Abstract
Automated medical diagnosis has become crucial and significantly supports medical doctors. Thus, there is a demand for inventing deep learning (DL) and convolutional networks for analyzing medical images. Dermatology, in particular, is one of the domains that was recently targeted by AI specialists to introduce new DL algorithms or enhance convolutional neural network (CNN) architectures. A significantly high proportion of studies in the field are concerned with skin cancer, whereas other dermatological disorders are still limited. In this work, we examined the performance of 6 CNN architectures named VGG16, EfficientNet, InceptionV3, MobileNet, NasNet, and ResNet50 for the top 3 dermatological disorders that frequently appear in the Middle East. An Image filtering and denoising were imposed in this work to enhance image quality and increase architecture performance. Experimental results revealed that MobileNet achieved the highest performance and accuracy among the CNN architectures and can classify disorder with high performance (95.7% accuracy). Future scope will focus more on proposing a new methodology for deep-based classification. In addition, we will expand the dataset for more images that consider new disorders and variations.
Keywords: Deep learning, convolutional neural networks (CNNs), classification, dermatological disorders, image classification, skin diseases
Introduction
Disease diagnosis and health care services have long been of high demands and faced significant challenges. Cost, time, and location barriers limit the quality of service, and patients cannot receive the services they acquire. Skin diseases affect approximately 1.9 billion people. 1 These kinds of diseases need follow-up with dermatologists, and most last for a long time, which makes the aforementioned barriers more challenging. Technology plays a crucial role in providing smarter and powerful solutions and can overcome the barriers. Artificial intelligence (AI), machine learning (ML), and deep learning (DL) offer a wide range of robust solutions.2-4 Specifically, dermatology conditions rely on morphological features and can be diagnosed throughout the patient’s images. This automated diagnosis is mainly based on recognizing visual patterns. Skin imaging tools and technology provide a variety of styles and designs that have become crucial for the clinical diagnosis of skin diseases.
AI has been developed in the last few decades for many different applications in medical science. It was developed as a rule-based induction for defined rules that are extracted from a set of observations and represent local patterns in the data. Medical image analysis is a crucial step in diagnosis. ML as a branch of AI has been utilized for the purposes of classification, object detection, segmentation, and image generation. Various methods and algorithms have been applied to medical image analysis, such as support vector machines (SVMs), decision trees, regression, and many other methods. 5 Moreover, artificial neural networks (ANNs), as AI domains, have been utilized in the last few decades for various areas of medical science.4,6 Although their use in the field of dermatology remains relatively limited, ANNs and other ML models were mainstream for quite a long time until the emergence of DL.7-13
Deep learning is a subset of machine learning methods that consists of multiple layers to extract higher-level features from the raw input data. It is based on networks that are capable of learning unsupervised data that are unstructured or unlabeled. Therefore, it is called deep neural learning or deep neural network. 14 It has been used for a variety of domains, including medicine. Litjens et al. 15 discussed DL methods for medical image analysis and covered its concepts, techniques, and architectures. Tian and Fu 16 reviewed 77 articles focused on DL in medical image classification, detection, segmentation, and generation. They divided it into supervised learning, weakly supervised learning, and unsupervised learning. They stated that the main difference between these 3 learning schemes is the proportion and granularity discrepancy of the annotated labels that drive the models. Common deep neural networks are mainly represented by convolutional neural networks (CNNs), recurrent neural networks (RNNs), and generative adversarial networks (GANs).
According to most of the literature on this subject, there are several challenges and limitations in current DL models applied to dermatology. Cullell-Dalmau et al. 17 mentioned the misclassification of images under process. A situation may occur if the image under process does not belong to any of the training classes; then, the model will classify the image into one of the other categories. They also discussed the importance of considering detailed metadata of images, which adds more challenges if a large number of images are used for training. As they stated, this further imposes the development of a quality test to automatically assess whether an image respects such quality standards; this is also supported by Haenssle et al. 18 Moreover, the quality of images and standardization of dermatological images affect the DL model performance since most dermatological diseases show common features that affect the quality of classification.
The rest of this paper is structured as follows. In Section 2, we present the DL techniques that have been used for skin cancer and some limited research on dermatological disorders. Section 3 describes the contributions of this work as we discuss the 6 main CNN architectures included in this analysis. Additionally, we describe our data used with the 6 CNN architectures. The method used for image enhancement and filtering is also discussed in this section. Section 4 discusses the results obtained. Section 5 discusses the limitations and challenges and analyzes the results. Finally, Section 6 concludes the paper and presents an outlook for future research.
