Summary
Satellite viruses are present across all domains of life, defined as sub-viral parasites that require infection by another virus for satellite progeny production. Phage satellites exhibit various regulatory mechanisms to manipulate phage gene expression to the benefit of the satellite, redirecting resources from the phage to the satellite, and often inhibiting phage progeny production. While small RNAs (sRNAs) are well documented as regulators of prokaryotic gene expression, they have not been shown to play a regulatory role in satellite-phage conflicts. Vibrio cholerae encodes the phage inducible chromosomal island-like element (PLE), a phage satellite, to defend itself against the lytic phage ICP1. Here we use Hi-GRIL-seq to identify a complex RNA-RNA interactome between PLE and ICP1. Both inter- and intra-genome RNA interactions were detected, headlined by the PLE sRNA, SviR. SviR is involved in regulating both PLE and ICP1 gene expression uniquely, decreasing ICP1 target translation and affecting PLE transcripts. The striking conservation of SviR across all known PLEs suggests the sRNA is deeply rooted in the PLE-ICP1 conflict and implicates sRNAs as unidentified regulators of gene expression in phage-satellite interactions.
Keywords: Bacteriophage, Vibrio cholerae, Regulatory RNA, Bacterial RNA, Satellite viruses
Introduction
The conflict between lytic bacteriophages and their hosts imposes tremendous selective pressure on both the virus and host to outcompete each other, resulting in a dynamic co-evolutionary arms race (Koskella and Brockhurst, 2014). To overcome phage predation, bacteria have acquired vast immune systems capable of blocking phages through unique mechanisms (Bernheim and Sorek, 2020). The interactions between the diarrheal pathogen Vibrio cholerae and its phages have been proposed to drive the seasonality of cholera outbreaks (Faruque et al., 2005). Two decades of surveillance of cholera patient stool samples from Bangladesh has brought to light the ongoing conflict between V. cholerae and the lytic bacteriophage ICP1 (Seed et al., 2011; Angermeyer et al., 2018; Boyd et al., 2021), providing a tractable model system to interrogate the molecular mechanisms employed by both phage and host to antagonize each other.
To defend itself against ICP1 predation, V. cholerae encodes a remarkable family of phage satellites called phage-inducible chromosomal island-like elements (PLEs) (O’Hara et al., 2017). Phage satellites are mobile genetic elements (MGEs) that lack the full suite of genes to autonomously produce progeny, instead leveraging host- and viral-encoded features to facilitate satellite spread (Penadés and Christie, 2015). Unlike many other generalized anti-phage immune systems, such as restriction-modification (Tock and Dryden, 2005) and CRISPR-Cas (Barrangou et al., 2007), the immunity provided by each phage satellite is typically specific to a single helper phage; in the case of PLE, that helper phage is ICP1. Ten unique PLEs have been identified, each of which is approximately 19 kb in length and encodes a suite of core genes, shared between all PLEs, and non-conserved accessory genes (Angermeyer et al., 2022). All identified PLE(+) V. cholerae contain a single PLE, which lays dormant in the V. cholerae chromosome until ICP1 infection. Upon infection, the PLE excises from the chromosome (McKitterick and Seed, 2018), replicates to high copy number (Barth, Silvas, et al., 2020), and deploys multiple anti-ICP1 factors, which collectively eliminate the production of ICP1 progeny, reducing the burst of released ICP1 virions from approximately 100 per infected cell to 0 (O’Hara et al., 2017; Hays and Seed, 2019; LeGault et al., 2022). Upon cell lysis, PLE is spread to the neighboring PLE(−) V. cholerae in modified ICP1 virions through transduction (Netter et al., 2021), resulting in population-wide immunity to ICP1 and the preservation of PLE. Transcriptomic profiling of PLE and ICP1 during phage infection revealed that the ICP1 transcriptome is nearly unperturbed in the presence of PLE, with only the capsid operon being downregulated by a PLE transcriptional repressor, CapR (Barth, Netter, et al., 2020; Netter et al., 2021). Additionally, RNA-seq identified two intergenic transcripts, a putative PLE small RNA (sRNA) and an ICP1 long non-coding RNA (lncRNA), to be the two most abundant transcripts late in phage infection (Barth, Netter, et al., 2020) (Figure 1A), raising the question of whether these transcripts may act as regulatory RNAs in the PLE-ICP1 conflict.
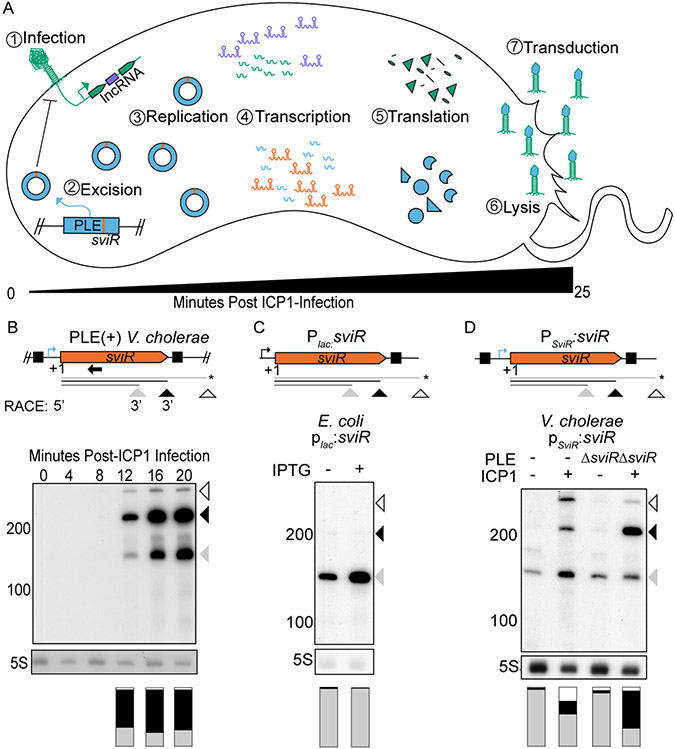
Figure 1). The PLE putative regulatory small RNA, SviR, is induced in response to ICP1 infection.
(A) A representative model of ICP1 phage infection of a PLE(+) V. cholerae host. The steps involved in PLE (blue) activation following ICP1 (green) infection are temporally numbered. Upon ICP1 infection, PLE excises from the chromosome (McKitterick and Seed, 2018), replicates to high copy (Barth, Silvas, et al., 2020), and undergoes its transcriptional (Barth, Netter, et al., 2020) and translational programs, resulting in lysis of host cells (Hays and Seed, 2019) and transduction of PLE to neighboring cells in modified ICP1 particles (Netter et al., 2021). ICP1 and PLE non-coding RNAs are shown in purple and orange, respectively.
(B) SviR 5’-/3’-RACE products (top, under model) and Northern blot analysis of SviR during a time-course of ICP1 infection of PLE(+) V. cholerae (bottom). Triangles under gene graphs and on the side of the blots represent the three most abundant species of SviR, shaded according to relative abundance. Annotations below the gene graph represent the sviR TSS, as determined by 5’-RACE and putative stop sites inferred from 3’-RACE data. The species marked with an asterisk correspond to a stop site not confirmed by 3’-RACE. For the full 5’/3’-RACE analysis, see Figure S1. The black boxes on the gene graph represent terminal inverted repeats flanking sviR. The native sviR promoter prediction is represented by the blue bent arrow icon. The stacked bar graph under each lane represents the ratio of each SviR transcript species abundance normalized to 5S rRNA for each lane, with each color of the stacked bar graph representing the corresponding SviR species above. The bar graphs are an average of two biological replicate Northern blot experiments. The Northern blot probe used to detect SviR is indicated by a black arrow.
(C) Northern blot analysis of ectopic sviR expression from a plac0-1:sviR construct in E. coli. The Plac promoter, represented by the black bent arrow icon, was induced with 1 mM IPTG to induce the expression of SviR. The different SviR species present in V. cholerae are indicated by the arrows present below the gene graph, as indicated in (B). The stacked bar graph under each lane represents SviR species abundance as described in (B).
(D) Northern blot analysis showing ectopic sviR expression from a PSviR:sviR construct in V. cholerae. The upstream 100 bp relative to the sviR TSS was inserted upstream of the insert used in (C), which includes the predicted SviR promoter sequence (blue bent arrow icon) and the left inverted repeat. RNA was isolated from mock (−) or ICP1 (+) infected samples 16 minutes post-infection. The PLE genotype of the V. cholerae host strain is indicated above each lane by either (−) for strains lacking PLE or ΔsviR for PLE(+) strains with the sviR deletion. The stacked bar graph under each lane represents the SviR species abundance as described in (B).
Bacterial sRNAs are a well-documented class of regulatory RNAs that respond to diverse cellular stresses through base pairing to target transcripts, leading to target regulation (Waters and Storz, 2009). sRNA base pairing is often initiated with a short seed region of imperfect complementarity to the target transcript (Bandyra et al., 2012), an interaction often, but not always, facilitated by RNA chaperones, such as Hfq and ProQ (Melamed et al., 2020). The result of sRNA regulation varies considerably, with a single sRNA capable of regulating multiple targets with different outcomes (Feng et al., 2015). The most frequently documented outcome for sRNA regulation is translational inhibition by blocking ribosome association with target transcripts (Storz, Vogel and Wassarman, 2012). However, many unique outcomes of sRNA regulation have been documented, and novel mechanisms surely remain to be discovered.
While many sRNAs have been shown to regulate various processes in V. cholerae (Hammer and Bassler, 2007; Richard et al., 2010; Peschek et al., 2020), less is known about sRNA regulation between MGEs, phages, and their host bacteria (Fröhlich and Papenfort, 2016; Altuvia, Storz and Papenfort, 2018). Both MGEs and prophages have been shown to use cis- and trans-encoded sRNAs to regulate transcripts encoded on the core host genome, as well as on the MGEs themselves (Tree et al., 2014; Wachter et al., 2018; Bloch et al., 2021). However, MGE- or host-encoded sRNAs regulating lytic phage transcripts have only been predicted from transcriptomic data sets and have not yet been experimentally validated (Chevallereau et al., 2016). Despite the impressive characterization of other phage satellites, such as P4 in Escherichia coli (Lindqvist, Dehò and Calendar, 1993) and the Staphylococcus aureus pathogenicity islands (Penadés and Christie, 2015), sRNAs have not yet been identified as regulators in satellite-phage conflict.
Recently, multiple techniques to identify the targets of bacterial sRNAs in vivo have emerged (Tree et al., 2014; Han, Tjaden and Lory, 2016; Melamed et al., 2016; Waters et al., 2017), each unique in its approach and limitations. Here, we apply High-throughput Global sRNA target Identification by Ligation and sequencing (Hi-GRIL-seq) (Zhang et al., 2017), to investigate the role of PLE and ICP1 regulatory RNAs during phage infection of V. cholerae under native expression conditions. In doing so, we generated the most thorough view to date of the RNA-RNA interactome between a lytic phage and a host phage defense element, capturing interactions between phage and satellite transcripts. Characterization of the PLE-encoded sRNA, SviR, identified the regulon of the sRNA, which includes both PLE and ICP1 transcripts. SviR’s differential regulation of PLE and ICP1 targets positions this sRNA as a key regulator in PLE-ICP1 conflict. Our findings suggest that SviR-target regulation is a conserved feature of PLEs’ manipulation of ICP1, unveiling a complex RNA-RNA landscape between the phage and its satellite.
Results
Induction of the PLE regulatory RNA, SviR, is reliant on both PLE and ICP1 factors
To begin the characterization of the satellite-encoded viral-induced sRNA (SviR), we first sought to confirm the previous transcriptomic predictions that PLE encodes an sRNA (Barth, Netter, et al., 2020). RNA-seq suggested SviR is expressed from a PLE intergenic region that contains no predicted open reading frames (ORFs), supporting that the transcript could function as a regulatory sRNA. For all experiments herein, we chose to use a co-isolated pair of PLE1(+) V. cholerae and ICP1 2011_Dha_A phage (Seed et al., 2013; Boyd et al., 2021), referred to simply as PLE and ICP1. Northern blot analysis of SviR showed that the sRNA is robustly detected 12 minutes following ICP1 infection and continues to increase for the duration of the approximately 25-minute ICP1 infection cycle of PLE(+) V. cholerae (Figure 1B). sviR expression peaking late in ICP1 infection is consistent with previous RNA-seq data (Barth, Netter, et al., 2020) and supports the induction of the putative sRNA in response to ICP1 infection.
