Abstract
The outer envelope of the extracellular form of vaccinia virus is derived from Golgi membranes that have been modified by the insertion of specific viral proteins, of which the major component is the 37-kDa, palmitylated, nonglycosylated product of the F13L gene. The F13L protein contains a variant of the HKD (His-Lys-Asp) motif, which is conserved in numerous enzymes of phospholipid metabolism. Vaccinia virus mutants with a conservative substitution of either the K (K314R) or the D (D319E) residue of the F13L protein formed only tiny plaques similar to those produced by an F13L deletion mutant, were unable to produce extracellular enveloped virions, and failed to mediate low-pH-induced fusion of infected cells. Membrane-wrapped forms of intracellular virus were rarely detected in electron microscopic images of cells infected with either of the mutants. Western blotting and pulse-chase experiments demonstrated that the D319E protein was less stable than either the K314R or wild-type F13L protein. Most striking, however, was the failure of either of the two mutated proteins to concentrate in the Golgi compartment. Palmitylation, oleation, and partitioning of the F13L protein in Triton X-114 detergent were unaffected by the K314R substitution. These results indicated that the F13L protein must retain the K314 and D319 for it to localize in the Golgi compartment and function in membrane envelopment of vaccinia virus.
Vaccinia virus (VV), the most intensively studied member of the Orthopoxvirus genus of the family Poxviridae, was previously used as a smallpox vaccine and is presently a vector in recombinant vaccine strategies to prevent infectious diseases and to treat cancer (15, 30). The characteristic features of orthopoxviruses are a double-stranded DNA genome of approximately 200,000 bp, a cytoplasmic site of replication, temporally regulated gene expression, and a complex process of morphogenesis involving multiple viral membranes (29). VV morphogenesis is the subject of this study.
VV forms several types of morphogenetically related particles. The intracellular mature virions (IMV) have two closely apposed outer membranes (45) and are infectious when released by cell lysis. To exit from an intact cell, the IMV undergo a second wrapping event and acquire two additional membrane layers from the trans-Golgi cisternae (20, 22, 41). The four membrane intracellular enveloped virions (IEV) then migrate to the cell periphery, where the outermost viral membrane is lost during fusion with the plasma membrane (22, 28). The externalized particles, with three remaining membranes, may be released into the medium as extracellular enveloped virions (EEV) or remain attached as cell-associated extracellular enveloped virions (CEV) (5, 22, 28, 34, 41).
Since enveloped virus particles are primarily responsible for the cell-to-cell spread of infection (1, 4, 7, 33, 47), the wrapping of virus particles is of considerable interest. The following six genes have been identified as encoding proteins that are specifically incorporated into the outer envelope: A56R (35, 44), F13L (21), B5R (13, 24), A36R (31), A34R (11), and A33R (38). Information regarding the roles of the individual envelope proteins was obtained by deletion of the cognate genes. Mutants with deleted A33R, F13L, B5R, A36R, or A34R genes produced plaques that were much smaller than those of wild-type (WT) virus, and those tested were attenuated in vivo (4, 23, 27, 31, 39, 48). The block in virus spread may be attributed to (i) a block in the wrapping of IMV to form IEV (e.g., the F13L and B5R genes) (4, 14, 48), (ii) a reduction in the amount of EEV released (e.g., the A36R gene) (31), (iii) a decrease in the infectivity of the EEV (A34R gene) (27), or (iv) a lack of formation of specialized actin containing microvilli that propel virus particles to the surface of infected cells (the A33R, A34R, and A36R genes) (39, 40, 49, 50).
The WT F13L gene encodes a 37-kDa polypeptide that localizes to the trans-Golgi membranes where the IMV are wrapped to form IEV (41). The membrane association is mediated via palmitylation of one or both of two adjacent cysteine residues (18, 43). The F13L gene is highly conserved in poxviruses, and a homolog is present in the distantly related molluscum contagiosum virus (3, 9), consistent with its important role in the virus life cycle. Interestingly, the F13L protein contains two 16-amino-acid HKD motifs (named for the most highly conserved residues), conserved in phospholipases and phospholipid synthases (9, 26, 37, 46), and possesses a broad-spectrum lipase activity (2). We reported previously that virus mutants containing F13L protein with a substitution of lysine to arginine at position 314 (K314R) or aspartic acid to glutamic acid at position 319 (D319E) produced tiny plaques similar to those of the F13L deletion (F13LΔ) mutant (46). Here, we have described the effects of these two independent mutations in the HKD motif of the VV F13L protein on the VV life cycle. The small-plaque phenotype was due to a block in the wrapping of IMV to form IEV. The data suggested that mutations of the HKD motif inhibited morphogenesis due to changes in membrane localization of the F13L protein.
MATERIALS AND METHODS
Cells and antibodies.
VV stocks were prepared in HeLa cells as described previously (12). BS-C-1 cells served for plaque assays, immunoprecipitation, and Western blotting experiments, and RK13 cells were used to propagate VV for CsCl purification. Cells were grown in Eagle’s minimal essential medium with 10% fetal bovine serum (FBS), and infections were carried out with 2.5% FBS. For titration of infectious VV and analysis of plaque size, monolayers were fixed and stained with 0.1% crystal violet in 20% ethanol. Mouse monoclonal antibody (MAb) 20 (B5R specific) and MAb 4 (A33R specific) were previously described (32). The F13L peptide RLVETLPENMDFRSDHLTTFEC (representing amino acids 14 to 35 of the F13L protein) conjugated to keyhole limpet hemocyanin was injected into rabbits to produce anti-F13L antibody.
Construction of viruses with F13L point mutations.
The construction and purification of viruses with K314R or D319E point mutations in the F13L gene and of a control virus with WT F13L sequence (F13L+) have been previously described (46).
Immunoprecipitation and Western blotting.
Immunoprecipitates and Western blots were made essentially as described previously (38).
Triton X-114 partitioning.
