ABSTRACT
Host genetic polymorphisms are recognized as a critical determinant of diversity in clinical symptoms of Coronavirus disease 2019 (COVID-19). Accordingly, this study aimed to determine possible associations between single nucleotide polymorphisms (SNPs) in 37 candidate genetic variants and clinical consequences of COVID-19 – especially long-term symptoms, Long COVID. A total of 260 COVID-19 patients, divided into mild (n = 239) and severe (n = 21) and further categorized based on the presence of Long COVID (no, n = 211; yes, n = 49), were recruited. Genotyping of selected polymorphisms responsible for viral entry, immune response, and inflammation was performed using MassARRAY system. Out of 37 SNPs, 9 including leucine zipper transcription factor like-1 (LZTFL1) rs10490770 C allele, LZTFL1 rs11385942 dupA allele, nicotinamide adenine dinucleotide synthetase-1 (NADSYN1) rs12785878 TT genotype, plexin A-4 (PLXNA4) rs1424597 AA genotype, LZTFL1 rs17713054 A allele, interleukin-10 (IL10) rs1800896 TC genotype and C allele, angiotensin converting enzyme-2 (ACE2) rs2285666 T allele, and plasmanylethanolamine desaturase-1 (PEDS1) rs6020298 GG genotype and G allele were significantly associated with an increased risk of developing Long COVID, whereas interleukin-10 receptor subunit beta (IL10RB) rs8178562 GG genotype was significantly associated with a reduced risk of Long COVID. Kaplan-Meier curve displayed that the above gene polymorphisms were significantly associated with cumulative rate of Long COVID occurrence. Polymorphisms in LZTFL1 rs10490770, LZTFL1 rs11385942, LZTFL1 rs17713054, NADSYN1 rs12785878, PLXNA4 rs1424597, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 appear to be genetic factors involved in development of Long COVID.
KEYWORDS: Genetic polymorphism, SARS-CoV-2, COVID-19, Long COVID, COVID-19 severity
Introduction
Coronavirus disease 2019 (COVID-19), a novel infectious disease initially detected in China near the end of 2019, is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which remains a major global public health problem because of its social and economic burdens [1]. Although SARS-CoV-2 is extremely infectious, the severity of its clinical manifestations varies considerably, ranging from asymptomatic or moderately symptomatic to life-threatening complications and death [2]. It has been reported that approximately 15% of COVID-19 patients develop severe form, which can proceed to pneumonia, respiratory failure, kidney injury, multiorgan dysfunction, and eventual death [3, 4]. In addition to this, more than 200 million COVID-19 patients worldwide reportedly present with long-term symptoms, commonly known as Long COVID defined as the persistence or emergence of new symptoms that manifest three months following the initial SARS-CoV-2 infection and endure for a minimum of two months without any other identifiable cause [5]. This condition results in a broad range of clinical manifestations including chest pain or tightness, cough, fatigue or breathlessness, ageusia and anosmia, headache, insomnia, anxiety, and depression, which are highly heterogeneous [6]. It is well recognized that variations in severity and long-term symptoms of COVID-19 can be entirely attributed to established risk factors, such as advanced age, male gender, and the presence of comorbidities including diabetes, obesity, hypertension, and cardiovascular disease [7]. However, severe outcomes and Long COVID have also been observed in young patients without comorbidities [8], thereby indicating that additional risk factors, in particular genetic predisposition, may potentially contribute to variations in clinical consequences of COVID-19.
Genetic variants are well recognized as a contributor to susceptibility to a variety of infectious diseases [9,10]. In the context of viral respiratory infection, multiple genetic polymorphisms, especially single nucleotide polymorphisms (SNPs), have been reportedly associated with susceptibility or resistance to SARS-CoV-2 infection [11–14]. More specifically, polymorphisms in genes responsible for viral binding and entry into host cells (ACE2, angiotensin converting enzyme-2 and TMPRSS, transmembrane serine protease) have been extensively reported to be associated with susceptibility to SARS-CoV-2 infection and COVID-19 severity [15,16], in addition to polymorphisms in genes relevant to innate and adaptive immune systems (TLRs, toll-like receptors; HLA, human leukocyte antigen class I and II; and cytokines/chemokines) [16,17]. Based on these premises, it is important to note that identification of genetic variations associated with severity and long-term symptoms of COVID-19 may pave the way for developing novel antiviral paradigms and provide insights into the ability to predict response to treatment and vaccination.
While the vast majority of previous studies focused on genetic determinants involved in susceptibility to SARS-CoV-2 infection and COVID-19 severity, influences of host genetic factors on long-term symptoms of COVID-19 are still relatively nascent and poorly understood. Accordingly, the objective of this study was to examine associations of selected 37 polymorphisms in genes responsible for viral entry as well as immune and inflammatory responses with clinical consequences of COVID-19 – especially long-term symptoms.
Materials and methods
The study protocol was approved by the ethical committee of the Faculty of Medicine Ramathibodi Hospital, Mahidol University (COA. MURA2021/264 Ref.2404) and carried out in compliance with the ethical standards outlined in the Declaration of Helsinki, The Belmont Report, CIOMS Guidelines and the International Conference on Harmonization in Good Clinical Practice (ICH-GCP). Prior to participation, all study subjects provided written informed consents.
Search strategy
To identify candidate SNPs for genotyping, systematic review search was undertaken. All relevant studies on associations between genetic polymorphisms and clinical consequences of COVID-19, published from inception and up to January 2022, were selected from the electronic databases: PubMed, Scopus, Cochrane, and Google Scholar websites. Public databases were examined with the following keywords: “coronaviruses,” “COVID-19,” “2019-nCoV,” “severe acute respiratory syndrome coronavirus 2,” “SARS-CoV-2,” “genetic polymorphism,” “mutation,” “single nucleotide polymorphism,” and “polymorphism.” The inclusion criteria included: (1) SNP was reported to be substantially related to susceptibility, severity, or mortality of COVID-19 in at least two studies, and (2) studies conducted on human subjects were selected with a higher priority. Exclusion criteria were as follows: (1) studies were conducted on non-human subjects, (2) no significant association between the disease severity and genetic polymorphism was reported, and (3) less than two studies reported on the importance of SNP in susceptibility to SARS-CoV-2 infection and COVID-19 severity. Both title and abstract were screened by two independent researchers. In the event of any dispute, a reciprocal agreement has been reached via discussion. Afterwards, the primary result was accomplished with data on genotypic and allelic frequencies in Thais for each SNP.
Study participants and sample collection
A total of 260 COVID-19 patients who had a positive real-time reverse transcription polymerase chain reaction (RT–PCR) test on a nasopharyngeal swab were recruited from Prachatipat Hospital. All COVID-19 patients were divided into mild (n = 239) and severe (n = 21) groups, as per World Health Organization (WHO) COVID-19 disease severity classification [18]. In the context of COVID-19 severity, COVID-19 patients with respiratory rate less than 24 per minute and oxygen saturation (SpO2) ≥ 94% were considered mild, while severe COVID-19 was evidenced by SpO2 < 94 mmHg, the need of invasive mechanical ventilation, and the presence of emergency signs, such as pneumonia, shock, or multiorgan failure. In terms of long-term symptoms of COVID-19, Long COVID, COVID-19 patients were further categorized into no Long COVID (n = 211) and Long COVID (n = 49) groups, according to the National Institute for Health and Care Excellence (NICE) definition of Long COVID [19]. In detail, Long COVID was defined as the presence of persistent/prolonged symptoms (constant, fluctuating, or relapsing) and/or functional disability following SARS-CoV-2 infection for at least four weeks after the onset of symptoms or the time of diagnosis in individuals in whom the infection has been self-reported, clinically diagnosed, and/or laboratory-confirmed.
Peripheral venous blood samples were drawn from all participants into ethylenediaminetetraacetic acid and kept instantly at −20°C till utilized. Clinical and demographic data including age, gender, body mass index (BMI), virus strains, and comorbidities were obtained from hospital records under the supervision of qualified medical professionals.
DNA extraction and SNPs genotyping
Genomic DNA was extracted from peripheral venous blood using QuickGene DNA Extraction Whole Blood Kit L (ADS Biotec Inc., USA), according to the manufacturer’s protocol. The quality and quantity of extracted DNA were both assessed by a nanodrop spectrometer (Thermo Fisher Scientific, Sunnyvale, CA, USA). The genomic DNA samples were stored at −20°C until genotyping. A total of 37 SNPs were selected from a systematic literature review and genotyped using the single nucleotide polymorphism detection with the iPLEX® assay on MassARRAY® system (Agena Bioscience, Inc., San Diego, CA, USA). Of 37 SNPs, 5 (13.51%) were in missense regions, 3 (8.11%) in coding regions, 3 (8.11%) in non-coding regions or upstream of gene regions, 3 (8.11%) in UTRs, 2 (5.41%) in downstream of gene regions, and 21 (56.76%) were intronic. The identity and origin of the DNA sample were concealed from laboratory staff.
Statistical analysis
All statistical analyses were executed using SPSS Statistics version 26.0 (SPSS Inc., Chicago, IL, USA). Comparisons in continuous variables represented as mean ± standard deviation (SD) were executed by Student’s t-test, while comparisons in categorical variables represented as numbers and percentages were accomplished using Chi-square test (χ2). Besides this, differences in genotypic and allelic frequencies between two groups were assessed using Chi-square test (χ2), in which odds ratio (OR) at a 95% confidence interval (CI) was employed to determine the strength of associations between genetic polymorphisms and susceptibility to severity and long-term symptoms of COVID-19. For multiple comparisons, the Bonferroni correction method was employed to adjust P-values. Furthermore, multivariate logistic regression analysis was undertaken to determine the independent associations, with adjustments for confounding factors including age, gender, BMI, and the presence of comorbidities. Additionally, Kaplan-Meier curves were constructed to determine whether polymorphisms in significant genes were associated with a cumulative incidence of Long COVID occurrence. For all analyses, a P-value <0.05 (based on a two-tailed test) was considered statistically significant.
