Abstract
Simian virus 40 (SV40) large tumor (T) antigen is the major regulatory protein that directs the course of viral infection, primarily by interacting with host cell proteins and modulating their functions. Initiation of viral DNA replication requires specific interactions of T antigen bound to the viral origin of DNA replication with cellular replication proteins. Transcription factors are thought to stimulate initiation of viral DNA replication, but the mechanism of stimulation is poorly understood. Since the transcription factor TATA-binding protein (TBP) binds to sequences within the origin of replication and interacts specifically with T antigen, we examined whether TBP complexes stimulate SV40 DNA replication in vitro. On the contrary, we found that depletion of TBP complexes from human cell extracts increased their ability to support viral DNA replication, and readdition of TBP complexes to the depleted extracts diminished their activity. We have mapped the sites of interaction between the proteins to residues 181 to 205 of T antigen and 184 to 220 of TBP. Titration of fusion proteins containing either of these peptides into undepleted cell extracts stimulated their replication activity, suggesting that they prevented the T antigen-TBP interaction that interfered with replication activity. TBP complexes also interfered with origin DNA unwinding by purified T antigen, and addition of either the T antigen or the TBP fusion peptide relieved the inhibition. These results suggest that TBP complexes associate with a T-antigen surface that is also required for origin DNA unwinding and viral DNA replication. We speculate that competition among cellular proteins for T antigen may play a role in regulating the course of viral infection.
Simian virus 40 (SV40) large tumor (T) antigen is a highly complex protein which performs a wide variety of functions during the viral life cycle (reviewed in references 28 and 29). Early after infection, T antigen begins to accumulate and alters cell growth regulation, in part by direct binding to the tumor suppressor proteins Rb, p107, p130, and p53 (reviewed in references 13 and 63) and to cellular chaperone proteins (reviewed in reference 7), thereby modulating their activities. T antigen also modulates patterns of viral and cellular gene expression. At the transition from the early to the late phase of infection, binding of accumulated T antigen to specific DNA sequences in the early promoter region leads to repression of early transcription (66; reviewed in reference 29). T antigen also serves as a promiscuous transcriptional activator of both cellular and viral promoters, including the SV40 late promoter (5, 43). Transactivation of the viral late promoter appears to be mediated in part by T-antigen-induced viral DNA replication; the increased number of viral genomes titrates cellular repressors of late transcription, resulting in increased late gene expression (reference 78 and references therein). T antigen also interacts directly with components of the transcription machinery to stimulate transcription. T antigen binds specifically to the TATA box-binding protein TBP (2, 14, 32, 41, 53, 83; reviewed in 38), several TBP-associated factors (TAFs) (14, 17, 55, 83), TEF-1 (2, 22, 32, 41), TFIIB (41), human TFIIB-related factor (17), TFIIA (16), Sp1 (41), and AP2 (59). These interactions appear to be essential for transcriptional activation of polymerase I, polymerase II, and polymerase III promoters by T antigen.
Another major function of T antigen is to direct the process of viral DNA replication (reviewed in reference 8). In the presence of ATP, binding of T antigen to a specific palindromic binding site in the viral origin of DNA replication leads to assembly of a T-antigen double hexamer on the DNA and causes a local distortion in the origin DNA sequences flanking its binding site (3, 18, 23, 54, 64). Driven by ATP hydrolysis, T antigen then functions as a DNA helicase, unwinding the two parental strands bidirectionally by reeling the DNA through the double-hexamer complex (19, 56, 57, 60, 69, 77). Specific interactions of T antigen with replication protein A (RPA) (6, 25, 47, 58, 76), DNA polymerase α-primase (12, 24, 25, 26, 61, 62), and topoisomerase I (68) are essential for initiation and elongation.
The essential core origin of SV40 DNA replication consists of the palindromic T-antigen binding site flanked by an AT-rich sequence on one side, which contains the TBP binding site of the early SV40 promoter, and an easily denatured sequence on the other side (reviewed in references 20 and 21). The core origin in turn is flanked by two auxiliary elements, termed aux-1 and aux-2, that are not essential but stimulate origin core activity. These elements consist of binding sites for T antigen and Sp1. In addition, the SV40 enhancer, located adjacent to aux-2, further stimulates replication. Since binding sites for several transcription factors, including Sp1 (34), AP1 (34), and NF1 (10), have been shown to stimulate replication when located in cis with the core origin, the auxiliary elements are thought to enhance activation of the SV40 origin by binding to transcription factors 33, 35, 39, 81; reviewed in references 20 and 21). However, the mechanism by which the auxiliary elements and the enhancer stimulate initiation is poorly understood.
Several possible mechanisms for origin activation by transcription factor binding sites have been proposed. Transcription factors such as Sp1, which binds to T antigen (41) and to an auxiliary element, have been proposed to promote association of T antigen with the origin through specific protein-protein interactions (20, 21, 34), whereas other transcription factors might recruit other replication proteins such as RPA. Other proposed mechanisms are that transcription factors stimulate initiation of DNA replication by stabilizing a partially unwound origin DNA structure (35) or by altering the viral chromatin structure so that the origin becomes more accessible to the proteins involved in the initiation of DNA replication (10, 11). However, the potential of a transcription factor binding site to stimulate viral replication does not appear to correlate with the transcriptional activation potential of the cognate factor (39). Moreover, in the absence of clear biochemical evidence that a transcription factor stimulates activation of the SV40 origin in a defined system, it remains possible some auxiliary elements enhance SV40 origin activation, at least in part, by effects on local DNA structure rather than through binding of the cognate transcription factor. Finally, despite compelling evidence that these auxiliary sequence elements stimulate SV40 DNA replication in vivo, there is still controversy as to whether this effect is also exerted during replication in vitro (9).
Stimulation of replication by transcription factors is common among the papovaviruses and adenoviruses (reviewed in references 20 and 21) but is not limited to viral replicons. Acidic transcription factors have been reported to activate replication from chromosomal origins of replication in budding yeast (49), and TBP has been implicated in replication initiation in yeast (51). Since TBP binds within the AT-rich element of the SV40 core origin and also physically interacts with T antigen, we examined whether TBP complexes might stimulate SV40 DNA replication in vitro. On the contrary, we found that removal of TBP complexes from cell extracts enhanced their ability to support SV40 DNA replication in vitro, while readdition of TBP complexes to the extracts reduced their replication activity. This interference appears to arise through physical interactions between T antigen and TBP that are mediated by a 24-amino-acid region in T antigen and a 36-amino-acid region in TBP. Interaction of purified T antigen with TBP complexes impaired its activity in bidirectional unwinding of the viral origin DNA. The inhibitory effect of TBP complexes on SV40 DNA replication in cell extracts and on origin unwinding activity was relieved by T antigen or TBP fusion peptides encompassing the interaction sites between the two proteins, which compete for the protein partner. The results suggest that TBP complexes associate with a T-antigen surface required for origin DNA unwinding and DNA replication, thereby inhibiting or interfering with them. We speculate that competition among cellular proteins for T antigen may play a role in its ability to regulate events in infected cells.
MATERIALS AND METHODS
Construction of recombinant pGEX plasmids.
