Comprehensive characterization of genomic and transcriptomic alterations in breast tumors from Hispanic/Latina patients reveals distinct genetic alterations and signatures, demonstrating the importance of inclusive studies to ensure equitable care for patients.
Abstract
Somatic mutational profiling is increasingly being used to identify potential targets for breast cancer. However, limited tumor-sequencing data from Hispanic/Latinas (H/L) are available to guide treatment. To address this gap, we performed whole-exome sequencing (WES) and RNA sequencing on 146 tumors and WES of matched germline DNA from 140 H/L women in California. Tumor intrinsic subtype, somatic mutations, copy-number alterations, and expression profiles of the tumors were characterized and compared with data from tumors of non-Hispanic White (White) women in The Cancer Genome Atlas (TCGA). Eight genes were significantly mutated in the H/L tumors including PIK3CA, TP53, GATA3, MAP3K1, CDH1, CBFB, PTEN, and RUNX1; the prevalence of mutations in these genes was similar to that observed in White women in TCGA. Four previously reported Catalogue of Somatic Mutations in Cancer (COSMIC) mutation signatures (1, 2, 3, 13) were found in the H/L dataset, along with signature 16 that has not been previously reported in other breast cancer datasets. Recurrent amplifications were observed in breast cancer drivers including MYC, FGFR1, CCND1, and ERBB2, as well as a recurrent amplification in 17q11.2 associated with high KIAA0100 gene expression that has been implicated in breast cancer aggressiveness. In conclusion, this study identified a higher prevalence of COSMIC signature 16 and a recurrent copy-number amplification affecting expression of KIAA0100 in breast tumors from H/L compared with White women. These results highlight the necessity of studying underrepresented populations.
Significance:
Comprehensive characterization of genomic and transcriptomic alterations in breast tumors from Hispanic/Latina patients reveals distinct genetic alterations and signatures, demonstrating the importance of inclusive studies to ensure equitable care for patients.
Introduction
Sequencing studies of breast cancer have identified recurrently mutated genes and somatic copy-number alterations (SCNA) affecting tumor suppressors and oncogenes (1–3). Both somatic mutations and CNAs may be useful in determining prognosis. Currently, therapies for breast cancer can be selected on the basis of particular somatic mutations [i.e., alpelisib for PIK3CA (4)], SCNAs (i.e., trastuzumab for HER2), and germline mutations in genes in the homologous recombination repair (HRR) pathway (PARP inhibitors).
Genetic ancestry is associated with specific somatic mutations in many cancer types. EGFR mutations are approximately 4-fold more common in lung cancer from women and men of East-Asian ancestry compared with lung cancer from women and men of other populations (5) with self-reported Hispanic/Latinos (H/L) representing an intermediate group (6, 7). FOXA1 mutations in prostate cancer also are substantially more common in East-Asian ancestry populations compared with European and African ancestry populations (8). Comprehensive analyses of The Cancer Genome Atlas (TCGA) have demonstrated that many mutations and CNAs are more common in specific ancestral populations (9, 10). In breast cancer, previous studies have demonstrated that women of African ancestry have higher rates of TP53 mutations and lower rates of PIK3CA mutations, likely related to a higher incidence of a basal-like breast cancer subtype in African-American women (11, 12). However, the genomic landscape of breast cancer has not been well characterized in H/L groups.
H/L represent the largest minority population in the United States and have diverse origins, with the largest subpopulations including Mexican Americans and Puerto Ricans. Genetically, H/L are a population of mixed European, Indigenous American (IA), and African ancestries with those ancestry proportions varying widely depending on country of origin and regions within a country. Although breast cancer is less common overall among H/L compared with self-reported non-Hispanic White (White) women due to both environmental (13) and genetic factors (14), there is a higher proportion of breast cancers diagnosed under age 50 years than in Whites (15). Moreover, outcomes are usually worse among H/L compared with White women (16). In some studies, IA ancestry was associated with poorer outcomes among H/L with breast cancer (17). HER2 amplifications are overrepresented among H/L and are more common among H/L with more IA ancestry compared with those with more European ancestry (18). Few studies have investigated the distribution of somatic mutations and SCNAs in breast tumors from H/L. In TCGA, out of 1,096 breast cancer cases, only 39 are self-reported H/L. A recent study analyzed data including whole-exome sequencing (WES) and gene expression data from 109 Mexican women living in Mexico (19). However, no similar size study has been conducted in H/L in the United States. To investigate the somatic mutational spectrum in breast cancer among H/L, we generated WES and RNA sequencing (RNA-seq) data from 146 tumors from 140 H/L from Southern California and performed analyses of somatic mutations, SCNAs, and gene expression.
Materials and Methods
Participants
One hundred and forty patients with breast cancer seen at City of Hope (COH) in Duarte, California were included in this study. All participants signed a written informed consent approved by the COH Institutional Review Board. Inclusion criteria were: (i) self-identified as H/L; (ii) tumor tissue from surgery was available and the sample contained more than 40% tumor based on examination by a single breast pathologist (D. Schmolze). The percentage tumor ranged from 40% to 90% with an average of 64% and a median of 65% tumor. An exclusion criterion was neoadjuvant therapy as treatment could change the mutation profile. Clinical data were abstracted from medical records including date at diagnosis, date at surgery, tumor stage, grade, histologic estrogen receptor (ER), progesterone receptor (PR), and HER2 status, second cancers, breast cancer recurrence, parity, history of breast feeding, age at menarche, and cause of death, if applicable. Six of the 140 patients with breast cancer had two primary contralateral breast cancers with tissue available for study for a total of 146 tumors.
DNA and RNA-seq
DNA extraction
Germline DNA was extracted from peripheral blood cells or from formalin-fixed paraffin-embedded (FFPE) normal breast tissue adjacent to tumor tissue from surgery. Peripheral blood cell DNA was extracted using a standard phenol chloroform method. For FFPE tissue, DNA and RNA were extracted from 10 30 μm sections from each tumor using the QIAmp DNA FFPE Tissue Kit (Qiagen) and miRNeasy Kit (Qiagen) according to manufacturer's instructions. DNA was quantified with the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific). After extraction and quantification, DNA was sent to The NCI Cancer Genomics Research Laboratory (CGR) for WES. For RNA-seq, 500 ng total RNA was sent to the COH Integrative Genomics Core (IGC).
DNA library construction, hybridization, and massively parallel sequencing
Library production and sequencing for 146 tumors and 140 matching normal samples was performed at CGR. The KAPA HyperPlus Kit (Kapa Biosystems, Inc.) was used to generate libraries from 300 ng DNA according to the KAPA-provided protocol. Libraries were pooled and sequence capture was performed with NimbleGen's SeqCap EZ exome v3 (Roche NimbleGen, Inc.), according to the manufacturer's protocol. The resulting postcapture enriched multiplexed sequencing libraries were used in cluster formation on an Illumina cBOT (Illumina) and paired-end sequencing was performed using an Illumina HiSeq 4000 following Illumina-provided protocols for 2 × 100 bp paired-end sequencing to an average-fold coverage of 80X for the tumors and 30X for the germline samples. Paired‐end reads from each sample were aligned to human reference genome (hg19) using Novoalign (v3.00.05), and the aligned binary format sequence (BAM) files were sorted and indexed using SAMtools (1, 2). The sorted and indexed BAMs were processed by Picard (v1.126, https://broadinstitute.github.io/picard/) to remove duplicate sequencing reads. Local realignment around suspected sites of indels was performed using Genome Analysis Toolkit (GATK) IndelRealigner (v3.3-0-g37228af). These mapped sequence reads were then base recalibrated before being used for somatic mutation calling by MuTect2 in GATK (v4.0.11.0).
RNA-seq
In the COH IGC, sequencing libraries were prepared with Kapa RNA HyperPrep Kit with RiboErase (Roche) and sequenced on a HiSeq 2500 (Illumina) with 40 million reads per sample. The RNA-seq reads were aligned to hg19 genome assembly using Tophat2 (v2.0.8) with default settings. The gene expression levels were counted by obtaining raw counts with HTSeq (v0.6.1p1) against Ensembl v86 annotation. The counts data were normalized using the trimmed mean of M values method implemented in R package edgeR (20). Log2-transformed counts were used to assign PAM50 subtypes based on the subgroup-specific gene centering method developed by Zhao and colleagues (21). We estimated Z-scores based on the corrected median absolute deviation (MAD) implemented by the robStandardize R function in the robustHD R package and defined expression outliers as gene-sample datapoints with robust Z-scores greater than three. Raw counts of RNA-seq data for 1,189 TCGA samples (including both tumor and matched normal samples) were downloaded from the Genomic Data Commons (GDC) using the GDCRNATools (22) R package. RNA-seq data for H/L tumor samples and TCGA samples were processed and analyzed separately.