Northern blot analysis revealed that multiple SviR species are present during ICP1 infection (Figure 1B). To identify the sequence of SviR and the origin of these transcript species, we used a combination of 5’- and 3’-RACE. Analysis of the SviR 5’-RACE transformants identified a single 5’ TSS unique to the TAP-treated RNA, which corroborates the increase in transcription detected by RNA-seq (Barth, Netter, et al., 2020). In the absence of TAP treatment, we detected infrequent 5’ start sites approximately 40 nucleotides into the predicted SviR sequence, suggesting there may be 5’ processing of SviR post-transcriptionally (Figure S1). Conversely, transformants of 3’-RACE products showed two unique regions containing 3’ termination products, one consistent with the shorter 160 nucleotide (nt) SviR species, and another consistent with the longer and more abundant 210 nt SviR species (Figure 1B, Figure S1). No transformants contained a 3’ termination product corresponding to the longer 300 nt species detected by Northern blot, which is consistent with the lower relative abundance of this species compared to the two dominant species. The 3’ termination product corresponding to the most abundant 210 nt species occurs shortly after a predicted rho-independent terminator, which we hypothesize is responsible for transcript termination. No predicted terminator was found corresponding to the shorter or longer species, from which we infer these species are likely the result of post-transcriptional processing of SviR. From the combination of RNA-seq and 5’/3’-RACE, we conclude that the expression of SviR is driven by a single promoter and that the SviR isoforms detected by Northern Blot are the result of either post-transcriptional processing of the sRNA or transcript termination at distinct sites.
Having determined the bounds of the SviR gene, we next generated plasmid-based constructs aiming to recapitulate the expression of the SviR species observed during ICP1 infection of PLE(+) V. cholerae. First, we inserted the SviR gene, informed by 5’-/3’-RACE, into the well-characterized PLac0-1 vector used for expressing sRNAs in E. coli (Corcoran et al., 2012). Induction of this construct in E. coli resulted in aberrant sviR expression (Figure 1C) compared to ICP1-infected PLE(+) V. cholerae (Figure 1B). While the minor 160 nt SviR species was present, the predominant 210 nt species was nearly undetectable, and the larger 300 nt species was not detected. The absence of accumulation of the 210 and 300 nt products upon promoter induction in E. coli suggests that the longer SviR isoforms are less stable out of the native context of sviR expression. The lack of SviR base pairing with target transcripts in E. coli could lead to decreased stability of the longer SviR transcripts in E. coli. Alternatively, there may be PLE, ICP1, or V. cholerae factors that are necessary for transcript maturation which are absent when expressing SviR in E. coli.
To gain insight into the factors necessary for wild type sviR expression, we next generated a similar plasmid-based construct in V. cholerae, allowing us to assess the impact of the host, the phage, and the satellite factors involved in sviR expression. In the context of PLE, sviR is flanked by inverted repeat sequences, with the left inverted repeat upstream of the sviR TSS and the right repeat after the predicted rho-independent terminator. We hypothesized that these inverted repeat sequences and the region upstream of the sviR TSS may be involved in sviR expression. To this end, we inserted the sviR gene and the 100 base pairs (bp) upstream from the sviR TSS into a promoterless plasmid and evaluated sviR expression during ICP1 infection (Figure 1D). Expression of SviR from this construct was induced by ICP1 infection, suggesting the 100 bp upstream of SviR encodes the native PSviR promoter. Interestingly, these findings suggest there may be an ICP1-encoded factor that has been co-opted to activate the SviR promoter.
Although PSviR:sviR expression was induced by ICP1 infection, the relative abundance of the SviR species when infecting a PLE(−) host did not match the relative intensity of the SviR species observed in the native context of expression. Given that sviR expression begins to increase after the degradation of the V. cholerae chromosomes by ICP1 (Barth, Silvas, et al., 2020), we reasoned that the factor(s) responsible for the accumulation of the expected relative band intensities of SviR are likely encoded by PLE or ICP1, and not the core V. cholerae genome. To assess the role of other PLE and ICP1 factors in sviR expression, we introduced the PSviR:sviR construct to a PLE(+)ΔsviR background and measured the relative abundance of each SviR species during ICP1 infection by Northern blot. In the presence of both PLE and ICP1 factors, the relative abundance of each species of SviR was highly similar to that of native sviR expression (Figure 1D). Together, these data support that ICP1 infection induces expression of the PSviR promoter, and that wild type SviR species accumulation is dependent on the presence of PLE factors. Whether this induction is directly due to an ICP1-encoded factor, or due to the change in the host cell in response to ICP1 infection is unknown. The exact nature of the PLE-dependent change in species abundance is also unknown, but we hypothesize that either SviR’s interaction with PLE target transcripts changes isoform stability, or that a PLE-encoded factor interacts with SviR transcripts, resulting in the accumulation of the larger transcript species.
To begin to address the role of SviR in PLE-ICP1 dynamics, we evaluated the response of PLEΔsviR to ICP1 infection. The deletion of sviR did not impair PLE’s capacity to block ICP1 progeny production, consistent with the previous observations that none of the 27 PLE ORFs are necessary for PLE’s inhibition of ICP1 (Hays and Seed, 2019). We detected decreased genome replication of both PLE and ICP1 (Figure S2A) and a delay in the timing of cell lysis (Figure S2B) when comparing PLEΔsviR infection to PLE(+) infection. However, both the host cell lysis (Hays and Seed, 2019) and genome replication (Barth, Silvas, et al., 2020; LeGault et al., 2022) are dependent on multiple PLE or ICP1 products, providing no clear candidate genes as targets of SviR regulation. Therefore, we next pursued identifying target transcripts of SviR during ICP1 infection of PLE(+) V. cholerae.
Hi-GRIL-seq reveals a complex RNA-RNA interactome between PLE and ICP1
To identify putative target transcripts regulated by SviR during ICP1 infection, we turned to Hi-GRIL-Seq (Zhang et al., 2017) to capture RNA-RNA interactions 16 minutes post-ICP1 infection, during peak sviR expression. RNA-seq of T4 RNA ligase treated samples and subsequent bioinformatic analysis (See Methods for further details) identified the presence of chimeric transcripts late in ICP1 infection of PLE(+) cells (Figure 2A). PLE-ICP1 cross-genome chimeric transcripts were identified as any transcript where the ends of the transcript mapped to these two distinct genomes. For reads that mapped to a single genome, transcripts were assumed to be chimeric only if the two ends of the reads mapped greater than 1 kb apart, comparable to prior applications of Hi-GRIL-seq (Zhang et al., 2017). This approach revealed that chimeras formed repeatably between each of the three biological replicates (Figure S3A, S3B), generating a data set from which we could identify candidate target transcripts of SviR regulation. While some chimeric transcripts were detected between PLE transcripts and transcripts on the core genome, nearly all of these interactions were between ribosomal RNAs and PLE transcripts, despite rRNA depletion during RNA isolation (Figure S3C). There are likely biologically relevant interactions occurring in this smaller PLE-chromosomal transcript dataset, however, they were not investigated further.
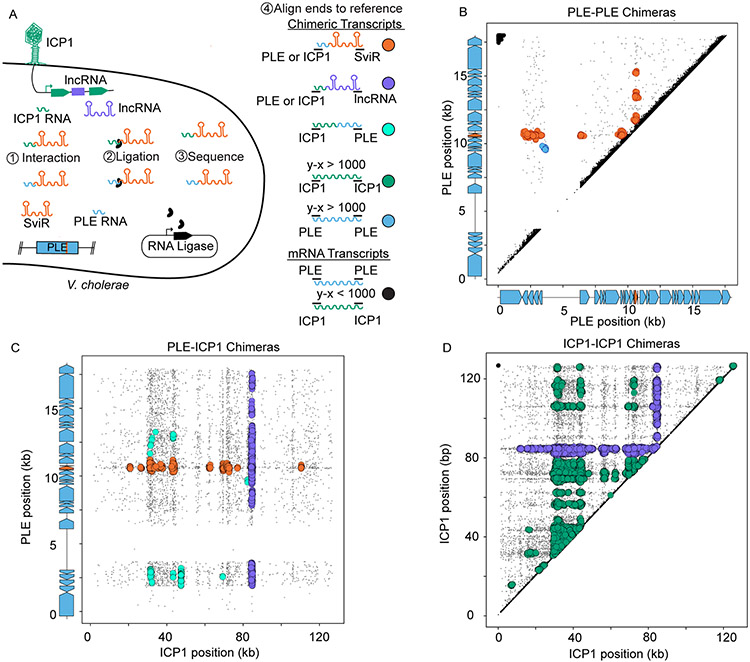
Figure 2). Hi-GRIL-seq during ICP1 infection detects a complex RNA-RNA interactome between PLE and ICP1 transcripts.
(A) A schematic of the Hi-GRIL-seq experiment carried out during ICP1 infection of PLE(+) V. cholerae. Assigning the genomic coordinates of the first and last 25 bp of each transcript provides an x-y coordinate pairing, which was then plotted for each read. Reads that map to the same genome with less than 1 kb between the ends of the transcript were treated as background reads (black), likely representing the ends of a single, non-chimeric transcript. Different classifications of transcript species, both chimeric (colors) and non-chimeric (black), are indicated to the right of the model.
(B, C, and D) The first and last 25 nt of Hi-GRIL-seq reads were mapped to the reference genomes of PLE-PLE transcripts (B), PLE-ICP1 transcripts (C), and ICP1-ICP1 transcripts (D). For the axes corresponding to PLE reads, the gene graphs on the graph axes demarcate protein-encoding ORFs in blue and sviR in orange. For PLE-PLE (B) and ICP1-ICP1 (D) interactions, upper triangle graphs were used to limit redundant mapping. Plotted reads were clustered using DBScan, where greater than 20 reads mapping to an area of diameter 250 bp (B), 350 bp (C), and 1000 bp (D), to scale for differences in genome size between PLE and ICP1, are indicated as a larger colored circle. Clusters of chimeras were colored according to the legend in (A). Reads not passing the threshold of clustering were colored gray and sized smaller than reads falling into a cluster. For the unclustered graphs and replicates, see Figure S3.
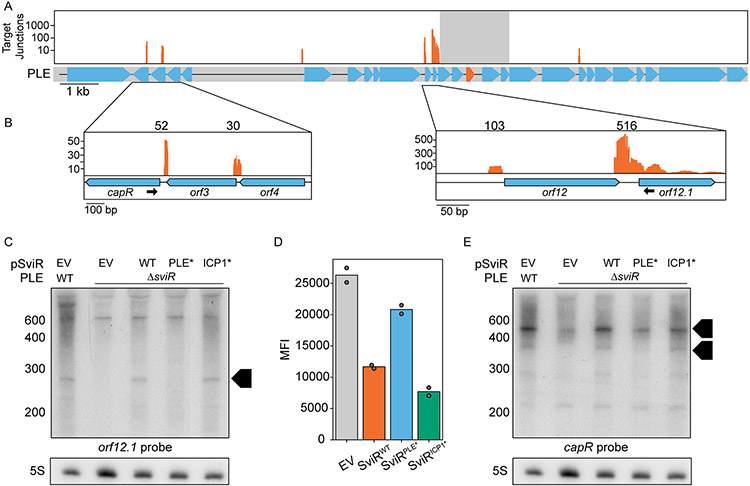
First, we analyzed the chimeric reads formed between PLE-PLE transcripts (n=1,289 ±196). After DBScan clustering to separate background noise from clusters of interactions, all but one cluster of PLE-PLE chimeric reads corresponded to a SviR-PLE transcript interaction, suggesting SviR regulates targets across multiple PLE operons (Figure 2B). Primarily, these interactions occurred between SviR and the 5’-UTR of target transcripts, consistent with canonical sRNA regulation. To our surprise, the only cluster of PLE-PLE chimeric reads which was not a SviR-PLE interaction occurred between the 5’-UTR of the operon for PLE orfs2-5 and PLE orf12 (Figure 2B), supporting this 5’-UTR as a potential second PLE non-coding RNA. Previously, RNA-seq identified this 5’-UTR as being transcribed during ICP1 infection, albeit to a much lower level than that of SviR (Barth, Netter, et al., 2020). The same promoter responsible for driving sviR expression is conserved upstream of this putative regulatory RNA (Figure S4A), suggesting that both SviR and the orfs2-5 5’-UTR transcript may be expressed in response to the same factors during ICP1 infection. This putative PLE promoter sequence shares a high level of identity with the E. coli sigma-70 promoter and is conserved in multiple loci across each of the 10 PLEs (Figure S4B), potentially serving as the primary PLE promoter for transcriptional activation upon ICP1 infection. Both SviR and the orfs2-5 5’-UTR transcript were found by Hi-GRIL-seq to interact with the orf12-12.1 operon, placing this target operon at the center of the PLE RNA-RNA interactome and making characterization of this locus of interest for furthering the understanding of PLE-ICP1 conflict.
