 10506). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, Phase 3 trial. Lancet Oncol 2018;19:1315–27. [DOI] [PubMed] [Google Scholar]
- 42. Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised Phase 3 trial. Lancet Oncol 2018;19:603–15. [DOI] [PubMed] [Google Scholar]
- 43. Dummer R, Flaherty KT, Robert C, Arance A, de Groot JWB, Garbe C, et al. COLUMBUS 5-year update: a randomized, open-label, Phase III trial of encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF V600-mutant melanoma. J Clin Oncol 2022;40:4178–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. New Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 45. Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino T, et al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol 2021;39:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Cutsem E, Huijberts S, Grothey A, Yaeger R, Cuyle PJ, Elez E, et al. Binimetinib, encorafenib, and cetuximab triplet therapy for patients with BRAF V600E-mutant metastatic colorectal cancer: safety lead-in results from the Phase III BEACON colorectal cancer study. J Clin Oncol 2019;37:1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ascierto PA, Dreno B, Larkin J, Ribas A, Liszkay G, Maio M, et al. 5-year outcomes with cobimetinib plus vemurafenib in BRAFV600 mutation-positive advanced melanoma: extended follow-up of the coBRIM study. Clin Cancer Res 2021;27:5225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ascierto PA, McArthur GA, Dreno B, Atkinson V, Liszkay G, Di Giacomo AM, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, Phase 3 trial. Lancet Oncol 2016;17:1248–60. [DOI] [PubMed] [Google Scholar]
- 49. Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. New Engl J Med 2014;371:1867–76. [DOI] [PubMed] [Google Scholar]
- 50. Laquerre S, Amone M, Moss K, Yang J, Fisher K, Kane-Carson LS, et al. A selective raf kinase inhibitor induces cell death and tumor regression of human cancer cell lines encoding B-raf(V600E) mutation. Mol Cancer Ther 2009;8(12 Suppl):B88. [Google Scholar]
- 51. Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Brit J Cancer 2010;102:1724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. New Engl J Med 2012;366:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. TAFINLAR® (dabrafenib) capsules, for oral use: FDA Packaging Insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/202806s022lbl.pdf.
- 54. Mullard A. BRAF plus MEK inhibitor combo secures tumour-agnostic FDA approval. Nat Rev Drug Discov 2022;21:548. [DOI] [PubMed] [Google Scholar]
- 55. U.S. Food and Drug Administration. FDA grants accelerated approval to dabrafenib in combination with trametinib for unresectable or metastatic solid tumors with BRAF V600E mutation. Available from:https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-dabrafenib-combination-trametinib-unresectable-or-metastatic-solid.
- 56. Drilon A, Hu ZI, Lai GGY, Tan DSW. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol 2018;15:151–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev 2005;16:441–67. [DOI] [PubMed] [Google Scholar]
- 58. Jhiang SM. The RET proto-oncogene in human cancers. Oncogene 2000;19:5590–7. [DOI] [PubMed] [Google Scholar]
- 59. Donis-Keller H, Dou S, Chi D, Carlson KM, Toshima K, Lairmore TC, et al. Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet 1993;2:851–6. [DOI] [PubMed] [Google Scholar]
- 60. Hofstra RM, Landsvater RM, Ceccherini I, Stulp RP, Stelwagen T, Luo Y, et al. A mutation in the RET proto-oncogene associated with multiple endocrine neoplasia type 2B and sporadic medullary thyroid carcinoma. Nature 1994;367:375–6. [DOI] [PubMed] [Google Scholar]
- 61. Mulligan LM, Kwok JB, Healey CS, Elsdon MJ, Eng C, Gardner E, et al. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature 1993;363:458–60. [DOI] [PubMed] [Google Scholar]
- 62. Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 1985;42:581–8. [DOI] [PubMed] [Google Scholar]
- 63. Li AY, McCusker MG, Russo A, Scilla KA, Gittens A, Arensmeyer K, et al. RET fusions in solid tumors. Cancer Treat Rev 2019;81:101911. [DOI] [PubMed] [Google Scholar]
- 64. Ishizaka Y, Itoh F, Tahira T, Ikeda I, Sugimura T, Tucker J, et al. Human ret proto-oncogene mapped to chromosome 10q11.2. Oncogene 1989;4:1519–21. [PubMed] [Google Scholar]
- 65. Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Faivre S, Djelloul S, Raymond E. New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin Oncol 2006;33:407–20. [DOI] [PubMed] [Google Scholar]
- 67. Qian Y, Chai S, Liang Z, Wang Y, Zhou Y, Xu X, et al. KIF5B-RET fusion kinase promotes cell growth by multilevel activation of STAT3 in lung cancer. Mol Cancer 2014;13:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Phay JE, Shah MH. Targeting RET receptor tyrosine kinase activation in cancer. Clin Cancer Res 2010;16:5936–41. [DOI] [PubMed] [Google Scholar]
- 69. Alberti L, Carniti C, Miranda C, Roccato E, Pierotti MA. RET and NTRK1 proto-oncogenes in human diseases. J Cell Physiol 2003;195:168–86. [DOI] [PubMed] [Google Scholar]
- 70. Subbiah V, Cassier PA, Siena S, Garralda E, Paz-Ares L, Garrido P, et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the Phase 1/2 ARROW trial. Nat Med 2022;28:1640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. U.S. Food and Drug Administration. RETEVMO™ (selpercatinib) capsules, for oral use: FDA Packaging Insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213246s008lbl.pdf.