Triton X-114 partitioning was performed as previously described (38). After partitioning, the detergent and aqueous phases were made equal in volume, detergent, and salt concentration and analyzed by Western blotting or immunoprecipitation and autoradiography.
Palmitate and oleate labeling of VV proteins.
Subconfluent monolayers of BS-C-1 cells were infected with virus at a multiplicity of 10 for 5 h. Medium was then replaced with Eagle’s minimal essential medium (no serum) with 200 μCi of [9, 10-3H(N)]palmitic acid in 10 μl of dimethyl sulfoxide or 100 μCi of [9, 10-3H(N)]oleaic acid in 20 μl of ethanol per well. After 24 h, medium was removed and centrifuged to pellet cells, which were then combined with cells scraped from wells. The cells were lysed, and F13L proteins were immunoprecipitated, solubilized in buffer containing 4% sodium dodecyl sulfate (SDS), 15 mM dithiothreitol, 0.05 M Tris base, 10% glycerol, and 0.1% bromphenol blue, heated at 80°C for 3 min, analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon-P membrane (Millipore, Bedford, Mass.), and exposed to Kodak Biomax MS film by using the Biomax TranScreen Low Energy intensifying screen (Kodak, Rochester, N.Y.).
Syncytium formation.
Confluent BS-C-1 cell monolayers were infected with virus at a multiplicity of 10 for 2 h, washed, and incubated in medium for an additional 10 h as described previously (4, 39). Cells were washed and treated with fusion buffer [phosphate-buffered saline with 10 mM 2-(N-morpholino)ethanesulfonic acid and 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] at pH 5.5 or 7.4 for 2 min at 37°C. Afterwards, fusion buffer was replaced with medium, and the cells were incubated at 37°C and then observed by phase-contrast microscopy.
Virus purification.
Wrapped and unwrapped virus particles were purified on the basis of their buoyant densities in CsCl gradients as described previously (38). The refractive index of the CsCl from samples was measured to verify correct collection of wrapped and unwrapped virus particles.
Electron microscopy.
For transmission electron microscopy, RK13 cells in 60-mm-diameter dishes were infected with VV at a multiplicity of 10 for 24 h, fixed in 2% glutaraldehyde, and embedded in Embed-812 (Electron Microscopy Sciences, Fort Washington, Pa.) or fixed with increasing concentrations of paraformaldehyde and prepared for immunoelectron microscopy as previously described (49). Thawed cryosections were incubated with either rabbit antibody to an F13L peptide or MAb 20, which recognizes the B5R protein (32). The samples were washed, incubated with gold particles conjugated to protein A (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands), and viewed with a Philips CM 100 electron microscope.
Immunofluorescence microscopy.
Fluorescence microscopy was performed as described previously (49). Infected HeLa cell monolayers on coverslips were washed with phosphate-buffered saline, fixed in 3% paraformaldehyde, and permeabilized with 0.05% saponin (Calbiochem, San Diego, Calif.). Samples were stained with rabbit anti-F13L antibody or MAb 4 to the VV A33R protein (32) as a control, and then with rhodamine-conjugated swine anti-rabbit or anti-mouse antibody (Dako Corporation, Carpinteria, Calif.). Samples were viewed and images were collected using a Zeiss Axiophot, and photographs were taken at identical exposure times.
RESULTS
Mutations of the F13L HKD motif cause a small-plaque phenotype.
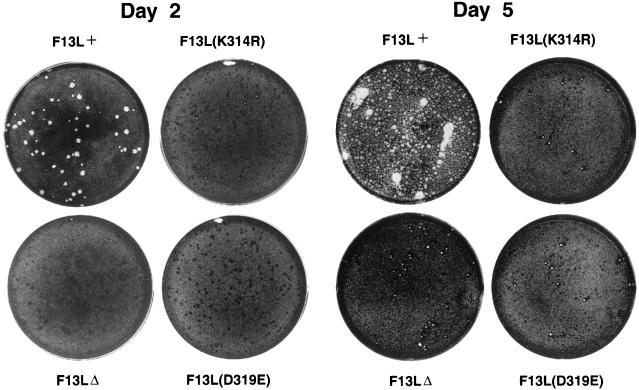
The VV F13L protein contains two previously identified 16-amino-acid HKD motifs at positions 118 to 133 and 312 to 327 (9, 26, 37). The second VV F13L HKD motif is more conserved than the first and is the focus of this study. This sequence has an N replacing the H in position 1 of the HKD motif. Therefore, because the K at amino acid 314 and D at 319 are the most highly conserved residues, we decided to mutate these two amino acids (46) to determine if this motif might be important for the function of the F13L protein in the VV life cycle. Viruses containing F13L point mutations, K314R and D319E, made barely visible plaques after 48 h (46). Here we compared the sizes of the plaques made by F13L(K314R) and F13L(D319E) mutants to those of a control virus engineered for these studies, F13L+ (with a WT copy of the F13L gene and the lacZ gene which is also present in the mutants), and the F13LΔ mutant (Fig. 1). On day 2, mutant virus infection was observed macroscopically as tiny foci, probably due to VV growth factor-induced cell proliferation (8). By day 5, tiny plaques were visible. Significantly, the plaques of the K314R and D319E mutants were similar to those of F13LΔ, suggesting that the function of the F13L protein was completely abrogated. In contrast, the plaques of F13L+ were quite large on day 2 and had spread extensively by day 5. It is apparent that the F13L HKD mutants do not form the diffuse elongated plaques, or comets, that some viruses with mutated EEV proteins produce (even though the parental WR [Western Reserve] virus does not form comets) and which are associated with increased release of wrapped virus particles into the medium (5, 27, 39). The results demonstrated the importance of the F13L HKD motif for the function of the F13L protein in cell-to-cell spread of VV.
FIG. 1.