A parallel analysis of chimeric transcripts between PLE and ICP1 (n=5,399±1,407) revealed a complex interactome between PLE and ICP1 (Figure 2C). Consistent with the PLE-PLE chimeras, SviR was the PLE transcript most often ligated with ICP1 transcripts, and chimeras were most frequently detected between 5’-UTRs of ICP1 transcripts and SviR. SviR-ICP1 chimeras were detected across multiple ICP1 operons, identifying a list of ICP1 candidate target genes for SviR regulation. Unlike PLE-PLE chimeras, where only one cluster did not include SviR, we identified multiple clusters of PLE-ICP1 chimeras which did not include the sRNA. The majority of these non-SviR clusters were formed between PLE transcripts and the putative ICP1-encoded lncRNA, supporting the lncRNA as a potential base pairing RNA encoded by ICP1. In agreement with this, we also observed ICP1-lncRNA chimeras above the detection threshold across ICP1’s genome, indicating RNA-RNA interactions between the lncRNA and both PLE and ICP1 transcripts (Figure 2D). Northern blot analysis of the lncRNA showed a similar expression pattern to that of SviR (Figure S5A), showing the lncRNA is robustly transcribed late in ICP1 infection. We hypothesized that this lncRNA may be functioning as an RNA sponge, sequestering SviR from target transcripts. Consistent with this hypothesis, both our Hi-GRIL-seq data and IntaRNA predictions identified the same base pairing region between the SviR and the lncRNA (Figure S5B). To test if the lncRNA affected the stability of SviR, or the SviR isoforms expressed, we generated a deletion of the lncRNA in ICP1 and monitored SviR abundance during infection by Northern blot. We were unable to detect a difference in the size or intensity of SviR species compared to infection by wild type phage (Figure S5C), suggesting that the stability of the two transcripts is not co-dependent, despite the predicted RNA-RNA base pairing between the two transcripts. While this does not rule out the lncRNA functioning as a sponge transcript, further analysis is required to determine the outcome of lncRNA-SviR interactions.
SviR interacts with PLE and ICP1 transcripts with unique seed regions
The RNA-RNA chimeras detected between SviR and both PLE and ICP1 transcripts strongly support SviR functioning as a base pairing sRNA. We next set out to identify the seed region that SviR uses to initiate base pairing with PLE and ICP1 transcripts by analyzing the junctions of the chimeras from the Hi-GRIL-seq dataset. We reasoned that the in vivo T4 RNA ligase-mediated junctions between SviR and target transcripts would allow for the detection of interacting base pairs between the SviR seed region and target transcript, as ligation occurs under native interacting conditions. For each chimeric read, we identified the first 25 nt of SviR proximal to the chimeric junction and mapped this 25mer back to the SviR sequence. These 25mers mapped to SviR primarily in three peaks. Of these three peaks of junctions, one near the 5’ end of SviR primarily corresponded to ICP1 target junctions, and one peak near the 3’ end of SviR largely corresponded to PLE target junctions, suggesting that discrete regions of SviR interact with PLE and ICP1 targets (Figure 3A). A third peak comprised of both PLE and ICP1 target junctions was observed near the TSS, which we expect likely represents non-specific ligation between the 5’ phosphoryl terminus of SviR and target transcripts due to the in vivo ligation step of Hi-GRIL-seq and does not likely represent biologically relevant RNA-RNA interactions.
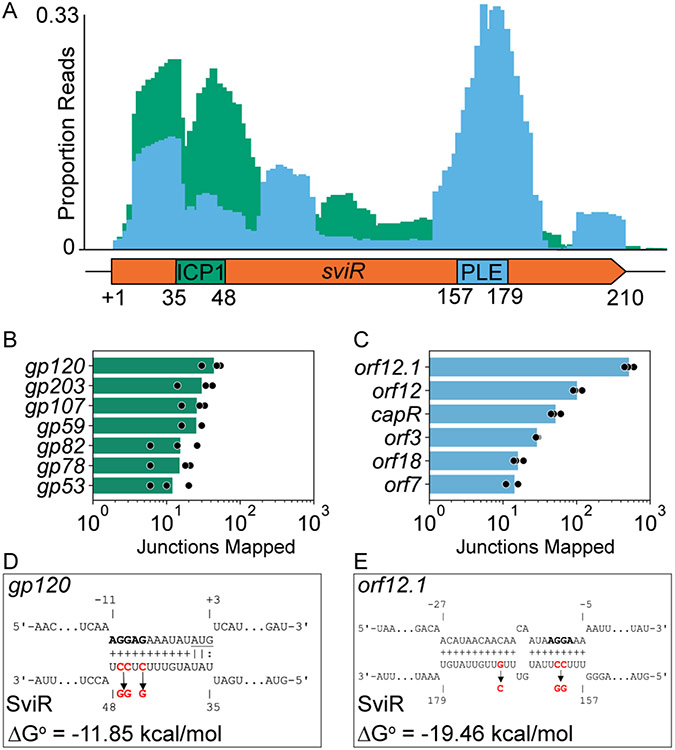
Figure 3). SviR has two unique seed regions to regulate ICP1 and PLE transcripts.
(A) The first 25 bp of SviR flanking the SviR-target junction were mapped to SviR for all SviR-PLE (blue) and SviR-ICP1 (green) chimeras detected by Hi-GRIL-seq. Mapped read junctions across SviR were graphed as a proportion of total SviR-target chimeras in each genome (PLE or ICP1) to normalize the number of chimeras. Annotations on the SviR gene graph below the histogram represent the seed regions predicted by IntaRNA for ICP1 gp120 (D) and PLE orf12.1 (E), which are representative of seed regions predicted for most PLE and ICP1 targets (Table 1).
(B and C) Quantification of SviR target ORFs in ICP1 (B) and PLE (C) detected by Hi-GRIL-seq. Mapping of the 25 bp flanking each SviR junction shows the target transcript for each SviR chimera. The number of reads mapping to a position on the target transcripts is reported for each target, requiring a minimum average value of 10 chimeras for inclusion. Reads mapping to intergenic loci were assigned to the downstream ORF if the mapping was within 50 bp of the ORF start codon. Presented values (bars) are averages of reads mapped across three biological replicates (replicates shown in black circles).
(D and E) IntaRNA predicted interactions between the most abundant SviR targets identified by Hi-GRIL-seq in ICP1, gp120 (D), and PLE, orf12.1 (E). Each predicted ribosome binding site is bolded and the gp120 start codon is underlined. Numbering on target transcripts is relative to the start codon and numbering on SviR is relative to the sviR 5’ TSS as determined by 5’-RACE. Nucleotides highlighted in red are predicted by CopomuS as important for sRNA-target base pairing and were mutated for the subsequent analyses using SviRICP1* and SviRPLE* alleles.
Leveraging the same SviR-target junctions, we mapped the adjacent, target transcript-mapping 25mer to its respective genome and quantified the number of chimeras mapping to each PLE and ICP1 target. Both PLE (n=6, Figure 3B) and ICP1 (n=7, Figure 3C) ORFs were identified to be targets of SviR from this analysis, as defined by having an average of greater than ten detected interactions across biological replicates. To provide support to this analysis in an unbiased manner, IntaRNA predictions were calculated between SviR and all PLE and ICP1 transcripts. From these predictions, we identified two putative SviR seed regions; one interacting with ICP1 targets (Figure 3D, Table 1) and a second interacting with PLE targets (Figure 3E, Table 1). Together, the analysis of the observed junctions of chimeric reads generated by Hi-GRIL-seq and the predicted seed regions by IntaRNA identified similar, conserved interactions between SviR and target transcripts (Figure 3A). These data identify two sets of candidate genes that are regulated by SviR and suggest that SviR employs two seed regions, one region for interacting with PLE transcripts and a second for interacting with ICP1 transcripts.
Table 1).
Chimeric junctions mapped from three Hi-GRIL-Seq replicates and IntaRNA interactions
| Target Transcript |
Genome | Hi-GRIL-Seq Chimeras |
SviR Range |
Predicted Interaction ΔG° |
Target Range |
|---|---|---|---|---|---|
| orf12.1 | PLE | 516±77.25 | 157 to 179 | −19.46 kcal/mol | −27 to −5 |
| orf12 | PLE | 101.33±15.5 | 155 to 179 | −10.42 kcal/mol | −28 to −4 |
| capR (orf2) | PLE | 51.66±8.3 | N/A | N/A | N/A |
| gp120 | ICP1 | 44±12.5 | 35 to 48 | −11.85 kcal/mol | −11 to +3 |
| gp203 | ICP1 | 30±14.4 | 37 to 64 | −21.91 kcal/mol | −35 to −2 |
| orf3 | PLE | 29±1 | N/A | N/A | N/A |
| gp107 | ICP1 | 25.6±8.7 | N/A | N/A | N/A |
| gp59 | ICP1 | 25.3±8.1 | 41 to 63 | −17.83 kcal/mol | −20 to +2 |
| orf18 | PLE | 19±2.64 | N/A | N/A | N/A |
| gp82 | ICP1 | 15.3±10 | 20 to 48 | −12.86 kcal/mol | −12 to +22 |
| gp78 | ICP1 | 15±7.9 | N/A | N/A | N/A |
| orf7 | PLE | 14.33±2.8 | 150 to 177 | −16.85 kcal/mol | −2 to −21 |
| gp53 | ICP1 | 12±7.2 | 33 to 52 | −23.14 kcal/mol | −7 to −27 |
The number of SviR-Target chimeric junctions with at least 10 junctions and the predicted IntaRNA interactions for each target. Junctions that mapped to within 50 bp upstream of the start codon of an ORF were attributed to the downstream ORF. Junctions that mapped to targets not predicted to interact with SviR by IntaRNA are qualified with N/A. Target ranges are relative to the start of translation, when applicable, and refer to predicted base pairing interactions as determined by IntaRNA.
SviR is necessary for the translational downregulation of ICP1 Gp120, a putative ICP1 structural protein
To investigate the outcome of SviR regulation of an ICP1 target, we focused on the most abundant SviR-ICP1 chimera, gp120 (Figure 4A, Table 1), a gene of unknown function expressed late in ICP1 infection (Barth, Netter, et al., 2020). Hi-GRIL-seq junction mapping detected SviR interactions with the 5’-UTR of gp120, just downstream of the TSS identified by 5’-RACE (Figure 4B). Northern blot analysis of gp120 during ICP1 infection showed equivalent transcript levels in the presence and absence of SviR (Figure 4C). Because SviR is predicted by IntaRNA to occlude the gp120 ribosome binding site (Figure 3D), potentially inhibiting translation, we next engineered a C-terminal 3xFLAG-tagged gp120 under the control of the endogenous gp120 promoter. Using this engineered phage, we monitored Gp120::3xFLAG levels during phage infection by Western blot analysis in the presence and absence of SviR. Gp120 protein levels increased in the PLEΔsviR background (Figure 4D, Figure S7), consistent with SviR downregulating Gp120 post-transcriptionally, potentially by occluding the ribosome binding site. Complementation of SviR from PSviR:sviR decreased Gp120::3xFLAG to wild type abundance, confirming the role of SviR in the downregulation of Gp120 translation. To assess if this downregulation is the result of direct SviR base pairing to the gp120 transcript, we introduced mutations to disrupt each SviR seed region individually (Figures 3D and 3E), as informed by CopomuS. Surprisingly, complementation of PLEΔsviR with either PSviR:sviRICP1* or PSviR:sviRPLE* resulted in wild type Gp120 levels when compared to the levels of PLEΔsviR background, suggesting that both mutant alleles were no longer functional in the regulation of gp120 translation. While this outcome is expected for the ICP1 seed region mutant, as SviR should be unable to interact with gp120, the PLE seed region is not predicted to interact with gp120 and therefore is not expected to be involved with gp120 regulation. Both mutant seed regions were expressed to levels comparable to the expression of the wild type allele (Figure S8A), ruling out differences in SviR abundance as limiting factors for Gp120 regulation. However, predictions of the secondary structure of the SviR with the mutated PLE seed region suggest some changes to the secondary structure, occluding both the PLE and ICP1 seed regions (Figure S8B). This disrupted secondary structure may lead to inhibition of direct base pairing between SviRPLE* and gp120, as the ICP1 seed region may no longer be capable of interacting with the target transcript, despite the wild type ICP1 seed region sequence being present.