Related Works
Recently, deep learning has made considerable progress in skin tumor and skin cancer detection and classification.19,20 Brinker et al. 21 reviewed CNNs that classify images of skin cancer and showed that the most common approach is to use a CNN pretrained by means of another large dataset and then optimize its parameters to the classification of skin cancers, which usually achieves the best performance with the currently available limited datasets. A significantly high proportion of studies in the field are concerned with skin cancer.14,22-24 Esteva et al. 19 first called attention to DCNN and proved that it could be significant in classifying skin disease, especially Keratinocyte carcinoma and melanoma with a level of competence comparable with that of board-certified dermatologists. This extensive interest is due to the availability of large datasets of skin cancer that are available publicly. Additionally, superior performance is achieved in the detection of skin cancer because the lesion area can easily be detected in images.
However, there is limited interest in dermatological diseases such as psoriasis, eczema, alopecia, and vitiligo. Most automated classification methods developed for dermatological diseases are based on supervised techniques. These methods are well known for ground-truth data and difficult to generalize since skin images vary based on skin tone, type, color, image lighting, and image contouring. 5 These difficulties increase the limitations of adopting it for dermatological imaging diseases. Moreover, the image features vary in this type of image, and the existing methods and techniques require different levels of engineering and filtering to reach a well-fit model. 25 However, convolutional deep learning methods have outperformed classical machine learning techniques for dermatological image classification. Yasir et al. 26 proposed a method based on computer vision techniques and image processing algorithms for feature extraction. They used an ANN to identify the diseases after training and testing. This method detected 9 different types of skin diseases with an accuracy rate of 90%. Kumar Patnaik et al. 27 examined the effect of InceptionV3, Inception-ResNet-v2 and MobileNet on colored skin images, and the architectures successfully predicted skin disease based on maximum voting from the 3 networks. Sae-Lim et al. 28 used MobileNet for skin lesion classification and compared it with the proposed modified MobileNet. DenseNet and ResNet architectures have also been examined with images collected using mobile phone cameras, and the accuracy with the models reached 80%. Facial disorder detection has been examined by Goceri. 25 Dermatological disease classification problems using CNNs have been discussed recently in Liu et al. 1 and Göçeri. 8
Material and Method
Dataset
The dataset was collected from different sources. The first source is Dermnet, which was founded by Thomas Habif, MD, in 1998 in Portsmouth, NH. It is one of the largest independent photodermatology sources for the purpose of medical education. It also provides descriptions of a wide range of skin conditions through innovative media. The second source is offered by the Department of Dermatology at the University of Iowa. They offer a repository of several disorders, such as cosmetic dermatology, skin remedies, and both photo and retinoid therapies. Images with the following diagnosis were included in this study: eczema, atopic, and psoriasis. We focus on these diagnoses since they are most common in the Middle East and Saudi Arabia.29-32
Image Filtering and noise reduction
The dataset contains a total of 6723 images, and 1934 are used for testing. The images were labeled with 3 classifications: eczema, atopic, and psoriasis. The image filtering process is important to note in image processing; therefore, the use of the original images affects the final results. Thus, image filtering is a crucial step in image processing, which may include modifying or enhancing the original images. In particular, filtering an image aims to emphasize certain features or remove other features. Image processing operations implemented with filtering include smoothing, sharpening, and edge enhancement. In this work, a block-matching and 3D filtering algorithm (BM3D) was adopted to enhance image dimensions. It is considered one of the current state-of-the-art methods for image denoising. This algorithm has a high capacity to achieve better noise removal results than other existing algorithms.33,34 Several studies have applied BM3D to deep learning for image classification.35,36 Figure 1 shows the result of applying BM3D filtering on a sample image. The images in the dataset were regenerated and resized to a scale of 224 224. The images have the same ratio scale and therefore do not need padding.
Figure 1.
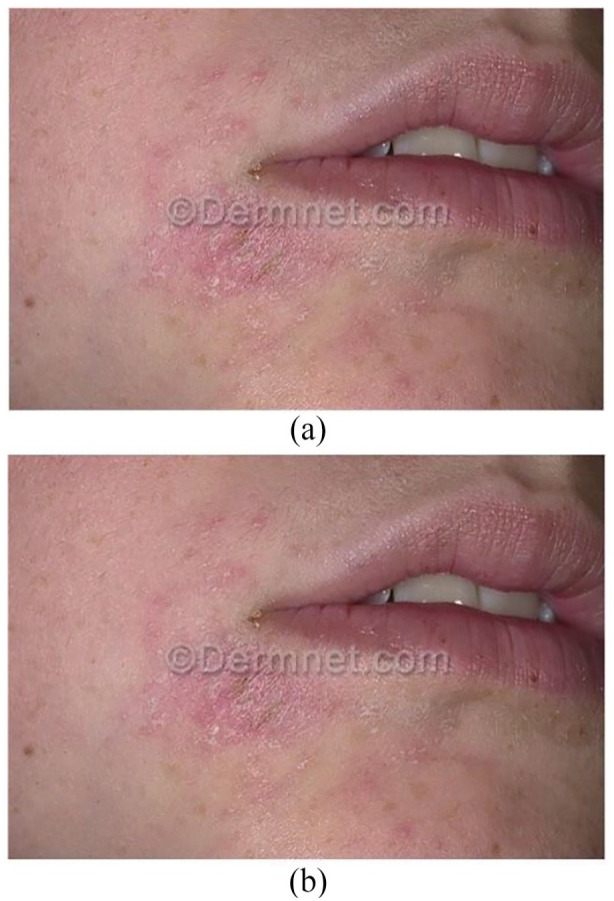
Result of applying BM3D filtering: (a) image before applying BM3D and (b) image after applying BM3D.