Results
Systematic review
In the light of our systematic review, 26 out of 37 SNPs were reportedly associated with severity or mortality of COVID-19, including IL6-AS1 (interleukin-6 antisense RNA-1) rs1800796, IL6 rs1524107, OAS3 (2'-5'-oligoadenylate synthetase-3) rs10735079, DDR1 (discoidin domain receptor tyrosine kinase-1) rs4618569, VEPH1 (ventricular zone expressed PH domain containing-1) rs1840680, TMPRSS2 (transmembrane serine protease-2) rs2070788, VEPH1 rs2305619, IFITM3 (interferon-induced transmembrane protein-3) rs12252, ACE2 (angiotensin converting enzyme-2) rs2285666, PEDS1 (plasmanylethanolamine desaturase-1) rs6020298, KDR (kinase insert domain receptor) rs1870377, IL17A rs2275913, TNFRSF1B (tumour necrosis factor receptor superfamily member-1B) rs1061624, ACE2 rs2074192, IFNAR2 (interferon alpha and beta receptor subunit-2) rs2236757, IL10RB (interleukin-10 receptor subunit beta) rs8178562, CXCR2 (CXC motif chemokine receptor-2) rs1126579, FOXP4-AS1 (forkhead box P4 antisense RNA-1) rs1853837, TMPRSS2 rs2298659, IFIH1 (interferon-induced with helicase C domain-1) rs1990760, DPP9 (dipeptidyl peptidase-9) rs2109069, XCR1 (X-C motif chemokine receptor-1) rs35951367, LZTFL1 (leucine zipper transcription factor like-1) rs11385942, LZTFL1 rs10490770, NADSYN1 (nicotinamide adenine dinucleotide synthetase-1) rs12785878, IFI44 (interferon-induced protein-44) rs544893099. In addition to this, 11 SNPs were shown to be significantly associated with susceptibility to SARS-CoV-2 infection, including TMPRSS2 rs12329760, GC (GC vitamin D binding protein) rs7041, TLR3 (toll like receptor-3) rs3775290, RAPGEF1 (Rap guanine nucleotide exchange factor-1) rs12551879, IL17A rs3819025, PLXNA4 (plexin A-4) rs1424597, IL17F rs763780, TLL1 (tolloid like-1) rs17047200, IFNλ3 (interferon lambda-3) rs12979860, LZTFL1 rs17713054, and IL10 (interleukin-10) rs1800896. From this viewpoint, a total of 37 SNPs were selected as candidates for genotyping using MassARRAY system.
Demographic characteristics of study participants
Demographic characteristics of study subjects are summarized in Table 1. Of 260 COVID-19 patients, 239 (91.92%) were mild cases with a mean age of 46.67 ± 21.17 years, and 21 (8.08%) were severe cases with a mean age of 67.81 ± 20.90 years. A significant difference in mean age between mild and severe cases was observed, in which severe COVID-19 patients were older than mild COVID-19 patients (P < 0.001). In parallel with this, patients aged 60 years or over were significantly more common in the severe group (P = 0.001). Furthermore, male patients were significantly more common in the severe group and had a significantly greater risk of developing severe COVID-19 than female patients (P = 0.014). There were no significant differences in the prevalence of SARS-CoV-2 variants between mild and severe COVID-19 patients. In terms of vaccination status, unvaccinated COVID-19 patients showed a significantly higher risk of developing severe form than vaccinated patients (P < 0.001). Regarding the presence of comorbidities, including chronic diseases and diabetes were significantly more common in severe COVID-19 patients (P = 0.012, P < 0.001, respectively).
Table 1.
Baseline characteristics of COVID-19 patients based on COVID-19 severity.
| Variables | Severity | P-value | OR | 95% CI | |
|---|---|---|---|---|---|
| Mild | Severe | ||||
| Number | 239 (91.92%) | 21 (8.08%) | |||
| Age (years) | 46.67 ± 21.17 | 67.81 ± 20.90 | <0.001 | - | - |
| <60 | 156 (65.27%) | 52 (23.81%) | Reference | ||
| ≥60 | 82 (34.31%) | 16 (76.19%) | 0.001 | 6.088 | 2.154–17.209 |
| Gender | |||||
| Female | 148 (61.92%) | 7 (33.33%) | Reference | ||
| Male | 91 (38.08%) | 14 (66.67%) | 0.014 | 3.253 | 1.265–8.361 |
| BMI (kg/m2) | 25.07 ± 5.64 | 23 ± 7.33 | 0.061 | - | - |
| <25 | 107 (44.77%) | 6 (28.57%) | Reference | ||
| ≥25 | 132 (55.23%) | 15 (71.43%) | 0.158 | 2.027 | 0.760-5.402 |
| Virus strains | |||||
| Delta | 5 (2.09%) | 0 (0.00%) | Reference | ||
| Omicron | 234 (97.91%) | 21 (100.00%) | 0.999 | N/A | N/A |
| Omicron (BA.1) | 12 (5.02%) | 0 (0.00%) | - | - | - |
| Omicron (BA.2) | 137 (57.32%) | 12 (57.14%) | - | - | - |
| Omicron (BA.2.12.1) | 3 (1.26%) | 0 (0.00%) | - | - | - |
| Omicron (BA.2/BA.5) | 1 (0.42%) | 0 (0.00%) | - | - | - |
| Omicron (BA.4) | 5 (0.42%) | 1 (4.76%) | - | - | - |
| Omicron (BA.4/BA.5) | 2 (0.84%) | 0 (0.00%) | - | - | - |
| Omicron (BA.5) | 74 (30.96%) | 8 (38.10%) | - | - | - |
| Vaccination status | |||||
| No | 23 (9.62%) | 8 (38.10%) | <0.001 | 5.779 | 2.169–15.399 |
| Yes | 216 (90.38%) | 13 (61.90%) | Reference | ||
| Comorbidities | |||||
| Chronic diseases | |||||
| No | 140 (58.58%) | 6 (28.57%) | Reference | ||
| Yes | 99 (41.42%) | 15 (71.43%) | 0.012 | 3.535 | 1.325–9.430 |
| Diabetes mellitus | |||||
| No | 203 (84.94%) | 10 (47.62%) | Reference | ||
| Yes | 36 (15.06%) | 11 (52.38%) | <0.001 | 6.203 | 2.455-15.671 |
| Hypertension | |||||
| No | 184 (15.06%) | 13 (61.90%) | Reference | ||
| Yes | 55 (23.01%) | 8 (38.10%) | 0.128 | 2.059 | 0.812-5.222 |
| Infectious diseases | |||||
| No | 239 (100.00%) | 20 (95.24%) | Reference | ||
| Yes | 0 (0.00%) | 1 (4.76%) | 0.999 | N/A | N/A |
P-values marked with bold indicate statistically significant differences between the groups.
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; N/A, not available; OR, odds ratio.
In the regard of long-term symptoms of COVID-19, COVID-19 patients were further divided into the patients without Long COVID (n = 211, 81.15%) and those with Long COVID (n = 49, 18.85%). Baseline characteristics of COVID-19 patients with and without Long COVID are presented in Table 2. Considering demographic characteristics of COVID-19 patients based on the presence of Long COVID, mean age of the non-Long COVID group was 46.36 ± 21.94 years, and mean age of Long COVID cases was 57.06 ± 19.59 years. This difference was statistically significant (P = 0.001). More specifically, COVID-19 patients older than 60 years showed a significantly higher risk of developing Long COVID than younger patients (P = 0.001). Besides this, COVID-19 patients with BMI <25 kg/m2 exhibited a significantly increased risk of developing Long COVID-19, compared with those with BMI ≥25 kg/m2 (P = 0.034). On the other hand, there was no significant association of gender with Long COVID risk. Likewise, no significant differences in the prevalence of SARS-CoV-2 variants between COVID-19 patients with and without Long COVID were observed. Besides this, there was no significant association between vaccination status and risk of developing Long COVID. Instead, associations between the presence of comorbidities and risk of Long COVID were detected, in which COVID-19 patients having comorbidities including chronic diseases (P < 0.001), diabetes (P = 0.001), and hypertension (P < 0.001) showed a significantly higher risk of developing Long COVID than those without comorbidities.
Table 2.
Baseline characteristics of COVID-19 patients based on the presence of Long COVID.
| Variables | Long COVID | P-value | OR | 95% CI | |
|---|---|---|---|---|---|
| No | Yes | ||||
| Number | 211 (81.15%) | 49 (18.85%) | |||
| Age (years) | 46.36 (21.94%) | 57.06 (19.59%) | 0.001 | - | - |
| <60 | 143 (67.77%) | 18 (8.53%) | Reference | ||
| ≥60 | 67 (31.75%) | 31 (14.69%) | <0.001 | 3.676 | 1.920–7.036 |
| Gender | |||||
| Female | 120 (56.87%) | 35 (71.43%) | 0.064 | 1.896 | 0.963–3.731 |
| Male | 91 (43.13%) | 14 (28.57%) | Reference | ||
| BMI (kg/m2) | 24.63 (5.84%) | 25.91 (5.66%) | 0.166 | - | - |
| <25 | 85 (40.25%) | 28 (57.14%) | Reference | ||
| ≥25 | 126 (59.72%) | 21 (42.86%) | 0.034 | 0.506 | 0.27–0.949 |
| Virus strains | |||||
| Delta | 0 | 5 (10.20%) | Reference | ||
| Omicron | 211 | 44 (89.80%) | 0.999 | N/A | N/A |
| Omicron (BA.1) | 1 (0.47%) | 11 (22.45%) | - | - | - |
| Omicron (BA.2) | 116 (54.98%) | 33 (67.35%) | - | - | - |
| Omicron (BA.2.12.1) | 3 (1.42%) | 0 (0.00%) | - | - | - |
| Omicron (BA.2/BA.5) | 1 (0.47%) | 0 (0.00%) | - | - | - |
| Omicron (BA.4) | 6 (2.84%) | 0 (0.00%) | - | - | - |
| Omicron (BA.4/BA.5) | 2 (0.95%) | 0 (0.00%) | - | - | - |
| Omicron (BA.5) | 82 (38.86%) | 0 (0.00%) | - | - | - |
| Vaccination status | |||||
| No | 26 (12.32%) | 5 (10.20%) | 0.681 | 0.809 | 0.294–2.224 |
| Yes | 185 (87.68%) | 44 (89.80%) | Reference | ||
| Comorbidities | |||||
| Chronic diseases | |||||
| No | 132 (62.56%) | 14 (28.57%) | Reference | ||
| Yes | 79 (37.44%) | 35 (71.43%) | <0.001 | 4.177 | 2.117–8.242 |
| Diabetes mellites | |||||
| No | 181 (85.78%) | 32 (65.31%) | Reference | ||
| Yes | 30 (14.22%) | 17 (34.69%) | 0.001 | 3.205 | 1.586–6.479 |
| Hypertension | |||||
| No | 177 (83.87%) | 20 (40.82%) | Reference | ||
| Yes | 34 (16.11%) | 29 (59.18%) | <0.001 | 7.549 | 3.833–14.865 |
| Infectious diseases | |||||
| No | 210 (99.53%) | 49 (100.00%) | Reference | ||
| Yes | 1 (0.47%) | 0 (0.00%) | 1.000 | N/A | N/A |
P-values marked with bold indicate statistically significant differences between the groups.