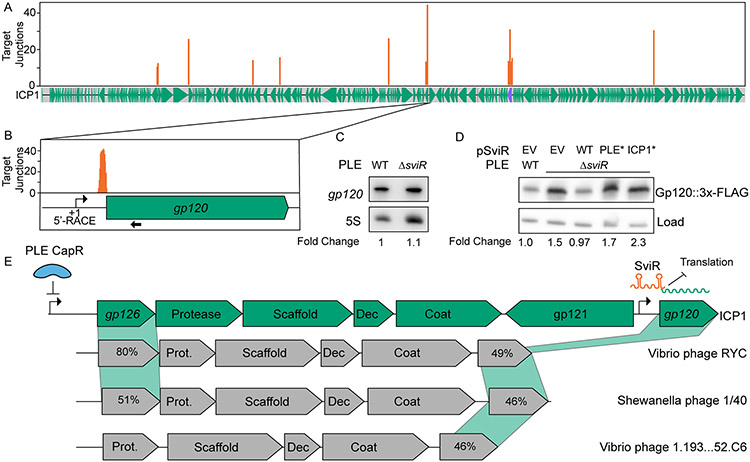
Figure 4). ICP1 Gp120 translation is decreased in the presence of SviR.
(A) Mapping of SviR-ICP1 chimeric junctions identified in Figure 3B to the ICP1 genome. The 25 bp following each SviR-ICP1 junction was mapped to the ICP1 reference genome. Mapping represents the average of three biological replicates and positions with an average number of reads less than 10 are not shown. The purple annotation on the ICP1 gene graph represents the ICP1 lncRNA. For individual replicates and unfiltered graphs, see Figure S6.
(B) Inset of SviR chimeras mapping to gp120, the target with the greatest average number of interactions of the SviR-ICP1 chimeras identified by Hi-GRIL-seq. The approximate position of the gp120 promoter is represented by a bent arrow icon, as determined by 5’ RACE and RNA-seq. The probe used for gp120 Northern blot in (C) is indicated by a black arrow under the gene graph.
(C) Northern blot analysis using a probe complementary to gp120. All RNA samples were isolated 16 minutes post-infection by ICP1, either in a PLE(+) or PLEΔsviR background. The values under each lane represent the gp120 transcript abundance, normalized to the 5S rRNA, compared to the wild type background between two biological replicates.
(D) Western blot analysis of Gp120::3x-FLAG ICP1 infection in a PLE(+) or PLEΔsviR background with the sviR expression construct as indicated (EV is the empty vector control, WT indicates expression of wild type SviR as in Figure 1D). SviRICP1* and SviRPLE* mutant alleles contain mutations highlighted in Figures 3D and 3E, respectively. Total protein input was normalized between lanes. The load control represents a non-specific FLAG background band present at approximately equal abundance across samples. The values under each lane represent the Gp120::3x-FLAG intensity, normalized to the loading control, compared to the wild type background, and averaged between three individual biological replicates. Each of the three biological replicates with the unaveraged normalized values as well as complementary statistical analyses, are shown in Figure S7.
(E) Gene graphs of ICP1 and non-related phages showing synteny of the capsid operon and a model of SviR-based regulation of gp120. CapR, PLE’s transcriptional repressor, binds upstream of gp126, downregulating the expression of ICP1’s capsid operon (Netter et al., 2021). gp121 encodes a putative mobile homing endonuclease gene, hypothesized to de-couple the regulation of gp120 from the rest of the capsid operon. Functional annotations of gene products from related phages are based on Pfam domain predictions using the HHPRED webserver. For gene products without annotation, percent amino acid similarity to ICP1 Gp126 or Gp120 was shown instead. Dec = capsid decoration protein; Prot. = scaffold protease, Coat = major capsid protein, Scaffold = predicted capsid maturation protease.
To further investigate if SviR and gp120 directly interact or if the regulation of Gp120 may be indirect, we generated a gp120::GFP translational fusion reporter and monitored GFP levels in the presence of sviR expression in E. coli. The expression of the gp120::GFP fusion construct was only mildly affected by sviR expression, and this decrease was not restored by the expression of either mutant allele (Figure S9). These data indicate that direct regulation of Gp120::GFP by SviR is unlikely to occur in E. coli. However, the expression of SviR in E. coli differs from that in V. cholerae (Figures 1C and 1D), which limits the interpretation of these results. While we were unable to validate direct interaction between SviR and gp120 in E. coli, the abundance of Hi-GRIL-seq chimeras formed between the two transcripts in V. cholerae is consistent with the predicted interactions by IntaRNA. Nonetheless, it is possible SviR’s regulation of Gp120 is indirect and SviR acts to recruit other factors, which are required for the post-transcriptional downregulation of Gp120. Regardless of the mechanism, these data support SviR-dependent downregulation of Gp120 translation during ICP1 infection under the native expression conditions of both the sRNA and target transcript.
The downregulation of Gp120 by SviR represents the second PLE-encoded mechanism of downregulating this region of the ICP1 genome, which includes the capsid operon. PLE-encoded CapR is a transcriptional repressor that binds to the promoter region of the capsid operon upstream of gp126 (Figure 4E) (Netter et al., 2021). However, in all sequenced ICP1 isolates, gp120 is transcriptionally de-coupled from the ICP1 capsid operon, due to gp121 encoded on the reverse strand. gp121 is a putative homing endonuclease gene, a type of selfish mobile gene which has been previously hypothesized to play a role in the co-evolution of PLE and ICP1 (Barth, Nguyen and Seed, 2021; Boyd et al., 2021; Netter et al., 2021). We identified multiple phages that lack a gp121 homolog but otherwise share gene synteny with ICP1’s capsid operon (Figure 4E), suggesting the gene architecture in an ancestral ICP1 may have included gp120 in the capsid operon. PSI-BLAST of these gp120 homologs detects similarity to the T4 inh protein, an inhibitor of the procapsid protease in coliphage T4, suggesting gp120 may play a role in regulating ICP1 capsid morphogenesis (Miller et al., 2003). Therefore, SviR-dependent downregulation of ICP1 gp120 may represent a secondary mechanism of downregulation of the ICP1 capsid components by PLE, in addition to the transcriptional repressor CapR.
Transcripts from multiple PLE operons are regulated by SviR
Next, we set out to determine the outcome of SviR’s interactions with putative PLE target transcripts. Mapping of SviR-target junctions across PLE revealed interactions specifically with the 5’-UTR of target sequences across multiple PLE operons (Figure 5A). To characterize the outcome of these interactions, we first focused on the most abundant chimeras detected by Hi-GRIL-Seq, those between SviR and orf12.1 (n=516) and orf12 (n=101) (Figure 5B, right and Table 1). PLE orf12 and orf12.1 are both small ORFs of 59 and 39 amino acids, respectively, each encoding proteins of unknown function with homologs only in other PLEs. We performed Northern blot analysis with a probe complementary to orf12.1 to assess the fate of the orfs12-12.1 operon transcript. In the presence of SviR, shorter transcript species, consistent with RNase degradation products, were observed for orf12.1 (Figure 5C), which were no longer present in PLEΔsviR. Complementation of SviR from PSviR:sviR restored the shorter transcripts, showing SviR regulation is necessary for the regulation of this operon. Importantly, complementation was successful with PSviR:sviRICP1*, but not PSviR:sviRPLE*, supporting the importance of base pairing of SviR’s PLE seed region for wild type regulation. The same transcript pattern was observed using a probe against orf12, showing that the outcome of transcript regulation is consistent across the operon (Figure S8C). Notably, the dispersed, longer transcripts detected with the orf12.1 probe were also detected with the orf12 probe (Figure S8C), and only detected upon phage infection (Figure S5C), consistent with the known kinetics of orf12.1 expression (Barth, Netter, et al., 2020). These data indicate that the longer transcripts are not a result of non-specific probe binding and indicate they are likely operon-length transcripts. Further supporting the direct regulation of orf12.1 by SviR, the expression of an orf12.1::GFP translational fusion reporter in E. coli was decreased in the presence of SviR in a PLE seed region-dependent manner (Figure 5D, Figure S10B). However, the introduction of compensatory mutations to restore the interaction between SviRPLE* and the orf12.1 5’-UTR necessitated mutation of the RBS, which reduced GFP expression significantly, precluding us from verifying the direct base pairing in a heterologous system (Figure S10A, S10C).
Figure 5). SviR regulation leads to altered levels of transcripts across PLE.
(A) Mapping of the SviR-PLE chimeras to the PLE reference genome. The mapping was performed as described in Figure 4A. The reads mapped here represent the average of three biological replicates. Positions with fewer than 10 interactions are filtered out to decrease noise. For each individual replicate and the unfiltered graphs, see Figure S6. Reads are mapped on a log scale in (A) due to the range of values mapped. The gray box overlaying the SviR annotation (orange) on the PLE gene graph represents the 1kb region flanking the center of SviR, where chimeric reads were unable to be discerned from non-chimeric transcripts due to proximity to the sRNA.
(B) Inset images of PLE target junction mapping. Zoomed insets show a linear scale of SviR interactions with capR (left) and orfs12 and 12.1 (right). The black arrows indicate the Northern blot probes used in (C) and (E). Scales in each inset differ based on local maxima.
(C and E) Northern blot analysis using a probe complementary to orf12.1 (C) or capR (E) in the 5’ region of the ORF as indicated in (B). All RNA samples were isolated 16 minutes post-ICP1 infection. SviRPLE* and SviRICP1* mutant alleles contain mutations highlighted in Figures 3E and 3D, respectively. Black boxed arrows highlight transcript species differentially regulated by SviR in a manner dependent on the PLE seed region. Blots were stripped and re-probed with each target probe, resulting in identical 5S images for both orf12.1 and capR blots.
(D) Flow cytometry analysis of orf12.1::GFP expression in the presence and absence of SviR alleles in E.coli. sviR expression was induced in all samples and mean population fluorescence was measured. Graphed values represent the mean fluorescent intensity of the GFP(+) population of 50,000 counted events, analyzed with FlowJo.
A second PLE operon detected by Hi-GRIL-seq to be targeted by SviR was the operon encoding CapR, where both capR and orf3 were observed as SviR targets by Hi-GRIL-Seq (Figure 5B, left and Table 1). To determine if the capR operon shared the same fate as the orf12.1 operon, we performed a similar Northern blot analysis using a probe complementary to capR. Consistent with SviR’s regulation of the orf12.1 operon, we observed the regulation of capR to be dependent on the PLE seed region of SviR (Figure 5E), with an accumulation of shorter transcript species, potentially RNase degradation products, when base pairing SviR alleles were expressed. However, complicating the model that these shorter transcripts represent RNase degradation products, the overall transcript abundance of the capR species appears to be higher in the presence of SviR (Figure 5E). Collectively, the PLE target transcripts (capR and orfs12-12.1) of SviR investigated here show varied transcript stability in a manner dependent on the PLE seed region, however the ultimate outcome of this regulation and the mechanism underlying it are not yet explicitly understood.
