Abstract
Aims
Epigenetic age is emerging as a personalized and accurate predictor of biological age. The aim of this article is to assess the association of subclinical atherosclerosis with accelerated epigenetic age and to investigate the underlying mechanisms mediating this association.
Methods and results
Whole blood methylomics, transcriptomics, and plasma proteomics were obtained for 391 participants of the Progression of Early Subclinical Atherosclerosis study. Epigenetic age was calculated from methylomics data for each participant. Its divergence from chronological age is termed epigenetic age acceleration. Subclinical atherosclerosis burden was estimated by multi-territory 2D/3D vascular ultrasound and by coronary artery calcification. In healthy individuals, the presence, extension, and progression of subclinical atherosclerosis were associated with a significant acceleration of the Grim epigenetic age, a predictor of health and lifespan, regardless of traditional cardiovascular risk factors. Individuals with an accelerated Grim epigenetic age were characterized by an increased systemic inflammation and associated with a score of low-grade, chronic inflammation. Mediation analysis using transcriptomics and proteomics data revealed key pro-inflammatory pathways (IL6, Inflammasome, and IL10) and genes (IL1B, OSM, TLR5, and CD14) mediating the association between subclinical atherosclerosis and epigenetic age acceleration.
Conclusion
The presence, extension, and progression of subclinical atherosclerosis in middle-aged asymptomatic individuals are associated with an acceleration in the Grim epigenetic age. Mediation analysis using transcriptomics and proteomics data suggests a key role of systemic inflammation in this association, reinforcing the relevance of interventions on inflammation to prevent cardiovascular disease.
Keywords: Subclinical atherosclerosis, Epigenetic age acceleration, Systemic inflammation, Transcriptomics, Proteomics, Methylomics
Structured Graphical Abstract
Structured Graphical Abstract.
The accelerated epigenetic age of individuals with subclinical atherosclerosis is mediated by a low-grade, chronic, systemic inflammation driven by key inflammatory cytokines and pathways.
See the editorial comment for this article ‘A Grim link: the association between subclinical atherosclerosis and epigenetic age’, by N. Velayutham and R.T. Lee, https://doi.org10.1093/eurheartj/ehad326.
Introduction
The total number of disabilities and deaths caused by cardiovascular diseases (CVD) are still increasing, mainly as a result of progressive aging in our societies. Atherosclerosis remains the main pathological mechanism underlying most life-threatening CVD events, especially myocardial infarction and stroke. Atherosclerotic plaques build progressively on arterial walls and can remain clinically silent (subclinical atherosclerosis, SA) for decades. In particular, the Progression of Early Subclinical Atherosclerosis (PESA) study previously showed that SA is highly prevalent in middle-aged individuals who were apparently healthy.1–3 While the reduced lifespan associated with atherosclerosis is thought to occur through the incidence of ischaemic events, the impact of SA on lifespan per se remains unclear.
The time elapsed since birth (chronological age, CA) is the leading risk factor for CVD.4 While CA is non-modifiable, several age-related functional parameters exhibit broad variability in individuals of similar CA, suggesting that biological aging, which refers to the decline in tissue/organismal function, proceeds at a different pace in different subjects. Indeed, there are several hallmarks of biological aging that vary substantially among individuals of the same CA, including genomic instability, telomere shortening, mitochondrial dysfunction, cellular senescence, and epigenetic changes.5,6 The best-known epigenetic mark is DNA methylation, a dynamic process consisting in the addition or removal of a methyl group (CH3), predominantly in CpG islands that are often located in the promoter region of genes.7 Methyl groups are added by DNA methyl transferases and removed by ten-eleven translocation methyl-cytosine dioxygenases. Importantly, these enzymes are regulated by genetic and environmental factors,8 including age and diet. In particular, specific DNA methylation patterns at multiple CpG sites across the genome have been demonstrated to change with age.9 This, together with the potential reversibility of epigenetic changes, has boosted the development of novel biological age measurements based on several epigenetic clocks that estimate the epigenetic age (EA) of an individual.10–13 Using distinct sets of epigenetic marks, the first generation of epigenetic clocks were derived to predict biological age and its departure from CA,10,11 while the second generation of clocks are predictors of lifespan, i.e. time to death.12,13 Importantly, epigenetic age acceleration (EAA), defined as the difference between EA and CA, has been associated with many physiological parameters (e.g. body mass index, and plasma levels of insulin, cholesterol, and C-reactive protein), lifestyle variables (e.g. exercise, smoking, and diet), psychosocial parameters (e.g. income and education), and several diseases.14,15
In this work, we estimated EAA based on different epigenetic clocks in 391 middle-aged, asymptomatic participants of the PESA study to assess the potential association between the presence, extension, and progression of SA and EA, as a proxy of their predicted time to CVD and time to death. Mediation analysis of transcriptomics and proteomics data was used to shed light on the molecular mechanisms underlying the interplay between SA and EAA. Our results strengthen the pathophysiological relevance of SA and highlight the need for its early detection before the life-threatening clinical manifestations of the disease are evident.
Methods
Study design
This study was conducted in a subset of participants from the PESA study, which enrolled between June 2010 and February 2014, 4184 healthy volunteers, employees of Santander Bank headquarters in Madrid, Spain, with ages ranging between 40 and 54 years.1 The study protocol was approved by the Carlos III Health Institute Ethics Committee, and all participants provided written informed consent.
For each participant, SA was assessed as the presence of plaques in any of the five territories evaluated (two carotids, two ilio-femorals, aorta) by 2D/3D vascular ultrasound or presence of coronary artery calcification (CAC, Agatston score ≥0.5) by computed tomography (CT). The extent of SA was evaluated according to the number of territories with disease by the PESA score: no SA, focal SA (one site affected), intermediate SA (two to three sites), or generalized SA (four to six sites). Multi-territory vascular ultrasound and CT were repeated 3 years after initial exams to evaluate atherosclerosis progression. Finally, atherosclerotic burden was calculated as the sum of all plaque volumes in different territories by 3D vascular ultrasound. The three-year progression of SA was defined as previously described.16 Two traditional cardiovascular (CV) risk scores that predict 10-year risk for a first atherosclerotic CVD event were used: European Society of Cardiology Systematic Coronary Risk Evaluation (SCORE)17 and the US pooled cohort risk score (ASCVD).18 Participants were classified according to their 10-year risk as having a low (<5%), borderline (≥5% and <7.5%), moderate (≥7.5% and <20%), and high (≥20%) risk of ASCVD event. The SCORE was used to estimate the CV age of the participants. The INFLA score19 is a measure of chronic, low-grade inflammation associated with a higher risk of mortality, derived from 20 337 adult individuals from the MOLI-SANI cohort. The score was calculated for the PESA cohort from the sum of 10-tiles of plasmatic (C-reactive protein) and cellular (leukocyte and platelet count, granulocyte/lymphocyte ratio) biomarkers of low-grade inflammation; higher levels indicated increased low-grade inflammation. Metabolic syndrome was defined as positive when a participant met at least three of the following conditions: central obesity (waist circumference ≥94.4 cm in men or ≥89.5 in women), elevated plasma triglycerides (≥150 mg/dL), low plasma HDL-cholesterol (<40 mg/dL in men or <50 mg/dL in women), elevated fasting plasma glucose (≥100 mg/dL), and high blood pressure (systolic ≥130 mmHg and/or diastolic ≥85 mmHg). Positron emission tomography/magnetic resonance imaging was performed in 130 participants from the PESA omics cohort as previously described.20
Progression of Early Subclinical Atherosclerosis omics cohort
For omics analyses, 240 PESA participants with extended SA and 240 without SA were matched by sex, age (± 3 years), and traditional CV risk factors (diabetes mellitus, current smoker, dyslipidemia, and hypertension). RNA-Seq and methylomics data were generated from whole blood samples from these individuals. Moreover, hypothesis-free proteomics were generated from plasma samples of these participants. A total of 391 samples fulfilled all quality control requirements in all three omics techniques. Baseline characteristics, CV risk factors, dietary patterns, and haematological parameters of PESA individuals in the omics cohort vs. those of the whole PESA cohort are shown in Supplementary data online, Tables S1 and S2.
