Abstract
Cryo–electron microscopy (cryo-EM) continues its remarkable growth as a method for visualizing biological objects,which has been driven by advances across the entire pipeline. Developments in both single-particle analysis and in situ tomography have enabled more structures to be imaged and determined to better resolutions, at faster speeds, and with more scientists having improved access. This review highlights recent advances at each stage of the cryo-EM pipeline and provides examples of how these techniques have been used to investigate real-world problems, including antibody development against the SARS-CoV-2 spike during the recent COVID-19 pandemic.
Keywords: cryo-EM, SARS-CoV-2, machine learning, in situ tomography, democratization, automation
INTRODUCTION
Major advances in the past decade in cryo–electron microscopy (cryo-EM) have driven the widespread success of cryo-EM as a method for visualizing biological objects. Better and faster microscopes, detectors, and data processing algorithms have accelerated data collection and processing speeds, thereby driving down total costs per micrograph. These innovations enabled more single-particle structures to be determined routinely at subnanometer resolution (reviewed in 1) and more biological objects to be imaged and identified in situ (2).One measure of results of these advances is the exponential growth in the number of cryo-EM entries in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) (Figure 1).
Figure 1.
Electron microscopy (EM) entries in data archives have been growing rapidly. In orange are released EM map entries in the Electron Microscopy Data Bank, in blue are released EM model coordinates in the Protein Data Bank, and in green are raw data sets contributed to the Electron Microscopy Public Image Archive (EMPIAR).
Today, cryo-EM continues to smash through barriers, with notable examples including the determination of the first truly atomic-resolution structures obtained by single-particle analysis (3–5) and the in situ visualization of antibiotic binding to ribosomes in bacterial cells imaged by cryo–electron tomography (cryo-ET) (6).These remarkable achievements highlight how the field continues to rapidly advance. This brief review is not intended to be a comprehensive overview of cryo-EM methods but instead describes some significant recent developments across the entire cryo-EM pipeline. Specifically, we focus our review on the general areas of samples, grids, instrumentation, data, processing, modeling, and dissemination and include discussions on facilities, in situ tomography, and microcrystal electron diffraction (MicroED). We highlight recent developments that have enabled cryo-EM to be a better, faster, and cheaper modern imaging tool for biological research; detail how these developments were applied in our efforts to combat the coronavirus disease 2019 (COVID-19) pandemic; and discuss potential future directions. We apologize to the authors whose work has not been cited in this review due to space constraints and for any potential oversights.
SAMPLES
Prior to imaging biological objects with cryo-EM, it is important to have biochemical and biophysical knowledge about a sample to complement and inform the imaging process. Information such as molecular weight, shape, and macromolecule composition guides the interpretation of cryo-EM micrographs and any troubleshooting required when dealing with a challenging sample. Conventional biochemical and biophysical characterization techniques can provide ample information regarding sample size, shape, molecular weight, purity, and homogeneity. Some useful techniques include native and denaturing gel electrophoresis, size exclusion chromatography, dynamic light scattering, differential scanning fluorimetry, and differential scanning calorimetry. In addition, negative stain electron microscopy (EM) is a useful tool for sample characterization prior to cryo-EM imaging. With negative stain EM, a sample can be rapidly screened for quality, avoiding the time and expense of vitrification and cryo-imaging, and negative staining also usually requires an order of magnitude less sample. An automated negative stain assay that could screen dozens of sample conditions would complement other biophysical characterization methods; some early attempts at this are underway (7).
A recently developed approach for characterizing macromolecule mass in solution is mass photometry (8, 9). Mass photometry detects changes in light scattering as molecules bind nonspecifically to a glass slide. This allows for the determination of a wide range of molecular masses with high accuracy, assessment of binding affinity and heterogeneity, and determination of oligomerization state. Additional advantages of mass photometry include the ability to use samples label-free; rapid results generation; and applicability to a wide range of macromolecules, including membrane proteins (10) and nucleic acids (11). These advantages make mass photometry a promising technology for initial sample characterization by providing useful preliminary information about a sample rapidly and at low cost.
One caveat for negative stain and mass photometry is that the protein concentration required for these methods is much lower than for cryo-EM. Complexes that might be stable at higher concentrations could disassociate at the concentrations required by these methods. One potential solution is to use chemical fixation to maintain complex stability at low concentrations. If there is sufficient sample volume, the presence of intact complexes may also be assessed using techniques like gel filtration and multi-angle light scattering.
Cryo-EM has become a powerful tool for imaging otherwise challenging samples such as large complexes and heterogeneous samples. One important class of samples on which cryo-EM has had a large impact is membrane proteins. These proteins need to be embedded in a membrane mimetic to maintain their structural integrity, which makes them especially challenging for alternative structural techniques like X-ray crystallography. Conversely, for cryo-EM, if the proteins can be extracted from the membrane, solubilized as individual particles, then vitrified in a thin layer on a grid, their structures can be solved with standard single-particle methods.