- 72. Brandhuber B, Haas J, Tuch B, Ebata K, Bouhana K, McFaddin E, et al. The development of a potent, KDRNEGFR2-sparing RET kinase inhibitor for treating patients with RET-dependent cancers. Eur J Cancer 2016;69:S144–SJ Clin Oncol 2020;38: 1209–21. [Google Scholar]
- 73. Subbiah V, Velcheti V, Tuch BB, Ebata K, Busaidy NL, Cabanillas ME, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol 2018;29:1869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. New Engl J Med 2020;383:813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, et al. Efficacy of selpercatinib in RET-altered thyroid cancers. New Engl J Med 2020;383:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998;18:184–7. [DOI] [PubMed] [Google Scholar]
- 77. Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016;1:e000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. U.S. Food and Drug Administration. VITRAKVI® (larotrectinib) capsules, for oral use: FDA packaging insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/210861s008lbl.pdf.
- 79. U.S. Food and Drug Administration. ROZLYTREK (entrectinib) capsules, for oral use: FDA packaging insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/212725s006lbl.pdf.
- 80. Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 2020;21:531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol 2020;21:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fusco MJ, West HJ, Walko CM. Tumor Mutation Burden and Cancer Treatment. JAMA Oncol 2021;7:316. [DOI] [PubMed] [Google Scholar]
- 83. Galuppini F, Dal Pozzo CA, Deckert J, Loupakis F, Fassan M, Baffa R. Tumor mutation burden: from comprehensive mutational screening to the clinic. Cancer Cell Int 2019;19:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Subbiah V, Solit DB, Chan TA, Kurzrock R. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) >/=10: a decision centered on empowering patients and their physicians. Ann Oncol 2020;31:1115–8. [DOI] [PubMed] [Google Scholar]
- 85. U.S. Food & Drug Administration. 2020 FDA approves pembrolizumab for adults and children with TMB-H solid tumors; 2020. Available from:https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors.
- 86. U.S. Food and Drug Administration. KEYTRUDA® (pembrolizumab) injection, for intravenous use: FDA Packaging insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/125514s133lbl.pdf.
- 87. U.S. Food and Drug Administration. JEMPERLI (dostarlimab-gxly) injection, for intravenous use: FDA Packaging Insert; 2022. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/761174s002lbl.pdf.
- 88. U.S. Food & Drug Administration. 2017 FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication; 2017. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication.
- 89. U.S. Food & Drug Administration: 2021 FDA grants accelerated approval to dostarlimab-gxly for dMMR advanced solid tumors; 2022. Available from:https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-dostarlimab-gxly-dmmr-advanced-solid-tumors.
- 90. Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. New Engl J Med 2022;386:2363–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Randen M, Rutqvist LE, Johansson H. Cancer patients without a known primary: incidence and survival trends in Sweden 1960–2007. Acta Oncol 2009;48:915–20. [DOI] [PubMed] [Google Scholar]
- 92. Kato S, Weipert C, Gumas S, Okamura R, Lee S, Sicklick JK, et al. Therapeutic actionability of circulating cell-free DNA alterations in carcinoma of unknown primary. JCO Precis Oncol 2021;5:PO.21.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pestana RC, Beal JR, Parkes A, Hamerschlak N, Subbiah V. Impact of tissue-agnostic approvals for patients with sarcoma. Trends Cancer 2022;8:135–44. [DOI] [PubMed] [Google Scholar]
- 94. Bhamidipati D, Subbiah V. Impact of tissue-agnostic approvals for patients with gastrointestinal malignancies. Trends Cancer 2023;9:237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wahida A, Buschhorn L, Frohling S, Jost PJ, Schneeweiss A, Lichter P, et al. The coming decade in precision oncology: six riddles. Nat Rev Cancer 2023;23:43–54. [DOI] [PubMed] [Google Scholar]
- 96. Meric-Bernstam F, J H, R B, H B, C F, R K, et al. MyPathway HER2 basket study: Pertuzumab (P) + trastuzumab (H) treatment of a large, tissueagnostic cohort of patients with HER2-positive advanced solid tumors. J Clin Oncol 39:15s, 2021 (abstr 3004). [Google Scholar]
- 97. Pestana RC, Sen S, Hobbs BP, Hong DS. Histology-agnostic drug development - considering issues beyond the tissue. Nat Rev Clin Oncol 2020;17:555–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes most frequently mutated in cases with tissue-agnostic targets in the overall altered population and top 5 cancers with highest number of profiled samples in each alteration
Frequency of tissue-agnostic targets in other additional different tumors (alphabetically arranged) in AACR GENIE v.13