DNA extraction and generation of methylomics data
Blood samples were collected in S-Monovette EDTA K3 tubes (Sarstedt). A total of 600 µL of blood were aliquoted in 0.75 mL 2D Barcoded Storage Tubes (Thermo Scientific) and stored at −80°C until processed. For DNA extraction, blood samples were thawed quickly in a 37°C water bath for 2 min to avoid degradation. Samples were then processed using the QIAsymphony DNA Mini Kit for the QiAsymphony SP automated station (QiAgen), following the manufacturer’s instructions. Briefly, blood samples were lysed and incubated with magnetic beads for DNA purification. After a wash, DNA was eluted in 200 µL of elution buffer, and DNA concentration and quality were measured by QuBit (Invitrogen) and Nanoquant (Tecan), respectively. One microgram of high-quality DNA (260/280 ratio over 1.8) was used for methylome analysis by MethylationEPIC BeadChip at the CEGEN (Madrid, Spain). DNA was bisulphite-converted using EZ DNA Methylation Kits (Zymo Research) following the manufacturer's recommendation. A hyper-methylated sample was included to ensure the correct conversion of the DNA, and a control sequence containing high CpG density was amplified and sequenced by Sanger to confirm the CpG bisulfite conversion of the samples. Converted DNAs were processed to be hybridized in the Infinium MethylationEPIC BeadChip BeadChip following the manufacturer's protocols (Illumina). Arrays were scanned using iScan System (Illumina).
Bioinformatics analysis of methylomics data, epigenetic age calculation, and deconvolution of methylomics samples
Array intensities were extracted from idat files, filtered, and normalized using the rnb.run.analysis function from Bioconductor package RnBeads21 version 2.0.1. Probes with single-nucleotide polymorphism overlap were removed, and the greedy cut algorithm implemented in RnBeads was applied with a detection P > .05 for probe detection assessment. Around 800 000 probes were retained after filtering and further normalized by the dasen method22 generating M-values and β-values for downstream analysis. Horvath’s,10Hannum’s,11Pheno,12 and Grim13 EA and EAA were inferred from the methylation state using the online DNA Methylation Age Calculator at https://dnamage.genetics.ucla.edu/new, and using β-values normalized by noob method implemented in the minfi23 Bioconductor package version 1.32.0. The quartiles of the distribution of Grim EAA calculated for the 391 PESA participants were used as cut-offs to define individuals with accelerated aging (Q3, with an EA 2.74 years above their CA) and with decelerated aging (Q1, with an EA 3.38 years below their CA). The DNA Methylation Age Calculator also provides estimations of the amount of different cellular types based on two deconvolution methods.24,25
RNA extraction and RNA-Seq data generation
Peripheral blood samples were collected from 391 PESA participants, transferred to PAXgene Blood RNA tubes (Preanalytix) to allow instant preservation of RNA, and stored at −80°C until use. Total cellular RNA, including miRNA, was automatically extracted from PAXgene tubes using a QIAsymphony SP liquid handling robot (Qiagen) and the QIAsymphony PAXgene blood RNA kit (Qiagen), following the manufacturer’s instructions. RNA quantity was measured by absorbance using a NanoQuant Plate (Tecan). The RNA integrity number (RIN) was calculated by automated electrophoresis in a 2100 Bioanalyzer (RNA6000 Nano LabChip, Agilent). Barcoded RNA-Seq libraries were generated using 200 ng of total RNA with RIN > 6 and the NEBNext Ultra RNA Library preparation kit (New England Biolabs). Briefly, polyA+ RNA was purified using polyT oligo-attached magnetic beads followed by fragmentation, and subsequent first and second cDNA strand synthesis. cDNA 3′-ends were adenylated, the adapters were ligated, and the library was amplified by PCR. The size and concentration of the libraries were checked using the Agilent 2100 Bioanalyzer DNA 1000 chip. Pools were prepared containing on average 11 libraries with a different barcode and equimolar concentrations of each library. Each pool concentration was determined using the Qubit® fluorometer (Life Technologies). Library pools were sequenced on a HiSeq2500 (Illumina) to generate 60 bases single reads. Fastq files for each sample were obtained using CASAVA v1.8 software (Illumina).
Bioinformatics analysis of RNA-Seq data
Single-end reads from different HiSeq2500 runs were processed with a pipeline implemented in work definition language for the Cromwell workflow engine. This pipeline includes sequenced reads trimming for adaptors with cutadapt26 v.1.1.1, and gene expression quantification from identified transcripts using RSEM27 v1.2.20, which internally made alignment of reads against human genome reference GRCh38.82 using bowtie228 v2.2.5. Expected counts from each sequencing run were then joined into a table with 43 186 genes. Genes with expression value in CPM >1 for almost 80% of participants were selected, and 12 062 genes were left for normalization with the TMM method implemented in EdgeR29 Bioconductor package for downstream analysis. Limma30 was used for differential expression analysis adjusting by batch and sex. Ingenuity Pathways Analysis software was used for enrichment of functional terms. The R package GOPlot31 was used for visualization of functional enrichment. Cytoscape software was used for network visualization.
Unbiased plasma proteomics
Unbiased plasma proteomics was performed and analysed as previously described.32 Briefly, plasma proteins were trypsin-digested using the filter-aided protocol, and the resulting peptides were subjected to 10-plex TMT isobaric labelling and fractionated by the high pH reverse phase method. Mass spectrometry was performed by nanoliquid chromatography coupled to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Peptides were identified with a 1% FDR criterion, and proteins were quantified using the WSPP model33 with the SanXoT software package.34 Protein abundance was expressed as standardized log2-ratios ( values). Protein quantifications by mass spectrometry of several proteins (C-reactive protein—CRP, Cystatin-C, and VCAM1) showed a very high correlation with those obtained by ELISA (P < .0001 in all the cases).
Basic statistical analysis
Continuous variables were expressed as mean ± SD [or median (interquartile range) if non-normally distributed] and categorical variables as n (%). Non-normal variables were log-transformed before analysis, and comparisons were performed using the Student’s t-test and the chi-square or Fisher exact test, as appropriate. For comparison across different levels of an ordinal variable (i.e. age acceleration), P-trend values were derived from Jonckheere-Terpstra tests for continuous variables and χ2 for categorical variables test for trend in proportions. Linear regression models were used to assess the univariate association between continuous variables. Logistic regression models were applied for binary outcomes. The models were adjusted for sex, hypertension (systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg, or use of antihypertensive medication), smoking (current smoking status or a lifetime consumption of >100 cigarettes), diabetes (fasting plasma glucose ≥126 mg/dL, or treatment with insulin or oral hypoglycemic medication), dyslipidemia (total cholesterol ≥240 mg/dL, LDL-cholesterol ≥160 mg/dL, HDL-cholesterol <40 mg/dL, or use of lipid-lowering medication), family history of CVD, and obesity (body mass index ≥30 kg/m2).35 Following the definition of the NIH, pack-years were calculated using the self-reported cigarettes smoked per day and the ages at which they started and ceased smoking, following the formula:
where the years smoking is calculated as the difference between the age of smoking cessation and the starting smoking age for past smokers, and the difference between the current age and the starting smoking age for current smokers. Statistical analyses were performed using R and STATA 15.0 (StataCorp, College Station, TX). For all endpoints, P < .05 was considered statistically significant.
Mediation analysis
Model-based causal mediation analysis was performed. In brief, two statistical models were specified: (i) the mediator model for the conditional distribution of the mediator Mi (differentially expressed genes) given the treatment Ti (SA); and (ii) the outcome model for the conditional distribution of the outcome Yi (EAA) given Ti and Mi, (Yi | Ti, Mi). These models were fitted separately, and the fitted objects were then used in the mediate function, which computes the estimated average causal mediation effect (ACME) and other parameters of interest obtained with these models under the sequential ignorability assumption. This supposition is plausible considering the design of the omics cohort, with cases and controls matched based on gender and age. Since the directionality of the association between SA and EAA is unknown, we fitted another model using EAA as treatment (Ti) and SA as outcome (Yi). The sequential ignorability assumption is more difficult to sustain in this case. All genes with a P ≤ .05 in the EAA and SA contrasts were used for the mediation analysis using this model. The ACME P-values were adjusted using Benjamini–Hochberg multiple correction. Genes with an ACME adjusted P ≤ .05 after adjustment for traditional CV risk factors (sex, dyslipidemia, hypertension, obesity, smoker, diabetes, and family history of CVD) were considered as potential mediators between SA and EAA.
