Abstract
Objective:
To develop and validate a radiomics nomogram based on CT for the pre-operative prediction of BRAF mutation and clinical outcomes in patients with colorectal cancer (CRC).
Methods:
A total of 451 CRC patients (training cohort = 190; internal validation cohort = 125; external validation cohort = 136) from 2 centers were retrospectively included. Least absolute shrinkage and selection operator regression was used to select radiomics features and the radiomics score (Radscore) was calculated. Nomogram was constructed by combining Radscore and significant clinical predictors. Receiver operating characteristic curve analysis, calibration curve and decision curve analysis were used to evaluate the predictive performance of the nomogram. Kaplan‒Meier survival curves based on the radiomics nomogram were used to assess overall survival (OS) of the entire cohort.
Results:
The Radscore consisted of nine radiomics features which were the most relevant to BRAF mutation. The radiomics nomogram integrating Radscore and clinical independent predictors (age, tumor location and cN stage) showed good calibration and discrimination with AUCs of 0.86 (95% CI: 0.80–0.91), 0.82 (95% CI: 0.74–0.90) and 0.82 (95% CI: 0.75–0.90) in the training cohort, internal validation and external validation cohorts, respectively. Furthermore,the performance of nomogram was significantly better than that of the clinical model (p < 0.05). The radiomics nomogram-predicted BRAF mutation high-risk group had a worse OS than the low-risk group (p < 0.0001).
Conclusion:
The radiomics nomogram showed good performance in predicting BRAF mutation and OS of CRC patients, which could provide valuable information for individualized treatment.
Advances in knowledge:
The radiomics nomogram could effectively predict BRAF mutation and OS in patients with CRC. High-risk BRAF mutation group identified by the radiomics nomogram was independently associated with poor OS.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and the leading cause of cancer-related deathin the world, 1 is a highly heterogeneous disease driven by a range of genetic and epigenetic events. 2 Genetic analysis of tumors is a powerful tool to personalize treatment by developing targeted therapies. 3,4 BRAF encodes a protein-dependent kinase, which is an important component of mitogen activated protein kinase pathway and plays a key role in regulating cell proliferation, differentiation and apoptosis. 5,6 BRAF mutation was a negative independent prognostic factor for survival and recurrence 7,8 in CRC. BRAF mutation predicts a lack of response to antiepidermal growth factor receptor (EGFR) monoclonal antibodies. 9 Several studies have also illustrated that anti-EGFR therapy is recommended for BRAF wild-type patients and that the combination of anti-EGFR therapy with BRAF inhibitors significantly improves the prognosis of patients with BRAF mutation. 10–13 The American Society of Clinical Oncology proposes to detect BRAF mutation in patients with CRC at the time of diagnosis for prognostic stratification and assessment of risk for Lynch syndrome. 14 In addition, the National Comprehensive Cancer Network guidelines have recommended that all patients suspected or diagnosed with metastatic CRC should be tested for BRAF mutation in tumor tissues. 9
To date, the detection of BRAF mutation requires the analysis of specimens obtained from surgery or biopsy, which is not only an invasive and expensive procedure but may also be limited by temporal and spatial heterogeneity of tumors, or poor DNA quality. Therefore, at pre-treatment, non-invasive and repeatable identification of BRAF mutation status is critical for predicting treatment effect and determining individual treatment strategies for patients with CRC.
In clinical practice, CT is a common imaging method for pre-operative evaluation of patients with CRC. Although traditional imaging characteristics, such as tumor enhancement modes, are related to BRAF mutation, 15 traditional imaging characteristics are highly subjective and non-specific, depending on the experience of the observer. Radiomics can objectively reflect the heterogeneity of tumors, and can obtain more information that is invisible to the naked eye and hidden in the tumor. 16–22 In several studies, researchers have proved that it is practicable to use radiomics methods to predict the KRAS/NRAS/BRAF mutation of CRC. 23,24 However, former studies had modest data sets (less than 200) and lacked independent external validation. Patients with any mutant KRAS/NRAS/BRAF were classified into the mutant group, which can confuse clinical application. Moreover, previous studies only focused on the evaluation of the value of radiomics in predicting KRAS/NRAS/BRAF mutation, and failed to provide further prognostic data.
Therefore, the purpose of this study was to explore the value of CT radiomics nomogram integrating Radscore and clinical independent predictors in predicting BRAFmutation and clinical outcomes in patients with CRC.
Methods and materials
Patients
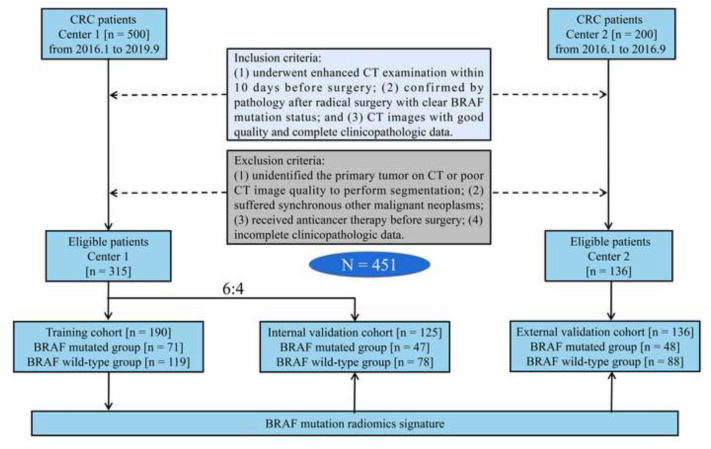
This retrospective study was approved by the ethics committees of the two centers, and waived the requirement for informed consent. We retrospectively recruited 451 patients with CRC who underwent radical resection in the two centers. Figure 1 shows the process of patient recruitment. The clinical characteristics of the patients were obtained from the hospital information system including age, sex, pre-operative carbohydrate antigen 199 (CA199), carcinoembryonic antigen (CEA), tumor location, tumor size, clinical T (cT) stage, and clinical N (cN) stage. Pathological information included BRAF mutation status, pathological TNM stage, tumor differentiation, lymphovascular invasion (LVI), and perineural invasion (PNI).