Data analysis
Germline variant calling
Germline variant calling from the BAM files was performed in the COH IGC using GATK HaplotypeCaller (https://software.broadinstitute.org/gatk). Variants with a call quality less than 20, read depth less than 10, or allele fraction ratio less than 20% were removed. Variants in variant call format files were evaluated for pathogenicity using Ingenuity Variant Analysis (IVA) version 4 (Qiagen) and American College of Medical Genetics and Genomics (ACMGG) guidelines were applied using the IVA ACMGG calling algorithm (23). Pathogenic or likely pathogenic variants were individually evaluated by the research team using the available literature and ClinVar to make a final determination (24).
We inferred germline SNP calls using the WES data. Low-pass whole-genome sequence data can be used to impute SNP data using reference human genomes and equivalent data from WES off-target reads can be leveraged with reference human genomes to infer common variants (25–27). To perform common variant inference, we used the BAM files from the germline WES datasets and performed common variant calling using STITCH (25) Version 1.6.6. We performed rigorous quality control on the data by excluding variants with info score <0.8, deviations from Hardy–Weinberg equilibrium P < 0.001, call rate < 0.95, minor allele frequency < 0.005. In addition, we performed a χ2 test for allele frequency differences between our sample and the 1000 Genomes AMR sample, which has a similar ancestry profile (mixture of mostly European and IA) and dropped variants that had a P value < 5 × 10−8. We used this additional filter to minimize SNPs with imputation errors because our goal was not to perform a genome-wide association study analysis but to obtain the best quality subset of SNPs to perform locus-specific ancestry estimates and conduct limited germline–somatic interaction and expression quantitative trait locus analysis (eQTL). Overall, 2,118,749 variants passed all the levels of quality control. Included in this set of SNPs was rs12628403 (imputation quality score = 0.97), which is a surrogate for the APOBEC3A/B deletion (r2 = 0.91; ref. 28).
Genetic ancestry analysis
We performed genetic ancestry estimation for each of the 140 women using the germline WES data. We used 90 European (1000 Genomes), 90 African (1000 Genomes), 90 East-Asian (1000 Genomes), and 71 IA ancestry (29) reference samples. We identified the SNPs that overlap all datasets (N = 9,935). We combined all SNPs and dropped SNPs that did not match based on reference and alternate alleles. To estimate the ancestry for each sample, we used ADMIXTURE 1.3.0 setting the K parameter to 4 and running the unsupervised algorithm (30). In addition, we used principal components analysis, calculated using PLINK 1.9 (31) as a complementary method to assess ancestry.
Locus-specific ancestry
The imputed data were phased using Beagle 5.4 (32). The phased data were combined with data from European, African, and IA reference samples as described above. We used RFMix version 2 [https://github.com/slowkoni/rfmix (33)] to estimate locus-specific ancestry on the overlapping 2,118,045 SNPs between our data and data from ancestral samples.
Somatic variant calling
We identified somatic single-nucleotide variants (SNV) using MuTect2 in GATK4 (v4.0.11.0) suite with default parameters (34) and indels using GATK Indelocator. Using the SNV and indel filtering method described in Pereira and colleagues (3), we focused on frameshift, nonsynonymous, canonical splicing site, and stop gain mutations. Briefly, somatic mutations were manually curated and considered true positives in a sample if the mutation was observed in >10% of reads or with a frequency of 5% to 10% if in frequently mutated breast cancer genes or seen in Catalogue of Somatic Mutations in Cancer (COSMIC) database (35). Because the tumors include both tumor and normal stromal cells, it is expected that the proportion of reads will have less than the expected 50% if 100% tumor. Mutations in <5% of reads, in segmental duplication regions, or indels that overlapped homopolymer stretches of six or more bases were considered false positives. We did visual checking using the Integrative Genomics Viewer (IGV) to assess the quality of all somatic mutations. We performed Sanger sequencing on a subset of samples to confirm specific mutations in AKT1, BARD1, MAP3K1, and MET. Using the filtered and annotated somatic mutations, we performed a somatic mutation significance analysis via MutSigCV (version 1.3.5; ref. 36) on Genepattern (https://www.genepattern.org/modules/docs/MutSigCV). Genes with FDR q < 0.05 are considered to be significantly mutated genes.
We compared the significant somatic mutations in our analysis with the mutations from the Romero-Cordoba dataset (19). Using the publicly available somatic mutation data from the Romero-Cordoba study of the Mexican patients, we combined our somatic mutation data and performed a MutSigCV analysis to identify the common significant genes. Similarly, to investigate whether these significantly mutated genes were associated with ancestry, we performed the same analysis on breast tumors from Whites in TCGA. Using 2% as the mutation frequency threshold, we counted the number of mutations and their corresponding mutation frequencies in each cohort, and performed Fisher exact test to investigate whether any gene was significantly more frequently mutated in either cohort. Furthermore, we tested whether common germline and somatic mutations (frequency > 5%) were associated with global and/or local ancestry using logistic regression models in which mutation (yes or no) was the outcome variable and numerical global ancestry (percentage) and local ancestry (the number of IA ancestry allele) were the explanatory variables.
CNA using FACETS
We used FACETS implemented in R package FACETS version 0.6.1 (37) to calculate CNAs. The counts of reads with the reference (ref) allele, alternate (alt) allele, errors (neither ref nor alt), and deletions at a specific genomic position were generated using BAM files from the 146 matched tumor-normal sample pairs using the application snp-pileup in the FACETS package. The segmentation of each tumor sample was then estimated with the critical value (cval) 150. The segmentation files generated by FACETS served as input files for the GISTIC2.0 (38) on the GenePattern server (https://genepattern.broadinstitute.org/gp) to identify significant SCNAs using a q-value cutoff < 0.05. A gene was considered as copy number altered with GISTIC2-thresholded scores of −2 (deep loss), −1 (shallow loss), 1 (low-level gain), and 2 (high-level gain). The GISTIC2 copy-number results and clinical data for 816 TCGA tumor samples were downloaded from the cBioPortal database (https://www.cbioportal.org; ref. 39). Expression outliers (defined by Z-scores greater than 3.0) were considered as driven by copy-number changes if greater than 90% expression outliers in a gene had a GISTIC2-thresholded copy-number score of 2 (high-level gain) or 1 (low-level gain). Fisher exact test was used to identify genes with frequency difference in expression outliers, driven by CNAs, between 146 tumor samples from H/L and 452 TCGA Whites (determined as having > 95% European ancestry as described below).
Mutation signature analysis
Using the previously called SNVs, we performed a mutational signature analysis via the MutationalPatterns R package (40). Hg19 was used as the reference genome. SNVs were parsed and classified into six mutation patterns (C>T, T>A, C>G, T>C, C>A and T>G) and 96 trinucleotide changes. Then a non-negative matrix factorization algorithm was implemented to extract mutation signatures. We compared the similarities of these mutation signatures with the COSMIC mutation signatures and each mutation signature could be treated as a linear combination of the 30 COSMIC mutation signatures. The 30 COSMIC mutation signature percentage contribution was then computed for each tumor and a contribution heatmap was generated. Within these tumor samples, we performed a signature contribution comparison using the two-sided Wilcoxon rank-sum tests among the five tumor subtypes (luminal A, luminal B, basal-like, HER2-enriched, and normal-like).
We also compared the mutation signature analysis with the breast tumors in the Romero-Cordoba dataset and the breast tumors from Whites in TCGA SNV dataset. For the significant COSMIC mutation signatures identified in our dataset, we performed two-sided Wilcoxon rank-sum tests among the three datasets to test whether the signature was enriched in Mexican patients.
