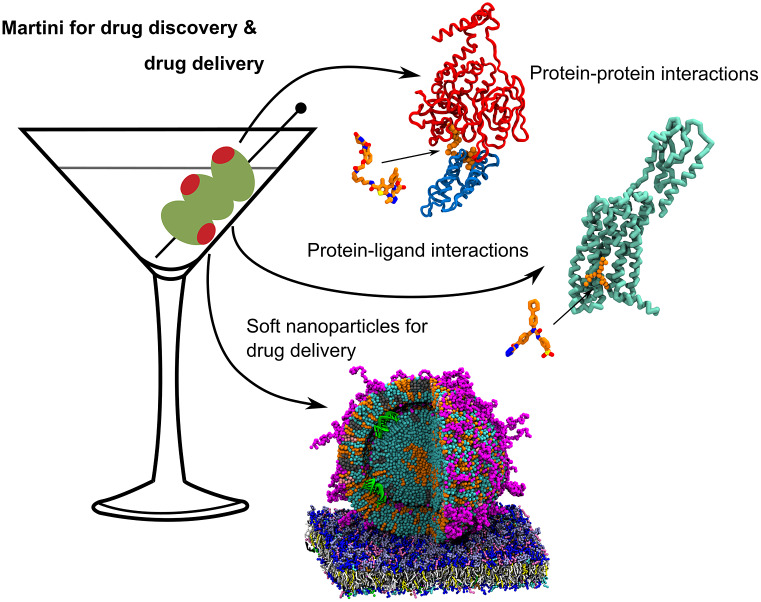
Key words: coarse-grained models, molecular dynamics, Martini, drug design, drug delivery, cryptic pockets, transmembrane proteins, protein-protein interactions, soft delivery systems, PROTACS, lipid nanoparticles
Abstract
Coarse-grained (CG) modelling with the Martini force field has come of age. By combining a variety of bead types and sizes with a new mapping approach, the newest version of the model is able to accurately simulate large biomolecular complexes at millisecond timescales. In this perspective, we discuss possible applications of the Martini 3 model in drug discovery and development pipelines and highlight areas for future development. Owing to its high simulation efficiency and extended chemical space, Martini 3 has great potential in the area of drug design and delivery. However, several aspects of the model should be improved before Martini 3 CG simulations can be routinely employed in academic and industrial settings. These include the development of automatic parameterisation protocols for a variety of molecule types, the improvement of backmapping procedures, the description of protein flexibility and the development of methodologies enabling efficient sampling. We illustrate our view with examples on key areas where Martini could give important contributions such as drugs targeting membrane proteins, cryptic pockets and protein–protein interactions and the development of soft drug delivery systems.
Introduction
Recent studies have shown that the cost of drug discovery and development is, on average, higher than several hundred million dollars (Mohs and Greig, 2017; Schlander et al., 2021). Moreover, several diseases such as Alzheimer, cancer, viral infections and cardiovascular diseases remain orphan of an effective, long-term and safe therapeutic protocol (Falzone et al., 2018; Nishiga et al., 2020; Brown and Wobst, 2021; Esang and Gupta, 2021). Current challenges in the development of novel therapeutic approaches include the unavailability of druggable binding pockets in the target structure (Weerakoon et al., 2022) and the lack of effective delivery systems, which can improve drug pharmacodynamics (Wang et al., 2021b).
Computational methodologies can speed up the drug discovery pipeline, decrease the associated costs and provide insight into the interactions between drugs and their targets, which is critical for rational drug design (Sliwoski et al., 2014; Lin et al., 2020). Computer modelling permeates both hit-identification and lead-optimisation stages of drug discovery pipelines. Computational methods have been used to predict protein–ligand binding modes (Śledź and Caflisch, 2018), binding affinities (Montalvo-Acosta and Cecchini, 2016), brain–blood barrier permeation (Crivori et al., 2000), compound activity against a given target (Pereira et al., 2018) or to identify and map potential binding sites (Yu and MacKerell, 2017; MacKerell et al., 2020). Some of these methods rely on atomistic molecular dynamics (MD) simulations to produce configurational ensembles (Siebenmorgen and Zacharias, 2020). However, converging on sampling the potential energy landscape of large biomolecular complexes is challenging and limits the application of atomistic MD to smaller systems. Nonetheless, numerical simulations and docking studies can still contribute to studies of protein–ligand interactions or identification of hit compounds (Jorgensen, 2009; Bollini et al., 2011; Frey et al., 2013). Alternatively, purpose-built hardware and software can help simulate large systems at atomistic resolution as shown by the DESRES team (Dror et al., 2011; Shaw et al., 2021).
Drug delivery has also seen an increase in usage of computational modelling, mainly because current development pipelines rest upon unpredictable trial and error experiments. Molecular modelling offers an attractive platform for understanding and optimising delivery systems in a biologically relevant context (Wang et al., 2021b). In this field, the limitations associated with system size and complexity are magnified. Several model systems exploring interaction with lipid bilayers have been constructed, with more realistic models mainly being of solid nanoparticles (NPs) such as gold NPs (Franco-Ulloa et al., 2021; Salassi et al., 2021). Limited studies have explored softer delivery systems like lipid-based NPs, mainly due to the lack of well-established computational protocols for constructing and studying these systems.
Coarse-grained (CG) modelling techniques alleviate sampling limitations of atomistic MD. The most widely used CG force field (FF) is the Martini FF (Marrink et al., 2007). The newly developed Martini 3 (Souza et al., 2021a) improves sampling efficiency by merging together two to four non-hydrogen atoms and corresponding associated hydrogens into one interaction bead, with the bonded and non-bonded parameters derived from a combination of bottom-up and top-down approaches, respectively. In parallel with the development of the Martini 3 FF, other CG approaches were pursued. For instance, some recent developments in protein CG models include the SIRAH2.0 FF (Machado et al., 2019), SPICA (Kawamoto et al., 2022) or the recently developed ProMPT, an alternative polarisable Martini model (Sahoo et al., 2022).
The Martini 2 FF currently supports a wide array of parameters for proteins, different lipid types, polymers, DNA and RNA (Monticelli et al., 2008; López et al., 2009; De Jong et al., 2013; Uusitalo et al., 2015, 2017; Grünewald et al., 2018; Salassi et al., 2018). Four main bead types were developed based on the polarity of chemical groups. These particles are further subdivided depending, for example, on their hydrogen-bonding capabilities (Marrink et al., 2007). Limitations of the Martini 2 model included overstabilisation of some biomolecular interactions, mainly noted for proteins and sugars (Alessandri et al., 2019) and the narrow range of chemical groups represented by the available beads (Kanekal and Bereau, 2019). The new version 3 (Souza et al., 2021a) addressed these issues and now provides promising solutions for drug design and delivery. New Martini 3 CG models allow simulations of more complex systems, facilitating the study of important biomolecular processes like ligand binding (Souza et al., 2020), fusion events (Bruininks et al., 2020), and the distribution of drugs within particle or carrier delivery systems (Casalini, 2021). This enables understanding of the forces behind encapsulation and drug release, which furthers the optimisation and development of delivery systems, as well as the interactions, which drive ligand binding, fundamental for drug design campaigns.
The Martini 2 and 3 FFs have been applied to study different biomolecular systems, among them proteins, membranes or vesicles, and is increasingly being used in the field of materials sciences (Marrink et al., 2019; Alessandri et al., 2021; Marrink et al., 2022). Examples exist of CG simulations studying fusion of delivery systems, such as lipoplexes or nanoemulsions, with lipid bilayers (Lee et al., 2012; Bruininks et al., 2020; Gupta et al., 2021; Machado et al., 2022). In 2020, Bruininks et al. (2020) used CG modelling to simulate the fusion of a cationic lipoplex containing DNA with a simple membrane model representing the endosomal membrane. This is one of the first stepping stones for using CG models to explore nucleic acid (NA) release. For drug binding, the potential of CG-Martini simulations in studies of protein–ligand binding was shown in the work of Negami et al. (2014) where they studied protein–ligand binding for two systems, levansucrase-glucose and LinB-1,2-dichloroethane. A more recent example is the application of Martini 3 FF to study protein–ligand binding in T4 Lysozyme with different small molecules and several pharmacologically relevant targets, such as G protein-coupled receptors (GPCRs), kinases and one example of nuclear receptor (Souza et al., 2020), achieving quantitative agreement with experimental binding affinities. Other examples are present in the literature (Delort et al., 2017; Ferré et al., 2019; Jiang and Zhang, 2019; Dandekar and Mondal, 2020; Negami et al., 2020). Furthermore, the application of Gō models (Poma et al., 2017) in the Martini 3 model leads to an improved description of protein flexibility while preserving computational efficiency. Combined with the new Martini 3 small molecule library (Souza et al., 2021b; Alessandri et al., 2022), CG Martini models now gather the conditions for successful applications in structure-based drug discovery campaigns.
In this perspective, we discuss potential applications of Martini 3 CG simulations to topics relevant for drug discovery and development pipelines, including design of innovative therapies, binding site identification and optimisation of soft delivery systems.
Protein conformation and cryptic pockets
Drug-binding sites are usually pockets or grooves located on the surface of the target protein (Vajda et al., 2018) accessible even in the absence of the drug (Vajda et al., 2018). However, since proteins are dynamic objects, ‘hidden’ binding sites may appear in the presence of an interacting compound (Oleinikovas et al., 2016; Vajda et al., 2018). These cryptic pockets are often not apparent on the unbound protein surface, only transiently opening up as rare events or shaping themselves in the presence of a ligand (Fig. 1). Cryptic sites can provide unforeseen tractable drug target sites, thus expanding the druggable proteome considerably (Vajda et al., 2018; Hopkins and Groom, 2002). On one hand, cryptic pockets offer the prospect to design allosteric drugs (Wenthur et al., 2014), a strategy that could be exploited as a therapeutic path towards treating cancer (Zhong et al., 2021), diabetes (Wang et al., 2021a), and more recently, SARS-CoV-2 infections (Zimmerman et al., 2021). On the other hand, cryptic pockets commonly occur at protein–protein interfaces [PPI; (see section ‘Drugs targeting protein–protein interactions’)]. Therefore, the ability to discover and target cryptic pockets would also enable the design of compounds targeting PPIs en route to new therapeutic formulations (Wells and McClendon, 2007; Shan et al., 2022).
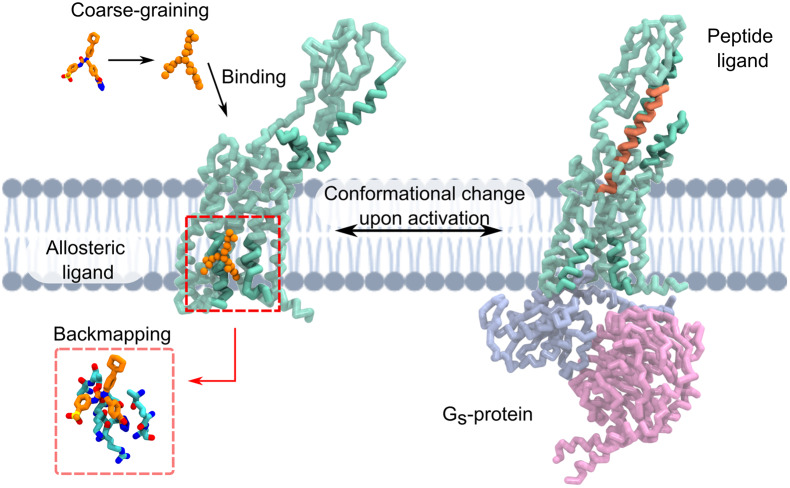
Fig. 1.
Schematic representation of a GPCR (PDB IDs 5XEZ & 6LMK) in inactive (left) and active (right) conformations with an allosteric and peptide ligand bound, respectively. Large conformational changes occur upon binding of the peptide ligand and Gs-protein binding intracellularly, which represent possible dynamics that could be observed with Martin combined with Gō-models. The allosteric pocket in the transmembrane domain exemplifies the possibility to use Martini models for identifying transmembrane pockets, allosteric or cryptic, in various complex membrane compositions. Once a ligand is bound, backmapping is a possibility to obtain higher resolution information for further ligand optimisation or design. All figures were rendered using VMD (Humphrey et al., 1996).