Appearance of plaques formed by F13L mutants. BS-C-1 cell monolayers were infected with F13L+, F13L(K314R), F13LΔ, or F13L(D319E) virus. After 2 or 5 days, media were removed, and the monolayers were fixed and stained with 0.1% crystal violet in 20% ethanol. At day 2, each well contained more than 50 foci of infection or plaques.
Expression of the mutated F13L proteins.
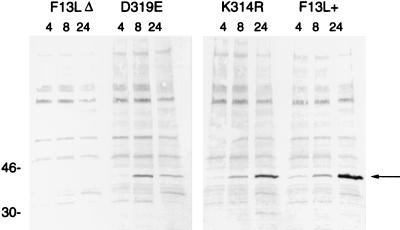
Western blot analysis showed that the two HKD point mutants made readily detectable F13L protein, although the D319E protein seemed reduced in amount. The accumulation of F13L protein during infection was analyzed at 4, 8, and 24 h (Fig. 2). WT F13L protein was barely detectable at 4 h, clearly visible at 8 h, and further increased at 24 h postinfection. The K314R protein showed a similar pattern, with slightly less protein present at 24 h. The D319E protein accumulated to 8 h, but then was reduced at 24 h, suggesting that this protein was less stable.
FIG. 2.
Synthesis and accumulation of F13L proteins. BS-C-1 cells were infected with F13L+, F13L(K314R), F13LΔ, or F13L(D319E) virus for 4, 8, and 24 h. Cell lysates were analyzed by SDS-PAGE, transferred to a membrane, and probed with anti-F13L protein antibodies. The molecular weights (thousands) of markers are shown to the left. The arrow points to the F13L protein.
Stability of the mutated F13L proteins.
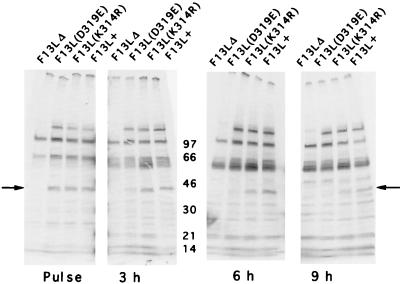
Pulse-chase experiments were carried out to compare the stabilities of the mutated and WT F13L proteins. Cells were infected with F13L+ or mutant viruses for 8 h, metabolically labeled for 90 min with [35S]methionine, and then harvested immediately or at 3-h intervals after the return of the cells to normal media. F13L protein was immunoprecipitated from the lysates and analyzed by SDS-PAGE and autoradiography. Although there were a number of background bands, a unique 37-kDa protein was present in all lanes, except that of F13LΔ (Fig. 3). (The F13LΔ lane was also missing a 120-kDa band which corresponds to the product of the Escherichia coli lacZ gene, which was not present in this recombinant virus [4]). The relative amounts of [35S]methionine-labeled 37-kDa protein were determined with a PhosphorImager. After the pulse, F13L(K314R) and F13L(D319E) proteins were present at 90 and 88%, respectively, of the WT level. After a 3-h chase incubation in unlabeled media, the amount of F13L(K314R) protein remained near the WT level (120% of the WT level), but the F13L(D319E) protein was reduced to 36% of the WT level. At 6 h, F13L(D319E) protein had degraded to 29% of the WT level, and at 9 h, it had degraded to 16% of the WT level. In comparison, F13L(K314R) protein was more stable, being reduced to 72% at 6 h and 51% at 9 h of the F13L+ levels. The persistence of the K314R protein, as determined by Western blotting and pulse-chase analysis, suggested that the profound effect on plaque formation was not solely due to protein instability. In the case of the D319E mutant, however, protein instability may play a significant role.
FIG. 3.
Stability of F13L proteins. BS-C-1 cells were infected with F13L+, F13L(K314R), F13LΔ, or F13L(D319E) virus as indicated. After 15 h of infection, the cells were incubated for 90 min with [35S]methionine, and the cells were either lysed immediately (pulse) or after an additional 3, 6, or 9 h of incubation in normal culture media. The F13L protein was immunoprecipitated, resolved by SDS-PAGE, and autoradiographed. The molecular weights (thousands) of markers are shown in the center. The arrows point to the F13L protein.
Triton X-114 phase partitioning of mutated F13L proteins.
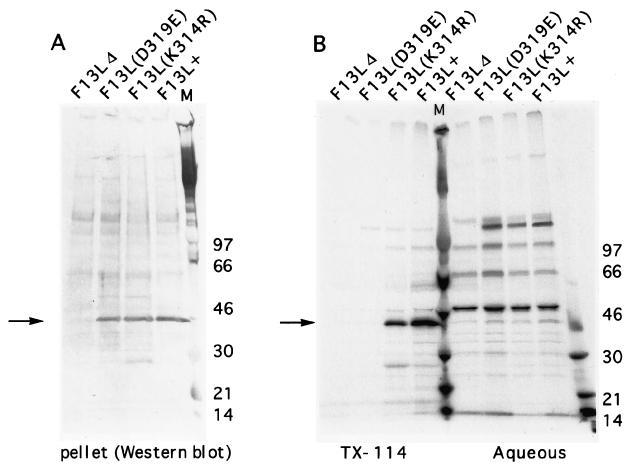
While the F13L protein contains several hydrophobic regions, of which one is predicted to be a transmembrane domain, the palmitylation state also affects its hydrophobicity and membrane association (18, 20, 43). To compare the hydrophobic properties of the mutated and WT F13L proteins, infected cells were metabolically labeled with [35S]methionine for 24 h and then lysed in buffer containing 2% Triton X-114. After extraction of proteins for 30 min, insoluble matter was pelleted and analyzed by Western blotting. The F13L+, K314R, and D319E pellets all contained detectable F13L protein (Fig. 4A). The Triton X-114 soluble material was then warmed to 37°C to allow separation of the detergent and aqueous phases. F13L proteins were immunoprecipitated, analyzed by SDS-PAGE, and autoradiographed (Fig. 4B). The WT F13L protein partitioned completely to the detergent phase, as did the K314R F13L protein, indicating their hydrophobicity and suggesting that the K314R protein was also palmitylated (18, 43). In contrast to the WT and K314R F13L proteins, the D319E F13L protein was not detected in either the detergent phase or the aqueous phase (Fig. 4B), even after extended autoradiography. This indicated that the D319E F13L protein was not extracted by Triton X-114 and was entirely within the insoluble pellet, as detected by Western blotting (Fig. 4A).