Germline–somatic interaction analyses: effect of germline SNP variants on somatic mutations, copy-number changes, and gene expression
To determine the potential effects of germline variants on somatic events and gene expression, we performed limited association test between the imputed SNP genotypes and a set of high-priority somatic events because we were severely underpowered in this dataset to perform genome wide searches for germline–somatic interactions. Specifically, we focused on SNPs that may be associated with (i) somatic mutations in the genes identified as significantly mutated by MutSigCV, (ii) the copy-number amplifications identified as significantly amplified by GISTIC, which have known driver genes, (iii) the top gene, KIAA0100, identified as significantly different for gene expression across datasets between White TCGA participants and our H/L dataset, (iv) the known APOBEC3A/B SNP, rs12628403, in linkage disequilibrium with the germline deletion associated with tumor mutational signatures. We restricted these analyses to the germline SNP variants in cis, defined as 1 MB upstream and downstream of the gene that is significantly mutated or the presumed target gene. Logistic regression models were used to test associations of somatic mutations (yes or no, outcome variable) in genes with mutation rates > 5% and germline SNP genotype (0, 1, and 2, primary predictor variable) adjusting for covariates including somatic mutation rate [the number of nonsilent mutations per megabase (Mb)], tumor stage, and PAM50 subtype (41, 42). Logistic regression models also were used to test associations between high-level gain (yes or no, outcome variable) in known genes in the region of gain and germline SNP genotype adjusting for covariates including somatic copy-number variation (CNV) rate (the number of segments each sample has genome wide), tumor stage, and PAM50 subtype. We also performed cis-eQTL analysis for a gene (s) with frequency difference in expression outliers between H/L and White tumors. We used a multivariate linear model in which gene expression in tumor was the outcome variable, germline SNP genotype was the primary predictor variable, and covariates were batch variable in RNA-seq, tumor stage, and PAM50 subtype (43). Benjamini–Horchberg (BH)-adjusted P values (FDR) < 0.05 were considered significant associations.
Data availability
All tumor/normal WES and RNA-seq data and accompanying phenotypic and clinical/histologic data are deposited in dbGAP (dbGaP Study Accession: phs003218). TCGA RNA-seq and clinical data are publicly available at http://firebrowse.org/?cohort=BRCA&download_dialog=true. The Romero-Cordoba dataset (19) was downloaded from their supplementary material publicly available at https://doi.org/10.1038/s41467-021-22478-5. All other raw data are available upon request from the corresponding author.
Results
Clinical/demographic data
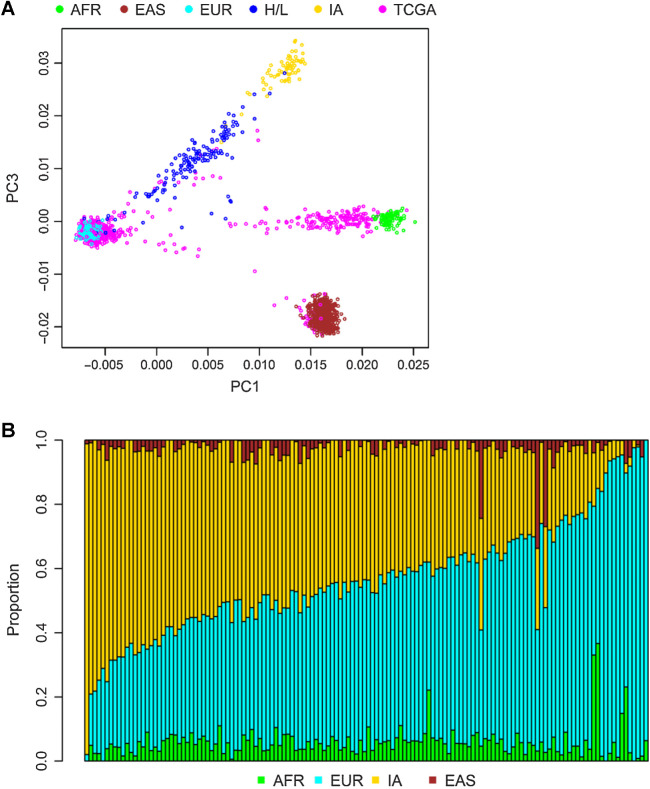
The mean age at diagnosis was 48.7 years with a range from ages 31 to 75 years (Table 1). Nearly all of the 140 H/L were of mixed European (Eur) and IA ancestry. The mean ancestry composition was 50.6% Eur, 40.8% IA, 5.9% African, and 2.7% Asian although the range of ancestry proportion varied widely from <1% to 96% IA at the extremes (Fig. 1). As shown in the principal component analysis plots in Fig. 1A, H/L samples are not well represented in TCGA. For the six individuals with two primary tumors (in the contralateral breasts), the tumors were considered independent tumors (Supplementary Table S1), which was borne out by different somatic mutation profiles. The majority of the women were diagnosed with stage I (44%) or II (43%) tumors (Table 1). There were 22 recurrences and 10 deaths during the time of follow-up. Of the 146 tumors, 83% were ER positive (ER+), 72% were PR positive, and 17% were HER2 positive (HER2+) and these proportions were similar to White women in TCGA (1).
Table 1.
Patient and tumor characteristics of 140 H/L breast cancer cases and their 146 breast tumors.
| Patient characteristics | Mean | Range | Median | |
|---|---|---|---|---|
| Age at diagnosis (years) | 48.7 | 31–75 | 48 | |
| Breastfeeding (months) | 7.2 | 0–84 | 2 | |
| Parity (number children) | 2.3 | 0–8 | 2 | |
| Age at menarche (years) | 12.6 | 9–18 | 12 | |
| Tumor characteristics | Positive | Negative | Unknown | Equivocal |
| Estrogen receptor | 120 (82%) | 25 (17%) | 1 (0.7%) | |
| Progesterone receptor | 104 (72%) | 41 (28%) | 1 (0.7%) | |
| HER2 | 25 (17%) | 116 (80%) | 1 (0.7%) | 4 (3%) |
| Stage at diagnosis | I | II | III | IV |
| 63 (44%) | 63 (43%) | 17 (12%) | 3(2%) |
Figure 1.
Ancestry of the cohort. Results of principal components analysis comparing the values for samples on principal component (PC) 1 (x-axis) and PC3 (y-axis) (A). Each dot represents the results from one individual. H/L, dark blue; TCGA, pink; and reference populations including African (AFR), Yoruban individuals from Nigeria from HapMap (light green); East Asians (EAS), Han Chinese from HapMap (brown); European American (EUR) from HapMap (light blue); and IA (yellow) from Mexico. PC2 (not shown) captures individuals of Asian and IA ancestry. B, Results from ADMIXTURE analysis. Each vertical bar represents estimate of ancestry from one individual. Ancestry is assigned for each individual as a fraction of either African (green), Asian (brown), European (light blue), or IA (yellow) ancestry.
Germline variant analyses
Germline pathogenic variants in breast cancer predisposition genes were identified in six participants including one BRCA1 exon 9–12 deletion, four CHEK2 L236P, and one NF1 Y408X variant, of which the BRCA1 and CHEK2 variants are of IA ancestry (44). Germline APOBEC3A/B deletions were observed in 56 of 140 women (43 heterozygous and 13 homozygous deletions). The APOBEC3A/B deletions were 3-fold more common in H/L than in TCGA White tumors (25.2% vs. 8.4%, linear trend P value = 1.76 × 10−11 from logistic regression). The high frequency of APOBEC3A/B deletion was significantly associated with both global IA ancestry (P = 0.017, logistic regression) and local IA ancestry (P = 0.016, Spearman rank correlation).