Several approaches have been proposed for the identification of cryptic sites. While some of them are entirely based on the analysis of protein crystallographic structures (Le Guilloux et al., 2009), the majority use MD for the identification of cryptic sites (Le Guilloux et al., 2009; Kokh et al., 2013; Laurent et al., 2015; Cimermancic et al., 2016; Kuzmanic et al., 2020; Zheng, 2021; Shan et al., 2022).
Cryptic sites are not usually captured in the 180,000+ tridimensional structures obtained by state-of-the-art experimental methods (Bank, 2021); their opening generally occurs on the microsecond-to-millisecond time timescale (Kuzmanic et al., 2020). These timescales are only accessible to all-atom (AA) MD simulations relying on specialised hardware, like the Anton3 (Dror et al., 2011; Shaw et al., 2021), or massive distributed computing, as in the Folding@home project (Zimmerman et al., 2021), but not yet for standard GPU-accelerated hardware (Schlick and Portillo-Ledesma, 2021). As a workaround, AA MD-based approaches involve the addition of hydrophilic (e.g. acetic acid, isopropanol) or hydrophobic (e.g. benzene) molecules in the simulation, the so-called mixed-solvent MD (Ghanakota and Carlson, 2016), the addition of the drug in high concentration, the so-called ‘flooding’ MD approach (Amaro and Li, 2010; Gray et al., 2017), or fragment-based screening (MacKerell et al., 2020). However, also in this case the opening of cryptic pockets may still require several microseconds (Kuzmanic et al., 2020). Enhanced sampling approaches have also been used (Bono et al., 2013; Herbert et al., 2013; Oleinikovas et al., 2016). For the collective variable (CV)-based approaches, the central challenge is choosing a suitable CV (Kuzmanic et al., 2020). For the CV-independent methods, running many simulations still entails high computational costs (Earl and Deem, 2005; Kokh et al., 2016), which constitutes the main limiting step.
The Martini 3 CG FF, with its increased accuracy and expanded coverage of the chemical space (Souza et al., 2021a; Alessandri et al., 2022), represents a competitive alternative for extracting and targeting druggable structures on such timescales and/or predicting ligand–target interactions (Souza et al., 2020, 2021b). So far, one of the limitations of the Martini models is the description of proteins’ conformational flexibility (Poma et al., 2017; Souza et al., 2019), which is fundamentally linked to biological function (Henzler-Wildman and Kern, 2007; Luo, 2012; Veesler and Johnson, 2012; Campaner et al., 2017; Hadden et al., 2018; Matthes et al., 2018; Maggi et al., 2020; Bolnykh et al., 2021; Noreng et al., 2021; Jackson et al., 2022) and pivotal to design new therapeutics (Hammes, 2002; Campaner et al., 2017; Sengupta and Udgaonkar, 2019; Schulz-Schaeffer et al., 2020; Gossen et al., 2021; Xiao et al., 2021; Zhao et al., 2021; Margreiter et al., 2022).
Commonly, Martini-based approaches implement an elastic network (EN), that is addition of a network of harmonic restraints to stabilise protein tertiary structure (Periole et al., 2009). The restraints are usually added based on a distance criterion, introducing a strong bias towards the starting conformation (Periole et al., 2009). Different strategies were devised to address this issue (Deplazes et al., 2012; Lelimousin et al., 2016; Poma et al., 2017): (i) localised distance-restraints on selected secondary structure elements, often driven by experimental information. For example, this approach was used to study the activation of the epidermal growth factor receptor, coupling Martini 2 CG simulations with enhanced sampling techniques, such as well-tempered metadynamics (Barducci et al., 2008) and distance-based restraints on transmembrane helices (Lelimousin et al., 2016). (ii) implementation of Gō-like models (GōMartini) by establishing a Lennard–Jones (LJ) potential based on the contact map of the native protein structure instead of the harmonic based potential (Poma et al., 2017; Souza et al., 2019; Mahmood et al., 2021). Different contact map definitions for GōMartini were tested on three protein systems (cohesin, titin and ubiquitin) and reproduced protein flexibility as observed in AA simulations. Two different contact map definitions were tested and compared against ENs. The first variant only considers van der Waals spheres (OV) overlaps, while the second builds on top of the OV approach and includes chemical information from the atoms in question. Here a contact between residues requires that the number of attractive contacts between atoms be larger than the number of repulsive ones (Wołek et al., 2015; Poma et al., 2017).
Other approaches also exist to characterise conformational transitions between two or more conformational states in the presence of ENs (Kim et al., 2002; Miyashita et al., 2003; Feng et al., 2009; Das et al., 2014), like gradually switching between two different types of EN connectivity through a switching parameter or the so-called ‘generalised elastic network’ (Poma et al., 2018) which, within a given cut-off, implements a canonical EN with harmonic potentials and above the chosen cutoff instead implements Gō-like contacts to the system. Recently, adaptive ENs have been developed as well (Kanada et al., 2022). These strategies represent a potential powerful development strategy for Martini models.
The Martini model can be coupled with strategies to introduce proteins’ dynamics, as discussed above, and combined with enhanced sampling approaches to ideally push the system towards the exploration of ‘rare’ events, like cryptic pocket opening. The use of artificial intelligence algorithms to identify and speed-up the slower modes, as done in Bonati et al. (2020 and 2021) could also be exploited to steer cryptic pockets’ opening. This represents a valid and computationally cheaper solution to identify and target cryptic pockets. Once possible pockets are identified at the CG level, the protein structure could be converted into atomistic resolution (Wassenaar et al., 2014; Vickery and Stansfeld, 2021) for further investigation and ligand design within a virtual screening (VS) workflow.
Protein binding pockets in membrane environments
A fast-growing area for Martini simulations is the analysis of protein–ligand interactions in membrane environments, which is experimentally and computationally rather challenging. Here, the ligand may be an endogenous lipid, that is, natively part of the physiological environment, or an exogenous compound targeting an allosteric pocket of a transmembrane protein or at the protein–lipid interface (Fig. 1). In this context, the structural characterisation of the ligand-binding sites in the transmembrane region and ranking based on binding energetics extracted by CG MD simulations is particularly attractive. This is even more so because standard computational approaches for protein–ligand binding like molecular docking, that do not account for the specificity of the membrane environment, that is, the strong hydrophobic character and the competition with native lipids, are prone to fail. Recently, several CG MD investigations of protein–ligand interactions in the transmembrane region of pharmacologically relevant targets have been reported. In general, the common computational strategy involves: (i) binding-site identification and structural characterisation of the protein–ligand complex using, among other methods, unbiased CG MD simulations and ligand-density maps (Ferraro et al., 2016; Dämgen and Biggin, 2021); (ii) ranking of binding modes by binding affinity calculations based on equilibrium MD (Souza et al., 2020), potential of mean force (PMF), alchemical transformations, metadynamics (Corey et al., 2019) or binding saturation curves (Ansell et al., 2021); and (iii) structural refinement of the protein ligand complex via backmapping to atomistic models (Wassenaar et al., 2014). Overall, the main advantage of CG modelling is the ability to converge on sampling the protein–ligand conformational space, currently out-of-reach by typical unbiased atomistic simulations. As a result, within the limits of the accuracy of the model, trends in dissociation constants (Kd) and rates (Koff) can potentially be accessed from unbiased MD (Souza et al., 2020, 2021b).
The vast majority of CG MD analyses of protein–ligand interactions in membrane environments involve protein–lipid binding based on Martini 2.2 simulations (De Jong et al., 2013). The use of the Martini 2.2 FF has allowed not only to discern specific versus nonspecific interactions but also to characterise the energetics involved in the binding reaction. Earlier efforts focused on the prediction of the binding site(s) for cholesterol, which is the most abundant endogenous steroid in mammalian cell membranes and was shown to modulate several membrane proteins including ion channels. Using multi-microsecond CG MD simulations of a homology model of the serotonin transporter embedded in a raft-like membrane, Ferraro et al (2016) provided evidence of the existence of specific binding sites for cholesterol, identifying a hotspot that largely overlaps with the cholesterol-binding site illuminated by X-ray crystallography of the closely related dopamine transporter (Ferraro et al., 2016). By combining CG MD simulations and PMF calculations, Ansell et al (2021) characterised the interaction between cholesterol and several membrane proteins including an ATP-dependent pump, a sterol receptor/transporter protein and a member of the TRP ion-channel family. A similar analysis of the chemokine receptor 3, a GPCR responsible for trafficking white blood cells, allowed for the identification of six cholesterol-binding sites, suggesting that recognition of cholesterol at these sites may modulate the affinity for agonists/antagonists allosterically via a rigidification of the protein structure (van Aalst et al., 2021). Using CG MD simulations and lipid-density maps, Damgen and Biggin (2021) explored the affinity of cholesterol and different lipid types for the glycine receptor channel in its active and resting states and found that lipids may act as allosteric modulators because their strength of binding strongly depend(s) on the physiological state of the receptor. In a similar study, protein–lipid interactions on the homologous nicotinic acetylcholine receptor were investigated using a complex quasi-neuronal membrane composed of 36 species of lipids, including cholesterol, in a binding competition assay (Sharp and Brannigan, 2021). Interestingly, the CG MD simulations suggested that cholesterol binds to concave inter-subunit sites and polyunsaturated fatty acids prefer convex sites at the outer transmembrane helix M4, while monounsaturated and saturated lipids are enriched at the protein–lipid interface (Sharp and Brannigan, 2021). Recently, the interaction of the anionic lipids cardiolipins with 42 inner membrane proteins from Escherichia. coli has been investigated by CG MD simulations. Overall, >700 independent cardiolipin binding sites were identified and structurally characterised, thus providing a molecular basis for protein–cardiolipin interactions (Corey et al., 2021). In the context of systematic comparative analyses, the method by Ansell et al. (2021) for protein–ligand binding affinities based on binding saturation curves appears particularly appealing as a high-throughput approach for binding-site comparison and ranking.
In addition to protein–lipid interactions, a potential area of development for CG simulations involves the exploration of modulatory ligand binding, such as agonists, antagonists and allosteric modulators, to the transmembrane region of proteins. In this case, and unlike for most lipid molecules, a serious difficulty is introduced by the lack of off-the-shelf CG parameters to model the ligand(s). As a result, examples of studies focusing on the allosteric modulation of transmembrane proteins via protein–ligand interactions are still rare in the literature. One of them focused on the investigation of the binding pathway of two orthosteric agonists of the μ-opioid receptor, that is, fentanyl and morphine (Sutcliffe et al., 2021). Using CG MD simulations and free energy calculations, Sutcliffe et al. (2021) compared the aqueous and lipophilic binding pathways to the orthosteric site and found that the synthetic opioid fentanyl prefers the lipophilic route, which might explain its lower susceptibility to overdose reversal. Since more and more high-resolution structures of relevant pharmacological targets highlight the existence of multiple allosteric sites in the transmembrane region of these proteins (Cerdan et al., 2020), the development of automatic parameterisation tools to facilitate the setup of CG MD simulations, similar to what is currently available for AA MD, is expected to leverage more exploratory analyses of protein–ligand interactions in the membrane environment and open to high-throughput screening powered by CG MD simulations. Additionally, the new Martini 3 FF offers an extended chemical space (Souza et al., 2020, 2021a, 2021b), providing an excellent platform for developing automatic parameterisation tools for ligands.
Drugs targeting protein–protein interactions
PPIs have been considered as promising drug targets since the early 2000s, with the hope to overcome the decline in the efficiency of conventional drug development. Three major types of PPI modulators currently described in the literature are small molecules, antibodies and peptides (Mabonga and Kappo, 2019; Lu et al., 2020; Martino et al., 2021). Small molecules typically require a prototypical binding site. The PPI interface is usually flat, shallow and hydrophobic, without an actual pocket where small-molecule ligands may bind (Lu et al., 2020). The natural alternative would be to increase the size of the modulator to maximise PPI interface coverage and establish many hydrophobic contacts (Lu et al., 2020). However, increasing small-molecule-based PPI modulator size may lead to undesirable pharmacokinetic profiles (An and Fu, 2018; Lu et al., 2020; Martino et al., 2021). Antibodies present an alternative therapeutic avenue, since these can fully cover the PPI interface due to their size (Bojadzic and Buchwald, 2018; Martino et al., 2021) and there is potential for the general application of antibody-based therapies when combined with novel drug delivery systems (Slastnikova et al., 2018). Peptides can also be used to modulate PPIs as they bind the PPI interface with high affinity (Cabri et al., 2021), but they may exhibit short half-lives and toxicity risks (Gupta et al., 2013; Nevola and Giralt, 2015; Mabonga and Kappo, 2019). Examples of small-molecule PPI modulators are venetoclax, to treat chronic lymphoblastic leukaemia (Lu et al., 2020), and pomalidomide to treat myeloma (Dimopoulos et al., 2014). ALRN-6924 is an α-helical peptide aimed at leukaemia therapy (Carvajal et al., 2018) while Bavencio is an antibody-based drug targeting Merkel cell carcinoma (Boyerinas et al., 2015).