Deep learning models for classifications
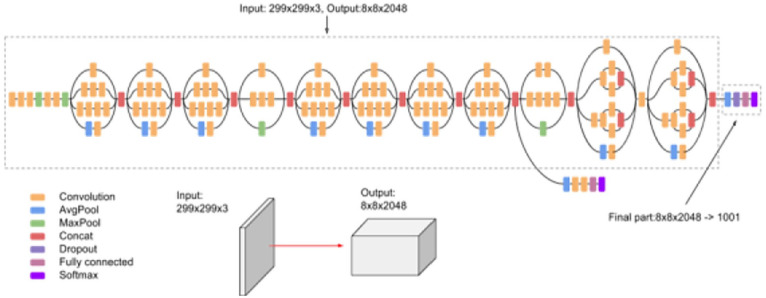
In this stage, classification was performed by employing different pretrained models for image classification. VGG is the first model considered in this study. It was proposed in 2014 by Simonyan and Zisserman 37 to test the efficiency and accuracy of layer depth on classification. The target was to evaluate the robustness of the model with layer weights up to 16. The model at that time was efficient and pretrained for other contexts of datasets. The second model is Inception-v3 (see Figure 2) introduced by Google, and it is the third version in the Inception family of deep convolutional architectures.38,39 It is based on parallel connected layers instead of a stacked layer architecture. 40 The Inception deep convolutional architecture was developed as GoogLeNet and later named Inception-v1. 38 Then, the Inception architecture was enhanced in Inception-v2 by including some batch normalization. Inception-v3 was launched after Inception-v2 with improvements in terms of factorization and fewer parameters that reached the 42-layer deep learning network. The third model applied in this study is residual networks or ResNet, which is presented as a network-in-network architecture. 41 The ResNet model is a block-based architecture where layers stacked on top of one another and form a block. The residual blocks (see Figure 3) are connected where each layer connects the input of a block with the output to ensure nonlinearity. The fourth model is MobileNets introduced by Howard et al. 42 as a class of CNNs with lightweight architectures. It has 2 layers, one for filtering and the other for combining. This architecture reduces the computation and model size in comparison with the standard convolution architecture. MobileNets, as stated in Howard et al. 42 performs depthwise convolution as a single filter on each input channel, and pointwise convolution performs a 1 × 1 convolution to combine the outputs of the depthwise convolution. We have included NasNet 43 as the fifth model in our evaluation. It stands for the neural architecture search network, which is designed to directly and automatically learn from the dataset to build the model architectures. This model is robust since it is applied in the search space where it looks for an architectural building block on a small dataset and then transfers the block to a larger dataset. The last model is EfficientNet, 44 which is based on scaling up CNNs using compound coefficients, where each dimension scales with fixed scaling coefficients. The optimized EfficientNet architecture relies on a baseline network built by performing a neural architecture search using the AutoML MNAS framework. 44
Figure 2.
Inception Model v3 source. 39
Figure 3.
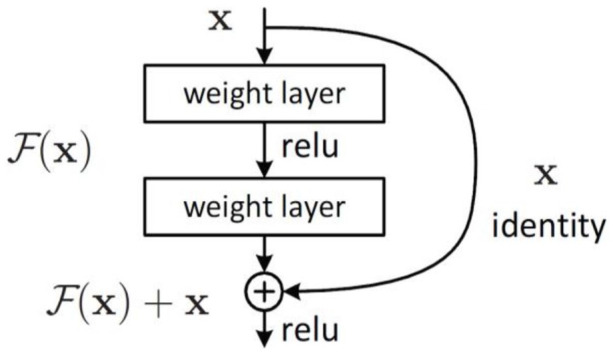
Residual learning: A building block. 41
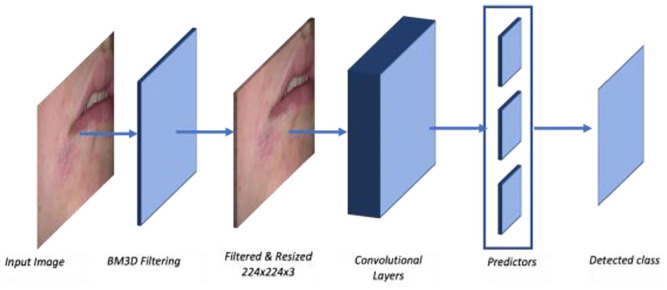
In summary, the classification process of CNN architectures for this work has passed through different stages. It is summarized in Figure 4. The process first takes the image for classification as input, and then BM3D filtering and denoising are applied. The filtered image is resized to 224 × 224 × 3 to prepare it for the CNN architecture model. Here, the architecture, the convolution layers in Figure 4, is one of the 6 CNN architectures included in this study. The results of the CNN architecture are sent to predictors to predict the image class. The last step is the result of the detected class.