Somatic mutations
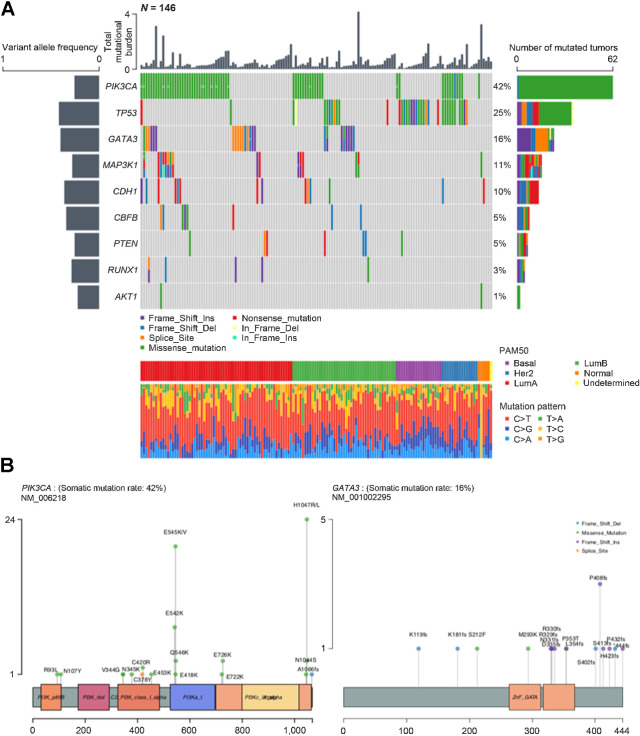
We observed a total of 4,510 true somatic mutations in 3,391 genes in the 146 primary breast tumors (Supplementary Table S2). The number of mutations per individual varied from 2 to 225. Using MutSigCV, we found that mutations in PIK3CA, TP53, GATA3, MAP3K1, CDH1, CBFB, PTEN, and RUNX1 were significant (FDR < 0.05) cancer driver mutations. To identify additional, potentially significantly mutated genes in H/L, we merged the mutation data from our cohort with a previously published study of Mexican patients with breast cancer (N = 135; ref. 19). Within the aggregated mutation data of this combined cohort (N = 281), we re-ran MutSigCV and identified one more significantly mutated gene, AKT1, which only occurred twice in our 146 primary breast tumors. Using the statistically significantly mutated genes obtained from the aggregated cohort, we visualized the mutational profiles within our cohort (Fig. 2A) and the variant locations for PIK3CA and GATA3 (Fig. 2B). For MAP3K1 and RUNX1, at least one tumor harbored multiple mutations in the same gene. Furthermore, in GATA3, seven tumors had the identical splice mutation (NM_001002295.2:c.925-3_925-2delCA) that affected expression (data not shown). Other genes of interest that did not meet the significance threshold (FDR < 0.05) but that have been identified as significant in prior studies and were mutated in our dataset included MLL3 (a.k.a KMTC2; 6%), PTPRD (3%), MAP2K4 (2%), PIK3R1 (2%), NF1 (1%), RB1 (1%), TBX3 (1%), FOXA1 (1%), PADI4 (1%), CDKN1B (1%), CTCF (1%), and NCOR1 (1%). In addition, we found mutations in MET (4.1%), which is not generally considered a breast cancer gene but is a known driver in other cancer types (45).
Figure 2.
Tumor mutational burdens and somatic mutational profiles. A, Mutation plot of nine significantly mutated genes in the 146 tumors. Different mutation classifications are color coded. Numbers are shown where multiple mutations of the same classification were detected. Total mutational burden for each tumor is shown as a bar chart on top. The mean variant allelic frequency is shown for each gene on the left. PAM50 subtype and mutation pattern for each tumor are shown at the bottom. B, Lollipop plots of PIK3CA and GATA3 mutations within the 146 tumors. Mutation classifications are color coded and amino acid changes are specified for each mutation.
The frequency of mutations in genes known to be significantly mutated in breast cancer, including PIK3CA, MAP3K1, GATA3, CBFB, and MLL3/KMT2C, were not significantly different in tumors from H/L compared with tumors from White women in TCGA (FDR q > 0.05; Supplementary Table S3). Similar to tumors from Whites, PIK3CA and TP53 were the most commonly mutated genes. We identified AKT1 mutations in two of 146 tumors (1.4%), including the E17K hotspot mutation, which was found to be mutated in 8% of patients among Mexican women (19). For the seven genes with mutation frequency >5% (Fig. 2A), there were no significant associations with either global or locus-specific ancestry (Supplementary Table S4). Of the seven individual mutations observed in more than 2% of tumors, we observed nominally significant higher frequency of the E545K hotspot mutation in PIK3CA (13.7% vs. 7.5%, Fisher exact test P value = 0.03) and a nonsignificant trend toward higher frequency of the splice variant in GATA3 (4.8% vs. 1.9%, Fisher exact test P value = 0.07) in H/L compared with TCGA Whites (Supplementary Table S5).
Mutational signature analysis
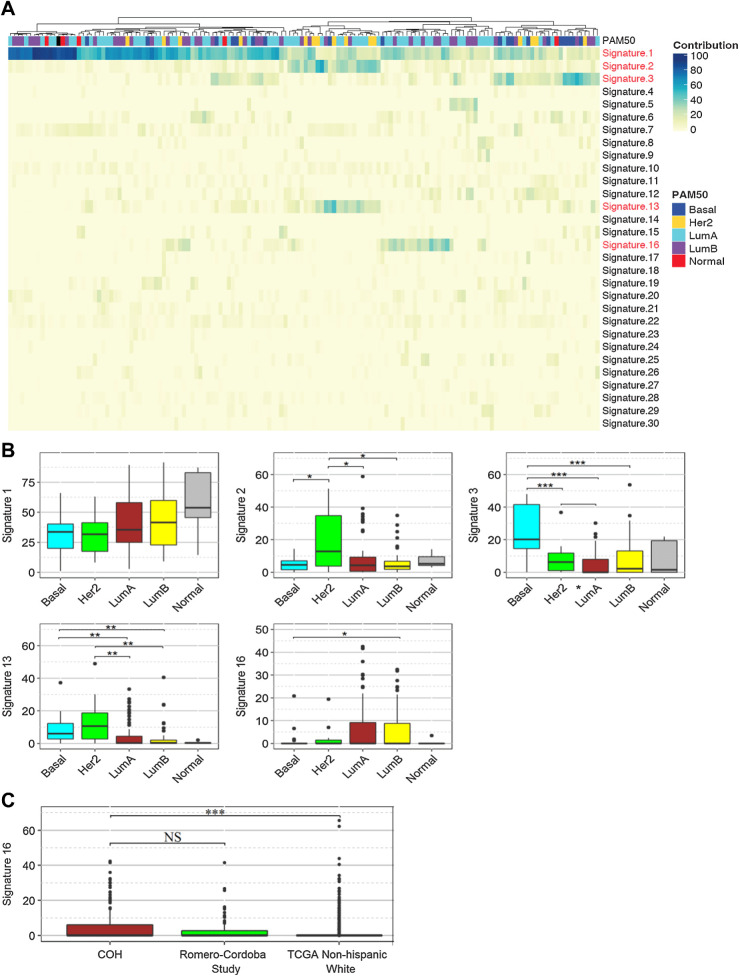
To investigate the mutational processes in H/L breast cancer tumors and the association between PAM50 subtypes and mutational patterns, we adopted the non-negative matrix factorization approach as proposed by Alexandrov and colleagues (46) for mutational signature analysis of tumors. Signature calling revealed five major contributing signatures in the 146 tumors corresponding to the COSMIC signatures 1, 2, 3, 13, and 16 (Fig. 3A; Supplementary Table S6). Signature 1 was detected in all 146 tumors. The contribution of COSMIC signature 1 was greater in luminal A and B subtypes than HER2 and basal subtypes (P < 0.05, two-sided Wilcoxon rank-sum test; Fig. 3B). Signatures 2 and 13, attributed to activity of the AID/APOBEC family of cytidine deaminases, were found in tandem in 16% (n = 23) of the tumors and were more common in tumors with HER2 subtype compared with luminal A and B subtypes (Fig. 3B). We found that 13 tumors were homozygous and 43 tumors were heterozygous for a common 29.5 kbp germline deletion spanning most of APOBEC3. Tumors with the deletion had a higher proportion of COSMIC signatures 2 (P = 0.0005, Wilcoxon rank-sum test) and 13 (P = 0.0008, Wilcoxon rank-sum test). Signature 3, attributed to defects of homologous recombination double-stranded DNA break repair, was found significantly more often in basal subtypes than the other PAM50 subtypes (P < 0.05, two-sided Wilcoxon rank-sum test) including the tumor with the germline BRCA1 exon 9–12 deletion. We observed a group of tumors (N = 40, 27.4%) with more than 5% COSMIC signature 16 contributions. There was no association of signature 16 with global IA ancestry. Because this was not previously reported in other breast tumor studies, we re-examined other datasets, using the same analytic pipeline used herein. We found that signature 16 was present in 20 (19.6%) tumors in a previous study of Mexican patients with breast cancer (19), which was not significantly different than the proportion in our dataset (P = 0.18, Fisher exact test). The proportion with this signature in tumors from TCGA White women (N = 75; 8.9%) was significantly lower than in our dataset (P < 0.001, Fisher exact test; Fig. 3C) and in the Romero-Cordoba and colleagues dataset (P < 0.0001, Fisher exact test). The percentage of this signature was significantly higher in luminal A and B subtypes compared with HER2 and basal tumors (P < 0.05, two-sided Wilcoxon rank-sum test; Fig. 3B).