Figure 1.
The process of patient recruitment. CRC, colorectal cancer.
BRAF mutation analysis
The tumor tissue was routinely treated after surgical resection, and the pathological tissue was embedded in paraffin after tissue fixation, dehydration and embedding. DNA was extracted from paraffin-embedded pathological tissue by nucleic acid extraction reagent, and then the mutation status of BRAF (V600E) gene was detected by gene detection kit based on amplification-refractory mutation system (ARMS) and fluorescent PCR technology.
CT image analysis
Supplementary Table 1 shows the CT scanning protocol used by the two centers. The pre-operative CT images were interpreted independently by two imaging diagnosis doctors of abdominal tumor with 15 and 25 years of experience who were blinded to the patients’ clinicopathological information.The criteria for cT stage and cN stage on contrast-enhanced CT are shown in Supplementary Material 1. Any differences between them will be discussed before a final consensus is reached.
Tumor delineation
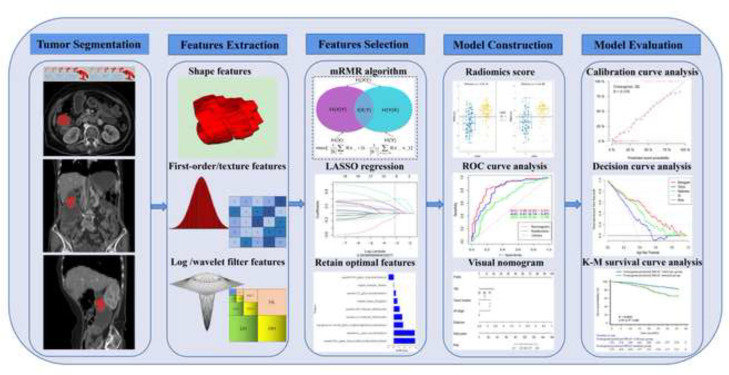
The workflow of radiomics is shown in Figure 2. Using ITK-SNAP (v. 3.8.0, https://www.itksnap.org) to manually delineate the whole tumor at the portal venous phase. Interclass and intraclass correlation coefficients (ICCs) were used to evaluate the inter- and intraobserver reproducibility of radiomics features extraction. Another two diagnostic radiologists with 8 years (Reader 1) and 16 years (Reader 2) of experience delineated the tumors for 40 randomly selected patients. Reader 1 repeated the delineation a month later. ICCs > 0.75 indicated that the consistency of feature extraction was good.
Figure 2.
Workflow of radiomics analysis.
Radiomics feature extraction
All CT images were pre-processed before feature extraction. 25,26 Spline interpolation was used to resample all CT images to 1 mm× 1 mm × 1 mm voxel size; a bin width of 25 was used for grayscale discretization to reduce the effect of imaging noise; the image intensity was normalized using the μ ± 3 σ method. Pyradiomics software (https://pyradiomics.readthedocs.io/en/latest/) was used to extract radiomics features. The shape features, first-order features, gray level dependence matrix (gldm) features, gray level co-occurrence matrix (glcm) features, gray level size zone matrix (glszm) features, and gray level run length matrix (glrlm) features were extracted.
Radiomics feature selection and model construction
The radiomics features of ICCs > 0.75 were retained. Then, the Max-relevance and min-redundancy (mRMR) is an effective and reliable method for radiomics feature selection. mRMR can seek the optimal feature subset by considering the importance of features and the relevance between features, i.e. maximizing the relevance between features and classification variables, while minimizing the redundancy between features. 27–29 In this study, mRMR can maximize the distinction between the features of BRAF mutated and BRAF wild-type genes. mRMR can reduce the dimension and improve the modeling efficiency and generalization ability of the model in the later stage (Supplementary Material 2). The least absolute shrinkage and selection operator (LASSO) logistic regression method was used to select the best radiomics features with non-zero coefficients. Finally, the radiomics score (Radscore) of each patient was calculated through a linear combination of selected radiomics features weighted by their respective coefficients. The radiomics model was built based on the Radscore to predict BRAF mutation of CRC.
Nomogram construction
The statistically significant predictors (clinical features and Radscore) obtained by univariate logistic regression analysis were entered into multivariate logistic analysis to establish a clinical model and a combined model. The combined model was visualized in the form of a nomogram.The Hosmer-Lemeshow test was used to analyze the goodness of fit of the nomogram. Decision curve analysis (DCA) was used to evaluate the clinical net benefit of the nomogram.
Follow-up and survival analysis
Follow-up every 3 months in the first 2 years and every 6 months in the third and subsequent years. Follow-up lasted at least 3 years or until the patient died. The end point of this study was overall survival (OS). Kaplan–Meier survival curves were plotted and compared using log-rank test. The statistically significant variables in univariate Cox regression analysis were included in multivariate Cox regression analysis to determine the independent predictors of OS (Supplementary Table 1).
Statistical analysis
Data processing and modeling were performed using R software (v. 4.2.1, https://www.R-project.org). Supplementary Table 2 shows the R software package used. Continuous variables were tested using an independent sample t test or Mann–Whitney U test. Classified variables were tested using the χ2 test or Fisher’s exact test. Using receiver operating characteristic (ROC) curves to evaluate the diagnostic efficiency of each model. DeLong test was used to compare the prediction performance of each model. P<0.05 was considered statistically significant.
Results
Patient characteristics
Overall, 451 CRC patients were included in the analysis. Characteristics of clinical baseline data in this study are summarized in Table 1. The rates of BRAF mutation were 37.4% (71 of 190), 37.6% (47 of 125) and 35.3% (48 of 136) in the training cohort, internal validation cohort and external validation cohort, respectively. There were no significant differences between the mutated group and the wild-type group in the three cohorts in terms of sex, CEA, CA199, cT stage, cN stage, tumor size, tumor location, tumor differentiation, pTNM stage, LVI, and PNI. There were significant differences in age between the mutated group and the wild-type group in the three cohorts (p < 0.05).
Table 1.