Validation cohort: omics data from Multi-Ethnic Study of Atherosclerosis
Publicly available methylomics and transcriptomics data from the Multi-Ethnic Study of Atherosclerosis (MESA)36 were used as external validation. Publicly available methylomics and transcriptomics data from CD14+ cells were retrieved from the GEO database (GSE56045 and GSE56046) for a total of 1193 individuals ranging 44–83 years of age.37 Complete imaging information to quantify SA was available for 1065 of them. Methylomics data were processed as described in the PESA study. The MESA individuals were categorized as having a decelerated, normal, or accelerated Grim EA using the first and third quartile of the distribution of EAA on the whole cohort. Statistical tests were adjusted by a composite variable of the participant’s race, site, and gender.
Results
The presence and extension of subclinical atherosclerosis is associated with epigenetic clocks predictive of lifespan
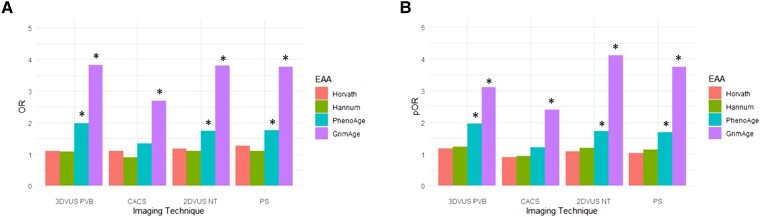
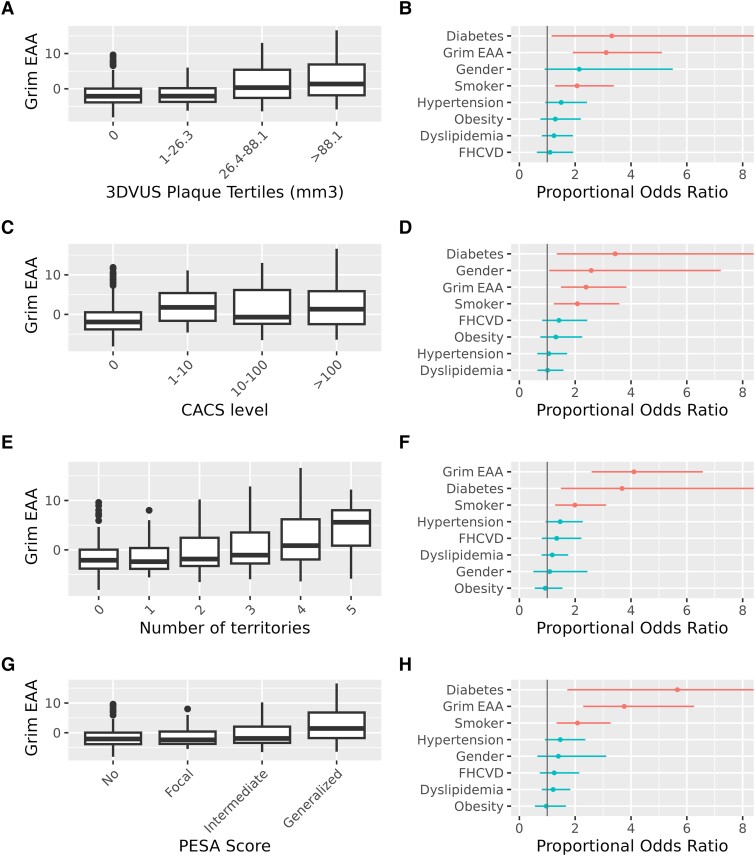
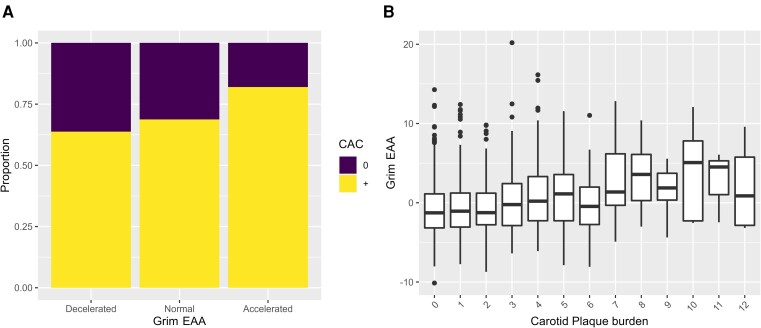
The EA and EAA (i.e. EA adjusted by CA) were estimated from methylomics data generated for 391 PESA participants from the omics cohort (see Supplementary data online, Tables S1 and S2). In particular, the EA of the participants was calculated based on four different epigenetic clocks: Grim13 and Pheno,12 which predict lifespan, vs. Horvath’s10 and Hannum’s11 clocks, which predict CA. The SA presence and extension were associated with Grim EAA and with Pheno EAA based on 2DVUS and 3DVUS plaque measurements (Figure 1) independent of traditional CV risk factors. Moreover, Grim EAA but not Pheno EAA was associated with positive coronary artery calcium score (CACS) and with the extension of CACS, regardless of traditional CV risk factors (Figure 1). The EAA based on the Horvath’s and Hannum’s epigenetic clocks, which are predictive of CA, was not significantly associated either with the presence or with the extension of SA based on the three different imaging techniques. Based on these results, and since Grim EA has been shown to outperform other epigenetic clocks in terms of health and lifespan prediction,13 we further explored its association with different imaging modalities and different ranges of SA. The results confirm an acceleration of Grim EA as SA increase, independent of traditional CV risk factors (Figure 2 and Supplementary data online, Figure S1). Likewise, MESA cohort participants37 with accelerated Grim EA (EAA higher than the third quantile of the distribution) had a two-fold increased risk of having a positive CAC compared with normal aging individuals (Figure 3A, P < .001). An increasing trend in EAA was also observed for carotid plaque score in the MESA study (Figure 3B, P-trend <.001). Interestingly, an analysis of the association of SA with the predicted levels of the eight biomarkers that conform the Grim Epigenetic Clock showed an association of SA with all markers using the 3DVUS plaque burden (see Supplementary data online, Figure S2), with all but Leptin using the PESA score (see Supplementary data online, Figure S3) and with all but ADM, B2M, and leptin using the CAC score (see Supplementary data online, Figure S4). These results confirm the multifaceted association between SA and Grim EAA.
Figure 1.
The presence and extension of subclinical atherosclerosis in Progression of Early Subclinical Atherosclerosis participants is associated with epigenetic clocks predictive of time-to-death. (A) Odds ratio of the logistic regression models for the presence of subclinical atherosclerosis based on different imaging techniques: 3DVUS PVB (positive global plaque burden measured by 3DVUS); coronary artery calcium score (positive calcium score measured using computed tomography, Agatson); 2DVUS NT (at least one territory with plaque detected by 2DVUS) and PS (Progression of Early Subclinical Atherosclerosis score focal, intermediate or generalized vs. No disease). (B) Proportional odds-ratio of the ordinal logistic regression for the following categories of each imaging technique: 3DVUS PVB: Tertiles of 3DVUS global plaque volume as previously described3; CACS (0, 1–10, 10–100, >100); 2DVUS NT (number of territories with 2DVUS atherosclerotic plaque); PS (Progression of Early Subclinical Atherosclerosis Score: no disease, focal, intermediate, and generalized disease). * P < .05. All models were adjusted by diabetes, gender, smoking, dyslipidemia, obesity, family history of CVD, and hypertension. CACS, coronary artery calcium score; EAA, epigenetic age acceleration; OR, odds ratio; pOR, proportional odds-ratio.