Recently, a new type of PPI modulator technology gained momentum: the Proteolysis Targeting Chimeras (PROTACs) (Sakamoto et al., 2001). These bivalent molecules consist of a linker connecting a small molecule binding the target (i.e. ‘warhead’) and a second small molecule binding an E3 ligase (the ‘recruiter’), acting as a PPI enhancer like molecular glues (Wang et al., 2020; Alabi and Crews, 2021; Bond and Crews, 2021; Békés et al., 2022). Simultaneous binding of both proteins by the PROTAC brings them into proximity, provoking target ubiquitination and posterior degradation by proteasome machinery (Wang et al., 2020; Alabi and Crews, 2021; Bond and Crews, 2021; Békés et al., 2022). Compared to small-molecule inhibitors, PROTACs work catalytically, requiring less compound concentration, having fewer off-target effects and exhibiting improved target selectivity (Troup et al., 2020; Alabi and Crews, 2021; Békés et al., 2022). In the last years, PROTACs attracted the interest of academic and pharmaceutical companies and currently two molecules developed by Arvinas were forwarded to Phase II clinical trials (Petrylak et al., 2020; Békés et al., 2022). Key steps in PROTAC development include the selection of the E3 ligase to pair with the target of interest (Cecchini et al., 2021), the accurate prediction of the ternary complex structure (Zaidman et al., 2020) and linker design (Troup et al., 2020; Bemis et al., 2021).
Computational modelling and simulations can help the rational design of PROTACs (Fig. 2). In the absence of ternary complex crystal structures, which must contain the target, the PROTAC and the ligase, one of the first steps of in silico design is sampling the conformational landscape of the complex, which is achievable by protein–protein docking (Hayashi et al., 2018; Drummond and Williams, 2019; Drummond et al., 2020; Rosell and Fernández-Recio, 2020; Zaidman et al., 2020; Bluntzer et al., 2021; Bai et al., 2021, 2022; Weng et al., 2021) and/or MD simulations at various levels of detail (Rakers et al., 2015; Yu et al., 2019; Perez et al., 2021). For very large systems however, AA MD can become prohibitively expensive (Durrant and McCammon, 2011; Amaro et al., 2018; Jung et al., 2021). This is particularly true when considering a VS campaign applied to ternary complexes in explicit solvent, due to system size and complexity. As an alternative, docking and MD simulations based on the Martini 2 and 3 CG framework (Roel-Touris et al., 2019; Roel-Touris and Bonvin, 2020; Souza et al., 2021b) may be used to facilitate the study of these large macromolecular systems. In a first stage, CG protein–protein docking can be used to capture the most important features of the interaction complex, providing many potential binding modes. It can then be combined with long and affordable CG MD simulations to probe complex stability, which is critical for PPI drug discovery. One example of CG-docking methods is HADDOCK (Roel-Touris et al., 2019; Roel-Touris and Bonvin, 2020). A limitation of some docking approaches is the treatment of proteins as rigid bodies (Vakser, 2020; Harmalkar and Gray, 2021). Recently, docking approaches including protein flexibility have been developed, including ‘divide-and-conquer’ (Karaca and Bonvin, 2011) and normal mode analysis-based strategies (May and Zacharias, 2008; Moal and Bates, 2010; Jiménez-García et al., 2018; Diaz et al., 2021). Alternatively, GōMartini simulations (Poma et al., 2017) could be used to cheaply produce protein–protein conformations which, after a back-mapping procedure, could be used in ensemble docking (Amaro et al., 2018).
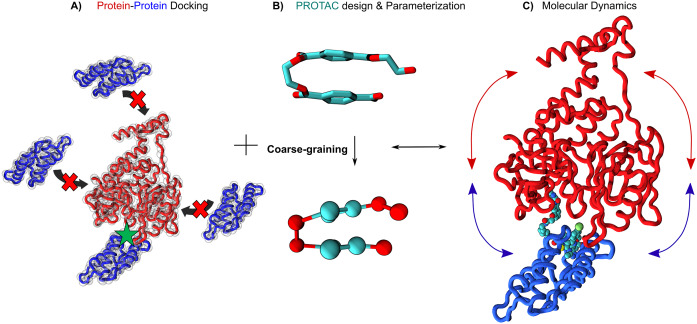
Fig. 2.
Important steps in PROTAC design for drug discovery campaigns. (a) Protein–protein docking either at the atomistic (ribbons) or coarse-grained level (red and cyan spheres). The E3 ligase is represented in red and the target protein in blue. (b) Coarse-graining of a small -molecule using the Martini 3 force field. (c) Dynamical motions of the ligase and the target (blue and red arrows, respectively) are important to query ternary complex stability in the presence of the PROTAC (represented as van der Waals spheres). All figures were rendered using VMD (Humphrey et al., 1996). The ternary complex structure is from Nowak et al. (2018) with the PDB ID code 6BN7.
Massive protein–protein docking for target identification (Zhang et al., 2014) of other PPI modulators can also greatly benefit from the use of CG approaches. In the case of PROTACS, not only the target but also the choice of E3 Ligase is fundamental for the stability of the ternary complex and cell-specific target degradation (Békés et al., 2022). Only a limited number of E3 ligases have been explored towards PROTAC development (Burslem and Crews, 2020; Troup et al., 2020; Alabi and Crews, 2021). Examples are the von Hippel-Landau or Cereblon E3 ligases (He et al., 2020; Bricelj et al., 2021). However, some ligases, for which there is currently no crystal PROTAC ternary complex available, are known to be enriched in specific cell types (Békés et al., 2022). Combining CG docking between a PROTAC-containing target and several candidate ligases separately with subsequent CG MD simulations could help to identify the most suitable target-ligase pair, enabling cell-type-based therapeutic PROTAC approaches.
Another PROTAC-specific challenge is the design of the linker portion (Alabi and Crews, 2021) as there exist no common practices or guidelines, and linker size and flexibility affect the degradation efficiency of PROTACs (Cyrus et al., 2011; Crew et al., 2018; Troup et al., 2020). Optimal linkers should be long and flexible enough to promote a ternary complex orientation that allow ubiquitin transfer to the lysines on the target surface. However, overly flexible linkers may hamper target degradation efficiency (Cecchini et al., 2021). CG-based approaches can help linker optimisation. For example, PROTAC CG docking simulations could be used to evaluate the possibility of other PROTAC molecules fitting into the available volume at the binding interface of a ligase/target complex. One route would be by harnessing structural data like the warhead-recruiter distances, extracted from ternary complexes from the Protein Data Bank (Burley et al., 2021) or from protein–protein docking experiments, as constraints. Filtering the predicted complexes using this information in combination with the docking score and other observables would allow retrieval of the best binding poses per system (Zaidman et al., 2020). From the most stable complexes, probed by CG MD, a linker template could then be designed and subsequently used in VS campaigns targeting chemically diverse linker libraries. Further, chemical modifications around the linker template would enable fine tuning of PROTAC properties like solubility, lipophilicity or toxicity effects (Troup et al., 2020). Recently, the group of Kihlberg illustrated that PROTACs cell permeability is deeply related to the linkers’ conformational flexibility. Although these compounds do not conform to oral bioavailability defined by the Lipinski rule-of-5 (Lipinski et al., 2001), by acting as ‘molecular chameleons’ they are able to fold-in on themselves in aqueous solution and reduce their solvent-accessible polar surface area to increase cell permeability and then unfurl after crossing the membrane (Atilaw et al., 2021). Thus, some of the key factors playing a role in PROTAC cell permeability are linker size (Klein et al., 2020), polarity and rigidity (Atilaw et al., 2021), further highlighting the importance of a rational linker design strategy. Similar concerns related to polarity and membrane permeability are also prevalent in PPI-targeting peptide design (Sugita et al., 2021). As such, transfer free energy calculations carried out at the CG level could enable the direct investigation of the ability of different PPI modulators to cross biological membranes in an efficient and affordable manner while still achieving a high degree of accuracy.
Tuning soft nanoparticles with Martini
Drug efficacy correlates with the ability of the drug to reach the target site in sufficient quantities. A high percentage of approved drugs display low aqueous solubility and are fast degraded. To tackle these problems, delivery systems have been developed (Malmsten, 2006; Wang et al., 2021b). Different physicochemical properties of the delivery system, such as morphology, composition and stiffness, can contribute to the drug solubility, targeting efficiency and stability (Zhang et al., 2015; Yu et al., 2018). As drug carrier rigidity affects physiological membrane crossing, developing soft nanoparticle (SN) systems that can easily deform appears attractive.
SNs include carriers consisting of lipids, polymers or surfactants. Lipid-based carriers are generally biocompatible and highly permeable; however, they exhibit low mechanical stability (Sercombe et al., 2015). Polymer-based carriers, on the other hand, have higher mechanical stability but lower biocompatibility and permeability (Jana et al., 2021). It is also possible to combine lipids and polymers and to harness the advantages of each component (Reimhult and Virk, 2021). Studies have shown that the mechanism of delivery for SNs depends on their morphology and composition, which is in turn correlated with the distribution of the drug within the carrier (El Maghraby et al., 2008). However, little is known about the morphology and mechanism of delivery for these hybrid systems (Reimhult and Virk, 2021).
CG modelling is a valuable tool for investigating the formation of SNs, their morphology, drug distribution within the carrier and the mechanism of delivery, including the interaction with different biological membranes (Yang et al., 2021; Parchekani et al., 2022). The first obstacle for using CG models is constructing the system. Fortunately, an increasing number of tools have been developed for building such CG models, examples being TS2CG, Charmm-GUI and Nano Disc builder, allowing the construction of vesicles and other SNs for drug delivery (Qi et al., 2015; Hsu et al., 2017; Kjølbye et al., 2020; Pezeshkian et al., 2020), the Polyply package for constructing polymer-based systems (Grünewald et al., 2022) or the Insane.py script for bilayers (Wassenaar et al., 2015). In combination, protocols for simulating soft delivery systems have also started to appear in the literature (Bruininks et al., 2019). Several CG studies have been performed using the Martini 2 model, investigating the morphology, size and internal organisation of the different components in lipid and polymer-based carriers (Hashemzadeh et al., 2020; Bono et al., 2021; Gao et al., 2021). Among the first described SNs are liposomes, consisting of a lipid bilayer surrounding a hydrophilic core, capable of trapping both hydrophobic and hydrophilic drugs. Liposomes were the first delivery system to reach clinical application (Doxil) (James et al., 1994) and have been widely used and characterised for many different therapeutics (Allen and Cullis, 2013). Further development of liposomes resulted in cationic lipids and subsequently cationic polymers for delivery of NAs, which proved invaluable at the outbreak of the COVID-19 pandemic (Polack et al., 2020; Baden et al., 2021). Cationic lipids or polymers can condense NA efficiently, thanks to the electrostatic interaction with the negatively charged NA to form lipoplexes and polyplexes, respectively (Li and Szoka, 2007; Schlich et al., 2021). Accurate description of the electrostatic interactions is a major challenge in the case of highly charged lipo- or polyplexes. The challenge could be tackled by developing polarisable Martini models, so far only available for water, ions and proteins with the Martini 2 FF (Yesylevskyy et al., 2010; De Jong et al., 2013; Michalowsky et al., 2017, 2018; Sahoo et al., 2022).
The main drawback of permanently charged cationic components is their toxicity and rapid elimination from circulation (Li and Szoka, 2007; Schlich et al., 2021). To avoid the toxicity and increase the circulation time and stability, particles can be covered by a PEGylated lipid shield (Li and Szoka, 2007), although PEGylation has shown to diminish particle uptake in target cells (PEG dilemma) (Gjetting et al., 2010). The Martini model has eased the way to study polymer coating in membranes (Grünewald et al., 2018; Lemaalem et al., 2020) and NPs (Pannuzzo et al., 2020).