Figure 4.
The process of image classification using the CNN architecture.
Experiment and Result
In this section, quantitative evaluations of the results obtained from CNN-based architectures are presented. In this experiment, we performed hyperparameter tuning to obtain a high-performing model, as shown in Table 1, which has 50 epochs, 10 steps per epoch and 5 validation steps, and Table 2, which has 50 epochs, 10 steps per epoch, and 10 validation steps. Finally, in Table 3, we increased the number of epochs to 100 with 10 steps per epoch and 5 steps to the validation. Figure 5 shows a sample of the resulted lesions for the 3 diseases.
Table 1.
Accuracy obtained by the CNN-based architectures applied in dermatology dataset at epochs = 50, val_step = 5, steps = 10.
| Models | Accuracy values | |
|---|---|---|
| Training (%) | Testing (%) | |
| VGG16 | 69.1 | 67.6 |
| EfficientNet | 84.6 | 88.1 |
| InceptionV3 | 76.1 | 69.9 |
| MobileNet | 95.4 | 92.8 |
| NasNet | 91.7 | 72.1 |
| ResNet50 | 82.5 | 85.1 |
Table 2.
Accuracy obtained by the CNN-based architectures applied in dermatology dataset at epochs = 50, val_step = 10, steps = 10.
| Models | Accuracy values | |
|---|---|---|
| Training (%) | Testing (%) | |
| VGG16 | 65.1 | 69.6 |
| EfficientNet | 81.8 | 89.2 |
| InceptionV3 | 68.2 | 68.4 |
| MobileNet | 96.1 | 93.2 |
| NasNet | 86.3 | 78.1 |
| ResNet50 | 78.9 | 88.5 |
Table 3.
Accuracy obtained by the CNN-based architectures applied in dermatology dataset at epochs = 100, val_step = 5, steps = 10.
| Models | Accuracy values | |
|---|---|---|
| Training (%) | Testing (%) | |
| VGG16 | 71.4 | 76.3 |
| EfficientNet | 84.2 | 91.3 |
| InceptionV3 | 71.7 | 72.1 |
| MobileNet | 99.3 | 95.7 |
| NasNet | 96.7 | 80.3 |
| ResNet50 | 70.9 | 91.1 |
Figure 5.
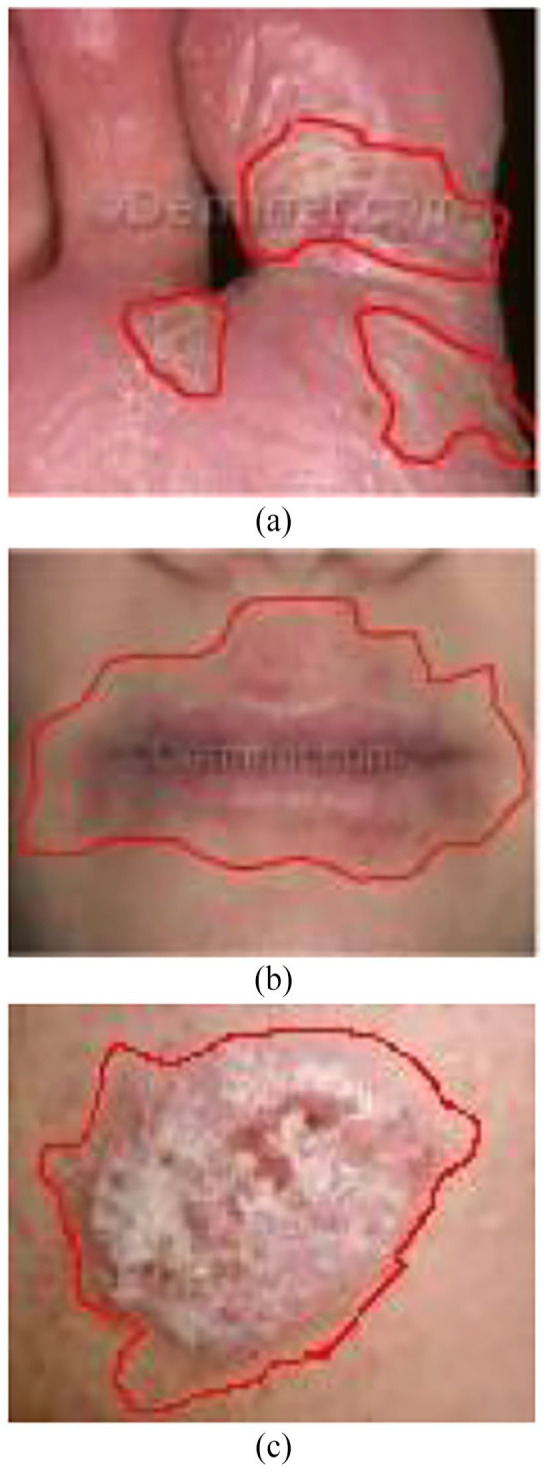
Sample of the resulted lesions for the 3 diseases: (a) eczema lesion, (b) atopic lesion, and (c) psoriasis lesion.