Baseline characteristics of the study population
| Characteristic | Training cohort (n = 190) | Internal validation cohort (n = 114) | Externalvalidation cohort (n = 136) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild-type group (n = 119) | Mutated group (n = 71) | p-value | Wild-type group (n = 78) | Mutated group (n = 47) | p- value | Wild-type group (n = 88) |
Mutated group (n = 48) | p-value | |
| Sex(%) | 0.549 | 0.792 | 0.704 | ||||||
| Female | 52 (43.7) | 35 (49.3) | 30 (38.5) | 20 (42.6) | 41 (46.6) | 24 (50.0) | |||
| Male | 67 (56.3) | 36 (50.7) | 48 (61.5) | 27 (57.4) | 47 (53.4) | 24 (50.0) | |||
| Age (mean ± SD, years) | 62.8 ± 12.2 | 67.3 ± 8.9 | 0.007 | 60.4 ± 13.5 | 67.3 ± 10.3 | 0.003 | 58.3 ± 11.6 | 64.2 ± 11.2 | 0.001 |
| CEA (%) | 0.115 | 0.091 | 0.197 | ||||||
| Normal | 69 (58.0) | 32 (45.1) | 53 (67.9) | 24 (51.1) | 43 (48.9) | 29 (60.4) | |||
| Abnormal | 50 (42.0) | 39 (54.9) | 25 (32.1) | 23 (48.9) | 45 (51.1) | 19 (39.6) | |||
| CA199 (%) | 0.702 | 0.126 | 0.107 | ||||||
| Normal | 81 (68.1) | 51 (71.8) | 58 (74.4) | 28 (59.6) | 56 (63.6) | 37 (77.1) | |||
| Abnormal | 38 (31.9) | 20 (28.2) | 20 (25.6) | 19 (40.4) | 32 (36.4) | 11 (22.9) | |||
| Tumor size (mean ± SD, cm) | 4.7 ± 2.0 | 4.5 ± 1.7 | 0.432 | 5.1 ± 1.8 | 4.6 ± 1.8 | 0.151 | 5.1 ± 1.9 | 5.0 ± 1.8 | 0.759 |
| Tumor location(%) | 0.095 | 0.992 | 0.268 | ||||||
| Right colon | 34 (28.6) | 31 (43.7) | 28 (35.9) | 17 (36.2) | 34 (38.6) | 25 (52.1) | |||
| Left colon | 58 (48.7) | 29 (40.8) | 34 (43.6) | 20 (42.6) | 39 (44.3) | 15 (31.3) | |||
| Rectum | 27 (22.7) | 11 (15.5) | 16 (20.5) | 10 (21.3) | 15 (17.0) | 8 (16.7) | |||
| cT stage (%) | 0.173 | 0.412 | 0.634 | ||||||
| cT1+cT2 | 12 (10.1) | 2 (2.8) | 11 (14.1) | 5 (10.6) | 5 (5.7) | 4 (8.3) | |||
| cT3 | 44 (37.0) | 27 (38.0) | 25 (32.1) | 11 (23.4) | 40 (45.5) | 24 (50.0) | |||
| cT4 | 63 (52.9) | 42 (59.2) | 42 (53.8) | 31 (66.0) | 43 (48.9) | 20 (41.7) | |||
| cN stage (%) | 0.095 | 0.187 | 0.557 | ||||||
| cN0 | 58 (48.7) | 25 (35.2) | 39 (50.0) | 17 (36.2) | 30 (34.1) | 14 (29.2) | |||
| cN1+cN2 | 61 (51.3) | 46 (64.8) | 39 (50.0) | 30 (63.8) | 58 (65.9) | 34 (70.8) | |||
| pTNM stage(%) | 0.342 | 0.144 | 0.235 | ||||||
| I | 12 (10.1) | 3 (4.2) | 11 (14.1) | 5 (10.6) | 2 (2.3) | 5 (10.4) | |||
| II | 50 (42.0) | 26 (36.6) | 32 (41.0) | 20 (42.6) | 38 (43.2) | 18 (37.5) | |||
| III | 46 (38.7) | 34 (47.9) | 32 (41.0) | 15 (31.9) | 44 (50.0) | 22 (45.8) | |||
| IV | 11 (9.2) | 8 (11.3) | 3 (3.8) | 7 (14.9) | 4 (4.5) | 3 (6.3) | |||
| pT stage(%) | 0.433 | 0.225 | 0.051 | ||||||
| 1 | 2 (1.7) | 0 (0.0) | 1 (1.3) | 2 (4.3) | 0 (0.0) | 1 (2.1) | |||
| 2 | 13 (10.9) | 5 (7.0) | 10 (12.8) | 4 (8.5) | 3 (3.4) | 4 (8.3) | |||
| 3 | 33 (27.7) | 25 (35.2) | 25 (32.1) | 9 (19.1) | 31 (35.2) | 23 (47.9) | |||
| 4 | 71 (59.7) | 41 (57.7) | 42 (53.8) | 32 (68.1) | 54 (61.4) | 20 (41.7) | |||
| pN stage(%) | 0.271 | 0.747 | 0.892 | ||||||
| 0 | 63 (52.9) | 29 (40.8) | 43 (55.1) | 26 (55.3) | 42 (47.7) | 24 (50.0) | |||
| 1 | 35 (29.4) | 26 (36.6) | 18 (23.1) | 13 (27.7) | 31 (35.2) | 15 (31.3) | |||
| 2 | 21 (17.6) | 16 (22.5) | 17 (21.8) | 8 (17.0) | 15 (17.0) | 9 (18.8) | |||
| pM stage(%) | 0.484 | 0.062 | 0.452 | ||||||
| 0 | 109 (91.6) | 62 (87.3) | 75 (96.2) | 40 (85.1) | 84 (95.5) | 44 (91.7) | |||
| 1 | 10 (8.4) | 9 (12.7) | 3 (3.8) | 7 (14.9) | 4 (4.5) | 4 (8.3) | |||
| Tumor differentiation (%) | 0.790 | 0.304 | 0.489 | ||||||
| Well | 3 (2.5) | 3 (4.2) | 3 (3.8) | 3 (6.4) | 0 (0.0) | 0 (0.0) | |||
| Moderate | 94 (79.0) | 56 (78.9) | 61 (78.2) | 40 (85.1) | 71 (80.7) | 41 (85.4) | |||
| Poor | 22 (18.5) | 12 (16.9) | 14 (17.9) | 4 (8.5) | 17 (19.3) | 7 (14.6) | |||
| Lymphovascular invasion (%) | 0.169 | 0.375 | 0.791 | ||||||
| Negative | 88 (73.9) | 45 (63.4) | 57 (73.1) | 30 (63.8) | 66 (75.0) | 35 (72.9) | |||
| Positive | 31 (26.1) | 26 (36.6) | 21 (26.9) | 17 (36.2) | 22 (25.0) | 13 (27.1) | |||
| Perineural invasion (%) | 0.121 | 0.246 | 0.753 | ||||||
| Negative | 101 (84.9) | 53 (74.6) | 66 (84.6) | 35 (74.5) | 81 (92.0) | 43 (89.6) | |||
| Positive | 18 (15.1) | 18 (25.4) | 12 (15.4) | 12 (25.5) | 7 (8.0) | 5 (10.4) | |||
CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen.