Figure 3.
Mutational signatures. A, Unsupervised clustered heatmap of contributions from each mutational signature for the 146 tumors. Significant signatures are highlighted in red. PAM50 subtype for each tumor is shown on top of the heatmap. B, Box plot comparisons of the contributions of the five significant mutational signatures (Signature 1, 2, 3, 13, 16) across the PAM50 subtypes. Statistical significance levels are indicated within the box plots. C, Box plot of signature 16 contributions in the 146 tumors from the Hispanic-Mexican cohort (COH), Romero-Cordoba study, and the non-Hispanic White tumors in TCGA dataset. Statistical significance levels are indicated within the box plot. NS, not significant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Wilcoxon rank-sum test.
SCNAs
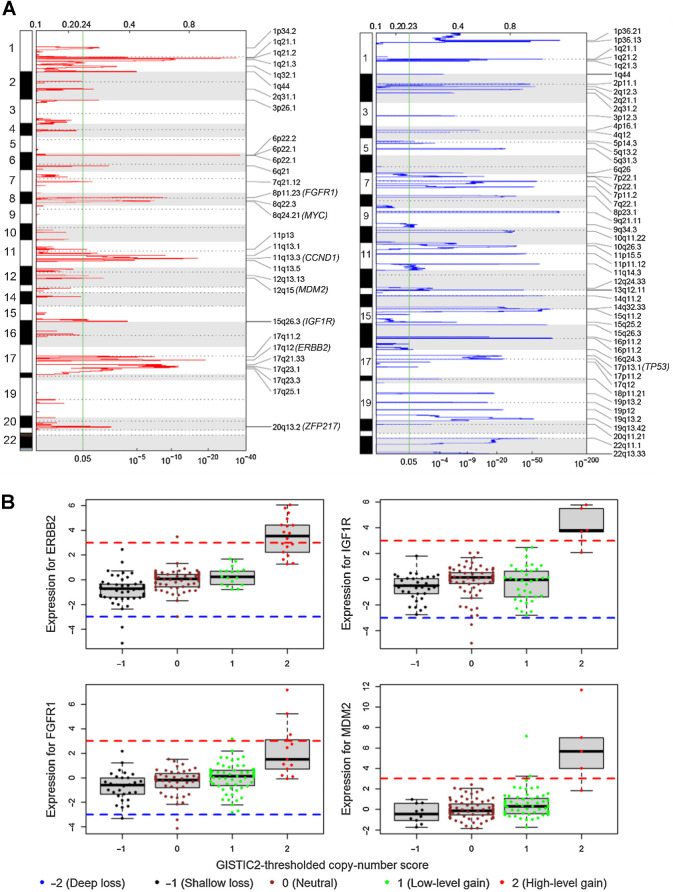
Using GISTIC2, we identified chromosome arm-level SCNAs that were significantly (q < 0.05) amplified at 1q, 8q, 6p, 1p, 6q, 16p, 20q, 8p, 12q and deleted at 22q, 16p, 17p, 8p (Supplementary Table S7). In addition to these broad SCNAs, we identified significantly (q < 0.05) amplified or deleted focal regions including 29 peak regions of amplification and 48 regions of deletion (Fig. 4A). Seven recurrently amplified regions contain common oncogenes (FGFR1, MYC, CCND1, MDM2, IGF1R, ERBB2, and ZNF217); one recurrently deleted region contains TP53 (Fig. 4A). There were no significant associations with either global or locus-specific ancestry (Supplementary Table S4). By integrative analysis of RNA-seq gene expression data and copy-number data, we observed that greater than 90% of expression outliers (defined by robust Z-score greater than 3.0) in ERBB2, FGFR1, IGF1R, and MDM2 were associated with copy-number gain (Fig. 4B). Therefore, we sought to identify expression outliers from 1,121 genes contained in the 29 copy-number amplification peak regions for the 146 H/L breast tumor samples and 452 White TCGA breast tumor samples. Of 1,121 genes in the 29 regions, over 90% of expression outliers were associated with copy-number gain in 214 genes, including 88 genes from the 146 H/L samples, 62 genes from the 452 TCGA White samples, and 64 genes from both sample groups (Supplementary Table S8). Driven by copy-number gains, 18 of 214 genes had significant (FDR < 0.05) difference in frequency of expression outliers between the 146 H/L and 452 TCGA White tumor samples (Table 2 and the top 18 rows in Supplementary Table S8). Expression outliers from those genes were more prevalent in the 146 H/L than in the 452 White tumors because we focused on the 29 copy-number regions (Fig. 4A) found in H/L (Table 2; Supplementary Table S8).
Figure 4.
CNAs. A, Genomic regions of significant copy-number gain (left) and loss (right) identified by GISTIC2. Common oncogenes and tumor suppressor genes are in parentheses next to the corresponding cytobands. The green vertical line marks the GISTIC2 q value of 0.05 (bottom x-axis). B, Outlying gene expression and copy-number gain in four genes in 146 H/L breast tumor samples. Gene expression values on the y-axis are Z-scores estimated by robust standardization; the red dash line of Z-score = 3 and blue dash line of Z-score = −3 are cut-off values for outliers of overexpression and underexpression, respectively.
Table 2.
Frequency difference in expression outliers driven by copy-number gain between 146 H/L tumors and 452 TCGA White tumors.
| Gene | GISTIC2 gain region | Specific to H/La | GISTIC2 q value | No. of outliers in 146 H/L | Frequency of outliers in 146 H/L | No. of outliers in 452 White | Frequency of outliers in 452 White | Fisher exact P valueb | BH-adjusted P value |
|---|---|---|---|---|---|---|---|---|---|
| KIAA0100 | 17q11.2 | yes | 7.85E-08 | 11 | 0.08 | 0 | 0 | 1.37E-07 | 2.93E-05 |
| DSCC1 | 8q24.21 | yes | 1.18E-06 | 7 | 0.05 | 0 | 0 | 4.63E-05 | 4.95E-03 |
| C4BPA | 1q32.1 | yes | 6.24E-04 | 10 | 0.07 | 4 | 0.01 | 2.31E-04 | 9.88E-03 |
| C4BPB | 1q32.1 | yes | 6.24E-04 | 6 | 0.04 | 0 | 0 | 1.96E-04 | 9.88E-03 |
| RNF169 | 11q13.5 | yes | 1.90E-05 | 12 | 0.08 | 6 | 0.01 | 1.41E-04 | 9.88E-03 |
| POLDIP2 | 17q11.2 | yes | 7.85E-08 | 10 | 0.07 | 5 | 0.01 | 5.48E-04 | 1.95E-02 |
| FOXJ3 | 1p34.2 | yes | 8.16E-03 | 7 | 0.05 | 2 | 0 | 1.05E-03 | 2.94E-02 |
| MIR4728 | 17q12 | no | 1.02E-19 | 12 | 0.08 | 9 | 0.02 | 1.10E-03 | 2.94E-02 |
| MYBPH | 1q32.1 | yes | 6.24E-04 | 8 | 0.05 | 4 | 0.01 | 2.13E-03 | 3.95E-02 |
| SAP30BP | 17q25.1 | yes | 2.60E-04 | 10 | 0.07 | 7 | 0.02 | 2.22E-03 | 3.95E-02 |
| SDF2 | 17q11.2 | yes | 7.85E-08 | 8 | 0.05 | 4 | 0.01 | 2.13E-03 | 3.95E-02 |
| UBE2O | 17q25.1 | yes | 2.60E-04 | 12 | 0.08 | 10 | 0.02 | 1.91E-03 | 3.95E-02 |
| AHCTF1 | 1q44 | no | 1.16E-05 | 5 | 0.03 | 1 | 0 | 3.96E-03 | 4.71E-02 |
| GSDMC | 8q24.21 | yes | 1.18E-06 | 10 | 0.07 | 8 | 0.02 | 3.95E-03 | 4.71E-02 |
| MTF1 | 1p34.2 | yes | 8.16E-03 | 4 | 0.03 | 0 | 0 | 3.44E-03 | 4.71E-02 |
| PIGS | 17q11.2 | yes | 7.85E-08 | 6 | 0.04 | 2 | 0 | 3.49E-03 | 4.71E-02 |
| QSER1 | 11p13 | no | 3.38E-02 | 9 | 0.06 | 6 | 0.01 | 3.13E-03 | 4.71E-02 |
| UNC13D | 17q25.1 | yes | 2.60E-04 | 5 | 0.03 | 1 | 0 | 3.96E-03 | 4.71E-02 |
Abbreviations: GISTIC2, GISTIC2 algorithm for copy-number analysis; H/L, Hispanic/Latino; White, non-Hispanic White.