Evaluation measures
The evaluation process of the pretrained models designed in this work considered some evaluation measures that have been extensively applied to measure the quality of the models. The results obtained from the pretrained models were evaluated according to accuracy, precision and sensitivity values. Equations (1)-(3) show how to calculate the values of these measures:
| (1) |
| (2) |
| (3) |
Where TP (true positive) is the number of images correctly classified with the correct disease, FP (false positive) is the number of images classified with the incorrect disease, TN (true negative) is the number of images correctly classified with no disease, and FN (false negative) is the number of images falsely classified with the correct disease. We included the F1 score, which is the harmonic mean of the precision and recall, to measure the performance of the classification. Matthew’s correlation coefficient (MCC) was also included to evaluate the performance of the models. The F1 score and MCC were computed with equations (4) and (5):
| (4) |
| (5) |
Evaluations for CNN-based architectures
In this work, we performed an image classification problem for dermatology diseases. A number of pretrained CNN-based architectures have been used. To measure the efficiency and compare the performance of the architectures, we performed some evaluations. The 6 models were evaluated using different hyperparameters and the same image sets in the training and testing stages. All networks were tested using the Python Keras interface for ANNs with TensorFlow at the backend. The accuracy of all tested hyperparameters is shown in Tables 1 to 3. The results include the comparison of these networks in terms of accuracy, precision, F1 score, MCC and sensitivity values at different hyperparameters, as shown in Tables 4 to 6. The networks were also tested considering each dermatological disease included in this study, atopic, psoriasis, and eczema. The result is shown in Table 7.
Table 4.
Average accuracy, precision, F1 score, MCC, and specificity values obtained by the results of the CNN-based architectures applied at epochs = 50, val_step = 5, steps = 10.
| Models | Accuracy (%) | Precision (%) | F1 score (%) | MCC (%) | Sensitivity (%) |
|---|---|---|---|---|---|
| VGG16 | 67.6 | 67.6 | 68.1 | 66.4 | 67.1 |
| EfficientNet | 88.1 | 89.3 | 87.3 | 87.6 | 87.3 |
| InceptionV3 | 69.9 | 69.4 | 70.4 | 68.5 | 68.7 |
| MobileNet | 92.8 | 92.2 | 92.6 | 93.3 | 91.5 |
| NasNet | 72.1 | 73.2 | 74.1 | 71.1 | 73.2 |
| ResNet50 | 85.1 | 85.2 | 84.6 | 83.9 | 84.9 |
Table 6.
Average accuracy, precision, F1 score, MCC, and specificity values obtained by the results of the CNN-based architectures applied at epochs = 100, val_step = 10, steps = 10.
| Models | Accuracy (%) | Precision (%) | F1 score (%) | MCC (%) | Sensitivity (%) |
|---|---|---|---|---|---|
| VGG16 | 76.3 | 78.1 | 77.2 | 76.3 | 77.1 |
| EfficientNet | 91.3 | 92.1 | 90.5 | 91.2 | 91.6 |
| InceptionV3 | 72.1 | 71.2 | 70.1 | 71.4 | 71.8 |
| MobileNet | 95.7 | 94.3 | 94.8 | 95.4 | 96.1 |
| NasNet | 80.3 | 82 | 82.4 | 84.1 | 83.6 |
| ResNet50 | 91.1 | 90.3 | 91.2 | 90.3 | 92.4 |
Table 7.
Precision, F1 score and Sensitivity values obtained from each CNN-based architectures for each disease.
| Model | Disease | Measures | ||
|---|---|---|---|---|
| Precision (%) | F1 score (%) | Sensitivity (%) | ||
| VGG16 | Atopic | 78 | 71 | 49 |
| Eczema | 76 | 82 | 83 | |
| Psoriasis | 56 | 61 | 62 | |
| EfficientNet | Atopic | 98 | 77 | 59 |
| Eczema | 86 | 86 | 97 | |
| Psoriasis | 68 | 69 | 65 | |
| InceptionV3 | Atopic | 73 | 71 | 69 |
| Eczema | 68 | 79 | 88 | |
| Psoriasis | 83 | 62 | 50 | |
| MobileNet | Atopic | 82 | 86 | 80 |
| Eczema | 86 | 83 | 79 | |
| Psoriasis | 86 | 84 | 95 | |
| NasNet | Atopic | 70 | 74 | 78 |
| Eczema | 86 | 89 | 92 | |
| Psoriasis | 88 | 78 | 70 | |
| ResNet50 | Atopic | 62 | 69 | 77 |
| Eczema | 67 | 64 | 62 | |
| Psoriasis | 75 | 60 | 50 | |
Table 5.