FIG. 4.
Triton X-114 (TX-114) partitioning of F13L proteins. BS-C-1 cells were infected with F13L+, F13L(K314R), F13LΔ, or F13L(D319E) virus as indicated. After 6 h of infection, the cells were incubated for 16 h with [35S]methionine and extracted in 2% Triton X-114. Insoluble material was centrifuged and analyzed by Western blotting (A). The Triton X-114 solution was warmed, and the phases were separated. The F13L protein from each phase was immunoprecipitated under identical salt and detergent conditions, resolved by SDS-PAGE, and autoradiographed (B). The molecular weights (thousands) (M) of markers are shown. The arrows point to the F13L protein.
Since the D319E protein possessed altered solubility properties in Triton X-114, we considered that the apparent instability determined in the previous section might be due in part to poor solubility in immunoprecipitation buffer containing 1% sodium deoxycholate and 1% Triton X-100. To evaluate this, cells were infected for 24 h and lysed in immunoprecipitation buffer. Western blotting was performed with the soluble and insoluble materials, and the proteins were quantitated (data not shown). In the case of the F13L+ virus, 87% of the F13L protein was present in the supernatant. For the F13L(K314R) and F13L(D319E) mutants, 78 and 53% of the F13L protein were soluble, respectively. Therefore, the reduced solubility of the mutated proteins may have led to a small underestimate of their stability as determined by immunoprecipitation.
Acylation of F13L proteins.
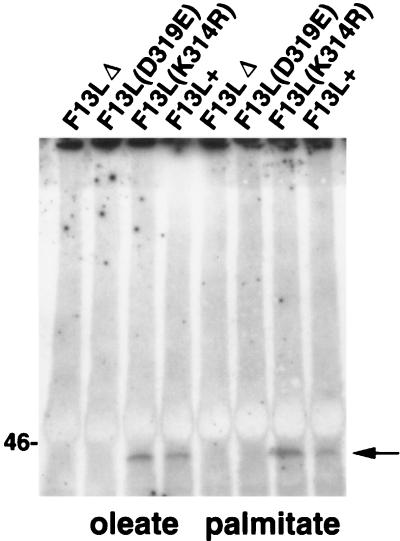
Since the mutant F13L proteins were not functional, we were interested in determining if they were posttranslationally modified as is WT F13L protein. The F13L protein has been shown to be oleated and palmitylated (20, 32). Since the acylation of a protein can determine its activation or interaction with other proteins or membranes (18, 43), the acylation status of the mutated F13L proteins was important to determine. The WT F13L protein is palmitylated on cysteine residues 185 and 186 (18). Although this is 125 amino acids from the location of the mutated HKD motif (Fig. 1), the effects of the point mutations on protein folding or localization might affect processing. Therefore, cells were infected and labeled with [9, 10-3H(N)]palmitic acid or [9, 10-3H(N)]oleaic acid, lysed with immunoprecipitation buffer containing 0.1% SDS, immunoprecipitated, electrophoresed, and autoradiographed. WT and K314R 37-kDa F13L proteins were both palmitylated and oleated (Fig. 5). However oleation or palmitylation of the F13L D319E protein was not detected, even after quadrupling of the exposure time of the gel to X-ray film. The immunoprecipitation buffer-insoluble material was also analyzed, and distinct tritiated 37-kDa bands (data not shown) were detected for F13L+ and the K314R mutant, but not for the D319E or F13LΔ mutant, suggesting that the inability to detect acylated D319E protein was not due to its altered solubility.
FIG. 5.
Palmitate and oleate labeling of VV proteins. BS-C-1 cells were infected with virus at a multiplicity of 10 for 5 h. Medium was then replaced with medium containing 200 μCi of [9, 10-3H(N)]palmitic acid in 10 μl of dimethyl sulfoxide or 100 μCi of [9, 10-3H(N)]oleaic acid in 20 μl of ethanol per well. After an additional 24 h, cells were lysed, and F13L proteins were immunoprecipitated, analyzed by SDS-PAGE, and exposed to film. The position of the 46-kDa marker is shown on the left. The arrow points to the 37-kDa F13L protein.
Because the K314R protein partitioned into the Triton X-114 phase and was acylated, it was expected to be membrane associated (18, 43). This association was confirmed by sucrose density flotation experiments (51) (data not shown). The amount of labeled D319E protein, however, was insufficient to determine membrane association. Although the K314R mutation does not appear to alter the Triton X-114 partitioning, acylation, or membrane association of the F13L protein, the mutant virus nonetheless formed tiny plaques.
Fusion of cells infected with F13L mutant virus.