Feature selection andradiomics model construction
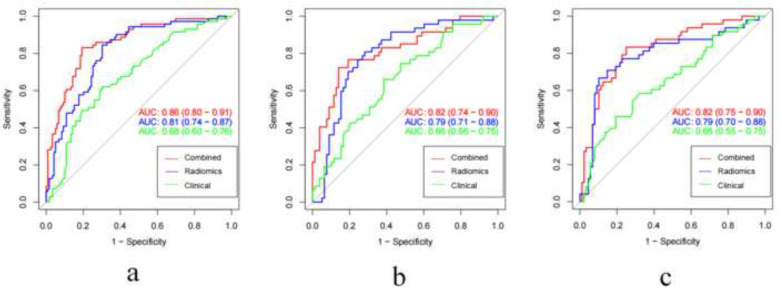
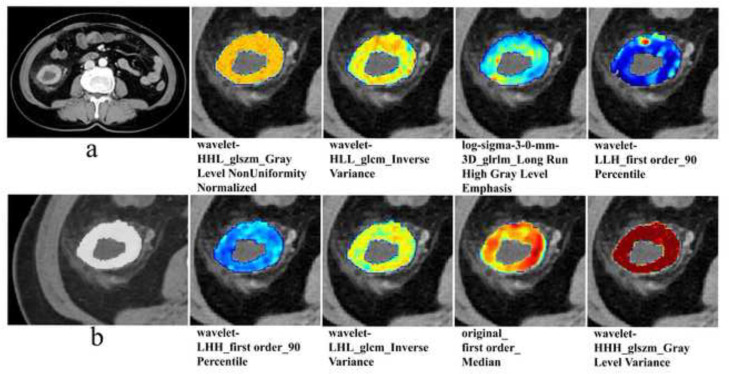
First, 1011 radiomics features were retained (ICCs > 0.75). Second, using mRMR algorithm to reduce the number of features to 30. Finally, LASSO regression with the best λ of 0.044 determined the radiomics features of nine non-zero coefficients, which can be used to develop the Radscore (Supplementary Figure 1). The Radscore was calculated using these nine features weighted by their respective coefficients. The Radscore formula was shown in Supplementary Material 3. The Wilcoxon test showed that the Radscore of BRAF mutated group was significantly higher than that BRAF wild-type group (p < 0.001) (Supplementary Figure 2). The AUCs of the radiomics model based on the Radscore were 0.81 (95% CI: 0.74–0.87), 0.79 (95% CI: 0.71–0.88) and 0.79 (95% CI: 0.70–0.88) in the training cohort, internal validation cohort and external validation cohort, respectively (Figure 3). Figure 4 provided a detailed visualization of each radiomics feature used over the CT image of a patient with BRAF mutation.
Figure 3.
The ROC curves of the radiomics model, clinical model, and combined model for predicting BRAF mutation in CRC in the training cohort (a), internal validation cohort (b) and external validation cohort (c). CRC, colorectal cancer; ROC, receiver operating characteristic.
Figure 4.
Detailed visualization of each used radiomics feature over the CT image. (a) The CT image of a 70-year-old female colorectal cancer patient with BRAF mutation. (b) Enlarged view of tumor region of interest. On the right side of figure a and b, detailed visualization of each used radiomics feature over the CT image, the name of each radiomics feature used was displayed below the corresponding figure. (glcm, gray level co-occurrence matrix; gldm, gray level dependence matrix; glrlm, gray level run length matrix;glszm, gray level size zone matrix)
Development and performance of the nomogram
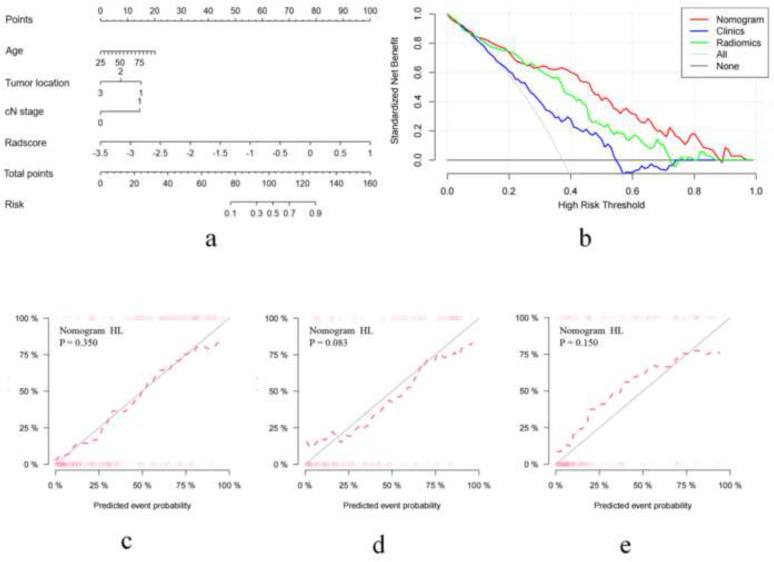
For the potential clinical features, univariate logistic regression analysis showed that age, tumor location and cN stage were correlated to BRAF mutation (p < 0.05). Multivariate logistic regression analysis revealed that age (odds ratio [OR] = 1.05; 95% CI: 1.02–1.08; p = 0.002), tumor location (OR = 0.64; 95% CI: 0.41–0.99; p = 0.045), and cN stage (OR = 1.99; 95% CI: 1.04–3.80; p = 0.037) were clinical independent predictors of BRAF mutation (Supplementary Table 3). The clinical independent predictors (age, tumor location and cN stage) were used to construct the clinical model. The AUCs of the clinical model in the training cohort, internal validation cohort, and external validation cohort were 0.68 (95% CI: 0.60–0.76), 0.66 (95% CI: 0.56–0.75) and 0.65 (95% CI: 0.55–0.75), respectively. For the potential clinical features and the Radscore, univariate logistic regression analysis showed that Radscore, age, tumor location, and cN stage were correlated to BRAF mutation (p < 0.05). Multivariate logistic regression analysis revealed that Radscore (OR = 6.91; 95% CI: 3.74–12.76; p < 0.001), tumor location (OR = 0.53; 95% CI: 0.31–0.90; p = 0.019), and cN stage (OR = 3.70; 95% CI: 1.68–8.12; p = 0.001) were independent predictors of BRAF mutation (Table 2). Correlation coefs between Radscore and each clinical feature were showed in Supplementary Table 4. When the absolute value of the correlation coef was less than 0.2, it could be considered that there was a very weak correlation or no correlation between Radscore and each clinical feature. By integrating Radscore, tumor location, cN stage and age, a combined model was successfully constructed. The AUCs of the