Figure 2.
Progression of Early Subclinical Atherosclerosis participants with larger extension of subclinical atherosclerosis have accelerated grim epigenetic age. a–h boxplots (A, C, E, G) and forest plots (B, D, F, H) for the ordinal logistic regression of the extension of subclinical atherosclerosis and Grim epigenetic age acceleration. Subclinical atherosclerosis was assessed using different metrics and imaging techniques: 3DVUS plaque volume (A, B), calcification of coronary arteries by computed tomography (C, D); number of territories with plaques detected by 2DVUS (E, F), and Progression of Early Subclinical Atherosclerosis score (G, H). Models were adjusted by diabetes, gender, smoking, dyslipidemia, obesity, family history of CVD, and hypertension. Jonckheere-Terpstra test for trend <0.001 for all comparisons in panels (A, C, E, G). Forest plots in panels (B, D, F, H) display the proportional Odds Ratio of the ordinal logistic regression model and its 95% Confidence Interval. EAA, epigenetic age acceleration; CACS, coronary artery calcium score; PESA, Progression of Early Subclinical Atherosclerosis.
Figure 3.
Subclinical atherosclerosis association with Grim epigenetic age acceleration in the Multi-Ethnic Study of Atherosclerosis study. (A) Distribution of the proportions of participants with coronary artery calcium score <0.5 (0) or > 0.5 (+) across Grim epigenetic age acceleration categories. (B) Distribution of Grim epigenetic age acceleration across different levels of carotid plaque burden measured as previously described.37 EAA, epigenetic age acceleration; CAC, coronary artery calcium.
Individuals with accelerated grim epigenetic age have an increased cardiovascular age and a higher risk of subclinical atherosclerosis progression
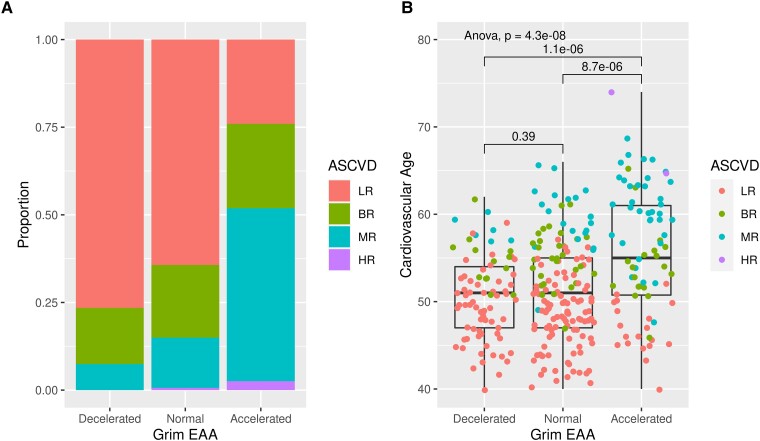
To confirm the prediction of reduced health and lifespan estimated by Grim EAA, we assessed the SA progression and CV risk of individuals with accelerated EA. The PESA participants were categorized in three groups based on their Grim EAA: (i) individuals with accelerated EA (top 25% EAA, i.e. individuals with an EA 2.74 years older than their CA, EAA ≥2.74); (ii) individuals with decelerated Grim EA (bottom 25%, i.e. individuals with their EA 3.38 years younger than their CA, EAA ≤ −3.38); and (iii) individuals with an EA similar to their CA (normal group, −3.38 < EAA < 2.74). Trend tests showed an increasing SA burden as EA accelerates by all imaging techniques (see Supplementary data online, Table S3). Importantly, the percentage of 3-year CAC progressors16 among the individuals with accelerated EA (43.9%) was almost double than in the group of normally aging individuals (24.1%) and almost three times more than among the decelerated individuals (14.3%, Supplementary data online, Table S3, P < .001). The association of EAA with global and CAC progression remained significant after adjustment by CV risk factors, including smoking (see Supplementary data online, Table S4). In general, individuals with an accelerated Grim EA had more CV risk factors than normal and decelerated individuals (Table 1), with a significant increase in the prevalence of diabetes, hypertension, high triglycerides, and low HDL, but not of high LDL and total cholesterol levels (Table 1). Importantly, they also had an increased prevalence of metabolic syndrome (P = .011). Consequently, individuals with accelerated Grim EA showed a significantly increased 10-year risk of CV events based on the ASCVD risk score18 and an increased CV age38 (Figure 4). This association remained significant after adjustment by traditional CV risk factors (see Supplementary data online, Table S5).
Table 1.
Demographics and CVRF of PESA participants comparing grim EAA categories
| Decelerated (n = 98) | Normal (n = 195) | Accelerated (n = 98) | P-trend | |
|---|---|---|---|---|
| Baseline characteristics | ||||
| Age, years | 49 (47–52) | 49 (45–52) | 50 (45–52) | .773 |
| Male, % | 90 (91.8) | 179 (91.8) | 95 (96.9) | .159 |
| BMI, kg/m2 | 26.6 ± 2.852 | 27.1 ± 3.371 | 27.3 ± 3.018 | .075 |
| Obesity (BMI 30 kg/m2), n (%) | 15 (15.3) | 31 (15.9) | 16 (16.3) | .845 |
| SBP, mm Hg | 120.0 ± 12.316 | 121.2 ± 12.406 | 122.7 ± 13.106 | .117 |
| DBP, mm Hg | 74.7 ± 9.352 | 76.2 ± 9.055 | 76.7 ± 9.553 | .256 |
| Total cholesterol, mg/dL | 204.7 ± 31.594 | 204.4 ± 33.21 | 208.1 ± 37.099 | .696 |
| LDL-C, mg/dL | 139.3 ± 28.031 | 138.8 ± 29.437 | 140.5 ± 32.018 | .935 |
| HDL-C, mg/dL | 46.1 ± 11.564 | 44.7 ± 10.53 | 40.8 ± 10.177 | .002 |
| Triglycerides, mg/dL | 79.5 (61.25–118.75) | 92 (68.5–123) | 114 (83.25–152.5) | .001 |
| Glucose, mg/dL | 91 (85–97) | 92 (87.5–99) | 95 (88–101) | .011 |
| Insulin, uU/mL | 5 (3.825–7.775) | 6 (4.15–8.2) | 6.4 (4.2–9.575) | .062 |
| Hemoglobin A1c, % | 5.5 (5.225–5.7) | 5.5 (5.2–5.7) | 5.6 (5.4–5.8) | .002 |
| GOT | 21 (18–24.75) | 20 (17–23) | 19 (17–22) | .029 |
| GPT | 21 (17–32) | 23 (17–30) | 21 (17–30.75) | .991 |
| GGT | 24.5 (17–34.75) | 26 (19–35.5) | 28 (20.25–43.62) | .058 |
| Anti-inflammatory therapy, n (%) | 31 (31.6) | 61 (31.3) | 29 (29.6) | .757 |
| Lipid-lowering therapy, n (%) | 14 (14.3) | 28 (14.4) | 14 (14.3) | 1 |
| Antihypertensive therapy, n (%) | 10 (10.2) | 32 (16.4) | 24 (24.5) | .008 |
| Antidiabetic therapy, n (%) | 1 (1) | 7 (3.6) | 5 (5.1) | .111 |
| Metabolic Syndrome (EU) | 18 (18.4) | 40 (20.5) | 33 (33.7) | .011 |
| Cardiovascular risk factors | ||||
| Dyslipidemia | 61 (62.2) | 121 (62.1) | 75 (76.5) | .035 |
| Total cholesterol ≤240 mg/dL | 15 (15.3) | 29 (14.9) | 18 (18.4) | .557 |
| LDL-C ≤160 mg/dL | 24 (24.5) | 46 (23.6) | 21 (21.4) | .612 |
| HDL-C < 40 mg/dL | 31 (31.6) | 69 (35.4) | 55 (56.1) | <.001 |
| Current smoker | 3 (3.1) | 36 (18.5) | 68 (69.4) | <.001 |
| Past smoker | 34 (34.7) | 90 (46.2) | 25 (25.5) | <.001 |
| Family history of CV disease | 10 (10.2) | 33 (16.9) | 22 (22.4) | .021 |
| Hypertension | 17 (17.3) | 43 (22.1) | 28 (28.6) | .06 |
| Diabetes mellitus | 1 (1) | 8 (4.1) | 7 (7.1) | .031 |
| Cardiovascular Risk Scores | ||||
| FHS 10y | 9.1 ± 4.2 | 10.6 ± 6.1 | 17.3 ± 9.5 | .001 |
| FHS 30y | 21.8 ± 8.9 | 24.9 ± 11.7 | 34.9 ± 13.6 | .001 |
| EU score | 0.8 ± 0.5 | 0.9 ± 0.60 | 1.5 ± 0.9 | .001 |
| ASCVD | 3.8 ± 2.0 | 4.7 ± 3.36 | 8.3 ± 4.7 | .001 |
Values are mean +/− SD, n (%), or median (interquartile range). P trend values are derived from Jonckheere-Terpstra tests for continuous variables and χ2 for categorical variables. Variables showing statistically significant differences are highlighted in bold.