Plasmids pGEX-T.1-249 and pGEX-T.101-249 were generously provided by A. Wildeman (2). pGEX-T.1-83 and pGEX-T.1-130 were kindly provided by A. Arthur and R. Weber (1, 75). pGEX-2T-T.101-164, pGEX-2T-T.181-205, pGEX-2T-T.164-205, pGEX-2T-T.164-249, and pGEX-2T-T.181-249 were generated by inserting the corresponding PCR amplification products into pGEX-2T (70). Each upstream primer for the PCR was designed to include a BamHI site, while the downstream primer included an EcoRI site to facilitate cloning into pGEX-2T. The template for the PCR was pT7-T antigen containing the full-length cDNA of T antigen (82). The following primers were used for PCR amplification: upstream and downstream primers for pGEX-T.101-164, 5′-TTGGATCCGAAAACCTGTTTTG-3′ and 5′-TTGAATTCTGTGGTGTAAATAGC-3′; upstream and downstream primers for pGEX-T.181-205, 5′-TTGGATCCGTAACCTTTATAAGT-3′ and 5′-TTGAATTCCACTCTATGCCTGTG-3′; upstream and downstream primers for pGEX-T.164-205, 5′-TTGGATCCACAAAGGAAAAAGCTGCACTG-3′ and 5′-TTGAATTCACTCTATGCCTGTGTGGAGT-3′; upstream and downstream primers for pGEX-T.164-249, 5′-TTGGATCCACAAAGGAAAAAGCTGCACTG-3′ and 5′-TTGAATTCTGGCAAACTTTCCTCAATAACAG-3′; upstream and downstream primers for pGEX-T.181-249, 5′-TTGGATCCGTAACCTTTATAAGTAGGCATAA-3′ and 5′-TTGAATTCTGGCAAACTTTCCTCAATAACAG-3′. After 30 cycles, the DNA was digested with BamHI and EcoRI, inserted into BamHI/EcoRI-digested plasmid pBluescript KSII+, and sequenced as specified by the manufacturer (Pharmacia, Piscataway, N.J.) to verify the sequence of each amplified fragment. The fragments were then subcloned into pGEX-2T for expression in Escherichia coli as glutathione S-transferase (GST) fusion peptides, using the restriction enzymes BamHI and EcoRI.
pGEX-2T-TBP was generously provided by R. Weber (75). GST-2T-TBP deletion constructs were generated as described above, using the following oligonucleotides as PCR primers: upstream and downstream primers for TBP1-159, 5′-TTGGATCCATGGATCAGAACAACAGCCT-3′ and 5′-CCGAATTCAGAACTCTCCGAAGCTGGC-3′; upstream and downstream primers for TBP156-339, 5′-TTGGATCCTCGGAGAGTTCTGGGATTG-3′ and 5′-TTGAATTCTTACGTCGTCTTCCTGAAT C-3′; upstream and downstream primers for TBP156-239, 5′-TTGGATCCTCGGAGAGTTCTGGGATTG-3′ and 5′-TTGAATTCTAGCATATTTCTTGCTGCCAG-3′; upstream and downstream primers for TBP242-339, 5′-TTGGATCCCAGAAGTTGGGTTTTCCAGC T-3′ and 5′-TTGAATTCTTACGTCGTCTTCCT GAATC-3′; upstream and downstream primers for TBP156-184, 5′-TTGGATCCTCGGAGAGTTCTGGGATTG-3′ and 5′-TTGAATTCCAATGGTCTTTAGGTCAAGTTTACAAC-3′; upstream and downstream primers for TBP184-220, 5′-TTGGATCCGCACTTCGTGCCCGAAAC-3′ and 5′-TTGAATTCCCATTTTCCCAGAACTGAAAAT-3′; upstream and downstream primers for TBP220-239, 5′-TTGGATCCGTGTGCACAGGAGCCAAGA-3′ and 5′-TTGAATTCTAGCATATTTCTTGCTGCCA G-3′. The template for the PCR was pBS.TBP, which was generated by inserting the BamHI/EcoRI fragment of full-length TBP from pGEX-2T-TBP into pBluescript KSII+ that had been digested with BamHI/EcoRI.
Purification of GST fusion proteins.
GST-TBP and GST-T antigen fusion proteins were expressed and purified as described previously (70), with the following modifications. An overnight culture of E. coli BL21(pLysS), transformed with the expression construct, was diluted 1:50 into Luria-Bertani broth containing ampicillin (100 μg/ml) and grown at 37°C for 3 h. Isopropylthiogalactoside was added to a final concentration of 0.5 mM, and growth was continued for 2 h. The cells were harvested by centrifugation and lysed by freezing and thawing in phosphate-buffered saline (PBS) containing freshly prepared protease inhibitors leupeptin (0.05 mM), aprotinin (0.02 mg/ml), and phenylmethylsulfonyl fluoride (0.5 mM). The lysate was sonicated to decrease the viscosity and then centrifuged for 10 min at 20,000 × g at 4°C to remove debris. The clarified cell lysates were rocked for 1 h at 4°C with glutathione-agarose beads (Sigma, St. Louis, Mo.) which had been equilibrated and resuspended 1:1 (vol/vol) in PBS. To recover the native fusion proteins from glutathione-agarose, the beads were washed with PBS and then eluted several times with 20 mM reduced glutathione (Sigma) in 50 mM Tris-HCl (pH 8.0). Proteins to be used in SV40 DNA replication assays were dialyzed against 2 liters of 20 mM HEPES-KOH (pH 7.8)–0.1 mM EDTA–100 mM NaCl–10% glycerol and stored at −80°C until use. TBP was released from GST-2T-TBP bound to glutathione-agarose by digestion with thrombin (Pharmacia) according to the manufacturer’s instructions. Protein concentrations were determined by the Bradford (4) assay (Bio-Rad, Hercules, Calif.), with bovine serum albumin (BSA; Sigma) as the standard.
To characterize fusion proteins bound to the beads, 10 μl of glutathione-agarose beads (1:1, vol/vol) was boiled with 10 μl of 4× sample buffer (44), and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (44). Proteins were visualized by staining the gels with Coomassie brilliant blue.
Antibodies.
Hybridoma Pab419 against SV40 T antigen (36), 4C8 against TBP (65), and 12CA5 against the hemagglutinin (HA) epitope of influenza virus (79) were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (Life Technologies, Gaithersburg, Md.). Immunoglobulins were purified by ammonium sulfate precipitation and affinity chromatography using a commercial kit (MAPS; Bio-Rad).
Other proteins.
SV40 T antigen was purified from Hi 5 insect cells infected with a recombinant baculovirus (46) as described previously (40). HA epitope-tagged TFIID complex (eTFIID) was purified from LTRα3 cells as described previously (84) except that the immunosorbent was covalently coupled 12CA5-Sepharose prepared with CNBr-activated Sepharose 4B (Pharmacia). Since the immunopurified eTFIID was washed with 0.4 M KCl, it contains TAFs and possibly associated general transcription factors (84). E. coli single-stranded DNA-binding protein (SSB) was purified as described previously (50) and was the kind gift of V. Podust. Human RPA was purified from recombinant E. coli extracts as described elsewhere (37). Calf thymus topoisomerase I was purified as described previously (72) and was the kind gift of I. Moarefi. Human DNA polymerase α-primase was purified from insect cells infected with recombinant baculoviruses as described elsewhere (71). All protein concentrations were determined by the Bradford (4) assay (Bio-Rad), with BSA (Sigma) as the standard.
Fusion protein binding assay.
GST fusion proteins immobilized on glutathione-agarose beads were washed twice with 500 μl of 30 mM HEPES-KOH (pH 7.8)–10 mM KCl–7 mM MgCl2 and incubated for 1 h at 4°C with either 0.5 μg of TBP or 0.5 μg of SV40 T antigen in 250 μl of the same buffer containing 2% nonfat dry milk powder. All reactions except that represented in Fig. 1 were performed in the presence of Benzonase endonuclease (0.1 U/μl; EM Science, Gibbstown, N.J.) to eliminate the possibility of protein-nucleic acid interactions. The beads were recovered by centrifugation and washed three times with 1 ml of 30 mM HEPES-KOH (pH 7.8)–25 mM KCl–7 mM MgCl2–0.25% inositol–0.25 mM EDTA–0.1% Nonidet P-40. The bound proteins were eluted by boiling in 1 volume of 4× sample buffer, separated by SDS-PAGE, and detected by Western blot analysis (27).