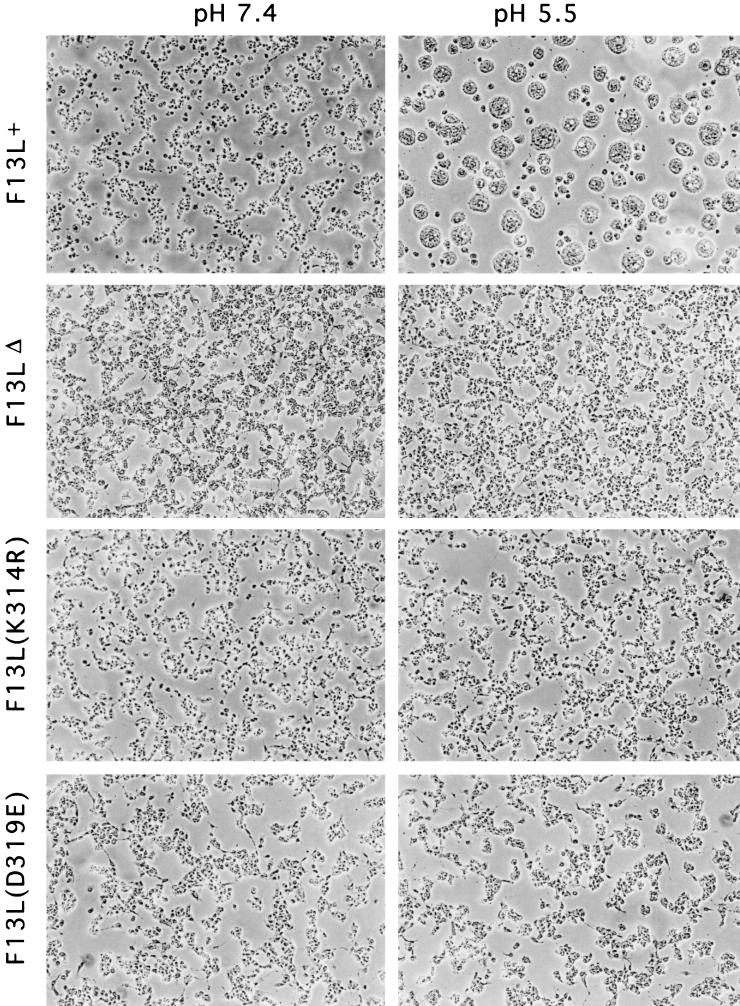
Cells infected with the F13LΔ do not undergo low-pH-induced fusion (4). This process is believed to require wrapping and display of virus particles on the cell surface (4, 10, 17, 39, 48). Therefore, we evaluated the ability of the F13L point mutant viruses to mediate acid fusion. Cells were infected for 12 h with F13L+, F13LΔ, F13L(K314R), or F13L(D319E) virus, incubated for 2 min in buffer at pH 5.5, returned to growth media, and, after 3 h, examined microscopically. While infected cells maintained at neutral pH did not fuse, brief acid treatment induced syncytium formation of F13L+ virus-infected cells. In contrast, none of the F13L mutants induced polykaryon formation (Fig. 6), even though the infected cells were observed for 12 h after the acid treatment. As controls, we confirmed the inability of the A34R deletion mutant to induce fusion and the ability of the A33R deletion mutant to do so (39, 49). These data suggested that the F13L HKD motif was required for cell fusion or that the HKD point mutants did not make wrapped virus particles that were associated with the cell surface.
FIG. 6.
Induction of syncytia by F13L+ or F13L mutants. BS-C-1 cells were infected for 12 h, treated for 2 min with buffer at pH 5.5 or 7.4, and then returned to growth medium for 3 h and examined by phase-contrast microscopy.
CsCl gradient centrifugation of virus particles.
Membrane-wrapped virions (IEV, CEV, and EEV) can be distinguished from unwrapped IMV by their lower buoyant density in CsCl. To determine whether the F13L mutations affected wrapping, cells were infected with WT or mutant viruses, and the particles from the medium and cell lysates were analyzed by CsCl centrifugation. While the production of infectious unwrapped IMV by the HKD mutants was similar to that of controls (WR and F13L+), the production of wrapped virus forms, both cell-associated (IEV or CEV) and free (EEV), was reduced by 89 to 98% (Table 1). These results were similar to those found with F13LΔ (Table 1). The titer of virus infectivity was determined before and after freeze-thawing and sonication (a procedure used to increase infectivity of defective EEV by releasing IMV [27]), with no significant difference in the result. Consequently, the mutation of the K or the D in the HKD motif of the F13L protein severely inhibited the ability of VV to form intracellular wrapped virus particles or to release wrapped particles into the medium (Table 1). This was consistent with the lack of comet formation in infected-cell monolayers (Fig. 1) and the absence of low-pH-induced cell fusion (Fig. 6).
TABLE 1.
Effects of F13L mutations on the formation of wrapped and unwrapped virus particles
| Virus | No. of PFU (106)a
|
||
|---|---|---|---|
| Enveloped (media) | Cell associated
|
||
| Enveloped | Unwrapped | ||
| WR | 2.4 | 40 | 26 |
| F13L+ | 1.2 | 44 | 42 |
| F13LΔ | 0.07 | 3.6 | 14 |
| F13L(K314R) | 0.04 | 3.6 | 60 |
| F13L(D319E) | 0.2 | 3.1 | 32 |
Total PFU from CsCl gradient fractions.
Electron microscopic examination of cells infected with F13L mutant virus.