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; N/A, not available; OR, odds ratio.
Genetic and allelic distributions of 37 SNPs in COVID-19 patients with different subgroups based on COVID-19 severity and Long COVID are detailed in Supplementary Tables 1 and 2.
SNPs associations with severe COVID-19
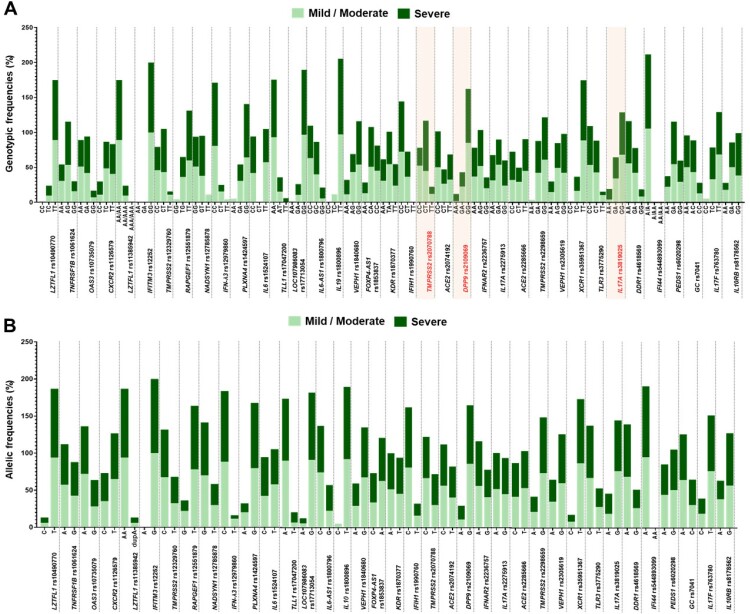
Overall, DPP9 rs2109069 showed a significant difference in genotypic distribution between mild and severe COVID-19 patients (P = 0.030), whereas multiple comparisons with adjustment of P-values by Bonferroni correction method revealed no significant difference in genotypic distribution of DPP9 rs2109069 between groups (Supplementary Table 3). We further stratified analyses based on different genetic models and also found that genotypic frequencies of 3 out of 37 SNPs were observed to be significantly different between mild and severe COVID-19 patients, including TMPRSS2 rs2070788, DPP9 rs2109069, and IL17A rs3819025 (Figure 1). Associations of genetic polymorphisms in TMPRSS2 rs2070788, DPP9 rs2109069, and IL17A rs3819025 with COVID-19 severity are detailed in Table 3.
Figure 1.
Genetic and allelic distributions between mild and severe COVID-19 patients. (A) Genetic distribution. (B) Allelic distribution. Fonts marked in red indicate statistically significant differences in genetic distribution between mild and severe COVID-19 patients.
Table 3.
Associations between polymorphisms in three particular genes and susceptibility to severe COVID-19.
| Genes | SNPs | Genotype/allele | Severity | Genetic models | Unadjusted model | Adjusted model | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mild | % | Severe | % | P-value | OR | 95% CI | P-value | aOR | 95%CI | ||||
| TMPRSS2 | rs2070788 | CC | 115 | 48.12 | 5 | 23.81 | CC vs. CT + TT | 0.040 | 0.337 | 0.120-0.949 | 0.197 | 0.499 | 0.174-1.434 |
| CT | 98 | 41.00 | 14 | 66.67 | CT vs. CC + TT | 0.028 | 2.878 | 1.120-7.390 | 0.278 | 1.723 | 0.645-4.604 | ||
| TT | 26 | 10.88 | 2 | 9.52 | TT vs. CC + CT | 0.848 | 0.862 | 0.190-3.915 | - | - | - | ||
| C | 328 | 68.62 | 24 | 57.14 | CT vs. CC | 0.027 | 3.286 | 1.143-9.447 | 0.205 | 0.499 | 0.170-1.462 | ||
| T | 150 | 31.38 | 18 | 42.86 | CC vs.TT | 0.509 | 0.565 | 0.104-3.076 | - | - | - | ||
| CT vs. TT | 0.432 | 1.857 | 0.397-8.692 | - | - | - | |||||||
| C vs. T | 0.13 | 0.61 | 0.321-1.157 | - | - | - | |||||||
| DPP9 | rs2109069 | AA | 3 | 1.26 | 2 | 9.52 | AA vs. GA + GG | 0.025 | 8.281 | 1.303-52.624 | 0.156 | 5.793 | 0.512-65.517 |
| GA | 49 | 20.50 | 4 | 19.05 | GA vs. AA + GG | 0.874 | 0.912 | 0.294-2.834 | - | - | - | ||
| GG | 187 | 78.24 | 15 | 71.43 | GG vs. AA + GA | 0.474 | 0.695 | 0.257-1.881 | - | - | - | ||
| A | 55 | 11.51 | 8 | 19.05 | AA vs. GA | 0.046 | 8.167 | 1.042-64.019 | 0.246 | 49.786 | 0.068-36,659.615 | ||
| G | 423 | 88.49 | 34 | 80.95 | AA vs. GG | 0.026 | 8.311 | 1.287-53.651 | 0.175 | 5.405 | 0.473-61.815 | ||
| GA vs. GG | 0.976 | 1.018 | 0.323-3.204 | - | - | - | |||||||
| A vs. G | 0.156 | 1.81 | 0.797-4.108 | - | - | - | |||||||
| IL17A | rs3819025 | AA | 9 | 3.77 | 3 | 14.29 | AA vs. GA + GG | 0.041 | 4.259 | 1.059-17.133 | 0.152 | 3.391 | 0.639-17.990 |
| GA | 77 | 32.22 | 6 | 28.57 | GA vs. AA + GG | 0.731 | 0.842 | 0.314-2.253 | - | - | - | ||
| GG | 153 | 64.02 | 12 | 57.14 | GG vs. AA + GA | 0.532 | 0.749 | 0.304-1.850 | - | - | - | ||
| A | 95 | 19.87 | 12 | 28.57 | AA vs. GA | 0.066 | 4.278 | 0.909-20.122 | - | - | - | ||
| G | 383 | 80.13 | 30 | 71.43 | AA vs. GG | 0.048 | 4.25 | 1.014-17.807 | 0.189 | 3.21 | 0.563-18.298 | ||
| GA vs. GG | 0.990 | 0.994 | 0.359-2.748 | - | - | - | |||||||
| A vs. G | 0.185 | 1.613 | 0.796-3.267 | - | - | - | |||||||
P-values marked with bold indicate statistically significant associations between genetic polymorphisms and susceptibility to severe COVID-19.
OR was adjusted for age, gender, vaccination status, and comorbidities.
Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; DPP9, dipeptidyl peptidase-9; IL17A, interleukin-17A; N/A, not available; OR, odds ratio; SNP, single nucleotide polymorphism; TMPRSS2, transmembrane serine protease-2.
Considering genetic polymorphism in TMPRSS2 rs2070788, CC genotype was significantly associated with a lower risk of severe COVID-19, compared with CT-plus-TT genotypes (OR = 0.34, 95% CI: 0.12, 0.95, P = 0.040). In contrast to this, CT genotype of TMPRSS2 rs2070788 was significantly associated with a higher risk of severe COVID-19, compared with CC-plus-TT genotypes and CC genotype (OR = 2.88, 95% CI: 1.12, 7.40, P = 0.028; OR = 3.29, 95% CI: 1.14, 9.45, P = 0.027, respectively).
Of genetic polymorphism in DPP9 rs2109069, AA genotype was significantly associated with an increased risk of severe COVID-19, compared with GA-plus-GG genotypes and GG genotype (OR = 8.28, 95% CI: 1.30, 52.62, P = 0.025; OR = 8.31, 95% CI: 1.29, 53.65, P = 0.026; respectively).
In the context of IL17A rs3819025 polymorphism, AA genotype was significantly associated with a greater risk of severe COVID-19 than GA-plus-GG genotypes (OR = 4.26, 95% CI: 1.06, 17.13, P = 0.041).
Given that age, gender, vaccination status, and the presence of comorbidities were all detected as contributors to severe COVID-19, multivariate logistic regression analysis with adjustments for the confounders was further performed to determine independent associations between genetic polymorphisms in particular SNPs and susceptibility to severe COVID-19. After adjusting for age, gender, and comorbidities, no significant associations of genetic polymorphisms in TMPRSS2 rs2070788, DPP9 rs2109069, and IL17A rs3819025 with susceptibility to severe COVID-19 were observed in COVID-19 patients, as revealed in Table 3.