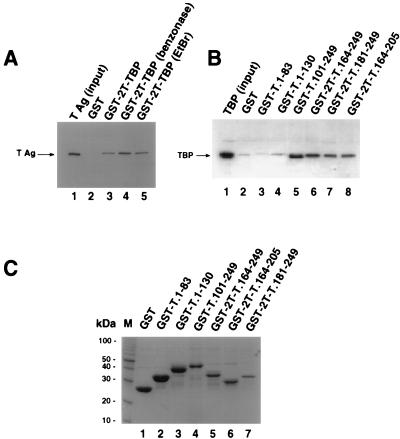
FIG. 1.
TBP binds within the DNA binding domain of T antigen. (A) Full-length T antigen (TAg; 0.5 μg) was incubated with 10 μl of glutathione-agarose containing GST (lane 2) or GST-TBP (lanes 3 to 5), either in buffer alone (lane 3) or in the presence of 1 U of benzonase (lane 4) or 200 μg of ethidium bromide (EtBr) per ml (lane 5). After washing, the beads were boiled in sample buffer, and the eluted proteins resolved by SDS-PAGE (12.5% polyacrylamide gel). T antigen was detected by immunoblotting using the T-antigen-specific monoclonal antibody Pab419. Lane 1 shows 0.1 μg of the input T antigen. (B) Full-length TBP was released from purified GST-2T-TBP by thrombin cleavage; 0.5 μg of cleaved TBP was incubated with 10 μl of glutathione-agarose containing GST (lane 2) or the indicated GST-T antigen fusion proteins in the presence of 1 U benzonase (lanes 3 to 8). After the beads were washed, the proteins were eluted by boiling in sample buffer and analyzed by SDS-PAGE (12.5% polyacrylamide gel). TBP was detected by immunoblotting using the monoclonal anti-TBP antibody 4C8. As a control, 0.1 μg of the input TBP was analyzed in parallel (lane 1). (C) Beads (10 μl) containing the GST fusion proteins used for panel B were analyzed by SDS-PAGE (12.5% polyacrylamide gel) and staining with Coomassie brilliant blue. M, 10-kDa marker protein ladder.
In vitro SV40 DNA replication.
Cytoplasmic extracts of the human embryonic kidney cell line 293S grown in monolayer were prepared as described previously (48). The extracts were adjusted to 100 mM NaCl and then centrifuged at 4°C for 1 h at 100,000 × g. The supernatant, designated S100, was depleted of TBP at 4°C by adjusting the salt concentration to 350 mM NaCl and incubating it twice for 30 min each with monoclonal anti-TBP antibody 4C8 covalently linked to CNBr-activated Sepharose. To generate the mock-depleted extract, the S100 extract at 4°C was adjusted to 350 mM NaCl and incubated twice for 30 min with the monoclonal anti-HA antibody 12CA5 covalently bound to CNBr-activated Sepharose. After depletion, the extracts were dialyzed against 20 mM HEPES-KOH (pH 7.8)–5 mM KCl–1.5 mM MgCl2–0.1 mM dithiothreitol (DTT)–100 mM NaCl–10% glycerol for 6 h, shock-frozen in liquid nitrogen, and stored at −80°C.
The template for the replication reaction was supercoiled pUC-HS plasmid DNA, which carries the HindIII-SphI fragment of SV40 DNA in pUC18 (74). The in vitro replication reaction was performed as described previously (60) except that the reaction mixture contained 190 μg of S100 extract or 250 μg of depleted or mock-depleted S100 extract, 100 ng of pUC-HS DNA, 1,000 ng of SV40 T antigen, 30 mM HEPES-KOH (pH 7.8), 7 mM magnesium acetate, 0.5 mM DTT, 1 mM EGTA, 4 mM ATP, 0.3 mM each GTP, CTP, and UTP, 0.1 mM each dATP and dGTP, 0.05 mM each dCTP and dTTP, 80 μg of creatine kinase (Boehringer Mannheim, Indianapolis, Ind.), 40 mM creatine phosphate (Boehringer Mannheim), and 5 μCi each of [α-32P]dCTP and [α-32P]dTTP (3,000 Ci/mmol; Amersham, Life Science Inc., Cleveland, Ohio). Where stated, GST fusion proteins in 20 mM HEPES-KOH (pH 7.8)–0.1 mM EDTA–100 mM NaCl–10% glycerol, or the same buffer alone, were added together with T antigen to the S100 extracts on ice. The mixture was incubated at 37°C for 90 min, the reaction was terminated with 30 μl of stop buffer (2% SDS, 60 mM EDTA-NaOH [pH 7.8]) and 20 ng of proteinase K (Sigma) and then the mixture was incubated for 30 min at 37°C. The mixture was extracted with phenol-chloroform, and DNA was precipitated with ammonium acetate-ethanol in the presence of 10 μg of tRNA. The replication products were resuspended in 20 μl of Tris-HCl (pH 7.8)–1 mM EDTA, and the incorporation of radiolabeled nucleotides into DNA in 5 μl of this solution was determined in a scintillation counter. To analyze the replication products, 5 μl of the solution was digested with EcoRI and EcoRI/DpnI, electrophoresed on a 1% agarose gel in Tris-borate buffer, and visualized by autoradiography (not shown).
Unwinding of SV40 origin DNA.
Unwinding reaction mixtures contained 200 ng of supercoiled closed circular pUC-HS DNA, 40 mM HEPES-KOH (pH 7.8), 0.5 mM DTT, 8 mM MgCl2, 4 mM ATP, 40 mM creatine phosphate, 0.5 μg of creatine kinase, 2 μg of BSA, 600 ng of SV40 T antigen, 120 ng of topoisomerase I, and 250 ng of E. coli SSB in a total volume of 20 μl. The mixture was incubated for 1 h at 37°C, and the reaction was stopped by addition of 0.2% SDS and 400 ng of proteinase K for 30 min at 37°C. After ethanol precipitation, the samples were redissolved in 10 mM EDTA–2% Ficoll–2% sucrose–0.01% bromophenol blue–0.1% SDS and electrophoresed in 1.5% agarose gels. The gels were stained with ethidium bromide and photographed.
RESULTS
TBP binds to a 24-amino-acid region within the DNA binding domain of T antigen.
Several independent studies have established that TBP binds specifically to SV40 T antigen in vitro and in vivo (2, 14, 32, 41, 53, 83). Taken together, these studies suggest that T antigen may contain two distinct binding sites for TBP, one between amino acids 5 and 172 and the other between amino acids 133 and 249. To further map the TBP binding domain of T antigen, we performed protein affinity pull-down experiments using T antigen, GST-T antigen fusion proteins, and TBP.
Since both T antigen and TBP bind to DNA, we wanted to first ensure that our assay measured direct interactions between the proteins rather than binding of both proteins to a common nucleic acid molecule. For this reason, the proteins used in this study were purified in the presence of benzonase to minimize nucleic acid contamination of the proteins. T antigen purified in this way bound to GST-TBP immobilized on glutathione-agarose (Fig. 1A, lane 3) but not to GST (lane 2). The interaction between T antigen and TBP was also observed in the presence of benzonase (lane 4) or ethidium bromide (lane 5), which disrupts DNA-protein interactions (45). These results indicate that the interaction between T antigen and TBP detected under these conditions is specific and direct.
GST-T antigen fusion proteins immobilized on glutathione-agarose were then tested for the ability to interact directly with TBP. Coarse mapping indicated that TBP bound to the N-terminal 259 amino acids of T antigen, while no interaction was observed with fusion peptides containing T-antigen amino acids 272 to 447, 336 to 537, 367 to 682, or 447 to 708 (data not shown). More detailed mapping of the N-terminal 259 residues of T antigen revealed that TBP bound well to T-antigen residues 101 to 249, 164 to 249, 181 to 249, and 164 to 205 (Fig. 1B, lanes 5 to 8), but interactions with GST alone or T-antigen residues 1 to 83 or 1 to 130 were barely detectable (Fig. 1B, lanes 2 to 4). Coomassie blue staining of the T antigen fusion proteins used in these experiments demonstrated that all of them were soluble and present in ample amounts, predominantly as a single polypeptide band (Fig. 1C).