To identify what other factors may be involved in SviR regulation of PLE transcripts, we tested whether the chaperones Hfq and ProQ, as well as the ICP1 lncRNA, played a role in SviR-based regulation of PLE targets. Interestingly, the regulation of all tested PLE target transcripts occurs independently of either of the RNA chaperones Hfq or ProQ, although the transcript abundance of SviR may be slightly decreased in the absence of Hfq (Figure S8). V. cholerae, PLE, or ICP1 may encode a novel RNA chaperone, which facilitates SviR-target interactions, or regulation of these targets occurs independently of chaperones. The regulation of PLE orf12.1 was unchanged in the absence of the ICP1 lncRNA (Figure S5C), indicating that, while the lncRNA is predicted to interact with SviR, SviR regulation of other transcripts is consistent in the presence or absence of the lncRNA.
SviR regulation is predicted to be a conserved feature of all known PLEs
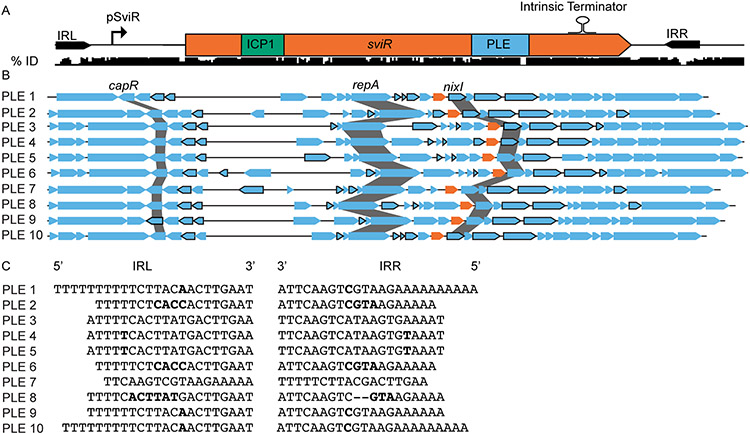
Each of the ten identified PLEs is distinct from another, containing a suite of core genes, including SviR, and variable genes. To predict whether SviR regulation is a conserved feature across the PLEs, we constructed a nucleotide alignment of SviR and the surrounding sequence from each of the ten PLEs, showing a high level of conservation between SviR alleles (Figure 6A). The regions of the highest conservation across the ten SviR alleles are those with functional relevance, including the promoter region, both seed regions, and an intrinsic terminator predicted to terminate SviR transcription to generate the most abundant 210 nt species (Figure 1B). Consistent with these predictions, RNA-seq profiling of PLEs 1-5 during ICP1 infection detected sviR expression consistent with the pattern observed for PLE 1 (Barth, Netter, et al., 2020). Because SviR is highly conserved across all PLEs, we next questioned if the SviR targets in other PLEs had predicted base pairing with their cognate SviR allele (Figure 6B). The bounds of the different SviR alleles were predicted based on the conserved sequence features and the interactions between each of the putative sRNA alleles and the cognate PLE ORFs were predicted using IntaRNA. Each SviR allele was predicted to interact with the 5’-UTR of PLE target transcripts using a homologous sequence to the seed region identified for PLE 1 (Table S5). Some conserved loci, such as the capR operon and the cluster of genes downstream of SviR, were consistently predicted to be regulated by SviR across all PLEs. However, the region between PLE repA and sviR, a highly variable gene neighborhood of the PLEs, was also predicted to be regulated by SviR. This suggests that SviR regulation of PLE transcripts is central to the PLE gene expression program, as SviR targets include core and accessory ORFs in multiple PLE operons.
Figure 6). SviR is a core regulatory component of all PLEs discovered to date.
(A) SviR and its flanking repeats sequences are conserved across all ten PLEs. A nucleotide alignment of SviR and the flanking regions (represented by the black bars below the gene graph) shows the percent identity of the region across all ten PLEs. The ICP1 and PLE seed regions are highlighted in green and blue, respectively. The inverted repeats flanking SviR are marked by black arrows and the relative location of the SviR promoter and intrinsic terminator are annotated above the sequence, as determined by RACE and bioinformatic predictions. IRL = inverted repeat left, IRR = inverted repeat right.
(B) SviR is predicted to be a regulatory feature of all ten PLEs. Each of the ten PLEs is shown by a representative gene graph with each SviR allele in orange. ORFs outlined in black are predicted by IntaRNA to interact with the SviR PLE seed region with a ΔG° < −10.
(C) The inverted repeats flanking SviR from each of the ten PLEs. The mismatched bases between repeat sequences are highlighted in bold, and gaps in the repeat sequences are represented by hyphens. Despite the highly variable sequences between the 10 PLEs, the presence of inverted repeats is consistent across all PLEs.
Finally, every SviR allele is flanked by an imperfect inverted repeat sequence, one upstream of the SviR promoter and another following the predicted Rho-dependent terminator (Figure 6A). Between the ten PLEs, the sequence of the inverted repeats is variable, but mutations present in one inverted repeat are routinely compensated for in the opposing repeat, suggesting selection for repeat complementarity (Figure 6C). While the function of these repeats is unknown, their inclusion on the plasmid constructs for sviR expression replicated the wild type expression patterns for SviR (Figure 1D), suggesting the potential role in sviR expression may lead to selection for conservation between repeats. Together, these bioinformatic analyses support SviR as a key factor in PLE-ICP1 co-evolution, regulating PLE and ICP1 targets throughout decades of MGE-phage pairings.
Discussion
This work describes the role of SviR, a satellite-encoded sRNA, in the differential regulation of both satellite and phage gene expression during infection of PLE(+) V. cholerae by the phage ICP1. The ability of SviR to regulate both PLE and ICP1 transcripts places the sRNA at the interface between the satellite and phage gene regulation. Satellite parasitism of phages requires a delicate balance of permitting and inhibiting select aspects of phage infection to proceed. Either premature or delayed inhibition of phage gene expression is deleterious to the satellite, which requires temporally controlled expression of phage gene products for satellite transmission in modified phage virions. To obtain this necessary balance, satellite genes encode diverse mechanisms to regulate the expression of both satellite and phage gene expression. PLEs (Netter et al., 2021), SaPIs (Ram et al., 2014), and P4 (Lindqvist, Dehò and Calendar, 1993) all downregulate phage late gene expression through unique mechanisms, now including a regulatory RNA, SviR. It is an attractive hypothesis to speculate that late genes are often the targets of satellite-mediated downregulation because the modified virions of satellites require fewer structural protein monomers than their phage prey, however, this has not been shown experimentally. SviR not only regulates ICP1 late gene expression but also differentially regulates PLE transcripts through a unique mechanism, highlighting the versatility of sRNAs as regulators. While PLE gene regulation remains mysterious, the characterization of SviR has shown that the sRNA is a pivotal piece of the puzzle. Broadly, the RNA-RNA interactome identified in this study brings to question the role of regulatory RNAs in phage-satellite conflicts, which have otherwise not been interrogated, and highlights the potential for discovery in such systems with global RNA interactome studies.
We demonstrated that Hi-GRIL-seq is a powerful tool to detect RNA-RNA interactions during phage infection, doing so without enrichment for specific types of interactions. The global view of interactions obtained from Hi-GRIL-seq certainly has benefits. Our data set contains robust interactions detected between multiple non-coding RNAs and other transcripts. However, the breadth of data obtained from Hi-GRIL-seq comes at a cost when compared to global RNA interactome techniques. First, Hi-GRIL-seq does not enrich for specific interactions, instead relying on increased sequencing depth to identify RNA-RNA interactions. Comparable RNA interactome approaches rely on co-immunoprecipitation of interacting transcripts with a cellular bait, such as Hfq (Melamed et al., 2016), RNase E (Waters et al., 2017), or a transcript itself (Lalaouna et al., 2017). Co-immunoprecipitated transcripts are then sheared and ligated in vitro, covalently joining the two transcripts at an artificial junction. In the absence of shearing and ligation, some interactions may not be captured, as the necessary 5’-PO4 and 3’-OH groups for ligation are not proximal to one another. Consistent with this limitation, many PLE ORFs predicted to interact with SviR using the PLE seed region were not detected by Hi-GRIL-seq (Table S4). Deeper sequencing of Hi-GRIL-seq libraries, or parallel analysis with another global RNA-RNA interactome technique, may reveal interactions between these transcripts.
Hi-GRIL-seq can detect interacting transcripts genome-wide under the conditions of interest, making it an optimal technique for the de novo discovery of regulatory RNAs independently of any co-associated protein. However, the lack of specificity in the interactions detected leads to high levels of noise, which complicates downstream analyses. Accordingly, we detected a high level of both inter- and intra-genomic ICP1 RNA-RNA interactions. These may be bona fide interactions detected by Hi-GRIL-seq that serve an unidentified role in the ICP1 infection program, perhaps by increasing RNA turnover to accommodate the rapid transcriptional takeover of the host cell. Alternatively, the abundance of phage transcripts expressed upon phage infection may result in non-specific ligation upon the expression of T4 RNA ligase. Previous Hi-GRIL-seq experiments show that the ligation efficiency of two transcripts is not directly correlated with transcript abundance (Zhang et al., 2017). However, these studies were not done in a phage-infected host, where cellular transcription and RNA decay are hijacked by phages, potentially affecting ligation dynamics. Future studies interrogating the RNA-RNA interactome of other phage-host pairings, by either Hi-GRIL-seq or other techniques, would provide further insight into whether these observations are specific to ICP1 or are observed across diverse phages.
The SviR-mediated regulation of PLE and ICP1 transcripts reveals a role for sRNAs in phage-satellite conflicts. However, much of the mechanism by which SviR regulates target transcripts remains unknown. While sRNAs are not well characterized in phage-satellite conflict, the similarities between satellites and prophages provide some grounds on which comparisons can be drawn. Prophage-encoded regulatory RNAs have primarily been shown to be involved in the lysis-lysogeny decision, triggering the switch from a lysogenic state to a lytic state following sRNA-mediated changes in the target transcript stability (Wagner, Altuvia and Romby, 2002). Most of these prophage regulatory RNAs, such as the OOP RNA of phage lambda, are encoded as cis-antisense regulatory RNAs. The OOP RNA employs long regions of perfect complementarity to direct RNase III-mediated cleavage of the cII RNA, driving the initiation of the lambda lytic cycle (Krinke and Wulff, 1990). The interaction between SviR and PLE target transcripts is likely not of the length necessary to recruit RNase III, making regulation by RNase III unlikely. Examples of trans-acting sRNAs encoded by MGEs are rare but have been documented. Characterization of the Hfq-associated sRNAs in enterohemorrhagic E. coli revealed an abundance of putative sRNAs, which act in trans to antagonize core genome and MGE transcripts, at least in part due to recruitment of RNase E (Tree et al., 2014; Waters et al., 2017). Other core genome sRNAs in V. cholerae are often found to regulate targets through the recruitment of RNase E and subsequent target degradation, resulting in the accumulation of short degradation products (Davis and Waldor, 2007; Venkat et al., 2021). This pattern is consistent with our Northern blot analysis of PLE-encoded targets of SviR, such as orfs12-12.1 and capR. However, there is potentially a slight increase in capR transcript abundance in the presence of SviR, which is more consistent with transcript stabilization than RNase recruitment. It is possible, albeit unlikely, that SviR regulates the capR and orfs12-12.1 transcripts differently, stabilizing capR transcripts while degrading orfs12-12.1 transcripts. SviR-capR RNA duplex formation could inhibit RNases from interacting with cleavage sites on RNA processing intermediates through a similar process as was previously documented for the V. cholerae sRNA SgrS (Papenfort et al., 2013). Finally, the impact ICP1 infection has on V. cholerae RNA decay has not been investigated. Other phages have been shown to disrupt the host RNA decay pathways (Uzan and Miller, 2010), which could further complicate our interpretations of these results.