aGISTIC2 gain regions are identified in the 146 H/L samples but not in the 663 TCGA Caucasian samples based on GISTIC2 results published by Romero-Cordoba and colleagues (19); the 8q24.21 region was identified in both groups; however, the wide-peak boundary for the 663 TCGA Caucasian samples (chr8:128657453-128779930) was narrower than that for the 146 HW samples (chr8:114449162-130760646), therefore, DSCC1 and GSDMC are included in 8q24.21 from the 146 H/L samples, but not in the 8q24.21 from the 663 TCGA Caucasian samples.
bFrequency difference in the number of expression outliers between H/L and White group was tested using the Fisher exact method.
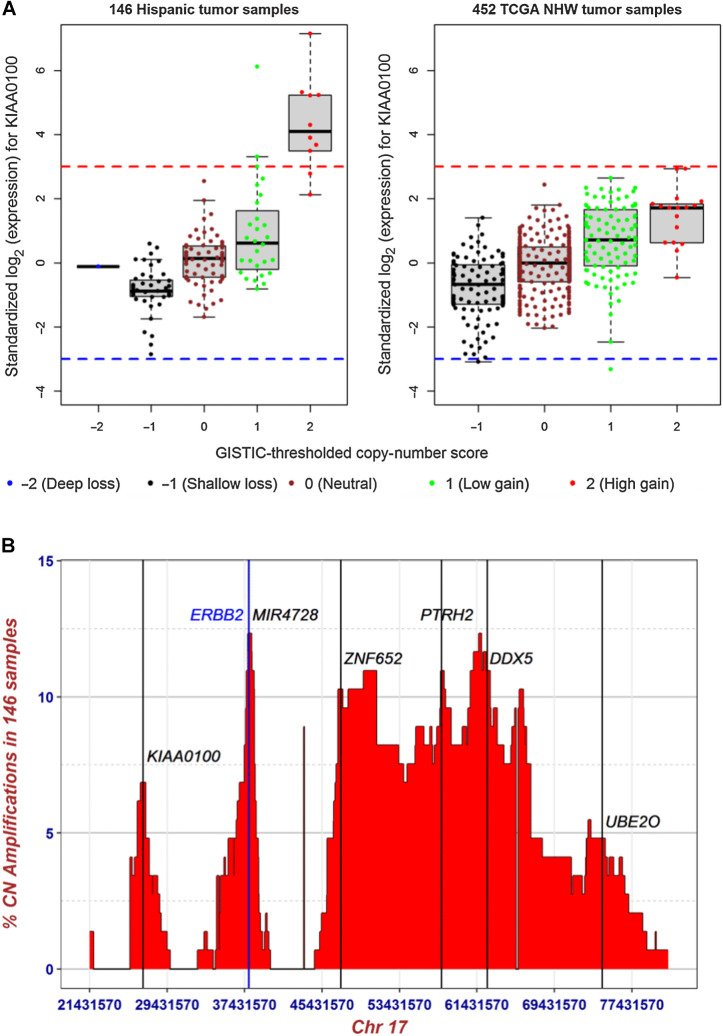
Using this combined copy-number and gene expression analysis approach, we identified KIAA0100, also known as breast cancer overexpressed gene 1 (BCOX1), as the top gene that was systematically different between Whites (TCGA) and our H/L cohort (Fig. 5A). Because this gene is within approximately 11 Mb of ERBB2 on chromosome 17q, we investigated whether it was part of the ERBB2 GISTIC amplification peak. The peaks for the copy-number amplifications (Fig. 5B) were distinct for KIAA0100 and ERBB2, located at 17q11.2 and 17q12, respectively. Of the 11 tumors with outlying expression of KIAA0100 in our H/L cohort, three also had outlying expression of ERBB2 (P = 0.04, Fisher exact test). To account for the partial correlation of overexpression between ERBB2 and KIAA0100, we removed the 11 of 142 H/L samples and 30 of 452 TCGA White tumor samples that were categorized as expression outliers in ERBB2 and re-did the robust standardization of KIAA0100 and compared the frequency difference. Eight of 135 H/L and two of 422 TCGA White tumors had overexpression of KIAA0100; the frequency difference is highly significant (P = 0.00029, Fisher exact test).
Figure 5.
Expression outliers and copy-number gain in KIAA0100. A, Distribution of gene expression and GISTIC2-thresholded copy-number scores in KIAA0100 for 146 breast tumor samples from H/L and 452 breast tumor samples from TCGA non-Hispanic Whites. The y-axis is standardized gene expression values (Z-scores) estimated robustly based on the corrected MAD. Red and blue dashed lines represent Z-score of 3 and −3, respectively. B, Distribution of proportion of high-level copy-number gain for 950 genes spanning the six amplified regions of 17q11.2, 17q12, 17q21.33, 17q23.1, 17q23.3, and 17q25.1. y-axis is the percentage of the 146 H/L samples with GISTIC2-thresholded copy-number score of 2; x-axis is genomic boundaries (Chr17: 21431570 – 81188573, hg19) for the six significantly amplified regions determined by GISTIC2. The vertical lines mark the genomic locations of KIAA0100 (BCOX1, 17q11.2) at Chr17:26941457 – 26972177, ERBB2 (17q12) at Chr17: 37844336 – 37873910, MIR4728 (microRNA 4728, 17q12) at Chr17: 37882747 – 37882814, ZNF652 (17q21.33) at Chr17: 47366567 – 47439476, PTRH2 (17q23.1) at Chr17: 57774666 – 57784959, DDX5 (17q23.3) at Chr17: 62494371 – 62503156, and UBE2O (17q25.1) at Chr17: 74385612 – 74449288.
To examine potential mechanisms underlying the outlying expression of KIAA0100, we first tested the association of KIAA0100 high-level copy-number gain and 920 germline SNP variants within ±1 Mb of this gene, adjusting for genome-wide CNV rate, tumor stage, and PAM50 subtype. We identified marginal evidence of association for a set of SNPs 3′ of KIAA0100 (lowest P value = 0.01 from logistic regression, FDR = 0.33). Then, we performed cis-eQTL analysis with the same 920 SNPs for KIAA0100 and observed significant associations (lowest P value = 8.61 × 10−5, beta coefficient = 0.245, and FDR = 0.017) for the same set of 3′ SNPs, while adjusting for RNA-seq batch, tumor stage, and PAM50 subtype as covariates (Supplementary Table S9). When including CNV as an additional covariate, the associations were attenuated (lowest P value = 1.86 × 10−3, beta coefficient = 0.137, and FDR = 0.40; Supplementary Table S9). There was no significant association of global or locus-specific IA ancestry with the cis-eQTL SNPs, KIAA0100 expression or copy-number gain.
Discussion
We analyzed tumor-germline sequencing data combined with RNA-seq data from 146 tumors from 140 self-identified H/L recruited from a single center in the Los Angeles region. As expected, the majority were of mixed European and IA ancestries. Because TCGA has extremely limited samples of breast cancer from H/L and particularly of H/L of mixed IA ancestry, our report fills a critical gap in the landscape of somatic mutations and CNAs in this increasing U.S. population. Together, our analyses and the recent article focused on Mexican women living in Mexico (19) substantially enhance the data in the public domain for women of H/L heritage.