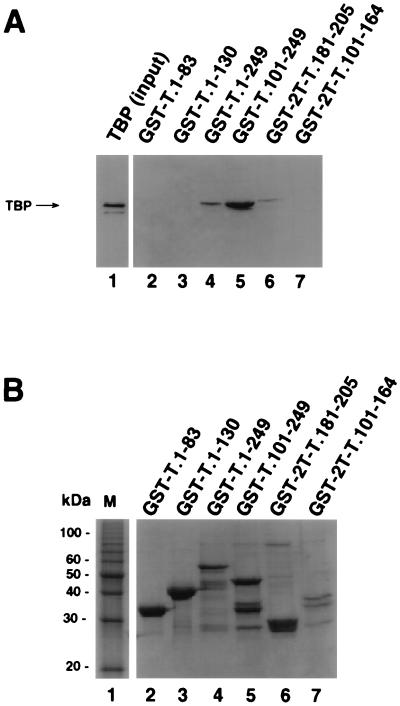
These data suggested that T-antigen residues 181 to 205 might be sufficient for direct interaction with TBP. To test this interpretation, two GST-T antigen fusion peptides, encompassing residues 101 to 164 and 181 to 205, were tested for TBP binding activity. The fusion proteins GST-T.1-249, GST-T.101-249, and GST-2T-T.181-205 were able to bind to TBP (Fig. 2A, lanes 4 to 6). Binding of GST-2T-T.181-205 to TBP was weaker than that of GST-T.101-249. The fusion protein GST-2T-T.101-164 (lane 7) and T-antigen fragments in the N-terminal domain GST-T.1-83 and GST-T.1-130 (lanes 2 and 3) showed little or no binding activity. A Coomassie blue-stained gel of the T antigen fusions used for this binding assay is shown in Fig. 2B. The fusion proteins were present in comparable amounts (lanes 2 to 7), except that GST-2T-T.101-164 was present at a level lower than those of the other fusion proteins (lane 7). These data demonstrate that a site sufficient for TBP binding resides within a 24-amino-acid region (residues 181 to 205) in the DNA binding domain of T antigen.
FIG. 2.
A 24-amino-acid region of T antigen is sufficient for specific interaction with TBP. (A) Purified full-length TBP was incubated with beads containing the indicated GST-T antigen fusion proteins (lanes 2 to 7) as described for Fig. 1. After the beads were washed, the bound proteins were analyzed by SDS-PAGE (12.5% gel), and TBP was detected by immunoblotting using the monoclonal anti-TBP antibody 4C8. Lane 1 contained 0.1 μg of the input TBP. (B) The GST-fusion proteins used for A (10 μl) were analyzed by SDS-PAGE (12.5% polyacrylamide gel) and staining with Coomassie brilliant blue. M, 10-kDa marker protein ladder.
T antigen binds to a 36-amino-acid region within the DNA binding domain of TBP.
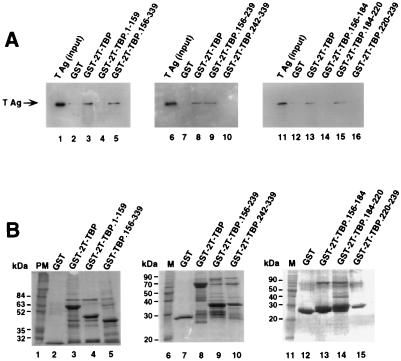
The T-antigen binding domain of TBP was reported to reside within the DNA binding domain of TBP (residues 204 to 275) (53). To map the T-antigen binding site of TBP in greater detail, GST-TBP fusion proteins containing small regions of TBP were tested for T-antigen binding in a pull-down assay. In agreement with previous results, only full-length TBP (Fig. 3A, lane 3) and the DNA binding domain of TBP (residues 156 to 339) (lane 5) were able to interact with T antigen under the conditions tested. No binding to GST (lanes 2, 7, and 12) or the TBP residues 1 to 159 (lane 4) was observed, indicating that the interaction was specific. Further analysis of this domain revealed that the residues involved in interactions with T antigen were located between amino acids 156 and 239 (Fig. 3A, lane 9), while residues at the carboxy terminus of TBP (amino acids 242 to 339) did not interact with T antigen (lane 10). To further narrow the T-antigen binding domain of TBP, GST-TBP fusion proteins containing overlapping peptides from the region from 156 to 239 were tested for the ability to bind T antigen in vitro. A 36-amino-acid fragment of TBP located between amino acids 184 and 220 was sufficient for interaction with T antigen (Fig. 3A, lane 15). Neither GST nor TBP fusion proteins containing residues 156 to 184 or 220 to 239 bound to T antigen under these conditions (lanes 12, 14, and 16), indicating that the interaction observed was specific. Coomassie blue staining of the GST-TBP fusion peptides revealed that all of them were soluble and present in similar amounts (Fig. 3B).
FIG. 3.
A 36-amino-acid region of TBP is sufficient for specific interaction with T antigen. (A) Purified full-length T antigen (TAg; 0.5 μg) was incubated with 10 μl of glutathione-agarose containing GST or the indicated GST-TBP fusion proteins in the presence of 1 U of benzonase (lanes 2 to 5, 7 to 10, and 12 to 16). After the beads were washed, the proteins were eluted by boiling in sample buffer and separated by SDS-PAGE (10% polyacrylamide gel). Bound T antigen was detected by immunoblotting using the monoclonal anti-T-antigen antibody Pab419. As a control, 0.1 μg of the input T antigen was analyzed in parallel (lanes 1, 6, and 11). (B) The beads containing the GST fusion proteins used for panel A (10 μl) were analyzed by SDS-PAGE (12.5% polyacrylamide gel) and staining with Coomassie brilliant blue. PM, prestained marker proteins; M, protein molecular weight markers.
TBP-associated protein complexes interfere with SV40 DNA replication in vitro.
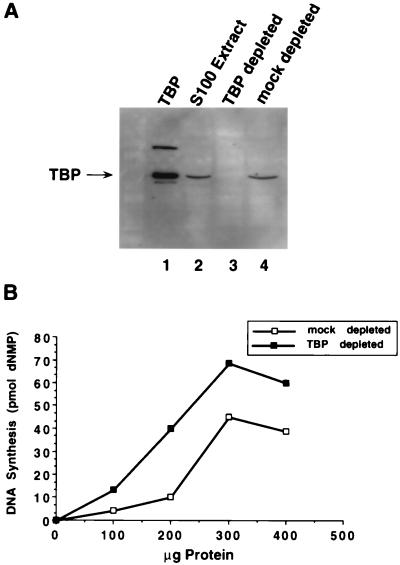
To investigate the possible effect of TBP on replication of SV40 origin-containing DNA, half of an S100 extract from human 293 cells was depleted of TBP by incubation with monoclonal anti-TBP (4C8) antibody beads and tested for its ability to support SV40 DNA replication in vitro. As a mock-depleted control, the other half of the S100 extract was incubated with monoclonal anti-HA (12CA5) antibody beads. Since 12CA5 recognizes the HA protein of influenza virus, it should not immunodeplete any proteins present in 293S extracts. Western blot analysis with anti-TBP antibody confirmed that TBP had been depleted from the extract incubated with 4C8-Sepharose (Fig. 4A, lane 3), while the mock-depleted extract contained approximately as much TBP as the untreated extract (Fig. 4A; compare lane 4 with lane 2).
FIG. 4.