A step further in the optimisation led to ionisable components, resulting in the formulation of lipid nanoparticles (LNPs) (Schlich et al., 2021) and dendrimers (Palmerston Mendes et al., 2017), branched polymers with well-defined molecular weights. The ionisable components are positively charged at low pH to encapsulate NA, and neutral at higher pH, for example, in the blood, thereby avoiding the drawbacks of lipo- and polyplexes. However, it has been shown that only 2–3% of the nucleotide drug load reaches the cytosol using LNPs (Gilleron et al., 2013). Once the LNP or dendrimer is endocytosed, endosomes eventually fuse with lysosomes and their cargo is degraded. For optimised release, the cargo needs to escape the endosome before fusion with the lysosome (Schlich et al., 2021). A general understanding of the delivery mechanism and its pH dependence is lacking. For investigating pH dependent release routes or interactions with NA, constant pH CG approaches are available (Grünewald et al., 2020; Aho et al., 2022). As a proof of concept, collective interactions between titratable sites in a G5 dendrimer poly(propylene imine) were simulated at different pH values, revealing how the particle expands in radius and increases in degree of protonation with decreasing pH, consistent with previous atomistic studies (Grünewald et al., 2020).
The delivery depends on the structural properties of the carrier. For LNPs, two different internal organisations have been proposed based on CG modelling with Martini 2 and cryo-transmission electron microscopy (Leung et al., 2012; Kulkarni et al., 2018). Understanding the structure–activity relationship is of paramount importance for the rational design of optimised LNPs and for cell-specific targeting. Cell specific LNPs can be built by changing one or more of the lipid components (Liu et al., 2021; Żak and Zangi, 2021), but models of synthetic lipids are not always available. The combination of the Martini 3 FF, with its extended chemical space, and building tools enables the prediction of properties of both empty structures of SNs, such as LNPs, and complexes with cargo of various sizes, from small interfering RNA (siRNAs) to large messenger RNA (mRNA) molecules, enabling studies of the internal organisation and interactions.
Cell specificity and drug efficacy can, in principle, be optimised in terms of interaction and fusion with the endosomal membrane. To this end, Martini 2 models of complex membranes have previously been constructed (Ingólfsson et al., 2014, 2020), which demonstrates the possibility of studying the interaction between various SNs formulations (Lee et al., 2021) and cell-specific plasma and endosomal membranes. However, this field remains to be explored.
In perspective, there is a general need to implement alternative design strategies to further optimise SNs. Tuning the chemical groups of the lipids or polymer, along with ratios of components, is key for altering the properties of SNs. However, synthesising and testing combinations of lipid or polymer variables are costly and time consuming. In silico screening of promising formulations is a viable alternative to study the role that each component plays in the SN morphology and delivery process (Fig. 3). One drawback when studying lipid-based systems using CG approaches is the loss of resolution compared to AA representations. For instance, changes in tail composition are not always well captured. Nevertheless, in the new version of Martini different tail chemistries can be easier to represent in the future as a result of the use of small and tiny beads.
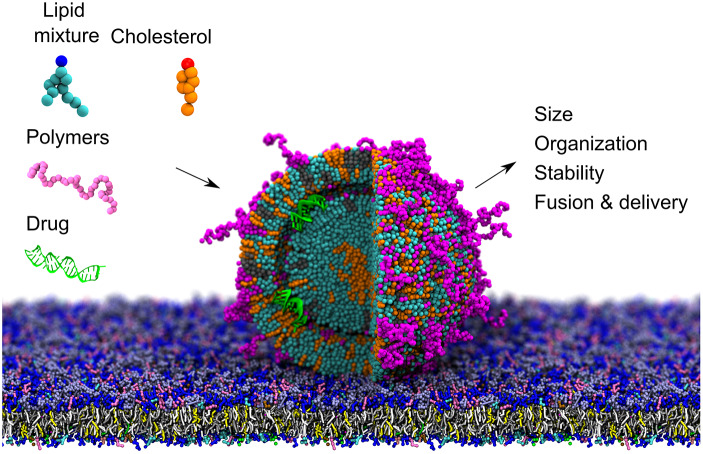
Fig. 3.
CG modelling enables predictions of organisation, size and stability of SNs containing various building blocks and cargo. Moreover, it can be used to study the interaction between various SN formulations and biological barriers, such as plasma and endosomal membranes. All figures were rendered using VMD (Humphrey et al., 1996).
Summary and future directions
For in silico studies of large complex systems, aiming at identifying possible druggable sites, predicting and optimising protein ligand binding for drug design or studying drug delivery systems, the Martini model provides an efficient approach relative to AA MD. Due to the timescales reachable by CG Martini simulations, it is possible to probe systems with respect to pockets formed transiently, interesting for drug discovery campaigns. However, further benchmarking is required to assess the accuracy of the FF. Furthermore, maintaining proteins’ tertiary structure in Martini models requires inclusion of EN or GōMartini potentials. Thus, a reasonable definition of the contact map, from which to draw the network of potentials, is critical. Improvements to the definition of contact maps may consider an ensemble of conformations and use knowledge of hydrogen bonds and residue protonation. Additionally, the use of LJ potentials in GōMartini enables the development of multi-basin models, as previously shown for AA MD (Okazaki et al., 2006), combining Gō-models of different protein conformations to promote conformational transitions.
Currently, a major challenge towards the use of the Martini CG model in drug design and drug delivery is the automatic parameterisation of ligands and components of delivery vectors, including AA-to-CG mapping, construction of the bonded parameters and bead-type assignment. To address this issue, tools like Swarm-CG (Empereur-Mot et al., 2020) or PyCGTOOL (Graham et al., 2017) have been developed. However, these approaches focus solely on optimising the bonded parameters. Automated parameterisation workflows for Martini 2 models of small molecules are available (Bereau and Kremer, 2015; Potter et al., 2021), including mapping, bonded-parameter definition and bead type selection based on optimisation of oil–water partitioning free energies. However, since the covered chemical space is larger in Martini 3, adapting these codes to Martini 3 is not straightforward. Equally important will be the generation of curated and extended libraries of Martini models, such as MAD (the MArtini Database server – https://mad.ibcp.fr), which can be used as reference to access the accuracy of such automatic approaches (Hilpert et al., 2022). However, the current library of Martini 3 small-molecule models (Alessandri et al., 2022) may already allow initial benchmarks based on fragment-based strategies. Another challenge is the backmapping procedure from CG to AA resolution (Wassenaar et al., 2014; Vickery and Stansfeld, 2021), as protein side-chain directionality is kept, but the binding mode may not be accurate. A standard solution is to perform cycles of energy minimisation and equilibration on the backmapped structure to improve side-chain packing. Another option in this direction would involve the use of machine learning methods to optimise side-chain orientation (Misiura et al., 2022).
Backmapping and small-molecule automatic parameterisation are fundamental goals towards VS of molecules targeting PPI systems, like PROTACS. Additionally, available tools for CG protein–protein docking with the Martini 3 FF could efficiently provide researchers with reasonable starting structures for these large complexes, whose dynamics can be probed by MD. This is the premise of the currently in-development CG version of LightDock (Roel-Touris et al., 2020a, 2020b), implementing the Martini 3 FF. Coupling these tools with CG docking and MD simulations would allow to derive rules for PROTAC linker design and screening and/or to evaluate ternary complex stability when varying the Ligase protein. Within the field of drug delivery, the Martini 3 model combined with the implementation of tools and protocols available for constructing and simulating soft delivery systems, such as LNPs, will enable in silico screening of various formulations, permitting more efficient optimisation or rational design of delivery methods. However, for NA-containing drug delivery systems, the parameters for RNA/DNA are still under development in Martini 3 and the lack of experimentally resolved structures complicates FF parameter optimisation. While previous Martini 2 NA models were rather rigid (Uusitalo et al., 2015, 2017), improving the dynamics of the future NA Martini 3 models is of utmost importance for the simulations of NA delivery systems.
Overcoming these challenges is fundamental for broader applications of the Martini 3 model in biologically relevant systems like SNs, protein–protein interactions, membrane systems and efficient discovery of druggable cryptic pockets, enabling an even larger impact of CG models in fields of drug discovery and delivery. For validation of the CG modelling within drug discovery, one example is the technique of co-crystallisation or soaking macromolecular crystals, essentially replacing solvent with a ligand within the crystal, enabling the comparison to for example, flooding CG-MD simulations for pocket identification (Wienen-Schmidt et al., 2021). Within the drug delivery field, one could imagine correlating predicted structures and organisation of SNs based on CG-MD simulation with fusion and transfection efficacy (Miao et al., 2020) measured experimentally, combined with fluorescence studies (Chen et al., 2019) enhancing the understanding and development of such delivery methods.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/qrd.2022.16.
Financial support
BJG is employed by Zymvol Biomodeling on a project which received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie grant agreement No. 801342 (Tecniospring INDUSTRY) and the Government of Catalonia’s Agency for Business Competitiveness (ACCIÓ). AB and MC received funding from the French National Research Agency (Grant no. ANR-18-CE11–0015). LM is supported by the French National Institute of Health and Medical Research (INSERM). PCTS, JM, GPP, and LRK are supported by the French National Center for Scientific Research (CNRS). Further funding of LRK, GPP, PCTS and LM came from a research collaboration with PharmCADD. SA and GR acknowledge the grant from the Interdisciplinary Centre for Clinical Research within the faculty of Medicine at the RWTH Aachen University (IZKF TN1-1/IA 532001; TN1–4/IA 532004) and the Deutsche Forschungsgemeinschaft (DFG) via the Research Training Group RTG2416 MultiSenses-MultiScales (368482240/GRK2416). AM and GR acknowledge the Helmholtz European Partnering fundings for the project ‘Innovative high-performance computing approaches for molecular neuromedicine’. GR acknowledges the Federal Ministry of Education and Research (BMBF) and the state of North Rhine-Westphalia as part of the NHR Program, as well as the Joint Lab ‘Supercomputing and Modeling for the Human Brain’ of the Helmholtz Association, Germany and the two European Union’s Horizon 2020 MSCA Program under grant agreement 956314 [ALLODD].
Conflicts of interest
The authors declare no conflicts of interest.
References
- Aho N, Buslaev P, Jansen A, Bauer P, Groenhof G and Hess B (2022) Scalable Constant pH Molecular Dynamics in GROMACS. Journal of Chemical Theory and Computation 18, 6148–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi SB and Crews CM (2021) Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. The Journal of Biological Chemistry 296, 100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandri R, Barnoud J, Gertsen AS, Patmanidis I, de Vries AH, Souza PCT and Marrink SJ (2022) Martini 3 coarse-grained force field: Small molecules. Advanced Theory and Simulations 5, 2100391. [Google Scholar]
- Alessandri R, Grünewald F and Marrink SJ (2021) The martini model in materials science. Advanced Materials 33, e2008635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandri R, Souza PCT, Thallmair S, Melo MN, De Vries AH and Marrink SJ (2019) Pitfalls of the martini model. Journal of Chemical Theory and Computation 15, 5448–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM and Cullis PR (2013) Liposomal drug delivery systems: From concept to clinical applications. Advanced Drug Delivery Reviews 65, 36–48. [DOI] [PubMed] [Google Scholar]
- Amaro RE, Baudry J, Chodera J, Demir Ö, McCammon JA, Miao Y and Smith JC (2018) Ensemble docking in drug discovery. Biophysical Journal 114, 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro RE and Li WW (2010) Emerging methods for ensemble-based virtual screening. Current Topics in Medicinal Chemistry 10, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S and Fu L (2018) Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. eBioMedicine 36, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell TB, Curran L, Horrell MR, Pipatpolkai T, Letham SC, Song W, Siebold C, Stansfeld PJ, Sansom MSP and Corey RA (2021) Relative affinities of protein-cholesterol interactions from equilibrium molecular dynamics simulations. Journal of Chemical Theory and Computation 17, 6548–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilaw Y, Poongavanam V, Svensson Nilsson C, Nguyen D, Giese A, Meibom D, Erdelyi M and Kihlberg J (2021) Solution conformations shed light on PROTAC cell permeability. ACS Medicinal Chemistry Letters 12, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J and Zaks T (2021) Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. The New England Journal of Medicine 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai N, Miller SA, Andrianov GV, Yates M, Kirubakaran P and Karanicolas J (2021) Rationalizing PROTAC-mediated ternary complex formation using Rosetta. Journal of Chemical Information and Modeling 61, 1368–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai N, Riching KM, Makaju A, Wu H, Acker TM, Ou S-C, Zhang Y, Shen X, Bulloch D, Rui H, Gibson B, Daniels DL, Urh M, Rock B and Humphreys SC (2022) Modeling the CRL4A ligase complex to predict target protein ubiquitination induced by cereblon-recruiting PROTACs. The Journal of Biological Chemistry 298, 101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protein Data Bank (2021) PDB Statistics: Overall Growth of Released Structures Per Year. Available at www.rcsb.org/stats/growth/growth-released-structures (accessed April 2022).