SNPs associations with long-term symptoms of COVID-19
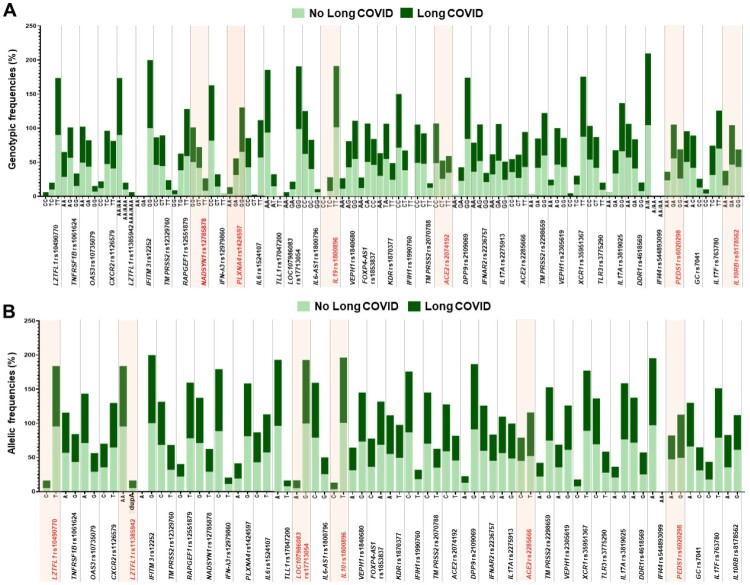
In terms of long-term symptoms of SARS-CoV-2 infection, 5 out of 37 SNPs were observed to have significant differences in genotypic distributions between COVID-19 patients with and without long-term symptoms of COVID-19, including LZTFL1 rs10490770 (P = 0.001), LZTFL1 rs11385942 (P = 0.001), LZTFL1 rs17713054 (P = 0.001), IL10 rs1800896 (P = 0.021), and PEDS1 rs6020298 (P = 0.045). After adjustment of P-values by the Bonferroni correction method, multiple comparison testing showed no significant differences in genotypic distributions of those SNPs between groups (Supplementary Table 3). In stratified analyses by seven genetic models, there were significant differences in genotypic and allelic distributions of 9 out of 37 SNPs between COVID-19 patients with and without long-term symptoms of COVID-19, including LZTFL1 rs10490770, LZTFL1 rs11385942, NADSYN1 rs12785878, PLXNA4 rs1424597, LZTFL1 rs17713054, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 (Figure 2). Associations of genetic polymorphisms in LZTFL1 rs10490770, LZTFL1 rs11385942, NADSYN1 rs12785878, PLXNA4 rs1424597, LZTFL1 rs17713054, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 with susceptibility to Long COVID are described in Table 4.
Figure 2.
Genetic and allelic distributions between COVID-19 patients with and without Long COVID. (A) Genetic distribution. (B) Allelic distribution. Fonts marked in red indicate statistically significant differences in genetic and allelic distributions between COVID-19 patients with and without Long COVID.
Table 4.
Associations between polymorphisms in nine particular genes and susceptibility to Long COVID.
| Genes | SNPs | Genotype/allele | Long COVID | Genetic models | Unadjusted model | Adjusted model | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | % | Yes | % | P-value | OR | 95% CI | P-value | aOR | 95% CI | ||||
| LZTFL1 | rs10490770 | CC | 0 | 0.00 | 3 | 6.12 | CC vs. TC + TT | 0.999 | NA | NA | - | - | - |
| TC | 21 | 9.95 | 5 | 10.20 | TC vs. CC + TT | 0.958 | 1.028 | 0.367-2.877 | - | - | - | ||
| TT | 190 | 90.05 | 41 | 83.67 | TT vs. CC + TC | 0.206 | 0.566 | 0.235-1.368 | - | - | - | ||
| C | 21 | 4.98 | 11 | 11.22 | CC vs. TC | 0.999 | N/A | N/A | - | - | - | ||
| T | 401 | 95.02 | 87 | 88.78 | CC vs. TT | 0.999 | N/A | N/A | - | - | - | ||
| TT vs. TC | 0.852 | 1.103 | 0.393-3.097 | - | - | - | |||||||
| C vs. T | 0.024 | 2.414 | 1.123-5.191 | 0.001 | 4.416 | 1.900-10.267 | |||||||
| LZTFL1 | rs11385942 | AA/AA | 190 | 90.05 | 41 | 83.67 | AA/AA vs. AA/dupA + dupA/dupA | 0.202 | 0.563 | 0.233-1.360 | - | - | - |
| AA/dupA | 21 | 9.95 | 5 | 10.20 | AA/dupA vs. AA/AA + dupA/dupA | 0.950 | 1.034 | 0.369-2.892 | - | - | - | ||
| dupA/dupA | 0 | 0.00 | 3 | 6.12 | dupA/dupA vs. AA/AA + AA/dupA | 0.999 | N/A | N/A | - | - | - | ||
| AA | 401 | 95.02 | 87 | 88.78 | AA/AA vs. AA/dupA | 0.844 | 1.109 | 0.395-3.113 | - | - | - | ||
| dupA | 21 | 4.98 | 11 | 11.22 | AA/AA vs. dupA/ dupA | 0.999 | N/A | N/A | - | - | - | ||
| AA/AA | AA/dupA vs. dupA/dupA | 0.999 | N/A | N/A | - | - | - | ||||||
| dupA vs. AA | 0.025 | 2.402 | 1.117-5.165 | 0.001 | 4.416 | 1.9-10.267 | |||||||
| NADSYN1 | rs12785878 | GG | 106 | 50.24 | 25 | 51.02 | GG vs. GT + TT | 0.945 | 1.022 | 0.549-1.903 | - | - | - |
| GT | 88 | 41.71 | 15 | 30.61 | GT vs. GG + TT | 0.162 | 0.622 | 0.319-1.210 | - | - | - | ||
| TT | 17 | 8.06 | 9 | 18.37 | TT vs. GT + GG | 0.034 | 2.581 | 1.074-6.202 | 0.039 | 2.612 | 1.048-6.510 | ||
| G | 300 | 71.09 | 65 | 66.33 | GG vs. GT | 0.377 | 1.371 | 0.681-2.759 | - | - | - | ||
| T | 122 | 28.91 | 33 | 33.67 | GG vs. TT | 0.081 | 0.441 | 0.176-1.105 | - | - | - | ||
| TT vs. GT | 0.023 | 3.106 | 1.17-8.242 | 0.026 | 3.727 | 1.171-11.860 | |||||||
| G vs. T | 0.354 | 0.801 | 0.501-1.280 | - | - | - | |||||||
| PLXNA4 | rs1424597 | AA | 7 | 3.32 | 5 | 10.20 | AA vs. GA + GG | 0.048 | 3.328 | 1.009-10.971 | - | - | - |
| GA | 66 | 31.28 | 12 | 24.49 | GA vs. AA + GG | 0.361 | 0.717 | 0.352-1.464 | - | - | - | ||
| GG | 138 | 65.40 | 32 | 65.31 | GG vs. AA + GA | 0.972 | 0.989 | 0.515-1.899 | - | - | - | ||
| A | 80 | 18.96 | 22 | 22.45 | AA vs. GA | 0.039 | 3.929 | 1.068-14.445 | 0.026 | 5.573 | 1.223-25.403 | ||
| G | 342 | 81.04 | 76 | 77.55 | AA vs. GG | 0.067 | 3.103 | 0.925-10.408 | - | - | - | ||
| GG vs. GA | 0.524 | 0.79 | 0.382-1.631 | - | - | - | |||||||
| A vs. G | 0.434 | 1.237 | 0.726-2.109 | - | - | - | |||||||
| LZTFL1 | rs17713054 | AA | 0 | 0.00 | 3 | 6.12 | AA vs. GA + GG | 0.999 | N/A | N/A | - | - | - |
| GA | 21 | 9.95 | 5 | 10.20 | GA vs. AA + GG | 0.950 | 1.034 | 0.369-2.892 | - | - | - | ||
| GG | 190 | 90.05 | 41 | 83.67 | GG vs. AA + GA | 0.202 | 0.563 | 0.233-1.360 | - | - | - | ||
| A | 21 | 4.98 | 11 | 11.22 | AA vs. GA | 0.999 | N/A | N/A | - | - | - | ||
| G | 401 | 95.02 | 87 | 88.78 | AA vs. GG | 0.999 | N/A | N/A | - | - | - | ||
| GA vs.GG | 0.844 | 1.109 | 0.395-3.113 | - | - | - | |||||||
| A vs. G | 0.024 | 2.414 | 1.123-5.191 | 0.001 | 4.416 | 1.900-10.267 | |||||||
| IL10 | rs1800896 | CC | 0 | 0.00 | 0 | 0.00 | CC vs. TC + TT | 0 | 0.231 | N/A | - | - | - |
| TC | 16 | 7.58 | 9 | 18.37 | TC vs. CC + TT | 0.025 | 2.756 | 1.138-6.676 | 0.035 | 2.984 | 1.078-8.258 | ||
| TT | 195 | 92.42 | 40 | 81.63 | TT vs. CC + TC | 0.025 | 0.363 | 0.150-0.879 | 0.035 | 0.335 | 0.121-0.927 | ||
| C | 16 | 3.79 | 9 | 9.18 | CC vs. TC | 0 | 0.231 | N/A | - | - | - | ||
| T | 406 | 96.21 | 89 | 90.82 | CC vs. TT | 0 | 0.231 | N/A | - | - | - | ||
| TC vs. TT | 0.025 | 2.756 | 1.138-6.676 | 0.035 | 2.984 | 1.078-8.258 | |||||||
| C vs. T | 0.029 | 2.566 | 1.099-5.993 | 0.042 | 2.708 | 1.038-7.062 | |||||||
| ACE2 | rs2285666 | CC | 71 | 33.65 | 9 | 18.37 | CC vs. CT + TT | 0.040 | 0.444 | 0.204-0.965 | 0.043 | 0.44 | 0.199-0.975 |
| CT | 54 | 25.59 | 16 | 32.65 | CT vs. CC + TT | 0.317 | 1.41 | 0.72-2.761 | - | - | - | ||
| TT | 86 | 40.76 | 24 | 48.98 | TT vs. CC + CT | 0.295 | 1.395 | 0.748-2.604 | - | - | - | ||
| C | 196 | 46.45 | 34 | 34.69 | CC vs. CT | 0.062 | 0.428 | 0.176-1.042 | - | - | - | ||
| T | 226 | 53.55 | 64 | 65.31 | CC vs. TT | 0.062 | 0.454 | 0.198-1.040 | - | - | - | ||
| CT vs. TT | 0.870 | 1.062 | 0.518-2.178 | - | - | - | |||||||
| T vs. C | 0.036 | 0.613 | 0.388-0.968 | 0.038 | 0.648 | 0.290-0.637 | |||||||
| PEDS1 | rs6020298 | AA | 47 | 22.27 | 6 | 12.24 | AA vs. GA + GG | 0.123 | 0.487 | 0.195-1.214 | - | - | - |
| GA | 112 | 53.08 | 23 | 46.94 | GA vs. AA + GG | 0.439 | 0.782 | 0.419-1.458 | - | - | - | ||