combined model in the training cohort, internal validation cohort, and external validation cohort were 0.86 (95% CI: 0.80–0.91), 0.82 (95% CI: 0.74–0.90) and 0.82 (95% CI: 0.75–0.90), respectively. Figure 3, Table 3 provide the predictive performance of the radiomics model, clinical model, and combined model in the three cohorts. DeLong’s test showed that there was a statistically significant difference in AUCs between the combined model and the clinical model (p = 0.0001, 0.0020 and 0.0006 in the training, internal and external validation cohorts, respectively). The difference of AUCs between radiomics model and clinical model was also statistically significant (p = 0.015, 0.044 and 0.038 in the training cohort, internal and external validation cohorts, respectively). The nomogram was used to visualize the combined model (Figure 5a). Supplementary Figure 3 showed an example of the clinical application of the developed nomogram. The Hosmer‒Lemeshow test showed that the nomogram fit well in the three cohorts (p = 0.350, 0.083 and 0.150 in the training cohort, internal and external validation cohorts, respectively) (Figure 5c-e). DCA showed that when the threshold probability was within the range of 10‒20 and 28‒90%, the nomogram was more beneficial than the “treat all”,“treat none”, the clinical model and the radiomics model (Figure 5b).
Table 2.
Univariate and multivariate logistic regression analysis in the training cohort
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | |
| Sex | 1.25 (0.70–2.26) | 0.454 | ... | ... |
| Age | 1.04 (1.01–1.07) | 0.009 | 1.03 (0.99–1.06) | 0.174 |
| CEA | 1.68 (0.93–3.04) | 0.085 | ... | ... |
| CA199 | 0.84 (0.44–1.59) | 0.586 | ... | ... |
| Tumor size | 0.94 (0.80–1.10) | 0.431 | ... | ... |
| Tumor location | 0.65 (0.42–0.98) | 0.041 | 0.53 (0.31–0.90) | 0.019 |
| cT stage | 1.44 (0.90–2.30) | 0.130 | ... | ... |
| cN stage | 1.86 (1.01–3.42) | 0.045 | 3.70 (1.68–8.12) | 0.001 |
| Radscore | 5.41 (3.15–9.29) | < 0.001 | 6.91 (3.74–12.76) | < 0.001 |
CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen.
Note. CI, confidence interval; cN stage, Clinical N stage; cT stage, Clinical T stage; OR, odds ratio; Radscore, radiomics score.
Table 3.
Comparison of predictive performance of the radiomics model, clinical model, and combined model in the three cohorts (training cohort, internal validation cohort, and external validation cohort)
| Model | Cut-off | Cohort | AUC (95% CI) | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Radiomics model | −0.50 | Training cohort | 0.81 (0.74–0.87) | 0.75 | 0.85 | 0.70 | 0.63 | 0.88 |
| Internal validation cohort | 0.79 (0.71–0.88) | 0.74 | 0.83 | 0.68 | 0.61 | 0.87 | ||
| External validation cohort | 0.79 (0.70–0.88) | 0.80 | 0.60 | 0.91 | 0.78 | 0.81 | ||
| Clinical model | −0.44 | Training cohort | 0.68 (0.60–0.76) | 0.67 | 0.62 | 0.70 | 0.55 | 0.75 |
| Internal validation cohort | 0.66 (0.56–0.75) | 0.61 | 0.51 | 0.67 | 0.48 | 0.69 | ||
| External validation cohort | 0.65 (0.55–0.75) | 0.65 | 0.56 | 0.69 | 0.50 | 0.74 | ||
| Combined model | −0.42 | Training cohort | 0.86 (0.80–0.91) | 0.82 | 0.83 | 0.81 | 0.72 | 0.89 |
| Internal validation cohort | 0.82 (0.74–0.90) | 0.76 | 0.65 | 0.84 | 0.77 | 0.76 | ||
| External validation cohort | 0.82 (0.75–0.90) | 0.79 | 0.90 | 0.60 | 0.81 | 0.76 |
Note. AUC, area under the receiver operating characteristic curve; CI, confidence interval; NPV, negative-predictive value; PPV, positive-predictive value.
Figure 5.
The developed radiomics nomogram to predict BRAF mutation (a); decision curve analysis for the nomogram (b); the calibration curves of the nomogram in the training cohort (c), internal validation cohort (d) and external validation cohort (e).