Conversely, the outcome of SviR regulation of gp120 does not impact the transcript abundance of gp120. Instead, SviR is necessary to regulate gp120 post-transcriptionally, reducing Gp120 translation without impacting gp120 transcript levels. While uncommon, this outcome of sRNA regulation is not unprecedented in V. cholerae. The MtlS sRNA, which regulates the mannitol transporter MtlA, does so post-transcriptionally without affecting mtlA transcript abundance (Mustachio et al., 2012). MtlS regulation also occurs independently of Hfq, consistent with our findings of SviR regulation of PLE transcripts, although gp120 regulation specifically was not assayed in the absence of Hfq. Neither the mechanism of MtlS regulation of MtlA nor that of SviR regulation of Gp120 has been fully resolved. As we only investigated RNA-RNA interactions between SviR and target transcripts, the SviR-gp120 interactions we observed may require the recruitment of another factor, which was absent from our reporter assays. We anticipate that SviR interacts with the ribosome binding site of gp120, disrupting the translation of the protein, consistent with both IntaRNA predictions and Hi-GRIL-seq data. However, we were unable to directly validate this hypothesis in vivo. Much stands to be learned about the inter-genome RNA-RNA interactions between phages and their satellites, and the mechanisms of regulation involved.
The role of the lncRNA in the PLE-ICP1 conflict is also of interest. It remains possible that the ICP1 lncRNA is a sponge transcript, sequestering SviR from interacting with targets. However, interactions between sponge transcripts and sRNAs often result in decreased sRNA levels due to coupled turnover of both transcripts (Denham, 2020), which we did not detect for the ICP1 lncRNA. Alternatively, the lncRNA could behave similarly to the anti-sRNA AgvB, mimicking sRNA target transcripts without resulting in coupled degradation (Tree et al., 2014). However, as we did not detect a difference in the regulation of target transcripts in the presence or absence of the lncRNA, the hypothesis of the lncRNA acting as a sponge transcript remains unsupported. The functional role of the lncRNA in ICP1-PLE dynamics remains elusive, despite attempts at identifying phenotypes related to the transcript. While much less is known about prokaryotic lncRNAs compared to their eukaryotic counterparts, there are multiple examples of lncRNAs encoded by other phages that have not been functionally characterized (Harris and Breaker, 2018). The GOLLD RNAs, lncRNAs found in Lactobacillales prophages, frequently are encoded proximal to tRNAs across diverse isolates and have been postulated to be involved with the phage lytic cycle (Weinberg et al., 2017). Interestingly, the ICP1 lncRNA is positioned upstream of the ICP1 lysis cassette, a locus that is occupied by tRNAs in phages with similar lysis cassettes to ICP1 (Boyd et al., 2021). The GOLLD RNAs are hypothesized to possess uncharacterized biochemical functions and, much like the ICP1 lncRNA, are not essential for phage virion production (Harris and Breaker, 2018). What the function of this family of lncRNAs is, and whether this function is conserved in other phage lncRNAs, remains to be understood. Determining the role of the lncRNA in the ICP1 infection cycle has the potential to reveal novel roles for lncRNAs in phages.
The profound conservation of the genomic locus of SviR, being flanked by variable repeat sequences, raises questions about the origins of the sRNA and what pressures continue to select for the maintenance of the locus architecture. The conservation of inverted repeats in each PLE with unique sequences between PLEs is of particular interest. Inverted repeats flanking an sRNA are hallmark characteristics of miniature inverted transposable elements (MITEs), a transposase-deficient class of MGEs found in prokaryotes and eukaryotes (Wachter et al., 2018). MITE-mediated sRNA mobilization occurs when a core-genome transposase mobilizes the MITE into a novel locus. Following mobilization, MITEs are thought to be domesticated by their host and have been shown to regulate adjacent transcripts to the integration site (Mazzone et al., 2001). MITE-encoded sRNA maturation can rely on an inverted repeat sequence (De Gregorio et al., 2005), possibly explaining the conservation of SviR repeat sequences within each PLE. The transposition of a regulatory RNA into an MGE involved in phage defense is consistent with the “guns-for-hire” model of genetic exchange between hosts, phages, and MGEs (Koonin et al., 2020), whereby SviR was domesticated into PLE to assist in the MGE’s anti-phage activity. Whether other viral satellites encode sRNAs that behave similarly to SviR remains to be seen, however, the rapid, specific regulation of target transcripts by sRNAs is in line with the requirements for satellite regulatory elements, which must rapidly respond to phage infection to enable their own spread. Collectively, SviR acts as a multi-functional regulator of gene expression in the PLE-ICP1 conflict, revealing a new mechanism of gene regulation to investigate in other satellite-phage conflicts.
Experimental Procedures
Bacterial strains and phage isolates
Bacterial strains
The genotypes of all Vibrio cholerae and Escherichia coli strains used in this study are listed in Table S1. V. cholerae strain KDS2 (a clinical isolate harboring PLE1) was used as the wild type strain for genetic manipulations. All overnight bacterial cultures were grown with aeration in a roller at 250 rpm at 37°C in LB medium. Larger volume cultures were grown shaking at 250 rpm at 37°C in LB medium. Ampicillin (100 μg/mL), streptomycin (100 μg/mL), and chloramphenicol (25 μg/mL for E. coli, 2.5 μg/mL for V. cholerae) were supplemented as necessary. 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was supplemented to induce cultures when indicated.
Phage isolates
All ICP1 listed in this study are derivatives of ICP1 strain 2011_Dha_A, detailed in Table S1. Bacteriophage stocks were stored in STE buffer (100 mM NaCl, 10 mM Tris-Cl – pH 8.0, 1 mM EDTA) at 4°C. All bacteriophage infections were initiated in a host cell culture grown to an OD600 of 0.3. Infections were either carried out at a multiplicity of infection (MOI) of 2.5 or 0.1, as indicated. V. cholerae harboring inducible expression constructs were induced at an OD600 of 0.2. Induced cultures were grown for 20 minutes before ICP1 infection and the MOI was calculated relative to the final culture OD600 value.
Cloning
Plasmid construction
The plasmids used in this study are available in Table S2 and the oligonucleotide sequences used for plasmid construction are detailed in Table S3. Oligonucleotides were purchased from Integrated DNA Technologies. All plasmid constructs, except CRISPR targeting constructs, were generated through Gibson Assembly. Plasmids were linearized by PCR with Q5 polymerase (New England Biolabs - NEB) using the primers indicated in Table S3. Gibson assembly reactions were carried out with a 3:1 molar ratio of insert to vector, with a total of 0.075 pmol DNA per reaction. 5 μL of the reaction was transformed into E. coli XL1-Blue chemically competent cells for further manipulation.
CRISPR targeting constructs for phage gene editing were generated through Golden Gate Assembly, as previously described (Box et al., 2016). Forward and reverse oligonucleotides complementary to the protospacer sequence with BsaI overhangs were mixed in 200 μL of water to a final concentration of 10 μM, boiled for 5 minutes, and cooled to room temperature. The annealed oligonucleotides were mixed with T4 Polynucleotide Kinase in 1x T4 DNA ligase buffer (NEB) and phosphorylated at 37°C for 30 min. Phosphorylated oligonucleotides were subsequently heat-inactivated for 20 minutes at 65°C. The heat-inactivated reactions phosphorylated oligonucleotides were diluted to a final concentration of 100 nM and the Golden Gate Assembly reaction was carried out with the BsaI restriction enzyme, 60 ng pCRISPR2.0 vector, and T4 DNA ligase at 37°C for 60 min.
Bacterial strain construction
V. cholerae strains containing plasmid constructs were generated by mating between recipient strains and plasmid-containing E. coli S17 donors. The donor E. coli and recipient V. cholerae strains were mixed at an equal ratio of 0.5 ODs each, incubated for 1 hour at 37°C, and plated on media selective for the recipient V. cholerae containing the plasmid of interest.
The markerless chromosomal deletion of sviR was generated using the allelic exchange vector pCVD422, encoding the sacB gene to enable sucrose counter-selection for recombinants. The deletion recombination template was generated through splicing by overlap extension PCR to include 1kb arms of homology upstream and downstream of the desired deletion to facilitate recombination into the V. cholerae chromosome. The PCR products were inserted into the pCVD442 multiple cloning site by Gibson assembly. Plasmids were mated into V. cholerae and exconjugants were grown overnight at 37°C in LB with antibiotic selection. Overnight cultures were plated on LB + 10% sucrose medium and incubated overnight at 30°C. The resulting colonies were screened for the deletion of interest by PCR and confirmed by sanger sequencing.
Phage mutant construction
Phage mutants were constructed as previously described (Box et al., 2016). V. cholerae harboring an inducible Ptac:CRISPR-Cas system was transformed with the pCRISPR2.0 plasmid containing spacer sequences to target sequences of interest. A homologous repair template containing the desired mutation of interest was inserted into the pCRISPR2.0 plasmid by Gibson assembly, facilitating repair of the targeted phage genome region by homologous recombination. Cells were grown to an OD600 of 0.2, induced for 20 minutes with 1 mM IPTG, and infected with phage in a soft agar overlay. Escape phages were isolated and purified by passaging three times under the selection of CRISPR-Cas targeting. Genomic DNA from purified phages was PCR amplified and Sanger sequenced to confirm the desired mutation was acquired.
RNA isolation
Bacterial strains were grown to an OD600 of 0.3 and, when applicable, infected with phage at an MOI of 2.5. The desired samples were then collected at the indicated time points and mixed 1:1 with ice-cold methanol. Methanol-treated samples were pelleted at 7,000 x g at 4°C, and the resulting pellets were washed in ice-cold 1X PBS. The washed pellets were resuspended in 200 μL TRI Reagent (Millipore/Sigma) and incubated for 5 minutes at room temperature. The samples were mixed with 40 μL chloroform, vortexed, and incubated for 10 minutes at room temperature. The chloroform-treated samples were then centrifuged at 12,000 x g for 10 min at 4°C. Following centrifugation, the upper (aqueous) phase was collected, promptly mixed with 110 μL 2-propanol and 11 μL pH 6.2 3M sodium acetate, and mixed vigorously. The samples were centrifuged at 12,000 x g for 15 min at 4°C, and the pellets were washed twice with 75% ethanol. The washed pellets were incubated at 65°C for 3 minutes to evaporate residual ethanol. The RNA was resuspended in 10-30 μL of diethyl dicarbonate (DEPC) water, and RNA integrity and concentration were analyzed by NanoDrop.
Hi-GRIL-Seq Sample Preparation
Hi-GRIL-seq was carried out based on previous protocols (Zhang et al., 2017), with slight modifications. All Hi-GRIL-seq replicates were generated in a V. cholerae PLE(+) host expressing T4 RNA ligase during ICP1 infection. Cells were grown to an OD600 of 0.2 and induced with 1 mM IPTG and 40 mM theophylline for 20 minutes to pre-induce t4rl1 expression. The induced V. cholerae culture was infected with ICP1 at an MOI of 2.5 and incubated at 37°C with aeration. 16 minutes after phage infection, 5 mL of the infected culture was mixed 1:1 with ice-cold methanol, and total RNA was isolated according to the RNA isolation protocol described above.
Following RNA isolation, each of the three biological replicates was submitted to the UC Berkeley QB3 functional genomics lab, where ribosomal RNA was depleted according to the RiboZero rRNA Depletion kit (Illumina), and cDNA was synthesized. An S220 focused-ultrasonicator (Covaris) was used to fragment the DNA, and library preparation was performed using the KAPA Hyper Prep kit for DNA (KK8504). Truncated universal stub adapters were ligated to each sample and adapter-ligated fragments were enriched by PCR amplification with indexed primers. Samples were quantified with Quant-iT dsDNA Assay Kit (Life Technologies), pooled equally by molarity, and submitted to the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, where libraries were sequenced on an Illumina HiSeq 4000 150 bp paired-end flow cell. RNA-seq data were analyzed as described below.