- Barducci A, Bussi G and Parrinello M (2008) Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Physical Review Letters 100, 020603. [DOI] [PubMed] [Google Scholar]
- Békés M, Langley DR and Crews CM (2022) PROTAC targeted protein degraders: The past is prologue. Nature Reviews. Drug Discovery 21, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis TA, La Clair JJ and Burkart MD (2021) Unraveling the role of linker design in proteolysis targeting chimeras. Journal of Medicinal Chemistry 64, 8042–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereau T and Kremer K (2015) Automated parametrization of the coarse-grained martini force field for small organic molecules. Journal of Chemical Theory and Computation 11, 2783–2791. [DOI] [PubMed] [Google Scholar]
- Bluntzer MTJ, O’Connell J, Baker TS, Michel J and Hulme AN (2021) Designing stapled peptides to inhibit protein-protein interactions: An analysis of successes in a rapidly changing field. Peptide Science 113, e24191. [Google Scholar]
- Bojadzic D and Buchwald P (2018) Toward small-molecule inhibition of protein-protein interactions: General aspects and recent Progress in targeting costimulatory and Coinhibitory (immune checkpoint) interactions. Current Topics in Medicinal Chemistry 18, 674–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini M, Domaoal R, Thakur V, Gallardo-Macias R, Spasov K, Anderson K and Jorgensen W (2011) Computationally-guided optimization of a docking hit to yield cathecol diethers as potent anti-HIV agents. Journal of Medicinal Chemistry 54, 8582–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnykh V, Rossetti G, Rothlisberger U and Carloni P (2021) Expanding the boundaries of ligand–target modeling by exascale calculations. Computational Molecular Science 11, e1535. [Google Scholar]
- Bonati L, Piccini G and Parrinello M (2021) Deep learning the slow modes for rare events sampling. Proceedings of the National Academy of Sciences of the United States of America 118, e2113533118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonati L, Rizzi V and Parrinello M (2020) Data-driven collective variables for enhanced sampling. Journal of Physical Chemistry Letters 11, 2998–3004. [DOI] [PubMed] [Google Scholar]
- Bond MJ and Crews CM (2021) Proteolysis targeting chimeras (PROTACs) come of age: Entering the third decade of targeted protein degradation. RSC Chemical Biology 2, 725–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono N, Coloma Smith B, Moreschi F, Redaelli A, Gautieri A and Candiani G (2021) In silico prediction of the in vitro behavior of polymeric gene delivery vectors. Nanoscale 13, 8333–8342. [DOI] [PubMed] [Google Scholar]
- Bono F, De Smet F, Herbert C, De Bock K, Georgiadou M, Fons P, Tjwa M, Alcouffe C, Ny A, Bianciotto M, Jonckx B, Murakami M, Lanahan AA, Michielsen C, Sibrac D, Dol-Gleizes F, Mazzone M, Zacchigna S, Herault JP, Fischer C, Rigon P, Ruiz de Almodovar C, Claes F, Blanc I, Poesen K, Zhang J, Segura I, Gueguen G, Bordes MF, Lambrechts D, Broussy R, van de Wouwer M, Michaux C, Shimada T, Jean I, Blacher S, Noel A, Motte P, Rom E, Rakic JM, Katsuma S, Schaeffer P, Yayon A, Van Schepdael A, Schwalbe H, Gervasio FL, Carmeliet G, Rozensky J, Dewerchin M, Simons M, Christopoulos A, Herbert JM and Carmeliet P (2013) Inhibition of tumor angiogenesis and growth by a small-molecule multi-FGF receptor blocker with allosteric properties. Cancer Cell 23, 477–488. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Jochems C, Fantini M, Heery CR, Gulley JL, Tsang KY and Schlom J (2015) Antibody-dependent cellular cytotoxicity activity of a novel anti-PD-L1 antibody Avelumab (MSB0010718C) on human tumor cells. Cancer Immunology Research 3, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricelj A, Steinebach C, Kuchta R, Gütschow M and Sosič I (2021) E3 ligase ligands in successful PROTACs: An overview of syntheses and linker attachment points. Frontiers in Chemistry 9, 707317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DG and Wobst HJ (2021) A decade of FDA-approved drugs (2010–2019): Trends and future directions. Journal of Medicinal Chemistry 64, 2312–2338. [DOI] [PubMed] [Google Scholar]
- Bruininks BMH, Souza PCT, Ingólfsson HI and Marrink SJ (2020) A molecular view on the escape of lipoplexed DNA from the endosome. eLife 9, e52012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruininks BMH, Souza PCT and Marrink SJ (2019) A practical view of the martini force field. Methods in Molecular Biology 2022, 105–127. [DOI] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, Christie CH, Dalenberg K, Di Costanzo L, Duarte JM, Dutta S, Feng Z, Ganesan S, Goodsell DS, Ghosh S, Green RK, Guranović V, Guzenko D, Hudson BP, Lawson CL, Liang Y, Lowe R, Namkoong H, Peisach E, Persikova I, Randle C, Rose A, Rose Y, Sali A, Segura J, Sekharan M, Shao C, Tao YP, Voigt M, Westbrook JD, Young JY, Zardecki C and Zhuravleva M (2021) RCSB protein data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Research 49, D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GM and Crews CM (2020) Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabri W, Cantelmi P, Corbisiero D, Fantoni T, Ferrazzano L, Martelli G, Mattellone A and Tolomelli A (2021) Therapeutic peptides targeting PPI in clinical development: Overview, mechanism of action and perspectives. Frontiers in Molecular Biosciences 8, 697586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner E, Rustighi A, Zannini A, Cristiani A, Piazza S, Ciani Y, Kalid O, Golan G, Baloglu E, Shacham S, Valsasina B, Cucchi U, Pippione AC, Lolli ML, Giabbai B, Storici P, Carloni P, Rossetti G, Benvenuti F, Bello E, D’Incalci M, Cappuzzello E, Rosato A and Del Sal G (2017) A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nature Communications 8, 15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, Narayanagari S-R, Wheat JC, Todorova TI, Mitchell K, Kenworthy C, Guerlavais V, Annis DA, Bartholdy B, Will B, Anampa JD, Mantzaris I, Aivado M, Singer RH, Coleman RA, Verma A and Steidl U (2018) Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Science Translational Medicine 10, eaa03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalini T (2021) Not only in silico drug discovery: Molecular modeling towards in silico drug delivery formulations. Journal of Controlled Release: Official Journal of the Controlled Release Society 332, 390–417. [DOI] [PubMed] [Google Scholar]
- Cecchini C, Pannilunghi S, Tardy S and Scapozza L (2021) From conception to development: Investigating PROTACs features for improved cell permeability and successful protein degradation. Frontiers in Chemistry 9, 672267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdan AH, Sisquellas M, Pereira G, Barreto Gomes DE, Changeux J-P and Cecchini M (2020) The glycine receptor allosteric ligands library (GRALL). Bioinformatics 36, 3379–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang D, Su N, Bao B, Xie X, Zuo F, Yang L, Wang H, Jiang L, Lin Q and Fang M (2019) Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nature Biotechnology 37, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Cimermancic P, Weinkam P, Rettenmaier TJ, Bichmann L, Keedy DA, Woldeyes RA, Schneidman-Duhovny D, Demerdash ON, Mitchell JC, Wells JA, Fraser JS and Sali A (2016) CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. Journal of Molecular Biology 428, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey RA, Song W, Duncan AL, Ansell TB, Sansom MSP and Stansfeld PJ (2021) Identification and assessment of cardiolipin interactions with inner membrane proteins. Science Advances 7(34), eabh2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey RA, Vickery ON, Sansom MSP and Stansfeld PJ (2019) Insights into membrane protein-lipid interactions from free energy calculations. Journal of Chemical Theory and Computation 15(10), 5727–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew AP, Raina K, Dong H, Qian Y, Wang J, Vigil D, Serebrenik YV, Hamman BD, Morgan A, Ferraro C, Siu K, Neklesa TK, Winkler JD, Coleman KG and Crews CM (2018) Identification and characterization of Von Hippel-Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. Journal of Medicinal Chemistry 61, 583–598. [DOI] [PubMed] [Google Scholar]
- Crivori P, Cruciani G, Carrupt PA and Testa B (2000) Predicting blood-brain barrier permeation from three-dimensional molecular structure. Journal of Medicinal Chemistry 43, 2204–2216. [DOI] [PubMed] [Google Scholar]
- Cyrus K, Wehenkel M, Choi E-Y, Han H-J, Lee H, Swanson H and Kim K-B (2011) Impact of linker length on the activity of PROTACs. Molecular BioSystems 7, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dämgen MA and Biggin PC (2021) State-dependent protein-lipid interactions of a pentameric ligand-gated ion channel in a neuronal membrane. PLoS Computational Biology 17, e1007856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar BR and Mondal J (2020) Capturing protein-ligand recognition pathways in coarse-grained simulation. Journal of Physical Chemistry Letters 11, 5302–5311. [DOI] [PubMed] [Google Scholar]
- Das A, Gur M, Cheng MH, Jo S, Bahar I and Roux B (2014) Exploring the conformational transitions of biomolecular systems using a simple two-state anisotropic network model. PLoS Computational Biology 10, e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schäfer LV, Periole X, Tieleman DP and Marrink SJ (2013) Improved parameters for the martini coarse-grained protein force field. Journal of Chemical Theory and Computation 9, 687–697. [DOI] [PubMed] [Google Scholar]
- Delort B, Renault P, Charlier L, Raussin F, Martinez J and Floquet N (2017) Coarse-grained prediction of peptide binding to G-protein coupled receptors. Journal of Chemical Information and Modeling 57, 562–571. [DOI] [PubMed] [Google Scholar]
- Deplazes E, Louhivuori M, Jayatilaka D, Marrink SJ and Corry B (2012) Structural investigation of MscL gating using experimental data and coarse-grained MD simulations. PLoS Computational Biology 8, e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz NC, Frezza E and Martin J (2021) Using normal mode analysis on protein structural models. How far can we go on our predictions? Proteins 89, 531–543. [DOI] [PubMed] [Google Scholar]
- Dimopoulos MA, Leleu X, Palumbo A, Moreau P, Delforge M, Cavo M, Ludwig H, Morgan GJ, Davies FE, Sonneveld P, Schey SA, Zweegman S, Hansson M, Weisel K, Mateos MV, Facon T and Miguel JFS (2014) Expert panel consensus statement on the optimal use of pomalidomide in relapsed and refractory multiple myeloma. Leukemia 28, 1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Young C and Shaw DE (2011) Anton, a special-purpose molecular simulation machine. In Padua D (ed.) Encyclopedia of Parallel Computing. Boston, MA: Springer US, pp. 60–71. [Google Scholar]
- Drummond ML, Henry A, Li H and Williams CI (2020) Improved accuracy for modeling PROTAC-mediated ternary complex formation and targeted protein degradation via new in silico methodologies. Journal of Chemical Information and Modeling 60, 5234–5254. [DOI] [PubMed] [Google Scholar]
- Drummond ML and Williams CI (2019) In silico modeling of PROTAC-mediated ternary complexes: Validation and application. Journal of Chemical Information and Modeling 59, 1634–1644. [DOI] [PubMed] [Google Scholar]
- Durrant JD and McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biology 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DJ and Deem MW (2005) Parallel tempering: Theory, applications, and new perspectives. Physical Chemistry Chemical Physics: PCCP 7, 3910–3916. [DOI] [PubMed] [Google Scholar]
- El Maghraby GM, Barry BW and Williams AC (2008) Liposomes and skin: From drug delivery to model membranes. European Journal of Pharmaceutical Sciences: Official Journal of the European Federation for Pharmaceutical Sciences 34, 203–222. [DOI] [PubMed] [Google Scholar]