Average accuracy, precision, F1 score, MCC, and specificity values obtained by the results of the CNN-based architectures applied at epochs = 50, val_step = 10, steps = 10.
| Models | Accuracy (%) | Precision (%) | F1 score (%) | MCC (%) | Sensitivity (%) |
|---|---|---|---|---|---|
| VGG16 | 69.6 | 69.1 | 68.7 | 69.3 | 67.8 |
| EfficientNet | 89.2 | 88.3 | 89.1 | 88.7 | 89.3 |
| InceptionV3 | 68.4 | 67.8 | 67.4 | 68.2 | 68.1 |
| MobileNet | 93.2 | 92.1 | 92.5 | 93.1 | 92.3 |
| NasNet | 78.1 | 77.4 | 78.4 | 77.1 | 78.2 |
| ResNet50 | 88.5 | 88.3 | 87.6 | 87.9 | 88.7 |
Discussion
This work aims to examine and better understand the effect of DL on dermatological disorders. Most current works are directed to skin cancer classification based on DL. The challenge in medical analysis and dermatology in particular is in the availability of data themselves. It has been noted that DL architectures are data-driven where data are crucial. In addition, local data for Middle Eastern dermatological disorders are also limited. Accordingly, we limited the analysis to photographs of 3 classes of dermatological disorders, atopic, eczema, and psoriasis, which commonly appear in the Middle East. Six CNN architectures were examined to classify these classes of dermatological disorders and test their performance. To measure the performance, (i) data were collected with predetermined classes, (ii) image filtering and noise reduction were applied using BM3D,33-36 (iii) 6 pretrained models with hyperparameter tuning were used, and (iv) evaluation and testing been performed.
Comparative evaluations of performance are shown in Tables 1 to 3. In these evaluations, we base our discussion on the hyperparameter at epochs = 100, val_step = 5, steps = 10 where it achieved the maximum performance. The comparative evaluations of performance are shown in Table 3, where the MobileNet architecture 42 produced high accuracy for training (99.3%) and testing (95/7%). The loss value of the MobileNet architecture reached the minimum loss (0.09). This indicates that the MobileNet architecture produced the highest performance against the other 5 architectures. Table 6 shows the results of precision, F1 score, MCC and sensitivity values of the 6 architectures. The MobileNet architecture also produced maximum values in the other evaluation metrics. The precision value of MobileNet was 94.3%, which indicates that 94.3% of all the patients actually suffering from dermatological disorders were correctly classified. In the same context, Table 7 shows the precision, F1 score and sensitivity values of each disease class for all 6 architectures. It clearly demonstrates that the MobileNet architecture produced the highest values on each dermatological disorder for all evaluation metrics. Only the EfficientNet architecture produced higher values in atopic, while NasNet produced values for eczema and psoriasis close to those of MobileNet. This is because of the compound coefficient used in EfficientNet in scaling up networks and the structure of atopic photographs. The second highest performance was obtained by the EfficientNet architecture.
The aforementioned results and discussion are significant; however, finding the correlation between the 3 classes for our classification is also crucial. The MCC correlation coefficient computes the similarity variables. The higher the correlation between true and predicted values, the better the prediction. This metric is perfect for symmetric classification, where no class has higher importance than the other. In our analysis, the results show positive correlations in all architectures for all classes (see Table 6). The MobileNet architecture reaches 95.4%, which is very close to 1 and indicates perfect positive correlation. EfficientNet comes after it with 91.2% correlation, which indicates that the predicted class and the true class are strongly correlated.
Conclusion and Future Works
The interest of DL and CNN in the field of medical sciences has increased recently. Several studies and experiments have been performed in different disciplines, including dermatology. While DL and CNN have been successfully adopted in skin cancer, they tend to be limited in dermatological disorders such as atopic, eczema, and psoriasis. In this work, we measured the performance of 6 CNN architectures on a dataset of dermatological disorder images. It is known that DL architectures are data-driven where data are crucial and image augmentations have been applied in some works 45 to increase the number of images. We have not used an image augmentation in this work. The results show that the MobileNet architecture outperformed the other 5 architectures. EfficientNet was next in terms of accuracy and other evaluation metrics. Despite the surpassing performance of CNN architectures in terms of medical analysis in dermatology, they face various challenges because of their data-driven nature. This is increasingly challenging since dermatological disorders are based on image analysis and computer vision. In addition, most dermatological disorder images have common features, which makes the process of preprocessing crucial. In this work, image filtering and denoising were applied throughout BM3D noise removal and edge enhancement.