| GG | 52 | 24.64 | 20 | 40.82 | GG vs. AA + GA | 0.024 | 2.109 | 1.101-4.040 | 0.037 | 2.062 | 1.046-4.066 | ||
| A | 206 | 48.82 | 35 | 35.71 | AA vs. GA | 0.332 | 0.622 | 0.238-1.625 | - | - | - | ||
| G | 216 | 51.18 | 63 | 64.29 | AA vs. GG | 0.030 | 0.332 | 0.123-0.897 | 0.050 | 0.347 | 0.120-0.998 | ||
| GA vs. GG | 0.072 | 0.534 | 0.270-1.058 | - | - | - | |||||||
| G vs. A | 0.020 | 1.717 | 1.089-2.706 | 0.046 | 1.608 | 1.008-2.565 | |||||||
| IL10RB | rs8178562 | AA | 32 | 15.17 | 10 | 20.41 | AA vs. GA + GG | 0.371 | 1.434 | 0.651-3.160 | - | - | - |
| GA | 93 | 44.08 | 27 | 55.10 | GA vs. AA + GG | 0.165 | 1.557 | 0.833-2.909 | - | - | - | ||
| GG | 86 | 40.76 | 12 | 24.49 | GG vs. AA + GA | 0.037 | 0.471 | 0.233-0.956 | 0.033 | 0.456 | 0.221-0.940 | ||
| A | 157 | 37.20 | 47 | 47.96 | AA vs. GA | 0.862 | 1.076 | 0.470-2.467 | - | - | - | ||
| G | 265 | 62.80 | 51 | 52.04 | AA vs. GG | 0.090 | 2.24 | 0.882-5.689 | - | - | - | ||
| GA vs. GG | 0.052 | 2.081 | 0.992-4.363 | - | - | - | |||||||
| A vs. G | 0.050 | 1.556 | 0.999-2.422 | - | - | - | |||||||
P-values marked with bold indicate statistically significant associations between genetic polymorphisms and susceptibility to Long COVID.
OR was adjusted for age, BMI, and comorbidities.
Abbreviations: ACE2, angiotensin converting enzyme-2; CI, confidence interval; COVID-19, coronavirus disease 2019; IL10, interleukin-10; IL10RB, interleukin-10 receptor subunit beta; LZTFL1, leucine zipper transcription factor like-1; N/A, not available; NADSYN1, NAD synthetase-1; OR, odds ratio; PEDS1, plasmanylethanolamine desaturase-1; PLXNA4, plexin A4; SNP, single nucleotide polymorphism.
Of LZTFL1 rs10490770, C allele showed a significant relationship with increased susceptibility to Long COVID, compared with T allele (OR = 2.414, 95% CI: 1.12, 5.19, P = 0.024).
Correspondingly, dupA allele of LZTFL1 rs11385942 was found to be significantly associated with a higher risk of Long COVID than AA allele (OR = 2.40, 95% CI: 1.12, 5.17, P = 0.025).
In addition to this, NADSYN1 rs12785878 GG genotype showed a substantial association with elevated risk of Long COVID when compared with GT-plus-GG genotypes and GT genotype (OR = 2.58, 95% CI: 1.07, 6.20, P = 0.034; OR = 3.11, 95% CI: 1.17, 8.24, P = 0.023; respectively).
Regardless of genetic polymorphism in PLXNA4 rs1424597, AA genotype was observed to be significantly associated with a greater susceptibility to Long COVID than GA genotype (OR = 3.93, 95% CI: 1.07, 14.45, P = 0.039).
Besides this, an increase in risk of Long COVID was significantly associated with G allele of LZTFL1 rs17713054, compared with A allele (OR = 2.41, 95% CI: 1.12, 5.19, P = 0.024).
For genetic polymorphism in IL10 rs1800896, TC genotype and C allele showed significant associations with higher susceptibility to Long COVID than CC-plus-TT genotypes, TT genotype, and T allele (OR = 2.76, 95% CI: 1.14, 6.68, P = 0.025; OR = 2.76, 95% CI: 1.14, 6.68, P = 0.025; OR = 2.57, 95% CI: 1.11, 5.99, P = 0.029; respectively), whereas TT genotype had a significant association with lower risk of Long COVID than CC-plus-TC genotypes (OR = 0.36, 95% CI: 0.15, 0.88, P = 0.025).
Of ACE2 rs2285666, CC genotype and C allele were found to be significantly related to a reduced risk of long COVID-19, compared with CT-plus-TT genotypes and T allele (OR = 0.44, 95% CI: 0.20, 0.97, P = 0.040; OR = 0.61, 95% CI: 0.39, 0.97, P = 0.036).
For genetic polymorphism in PEDS1 rs6020298, GG genotype and G allele both exhibited a significant relationship with higher risk of Long COVID than AA-plus-GA genotype and A allele (OR = 2.11, 95% CI: 1.10, 4.04, P = 0.024; OR = 1.72, 95% CI: 1.09, 2.71, P = 0.020, respectively).
Additionally, GG genotype of IL10RB rs8178562 was significantly associated with a reduced risk of long COVID-19, compared with AA-plus-GA genotype (OR = 0.47, 95% CI: 0.23, 0.96, P = 0.037).
Due to significant differences in age, BMI, and comorbidities between the patients with and without long COVID, multivariate logistic regression analysis was further undertaken to determine whether genetic polymorphisms in LZTFL1 rs10490770, LZTFL1 rs11385942, NADSYN1 rs12785878, PLXNA4 rs1424597, LZTFL1 rs17713054, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 were independently associated with susceptibility to long COVID-19. As demonstrated in Table 3, there still remain significant associations with susceptibility to Long COVID in genetic models of LZTFL1 rs10490770 (C vs. T: OR = 4.42, 95% CI: 1.90, 10.27, P = 0.001), LZTFL1 rs11385942 (dupA vs. AA: OR = 4.42, 95% CI: 1.90, 10.27, P = 0.001), NADSYN1 rs12785878 (TT vs. GT + GG: OR = 2.61, 95% CI: 1.05, 6.51, P = 0.039; TT vs. GT: OR = 3.73, 95% CI: 1.17, 11.86, P = 0.026), PLXNA4 rs1424597 (AA vs. GA + GG: OR = 5.02, 95% CI: 1.33, 18.97, P = 0.017; AA vs. GA: OR = 5.57, 95% CI: 1.22, 25.40, P = 0.026), LZTFL1 rs17713054 (A vs. G: OR = 4.42, 95% CI: 1.90, 10.27, P = 0.001), IL10 rs1800896 (TC vs. CC + TT: OR = 2.98, 95% CI: 1.08, 8.26, P = 0.035; TT vs. CC + TC: OR = 0.34, 95% CI: 0.12, 0.93, P = 0.035; TC vs. TT: OR = 2.98, 95% CI: 1.08, 8.26, P = 0.035; C vs. T: OR = 2.70, 95% CI: 1.04, 7.06, P = 0.042), ACE2 rs2285666 (CC vs. CT + TT: OR = 0.44, 95% CI: 0.20, 0.98, P = 0.043; C vs. T: OR = 0.65, 95% CI: 0.29, 0.64, P = 0.038), PEDS1 rs6020298 (GG vs. AA + GA: OR = 2.06, 95% CI: 1.05, 4.07, P = 0.037), and IL10RB rs8178562 (GG vs. AA + GA: OR = 0.46, 95% CI: 0.22, 0.94, P = 0.033), after adjusting for age, BMI, and comorbidities.
Population-based analysis of allelic frequencies
Whether there are differences in allelic frequencies of 9 SNPs observed in this study and those of other ethnic populations including Asians and Europeans was further determined using the Genome Aggregation Database (gnomAD) [20]. Comparisons in allelic frequencies of 9 SNPs between Thai COVID-19 patients and other ethnic populations are detailed in Table 5. When compared to general populations of Asia and Europe, allelic frequencies of LZTFL1 rs10490770 (Asians, P < 0.001), LZTFL1 rs11385942 (Asians, P < 0.001), NADSYN1 rs12785878 (Europeans, P < 0.001), PLXNA4 rs1424597 (Asians, P < 0.001; Europeans, P < 0.001), IL10 rs1800896 (Asians, P < 0.001), ACE2 rs2285666 (Europeans, P < 0.001), PEDS1 rs6020298 (Asians, P = 0.013), and IL10RB rs8178562 (Europeans, P < 0.001) in Thai COVID-19 patients were significantly different. Instead, there was no significant difference in allelic frequency of LZTFL1 rs17713054 between Thai COVID-19 patients and natural populations of Asia and Europe.
Table 5.