- Empereur-Mot C, Pesce L, Doni G, Bochicchio D, Capelli R, Perego C and Pavan GM (2020) Swarm-CG: Automatic parametrization of bonded terms in MARTINI-based coarse-grained models of simple to complex molecules via fuzzy self-tuning particle swarm optimization. ACS Omega 5, 32823–32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esang M and Gupta M (2021) Aducanumab as a novel treatment for Alzheimer’s disease: A decade of hope, controversies, and the future. Cureus 13, e17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone L, Salomone S and Libra M (2018) Evolution of cancer pharmacological treatments at the turn of the third millennium. Frontiers in Pharmacology 9, 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yang L, Kloczkowski A and Jernigan RL (2009) The energy profiles of atomic conformational transition intermediates of adenylate kinase. Proteins 77, 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro M, Masetti M, Recanatini M, Cavalli A and Bottegoni G (2016) Mapping cholesterol interaction sites on serotonin transporter through coarse-grained molecular dynamics. PLoS One 11, e0166196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré G, Louet M, Saurel O, Delort B, Czaplicki G, M’Kadmi C, Damian M, Renault P, Cantel S, Gavara L, Demange P, Marie J, Fehrentz J-A, Floquet N, Milon A and Banères JL (2019) Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain. Pro1ceedings of the National Academy of Sciences of the United States of America 116, 17525–17530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Ulloa S, Guarnieri D, Riccardi L, Pompa PP and De Vivo M (2021) Association mechanism of peptide-coated metal nanoparticles with model membranes: A coarse-grained study. Journal of Chemical Theory and Computation 17, 4512–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey K, Gray W, Spasov K, Bollini M, Gallardo-Macias R, Jorgensen W and Anderson K (2013) Structure-based evaluation of C5 derivatives in the catechol Diether series targeting HIV-1 reverse transcriptase. Chemical Biology & Drug Design 83, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Nicolas J and Ha-Duong T (2021) Supramolecular organization of polymer prodrug nanoparticles revealed by coarse-grained simulations. Journal of the American Chemical Society 143, 17412–17423. [DOI] [PubMed] [Google Scholar]
- Ghanakota P and Carlson HA (2016) Moving beyond active-site detection: MixMD applied to allosteric systems. The Journal of Physical Chemistry. B 120, 8685–8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M and Zerial M (2013) Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nature Biotechnology 31, 638–646. [DOI] [PubMed] [Google Scholar]
- Gjetting T, Arildsen NS, Christensen CL, Poulsen TT, Roth JA, Handlos VN and Poulsen HS (2010) In vitro and in vivo effects of polyethylene glycol (PEG)-modified lipid in DOTAP/cholesterol-mediated gene transfection. International Journal of Nanomedicine 5, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen J, Albani S, Hanke A, Joseph BP, Bergh C, Kuzikov M, Costanzi E, Manelfi C, Storici P, Gribbon P, Beccari AR, Talarico C, Spyrakis F, Lindahl E, Zaliani A, Carloni P, Wade RC, Musiani F, Kokh DB and Rossetti G (2021) A blueprint for high affinity SARS-CoV-2 Mpro inhibitors from activity-based compound library screening guided by analysis of protein dynamics. ACS Pharmacology & Translational Science 4, 1079–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JA, Essex JW and Khalid S (2017) PyCGTOOL: Automated generation of coarse-grained molecular dynamics models from atomistic trajectories. Journal of Chemical Information and Modeling 57, 650–656. [DOI] [PubMed] [Google Scholar]
- Gray GM, Ma N, Wagner CE and van der Vaart A (2017) Molecular dynamics simulations and molecular flooding studies of the retinoid X-receptor ligand binding domain. Journal of Molecular Modeling 23, 98. [DOI] [PubMed] [Google Scholar]
- Grünewald F, Alessandri R, Kroon PC, Monticelli L, Souza PCT and Marrink SJ (2022) Polyply; A python suite for facilitating simulations of macromolecules and nanomaterials. Nature Communications 13, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald F, Rossi G, de Vries AH, Marrink SJ and Monticelli L (2018) Transferable MARTINI model of poly(ethylene oxide). The Journal of Physical Chemistry. B 122, 7436–7449. [DOI] [PubMed] [Google Scholar]
- Grünewald F, Souza PCT, Abdizadeh H, Barnoud J, de Vries AH and Marrink SJ (2020) Titratable martini model for constant pH simulations. The Journal of Chemical Physics 153, 024118. [DOI] [PubMed] [Google Scholar]
- Gupta KM, Das S and Chow PS (2021) Molecular dynamics simulations to elucidate translocation and permeation of active from lipid nanoparticle to skin: Complemented by experiments. Nanoscale 13, 12916–12928. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Consortium OSDD and Raghava GPS (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS One 8, e73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden JA, Perilla JR, Schlicksup CJ, Venkatakrishnan B, Zlotnick A and Schulten K (2018) All-atom molecular dynamics of the HBV capsid reveals insights into biological function and cryo-EM resolution limits. eLife 7, e32478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes GG (2002) Multiple conformational changes in enzyme catalysis. Biochemistry 41, 8221–8228. [DOI] [PubMed] [Google Scholar]
- Harmalkar A and Gray JJ (2021) Advances to tackle backbone flexibility in protein docking. Current Opinion in Structural Biology 67, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemzadeh H, Javadi H and Darvishi MH (2020) Study of structural stability and formation mechanisms in DSPC and DPSM liposomes: A coarse-grained molecular dynamics simulation. Scientific Reports 10, 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Matsuzaki Y, Yanagisawa K, Ohue M and Akiyama Y (2018) MEGADOCK-web: An integrated database of high-throughput structure-based protein-protein interaction predictions. BMC Bioinformatics 19, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Khan S, Huo Z, Lv D, Zhang X, Liu X, Yuan Y, Hromas R, Xu M, Zheng G and Zhou D (2020) Proteolysis targeting chimeras (PROTACs) are emerging therapeutics for hematologic malignancies. Journal of Hematology & Oncology 13, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K and Kern D (2007) Dynamic personalities of proteins. Nature 450, 964–972. [DOI] [PubMed] [Google Scholar]
- Herbert C, Schieborr U, Saxena K, Juraszek J, De Smet F, Alcouffe C, Bianciotto M, Saladino G, Sibrac D, Kudlinzki D, Sreeramulu S, Brown A, Rigon P, Herault J-P, Lassalle G, Blundell TL, Rousseau F, Gils A, Schymkowitz J, Tompa P, Herbert JM, Carmeliet P, Gervasio FL, Schwalbe H and Bono F (2013) Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling. Cancer Cell 23, 489–501. [DOI] [PubMed] [Google Scholar]
- Hilpert C, Beranger L, Souza PCT, Vainikka PA, Nieto V, Marrink SJ, Monticelli L and Launay G (2022) Facilitating CG simulations with MAD: The MArtini Database Server. BiorXiv. 10.1101/2022.08.03.502585 [DOI] [PubMed]
- Hopkins AL and Groom CR (2002) The druggable genome. Nature Reviews. Drug Discovery 1, 727–730. [DOI] [PubMed] [Google Scholar]
- Hsu P-C, Bruininks BMH, Jefferies D, Souza PCT, Lee J, Patel DS, Marrink SJ, Qi Y, Khalid S and Im W (2017) CHARMM-GUI martini maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. Journal of Computational Chemistry 38, 2354–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A and Schulten K (1996) VMD: Visual molecular dynamics. Journal of Molecular Graphics 14, 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- Ingólfsson HI, Bhatia H, Zeppelin T, Bennett WFD, Carpenter KA, Hsu PC, Dharuman G, Bremer PT, Schiøtt B, Lightstone FC and Carpenter TS (2020) Capturing biologically complex tissue-specific membranes at different levels of compositional complexity. The Journal of Physical Chemistry. B 124, 7819–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP and Marrink SJ (2014) Lipid organization of the plasma membrane. Journal of the American Chemical Society 136, 14554–14559. [DOI] [PubMed] [Google Scholar]
- Jackson CB, Farzan M, Chen B and Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews. Molecular Cell Biology 23, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James ND, Coker RJ, Tomlinson D, Harris JR, Gompels M, Pinching AJ and Stewart JS (1994) Liposomal doxorubicin (Doxil): An effective new treatment for Kaposi’s sarcoma in AIDS. Clinical Oncology 6, 294–296. [DOI] [PubMed] [Google Scholar]
- Jana P, Shyam M, Singh S, Jayaprakash V and Dev A (2021) Biodegradable polymers in drug delivery and oral vaccination. European Polymer Journal 142, 110155. [Google Scholar]
- Jiang Z and Zhang H (2019) Molecular mechanism of S1P binding and activation of the S1P receptor. Journal of Chemical Information and Modeling 59, 4402–4412. [DOI] [PubMed] [Google Scholar]
- Jiménez-García B, Roel-Touris J, Romero-Durana M, Vidal M, Jiménez-González D and Fernández-Recio J (2018) LightDock: A new multi-scale approach to protein-protein docking. Bioinformatics 34, 49–55. [DOI] [PubMed] [Google Scholar]
- Jorgensen W (2009) Efficient drug lead discovery and optimization. Accounts of Chemical Research 42, 724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kobayashi C, Kasahara K, Tan C, Kuroda A, Minami K, Ishiduki S, Nishiki T, Inoue H, Ishikawa Y, Feig M and Sugita Y (2021) New parallel computing algorithm of molecular dynamics for extremely huge scale biological systems. Journal of Computational Chemistry 42, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada R, Terayama K, Tokuhisa A, Matsumoto S and Okuno Y (2022) Enhanced conformational sampling with an adaptive coarse-grained elastic network model using short-time all-atom molecular dynamics. Journal of Chemical Theory and Computation 18, 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekal KH and Bereau T (2019) Resolution limit of data-driven coarse-grained models spanning chemical space. The Journal of Chemical Physics 151, 164106. [DOI] [PubMed] [Google Scholar]
- Karaca E and Bonvin AMJJ (2011) A multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure 19, 555–565. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Liu H, Miyazaki Y, Seo S, Dixit M, DeVane R, MacDermaid C, Fiorin G, Klein ML and Shinoda W (2022) SPICA force field for proteins and peptides. Journal of Chemical Theory and Computation 18, 3204–3217. [DOI] [PubMed] [Google Scholar]
- Kim MK, Jernigan RL and Chirikjian GS (2002) Efficient generation of feasible pathways for protein conformational transitions. Biophysical Journal 83, 1620–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjølbye LR, De Maria L, Wassenaar TA, Abdizadeh H, Marrink SJ, Ferkingoff-Borg J and Schiøtt B (2020) A generic protocol for constructing molecular models of nanodiscs in silico. Journal of Chemical Information and Modeling 61, 2869–2883. [DOI] [PubMed] [Google Scholar]
- Klein VG, Townsend CE, Testa A, Zengerle M, Maniaci C, Hughes SJ, Chan K-H, Ciulli A and Lokey RS (2020) Understanding and improving the membrane permeability of VH032-based PROTACs. ACS Medicinal Chemistry Letters 11, 1732–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokh DB, Czodrowski P, Rippmann F and Wade RC (2016) Perturbation approaches for exploring protein binding site flexibility to predict transient binding pockets. Journal of Chemical Theory and Computation 12, 4100–4113. [DOI] [PubMed] [Google Scholar]
- Kokh DB, Richter S, Henrich S, Czodrowski P, Rippmann F and Wade RC (2013) TRAPP: A tool for analysis of transient binding pockets in proteins. Journal of Chemical Information and Modeling 53, 1235–1252. [DOI] [PubMed] [Google Scholar]
- Kulkarni JA, Darjuan MM, Mercer JE, Chen S, van der Meel R, Thewalt JL, Tam YYC and Cullis PR (2018) On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 12, 4787–4795. [DOI] [PubMed] [Google Scholar]
- Kuzmanic A, Bowman GR, Juarez-Jimenez J, Michel J and Gervasio FL (2020) Investigating cryptic binding sites by molecular dynamics simulations. Accounts of Chemical Research 53, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B, Chavent M, Cragnolini T, Dahl ACE, Pasquali S, Derreumaux P, Sansom MSP and Baaden M (2015) Epock: Rapid analysis of protein pocket dynamics. Bioinformatics 31, 1478–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guilloux V, Schmidtke P and Tuffery P (2009) Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Cheng Q, Yu X, Liu S, Johnson LT and Siegwart DJ (2021) A systematic study of unsaturation in lipid nanoparticles leads to improved mRNA transfection in vivo. Angewandte Chemie 60, 5848–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Schlesinger PH, Wickline SA, Lanza GM and Baker NA (2012) Simulation of fusion-mediated nanoemulsion interactions with model lipid bilayers. Soft Matter 8, 3024–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelimousin M, Limongelli V and Sansom MSP (2016) Conformational changes in the epidermal growth factor receptor: Role of the transmembrane domain investigated by coarse-grained MetaDynamics free energy calculations. Journal of the American Chemical Society 138, 10611–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaalem M, Hadrioui N, Derouiche A and Ridouane H (2020) Structure and dynamics of liposomes designed for drug delivery: Coarse-grained molecular dynamics simulations to reveal the role of lipopolymer incorporation. RSC Advances 10, 3745–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKK, Hafez IM, Baoukina S, Belliveau NM, Zhigaltsev IV, Afshinmanesh E, Tieleman DP, Hansen CL, Hope MJ and Cullis PR (2012) Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured Core. The Journal of Physical Chemistry. C, Nanomaterials and Interfaces 116, 18440–18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W and Szoka FC (2007) Lipid-based nanoparticles for nucleic acid delivery. Pharmaceutical Research 24, 438–449. [DOI] [PubMed] [Google Scholar]