TBP depletion of S100 extracts stimulates their activity in SV40 DNA replication in vitro. (A) A 100-μg aliquot of S100 extract from 293 cells (lane 2), TBP-depleted S100 extract (lane 3), or mock depleted S100 extract (lane 4) was analyzed by SDS-PAGE (10% gel) and immunoblotting with anti-TBP monoclonal antibody 4C8. As a marker, 0.25 μg of thrombin-cleaved GST-TBP was analyzed in parallel (lane 1). The upper band contains some uncleaved fusion protein. (B) SV40 DNA replication in vitro was assayed by using the indicated amounts of protein from TBP-depleted S100 extract or mock-depleted S100 extract. Replication activity was quantitated by measuring the incorporation of radiolabeled nucleotides into DNA.
Increasing amounts of these extracts were then titrated into an in vitro SV40 DNA replication assay mixture which contained a T-antigen concentration within the linear range of response. The replication activity of the TBP-depleted extract was reproducibly at least 60% greater than that of the mock-depleted extract (Fig. 4B). This stimulation was observed with three different preparations of the S100 extracts and with three different preparations of purified antibodies. These results suggest that TBP complexes in human cell extracts may inhibit SV40 DNA replication in vitro.
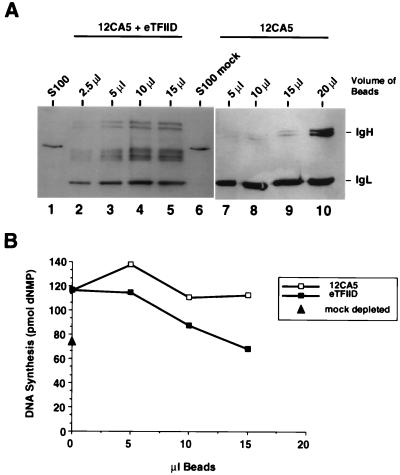
If this interpretation is correct, then readdition of purified TBP complexes to immunodepleted extracts should reduce their replication activity. TFIID was purified from LTRα3 cells, a HeLa cell line that constitutively expresses HA epitope-tagged TBP (84), by immunoprecipitation on anti-HA antibody 12CA5-Sepharose. To test the effect of eTFIID on SV40 DNA replication, increasing amounts of these beads were added to in vitro replication assay mixtures containing TBP-depleted extract. As a control to ensure that any effects observed did not arise from nonspecific effects of the beads, replication assays were also performed in the presence of equivalent amounts of 12CA5-Sepharose which had been preincubated with a nuclear extract from control HeLa cells that expressed no HA-tagged proteins. A Western blot of increasing amounts of the eTFIID beads revealed several TBP-related proteins that migrated slightly faster than intact TBP (Fig. 5A; compare lanes 2 to 5 with lanes 1 and 6), while no TBP was detected on the 12CA5 control beads (lanes 7 to 10). The faster migration of the TBP polypeptides in eTFIID was probably due to partial degradation during purification.
FIG. 5.
eTFIID inhibits SV40 DNA replication in vitro. (A) The indicated volumes of 12CA5-Sepharose beads containing epitope-tagged TBP complexes from LTRα3 cell extracts (lanes 2 to 5) or 12CA5-Sepharose beads which had been preincubated with HeLa control nuclear extract (lanes 7 to 10) were analyzed by SDS-PAGE (12.5% polyacrylamide gel) and immunoblotting with anti-TBP monoclonal antibody 4C8. Samples of undepleted 293S S100 extract (lane 1) and mock-depleted S100 extract (lane 6) (190 μg of each) were analyzed in parallel for comparison. Positions of immunoglobulin heavy and light chains (IgH and IgL) are shown on the right. (B) The indicated volumes of eTFIID bound to 12CA5-Sepharose beads or control 12CA5-Sepharose beads were added to an SV40 DNA replication reaction mixture containing 250 μg of S100 extract which had been depleted of TBP (Fig. 4A). Replication activity was quantitated by measuring the incorporation of radiolabeled nucleotides into DNA. The triangle on the ordinate indicates the SV40 DNA replication activity observed with 250 μg of mock-depleted S100 extract.
When increasing amounts of eTFIID beads were added to the TBP-depleted extract, replication activity decreased in a dose-dependent manner to the level observed with the mock-depleted extract (Fig. 5B). In contrast, the 12CA5 control beads did not significantly diminish the replication activity of the extracts. Consistent with the results in Fig. 5B, eTFIID beads also reduced the SV40 DNA replication activity of undepleted S100 extracts by 40% (data not shown). These data support the interpretation that eTFIID or other TBP-associated proteins interfered with SV40 DNA replication.
The fusion proteins GST-2T-TBP.184-220 and GST-2T-T.181-205 stimulate SV40 DNA replication in vitro by blocking the interaction between TFIID and T antigen.
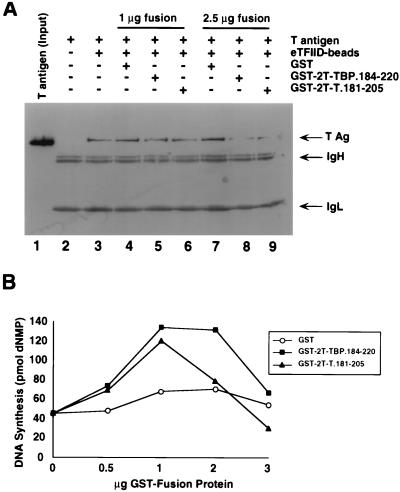
The ability of eTFIID to interfere with SV40 DNA replication suggested that physical interactions between TFIID and T antigen could be responsible for the interference. To test this possibility, we reasoned that a GST fusion peptide containing either the TBP binding site in T antigen or the T-antigen binding site in TBP might compete with T antigen for TFIID and relieve the inhibition of replication. To first establish whether either of the fusion peptides could compete with full-length T antigen for eTFIID, we tested T-antigen binding to eTFIID beads in the presence and absence of GST or GST fusion peptides. T antigen that had bound to the beads was detected by denaturing gel electrophoresis and immunoblotting with a monoclonal anti-T-antigen antibody (Fig. 6A). T antigen did not bind to 12CA5 beads preincubated with HeLa control extract (lane 2) but did bind to the eTFIID beads (lane 3). The presence of GST in the reaction mixture did not diminish T-antigen binding to the eTFIID beads (lanes 4 and 7). However, the interaction between T antigen and eTFIID was slightly reduced when 1 μg of either GST-2T-TBP.184-220 (lane 5) or GST-2T-T.181-205 (lane 6) was present in the binding reaction, and it was further reduced when 2.5 μg of either fusion peptide was present (lanes 8 and 9). The molar ratios of GST-T antigen fusion peptide to T antigen corresponded to 1.7 and 4.1 (lanes 6 and 9). These results demonstrate that the fusion peptides GST-2T-TBP.184-220 and GST-2T-T.181-205 competed for binding of eTFIID to T antigen.
FIG. 6.
Fusion proteins GST-2T-TBP.184-220, and GST-2T-T.181-205 stimulate SV40 DNA replication in vitro and inhibit the interaction between T antigen and TFIID. (A) One microgram of purified full-length T antigen was incubated with 10 μl of 12CA5-Sepharose beads preincubated with HeLa control extract (lane 2) or eTFIID immobilized on 12CA5-Sepharose beads in the absence (lane 3) or presence of the indicated amounts of GST fusion proteins (lanes 4 to 9). After the beads were washed, the bound proteins were eluted by boiling in sample buffer and separated by SDS-PAGE (12.5% polyacrylamide gel). T antigen was detected by immunoblotting with the monoclonal anti-T antigen antibody Pab419. Lane 1 shows 0.1 μg of the input T antigen as a marker. Positions of T antigen (T Ag) and immunoglobulin heavy and light chains (IgH and IgL) are shown on the right. (B) Increasing amounts of GST, GST-2T-TBP.184-220, or GST-2T-T.181-205 were added to an SV40 DNA replication reaction containing 190 μg of undepleted S100 extract from 293S cells. Replication activity was quantitated by measuring the incorporation of radiolabeled nucleotides into DNA.