The most commonly mutated gene in our population was PIK3CA, which is the most commonly mutated gene in TCGA White samples. For women with advanced ER+/HER2− breast cancers, alpelisib is a currently approved therapy, and our results suggest that this therapy should be useful in a large fraction of H/L women. The Romero-Cordoba and colleagues study identified a high frequency (8%) of the E17K-activating AKT1 mutation, indicating such women may benefit from AKT inhibitors. We only identified two tumors with mutations in AKT1 and only one with the E17K mutation. The difference between our results and those of Romero-Cordoba may be due to chance, differences in selection criteria between the two cohorts, and/or differences in environmental exposures between the two cohorts. Because the ancestry of our population is similar, it is unlikely that the differences we observed are due to germline-genetic differences between the two cohorts.
We performed analyses of the somatic mutational signatures and compared them to TCGA dataset. Our analysis identified COSMIC signature 16 (contribution > 5%) in a significant fraction of tumors (27.4%) in our dataset with similar rates in the data from Romero-Cordoba and colleagues who analyzed breast tumors from Mexican patients. Because Romero-Cordoba and colleagues used a contribution cutoff in their mutation-signature-analysis pipeline, they did not report this signature. However, in our analysis, we implemented the non-negative matrix factorization algorithm and no contribution cutoff was applied such that signature 16 was observed. There were significantly lower rates of this signature in TCGA White women (P < 0.001). We do not believe our finding is a technical artifact from FFPE because this signature was found in frozen tissue in the Romero-Cordoba and colleagues data. No known genetic or environmental exposures that predispose to this signature have been reported and prior studies have not found this mutational signature in breast cancer, although it has been reported to be common in liver cancers (46).
Other COSMIC signatures were the same as those previously reported in TCGA. We found signatures 2 and 13 associated with APOBEC loss as a relatively common finding, associated with HER2-amplified tumors and specifically with the germline APOBEC copy-number variant similar to previous reports (47). The common APOBEC3A/B 29.5 kbp germline deletion (3-fold more common in H/L breast cancer cases than White breast cancer cases) results in the fusion of APOBEC3A and the 3′-UTR (untranslated region) of APOBEC3B (48). Consistent with these results, we also found that presence of the deletion is more common in IA ancestry chromosomes among the H/L women in our study. This fusion generates a more stable APOBEC3A mRNA, resulting in increased expression of APOBEC3A, higher overall mutation burden, and a higher OR of developing breast cancer (49, 50). We also found signature 3, associated with defects in HRR as a common signature, which is overrepresented in basal-like tumors as reported previously (46, 51).
Our copy-number analyses identified copy-number gains, that is, 1q, 8q, 17q, which are common in breast cancer in other populations (1, 2). We also identified several known CNAs, which were recurrently gained in our dataset. In combined analysis of CNAs and gene expression, we identified KIAA0100 (BCOX1) as a recurrently amplified region with high gene expression, which was more common in tumors from H/L than tumors from White women in TCGA. KIAA0100 was originally identified in a screen for genes that were more frequently found in breast tumor than in normal breast tissue (52) and increased expression was associated with poor prognosis (52, 53). Knockdown of KIAA0100 by siRNA in the breast cancer cell line MDA-MB-231 reduced cell aggregation, reattachment, cell metastasis, and invasion (54). Thus, KIAA0100 may be of interest for further study in understanding the biology of tumors in H/L and stratifying women for risk of recurrence. We investigated whether genetic ancestry or SNPs at the locus were associated with expression and or genomic amplification. We found no association with ancestry, but we did identify some SNPs in cis associated with both expression and copy-number amplification. One interpretation of these results is that the alleles associated with higher expression are more likely amplified as that has a selective advantage for the tumor. Such cis interactions have been detected in prostate cancer, for example, where variants associated with increased TMPRSS2 expression are associated with TMPPRS2:ERG fusion (55). Testing these hypotheses in larger datasets should help clarify the role of KIAA0100 in breast cancer in H/L women.
The differences we observed between H/L and White breast tumors with higher prevalence of signature 16 and more KIAA0100 amplifications with high levels of gene expression may be due to differences in the genetic and/or environmental factors underlying breast cancer susceptibility between White and H/L women. However, we cannot rule out technical factors related to the sequencing or informatics or cohort selection effects. We did not identify any association between individual ancestry and either of these factors as might be expected if the effect were related to genetic variants that are more common in one ancestral population. However, we note that the number of individuals with signature 16 or KIAA0100 amplification and high expression is small and we were likely underpowered to see associations with individual ancestry. Associations with individual ancestry have been observed with Her2 amplification among Latinas (18) and with EGFR mutation in lung cancer among Latinos (7). However, the sizes in these studies are both greater than 1,000 individuals and the somatic events that they test are both common (>15%). Therefore, it is likely that testing associations with individual ancestry and signature 16 and KIAA0100 will require much larger sample sizes.
Expanding somatic profiling of breast cancer to H/L women identified new potentially important somatic events including signature 16 and KIAA0100 overexpression. Although these events were significantly more common in H/L women, the signature and/or specific somatic mutation or aberration should be the focus of future studies for effects on prognosis or targeted therapies. Similar to EGFR in lung cancer, where ancestry is strongly associated with mutations, the mutation is the key marker of response to EGFR targeted therapy rather than ancestry (56).
Our study has several limitations. We included only women who did not have neoadjuvant therapy prior to surgical resection. We chose this subset of women to avoid effects possibly induced by neoadjuvant chemotherapy such as new mutations and/or selection for resistant subclones. However, because neoadjuvant therapy is more likely to be given to patients with large tumors and/or tumors with poor prognosis (57), tumors included in our study may have some differences in comparison with prior studies due to these selection criteria. For example, because most triple-negative breast tumors are first treated with neoadjuvant therapy, the proportion of triple-negative tumors in our study was lower than reported previously (58). Our analysis of tumor CNAs was based on WES data. Although WES and other forms of targeted sequencing are used for CNA analysis, it makes it difficult to conduct one-to-one comparisons with array-based or whole genome sequencing–based analyses. Therefore, we limited our analyses to copy-number events that also demonstrated gene expression differences across populations. Finally, although our study substantially increases the number of tumors analyzed by WES in H/L, the overall numbers are still substantially lower than in White women. In particular, we are likely underpowered to discover low frequency, ethnic and/or ancestry-specific drivers that may be unique to this population. There also were too few recurrences and deaths for statistical analyses.
In summary, we conducted a comprehensive characterization of somatic mutations, CNAs, and gene expression in 146 breast tumors from 140 H/L from Los Angeles County, California. We found that COSMIC signature 16 was more common in our dataset and a recently published dataset of Mexican women living in Mexico, suggesting that this signature may be important in self-reported H/L women and potentially useful to understand differences at diagnosis and for outcome. The frequency of APOBEC3A/B germline deletions was significantly higher in H/L than Whites and associated with local IA ancestry. Finally, our combined CNA and gene expression analysis suggested that KIAA0100 may be a possible driver of breast cancer aggressiveness in a subset of our sample. These results provide a better understanding of the biology of breast cancer in H/L women.