Pre-operative predictors of survival
The median follow-up period of the whole cohort was 41 months (range, 1–84 months). The Kaplan–Meier survival curves showed that the pathological BRAF mutation status and nomogram-predicted BRAF mutation status could significantly distinguish the high death risk group from the low death risk group (HR = 2.42; 95% CI: 1.62–3.62; and HR = 2.33; 95% CI: 1.57–3.46; respectively, all p < 0.0001) (Figure 6a–b). The radiomics nomogram-predicted high-risk group had a worse OS than the low-risk group (p < 0.0001). The univariate and multivariate Cox regression analysis results of the predictors of OS in the whole cohort were showed in Table 4. The multivariate Cox regression analysis showed that the nomogram-predicted BRAF mutation was an independent predictor of OS (HR, 1.93; 95% CI: 1.31–2.85; p < 0.001). In the whole cohort, the C-index of pathological BRAF mutation and nomogram-predicted BRAF mutation was 0.63 (95% CI: 0.58–0.67) and 0.62 (95% CI: 0.57–0.66), respectively.
Figure 6.
Kaplan‒Meier survival curves according to pathological BRAF mutation (a) and nomogram-predicted BRAF mutation in the whole cohort (b).
Table 4.
Univariate and multivariate Cox regression analysis of predictors of OS in the whole cohort
| Variable | Univariateanalysis | Multivariate analysis (Pathological) | Multivariate analysis (Nomogram) | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Sex | 1.06 (0.73–1.56) | 0.750 | ... | ... | ... | ... |
| Age | 1.03 (1.01–1.05) | < 0.001 | 1.02 (1.01–1.04) | < 0.001 | 1.03 (1.01–1.04) | 0.005 |
| CEA | 1.87 (1.27–2.74) | 0.001 | 1.32 (0.89–1.96) | 0.167 | 1.35 (0.91–2.00) | 0.141 |
| CA199 | 2.16 (1.48–3.15) | < 0.001 | 1.63 (1.10–2.42) | 0.016 | 1.68 (1.13–2.49) | 0.011 |
| Tumor size | 1.04 (0.95–1.14) | 0.422 | ... | ... | ... | ... |
| Tumor location | 0.91 (0.70–1.19) | 0.488 | ... | ... | ... | ... |
| cT stage | 2.88 (1.93–4.29) | < 0.001 | 2.54 (1.69–3.83) | < 0.001 | 2.51 (1.66–3.79) | < 0.001 |
| cN stage | 1.82 (1.20–2.76) | 0.005 | 1.40 (0.91–2.17) | 0.129 | 1.40 (0.91–2.16) | 0.129 |
| Pathological BRAF mutation | 2.25 (1.54–3.29) | < 0.001 | 1.90 (1.28–2.81) | 0.001 | ... | ... |
| Nomogram-predicted BRAF mutation | 2.21 (1.51–3.23) | < 0.001 | ... | ... | 1.93 (1.31–2.85) | < 0.001 |
CA199, carbohydrate antigen 199; CEA, carcinoembryonic antigen.
Note. CI, confidence interval; cN stage, Clinical N stage; cT stage, Clinical T stage; HR, hazard ratio,
Discussion
Using relatively large data sets from two centers, we developed a CT-based radiomics nomogram to predict pre-operative BRAF mutation. The model had a good diagnostic significance for the state of BRAF, and its results were also validated in the internal and external cohorts, which indicated the reliability and reproducibility of the developed prediction model. Therefore, CT-based radiomics nomogram may provide additional information to determine BRAF status of CRC patients to guide targeted therapy. Additionally, the radiomics nomogram was significantly related to OS, which played an important role in predicting the prognosis of patients with CRC.
A previous study showed that CT radiomics features have potential value for evaluating KRAS mutation in patients with CRC. 22 Moreover, two recent studies showed that MRI or ultrasound radiomics features performed well in predicting BRAF mutation in patients with melanoma brain metastases or papillary thyroid microcarcinoma. 30,31 The above studies indicated that it was feasible to use radiomics to predict the BRAF mutation status of tumors. Shi et al 23 constructed a CT-based radiomics model to predict RAS/BRAF mutation of CRC liver metastasis and achieved moderate predictive efficiency (AUC = 0.74) in the validation set (159 cases). In addition, Yang et al 24 established a CT-based radiomics model to predict KRAS/NRAS/BRAF mutation inpatients with CRC and found good predictive efficiency (AUC = 0.83) in the validation set (117 cases). Our research has some advantages compared with previous research. First, our study not only included a relatively large number of patients (451 cases), but also had an independent external validation cohort. Second, in previous studies, researchers identified the positive group based on the mutation of KRAS/NRAS/BRAF; i.e. one or more gene mutations were defined as the positive group, which will undoubtedly complicate the clinical application of the results found in their studies. Finally, previous studies only focused on evaluating the predictive value of radiomics features for KRAS/NRAS/BRAF mutation, but did not provide further prognostic information.
1218 radiomics features were simplified into 9 key features, which had the best identification ability and the richest biological information. Interestingly, we found that wavelet-filtered features were the optimal radiomics features (6/9), which means that the wavelet transform filter can improve the efficiency of capturing BRAF mutation-related features in CRC. Wavelet features can quantify the heterogeneity of tumors at different scales and have the characteristics of multiresolution analysis. 32 Because it can mine rich texture information in images, related studies also show that wavelet features have strong prediction ability and are an important part of the optimal radiomics feature set to predict KRAS mutation in CRC. 22,24 We found that the wavelet-filtered feature “inverse variance” of the glcm played an important role in predicting BRAF mutation in this study. “Inverse variance” was the measurement of image spatial heterogeneity, and larger values of “inverse variance” reflect greater heterogeneity. That is, the greater the possibility of BRAF mutation. This observation is also in line with the results of previous studies. 22,24,33 All these results suggested that our radiomics features provide a wealth of information related to BRAF mutation.