GST-T antigen fusion peptides (GST-2T-T.181-205 and GST-2T-TBP.184-220) were then titrated into in vitro SV40 DNA replication assay mixtures containing undepleted human cell extract. GST alone served as a control to ensure that any effects observed did not arise from the GST portion of the peptide. Both fusion peptides clearly stimulated SV40 DNA replication, while GST alone had only a modest effect (Fig. 6B). The stimulation by GST-2T-T.181-205 reached a maximum of about threefold with 1 μg of peptide (peptide to T antigen molar ratio of 2.7). As more peptide was added, DNA synthesis dropped to the level observed without fusion peptide and with 3 μg of peptide (molar ratio of 8.3), DNA synthesis was slightly inhibited. The fusion peptide GST-2T-TBP.184-220 also stimulated DNA replication almost threefold; similarly, DNA synthesis declined when more peptide was present (Fig. 6B). Taken together, these results indicate that the fusion peptides, when present in appropriate amounts, competed out the interaction between T antigen and TBP, thereby relieving the inhibitory effect of TFIID or other TBP complexes on SV40 DNA replication.
eTFIID inhibits SV40 DNA replication in vitro at the step of origin DNA unwinding.
It is possible that eTFIID binding to the AT-rich element of the core origin DNA interfered with T-antigen assembly on the DNA (43a), thereby preventing replication. Alternatively, binding of eTFIID to T-antigen complexes on the origin may interfere with a later step in replication. To determine which step(s) of SV40 DNA replication is impaired by TBP complexes in cell extracts, we assayed origin DNA binding under replication conditions and unwinding by purified T antigen, as well as initiation of DNA replication by purified T antigen, RPA, topoisomerase I, and DNA polymerase α-primase, in the presence and absence of eTFIID beads. Band shift experiments using a labeled origin DNA fragment showed that T antigen bound equally well to the DNA in the presence and absence of eTFIID beads (data not shown), demonstrating that origin binding was not impaired. However, the ability of T antigen to unwind closed circular supercoiled DNA carrying the SV40 origin of replication was essentially abolished in the presence of eTFIID beads (Fig. 7, lane 4), while the control beads had no effect (lane 3). Consistent with this result, addition of eTFIID beads to an initiation reaction mixture containing SV40 DNA and purified T antigen and cellular replication proteins prevented primer formation on the origin DNA template (data not shown). These results suggest that eTFIID binding to T antigen may inhibit its ability to unwind supercoiled origin DNA and hence to initiate SV40 replication.
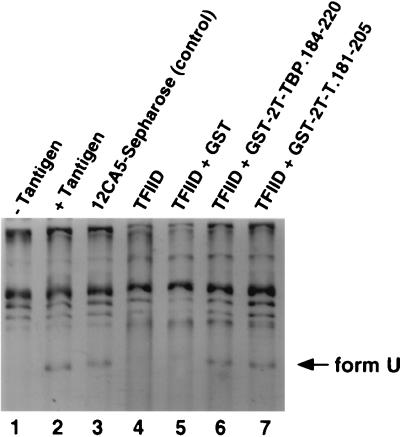
FIG. 7.
GST-2T-TBP.184-220 and GST-2T-T.181-205 relieve the inhibitory effect of eTFIID on unwinding of SV40 origin DNA. The unwinding assay contained closed circular supercoiled pUC-HS DNA, topoisomerase I, E. coli SSB (lanes 1 to 7), and SV40 T antigen (lanes 2 to 7). Reactions were performed in the presence of 12CA5-Sepharose preincubated with HeLa control extract (lane 3), eTFIID bound to 12CA5-Sepharose (lanes 4 to 7), and 1 μg of the indicated GST fusion proteins (lanes 5 to 7). The reaction products were analyzed by electrophoresis and ethidium bromide staining and then photographed. Form U indicates underwound DNA generated during the reaction.
Since the fusion peptides GST-2T-TBP.184-220 and GST-2T-T.181-205 were able to specifically relieve the inhibitory effect of eTFIID on in vitro SV40 DNA replication by competing for binding between TBP and T antigen in cell extracts (Fig. 6), one might also expect the peptides to relieve the inhibition of unwinding by eTFIID. Indeed, both GST-2T-TBP.184-220 and GST-2T-T.181-205 (Fig. 7, lanes 6 and 7), but not GST (lane 5), were able to restore the ability of purified T antigen to unwind the SV40 origin in the presence of eTFIID. The molar ratio of GST-T antigen peptide to T antigen corresponded to 2.7 (lane 7). The fusion peptides also relieved the eTFIID-mediated inhibition of initiation of SV40 DNA replication in an assay containing purified T antigen, RPA, topoisomerase I, and DNA polymerase α-primase (data not shown). We conclude that interaction of eTFIID with T antigen specifically interfered with its ability to unwind the viral origin and hence to initiate replication.
DISCUSSION
In this report, we have identified the sites in T antigen and in TBP that mediate specific interactions between the two proteins (Fig. 1 to 3). We have demonstrated that depletion of TBP complexes from cell extracts enhances their ability to support SV40 DNA replication in vitro, while readdition of TBP complexes restores replication activity to the lower level (Fig. 4 and 5). Addition of TBP complexes to purified T antigen impaired its ability to unwind SV40 origin DNA (Fig. 7), suggesting that TBP complexes interfere with replication in cell extracts by reducing origin unwinding. Evidence from competition experiments supports the interpretation that TBP interactions with T antigen are responsible for inhibition of both origin DNA unwinding and viral DNA replication (Fig. 6 and 7).
TBP binding to T antigen and the mechanism of inhibition of viral DNA replication.
TBP was shown previously to bind to two distinct regions of T antigen located between residues 84 and 172 (15, 32) and residues 133 and 249 (41). We observed specific interaction of TBP with T-antigen residues 164 to 205 (Fig. 1B) and 181 to 205 (Fig. 2A) but failed to detect interactions with regions N terminal to amino acid 164 or C terminal to amino acid 205 (Fig. 1B and 2A). These results are further supported by coimmunoprecipitation studies with monoclonal antibodies against T antigen. Monoclonal antibodies that recognize epitopes in the amino-terminal region of T antigen (residues 1 to 130) had no effect on the interaction with TBP, while antibodies with epitopes in the region from 131 to 259 nearly abolished TBP binding activity (data not shown). TBP binding to T-antigen residues 181 to 205 was weaker than binding to residues 164 to 205, suggesting that sequences on the amino-terminal side of residue 181 also contribute to TBP binding. Either there may be a second independent TBP binding site located between residues 164 and 181, as suggested by the TBP binding activity of residues 84 to 172 (15, 32), or peptide 181 to 205 represents a partial site that is sufficient to detect some TBP binding and to compete with intact T antigen for TBP complexes (Fig. 6A) but not at the level observed with the complete site.
The TBP binding site of T antigen resides within the origin DNA binding domain of the protein (residues 131 to 259) (1, 42, 52, 67). Genetic evidence indicates that this domain has multiple functions in addition to its sequence-specific DNA binding activity (80). Biochemical studies have shown that not only TBP but also several other transcription factors interact with the DNA binding domain of T antigen. Moreover, TBP, TFIIB, Sp1, RNA polymerase II, and TEF-1 compete with each other for T antigen, suggesting that their binding sites overlap (41). This observation raises the possibility that these other transcription factors also interfere with SV40 replication, a possibility which remains to be tested. However, these factors may not interact with identical residues in T antigen, since certain mutations in T antigen specifically impair binding of one transcription factor without affecting binding of the others. For example, substitution of serine 189 by asparagine strongly reduced T-antigen binding to TEF-1 (2, 22) but had no effect on binding to TBP (data not shown). On the other hand, a mutation at residues 173 and 174 (K173A,K174A) was reported to inhibit T-antigen binding to several transcription factors (41). The genetic evidence does not distinguish whether residues 173 and 174 comprise part of one binding site recognized by all of these transcription factors or whether the mutations may disrupt the structural integrity of multiple distinct binding sites.