Supplementary Material
Tables S1-S9
Acknowledgments
This work was funded by the NCI (R01CA184585, K24CA169004), the National Institute on Minority Health and Health Disparities Division of Intramural Research, and the California Initiative to Advance Precision Medicine (OPR18111). Research reported in this publication included work performed in the City of Hope Integrative Genomics Core and the Pathology Core supported by the NCI of the NIH under grant number P30CA033572. The content and views are solely the responsibility of the authors and should not be construed to represent the views of the NIH. S.L. Neuhausen and this research were partially funded by the Morris and Horowitz Families Professorship. C.D. Adams is supported by the National Heart, Lung, and Blood Institute (NHLBI T32HL007118) through the training Program in Molecular and Integrative Physiological Sciences at the Harvard T.H. Chan School of Public Health. L. Fejerman is supported by R01CA204797. J.N. Weitzel was supported by NIH RC4 CA153828; Breast Cancer Research Foundation (#20-172), and American Society of Clinical Oncology Conquer Cancer Research Professorship in Breast Cancer Disparities.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
S. Tao reports grants from NCI of NIH Cancer Center Support Grant (P30) awarded to Integrative Genomics and Bioinformatics Core of Beckman Research Institute of the City of Hope during the conduct of the study. J.N. Weitzel reports personal fees from Natera outside the submitted work. S.L. Neuhausen reports grants from NCI and grants from State of California during the conduct of the study. E. Ziv reports grants from NCI and California Initiative to Advance Precision Medicine during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
Y.C. Ding: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. H. Song: Formal analysis, writing–original draft. A.W. Adamson: Data curation, writing–original draft. D. Schmolze: Data curation, methodology. D. Hu: Formal analysis, writing–original draft. S. Huntsman: Data curation, formal analysis. L. Steele: Resources, data curation. C.S. Patrick: Data curation. S. Tao: Data curation. N. Hernandez: Data curation. C.D. Adams: Formal analysis. L. Fejerman: Data curation, formal analysis. K. Gardner: Data curation. A.M. Nápoles: Data curation. E.J. Perez-Stable: Data curation. J.N. Weitzel: Data curation. H. Bengtsson: Formal analysis. F.W. Huang: Formal analysis. S.L. Neuhausen: Conceptualization, resources, funding acquisition, methodology, writing–original draft, writing–review and editing. E. Ziv: Conceptualization, resources, formal analysis, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 5. Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 2004;10:8195–203. [DOI] [PubMed] [Google Scholar]
- 6. Arrieta O, Cardona AF, Martin C, Mas-Lopez L, Corrales-Rodriguez L, Bramuglia G, et al. Updated frequency of EGFR and KRAS mutations in NonSmall-cell lung cancer in Latin America: the Latin-American consortium for the investigation of lung cancer (CLICaP). J Thorac Oncol 2015;10:838–43. [DOI] [PubMed] [Google Scholar]
- 7. Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discov 2021;11:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020;580:93–9. [DOI] [PubMed] [Google Scholar]
- 9. Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell 2018;34:549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carrot-Zhang J, Chambwe N, Damrauer JS, Knijnenburg TA, Robertson AG, Yau C, et al. Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 2020;37:639–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin 2019;69:211–33. [DOI] [PubMed] [Google Scholar]
- 12. Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of breast cancer molecular features and survival by African and European ancestry in The Cancer Genome Atlas. JAMA Oncol 2017;3:1654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. John EM, Phipps AI, Davis A, Koo J. Migration history, acculturation, and breast cancer risk in Hispanic women. Cancer Epidemiol Biomarkers Prev 2005;14:2905–13. [DOI] [PubMed] [Google Scholar]
- 14. Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun 2014;5:5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hendrick RE, Monticciolo DL, Biggs KW, Malak SF. Age distributions of breast cancer diagnosis and mortality by race and ethnicity in US women. Cancer 2021;127:4384–92. [DOI] [PubMed] [Google Scholar]
- 16. Primm KM, Zhao H, Hernandez DC, Chang S. A contemporary analysis of racial and ethnic disparities in diagnosis of early-stage breast cancer and stage-specific survival by molecular subtype. Cancer Epidemiol Biomarkers Prev 2022;31:1185–94. [DOI] [PubMed] [Google Scholar]
- 17. Fejerman L, Hu D, Huntsman S, John EM, Stern MC, Haiman CA, et al. Genetic ancestry and risk of mortality among U.S. Latinas with breast cancer. Cancer Res 2013;73:7243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marker KM, Zavala VA, Vidaurre T, Lott PC, Vasquez JN, Casavilca-Zambrano S, et al. Human epidermal growth factor receptor 2-positive breast cancer is associated with indigenous American ancestry in Latin American women. Cancer Res 2020;80:1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romero-Cordoba SL, Salido-Guadarrama I, Rebollar-Vega R, Bautista-Pina V, Dominguez-Reyes C, Tenorio-Torres A, et al. Comprehensive omic characterization of breast cancer in Mexican-Hispanic women. Nat Commun 2021;12:2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Lun AT, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using rsubread and the edgeR quasi-likelihood pipeline. F1000Res 2016;5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao X, Rodland EA, Tibshirani R, Plevritis S. Molecular subtyping for clinically defined breast cancer subgroups. Breast Cancer Res 2015;17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li R, Qu H, Wang S, Wei J, Zhang L, Ma R, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018;34:2515–7. [DOI] [PubMed] [Google Scholar]
- 23. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davies RW, Flint J, Myers S, Mott R. Rapid genotype imputation from sequence without reference panels. Nat Genet 2016;48:965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rubinacci S, Ribeiro DM, Hofmeister RJ, Delaneau O. Publisher correction: efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat Genet 2021;53:412. [DOI] [PubMed] [Google Scholar]
- 27. Rubinacci S, Ribeiro DM, Hofmeister RJ, Delaneau O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat Genet 2021;53:120–6. [DOI] [PubMed] [Google Scholar]
- 28. Middlebrooks CD, Banday AR, Matsuda K, Udquim KI, Onabajo OO, Paquin A, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet 2016;48:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spear ML, Hu D, Pino-Yanes M, Huntsman S, Eng C, Levin AM, et al. A genome-wide association and admixture mapping study of bronchodilator drug response in African Americans with asthma. Pharmacogenomics J 2019;19:249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 2011;12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Browning BL, Tian X, Zhou Y, Browning SR. Fast two-stage phasing of large-scale sequence data. Am J Hum Genet 2021;108:1880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013;93:278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van der Auwera GA, O'Connor BD. Genomics in the cloud: using Docker, GATK, and WDL in Terra. 1st ed. Sebastopol (CA): O'Reilly Media; 2020. [Google Scholar]
- 35. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carter H, Marty R, Hofree M, Gross AM, Jensen J, Fisch KM, et al. Interaction landscape of inherited polymorphisms with somatic events in cancer. Cancer Discov 2017;7:410–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramroop JR, Gerber MM, Toland AE. Germline variants impact somatic events during tumorigenesis. Trends Genet 2019;35:515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li QY, Seo JH, Stranger B, McKenna A, Pe'er I, LaFramboise T, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 2013;152:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weitzel JN, Neuhausen SL, Adamson A, Tao S, Ricker C, Maoz A, et al. Pathogenic and likely pathogenic variants in PALB2, CHEK2, and other known breast cancer susceptibility genes among 1054 BRCA-negative Hispanics with breast cancer. Cancer 2019;125:2829–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lorenzato A, Olivero M, Patane S, Rosso E, Oliaro A, Comoglio PM, et al. Novel somatic mutations of the MET oncogene in human carcinoma metastases activating cell motility and invasion. Cancer Res 2002;62:7025–30. [PubMed] [Google Scholar]
- 46. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol 2016;17:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Komatsu A, Nagasaki K, Fujimori M, Amano J, Miki Y. Identification of novel deletion polymorphisms in breast cancer. Int J Oncol 2008;33:261–70. [PubMed] [Google Scholar]
- 49. Caval V, Suspene R, Shapira M, Vartanian JPandWain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun 2014;5:5129. [DOI] [PubMed] [Google Scholar]
- 50. Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, Shu XO, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis 2013;34:2240–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song J, Yang W, Shih Ie M, Zhang Z, Bai J. Identification of BCOX1, a novel gene overexpressed in breast cancer. Biochim Biophys Acta 2006;1760:62–9. [DOI] [PubMed] [Google Scholar]
- 53. Liu T, Zhang XY, He XH, Geng JS, Liu Y, Kong DJ, et al. High levels of BCOX1 expression are associated with poor prognosis in patients with invasive ductal carcinomas of the breast. PLoS One 2014;9:e86952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhong Z, Pannu V, Rosenow M, Stark A, Spetzler D. KIAA0100 modulates cancer cell aggression behavior of MDA-MB-231 through microtubule and heat shock proteins. Cancers 2018;10:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Emami NC, Kachuri L, Meyers TJ, Das R, Hoffman JD, Hoffmann TJ, et al. Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nat Commun 2019;10:3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 57. Thompson AM, Moulder-Thompson SL. Neoadjuvant treatment of breast cancer. Ann Oncol 2012;23:x231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zavala VA, Bracci PM, Carethers JM, Carvajal-Carmona L, Coggins NB, Cruz-Correa MR, et al. Cancer health disparities in racial/ethnic minorities in the United States. Br J Cancer 2021;124:315–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S9
Data Availability Statement
All tumor/normal WES and RNA-seq data and accompanying phenotypic and clinical/histologic data are deposited in dbGAP (dbGaP Study Accession: phs003218). TCGA RNA-seq and clinical data are publicly available at http://firebrowse.org/?cohort=BRCA&download_dialog=true. The Romero-Cordoba dataset (19) was downloaded from their supplementary material publicly available at https://doi.org/10.1038/s41467-021-22478-5. All other raw data are available upon request from the corresponding author.