- Lin X, Li X and Lin X (2020) A review on applications of computational methods in drug screening and design. Molecules 25, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW and Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews 46, 3–26. [DOI] [PubMed] [Google Scholar]
- Liu S, Cheng Q, Wei T, Yu X, Johnson LT, Farbiak L and Siegwart DJ (2021) Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nature Materials 20, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CA, Rzepiela AJ, de Vries AH, Dijkhuizen L, Hünenberger PH and Marrink SJ (2009) Martini coarse-grained force field: Extension to carbohydrates. Journal of Chemical Theory and Computation 5, 3195–3210. [DOI] [PubMed] [Google Scholar]
- Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R and Shi J (2020) Recent advances in the development of protein-protein interactions modulators: Mechanisms and clinical trials. Signal Transduction and Targeted Therapy 5, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M (2012) Influenza virus entry. Advances in Experimental Medicine and Biology 726, 201–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabonga L and Kappo AP (2019) Protein-protein interaction modulators: Advances, successes and remaining challenges. Biophysical Reviews 11, 559–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado M, Barrera E, Klein F, Soñora M, Silva S and Pantano S (2019) The SIRAH2.0 force field: Altius, Fortius, Citius. Journal of Chemical Theory and Computation 15, 2719–2733. [DOI] [PubMed] [Google Scholar]
- Machado N, Bruininks BMH, Singh P, Santos L d, Pizzol CD, Dieamant GC, Kruger O, Martin AA, Marrink SJ, Souza PCT and Favero PP (2022) Complex nanoemulsion for vitamin delivery: Droplet organization and interaction with skin membranes. Nanoscale 14, 506–514. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Jo S, Lakkaraju SK, Lind C and Yu W (2020) Identification and characterization of fragment binding sites for allosteric ligand design using the site identification by ligand competitive saturation hotspots approach (SILCS-hotspots). Biochimica et Biophysica Acta, General Subjects 1864, 129519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi L, Carloni P and Rossetti G (2020) Modeling the allosteric modulation on a G-protein coupled receptor: The case of M2 muscarinic acetylcholine receptor in complex with LY211960. Scientific Reports 10, 3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood MI, Poma AB and Okazaki K-I (2021) Optimizing Gō-MARTINI coarse-grained model for F-BAR protein on lipid membrane. Frontiers in Molecular Biosciences 8, 619381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmsten M (2006) Soft drug delivery systems. Soft Matter 2, 760–769. [DOI] [PubMed] [Google Scholar]
- Margreiter MA, Witzenberger M, Wasser Y, Davydova E, Janowski R, Metz J, Habib P, Sahnoun SEM, Sobisch C, Poma B, Palomino-Hernandez O, Wagner M, Carell T, Jon Shah N, Schulz JB, Niessing D, Voigt A and Rossetti G (2022) Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death. Computational and Structural Biotechnology Journal 20, 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Corradi V, Souza PCT, Ingólfsson HI, Tieleman DP and Sansom MSP (2019) Computational modeling of realistic cell membranes. Chemical Reviews 119, 6184–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Monticelli L, Melo MN, Alessandri R, Tieleman DP and Souza PCT (2022) Two decades of martini: Better beads, broader scope. WIREs Computational Molecular Science. 12, e1620. [Google Scholar]
- Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP and De Vries AH (2007) The MARTINI force field: Coarse grained model for biomolecular simulations. The Journal of Physical Chemistry. B 111, 7812–7824. [DOI] [PubMed] [Google Scholar]
- Martino E, Chiarugi S, Margheriti F and Garau G (2021) Mapping, structure and modulation of PPI. Frontiers in Chemistry 9, 718405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes F, Massari S, Bochicchio A, Schorpp K, Schilling J, Weber S, Offermann N, Desantis J, Wanker E, Carloni P, Hadian K, Tabarrini O, Rossetti G and Krauss S (2018) Reducing mutant huntingtin protein expression in living cells by a newly identified RNA CAG binder. ACS Chemical Neuroscience 9, 1399–1408. [DOI] [PubMed] [Google Scholar]
- May A and Zacharias M (2008) Energy minimization in low-frequency normal modes to efficiently allow for global flexibility during systematic protein-protein docking. Proteins 70, 794–809. [DOI] [PubMed] [Google Scholar]
- Miao L, Lin J, Huang Y, Li L, Delcassian D, Ge Y, Shi Y and Anderson DG (2020) Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nature Communications 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowsky J, Schäfer LV, Holm C and Smiatek J (2017) A refined polarizable water model for the coarse-grained MARTINI force field with long-range electrostatic interactions. The Journal of Chemical Physics 146, 054501. [DOI] [PubMed] [Google Scholar]
- Michalowsky J, Zeman J, Holm C and Smiatek J (2018) A polarizable MARTINI model for monovalent ions in aqueous solution. The Journal of Chemical Physics 149, 163319. [DOI] [PubMed] [Google Scholar]
- Misiura M, Shroff R, Thyer R and Kolomeisky AB (2022) DLPacker: Deep learning for prediction of amino acid side chain conformations in proteins. Proteins 90, 1278–1290. [DOI] [PubMed] [Google Scholar]
- Miyashita O, Onuchic JN and Wolynes PG (2003) Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proceedings of the National Academy of Sciences of the United States of America 100, 12570–12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moal IH and Bates PA (2010) SwarmDock and the use of normal modes in protein-protein docking. International Journal of Molecular Sciences 11, 3623–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohs RC and Greig NH (2017) Drug discovery and development: Role of basic biological research. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 3, 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Acosta JJ and Cecchini M (2016) Computational approaches to the chemical equilibrium constant in protein-ligand binding. Molecular Informatics 35, 555–567. [DOI] [PubMed] [Google Scholar]
- Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP and Marrink SJ (2008) The MARTINI coarse-grained force field: Extension to proteins. Journal of Chemical Theory and Computation 4, 819–834. [DOI] [PubMed] [Google Scholar]
- Negami T, Shimizu K and Terada T (2014) Coarse-grained molecular dynamics simulations of protein-ligand binding. Journal of Computational Chemistry 35, 1835–1845. [DOI] [PubMed] [Google Scholar]
- Negami T, Shimizu K and Terada T (2020) Coarse-grained molecular dynamics simulation of protein conformational change coupled to ligand binding. Chemical Physics Letters 742, 137144. [Google Scholar]
- Nevola L and Giralt E (2015) Modulating protein-protein interactions: The potential of peptides. Chemical Communications 51, 3302–3315. [DOI] [PubMed] [Google Scholar]
- Nishiga M, Wang DW, Han Y, Lewis DB and Wu JC (2020) COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nature Reviews. Cardiology 17, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreng S, Li T and Payandeh J (2021) Structural pharmacology of voltage-gated sodium channels. Journal of Molecular Biology 433, 166967. [DOI] [PubMed] [Google Scholar]
- Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, Zhang T, Mancias JD, Gray NS, Bradner JE and Fischer ES (2018) Plasticity in binding confers selectivity in ligand-induced protein degradation. Nature Chemical Biology 14, 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K-I, Koga N, Takada S, Onuchic JN and Wolynes PG (2006) Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proceedings of the National Academy of Sciences of the United States of America 103, 11844–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinikovas V, Saladino G, Cossins BP and Gervasio FL (2016) Understanding cryptic pocket formation in protein targets by enhanced sampling simulations. Journal of the American Chemical Society 138, 14257–14263. [DOI] [PubMed] [Google Scholar]
- Palmerston Mendes L, Pan J and Torchilin VP (2017) Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules 22(9), 1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuzzo M, Esposito S, Wu L-P, Key J, Aryal S, Celia C, di Marzio L, Moghimi SM and Decuzzi P (2020) Overcoming nanoparticle-mediated complement activation by surface PEG pairing. Nano Letters 20, 4312–4321. [DOI] [PubMed] [Google Scholar]
- Parchekani J, Allahverdi A, Taghdir M and Naderi-Manesh H (2022) Design and simulation of the liposomal model by using a coarse-grained molecular dynamics approach towards drug delivery goals. Scientific Reports 12, 2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Szwarc B, Mondragão MA, Lima PA and Pereira F (2018) A ligand-based approach to the discovery of lead-like potassium channel KV 1.3 inhibitors. ChemistrySelect 3, 1352–1364. [Google Scholar]
- Perez JJ, Perez RA and Perez A (2021) Computational modeling as a tool to investigate PPI: From drug design to tissue engineering. Frontiers in Molecular Biosciences 8, 681617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periole X, Cavalli M, Marrink SJ and Ceruso MA (2009) Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. Journal of Chemical Theory and Computation 5, 2531–2543. [DOI] [PubMed] [Google Scholar]
- Petrylak DP, Gao X, Vogelzang NJ, Garfield MH, Taylor I, Dougan Moore M, Peck RA and Burris HA (2020) First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). Journal of Clinical Orthodontics: JCO 38, 3500–3500. [Google Scholar]
- Pezeshkian W, König M, Wassenaar TA and Marrink SJ (2020) Backmapping triangulated surfaces to coarse-grained membrane models. Nature Communications 11, 2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU and Gruber WC (2020) Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. The New England Journal of Medicine 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poma AB, Cieplak M and Theodorakis PE (2017) Combining the MARTINI and structure-based coarse-grained approaches for the molecular dynamics studies of conformational transitions in proteins. Journal of Chemical Theory and Computation 13, 1366–1374. [DOI] [PubMed] [Google Scholar]
- Poma AB, Li MS and Theodorakis PE (2018) Generalization of the elastic network model for the study of large conformational changes in biomolecules. Physical Chemistry Chemical Physics: PCCP 20, 17020–17028. [DOI] [PubMed] [Google Scholar]
- Potter TD, Barrett EL and Miller MA (2021) Automated coarse-grained mapping algorithm for the martini force field and benchmarks for membrane–water partitioning. Journal of Chemical Theory and Computation 17, 5777–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Ingólfsson HI, Cheng X, Lee J, Marrink SJ and Im W (2015) CHARMM-GUI martini maker for coarse-grained simulations with the martini force field. Journal of Chemical Theory and Computation 11, 4486–4494. [DOI] [PubMed] [Google Scholar]
- Rakers C, Bermudez M, Keller BG, Mortier J and Wolber G (2015) Computational close up on protein-protein interactions: How to unravel the invisible using molecular dynamics simulations? Wiley Interdisciplinary Reviews. Computational Molecular Science 5, 345–359. [Google Scholar]
- Reimhult E and Virk MM (2021) Hybrid lipopolymer vesicle drug delivery and release systems. Journal of Biomedical Research 35, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roel-Touris J and Bonvin AMJJ (2020) Coarse-grained (hybrid) integrative modeling of biomolecular interactions. Computational and Structural Biotechnology Journal 18, 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roel-Touris J, Bonvin AMJJ and Jiménez-García B (2020a) LightDock goes information-driven. Bioinformatics 36, 950–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roel-Touris J, Don CG, Honorato RV, Rodrigues JPGLM and Bonvin AMJJ (2019) Less is more: Coarse-grained integrative modeling of large biomolecular assemblies with HADDOCK. Journal of Chemical Theory and Computation 15, 6358–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roel-Touris J, Jiménez-García B and Bonvin AMJJ (2020b) Integrative modeling of membrane-associated protein assemblies. Nature Communications 11, 6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell M and Fernández-Recio J (2020) Docking-based identification of small-molecule binding sites at protein-protein interfaces. Computational and Structural Biotechnology Journal 18, 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo A, Lee P and Matysiak S (2022) Transferable and polarizable coarse grained model for proteins─ProMPT. Journal of Chemical Theory and Computation 18, 5046–5055. [DOI] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM and Deshaies RJ (2001) Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences of the United States of America 98, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salassi S, Caselli L, Cardellini J, Lavagna E, Montis C, Berti D and Rossi G (2021) A martini coarse grained model of citrate-capped gold nanoparticles interacting with lipid bilayers. Journal of Chemical Theory and Computation 17, 6597–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salassi S, Simonelli F, Bartocci A and Rossi G (2018) A martini coarse-grained model of the calcein fluorescent dye. Journal of Physics D: Applied Physics 51, 384002. [Google Scholar]