BMI: body mass index; FHS: Framingham Heart Study Score; ASCVD: atherosclerotic cardiovascular disease risk score.
Figure 4.
Progression of Early Subclinical Atherosclerosis individuals with accelerated grim epigenetic age have a higher predicted risk of having a cardiovascular event in 10 years and an increased cardiovascular age predicted by traditional cardiovascular risk scores. (A) Proportion of individuals with low atherosclerotic cardiovascular disease risk, borderline risk, medium risk, and high risk among individuals with accelerated, normal, or decelerated Grim epigenetic age. Chi2 test P < .001. (B) Cardiovascular age derived from the Systematic Coronary Risk Evaluation algorithm calculated for individuals with accelerated, normal, and decelerated Grim epigenetic age (colour coded based on atherosclerotic cardiovascular disease risk score). ANOVA and t-test P-values are displayed. ASCVD, atherosclerotic cardiovascular disease; LR, low risk; BR, borderline risk; MR, medium; HR, high risk.
Accelerated grim epigenetic age is associated with increased systemic inflammation
Hematological analyses unveiled a systemic inflammatory profile in individuals with an accelerated Grim EA (Table 2), with a significant increase in the total number of platelets and white blood cells, including an increase in the number of neutrophils, lymphocytes, monocytes, eosinophils, and basophils. Importantly, biomarkers of systemic inflammation such as fibrinogen, high-sensitivity C-reactive protein, and the neutrophil/lymphocyte ratio also correlated with Grim EAA. A score of low-grade, chronic inflammation, the INFLA score,19 was also found to be significantly associated with Grim EAA (P < .001, Table 2). We next studied the potential association between Grim EAA and vascular inflammation, determined as the number of fluorine-18 fluorodeoxyglucose (18F-FDG) uptakes assessed by positron emission tomography.20 In spite of the low sample size, we found a trend in 18FDG uptakes, with the proportion of individuals with ≥3 uptakes being three-fold larger in the group of participants with accelerated Grim EA than in the normal EAA participants (Table 2, P-trend = .1). All significant associations identified in Table 2 remained significant after adjustment by smoking (see Supplementary data online, Table S6).
Table 2.
Haematological profile and inflammatory markers of PESA participants comparing grim EAA categories
| Decelerated (n = 98) | Normal (n = 195) | Accelerated (n = 98) | P-trend | |
|---|---|---|---|---|
| Erythrocytes | 4.88 (4.64–5.18) | 4.93 (4.68–5.16) | 4.85 (4.64–5.137) | .88 |
| Platelet count | 208.5 (186.25–24) | 216 (190–252) | 229 (202–261) | .001 |
| White blood cell count (num, 103/µL) | 5.28 (4.48–5.99) | 5.45 (4.77–6.32) | 6.98 (5.42–8.35) | .001 |
| Neutrophils (num, 103/µL) | 2.9 (2.52–3.36) | 3.11 (2.64–3.71) | 4.075 (3.09–4.97) | .001 |
| Lymphocytes (num, 103/µL) | 1.66 (1.37–2.01) | 1.71 (1.43–2) | 1.945 (1.56–2.438) | .001 |
| Monocytes (num, 103/µL) | 0.36 (0.322–0.48) | 0.41 (0.35–0.5) | 0.49 (0.39–0.57) | .001 |
| Eosinophils (num, 103/µL) | 0.12 (0.07–0.178) | 0.12 (0.08–0.2) | 0.16 (0.1–0.24) | .001 |
| Basophils (num, 103/µL) | 0.05 (0.04–0.06) | 0.05 (0.04–0.06) | 0.05 (0.05–0.08) | .002 |
| % neutrophils | 56.7 (52.3–60.725) | 56.5 (51.55–60.85) | 59.05 (54.2–64.4) | .012 |
| % monocytes | 7.35 (6.25–8.5) | 7.6 (6.45–8.9) | 7.1 (6–8.3) | .345 |
| % lymphocytes | 32.4 (27.25–36.22) | 31.3 (27.95–36.1) | 28.85 (26.12–34) | .006 |
| % eosinophils | 2.2 (1.4–3.2) | 2.3 (1.4–3.5) | 2.3 (1.6–3.175) | .668 |
| % basophils | 0.9 (0.7–1.2) | 0.9 (0.7–1.2) | 0.9 (0.7–1.1) | .216 |
| Neutrophile/Lymphocyte | 1.75 (1.45–2.28) | 1.805 (1.426–2.166) | 2.039 (1.61–2.48) | .006 |
| RDW | 14.1 (13.62–14.8) | 14.4 (13.8–14.95) | 14.5 (13.92–15.08) | .028 |
| Hemoglobin | 14.9 (14.3–15.6) | 15 (14.3–15.6) | 15.45 (14.7–15.97) | .003 |
| Hematocrit | 44.15 (42.05–46.2) | 44.3 (42.4–46.3) | 44.7 (42.925–46.55) | .1 |
| MCV | 90.55 (87.97–92.4) | 90.3 (88.2–92.75) | 92.1 (89.25–95.07) | .003 |
| Fibrinogen (mg/dL) | 254.3 (231.45–273.32) | 263.9 (238.7–284.2) | 281.85 (243.15–318.92) | .001 |
| Vascular Cell Adhesion Molecule-1 | 641.65 (531.55–803.02) | 609.7 (478.25–754.1) | 621.2 (479.12–747.6) | .079 |
| P-Selectin | 141.75 (109.92–163.5) | 135.2 (112.4–160.9) | 147.05 (111.4–177.75) | .245 |
| Oxidized low density lipoprotein | 51.355 (41.35–65.98) | 49.8 (41.23–61.31) | 56.51 (42.52–68.325) | .195 |
| Serum lipoprotein(a) | 14.4 (5.505–39.47) | 19.7 (8.305–39.9) | 17.25 (7.66–33.85) | .809 |
| INFLA SCORE PESA | −2 (−5–1.75) | −1 (−4–4) | 4 (1–9) | <.001 |
| Hs C-reactive protein | 0.08 (0.05–0.15) | 0.11 (0.06–0.18) | 0.16 (0.09–0.29) | .001 |
| FDG-PET uptakes ≥3 | 1 (1) | 7 (3.6) | 9 (9.2) | .109 |
Data are expressed as mean mean ± SD or median (IQR) when appropriate. P-trend values are derived from Jonckheere-Terpstra tests for continuous variables and χ2 for categorical variables. Variables with statistically-significant differences are highlighted in bold.
RDW: red cell distribution width; MCV: mean corpuscular volume.