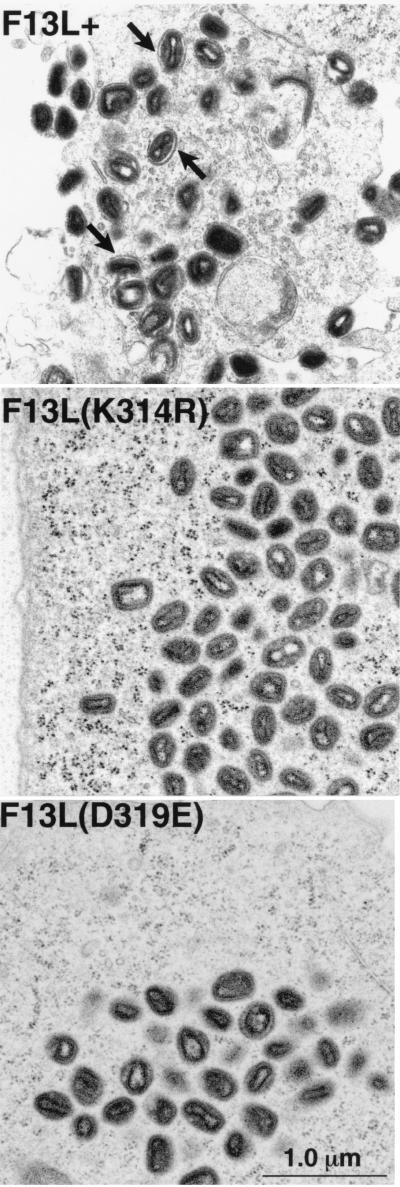
Since the F13LΔ mutant virus is blocked in morphogenesis at the stage of IMV wrapping to form IEV (4) and the F13L point mutants form plaques the same size as those formed by the deletion mutant, electron microscopy was used to analyze the effects of the mutations on morphogenesis. In cells infected with F13L+, numerous wrapped IEV were visible (Fig. 7). However, in the cells infected with the F13LΔ mutant or the F13L point mutants, K314R and D319E, IMV were common, but IEV were rare or absent, consistent with the result obtained by CsCl gradient centrifugation of virus particles. Interestingly, rare IEV could be seen in the D319E mutant virus, which makes the less stable F13L protein, but they were almost completely absent in the K314R mutant with the more stable F13L protein, indicating that the phenotypic changes did not correlate with protein levels. These data indicated that the F13L point mutants were blocked in membrane wrapping of IMV, similar to the phenotype of the F13LΔ mutant.
FIG. 7.
Electron micrographs of Epon-embedded infected cells. RK13 cells were infected with F13L+, F13L(K314R), or F13L(D319E) virus. After 24 h, cells were fixed in glutaraldehyde and embedded in Epon. In F13L+-infected cells, arrows point to examples of wrapped IEV particles.
Localization of mutated F13L proteins by immunofluorescence.
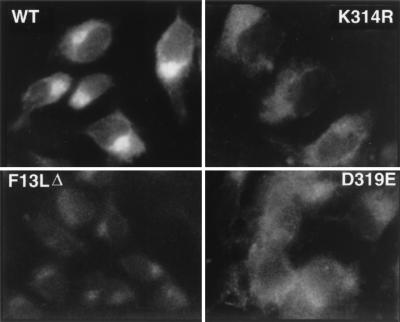
The WT F13L protein has been shown to localize to the trans-Golgi membranes which wrap IMV to form IEV (20, 41). Since efficient wrapping of the F13L HKD point mutant virus particles did not occur, we investigated the intracellular localization of the mutant F13L proteins. WT F13L protein showed the typical bright juxtanuclear staining pattern characteristic of Golgi localization (Fig. 8) (20). Both the F13L(K314R) and F13L(D319E) proteins showed levels of staining above that of uninfected cells or the F13LΔ mutant, but the staining appeared to be distributed throughout the cell rather than intensely localized in the Golgi compartment. In the experiments described above, the cells were also stained with antibody recognizing the surface- and Golgi-localized A33R protein as a control for virus gene expression (38, 41) (data not shown).
FIG. 8.
Immunofluorescence detection of F13L proteins. HeLa cells were infected with F13L+, F13L(K314R), F13LΔ, or F13L(D319E) virus. At 16 h, the cells were fixed, permeabilized, and incubated with antibodies to F13L protein and rhodamine-conjugated swine anti-rabbit immunoglobulin.
Localization of mutated F13L proteins by immunogold electron microscopy.
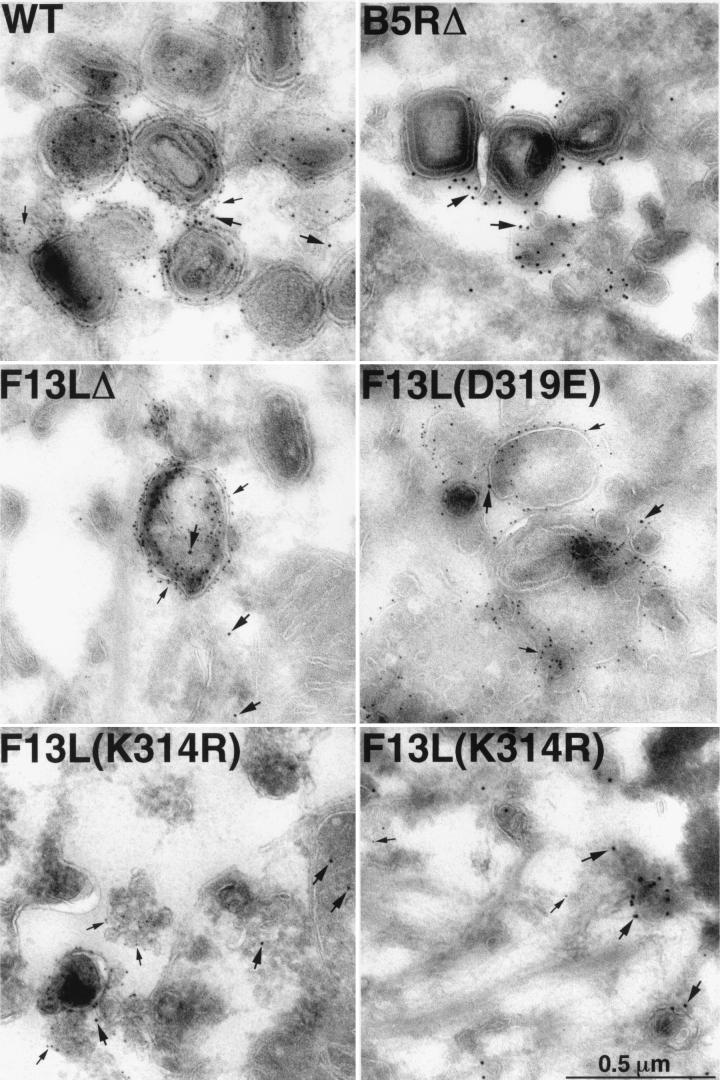
We investigated the localization of the F13L proteins by comparing them with the VV B5R EEV protein, which also localizes to the trans-Golgi network and wrapping membranes (41), by using specific antibodies and two sizes of protein A-gold spheres. The 10- and 5-nm particles target the F13L and B5R proteins, respectively. We found that in VV strain WR-infected cells, membranes were heavily labeled for both proteins, with 92% of the F13L protein closely juxtaposed with B5R protein (Fig. 9 and Table 2). Although wrapped IMV are rarely seen in cells infected with the B5R and F13L deletion mutants, virus protein-laden membranes may still be noted adjacent to IMV or localized in vesicular structures. In both of the HKD point mutant virus-infected cells, it was possible to see membranes heavily labeled for the B5R protein, while the F13L protein was distributed throughout the cell, rather than concentrated in particular areas. For this reason, it was difficult to find electron microscope fields with concentrated labeling of F13L protein. However, in the cells infected with the K314R mutant, the F13L protein could occasionally be seen concentrated in discrete membraneous structures, but these membranes were lacking B5R protein (Fig. 9, lower right panel). Quantification indicated a high level of K314R protein, of which only 24% was juxtaposed with the B5R protein (Table 2). This value is similar to the background value of 23% obtained in cells infected with F13LΔ (Table 2). For the D319E mutant, there was less total staining of F13L protein, and only 19% was adjacent to the B5R protein (Table 2). Thus the K314R and D319E F13L proteins did not localize with B5R protein above background levels. A B5R deletion mutant virus was also included as a control; background B5R staining was very low (Fig. 9), and only 1% juxtaposed with F13L protein. These data suggested that the block in IEV formation was due to improper membrane localization of both the K314R and D319E F13L proteins.