Population-based analysis of allelic frequencies.
| Genes | SNPs | Regions | General population | COVID-19 patients | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Reference alleles | Alternative alleles | P-value | Total | Reference alleles | Alternative alleles | P-value | |||||||
| LZTFL1 | rs10490770 | Present study | 520 | T | 488 (0.94) | C | 32 (0.06) | Reference | 520 | T | 488 (0.94) | C | 32 (0.06) | Reference |
| Asians | 702 | 689 (0.98) | 13 (0.02) | <0.001 | 7,047 | 6,075 (0.14) | 972 (0.14) | <0.001* | ||||||
| Europeans | 174,760 | 161,006 (0.92) | 13,754 (0.08) | 0.165 | ||||||||||
| LZTFL1 | rs11385942 | Present study | 520 | AA | 488 (0.94) | dupA | 32 (0.06) | Reference | 520 | AA | 488 (0.94) | dupA | 32 (0.06) | Reference |
| Asians | 112 | 112 (1.00) | 0 (0.00) | <0.001 | - | - | - | - | ||||||
| Europeans | 14,152 | 12,977 (0.92) | 1,175 (0.08) | 0.087 | 72 | 46 (0.64) | 26 (0.36) | <0.001 | ||||||
| NADSYN1 | rs12785878 | Present study | 520 | G | 365 (0.70) | T | 155 (0.30) | Reference | 520 | G | 365 (0.70) | T | 155 (0.30) | Reference |
| Asians | 3,374 | 2,240 (0.66) | 1,134 (0.34) | 0.089 | - | - | - | - | ||||||
| Europeans | 163,228 | 42,793 (0.26) | 120,435 (0.74) | <0.001 | 120 | 36 (0.30) | 84 (0.70) | <0.001 | ||||||
| PLXNA4 | rs1424597 | Present study | 520 | G | 102 (0.20) | A | 418 (0.80) | Reference | - | - | - | - | ||
| Asians | 160 | 124 (0.78) | 36 (0.23) | <0.001 | - | - | - | - | ||||||
| Europeans | 15,532 | 13,982 (0.90) | 1,550 (0.10) | <0.001 | - | - | - | - | ||||||
| LZTFL1 | rs17713054 | Present study | 520 | G | 488 (0.94) | A | 32 (0.06) | Reference | - | - | - | - | ||
| Asians | 168 | 164 (0.98) | 4 (0.02) | 0.071 | - | - | - | - | ||||||
| Europeans | 19,446 | 17,856 (0.92) | 1,590 (0.08) | 0.104 | - | - | - | - | ||||||
| IL10 | rs1800896 | Present study | 520 | T | 495 (0.95) | C | 25 (0.05) | Reference | 520 | T | 495 (0.95) | C | 25 (0.05) | Reference |
| Asians | 3,938 | 140,244 (0.53) | 125,934 (0.47) | <0.001 | 3,184 | 2,842 (0.89) | 342 (0.11) | <0.001 | ||||||
| Europeans | 266,178 | 3,690 (0.94) | 248 (0.06) | 0.206 | - | - | - | - | ||||||
| ACE2 | rs2285666 | Present study | 520 | C | 230 (0.44) | T | 290 (0.56) | Reference | 520 | C | 230 (0.44) | T | 290 (0.56) | Reference |
| Asians | 514 | 229 (0.45) | 285 (0.55) | 0.950 | - | - | - | - | ||||||
| Europeans | 100,440 | 79,975 (0.80) | 20,465 (0.20) | <0.001 | 481 | 354 (0.74) | 127 (0.26) | <0.001 | ||||||
| PEDS1 | rs6020298 | Present study | 520 | G | 279 (0.54) | A | 241 (0.46) | Reference | - | - | - | - | ||
| Asians | 202 | 87 (0.43) | 115 (0.57) | 0.013 | - | - | - | - | ||||||
| Europeans | 89,342 | 45,795 (0.51) | 43,547 (0.49) | 0.291 | - | - | - | - | ||||||
| IL10RB | rs8178562 | Present study | 520 | G | 316 (0.61) | A | 204 (0.39) | Reference | - | - | - | - | ||
| Asians | 624 | 402 (0.64) | 222 (0.36) | 0.219 | - | - | - | - | ||||||
| Europeans | 121,598 | 99,511 (0.82) | 22,087 (0.18) | <0.001 | - | - | - | - | ||||||
P-values marked with bold indicate statistically significant differences in allelic frequencies of SNPs between Thai and other ethic populations.
*P-value for comparison in allelic frequency of LZTFL1 rs10490770 among Thai, Asian, and European patients with COVID-19.
Abbreviations: ACE2, angiotensin converting enzyme-2; CI, confidence interval; COVID-19, coronavirus disease 2019; IL10, interleukin-10; IL10RB, interleukin-10 receptor subunit beta; LZTFL1, leucine zipper transcription factor like-1; N/A, not available; NADSYN1, NAD synthetase-1; OR, odds ratio; PEDS1, plasmanylethanolamine desaturase-1; PLXNA4, plexin A4; SNP, single nucleotide polymorphism.
In the context of SARS-CoV-2 infection, statistically significant differences in allelic frequencies of LZTFL1 rs10490770 [13], LZTFL1 rs11385942 [21], NADSYN1 rs12785878 [22], IL10 rs1800896 [23], and ACE2 rs2285666 [24–27] between Thai and other ethnic patients, particularly Europeans were observed (P < 0.001, P < 0.001, P < 0.001, P < 0.001, P < 0.001, respectively) (Table 5).
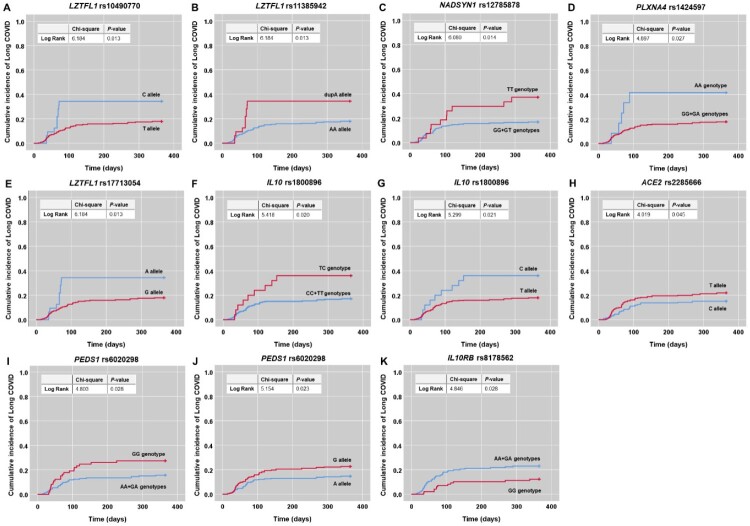
SNPs associations with cumulative incidence of Long COVID
The effect of polymorphisms in significant genes on cumulative incidence of Long COVID was further determined. Kaplan-Meier curves with log-rank analysis displayed that LZTFL1 rs10490770 C allele (χ2 = 6.184, P = 0.013), LZTFL1 rs11385942 dupA allele (χ2 = 6.184, P = 0.013), NADSYN1 rs12785878 TT genotype (χ2 = 6.080, P = 0.014), PLXNA4 rs1424597 AA genotype (χ2 = 4.897, P = 0.027), LZTFL1 rs17713054 A allele (χ2 = 6.184, P = 0.013), IL10 rs1800896 TC genotype and C allele (χ2 = 5.418, P = 0.020; χ2 = 5.299, P = 0.021, respectively), ACE2 rs2285666 T allele (χ2 = 4.019, P = 0.045), and PEDS1 rs6020298 GG genotype and G allele (χ2 = 4.803, P = 0.028; χ2 = 5.154, P = 0.023, respectively) were significantly related to a higher cumulative incidence of Long COVID occurrence than other alleles and genotypes (Figures 3). On the contrary, IL10RB rs8178562 GG genotype was found to be significantly associated with a lower cumulative incidence of Long COVID occurrence than AA-plus-GA genotypes (χ2 = 4.846, P = 0.028) (Figure 3).
Figure 3.
Kaplan-Meier curves demonstrating associations between polymorphisms in nine significant genes and cumulative incidence of Long COVID. (A) LZTFL1 rs10490770 C allele. (B) LZTFL1 rs11385942 dupA allele. (C) NADSYN1 rs12785878 TT genotype. (D) PLXNA4 rs1424597 AA genotype. (E) LZTFL1 rs17713054 A allele. (F) IL10 rs1800896 TC genotype and (G) C allele. (H) ACE2 rs2285666 T allele. (I) PEDS1 rs6020298 GG genotype and (J) G allele. (K) IL10RB rs8178562 GG genotype.
Discussion
Identifying host genetic factors involved in clinical consequences of COVID-19 are crucial for a better understanding of COVID-19 pathogenesis and interindividual heterogeneity in severity and long-term symptoms of COVID-19, which may be helpful for development of novel therapeutic strategies for prophylaxis and clinical surveillance of COVID-19. From this perspective, the present study attempted to investigate the influences of 37 SNPs known to be implicated in viral entry and immune as well as inflammatory responses on susceptibility to severe COVID-19 and long-term symptoms of SARS-CoV-2 infection. Out candidate genetic variants selected from data on systematic literature review, 9 including LZTFL1 rs10490770, LZTFL1 rs11385942, LZTFL1 rs17713054, NADSYN1 rs12785878, PLXNA4 rs1424597, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 were found to be significantly associated with Long COVID, whereas there were no significant associations between polymorphisms in 37 selected variants and susceptibility to severe COVID-19, after adjusting for age, gender, the presence of comorbidities, and vaccination status. More specifically, polymorphisms in LZTFL1 rs10490770, LZTFL1 rs11385942, LZTFL1 rs17713054, NADSYN1 rs12785878, PLXNA4 rs1424597, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 were shown to be significantly associated with a cumulative incidence of Long COVID occurrence. Collectively, the aforementioned findings lend support to the notion that host genetic factors may influence clinical consequences of COVID-19 – particularly Long COVID, which may open the door for personalized medicine against long-term symptoms of COVID-19.