- Schlander M, Hernandez-Villafuerte K, Cheng C-Y, Mestre-Ferrandiz J and Baumann M (2021) How much does it cost to research and develop a new drug? A Systematic Review and Assessment. PharmacoEconomics 39, 1243–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlich M, Palomba R, Costabile G, Mizrahy S, Pannuzzo M, Peer D and Decuzzi P (2021) Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles. Bioengineering and Translational Medicine 6, e10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T and Portillo-Ledesma S (2021) Biomolecular modeling thrives in the age of technology. Nature Computational Science 1, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Schaeffer WJ, Wemheuer WM and Wrede A (2020) Chapter 21 - prion diseases: Conformational changes of a protein create an unconventional infectious agent. In Ennaji MM (ed.), Emerging and Reemerging Viral Pathogens, Academic Press, pp. 479–488. [Google Scholar]
- Sengupta I and Udgaonkar J (2019) Monitoring site-specific conformational changes in real-time reveals a misfolding mechanism of the prion protein. eLife 8, e44698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK and Hua S (2015) Advances and challenges of liposome assisted drug delivery. Frontiers in Pharmacology 6, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Mysore VP, Leffler AE, Kim ET, Sagawa S and Shaw DE (2022) How does a small molecule bind at a cryptic binding site? PLoS Computational Biology 18, e1009817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp L and Brannigan G (2021) Spontaneous lipid binding to the nicotinic acetylcholine receptor in a native membrane. The Journal of Chemical Physics 154, 185102. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Adams PJ, Azaria A, Bank JA, Batson B, Bell A, Bergdorf M, Bhatt J, Butts JA, Correia T, Dirks RM, Dror RO, Eastwood MP, Edwards B, Even A, Feldmann P, Fenn M, Fenton CH, Forte A, Gagliardo J, Gill G, Gorlatova M, Greskamp B, Grossman JP, Gullingsrud J, Harper A, Hasenplaugh W, Heily M, Heshmat BC, Hunt J, Ierardi DJ, Iserovich L, Jackson BL, Johnson NP, Kirk MM, Klepeis JL, Kuskin JS, Mackenzie KM, Mader RJ, McGowen R, McLaughlin A, Moraes MA, Nasr MH, Nociolo LJ, O’Donnell L, Parker A, Peticolas JL, Pocina G, Predescu C, Quan T, Salmon JK, Schwink C, Shim KS, Siddique N, Spengler J, Szalay T, Tabladillo R, Tartler R, Taube AG, Theobald M, Towles B, Vick W, Wang SC, Wazlowski M, Weingarten MJ, Williams JM and Yuh KA (2021) Anton 3: Twenty microseconds of molecular dynamics simulation before lunch. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. New York, NY: Association for Computing Machinery, pp. 1–11. [Google Scholar]
- Siebenmorgen T and Zacharias M (2020) Computational prediction of protein–protein binding affinities. Computational Molecular Science 10, e1448. [DOI] [PubMed] [Google Scholar]
- Slastnikova TA, Ulasov AV, Rosenkranz AA and Sobolev AS (2018) Targeted intracellular delivery of antibodies: The state of the art. Frontiers in Pharmacology 9, 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P and Caflisch A (2018) Protein structure-based drug design: From docking to molecular dynamics. Current Opinion in Structural Biology 48, 93–102. [DOI] [PubMed] [Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J and Lowe EW (2014) Computational methods in drug discovery. Pharmacological Reviews 66, 334–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PCT, Alessandri R, Barnoud J, Thallmair S, Faustino I, Grünewald F, Patmanidis I, Abdizadeh H, Bruininks BMH, Wassenaar TA, Kroon PC, Melcr J, Nieto V, Corradi V, Khan HM, Domański J, Javanainen M, Martinez-Seara H, Reuter N, Best RB, Vattulainen I, Monticelli L, Periole X, Tieleman DP, de Vries AH and Marrink SJ (2021a) Martini 3: A general purpose force field for coarse-grained molecular dynamics. Nature Methods 18, 382. [DOI] [PubMed] [Google Scholar]
- Souza PCT, Limongelli V, Wu S, Marrink SJ and Monticelli L (2021b) Perspectives on high-throughput ligand/protein docking with martini MD simulations. Frontiers in Molecular Biosciences 8, 657222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PCT, Thallmair S, Conflitti P, Ramírez-Palacios C, Alessandri R, Raniolo S, Limongelli V and Marrink SJ (2020) Protein-ligand binding with the coarse-grained martini model. Nature Communications 11, 3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PCT, Thallmair S, Marrink SJ and Mera-Adasme R (2019) An allosteric pathway in copper, zinc superoxide dismutase unravels the molecular mechanism of the G93A amyotrophic lateral sclerosis-linked mutation. Journal of Physical Chemistry Letters 10, 7740–7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiyama S, Fujie T, Yoshikawa Y, Yanagisawa K, Ohue M and Akiyama Y (2021) Large-scale membrane permeability prediction of cyclic peptides crossing a lipid bilayer based on enhanced sampling molecular dynamics simulations. Journal of Chemical Information and Modeling 61, 3681–3695. [DOI] [PubMed] [Google Scholar]
- Sutcliffe KJ, Corey RA, Charlton SJ, Sessions RB, Henderson G and Kelly E (2021) Fentanyl binds to the μ-opioid receptor via the lipid membrane and transmembrane helices. BiorXiv. 10.1101/2021.02.04.429703 [DOI]
- Troup RI, Fallan C and Baud MGJ (2020) Current strategies for the design of PROTAC linkers: A critical review. Exploration of Targeted Anti-tumor Therapy 1, 273–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo JJ, Ingólfsson HI, Akhshi P, Tieleman DP and Marrink SJ (2015) Martini coarse-grained force field: Extension to DNA. Journal of Chemical Theory and Computation 11, 3932–3945. [DOI] [PubMed] [Google Scholar]
- Uusitalo JJ, Ingólfsson HI, Marrink SJ and Faustino I (2017) Martini coarse-grained force field: Extension to RNA. Biophysical Journal 113, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S, Beglov D, Wakefield AE, Egbert M and Whitty A (2018) Cryptic binding sites on proteins: Definition, detection, and druggability. Current Opinion in Chemical Biology 44, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakser IA (2020) Challenges in protein docking. Current Opinion in Structural Biology 64, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalst E, Koneri J and Wylie BJ (2021) In silico identification of cholesterol binding motifs in the chemokine receptor CCR3. Membranes 11, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D and Johnson JE (2012) Virus maturation. Annual Review of Biophysics 41, 473–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery ON and Stansfeld PJ (2021) CG2AT2: An enhanced fragment-based approach for serial multi-scale molecular dynamics simulations. Journal of Chemical Theory and Computation 17, 6472–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang X, Feng F, Liu W and Sun H (2020) Degradation of proteins by PROTACs and other strategies. Acta pharmaceutica Sinica. B 10, 207–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang D, Cheng X, Yang L, Wang Z, Dai A, Cai X, Zhang C, Yuliantie E, Liu Q, Jiang H, Liu H, Wang M-W and Yang H (2021a) Allosteric modulators enhancing GLP-1 binding to GLP-1R via a transmembrane site. ACS Chemical Biology 16, 2444–2452. [DOI] [PubMed] [Google Scholar]
- Wang W, Ye Z, Gao H and Ouyang D (2021b) Computational pharmaceutics - A new paradigm of drug delivery. Journal of Controlled Release: Official Journal of the Controlled Release Society 338, 119–136. [DOI] [PubMed] [Google Scholar]
- Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP and Marrink SJ (2015) Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. Journal of Chemical Theory and Computation 11, 2144–2155. [DOI] [PubMed] [Google Scholar]
- Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ and Tieleman DP (2014) Going backward: A flexible geometric approach to reverse transformation from coarse grained to atomistic models. Journal of Chemical Theory and Computation 10, 676–690. [DOI] [PubMed] [Google Scholar]
- Weerakoon D, Carbajo RJ, De Maria L, Tyrchan C and Zhao H (2022) Impact of PROTAC linker plasticity on the solution conformations and dissociation of the ternary complex. Journal of Chemical Information and Modeling 62, 340–349. [DOI] [PubMed] [Google Scholar]
- Wells JA and McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Weng G, Li D, Kang Y and Hou T (2021) Integrative modeling of PROTAC-mediated ternary complexes. Journal of Medicinal Chemistry 64, 16271–16281. [DOI] [PubMed] [Google Scholar]
- Wenthur CJ, Gentry PR, Mathews TP and Lindsley CW (2014) Drugs for allosteric sites on receptors. Annual Review of Pharmacology and Toxicology 54, 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienen-Schmidt B, Oebbeke M, Ngo K, Heine A and Klebe G (2021) Two methods, one goal: Structural differences between cocrystallization and crystal soaking to discover ligand binding poses. ChemMedChem 16, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wołek K, Gómez-Sicilia À and Cieplak M (2015) Determination of contact maps in proteins: A combination of structural and chemical approaches. The Journal of Chemical Physics 143, p243105. [DOI] [PubMed] [Google Scholar]
- Xiao J, Bondarenko V and Wang Y (2021) Regulation and drug modulation of a voltage-gated sodium channel: Pivotal role of the S4–S5 linker in activation and slow inactivation. Proceedings of the National Academy of Sciences of the United States of America 118, e2102285118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Guo Z, Tian H and Chen X (2021) Enhancers in polymeric nonviral gene delivery systems. Viewpoints on Digestive Diseases 2, 20200072. [Google Scholar]
- Yesylevskyy SO, Schäfer LV, Sengupta D and Marrink SJ (2010) Polarizable water model for the coarse-grained MARTINI force field. PLoS Computational Biology 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Jo S, Lakkaraju SK, Weber DJ and MacKerell AD (2019) Exploring protein-protein interactions using the site-identification by ligand competitive saturation methodology. Proteins 87, 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W and MacKerell AD (2017) Computer-aided drug design methods. Methods in Molecular Biology 1520, 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Xu L, Tian F, Su Q, Zheng N, Yang Y, Wang J, Wang A, Zhu C, Guo S, Zhang X, Gan Y, Shi X and Gao H (2018) Rapid transport of deformation-tuned nanoparticles across biological hydrogels and cellular barriers. Nature Communications 9, 2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman D, Prilusky J and London N (2020) PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. Journal of Chemical Information and Modeling 60, 4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żak MM and Zangi L (2021) Lipid nanoparticles for organ-specific mRNA therapeutic delivery. Pharmaceutics 13, 1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gao H and Bao G (2015) Physical principles of nanoparticle cellular endocytosis. ACS Nano 9, 8655–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tang B, Wang Q and Lai L (2014) Discovery of binding proteins for a protein target using protein-protein docking-based virtual screening. Proteins 82, 2472–2482. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Capelli R, Carloni P, Lüscher B, Li J and Rossetti G (2021) Enhanced sampling approach to the induced-fit docking problem in protein-ligand binding: The case of mono-ADP-Ribosylation hydrolase inhibitors. Journal of Chemical Theory and Computation 17, 7899–7911. [DOI] [PubMed] [Google Scholar]
- Zheng W (2021) Predicting cryptic ligand binding sites based on normal modes guided conformational sampling. Proteins 89, 416–426. [DOI] [PubMed] [Google Scholar]
- Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, Yang W, Tian C, Miao Z, Wang T and Yang S (2021) Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduction and Targeted Therapy 6, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MI, Porter JR, Ward MD, Singh S, Vithani N, Meller A, Mallimadugula UL, Kuhn CE, Borowsky JH, Wiewiora RP, Hurley MFD, Harbison AM, Fogarty CA, Coffland JE, Fadda E, Voelz VA, Chodera JD and Bowman GR (2021) SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nature Chemistry 13, 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]