FIG. 9.
Immunogold labeling of viral membranes. RK13 cells were infected with WR, F13L(K314R), F13LΔ, or F13L(D319E) virus for 24 h, fixed in paraformaldehyde, cryosectioned, and double labeled with antibodies specific to either the F13L (and 10-nm protein A-gold spheres) or B5R (and 5-nm protein A-gold spheres) protein. Thick and thin arrows point to representative 10- and 5-nm gold spheres, respectively. Occasional 10-nm gold particles in the F13LΔ are due to background staining as quantified in Table 2.
TABLE 2.
Juxtaposition of F13L and B5R proteins
| Virus | No. of F13L particles/1,000 B5R particlesa | % F13L proteins juxtaposed with B5R protein |
|---|---|---|
| WR | 534 | 92 |
| F13L(K314R) | 718 | 24 |
| F13L(D319E) | 253 | 19 |
| F13LΔ | 148 | 23 |
B5R and F13L proteins were detected with specific antibodies and 5- and 10-nm protein A-gold particles, respectively. The numbers of 10-nm particles per 1,000 5-nm particles are indicated.
DISCUSSION
We combined genetic, biochemical, and microscopic approaches to investigate the role of the HKD motif of the F13L protein. Conservative mutations of the K and D positions of the second VV F13L HKD motif caused a small-plaque phenotype, indicating the functional importance of this sequence. The F13L K314R and D319E phenotypes were as severe as that of F13LΔ, indicating that the function of the F13L protein was completely abrogated by these single-amino-acid mutations in the HKD motif. The specificity of these mutations is demonstrated by several other known amino acid changes not affecting plaque size (42, 16, 36).
Biochemical analyses showed that the properties of the K314R protein were similar to those of WT F13L, while those of the D319E protein were more severely altered. Thus the K314R mutant, which partitioned normally in Triton X-114, was acylated and membrane bound. In contrast, the D319E protein was less stable, was insoluble in Triton X-114, and was less soluble than WT F13L protein in immunoprecipitation buffer. Acylation of the D319E protein could not be detected, either because of the small amount of the protein or because the mutated protein was sequestered or degraded before it could undergo posttranslational modifications.
Small-plaque phenotypes have been associated with reductions in the quantity (5, 31, 48) or infectivity (27) of extracellular virus and with a defect in actin tail formation (39, 40, 49, 50). The F13L(K314R) and F13L(D319E) mutants mirror the F13LΔ mutant in their formation of small plaques, lack of virus released into the medium, and lack of comet formation. These data uphold the correlation between EEV in the medium and the formation of elongated comet-shaped plaques under liquid medium (4–6, 27, 39). Both F13L HKD mutant viruses were blocked in morphogenesis at the stage of membrane wrapping of IMV to form IEV. Very few examples of wrapped IEV were seen, and there was a log or more reduction in the amount of EEV released into the media by the K314R and D319E mutants compared to the F13L+ control viruses. The F13L point mutants did not mediate acid-induced fusion of infected cells, consistent with previous observations that formation of wrapping and display of virus on the surface are necessary for acid-induced polykaryon formation (4, 10, 48).
Although the F13L(K314R) and F13L(D319E) proteins could be detected, they did not localize properly to the Golgi compartment. These data indicated that the HKD motif may be important for the proper sorting of the F13L protein in intracellular membranes during VV infection. It has previously been shown that F13L localization is dependent on other VV protein(s) (25). In the case of the K314R mutant, hydrophobicity, oleation, and palmitylation alone were insufficient to direct the protein to its proper compartment in the membranes. Thus, the K314 of the HKD motif was required for proper localization. It is unknown whether the F13L HKD motif functions through an interaction with another VV protein or directly with membranes. The data indicate that there are three requirements for correct intracellular localization of the F13L protein: (i) palmitylation (18, 43), (ii) another VV-encoded protein (25), and (iii) amino acids in the HKD motif.