As LZTFL1 is one of candidate effector genes potentially implicated in the disease progression by viral entry or clearance and immune response, recent studies examined its functional significance in SARS-CoV-2 infection [28,29]. From this, selective spatial transcriptomic analysis of lung samples from COVID-19 patients uncovered the presence of epithelial–mesenchymal transition (EMT) signalling, a viral response pathway regulated by LZTFL1 [30]. In addition to this, it has been shown that LZTFL1 was highly expressed in pulmonary epithelial cells, particularly ciliated epithelial cells, one of the primary cellular targets for SARS-CoV-2 infection [31]. In view of the foregoing findings, it has been speculated that alterations in LZTFL1 expression involved in susceptibility to SARS-CoV-2 infection and COVID-19 severity may be governed by genetic polymorphisms. In support of this hypothesis, global data depicted a significant association between LZTFL1 rs10490770 polymorphism and COVID-19 severity [32]. Indeed, C allele of LZTFL1 rs10490770 has been reportedly associated with an increased susceptibility to severe COVID-19, compared with T allele [13]. Contrary to the previous findings, our result showed no significant association between LZTFL1 rs10490770 polymorphism and COVID-19 severity. In terms of long-term symptoms of COVID-19, our result further demonstrated that C allele was significantly associated with a higher risk of Long COVID than T allele. Besides LZTFL1 rs10490770 polymorphism, the present study revealed that dupA allele of LZTFL1 rs11385942 was significantly associated with a greater risk of Long COVID than AA allele. Our finding is partly supported by a genome-wide association study denoting a considerable association between LZTFL1 rs11385942 polymorphism and severe COVID-19 with respiratory failure [14]. In addition to LZTFL1 rs10490770 and rs11385942 polymorphisms, our additional finding uncovered that A allele of LZTFL1 rs17713054 was significantly associated with an increased risk of developing Long COVID-19, consistent with a previous study by Downes et al [30]. revealing a significant association of LZTFL1 rs17713054 with two-fold increased risk of respiratory failure. When compared to other ethnic populations with COVID-19, especially Europeans [13,21], the allelic frequencies of LZTFL1 rs10490770 and rs11385942 observed in our study were significantly different. All above-mentioned findings highlight the importance of LZTFL1 as a candidate effector gene in clinical consequences of COVID-19 – particularly long-term symptoms.
Given the involvement of vitamin D homeostasis and its metabolic pathway in COVID-19 severity [33], NADSYN1, a precursor for several cellular signalling and metabolic molecules including vitamin D, is gaining increasing interest as a candidate molecule for exploring its polymorphisms involved in COVID-19 severity. It has been suggested that the significance of vitamin D in host immunity against viral infection, in particular SARS-CoV-2 infection, might be partly explained by polymorphisms in relevant genes [33, 34]. Regarding this, a recent study by Kotur et al [22]. demonstrated a significant association between DHCR7/NADSYN rs12785878 polymorphism and susceptibility to severe COVID-19, in which TG-plus-GG genotypes showed a significant association with decreased risk of developing severe COVID-19. Consistent with this, a study by Freitas et al [35]. showed that polymorphism in DHCR7/NADSYN rs12785878 was significantly associated with COVID-19 severity. The above findings attest our result regarding a significant association between TT genotype of NADSYN rs12785878 and higher risk of developing Long COVID, compared with other genotypes, while after adjusting for confounders, no association of NADSYN rs12785878 polymorphism with susceptibility to severe COVID-19 was found. This may be due to existing risk factors like vaccination, possibly exerting the dominant susceptibility effect on severe COVID-19, rather than genetic factors. Comparison in the allelic frequency of NADSYN rs12785878 between Thai and European patients with COVID-19 uncovered significant difference [22].
The present study further identified a possible causative gene responsible for a three-fold greater risk of long-term symptoms of COVID-19, PLXNA4 rs1424597 polymorphism. Our additional finding is supported by genomic data derived from a recent study by Razzaq et al [36], denoting that AA genotype of PLXNA4 rs1424597 had an increased risk of developing pulmonary embolism in COVID-19 patients. As to its biological role, Plexin A4, encoded by PLXNA4 gene, is a component of a receptor complex involved in signal transduction of semaphorin 3 signals responsible for cytoskeletal reorganization, leading to inhibited integrin adhesion [37]. This highlights the importance of Plexin A4 in microvascular thrombosis, one of the critical complications observed in COVID-19 patients. In critically ill COVID-19 patients, it has been found that decreased plasma Plexin A4 levels were significantly linked to deteriorated respiratory function [36].
Since the pathophysiology of COVID-19 is characterized by an inflammatory response that activates a complex collection of mediators, one of which is a group of interleukins (IL) [38], genetic polymorphisms in ILs have attracted a great deal of scientific attention. Among others, genetic polymorphism in IL10 rs1800896 has been shown to have a strong association with the prevalence of COVID-19 [39]. In line with the previous finding, our study further depicted a significant relationship between IL10 rs1800896 polymorphism with long-term symptoms of COVID-19, in which TC genotype and C allele provided a two-fold increase in risk of Long COVID development. In comparison to COVID-19 patients in Europe [23], the allelic frequency of IL10 rs1800896 obtained in our study was significantly different.
As ACE2 acts as a predominant receptor through which the SARS-CoV-2 enters and infects cells, alterations in ACE2 expression mediated by its genetic variants have been demonstrated to contribute to severe outcomes in COVID-19 patients [40,41]. From that view, several studies focused on investigating possible association between genetic polymorphism in ACE2 and COVID-19 severity and demonstrated that TT genotype and T allele of ACE2 rs2285666 were substantially related to increased vulnerability to severe and critical COVID-19 [14,15]. The previous findings pointed out the consistency of our result regarding association between ACE2 rs2285666 polymorphism with Long COVID, where T allele was observed to be associated with a greater susceptibility to developing long-term symptoms of COVID-19. In the allelic frequency of ACE2 rs2285666 between our study and previous studies conducted in Europe [24–27], a significant difference was observed.
As PEDS1 is recognized as one of molecules involved in IL1 signalling pathway responsible for inflammatory and immune responses, genetic polymorphism in PEDS1 has gained considerable research interest recently. Notably, GWAS data showed that PEDS1 rs6020298, the most significant SNP, was associated with COVID-19 severity [42]. Supporting the previous finding, GG genotype and G allele of PEDS1 rs6020298 were shown to be significantly associated with an elevated risk of Long COVID in our study.
Aside from genetic polymorphisms in the aforementioned genes, IL10RB was recently identified as the top candidate gene target for COVID-19 host susceptibility [43]. Regardless of its primary role, IL10RB is an integral part of the IL10 receptor complex. It has been proved that coexpression of IL10RB with IL10RA is essential for IL10-induced signal transduction [44]. In the context of COVID-19, data from translational genomics approach revealed that upregulation of IL10RB expression was associated with worse outcomes and increased viral load [43]. In this regard, it has been assumed that COVID-19 progression may be linked to genetic variations in IL10RB possibly affecting its expression. To address this speculation, a recent result derived from a bioinformatics approach unveiled that genetic polymorphism in IL10RB rs8178562 was remarkably associated with COVID-19 severity [45], lending credence to our finding of significant relationship between GG genotype of IL10RB rs8178562 and decreased susceptibility to developing Long COVID.
In view of the foregoing, it is worth noting that genetic polymorphisms in 9 out of 37 variants, including LZTFL1 rs10490770, LZTFL1 rs11385942, LZTFL1 rs17713054, NADSYN1 rs12785878, PLXNA4 rs1424597, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562, might play a role in susceptibility to long-term symptoms of COVID-19, conceivably by altering their protein expression and subsequently activating the downstream effectors relevant to COVID-19 pathogenesis. In support of this postulation, LZTFL1 rs11385942 polymorphism has been reportedly associated with elevated plasma levels of complement component 5a (C5a) and soluble terminal complement complex C5b-9 (SC5b-9) during SARS-CoV-2 infection [21], indicating that enhanced activation of the immune system and complement pathways might contribute to the deleterious effect of this variant. Apart from this, it has been shown that ACE2 rs2285666 T allele was significantly associated with increased expression of ACE2 receptor in COVID-19 patients [46]. However, it is important to keep the inherent limitations in mind when interpreting the data presented herein. One of the most important limitations of this study is that the functional analysis of those identified 9 SNPs was not performed. In that context, it may be challenging to explain how genetic polymorphisms in 9 SNPs affect their gene functions implicated in Long COVID development and how they interact with each other. Another drawback of this study is lack of data on environmental factors such as smoking status, which may influence COVID-19 severity. Furthermore, given that MassArray system, a method employed for the identification of genetic polymorphisms, allows us to analyse up to 40 SNPs, we encountered difficulties in determining genetic polymorphisms of all reported SNPs with no significant associations with COVID-19 severity and unfavourable long-term outcomes. Additionally, no associations between genetic polymorphisms in 37 chosen host variants and severe COVID-19 were discovered in this study, which might be due to the fact that we were unable to comprehensively examine other genes implicated in COVID-19 development. Besides this, it might be attributable to the fact that over 80% of COVID-19 patients in Thailand are vaccinated, for which the disease infrequently progresses to a severe form among those who do have it. Vaccines against COVID-19 are generally accepted as a means of preventing life-threatening complications and death.
Conclusions
This study added to the growing body of evidence regarding the influences of host genetic factors on COVID-19 clinical consequences by revealing that polymorphisms in LZTFL1 rs10490770, LZTFL1 rs11385942, LZTFL1 rs17713054, NADSYN1 rs12785878, PLXNA4 rs1424597, IL10 rs1800896, ACE2 rs2285666, PEDS1 rs6020298, and IL10RB rs8178562 were significantly related to not only risk of long-term symptoms of COVID-19, but also cumulative incidence of Long COVID occurrence. Since genetic variants have been shown to increase susceptibility to SARS-CoV-2 infection and cause devastating outcomes, it is imperative to incorporate individual genetic data in order to implement personalized therapeutics and improve COVID-19 prognostication.