In addition to binding sites for transcription factors, the origin DNA binding domain of T antigen also harbors a binding site for replication protein A which was demonstrated to be essential for viral DNA replication (76). Although the RPA binding site mapped within residues 164 to 249, RPA did not compete with TBP for T-antigen binding (data not shown), indicating that these proteins recognize nonoverlapping sites. Moreover, mutations within the DNA binding domain of T antigen that did not affect origin DNA binding, double-hexamer assembly, local origin DNA distortion, or DNA helicase activity have been shown to disrupt unwinding of supercoiled origin DNA by T antigen (80). This result suggested that the DNA binding domain also plays a novel role in origin unwinding. Since unwinding of supercoiled origin DNA is thought to require interactions between the two hexamers of T antigen (56, 57, 60, 69, 76a, 77), and these interactions are defective in mutants that map in the DNA binding domain of T antigen (76a, 80), the DNA binding domain may mediate these interactions.
The solution structure of the DNA binding domain of T antigen was recently determined by nuclear magnetic resonance spectroscopy (52). The structure reveals that residues 181 to 205 reside in β strands B and C, which are only partially exposed in two patches (residues 186 to 193 and 200 to 204) on well-separated surfaces of the structure. Residues 173 and 174, implicated in binding to multiple transcription factors (41), protrude from a ridge located between these two surfaces (52). Residue 189, which is implicated in TEF-1 binding (2, 22), is exposed at the edge of the patch composed of residues 186 to 193 (52) and might specifically contact TEF-1. Based on these considerations, we suggest that TBP and the other transcription factors probably make contact with one of the exposed patches (residues 186 to 193) and the ridge.
Two of the mutations that specifically inhibit origin unwinding (80) target residues (K167R and F220Y) that are localized immediately adjacent to the 186–193 patch (52). The potential overlap between the TBP binding surface and this region required for origin unwinding suggests that TBP may block functional interactions between T-antigen hexamers. This speculation provides an attractive explanation for the ability of TBP to inhibit origin DNA unwinding by T antigen and for the ability of T antigen or TBP competitor peptides in appropriate concentrations to relieve the inhibition (Fig. 7). If unwinding requires that T-antigen hexamers interact in part through the surface that is bound by TBP, then it should also be possible to block origin unwinding by adding excess TBP or T antigen fusion peptide. Indeed, origin unwinding was inhibited by concentrations of T antigen or TBP competitor peptide threefold higher than those used for Fig. 7 (data not shown).
The T-antigen binding site of TBP.
The T-antigen binding site of TBP has been localized to within residues 184 to 220 (Fig. 3). Consistent with this result, deletion of TBP residues 186 to 377 or 186 to 208 abolished the T-antigen binding activity of TBP (15). Moreover, a truncated TBP mutant lacking residues 209 to 337 retained full T-antigen binding activity and a mutant lacking residues 1 to 196 retained partial activity (15), suggesting that residues 197 to 208 may be sufficient to bind to T antigen. These data appear to conflict with a previous report that T antigen binds to TBP residues 204 to 275 (53). However, the T-antigen binding observed with TBP residues 204 to 335 was much weaker than that observed with residues 1 to 275 (53).
The T-antigen binding site of TBP residues in the N-terminal repeated sequence of TBP in the first stirrup of the saddle structure elucidated by crystallography (reviewed in reference 38). It is composed of two separate surfaces directly adjacent to the TFIIA binding surface in TBP-TFIIA-DNA cocrystals (31, 73). The proximity of the T-antigen binding site to the TFIIA binding site may allow T antigen to contact both transcription factors, providing a possible mechanism for the recently reported stabilization of TBP-TFIIA complexes by T antigen and the resulting transcriptional activation (16).
Competition among cellular proteins for T antigen: implications for viral infection.
Our findings indicate that binding of TBP complexes to T antigen antagonizes viral DNA replication in vitro (Fig. 4 to 6), most likely by interfering with interactions between T-antigen hexamers in unwinding viral DNA (Fig. 7). Other transcription factors that compete for the TBP binding site on T antigen (41) may also interfere with replication. Similarly, binding of p53 to T antigen antagonizes viral DNA replication, apparently by blocking its interactions with DNA polymerase α-primase (30). These examples of competition between protein-protein interactions involving T antigen suggest that T antigen in infected cells may exist in multiple separate complexes with different and mutually exclusive functions. Accumulation of high concentrations of T antigen in the infected cell during the early phase of infection may be required to fulfill all of the functions ascribed to this protein, since each T-antigen complex could participate in only a subset of these functions. Moreover, balanced regulation of viral infection by T antigen may depend not only on the concentration of T antigen but also on the affinity of its interactions with cellular proteins and their concentrations during the infection. Finally, if T antigen is limiting due to competing protein-protein interactions, the role of the J domain of T antigen and cellular chaperone proteins that interact with it may be to remodel multiprotein complexes of T antigen formed early in infection into other T-antigen complexes needed later in the infection (7).
ACKNOWLEDGMENTS
We thank A. Arthur, A. Berk, I. Moarefi, V. Podust, H. Stunnenberg, R. Weber, A. Wildeman, and K. van Zee for cells, plasmids, and proteins. We especially thank J. Flint for monoclonal antibody 4C8 and J. Alwine for communication of unpublished data. The skillful technical assistance of A. Brunahl and L. O’Rear is gratefully acknowledged.
The support of the NIH (grant GM 52948) and Vanderbilt University is gratefully acknowledged. This work was begun at the University of Munich with the support of the German Science Foundation.
REFERENCES
- 1.Arthur A K, Höss A, Fanning E. Expression of simian virus 40 T antigen in Escherichia coli: localization of T-antigen origin DNA-binding domain to within 129 amino acids. J Virol. 1988;62:1999–2006. doi: 10.1128/jvi.62.6.1999-2006.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger L, Smith D B, Davidson I, Hwang J-J, Fanning E, Wildeman A G. Interaction between T antigen and TEA domain of the factor TEF-1 derepresses simian virus 40 late promoter in vitro: identification of T-antigen domains important for transcription control. J Virol. 1996;70:1203–1212. doi: 10.1128/jvi.70.2.1203-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of DNA replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brady J, Khoury G. trans-activation of the simian virus 40 late transcription unit by T antigen. Mol Cell Biol. 1985;5:1391–1399. doi: 10.1128/mcb.5.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun K A, Lao Y, He Z, Ingles C J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 9.Bullock P A, Joo W S, Sreekumar K R, Mello C. Initiation of SV40 DNA replication in vitro: analysis of the role played by sequences flanking the core origin on initial synthesis events. Virology. 1997;227:460–473. doi: 10.1006/viro.1996.8347. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Kelly T J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989;59:541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L, Workman J L, Kingston R E, Kelly T J. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4-VP16. Proc Natl Acad Sci USA. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins K L, Kelly T J. Effects of T antigen and replication protein A on the initiation of DNA synthesis of DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conzen S D, Cole C N. The transforming proteins of simian virus 40. Semin Virol. 1994;5:349–356. [Google Scholar]
- 14.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 15.Damania, B., and J. C. Alwine. Unpublished data.