In clinical practice, joint analysis of multiple markers can achieve better results than single analysis. Previous studies had reported the combination of CT-based radiomics features and clinical risk factors, such as pre-operative cN stage, age or tumor location, it was of great value in predicting lymph node metastasis, LVI, PNI and microsatellite instability in CRC. 17–20 Similarly, in our study, multivariate logistic regression analysis found that age, tumor location and cN stage were clinical independent predictors for BRAF mutation. Consistent with previous studies, 34–36 clinical factors are associated with genetic alterations in tumors. Our study showed that BRAF mutation in CRC predominantly occurs in older patients and on the right colon. This may be explained by the distinct embryologic origins of the right and left colon, leading to different biological characteristics. 37 While the clinical model based on independent predictors (age, tumor location and cN stage) was moderately effective, with an AUC of 0.68 in the training cohort, which was lower than that of the radiomics model. There may be two main reasons to explain this phenomenon. First, clinical models containing clinical risk factors only reflect specific tumor information. Even patients with the same features may exhibit a differential BRAF mutation status. Second, radiomics features extracted from the whole tumor are able to comprehensively quantify intratumor heterogeneity, which is considered a well-established predictor that is barely available from traditional clinical models. In addition, the good predictive power of radiomics models for BRAF mutation confirmed their predictive values. As shown,the predictive efficacy was further improved by integrating the radiomics model with the clinical model to construct a combined model. A nomogram was generated for the visualization of the combined model. This nomogram allows clinicians to make individualized predictions about the risk of BRAF mutation in CRC patients.
Previous studies have demonstrated that CT radiomics features can predict survival in patients with CRC. 19,21 Many studies have shown that pathological BRAF mutation are associated with poor prognosis in CRC patients. 5–7 We obtained a similar result that BRAF mutation could lead to poor OS in patients with CRC. Additionally, there was a significant difference between the survival outcomes of high- and low-risk group determined by the radiomics nomogram, and the OS of the patients in the high risk group was significantly lower than that in the low risk group. Through uni- and multivariate Cox regression analysis, nomogram scores was an independent prognostic factor for OS. Because of unsatisfactory outcomes in high-risk group, alternative intention to treat methods should be provided in time to avoid unnecessary toxicity and improve survival outcomes. Therefore, it is practical to use our nomogram model in guiding treatment plans and carrying out personalized treatment.
This study has some limitations. First, this was a retrospective study, which may have inevitably led to selection bias. A prospective study is needed to validate the generalizability and clinical applicability of this model. Second,in this study, the radiomics features of CT portal phase images were extracted. Plain scan and arterial phase images may also contain information to predict BRAF mutation.
In conclusion, the radiomics nomogram, which integrates CT radiomics features and clinical independent predictors, has a good performance in predicting BRAF mutation and clinical outcome of CRC patients. This approach allows for better risk stratification of CRC patients prior to treatment, and provide support for clinical decision-making and personalized treatment.
Footnotes
* Corresponding authors: Feng Feng and Yanfen Cui contributed equally to this work.
The authors Manman Li and Guodong Xu contributed equally to the work.
Contributor Information
Manman Li, Email: 380232471@qq.com.
Guodong Xu, Email: xgd20133@163.com.
Hui Zhou, Email: zh12345st@163.com.
Qiaoling Chen, Email: 1372431158@qq.com.
Qi Fan, Email: xinjie22@yeah.net.
Jian Shi, Email: sj9399@139.com.
Shaofeng Duan, Email: 18910063803@163.com.
Yanfen Cui, Email: yanfen210@126.com.
Feng Feng, Email: fengfeng@ntu.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet 2019; 394: 1467–80: S0140-6736(19)32319-0. doi: 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 3. Strickler JH, Wu C, Bekaii-Saab T. Targeting BRAF in metastatic colorectal cancer: maximizing molecular approaches. Cancer Treat Rev 2017; 60: 109–19: S0305-7372(17)30129-9. doi: 10.1016/j.ctrv.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 4. Sundar R, Hong DS, Kopetz S, Yap TA. Targeting BRAF-mutant colorectal cancer: progress in combination strategies. Cancer Discov 2017; 7: 558–60. doi: 10.1158/2159-8290.CD-17-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cicenas J, Tamosaitis L, Kvederaviciute K, Tarvydas R, Staniute G, Kalyan K, et al. Kras, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol 2017; 34: 26. doi: 10.1007/s12032-016-0879-9 [DOI] [PubMed] [Google Scholar]
- 6. Jiang D, Wang X, Wang Y, Philips D, Meng W, Xiong M, et al. Mutation in BRAF and SMAD4 associated with resistance to neoadjuvantchemoradiation therapy in locally advanced rectal cancer. Virchows Arch 2019;475:39-47. doi: 10.1007/s00428-019-02576-y. [DOI] [PubMed] [Google Scholar]
- 7. Margonis GA, Buettner S, Andreatos N, Kim Y, Wagner D, Sasaki K, et al. Association of BRAF mutations with survival and recurrence in surgically treated patients with metastatic colorectal liver cancer. JAMA Surg 2018; 153(): e180996. doi: 10.1001/jamasurg.2018.0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ducreux M, Chamseddine A, Laurent-Puig P, Smolenschi C, Hollebecque A, Dartigues P, et al. Molecular targeted therapy of BRAF -mutant colorectal cancer . Ther Adv Med Oncol 2019; 11: 175883591985649. doi: 10.1177/1758835919856494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen Y-J, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021; 19: 329–59. doi: 10.6004/jnccn.2021.0012 [DOI] [PubMed] [Google Scholar]