Future works for this analysis include different directions. Collaborations between computer science practitioners and dermatologists will open novel data-driven solutions toward dermatological disorder detection and diagnosis. This domain still requires further enhancements in terms of data availability, real clinical experiments, and robust automated diagnosis systems. One crucial point in adopting this kind of approach is the acceptance of patients and physicians. Patient privacy, ethical data, and trustworthiness are critical issues that lead to reluctance. Other enhancements may include powerful CNN architectures in computer vision for dermatological disorders. Communities of dermatologists and computer vision specialists must work together to achieve this goal. This kind of collaboration will further explore opportunities that are cost effective, remotely accessible, and accurate. DL and CNN have demonstrated the capability of achieving highly accurate diagnoses in the classification of dermatological disorders. However, real datasets are still limited, and most current data available are detected for medical student illustrations. To proceed and succeed in this direction, initiatives should take place to offer a large dermatological disorder dataset while preserving patient privacy. This will have a positive effect on the proposed algorithms and experiments that are tested in real life.
Acknowledgments
The author extends her appreciation to the Innovation and Emerging Technologies Center at Digital Government Authority for funding this work. She also would like to thank Ms. Fatima Alogayyel for her valuable support in the technical part and experiments.
Footnotes
Funding: This work was supported by the Innovation and Emerging Technologies Center at Digital Government Authority.
The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All work done by the single author Lulwah AlSuwaidan.
References
- 1. Liu Y, Jain A, Eng C, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. 2020;26:900-908. [DOI] [PubMed] [Google Scholar]
- 2. Goceri E, Songul C. Biomedical information technology: image based computer aided diagnosis systems. In: International Conference on Advanced Technologies, Antalaya, Turkey, 2018. [Google Scholar]
- 3. Goceri E. Formulas behind deep learning success. In: International Conference on Applied Analysis and Mathematical Modeling (ICAAMM2018), Istanbul, Turkey, 2018. [Google Scholar]
- 4. Goceri E, Goceri N. Deep Learning in Medical Image Analysis: Recent Advances and Future Trends. IADIS; 2017. [Google Scholar]
- 5. Thiyaneswaran B, Anguraj K, Kumarganesh S, Thangaraj K. Early detection of melanoma images using gray level co-occurrence matrix features and machine learning techniques for effective clinical diagnosis. Int J Imaging Syst Technol. 2021;31:682-694. [Google Scholar]
- 6. Goceri E, Martinez E. Artificial neural network based abdominal organ segmentations: a review. In: 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA). IEEE; 2015:1191-1194. [Google Scholar]
- 7. Goceri E. Skin disease diagnosis from photographs using deep learning. In: ECCOMAS Thematic Conference on Computational Vision and Medical Image Processing. Springer; 2019:239-246. [Google Scholar]
- 8. Göçeri E. Convolutional neural network based desktop applications to classify dermatological diseases. In: 2020 IEEE 4th International Conference on Image Processing, Applications and Systems (IPAS). IEEE; 2020:138-143. [Google Scholar]
- 9. Göçeri E. Impact of deep learning and smartphone technologies in dermatology: Automated diagnosis. In: 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA). IEEE; 2020:1-6. [Google Scholar]
- 10. Goceri E, Karakas AA. Comparative evaluations of cnn based networks for skin lesion classification. In: 14th International Conference on Computer Graphics, Visualization, Computer Vision and Image Processing (CGVCVIP), Zagreb, Croatia, 2020:1-6. [Google Scholar]
- 11. Goceri E. Capsule neural networks in classification of skin lesions. In: International Conferences Computer Graphics, Visualization, Computer Vision and Image Processing. 2021. [Google Scholar]
- 12. Goceri E. Diagnosis of skin diseases in the era of deep learning and mobile technology. Comput Biol Med. 2021;134:104458. [DOI] [PubMed] [Google Scholar]
- 13. Göçeri E. An application for automated diagnosis of facial dermatological diseases. İ.zmir Katip Çelebi Üniversitesi Sağlık Bilimleri Fakültesi Dergisi. 2021;6:91-99. [Google Scholar]
- 14. Fujisawa Y, Otomo Y, Ogata Y, et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. 2019;180:373-381. [DOI] [PubMed] [Google Scholar]
- 15. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [DOI] [PubMed] [Google Scholar]
- 16. Tian Y, Fu S. A descriptive framework for the field of deep learning applications in medical images. Knowl Based Syst. 2020;210:106445. [Google Scholar]