- 16.Damania B, Lieberman P, Alwine J C. Simian virus 40 large T antigen stabilizes the TATA-binding protein–TFIIA complex on the TATA element. Mol Cell Biol. 1998;18:3926–3935. doi: 10.1128/mcb.18.7.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damania B, Mital R, Alwine J C. Simian virus large T antigen interacts with human TFIIB-related factor and small nuclear RNA-activating protein complex for transcriptional activation of TATA-containing polymerase III promoters. Mol Cell Biol. 1998;18:1331–1338. doi: 10.1128/mcb.18.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 20.DePamphilis M L. Eucaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;63:29–62. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 21.DePamphilis M L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993;3:1161–1163. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 22.Dickmanns A, Zeitvogel A, Simmersbach F, Weber R, Arthur A K, Dehde S, Wildeman A G, Fanning E. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J Virol. 1994;68:5496–5508. doi: 10.1128/jvi.68.9.5496-5508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–968. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 24.Dornreiter I, Höss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the large subunit of DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dornreiter I, Copeland W C, Wang T S. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase alpha with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn S D. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem. 1986;157:144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- 28.Fanning E. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fanning E, Knippers R. Structure and function of simian virus large T antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 30.Gannon J V, Lane D P. p53 and DNA polymerase α compete for binding to SV40 T antigen. Nature. 1987;329:456–458. doi: 10.1038/329456a0. [DOI] [PubMed] [Google Scholar]
- 31.Geiger J H, Hahn S, Lee S, Sigler P B. TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 32.Gruda M C, Zablotny J M, Xiao J H, Davidson I, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z-S, Gutierrez C, Heine U, Sogo J M, DePamphilis M L. Origin auxiliary sequences can facilitate initiation of simian virus 40 DNA replication in vitro as they do in vivo. Mol Cell Biol. 1989;9:3593–3602. doi: 10.1128/mcb.9.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Z-S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez C, Guo Z S, Roberts J M, DePamphilis M L. Simian virus 40 origin auxiliary sequences weakly facilitate T-antigen binding but strongly facilitate DNA unwinding. Mol Cell Biol. 1990;10:1719–1728. doi: 10.1128/mcb.10.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harlow E, Crawford L, Pim D, Williamson N. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 38.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 39.Hoang A T, Weidong W, Gralla J D. The replication activation potential of selected RNA polymerase II promoter elements of the simian virus 40 origin. Mol Cell Biol. 1992;12:3087–3093. doi: 10.1128/mcb.12.7.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Höss A, Moarefi I, Scheidtmann K-H, Cisek L J, Corden J L, Dornreiter I, Arthur A, Fanning E. Altered phosphorylation pattern of simian virus 40 T antigen expressed in insect cells by using a baculovirus vector. J Virol. 1990;64:4799–4807. doi: 10.1128/jvi.64.10.4799-4807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston S D, Yu X-M, Mertz J E. The major transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J Virol. 1996;70:1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joo W X, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T-antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keller J M, Alwine J C. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell. 1984;36:381–389. doi: 10.1016/0092-8674(84)90231-9. [DOI] [PubMed] [Google Scholar]
- 43a.Kilwinski J, Baack M, Heiland S, Knippers R. Transcription factor Oct1 binds to the AT-rich segment of the simian virus 40 replication origin. J Virol. 1995;69:575–578. doi: 10.1128/jvi.69.1.575-578.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Lai J, Herr W. Ethidium bromide provides a simple tool for establishing genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanford R E. Expression of simian virus 40 T antigen in insect cells using a baculovirus vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 47.Lee S H, Kim D K. The role of the 34-kDa subunit of human replication protein A in simian virus 40 DNA replication in vitro. J Biol Chem. 1995;270:12801–12807. doi: 10.1074/jbc.270.21.12801. [DOI] [PubMed] [Google Scholar]
- 48.Li J J, Kelly T J. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Yu D S, Tanaka M, Zheng L, Berger S L, Stillman B. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol Cell Biol. 1998;18:1296–1302. doi: 10.1128/mcb.18.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lohman T M, Green J M, Beyer R S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 51.Lue N F, Kornberg R D. A possible role for the yeast TATA-element-binding protein in DNA replication. Proc Natl Acad Sci USA. 1993;90:8018–8022. doi: 10.1073/pnas.90.17.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo X, Sanford D G, Bullock P A, Bachovchin W W. Solution structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 53.Martin D W, Subler M A, Munoz R M, Brown D R, Deb S P, Deb S. p53 and SV40 T antigen bind to the same region overlapping the conserved domain of the TATA-binding protein. Biochem Biophys Res Commun. 1993;195:428–434. doi: 10.1006/bbrc.1993.2061. [DOI] [PubMed] [Google Scholar]
- 54.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 55.Mazzarelli J M, Atkins G B, Geisberg J V, Ricciardi R P. The viral oncoproteins Ad5 E1A, HPV16 E7 and SV40 Tag bind a common region of the TBP-associated factor-110. Oncogene. 1995;11:1859–1864. [PubMed] [Google Scholar]
- 56.McVey D, Ray S, Gluzman Y, Berger L, Wildeman A G, Marshak D R, Tegtmeyer P. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J Virol. 1993;67:5206–5215. doi: 10.1128/jvi.67.9.5206-5215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 59.Mitchell P J, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 60.Moarefi I F, Small D, Gilbert I, Hoepfner M, Randall S K, Schneider C, Russo A R R, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA binding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murakami Y, Hurwitz J. DNA polymerase alpha stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 62.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:1108–11017. [PubMed] [Google Scholar]
- 63.Nevins J R. Cell cycle targets of the DNA tumor viruses. Curr Opin Genet Dev. 1994;4:130–134. doi: 10.1016/0959-437x(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 64.Parsons R E, Stenger J E, Ray S, Walker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus T antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pruzan R, Chatterjee P K, Flint S J. Specific transcription from the adenovirus E2E promoter by RNA polymerase III requires a subpopulation of TFIID. Nucleic Acids Res. 1992;21:5705–5712. doi: 10.1093/nar/20.21.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed S I, Stark G R, Alwine J C. Autoregulation of simian virus 40 gene A by T antigen. Proc Natl Acad Sci USA. 1976;73:3083–3087. doi: 10.1073/pnas.73.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons D T, Loeber G, Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990;64:1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 69.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 71.Stadlbauer F, Voitenleitner C, Brückner A, Fanning E, Nasheuer H-P. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase α-primase. Mol Cell Biol. 1996;16:94–1104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strausfeld U, Richter A. Simultaneous purification of DNA topoisomerases I and II from eucaryotic cells. Prep Biochem. 1989;19:37–48. doi: 10.1080/10826068908544895. [DOI] [PubMed] [Google Scholar]
- 73.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 74.Traut W, Fanning E. Sequence-specific interactions between a cellular DNA-binding protein and the simian virus 40 origin of DNA replication. Mol Cell Biol. 1988;8:903–911. doi: 10.1128/mcb.8.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weber R. Doctoral dissertation. Munich, Germany: University of Munich; 1996. [Google Scholar]
- 76.Weisshart K, Taneja P, Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76a.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. Two regions of simian virus 40 T antigen determine cooperativity of double hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 77.Wessel R, Schweizer J, Stahl H. Simian virus 40 T antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wiley S R, Kraus R J, Zuo F, Murray E E, Loritz K, Mertz J E. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 1993;7:2206–2219. doi: 10.1101/gad.7.11.2206. [DOI] [PubMed] [Google Scholar]
- 79.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 80.Wun-Kim K, Upson R H, Young W, Simmons D T. The DNA binding domain of simian virus large T antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi M, DePamphilis M L. DNA binding site for a factor(s) required to initiate simian virus 40 DNA replication. Proc Natl Acad Sci USA. 1986;83:1646–1650. doi: 10.1073/pnas.83.6.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Zee K. Doctoral dissertation. Munich, Germany: University of Munich; 1991. [Google Scholar]
- 83.Zhai W, Tuan J A, Comai L. SV40 large T antigen binds to the TBP-TAFI complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 84.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]