- 10. Hong DS, Morris VK, El OB, Sorokin AV, Janku F, Fu S, et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab inPatients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer Discov 2016;6:1352-1365. doi: 10.1158/2159-8290.CD-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopetz S, Guthrie KA, Morris VK, Lenz H-J, Magliocco AM, Maru D, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406). J Clin Oncol 2021; 39: 285–94. doi: 10.1200/JCO.20.01994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanz-Garcia E, Argiles G, Elez E, Tabernero J. Braf mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol 2017; 28: 2648–57: S0923-7534(19)34586-7. doi: 10.1093/annonc/mdx401 [DOI] [PubMed] [Google Scholar]
- 13. Prasad V. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2020; 382: 876: 10.1056/NEJMc1915676#sa2. doi: 10.1056/NEJMc1915676 [DOI] [PubMed] [Google Scholar]
- 14. Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for clinical pathology, College of American pathologists, association for molecular pathology, and American Society of clinical oncology. J Mol Diagn 2017; 19: 187–225: S1525-1578(16)30224-0. doi: 10.1016/j.jmoldx.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eurboonyanun K, Lahoud RM, Kordbacheh H, Pourvaziri A, Promsorn J, Chadbunchachai P, et al. Imaging predictors of BRAF mutation in colorectal cancer. Abdom Radiol 2020; 45: 2336–44. doi: 10.1007/s00261-020-02484-9 [DOI] [PubMed] [Google Scholar]
- 16. Qi Y, Zhao T, Han M. The application of radiomics in predicting gene mutations in cancer. Eur Radiol 2022; 32: 4014–24. doi: 10.1007/s00330-021-08520-6 [DOI] [PubMed] [Google Scholar]
- 17. Huang Y-Q, Liang C-H, He L, Tian J, Liang C-S, Chen X, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016; 34: 2157–64. doi: 10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- 18. Li M, Jin Y, Rui J, Zhang Y, Zhao Y, Huang C, et al. Computed tomography-based radiomics for predicting lymphovascular invasion in rectal cancer. European Journal of Radiology 2022; 146: 110065. doi: 10.1016/j.ejrad.2021.110065 [DOI] [PubMed] [Google Scholar]
- 19. Ma J, Guo D, Miao W, Wang Y, Yan L, Wu F, et al. The value of 18f-FDG PET/CT-based radiomics in predicting perineural invasion and outcome in non-metastatic colorectal cancer. Abdom Radiol 2022; 47: 1244–54. doi: 10.1007/s00261-022-03453-0 [DOI] [PubMed] [Google Scholar]
- 20. Pei Q, Yi X, Chen C, Pang P, Fu Y, Lei G, et al. Pre-Treatment CT-based radiomics nomogram for predicting microsatellite instability status in colorectal cancer. Eur Radiol 2022; 32: 714–24. doi: 10.1007/s00330-021-08167-3 [DOI] [PubMed] [Google Scholar]
- 21. Xue T, Peng H, Chen Q, Li M, Duan S, Feng F. A CT-based radiomics nomogram in predicting the postoperative prognosis of colorectal cancer: a two-center study. Academic Radiology 2022; 29: 1647–60. doi: 10.1016/j.acra.2022.02.006 [DOI] [PubMed] [Google Scholar]
- 22. Xue T, Peng H, Chen Q, Li M, Duan S, Feng F. Preoperative prediction of KRAS mutation status in colorectal cancer using a CT-based radiomics nomogram. Br J Radiol 2022; 95(): 20211014. doi: 10.1259/bjr.20211014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi R, Chen W, Yang B, Qu J, Cheng Y, Zhu Z, et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res 2020; 10: 4513–26. [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol 2018; 28: 2058–67. doi: 10.1007/s00330-017-5146-8 [DOI] [PubMed] [Google Scholar]
- 25. Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 2017; 44: 1050–62. doi: 10.1002/mp.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018; 19: 1180–91: S1470-2045(18)30413-3. doi: 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 27. Remeseiro B, Bolon-Canedo V. A review of feature selection methods in medical applications. Comput Biol Med 2019; 112: 103375: S0010-4825(19)30252-5. doi: 10.1016/j.compbiomed.2019.103375 [DOI] [PubMed] [Google Scholar]
- 28. De Jay N, Papillon-Cavanagh S, Olsen C, El-Hachem N, Bontempi G, Haibe-Kains B. MRMRe: an R package for parallelized mrmr ensemble feature selection. Bioinformatics 2013; 29: 2365–68. doi: 10.1093/bioinformatics/btt383 [DOI] [PubMed] [Google Scholar]
- 29. Hong JH, Jung J-Y, Jo A, Nam Y, Pak S, Lee S-Y, et al. Development and validation of a radiomics model for differentiating bone islands and osteoblastic bone metastases at abdominal CT. Radiology 2021; 299: 626–32. doi: 10.1148/radiol.2021203783 [DOI] [PubMed] [Google Scholar]
- 30. Meißner A-K, Gutsche R, Galldiks N, Kocher M, Jünger ST, Eich M-L, et al. Radiomics for the noninvasive prediction of the BRAF mutation status in patients with melanoma brain metastases. Neuro Oncol 2022; 24: 1331–40. doi: 10.1093/neuonc/noab294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang J, Jiang S, Ma J, Xi X, Li H, Wang L, et al. Nomogram based on radiomics analysis of ultrasound images can improve preoperative BRAF mutation diagnosis for papillary thyroid microcarcinoma. Front Endocrinol (Lausanne) 2022; 13: 915135: 915135. doi: 10.3389/fendo.2022.915135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soufi M, Arimura H, Nagami N. Identification of optimal mother wavelets in survival prediction of lung cancer patients using wavelet decomposition-based radiomic features. Med Phys 2018; 45: 5116–28. doi: 10.1002/mp.13202 [DOI] [PubMed] [Google Scholar]
- 33. Zheng Y-M, Chen J, Zhang M, Wu Z-J, Tang G-Z, Zhang Y, et al. Ct radiomics nomogram for prediction of the Ki-67 index in head and neck squamous cell carcinoma. Eur Radiol 2023; 33: 2160–70. doi: 10.1007/s00330-022-09168-6 [DOI] [PubMed] [Google Scholar]
- 34. Atreya CE, Greene C, McWhirter RM, Ikram NS, Allen IE, Van Loon K, et al. Differential radiographic appearance of BRAF V600E-mutant metastatic colorectal cancer in patients matched by primary tumor location. J Natl Compr Canc Netw 2016; 14: 1536–43. doi: 10.6004/jnccn.2016.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tran B, Kopetz S, Tie J, Gibbs P, Jiang Z-Q, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011; 117: 4623–32. doi: 10.1002/cncr.26086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, et al. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/alliance N0147. J Natl Cancer Inst 2014; 106(. doi: 10.1093/jnci/dju106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roberto M, Marchetti P, Arrivi G, Di Pietro FR, Cascinu S, Gelsomino F, et al. The treatment paradigm of right-sided metastatic colon cancer: harboring BRAF mutation makes the difference. Int J Colorectal Dis 2020; 35: 1513–27. doi: 10.1007/s00384-020-03589-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.