The HKD motif is conserved in phospholipases and phospholipid synthases, leading to speculation that the VV F13L protein and the process of envelopment require phospholipid biosynthesis or cleavage (26, 37). Indeed, the VV F13L protein has now been characterized as a broad-specificity lipase, possessing triacylglycerol lipase and phospholipase A and C activities (2). The EEV is known to differ in phospholipid composition from the IMV (19). However, the direct involvement of the 16 amino acids of the VV F13L HKD motif in enzymatic activity seems unlikely for several reasons. (i) Although this motif is characteristic of the phospholipase D superfamily, phospholipase D activity of the VV F13L protein has not been demonstrated (2, 46). (ii) The H is believed to form part of a catalytic H, K, and D triad and is conserved in all phospholipase D enzymes, cardiolipin synthases, and phosphatidyl serine synthases, suggesting that it is essential for enzyme activity. (iii) None of the other enzymes in this family has an N replacing the H at position 1 of the motif, as is found in the VV F13L HKD, and mutation of the human phospholipase D HKD motif to NKD totally ablated phospholipase D function (2, 46). (iv) Finally, F13L phospholipase A and C activities persisted after mutation of D319 (which causes a small-plaque phenotype) of the VV HKD motif (2). Mutation of S327 of the VV F13L HKD motif (deemed possibly to be important for phospholipase A or C activity) also did not affect the reported phospholipase activity (2). One interpretation is that the VV F13L HKD motif is involved in an enzymatic activity not yet assayed, possibly even a phospholipid synthesis activity. Another possibility is that the motif mediates a nonenzymatic interaction of the F13L protein with phospholipids in membranes needed for wrapping. This hypothesis is supported by the loss of proper membrane localization of the VV HKD mutants.
ACKNOWLEDGMENTS
We thank N. Cooper for cells, E. Wolffe for electron microscopy and comments on the manuscript, A. Weisberg for electron microscopy and help with preparation of figures, and D. Hruby and D. Grosenbach for advice on palmitate labeling.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Baek S-H, Kwak J-Y, Lee S H, Lee T, Ryu S H, Uhlinger D J, Lambeth J D. Lipase activities of p37, the major envelope protein of vaccinia virus. J Biol Chem. 1997;272:784–790. doi: 10.1074/jbc.272.51.32042. [DOI] [PubMed] [Google Scholar]
- 3.Blake N W, Porter C D, Archard L C. Characterization of a molluscum contagiosum virus homolog of the vaccinia virus p37K major envelope antigen. J Virol. 1991;65:3583–3589. doi: 10.1128/jvi.65.7.3583-3589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulter E A, Appleyard G. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog Med Virol. 1973;16:86–108. [PubMed] [Google Scholar]
- 8.Buller R M L, Chakrabarti S, Cooper J A, Twardzik D R, Moss B. Deletion of the vaccinia virus growth factor gene reduces virus virulence. J Virol. 1988;62:866–874. doi: 10.1128/jvi.62.3.866-874.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J X, Koop B F, Upton C. A human homolog of the vaccinia virus HindIII K4L gene is a member of the phospholipase D superfamily. Virus Res. 1997;48:11–18. doi: 10.1016/s0168-1702(96)01422-0. [DOI] [PubMed] [Google Scholar]
- 10.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 13.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 14.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 15.Fenner F, Henderson D A, Arita I, Jezek Z, Ladnyi I D. Smallpox and its eradication. 1st ed. Geneva, Switzerland: World Health Organization; 1988. [Google Scholar]
- 16.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 17.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 18.Grosenbach D W, Ulaero D O, Hruby D E. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. J Biol Chem. 1997;272:1956–1964. doi: 10.1074/jbc.272.3.1956. [DOI] [PubMed] [Google Scholar]
- 19.Hiller G, Eibl H, Weber K. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J Virol. 1981;39:903–913. doi: 10.1128/jvi.39.3.903-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 23.Isaacs, S., R. Blasco, and B. Moss. Unpublished observations.
- 24.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz E, Wolffe E J, Moss B. The cytoplasmic and transmembrane domains of the vaccinia virus B5R protein target a chimeric human immunodeficiency virus type I glycoprotein to the outer envelope of nascent vaccinia virions. J Virol. 1997;71:3178–3187. doi: 10.1128/jvi.71.4.3178-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin E V. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem Sci. 1996;21:242–243. [PubMed] [Google Scholar]
- 27.McIntosh A A G, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 29.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. New York, N.Y: Lippincott-Raven Press; 1996. pp. 2637–2671. [Google Scholar]
- 30.Moss B. Vaccinia virus: a tool for research and vaccine development. Science. 1991;252:1662–1667. doi: 10.1126/science.2047875. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 32.Payne L G. Characterization of vaccinia virus glycoproteins by monoclonal antibody preparations. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 33.Payne L G. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol. 1980;31:147–155. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 34.Payne L G, Kristensson K. Mechanism of vaccinia virus release and its specific inhibition by N1-isonicatinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J Virol. 1979;32:614–622. doi: 10.1128/jvi.32.2.614-622.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne L G, Norrby E. Presence of haemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponting C P, Kerr I D. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roper R L, Wolffe E J, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson C M, Rischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 41.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmutz C, Payne L G, Gubser J, Wittek R. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol. 1991;65:3435–3442. doi: 10.1128/jvi.65.7.3435-3442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmutz C, Rindisbacher L, Galmiche M C, Wittek R. Biochemical analysis of the major vaccinia virus envelope antigen. Virology. 1995;213:19–27. doi: 10.1006/viro.1995.1542. [DOI] [PubMed] [Google Scholar]
- 44.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 45.Sodeik B, Doms R W, Ericcson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sung T-C, Roper R L, Zhang Y, Rudge S A, Temel R, Hammond S M, Morris A J, Moss B, Engebrecht J, Frohman M A. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 1997;16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner G S, Squires E J. Inactivated smallpox vaccine: immunogenicity of inactivated intracellular and extracellular vaccinia virus. J Gen Virol. 1971;13:19–25. doi: 10.1099/0022-1317-13-1-19. [DOI] [PubMed] [Google Scholar]
- 48.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolffe E J, Weisberg A S, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Lamb R A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]