Author contributions
WU and WC conceived and designed the study; WU and WS performed systematic review; BP, PJ, NT, SS, CR, and IS carried out the experiments; WU and IS analysed and interpreted the data; WU, JJ, and UC participated in statistical analyses; WC contributed reagents, materials, and analytical tools; NN examined all the patients and collected the clinical data; WU drafted and revised the manuscript; WU reviewed the manuscript; All approved the final version of the manuscript.
Ethics approval and consent to participate
The study protocol was approved by the ethical committee of the Faculty of Medicine Ramathibodi Hospital, Mahidol University (COA. MURA2021/264 Ref.2404) and carried out in compliance with the ethical standards outlined in the Declaration of Helsinki, The Belmont Report, CIOMS Guidelines and the International Conference on Harmonization in Good Clinical Practice (ICH-GCP). All study subjects provided written informed consents, prior to their participation.
Supplementary Material
Funding Statement
This research project was supported by the National Research Council of Thailand (NRCT) (วช.อว. (อ) (กบท2) 332/2564) and Mahidol University (Fundamental Fund: fiscal year 2023 by National Science Research and Innovation Fund (NSRF)) (FF-048/2566).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Guo G, Ye L, Pan K, et al. . New insights of emerging SARS-CoV-2: epidemiology, etiology, clinical features, clinical treatment, and prevention. Front Cell Dev Biol. 2020;8:410. doi: 10.3389/fcell.2020.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X, Yang R.. COVID-19 transmission through asymptomatic carriers is a challenge to containment. Influenza Other Respir Viruses. 2020;14:474–475. doi: 10.1111/irv.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alshoabi SA, Alhazmi FH, Abdulaal OM, et al. . Frequent clinical and radiological manifestations of the novel SARS-CoV-2: A review article. J Family Med Prim Care. 2021;10:122–126. doi: 10.4103/jfmpc.jfmpc_1985_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atzrodt CL, Maknojia I, McCarthy RDP, et al. . A guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020;287:3633–3650. doi: 10.1111/febs.15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Haupert SR, Zimmermann L, et al. . Global prevalence of post COVID-19 condition or long COVID: a meta-analysis and systematic review. J Infect Dis. 2022;226:1593–1607. doi: 10.1093/infdis/jiac136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Huang L, Wang Y, et al. . 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou F, Yu T, Du R, et al. . Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abou-Ghaida J, Foster A, Klein S, et al. . The world-wide adaptations of diabetic management in the face of COVID-19 and socioeconomic disparities: a scoping review. Cureus. 2022;14:e31911. doi: 10.7759/cureus.31911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patarčić I, Gelemanović A, Kirin M, et al. . The role of host genetic factors in respiratory tract infectious diseases: systematic review, meta-analyses and field synopsis. Sci Rep. 2015;5:16119. doi: 10.1038/srep16119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenney AD, Dowdle JA, Bozzacco L, et al. . Human genetic determinants of viral diseases. Annu Rev Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debnath M, Banerjee M, Berk M.. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34:8787–8795. doi: 10.1096/fj.202001115R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos-Lopez O, Daimiel L, Ramirez de Molina A, et al. . Exploring host genetic polymorphisms involved in SARS-CoV infection outcomes: implications for personalized medicine in COVID-19. Int J Genomics. 2020;2020:6901217. doi: 10.1155/2020/6901217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakanishi T, Pigazzini S, Degenhardt F, et al. . Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J Clin Invest. 2021;131:e152386. doi: 10.1172/JCI152386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, et al. . Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saengsiwaritt W, Jittikoon J, Chaikledkaew U, et al. . Genetic polymorphisms of ACE1, ACE2, and TMPRSS2 associated with COVID-19 severity: a systematic review with meta-analysis. Rev Med Virol. 2022;32:e2323. doi: 10.1002/rmv.2323 [DOI] [PubMed] [Google Scholar]
- 16.Grolmusz VK, Bozsik A, Papp J, et al. . Germline genetic variants of viral entry and innate immunity may influence susceptibility to SARS-CoV-2 infection: toward a polygenic risk score for risk stratification. Front Immunol. 2021;12:6534. doi: 10.3389/fimmu.2021.653489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anastassopoulou C, Gkizarioti Z, Patrinos GP, et al. . Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics. 2020;14:40. doi: 10.1186/s40246-020-00290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization 2021 . Clinical management of COVID-19: living guidance 2021:1–81.
- 19.NICE COVID-19 Rapid Guideline: Managing the Long-Term Effects of COVID-19. [cited 2021 Jan 13]; Available from: https://www.nice.org.uk/guidance/ng188. [PubMed]
- 20.The Genome Aggregation Database (gnomAD) v2.1. [cited 2022 May 22]; Available from: https://gnomad.broadinstitute.org.
- 21.Valenti L, Griffini S, Lamorte G, et al. . Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J Autoimmun. 2021;117:102595. doi: 10.1016/j.jaut.2021.102595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotur N, Skakic A, Klaassen K, et al. . Association of vitamin D, zinc and selenium related genetic variants with COVID-19 disease severity. Front Nutr. 2021;8:689419. doi: 10.3389/fnut.2021.689419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbood SJA, Anvari E, Fateh A.. Association between interleukin-10 gene polymorphisms (rs1800871, rs1800872, and rs1800896) and severity of infection in different SARS-CoV-2 variants. Hum Genomics. 2023;17(1):19. doi: 10.1186/s40246-023-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traets MJM, Nijhuis RHT, Morré SA, et al. . Association of genetic variations in ACE2, TIRAP and factor X with outcomes in COVID-19. PLoS One. 2022;17(1):e0260897. doi: 10.1371/journal.pone.0260897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Möhlendick B, Schönfelder K, Breuckmann K, et al. . ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenet Genomics. 2021;31(8):165–171. doi: 10.1097/FPC.0000000000000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jevnikar K, Lapajne L, Petrovič D, et al. . The role of ACE, ACE2, and AGTR2 polymorphisms in COVID-19 severity and the presence of COVID-19-related retinopathy. Genes (Basel). 2022;13(7):1111. doi: 10.3390/genes13071111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalilzadeh F, Sakhaee F, Sotoodehnejadnematalahi F, et al. . Angiotensin-converting enzyme 2 rs2285666 polymorphism and clinical parameters as the determinants of COVID-19 severity in Iranian population. Int J Immunogenet. 2022;49(5):325–332. doi: 10.1111/iji.12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Cai S, Feng H, et al. . Single-cell analysis reveals bronchoalveolar epithelial dysfunction in COVID-19 patients. Protein Cell. 2020;11:680–687. doi: 10.1007/s13238-020-00752-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borczuk AC, Salvatore SP, Seshan SV, et al. . COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York city. Mod Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downes DJ, Cross AR, Hua P, et al. . Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat Genet. 2021;53:1606–1615. doi: 10.1038/s41588-021-00955-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravindra NG, Alfajaro MM, Gasque V, et al. . Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19:e3001143. doi: 10.1371/journal.pbio.3001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Macdonald-Dunlop E, Chen J, et al. . Genetic landscape of the ACE2 coronavirus receptor. Circulation. 2022;145:1398–1411. doi: 10.1161/CIRCULATIONAHA.121.057888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant WB, Lahore H, McDonnell SL, et al. . Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients. 2020;12:988. doi: 10.3390/nu12040988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossein-nezhad A, Spira A, Holick MF.. Influence of vitamin D status and vitamin D3 supplementation on genome wide expression of white blood cells: a randomized double-blind clinical trial. PLoS One. 2013;8:e58725. doi: 10.1371/journal.pone.0058725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freitas AT, Calhau C, Antunes G, et al. . Vitamin D-related polymorphisms and vitamin D levels as risk biomarkers of COVID-19 disease severity. Sci Rep. 2021;11:20837. doi: 10.1038/s41598-021-99952-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razzaq M, Iglesias MJ, Ibrahim-Kosta M, et al. . An artificial neural network approach integrating plasma proteomics and genetic data identifies PLXNA4 as a new susceptibility locus for pulmonary embolism. Sci Rep. 2021;11:14015. doi: 10.1038/s41598-021-93390-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fard D, Tamagnone L.. Semaphorins in health and disease. Cytokine Growth Factor Rev. 2021;57:55–63. doi: 10.1016/j.cytogfr.2020.05.006 [DOI] [PubMed] [Google Scholar]
- 38.Chen G, Wu D, Guo W, et al. . Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karcioglu Batur L, Hekim N.. Correlation between interleukin gene polymorphisms and current prevalence and mortality rates due to novel coronavirus disease 2019 (COVID-2019) in 23 countries. J Med Virol. 2021;93:5853–5863. doi: 10.1002/jmv.27127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakhshandeh B, Sorboni SG, Javanmard AR, et al. . Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect Genet Evol. 2021;90:104773. doi: 10.1016/j.meegid.2021.104773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez-Gómez LE, Herrera-López B, Martinez-Armenta C, et al. . ACE and ACE2 gene variants are associated with severe outcomes of COVID-19 in men. Front Immunol. 2022;13:812940. doi: 10.3389/fimmu.2022.812940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Huang S, Gao R, et al. . Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6:83. doi: 10.1038/s41421-020-00231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voloudakis G, Vicari JM, Venkatesh S, et al. . A translational genomics approach identifies IL10RB as the top candidate gene target for COVID-19 susceptibility. NPJ Genom Med. 2022;7:52. doi: 10.1038/s41525-022-00324-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo S, Dong X, Guo S, et al. . Regulation of the human IL-10RB gene expression by Sp8 and Sp9. J Alzheimers Dis. 2022;88:1469–1485. doi: 10.3233/JAD-220321 [DOI] [PubMed] [Google Scholar]
- 45.Karakas Celik S, Cakmak Genc G, Dursun A.. A bioinformatic approach to investigating cytokine genes and their receptor variants in relation to COVID-19 progression. Int J Immunogenet. 2021;48:211–218. doi: 10.1111/iji.12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karakaş Çelik S, Çakmak Genç G, et al. . Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: a case study. J Med Virol. 2021;93(10):5947–5952. doi: 10.1002/jmv.27160 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.