Hi-GRIL-seq read mapping
Forward and reverse read fastq files were first trimmed with Trimmomatic (-leading 30 -trailing 30 -slidingwindow 5:30 -minlen 25) to remove low-quality bases from the ends of reads. Trimmed read pairs were merged with PEAR (-j 3 -u 0.1 -p 0.01 -v 10 -n 50), requiring merged read pairs to be a minimum of 50 bp in length with a minimum p-value of 0.01. The first and last 25 bp of each paired read were then mapped to reference Genbank files for PLE, ICP1, and both V. cholerae chromosomes with bowtie2 (-p 4 -D 1 -R 1 -N 0 -k 1 -I 0 -X 50 --ignore-quals --end-to-end --no-unal). The resulting bam file was parsed with samtools and any read that mapped to two distinct genomes was considered chimeric. Reads that mapped to the same genome but had at least 1kb of sequence separating the two ends of the read were also considered chimeric, predicted to be interactions between two distinct transcripts in the same genome. The terminal mapping positions of all chimeric reads were then recorded as x-y values and plotted on scatter plots against the reference genome position. Scatter plots were then analyzed with DBScan (-n 10 -eps [min distance]) with a variable minimum distance between reads based on genome size for PLE-PLE (250 bp), PLE-ICP1 (350 bp), and ICP1-ICP1 (1000 bp) chimeras, respectively. Clusters of 10 or more reads where the average x or y value of all reads in the cluster mapped to a locus of interest were colored according to the figure legends. The final clustered plots were generated with matplotlib.
SviR-chimera junction mapping
The trimmed and merged Hi-GRIL-seq chimeric reads were aligned to their respective reference genomes using BLAST with default settings. Any reads that mapped to both SviR and a non-SviR target transcript were split at the junction between the two uniquely mapped segments. The 25 bp closest to the junction site between SviR and the target transcript was extracted in fasta format for further analysis. Both the SviR and target junction sequences were then mapped to their respective reference Genbank files using Bowtie2 with default settings and sorted and indexed with samtools. The sorted, indexed bam files were used to calculate junction read coverage bedgraph files using bedtools genomecov with default settings.
The mean genome coverage of three replicates was filtered to remove any sites with fewer than 10 reads mapping to a single locus and the filtered mean coverage graphs were visualized with Integrative Genome Viewer (IGV). SviR-target interactions at a given gene of interest were calculated by averaging the number of chimeras mapping to the gene across three replicates. Reads mapping to the intergenic region between two coding sequences were binned to the nearest ORF if the read mapped to within 50 bp of the gene coding sequence. Values were reported as average ± SEM between replicates.
For the mapping of the SviR component of chimeric junctions, the read coverage was calculated with bedtools genomecov to average the coverage of chimeric reads across the SviR sequence. Coverage was normalized to the total number of junctions mapping to SviR in a given replicate, and the average relative frequency of a junction mapping to a given SviR position across three replicates was plotted across the SviR sequence with IGV.
IntaRNA and CopomuS sRNA-target predictions
IntaRNA was used to predict interactions between SviR and target transcripts. The 50 bp upstream through the stop codon of each PLE and ICP1 open reading frame were considered candidate transcripts for interaction with SviR. SviR-target interaction predictions were calculated between these extracted sequences and SviR using IntaRNA, requiring maximum free energy of −10 kcal/mol (--outMaxE −10). For predictions of non-PLE 1 SviR alleles with target transcripts, the SviR sequence was predicted based on homology to the PLE 1 allele. For mutational analysis of SviR-target base pairing, predictions were performed using CopomuS using default settings.
5’/3’-Rapid amplification of cDNA ends (RACE)
5’- and 3’-RACE were carried out on RNA isolated 16 minutes-post ICP1 infection of PLE(+) V. cholerae, as previously described (Argaman et al., 2001) with some modifications. 5’-RACE samples were prepared with 20 μg of RNA, with and without Tobacco Acid Pyrophosphatase (TAP) (Millipore/Sigma) treatment for 1 hour at 37°C to remove 5’ triphosphates. Reactions were quenched with TRI Reagent (Millipore/Sigma) and RNA was purified as described previously, with pellets resuspended in 20 μL DEPC water. RNA was dephosphorylated with Bacterial Alkaline Phosphatase (Thermo Fisher) in the presence of 20 U RNase inhibitor (Thermo Fisher) for one hour at 65°C. The reactions were inactivated with 1 μL of 0.5 M EDTA for 10 min at 50°C, and RNA was purified as described with TRI Reagent. Pelleted, dephosphorylated RNA was resuspended in 10 μL DEPC water, mixed with 500 pmol of 5’ RNA adaptor A3, and annealed with T4 RNA ligase (NEB) overnight at 16°C. Ligated RNA was purified as described, resuspended in 37 μL DEPC water, and reverse transcribed with SuperScript III RT (Thermo Fisher) per manufacturers guidelines using a gene-specific primer against the transcript of interest. First-strand synthesized cDNA was treated with RNAse H at 37°C for 20 min and subsequently amplified in a secondary reaction with primer A4 and a nested gene-specific primer to amplify 5’ ends. Products were PCR purified with the Monarch PCR & DNA cleanup kit (NEB), ligated into pCR2.1-TOPO vector (Thermo Fisher) per the manufacturer's guidelines, and transformed into XL1 Blue E. coli. At least 20 transformants were screened by colony PCR, and 5’ TSS were identified from Sanger sequencing of PCR products.
3’-RACE was performed with 20 μg of equivalent RNA to 5’-RACE. RNA was dephosphorylated as described in 5’-RACE and purified as described with TRI Reagent (Sigma-Aldrich). Pelleted RNA was dissolved in 25 μL DEPC water, mixed with 500 pmol of RNA adapter E1, denatured for 5 min at 95°C, and chilled on ice for 5 min. 7 μL of RNA-adapter mix was mixed with 50 units of T4 RNA ligase, incubated overnight at 16°C, purified as described, and resuspended in 9 μL DEPC water. 1 μL of primer E4 was added to the ligated transcripts, and cDNA synthesis was carried out with SuperScript III RT per the manufacturer's guidelines. cDNA was treated with RNAse H at 37°C for 20 min, and secondary PCR amplification was carried out with primer E4 and a gene-specific primer, resulting in PCR amplicons containing 3’-RACE products. Ligation, transformation, colony PCR, and sequencing were performed as described for 5’-RACE, identifying 3’ ends of transcripts of interest.
Northern blot analysis
For each sample, 5 μg of RNA was run on an 8% polyacrylamide urea gel (National Diagnostics SequaGel 1:4) in 1x TBE buffer (100mM Tris base, 100m boric acid, 2 mM EDTA, pH 7.0) at 300V for 90 min. RNA was transferred from the gel to a Zeta-Probe GT membrane (Bio-Rad) for 16 hours in 0.5X TBE at 20 V at 4°C. Transferred blots were UV-crosslinked on both sides, and the RiboRuler LR RNA Ladder (Thermo Fisher) was visualized and marked by UV illumination. Membranes were blocked for 2 hours at 45°C with ULTRAhyb-Oligo Hybridization Buffer (Invitrogen). Northern probes (Table S3) were 5’ 32P-end-labeled with 0.3 mCi of γ-32P ATP (Perkin Elmer) with 10 U of T4 Polynucleotide Kinase (NEB) for 1 hour at 37°C. Labeled probes were spun through G-50 purification columns (Cytiva) to remove the unincorporated ATP, incubated at 95°C for 3 minutes, and added to the blocked membrane. Membranes were hybridized for 16 hours at 45°C in a rolling hybridization oven (Analytic Jena). Hybridized membranes were washed twice with 2X SSC buffer (3M sodium chloride, 300 mM sodium citrate, pH 7.0), washed three times with 0.2X SSC, and dried for 5 minutes at room temperature. Membranes were sealed in plastic page protectors, covered with autoradiography film (Denville), and exposed at −80°C in an X-ray film cassette (Cole-Parmer) as required. All Northern blots shown are representative of at least two biological replicates. All the replicates are available under the link in the data availability section.
Western blot analysis
V. cholerae was infected with ICP1 at an MOI of 0.1 and incubated until the indicated time point. 2 mL of infected cells were mixed with 2 mL ice-cold methanol and centrifuged for 10 minutes at 5,000x g. Cell pellets were resuspended in ice-cold lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1X Pierce Protease Inhibitor Mini Tab (Thermo)). The protein concentration was calculated against a BSA standard curve using the Pierce BCA Protein Assay Kit. A total of 30 μg per sample was resuspended in 30 μL of 1X Laemmli buffer (Bio-Rad) supplemented with 5% 2-mercaptoethanol and boiled for 10 minutes. 12 μL of the sample was run on an Any-kD TGX-SDS-PAGE gel (Bio-Rad) in 1X TBS (200 mM Tris, 150 mM NaCl) buffer for 30 minutes at 250 V and transferred using the Trans-Blot turbo Mini 0.2 μm PDVF transfer kit (Bio-Rad) with a Transblot Turbo transfer system (Bio-Rad) for 10 min at 2.5 V. Primary rabbit anti-FLAG antibody (Sigma) was diluted 1:1,500 and applied to the membrane for 1 hour. The membrane was washed for 5 minutes three times with TBS-T (TBS with 0.1% Tween 20) and incubated with secondary goat anti-rabbit-HRP antibody (Bio-Rad) at 1:10,000 in TBS buffer. The membrane was washed three times for 5 minutes with TBS-T and developed with Clarity Western ECL (Bio-Rad) per the manufacturer's guidelines. The activated membranes were imaged using a Chemidoc XRS Imaging System (Bio-Rad). The Gp120-3xFLAG western blot in Figure 4D is representative of three biological replicates, which are available in Figure S7. Statistical analysis of the significance of fold changes was calculated by a two-sample unpaired t-test, comparing the normalized band intensity of each mutant to that of the wild type from each of the three replicates.
GFP reporter assay
GFP reporter assays were carried out as previously described (Corcoran et al., 2012). E. coli MG1655 was transformed with an orf12.1::GFP reporter plasmid and a Plac-sviR small RNA expression plasmid or empty vector control. Cells were grown from single colonies overnight at 37°C, and then diluted to an OD600 of 0.05 in the presence of 1 mM IPTG and incubated for 3 hours. Following incubation, a cell pellet was harvested from 1 mL of each culture by centrifugation at 5,000 x g for 3 minutes. Cells were washed with 1X PBS and the pellets were resuspended in 2 mL 1X PBS. The samples were analyzed using an LSR II cell analyzer (Beckman Coulter). Two biological replicates of 50,000 cell counts were analyzed with FlowJo, with the mean fluorescent intensity of the GFP(+) population of cells reported for each replicate.
Phage plaque assay
V. cholerae cells were grown to an OD600 of 0.2 and pre-induced for 20 minutes, as necessary. After a 20-minute pre-induction, or at OD600 of 0.3, 50 μL of the culture was mixed with 10 μL of 10-fold serial dilutions of phage. After an 8-minute incubation to allow for phage attachment, the infected cells were mixed with 3 mL molten soft agar (LB broth + 0.5% agar) in a 6-well tissue culture plate (Gennessee scientific) and incubated overnight at 37°C.
Lysis kinetic assay
Lysis kinetic assays were carried out as previously described (Hays and Seed, 2019). V. cholerae cultures were grown to an OD600 of approximately 1.0 and back diluted to an OD600 of 0.05. The diluted cultures were grown to an OD600 of 0.3, and 150 μL of culture was mixed with phage in a 96-well plate at an MOI of 2.5. Cell lysis was observed by measuring OD600 with an i3x SpectraMax plate reader (Molecular Devices) at 37°C over 60 minutes with readings every 90 seconds, shaking in between reads. Presented lysis kinetic curves are an average of values from three biological replicates.
Genome replication qPCR
Genome replication assays were carried out as previously described (O’Hara et al., 2017). V. cholerae cultures were grown to an OD600 of approximately 1.0 and back diluted to an OD600 of 0.05. The diluted cultures were grown to an OD600 0.3 and infected with ICP1 at an MOI of 0.1 (ICP1 replication) or an MOI of 2.5 (PLE replication). 100 μL of culture was taken immediately before (PLE replication) or immediately after (ICP1 replication) phage infection and boiled for 10 minutes. An endpoint sample was taken at 20 minutes post-infection and boiled for 10 minutes. The boiled samples were diluted at 1:50 (ICP1) or 1:1000 (PLE) in water, and 3.75 μL of each sample was mixed with 0.18 μL of 25 μM qPCR primers and 7.5 μL of iQ SYBR master mix (Bio-Rad). 15 μL reactions were analyzed with a CFX Connect Real-time PCR detection system (Bio-Rad) in technical duplicate. The fold change over time was calculated by normalizing the detected DNA at 20-minute time points to the input DNA calculated at 0-minute time points.