Transcriptomics and proteomics data reveal specific immune response pathways dysregulated in individuals with accelerated epigenetic age
In order to deepen our understanding of the mechanisms underlying the association between EAA and SA, RNA-Seq data from peripheral blood cells of the 391 PESA participants with methylomics data were analysed, to compare gene expression in individuals with accelerated EA vs. individuals with non-accelerated EA. We found 737 differentially expressed genes between both groups with an adjusted P ≤ .05 (see Supplementary data online, Table S7). The top canonical pathways associated with an accelerated EA were related to immune response (see Supplementary data online, Table S7), including the Th1 and Th2 activation pathways, STAT3 pathway, inflammasome, IL-10, and Toll-like receptor signalling pathway. Proteomics data on the same PESA sub-cohort revealed several inflammatory biomarkers also dysregulated between accelerated and non-accelerated individuals at the protein level (see Supplementary data online, Table S7), including HP (logFC = 1.66, adj-P = 7 × 10−6), CRP (logFC = 1.35, adj-P = .004) and VCAM1 (logFC = −0.35, adj-P = .06), consistent with the results of the hemogram. Other proteins involved in B-cell signalling (IGHA2) and adaptive and innate immune system responses (S100A8, S100A9, LRG1, and PTPRJ) were found to be up-regulated in accelerated compared with non-accelerated individuals using both omics techniques.
Deconvolution of methylomics data identified a dysregulation of cellular types consistent with these genes and proteins (see Supplementary data online, Figure S5) and with previous reports.13 We found a significant negative association of Grim EAA with CD8 naïve cells (P = .0027), with CD8+ T cells (P = .006), and with NK cells (P = .0004), and a decreasing trend with B-cells (P = .083). On the other hand, there was a very significant positive association with granulocytes (P = .015), with CD4 naïve (P = .015), and with plasma blasts (P = .0024). The latter express IGHA2, identified by transcriptomics and proteomics as one of the top molecules associated with EAA (logFC = 1.03 adj-P = 9.8 × 10−7 in transcriptomics and logFC = 0.70, adj-P = .097 in proteomics Supplementary data online, Table S7).
Innate and adaptive immune pathways are common underlying mechanisms between accelerated epigenetic age and subclinical atherosclerosis
We then investigated gene expression changes in peripheral blood cells associated with generalized SA. IGHA2, ORM1, MCEMP1, F5, MFHAS1, GPR55, SCART1, and LPCAT3 were significantly associated with SA extension at an adj-P ≤ .05 at the transcriptional level (see Supplementary data online, Table S8). These genes were also associated with Grim EAA. Moreover, IGHA2 was also identified as differentially expressed in plasma at the protein level (see Supplementary data online, Table S8) and among the top biomarkers associated with EAA both using transcriptomics and proteomics data. LRRN3, the most differentially expressed gene between accelerated and non-accelerated individuals, was among the top dysregulated genes in individuals with generalized SA (logFC = 0.47, adj-P = .07).
In fact, the top 100 most significant genes associated with generalized SA were enriched in some of the pathways linked to EAA, such as phagosome, B cell receptor signalling, IL-15 signalling, IL-10 signalling, and Toll-like receptor signalling. Similar results were obtained with the proteomics analysis, which identified IL-15 signalling and the overlapping B cell receptor signalling, and communication between innate and adaptive immune cell pathway as the three most enriched pathways within the top 100 proteins differentially expressed between PESA participants with generalized SA and participants without any disease (see Supplementary data online, Table S8). The genes and proteins associated with generalized SA were concordant with the results of the cellular types identified by the deconvolution of methylomics data (see Supplementary data online, Figure S6). Similar to the results for EAA, the number of CD8 naïve cells was also inversely correlated with SA extension (P = .0014). In contrast, there was a trend for more CD4 naïve cells (P = .14) and for more CD45RA-CD28-CD8+ (P = .17) in individuals with generalized subclinical atherosclerosis, who also had significantly more plasma blasts than individuals with no disease (P = .066). In sum, innate and adaptive immune pathways, and the molecules involved in their communication seem to be common underlying mechanisms between SA and accelerated EA.
Key pro-inflammatory cytokines mediate the association between subclinical atherosclerosis and accelerated epigenetic age
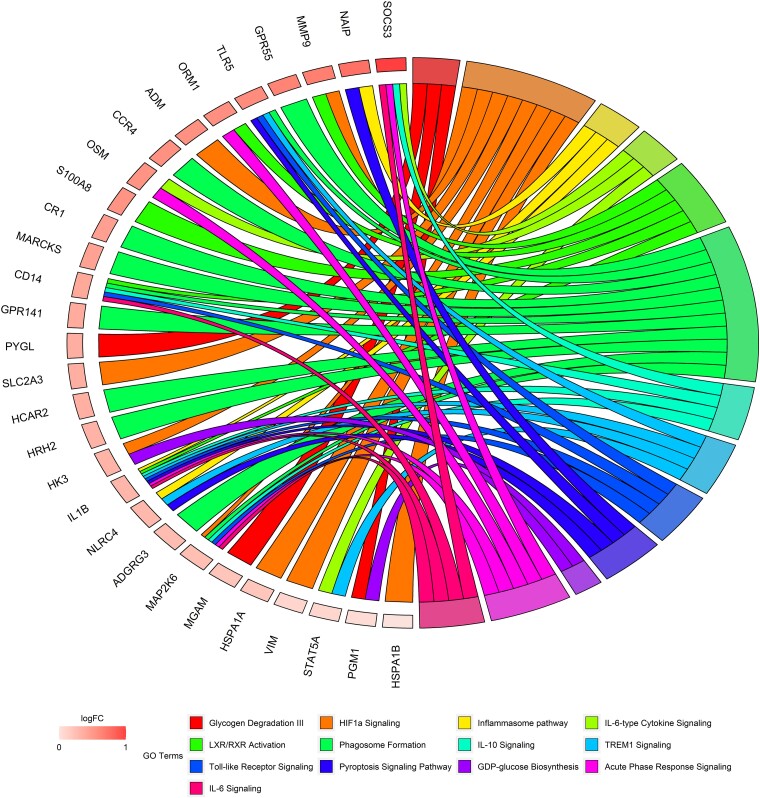
Mediation analysis was used to disentangle the molecular pathways potentially mediating the relationship between generalized SA and accelerated EA in PESA participants. Genes differentially expressed (P ≤ .05) between individuals with generalized SA and with no disease and also between individuals with accelerated vs. normal Grim EA (shared number of genes = 393) were tested as potential mediators between these two processes. Mediation analysis identified 114 genes as potential mediators of the effect of SA on EAA after adjustment by traditional CV risk factor (ACME adj-P ≤ .05, Supplementary data online, Table S9). Among them, we identified several pro-inflammatory markers associated with IL10 signalling (CD14, IL1B, MAP2K6, and SOCS3), the inflammasome pathway (IL1B, NAIP, and NLRC4), TREM1, and the Toll-like receptor pathway (IL1B, NLRC4, STAT5A, and TLR5) (Figure 5, Supplementary data online, Table S9). The same analysis using proteomics data identified several immune-related proteins (PIGR and IGHA2) and a key pro-inflammatory marker (HP) as mediators between SA and EAA. We then investigated the regulatory network controlling the expression of mediator genes. Among the upstream regulators identified, IL1B, CLEC10A, and OSM were also mediators themselves (see Supplementary data online, Table S9). We also tested the hypothesis that EAA could have an effect on subclinical atherosclerosis. Interestingly, only four genes (CD14, F5, LMNA, and ANXA11) and no protein were identified as significant mediators of the effect of EAA on SA. These findings were confirmed using the INFLA score.20 While there was no evidence of the INFLA score being a mediator of EAA on SA (P = .32), the same score was found to be a partial mediator between SA initiation and EAA (proportion explained = 12%; P = .04), suggesting that the chronic inflammation triggered by SA may have a detrimental role in health and lifespan.
Figure 5.
Transcriptomics analysis identifies inflammatory pathways as mediators between subclinical atherosclerosis and accelerated grim epigenetic age. ChordPlot31 of the most representative genes from the top 10 most enriched canonical pathways enriched in mediator genes after adjustment by cardiovascular risk factors. Data extracted from Supplementary data online, Table S9. logFC represents changes associated with EAA (online Table S7).