- 17. Cullell-Dalmau M, Otero-Viñas M, Manzo C. Research techniques made simple: deep learning for the classification of dermatological images. J Investig Dermatol. 2020;140:507-514. [DOI] [PubMed] [Google Scholar]
- 18. Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29:1836-1842. [DOI] [PubMed] [Google Scholar]
- 19. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goceri E. Automated Skin Cancer Detection: Where We Are and The Way to The Future. In: 2021 44th International Conference on Telecommunications and Signal Processing (TSP). IEEE; 2021:48-51. [Google Scholar]
- 21. Brinker TJ, Hekler A, Utikal JS, et al. Skin cancer classification using convolutional neural networks: systematic review. J Med Internet Res. 2018;20:e11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Magalhaes C, Mendes J, Vardasca R. The role of AI classifiers in skin cancer images. Skin Res Technol. 2019;25:750-757. [DOI] [PubMed] [Google Scholar]
- 23. Dascalu A, David EO. Skin cancer detection by deep learning and sound analysis algorithms: A prospective clinical study of an elementary dermoscope. EBioMedicine. 2019;43:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Demir A, Yilmaz F, Kose O. Early detection of skin cancer using deep learning architectures: resnet-101 and inception-v3. In: 2019 Medical Technologies Congress (TIPTEKNO). IEEE; 2019:1-4. [Google Scholar]
- 25. Goceri E. Deep learning based classification of facial dermatological disorders. Comput Biol Med. 2021;128:104118. [DOI] [PubMed] [Google Scholar]
- 26. Yasir R, Rahman MA, Ahmed N. Dermatological disease detection using image processing and artificial neural network. In: 8th International Conference on Electrical and Computer Engineering. IEEE; 2014:687-690. [Google Scholar]
- 27. Kumar Patnaik S, Singh Sidhu M, Gehlot Y, Sharma B, Muthu P. Utomated skin disease identification using deep learning algorithm. Biomed Pharmacol J. 2018;11:1429-1436. [Google Scholar]
- 28. Sae-Lim W, Wettayaprasit W, Aiyarak P. Convolutional neural networks using mobilenet for skin lesion classification. In: 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE). IEEE; 2019:242-247. [Google Scholar]
- 29. Alsuwaidan SN. Childhood psoriasis: analytic retrospective study in Saudi patients. J Saudi Soc Dermatol Dermatol Surg. 2011;15:57-61. [Google Scholar]
- 30. Binyamin ST, Algamal F, Yamani AN, et al. Prevalence and determinants of eczema among females aged 21 to 32 years in Jeddah city – Saudi Arabia. Our Dermatol Online. 2017;8:22-26. [Google Scholar]
- 31. Raddadi AA, Jfri A, Samarghandi S, et al. Psoriasis: Correlation between severity index (PASI) and quality of life index (DLQI) based on the type of treatment. J Dermatol Dermatol Surg. 2016;20:15-18. [Google Scholar]
- 32. Almohideb M. Epidemiological patterns of skin disease in Saudi Arabia: a systematic review and meta-analysis. Dermatol Res Pract. 2020;2020:5281957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abubakar A, Zhao X, Li S, Takruri M, Bastaki E, Bermak A. A block-matching and 3-D filtering algorithm for Gaussian noise in DoFP polarization images. IEEE Sens J. 2018;18:7429-7435. [Google Scholar]
- 34. Zheng Z, Chen W, Zhang Y, Chen S, Liu D. Denoising the space-borne high-spectral-resolution lidar signal with block-matching and 3D filtering. Appl Opt. 2020;59:2820-2828. [DOI] [PubMed] [Google Scholar]
- 35. Yang D, Sun J. Bm3d-net: A convolutional neural network for transform-domain collaborative filtering. IEEE Signal Process Lett. 2018;25:55-59. [Google Scholar]
- 36. Huang S, Zhou P, Shi H, Sun Y, Wan S. Image speckle noise denoising by a multi-layer fusion enhancement method based on block matching and 3D filtering. Imaging Sci J. 2019;67:224-235. [Google Scholar]
- 37. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 38. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2015:1-9. [Google Scholar]
- 39. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2016:2818-2826. [Google Scholar]
- 40. Brodzicki A, Jaworek-Korjakowska J, Kleczek P, Garland M, Bogyo M. Pre-Trained deep convolutional neural network for clostridioides difficile bacteria cytotoxicity classification based on fluorescence images. Sensors. 2020;20:6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2016:770-778. [Google Scholar]
- 42. Howard AG, Zhu M, Chen B, et al. Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704-04861. [Google Scholar]
- 43. Zoph B, Vasudevan V, Shlens J, Le QV. Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2018:8697-8710. [Google Scholar]
- 44. Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning. PMLR; 2019:6105-6114. [Google Scholar]
- 45. Goceri E. Image augmentation for deep learning based lesion classification from skin images. In: 2020 IEEE 4th International Conference on Image Processing, Applications and Systems (IPAS). IEEE; 2020:144-148. [Google Scholar]