Supplementary Material
Acknowledgments
We thank Dr. Gigi Storz, Dr. Philip Adams, and Dr. Sahar Melamed for their training on Northern blot analysis, general advice, and for providing the E. coli reporter plasmid constructs. We acknowledge the UC Berkeley CRL-FACS Core Facility and Dr. Kartoosh Heydari’s support with flow cytometry experiments. We also thank the members of the Seed Lab for their feedback, in particular, Dr. Reid Oshiro and Caroline Boyd for help during manuscript revision.
Funding
This work was supported by a National Science Foundation Graduate Research Fellowship [2018257700 to D.T.D.] and by the National Institutes of Health [R01AI127652 to K.D.S, R01AI153303 to K.D.S]. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or NIH. K.D.S. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
Hi-GRIL-seq RNA sequencing reads for all three biological replicates are available online at the NCBI SRA (SRA#PRJNA824536). Uncropped replicate gels are available on Mendeley Data, DOI:10.17632/xsx6wv435b.1 https://data.mendeley.com/datasets/xsx6wv435b/1
References
- Altuvia S, Storz G and Papenfort K (2018) ‘Cross-Regulation between Bacteria and Phages at a Posttranscriptional Level’, Microbiology Spectrum, 6(4). doi: 10.1128/microbiolspec.RWR-0027-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermeyer A. et al. (2018) ‘Analysis of 19 highly conserved Vibrio cholerae bacteriophages isolated from environmental and patient sources over a twelve-year period’, Viruses, 10(6). doi: 10.3390/v10060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermeyer A. et al. (2022) ‘Evolutionary sweeps of subviral parasites and their phage host bring unique parasite variants and disappearance of a phage CRISPR-Cas system’, mBio, 13(1). doi: 10.1128/mbio.03088-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L. et al. (2001) ‘Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis’, Current Biology, 11(12), pp. 941–951. doi: 10.1016/j.gene.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Bandyra KJ et al. (2012) ‘The Seed Region of a Small RNA Drives the Controlled Destruction of the Target mRNA by the Endoribonuclease RNase E’, Molecular Cell, 47(6), pp. 943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. et al. (2007) ‘CRISPR provides acquired resistance against viruses in prokaryotes’, Science, 315(5819), pp. 1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Barth ZK, Netter Z, et al. (2020) ‘A Family of Viral Satellites Manipulates Invading Virus Gene Expression and Can Affect Cholera Toxin Mobilization’, mSystems, 5(5), pp. 1–18. doi: 10.1128/msystems.00358-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ZK, Silvas TV, et al. (2020) ‘Genome replication dynamics of a bacteriophage and its satellite reveal strategies for parasitism and viral restriction’, Nucleic acids research, 48(1), pp. 249–263. doi: 10.1093/nar/gkz1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ZK, Nguyen MHT and Seed KD (2021) ‘A chimeric nuclease substitutes a phage CRISPR-Cas system to provide sequence-specific immunity against subviral parasites’, eLife, 10. doi: 10.7554/eLife.68339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim A and Sorek R (2020) ‘The pan-immune system of bacteria: antiviral defence as a community resource’, Nature Reviews Microbiology, 18(2), pp. 113–119. doi: 10.1038/s41579-019-0278-2. [DOI] [PubMed] [Google Scholar]
- Bloch S. et al. (2021) ‘Bacteriophages as sources of small non-coding RNA molecules’, Plasmid, 113(August 2020). doi: 10.1016/j.plasmid.2020.102527. [DOI] [PubMed] [Google Scholar]
- Box AM et al. (2016) ‘Functional Analysis of Bacteriophage Immunity through a Type I-E CRISPR-Cas System in Vibrio cholerae and Its Application in Bacteriophage Genome Engineering’, Journal of Bacteriology, 198(3), pp. 578–590. doi: 10.1128/JB.00747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd CM et al. (2021) ‘Bacteriophage ICP1: A Persistent Predator of Vibrio cholerae’, Annual Review of Virology, 8, pp. 285–304. doi: 10.1146/annurev-virology-091919-072020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallereau A. et al. (2016) ‘Next-Generation “-omics” Approaches Reveal a Massive Alteration of Host RNA Metabolism during Bacteriophage Infection of Pseudomonas aeruginosa’, PLoS Genetics, 12(7). doi: 10.1371/journal.pgen.1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran CP et al. (2012) ‘Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA’, Molecular Microbiology, 84(3), pp. 428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- Davis BM and Waldor MK (2007) ‘RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA’, Voenno-meditsinskii zhurnal, 65(2), pp. 373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham EL (2020) ‘The Sponge RNAs of bacteria – How to find them and their role in regulating the post-transcriptional network’, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms, 1863(8). doi: 10.1016/j.bbagrm.2020.194565. [DOI] [PubMed] [Google Scholar]
- Faruque SM et al. (2005) ‘Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages’, Proceedings of the National Academy of Sciences, 102(5), pp. 1702–1707. doi: 10.1073/PNAS.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L. et al. (2015) ‘A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics’, Cell, 160(1–2), pp. 228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS and Papenfort K (2016) ‘Interplay of regulatory RNAs and mobile genetic elements in enteric pathogens’, Molecular Microbiology, 101(5), pp. 701–713. doi: 10.1111/mmi.13428. [DOI] [PubMed] [Google Scholar]
- De Gregorio E. et al. (2005) ‘Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: Genomic organization and functional properties’, Journal of Bacteriology, 187(23), pp. 7945–7954. doi: 10.1128/JB.187.23.7945-7954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK and Bassler BL (2007) ‘Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae’, Proceedings of the National Academy of Sciences of the United States of America, 104(27), pp. 11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Tjaden B and Lory S (2016) ‘GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation’, Nature Microbiology, 2. doi: 10.1038/nmicrobiol.2016.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KA and Breaker RR (2018) ‘Large Noncoding RNAs in Bacteria’, Regulating with RNA in Bacteria and Archaea, 6(4), pp. 515–526. doi: 10.1128/microbiolspec.rwr-0005-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SG and Seed KD (2019) ‘Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite’, eLife, 9. doi: 10.1101/790493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV et al. (2020) ‘Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire’, Nature Reviews Genetics, 21(2), pp. 119–131. doi: 10.1038/s41576-019-0172-9. [DOI] [PubMed] [Google Scholar]
- Koskella B and Brockhurst MA (2014) ‘Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities’, FEMS Microbiology Reviews, 38(5), pp. 916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke L and Wulff DL (1990) ‘RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA’, Genes and Development, 4, pp. 2223–2233. [DOI] [PubMed] [Google Scholar]
- Lalaouna D. et al. (2017) ‘Identification of unknown RNA partners using MAPS’, Methods, 117, pp. 28–34. doi: 10.1016/j.ymeth.2016.11.011. [DOI] [PubMed] [Google Scholar]
- LeGault KN et al. (2022) ‘A phage parasite deploys a nicking nuclease effector to inhibit viral host replication’, Nucleic acids research, 50(15), pp. 8401–8417. doi: 10.1093/nar/gkac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist BH, Dehò G and Calendar R (1993) ‘Mechanisms of genome propagation and helper exploitation by satellite phage P4’, Microbiological Reviews, 57(3), pp. 683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M. et al. (2001) ‘Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae’, Gene, 278, pp. 211–222. [DOI] [PubMed] [Google Scholar]
- McKitterick AC and Seed KD (2018) ‘Anti-phage islands force their target phage to directly mediate island excision and spread’, Nature Communications, 9(1). doi: 10.1038/s41467-018-04786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed S. et al. (2016) ‘Global Mapping of Small RNA-Target Interactions in Bacteria’, Molecular Cell, 63(5), pp. 884–897. doi: 10.1016/j.molcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed S et al. (2020) ‘RNA-RNA Interactomes of ProQ and Hfq Reveal Overlapping and Competing Roles’, Molecular Cell, 77(2), pp. 411–425. doi: 10.1016/j.molcel.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ES et al. (2003) ‘Bacteriophage T4 Genome’, Microbiology and Molecular Biology Reviews, 67(1), pp. 86–156. doi: 10.1128/MMBR.67.1.86.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustachio LM et al. (2012) ‘The Vibrio cholerae mannitol transporter is regulated posttranscriptionally by the MtlS small regulatory RNA’, Journal of Bacteriology, 194(3), pp. 598–606. doi: 10.1128/JB.06153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netter Z. et al. (2021) ‘A phage satellite tunes inducing phage gene expression using a domesticated endonuclease to balance inhibition and virion hijacking’, Nucleic Acids Research, 49(8), pp. 4386–4401. doi: 10.1093/nar/gkab207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara BJ et al. (2017) ‘A highly specific phage defense system is a conserved feature of the Vibrio cholerae mobilome’, PLoS Genetics, 13(6). doi: 10.1371/journal.pgen.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K. et al. (2013) ‘Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis’, Cell, 153(2), pp. 426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penadés JR and Christie GE (2015) ‘The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites’, Annual Review of Virology, 2(1), pp. 181–201. doi: 10.1146/annurev-virology-031413-085446. [DOI] [PubMed] [Google Scholar]
- Peschek N. et al. (2020) ‘RNA-mediated control of cell shape modulates antibiotic resistance in Vibrio cholerae’, Nature Communications. doi: 10.1038/s41467-020-19890-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G. et al. (2014) ‘Precisely modulated pathogenicity island interference with late phage gene transcription’, Proceedings of the National Academy of Sciences of the United States of America, 111(40), pp. 14536–14541. doi: 10.1073/pnas.1406749111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. et al. (2010) ‘The Vibrio cholerae virulence regulatory cascade controls glucose update through activation of TarA, a small regulatory RNA’, Molecular Microbiology, 78(5), pp. 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD et al. (2011) ‘Evidence of a Dominant Lineage of Vibrio cholerae-Specific Lytic Bacteriophages Shed by Cholera Patients over a 10-Year Period in Dhaka, Bangladesh’, mBio, 2(1), pp. e00334–10. doi: 10.1128/mBio.00334-10.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD et al. (2013) ‘A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity’, Nature, 494(7438), pp. 489–491. doi: 10.1038/nature11927.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J and Wassarman KM (2012) ‘Regulation by Small RNAs in Bacteria: Expanding Frontiers’, 40(6), pp. 1301–1315. doi: 10.1007/s10439-011-0452-9.Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR and Dryden DTF (2005) ‘The biology of restriction and anti-restriction’, Current Opinion in Microbiology, 8(4), pp. 466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Tree JJ et al. (2014) ‘Identification of Bacteriophage-Encoded Anti-sRNAs in Pathogenic Escherichia coli’, Molecular Cell, 55(2), pp. 199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan M and Miller ES (2010) ‘Post-transcriptional control by bacteriophage T4: MRNA decay and inhibition of translation initiation’, Virology Journal, 7(1), p. 360. doi: 10.1186/1743-422X-7-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat K. et al. (2021) ‘A dual-function RNA balances carbon uptake and central metabolism in Vibrio cholerae’, The EMBO Journal, 40(24). doi: 10.15252/embj.2021108542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter S. et al. (2018) ‘Identification of novel MITEs (miniature inverted-repeat transposable elements) in Coxiella burnetii: Implications for protein and small RNA evolution’, BMC Genomics, 19(247). doi: 10.1186/s12864-018-4608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EGH, Altuvia S and Romby P (2002) ‘Antisense RNAs in bacteria and their genetic elements’, Advances in Genetics, 46, pp. 361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- Waters LS and Storz G (2009) ‘Regulatory RNAs in Bacteria’, Cell, 136(4), pp. 615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SA et al. (2017) ‘Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E’, The EMBO Journal, 36(3), pp. 374–387. doi: 10.15252/embj.201694639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. et al. (2017) ‘Detection of 224 candidate structured RNAs by Comparative analysis of specific subsets of intergenic regions’, Nucleic Acids Research, 45(18), pp. 10811–10823. doi: 10.1093/nar/gkx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-F et al. (2017) ‘Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility’, Molecular Microbiology, 106(6), pp. 919–937. doi: 10.1111/mmi.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-GRIL-seq RNA sequencing reads for all three biological replicates are available online at the NCBI SRA (SRA#PRJNA824536). Uncropped replicate gels are available on Mendeley Data, DOI:10.17632/xsx6wv435b.1 https://data.mendeley.com/datasets/xsx6wv435b/1