Discussion
In this study, we used several epigenetic clocks to investigate the interplay between EAA and subclinical atherosclerosis. Importantly, epigenetic clocks predictive of health and lifespan12,13 (i.e. time-to-disease or time-to-death) were strongly associated with the presence, extension, and progression of SA (Structured Graphical Abstract). In contrast, the epigenetic predictors of CA10,11 did not add any further information on the association between SA and CA. The association between SA and Grim EAA was confirmed using three different imaging modalities to characterize SA and validated in CD14+ methylomics data from the MESA study.37 Moreover, we found that individuals with accelerated Grim EA have an increased CV age38 and a higher risk of 3-year CAC progression even after adjustment for CV risk factors, in line with previous findings in an independent cohort.39
Interestingly, we found that individuals with an accelerated EA had a pro-inflammatory profile, characterized by elevated plasma C-reactive protein level, neutrophil/lymphocyte ratio, white blood cells amount—previously identified as a key predictor of SA extension40—and an increased INFLA score.19 Importantly, mediation analysis showed that the INFLA score partly mediates the effect of SA on EAA (12%), confirming recent reports about the key role of inflammation not only in the residual risk of atherosclerosis in individuals who have suffered a CV event41 but also in plaque formation in healthy individuals with bone marrow activation in response to metabolic syndrome.42 In contrast with previous reports,43 we found no evidence of increased lymphopenia with accelerated EA or with increased SA burden, probably due to the age and fitness of the participants recruited for the PESA study.
Mediation analysis using transcriptomics and proteomics data showed that key pro-inflammatory molecules such as IL1B and OSM regulate other mediator genes involved in the IL-10 and inflammasome pathways such as NAIP, NLRC4, STAT5A, and TLR5. A study using the same anti-IL1β treatment to that used in the CANTOS trial41 showed an improvement in peripheral artery disease with little impact on plaque burden,44 suggesting that inflammation might be a mediator of the pathological effects of atherosclerosis. Additionally, OSM has been reported to be expressed in atherosclerotic lesions45 and to play a role in atherosclerosis progression and survival in humans.46 These findings, together with the down-regulation in our data of NR4A1 in individuals with SA (logFC = −0.27, adj-P = .24) and in those with EAA (logFC = −0.39, adj-P < .001), might indicate an imbalance in the Th17/Tregs ratio, which has been previously associated with the development of atherosclerosis.47
Consistent with these results, deconvolution of the methylomics data revealed changes in the proportions of different types of T and B cells. Interestingly, we observed an increased number of plasma blast cells in individuals with accelerated EA and in those with SA. One of the main mediators identified both by proteomics and transcriptomics analysis (IGHA2) is secreted by plasma blasts. This is an interesting finding since plasma B cells play a crucial role in the maintenance of intestinal homeostasis, and the role of an individual’s microbiome in atherosclerosis development is being currently explored.
In summary, we have identified a robust association between the presence, extension, and progression of SA and an acceleration in the Grim EA, partly mediated by chronic, low-grade inflammation, triggered by inflammatory cytokines. Although these findings suggest a relevant role of inflammation in the detrimental impact of SA on health, in line with previous reports,41,44 we cannot rule out the existence of confounders or of other mediators, in view of the percentage of mediation explained by the INFLA score and due to the use of blood and plasma samples in the omics experiments. Moreover, the nature of this study does not allow to establish a causal effect between SA and EAA. While the mediation analysis suggests a directionality, more longitudinal studies with a longer follow-up and more experimental data are needed to further characterize the actual effect of SA on health and lifespan and to elucidate the directionality between both processes.
Supplementary Material
Acknowledgements
We would like to thank Jesús Molina, Sergio Cardenas and Iñaki Polo for database development and maintenance and all Bioinformatics Unit for support. We would also like to thank Almudena R. Ramiro and Pilar Martín for insightful discussions on the deconvolution data.
Contributor Information
Fátima Sánchez-Cabo, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Valentín Fuster, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; The Zena and Michael A. Wiener Cardiovascular Institute/Marie-Josée and Henry R. Kravis Center for Cardiovascular Health, Mount Sinai School of Medicine, One Gustave L. Levy. Place, New York, NY 10029, USA.
Juan Carlos Silla-Castro, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Gema González, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Erika Lorenzo-Vivas, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Rebeca Alvarez, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Sergio Callejas, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Alberto Benguría, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Eduardo Gil, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Estefanía Núñez, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Belén Oliva, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
José María Mendiguren, Banco de Santander, Av. de Cantabria, s/n, 28660 Boadilla del Monte, Madrid, Spain.
Marta Cortes-Canteli, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Cardiology, IIS-Fundación Jiménez Díaz Hospital, Av. de los Reyes Católicos, 2, 28040 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Neurodegenerativas (CIBERNED), Madrid, Spain.
Héctor Bueno, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Cardiology Department, Hospital Universitario 12 de Octubre and Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), Avda. de Córdoba, s/n 28041 Madrid, Spain.
Vicente Andrés, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Jose María Ordovás, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Precision Nutrition and Obesity Research Program, IMDEA Food Institute, CEI UAM + CSIC, Carr. de Canto Blanco, nº 8 E, 28049 Madrid, Spain; U.S. Department of Agriculture Human Nutrition Research Center on Aging, Tufts University, 711 Washington Street, Boston, MA 02111, USA.
Leticia Fernández-Friera, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain; HM Hospitales–Centro Integral de Enfermedades Cardiovasculares HM CIEC, Av. de Montepríncipe, 25, 28660 Boadilla del Monte, Madrid, Spain.
Antonio J Quesada, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain.
Jose Manuel Garcia, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Hospital Universitario Central de Oviedo, Av. Roma, s/n, 33011 Asturias, Spain.
Xavier Rossello, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain; Hospital Universitari Son Espases-IDISBA, Carretera de Valldemossa, 79, 07120 Palma de Mallorca, Mallorca, Islas Baleares (Balearic Islands), Spain.
Jesús Vázquez, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Ana Dopazo, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Antonio Fernández-Ortiz, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain; Hospital Clínico San Carlos, Calle del Prof Martín Lagos, S/N, 28040 Madrid, Spain.
Borja Ibáñez, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain; Cardiology, IIS-Fundación Jiménez Díaz Hospital, Av. de los Reyes Católicos, 2, 28040 Madrid, Spain.
Jose Javier Fuster, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Enrique Lara-Pezzi, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Melchor Fernández Almagro, 3, 28029 Madrid, Spain; Centro de Investigacion Biomedica en Red en Enfermedades Cardiovasculares (CIBERCV), Spain.
Supplementary data
Supplementary data is available at European Heart Journal online.
Pre-registered clinical trial number
Pre-registered clinical trial number: NCT01410318.
Ethical approval
The study protocol was approved by the Carlos III Health Institute Ethics Committee, and all participants provided written informed consent.
Data availability
The data underlying this article are available in Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/ and can be accessed with accession numbers GSE220622 and GSE221615.
Funding
The PESA study is co-funded by the Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid, Spain, and Banco Santander, Madrid, Spain. The study also receives funding from the Instituto de Salud Carlos III (PI15/02019, PI17/00590, and PI20/00819) and the European Regional Development Fund (ERDF) ‘Una manera de hacer Europa’. The CNIC is supported by the Instituto de Salud Carlos III (ISCIII), the Ministerio de Ciencia e Innovación (MCIN) and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S funded by MCIN/AEI/10.13039/501100011033).
References
- 1. Fernández-Ortiz A, Jiménez-Borreguero LJ, Peñalvo JL, Ordovás JM, Mocoroa A, Fernández-Friera L, et al. The Progression and Early detection of Subclinical Atherosclerosis (PESA) study: rationale and design. Am Heart J 2013;166:990–998. 10.1016/j.ahj.2013.08.024 [DOI] [PubMed] [Google Scholar]
- 2. Fernández-Friera L, Peñalvo JL, Fernández-Ortiz A, Ibañez B, López-Melgar B, Laclaustra M, et al. Prevalence, vascular distribution, and multiterritorial extent of subclinical atherosclerosis in a middle-aged cohort: the PESA (Progression of Early Subclinical Atherosclerosis) study. Circulation 2015;131:2104–2113. 10.1161/CIRCULATIONAHA.114.014310 [DOI] [PubMed] [Google Scholar]
- 3. López-Melgar B, Fernández-Friera L, Oliva B, García-Ruiz JM, Peñalvo JL, Gómez-Talavera S, et al. Subclinical atherosclerosis burden by 3D ultrasound in mid-life: the PESA study. J Am Coll Cardiol 2017;70:301–313. 10.1016/j.jacc.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 4. Sniderman AD, Furberg CD. Age as a modifiable risk factor for cardiovascular disease. Lancet 2008;371:1547–1549. 10.1016/S0140-6736(08)60313-X [DOI] [PubMed] [Google Scholar]
- 5. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 2013;153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamczyk MR, Nevado RM, Barettino A, Fuster V, Andrés V. Biological versus chronological aging: JACC focus seminar. J Am Coll Cardiol 2020;75:919–930. 10.1016/j.jacc.2019.11.062 [DOI] [PubMed] [Google Scholar]
- 7. Weinhold B. Epigenetics: the science of change. Environ Health Perspect 2006;114:A160–A167. 10.1289/ehp.114-a160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schübeler D. Function and information content of DNA methylation. Nature 2015;517:321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 9. Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell 2015;14:924–932. 10.1111/acel.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda SV, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49:359–367. 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–591. 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11:303–327. 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levine ME. Assessment of epigenetic clocks as biomarkers of aging in basic and population research. J Gerontol A Biol Sci Med Sci 2020;75:463–465. 10.1093/gerona/glaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018;19:371–384. 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 16. López-Melgar B, Fernández-Friera L, Oliva B, García-Ruiz JM, Sánchez-Cabo F, Bueno H, et al. Short-term progression of multiterritorial subclinical atherosclerosis. J Am Coll Cardiol 2020;75:1617–1627. 10.1016/j.jacc.2020.02.026 [DOI] [PubMed] [Google Scholar]
- 17. Conroy RM, Pyörälä K, Fitzgerald AP, Sans S, Menotti A, de Backer G, et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J 2003;24:987–1003. 10.1016/S0195-668X(03)00114-3 [DOI] [PubMed] [Google Scholar]
- 18. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines. Circulation 2019;140:e596–e646. 10.1161/CIR.0000000000000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonaccio M, di Castelnuovo A, Pounis G, de Curtis A, Costanzo S, Persichillo M, et al. A score of low-grade inflammation and risk of mortality: prospective findings from the Moli-sani study. Haematologica 2016;101:1434–1441. 10.3324/haematol.2016.144055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández-Friera L, Fuster V, López-Melgar B, Oliva B, Sánchez-González J, Macías A, et al. Vascular inflammation in subclinical atherosclerosis detected by hybrid PET/MRI. J Am Coll Cardiol 2019;73:1371–1382. 10.1016/j.jacc.2018.12.075 [DOI] [PubMed] [Google Scholar]
- 21. Müller F, Scherer M, Assenov Y, Lutsik P, Walter J, Lengauer T, et al. Rnbeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol 2019;20:55. 10.1186/s13059-019-1664-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450 K methylation array data. BMC Genomics 2013;14:293. 10.1186/1471-2164-14-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–1361369. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA Methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86. 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 2016;17:171. 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 27. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods 2012;9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson MD, McCarthy DJ, Smyth GK. Edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-Seq read counts. Genome Biol 2014;15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912–2914. 10.1093/bioinformatics/btv300 [DOI] [PubMed] [Google Scholar]
- 32. Núñez E, Fuster V, Gómez-Serrano M, Valdivielso JM, Fernández-Alvira JM, Martínez-López D, et al. Unbiased plasma proteomics discovery of biomarkers for improved detection of subclinical atherosclerosis. EBioMedicine 2022;76:103874. 10.1016/j.ebiom.2022.103874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Navarro P, Trevisan-Herraz M, Bonzon-Kulichenko E, Núñez E, Martínez-Acedo P, Pérez-Hernández D, et al. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res 2014;13:1234–1247. 10.1021/pr4006958 [DOI] [PubMed] [Google Scholar]
- 34. Trevisan-Herraz M, Bagwan N, García-Marqués F, Rodriguez JM, Jorge I, Ezkurdia I, et al. Sanxot: a modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics 2019;35:1594–1596. 10.1093/bioinformatics/bty815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rossello X, Fuster V, Oliva B, Sanz J, Fernández Friera LA, López-Melgar B, et al. Association between body size phenotypes and subclinical atherosclerosis. J Clin Endocrinol Metab 2020;105:3734–3744. 10.1210/clinem/dgaa620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–881. 10.1093/aje/kwf113 [DOI] [PubMed] [Google Scholar]
- 37. Liu Y, Reynolds LM, Ding J, Hou L, Lohman K, Young T, et al. Blood monocyte transcriptome and epigenome analyses reveal loci associated with human atherosclerosis. Nat Commun 2017;8:393. 10.1038/s41467-017-00517-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cuende JI, Cuende N, Calaveras-Lagartos J. How to calculate vascular age with the SCORE project scales: a new method of cardiovascular risk evaluation. Eur Heart J 2010;31:2351–2358. 10.1093/eurheartj/ehq205 [DOI] [PubMed] [Google Scholar]
- 39. Joyce B, Gao T, Zheng Y, Ma J, Hwang SJ, Liu L, et al. Epigenetic age acceleration reflects long-term cardiovascular health. Circ Res 2021;129:770–781. 10.1161/CIRCRESAHA.121.318965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sánchez-Cabo F, Rossello X, Fuster V, Benito F, Manzano JP, Silla JC, et al. Machine learning improves cardiovascular risk definition for young, asymptomatic individuals. J Am Coll Cardiol 2020;76:1674–1685. 10.1016/j.jacc.2020.08.017 [DOI] [PubMed] [Google Scholar]
- 41. Matsuura Y, Kanter JE, Bornfeldt KE. Highlighting residual atherosclerotic cardiovascular disease risk. Arterioscler Thromb Vasc Biol 2019;39:e1–e9. 10.1161/ATVBAHA.118.311999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Devesa A, Lobo-González M, Martínez-Milla J, Oliva B, García-Lunar I, Mastrangelo A, et al. Bone marrow activation in response to metabolic syndrome and early atherosclerosis. Eur Heart J 2022;43:1809–1828. 10.1093/eurheartj/ehac102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zidar DA, Al-Kindi SG, Liu Y, Krieger NI, Perzynski AT, Osnard M, et al. Association of lymphopenia with risk of mortality among adults in the US general population. JAMA Netw Open 2019;2:e1916526. 10.1001/jamanetworkopen.2019.16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Russell KS, Yates DP, Kramer CM, Feller A, Mahling P, Colin L, et al. A randomized, placebo-controlled trial of canakinumab in patients with peripheral artery disease. Vasc Med 2019;24:414–421. 10.1177/1358863X19859072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Albasanz-Puig A, Murray J, Preusch M, Coan D, Namekata M, Patel Y, et al. Oncostatin M is expressed in atherosclerotic lesions: a role for Oncostatin M in the pathogenesis of atherosclerosis. Atherosclerosis 2011;216:292–298. 10.1016/j.atherosclerosis.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 46. van Keulen D, Pouwer MG, Emilsson V, Matic LP, Pieterman EJ, Hedin U, et al. Oncostatin M reduces atherosclerosis development in APOE*3Leiden.CETP mice and is associated with increased survival probability in humans. PLoS One 2019;14:e0221477. 10.1371/journal.pone.0221477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsilingiri K, de la Fuente H, Relaño M, Sánchez-Díaz R, Rodríguez C, Crespo J, et al. Oxidized low-density lipoprotein receptor in lymphocytes prevents atherosclerosis and predicts subclinical disease. Circulation 2019;139:243–255. 10.1161/CIRCULATIONAHA.118.034326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/ and can be accessed with accession numbers GSE220622 and GSE221615.