Many membrane solubilization methods are in use (recently reviewed in 12, 13), and we summarize only a few here. The extraction of membrane proteins from a native bilayer traditionally has been done with detergents. While harsh detergents such as Triton X-100 work for some complexes (14), the trend in cryo-EM is to use gentler detergents such as digitonin or glyco-diosgenin to keep complexes intact (12). An alternative popular membrane mimetic used in cryo-EM is amphipols, protein polymers that wrap around the hydrophobic portion of the membrane protein and displace the detergent. These have been used very successfully with TRP ion channels (13). In addition, nanodiscs are becoming increasingly popular in cryo-EM (Figure 2). They function in a similar way as amphipols, except that they are composed of amphipathic protein helices. While amphipols and nanodiscs are usually applied to samples after detergent solubilization, another class of mimetics, saposins and SMALPs/DIBMA (13), attempts to excise the protein directly from the membrane, skipping the step of detergent solubilization entirely. Finally, it is possible to preserve membrane proteins in a native liposome, which can be imaged directly (reviewed in 15). This membrane mimetic has the benefit of allowing for manipulation of bilayer-specific properties like an electrochemical gradient to probe the mechanism of action of membrane proteins like ion channels (16, 17). As membrane proteins are important drug targets, being able to determine more such structures with cryo-EM at better resolutions has accelerated efforts to develop better drugs to treat disease (18).
Figure 2.
Advances in membrane protein preparation for cryo–electron microscopy (cryo-EM) include the use of various membrane mimetics, such as nanodiscs (green), to prepare membrane proteins, for example, the sugar transferase AftD (red), for cryo-EM imaging (19).
Sample preparation and grid-making for cryo-EM present the largest hurdles for most projects. It remains unpredictable as to which samples will be amenable to imaging by single-particle cryo-EM. Akin to the problems of making crystals for X-ray crystallography, in cryo-EM it is unknown whether a novel sample will survive current grid-making methods until it is attempted. However, we anticipate that obtaining more prior knowledge about the sample of interest by using complementary biochemical and biophysical assays may improve success and predictability in the future.
GRIDS
The goal of grid preparation for cryo-EM is to suspend proteins in a well-vitrified layer of ice as thin as possible on a substrate supported by an EM grid. On an ideal grid, the ice will be vitreous and uniform across the entire grid, just slightly thicker than the molecule of interest, and contain well-distributed particles at sufficient concentration. The ability to reproducibly make good-quality grids means more high-quality images can be collected faster, making the entire process cheaper. However, even with robotic plunge freezers like the Vitrobot (Thermo Fisher Scientific) and Leica GP2 (Leica Microsystems), vitrification is a nontrivial optimization task that requires good fine motor skills and can still be difficult to replicate from grid to grid.
To make a cryo-EM grid, a sample volume (~3 μL) is applied to a grid, then reduced to a thin film of ideally 10–100 nm by blotting with filter paper, followed by plunging the grid rapidly into a cryogen. In the process of making the thin film, the surface area to volume ratio of the sample increases dramatically, with the result that proteins collide with the air-water interface (AWI) on the order of 100–1,000 times during the time interval between blotting and vitrifying (20, 21). These collisions with the AWI can result in preferred orientation, denaturation, complex dissociation, and compositional heterogeneity (22, 23). While the AWI causes a variety of problems, adherence to the AWI is also the reason we observe a high number of particles in micrographs despite using low protein concentrations (24, 25). For example, for our apoferritin test sample we typically prepare grids at a concentration of 8 mg/mL.At this concentration, in a micrograph with 4,092 × 5,760 pixels at 1.1 Å per pixel, and assuming an ice thickness of 35 nm, approximately 100 particles of apoferritin would be expected to be observed. In practice, approximately 1,300 particles per micrograph are observed, presumably because they are concentrated by sticking to the AWI (25). The overall effects of the AWI are still poorly understood and require further investigation.
There have been several approaches toward solving the problems caused by the AWI, including using a non-grid support such as layering graphene or graphene oxide onto holey grids to sequester proteins away from the AWI (23, 26, 27); using films made of streptavidin monolayer crystals (28); using films functionalized with Ni-NTA, Protein G, oligonucleotides (29),or antibodies (30); using detergents such as CHAPSO that protect the protein from the AWI (27); and using chemical fixation to preserve protein structural integrity (31). Alternatively, instead of changing the behavior of proteins on the grids, modifications to the method of vitrification have also been made. Devices utilizing microfluidic spray plungers (32), surface acoustic waves (33), microcapillary writing (34), pin printing and jet cooling (35), and piezoelectric transducers (36, 37) have the potential for reducing the blot-to-plunge times or eliminating the need for blotting entirely. Reducing blotto-vitrification times to ~100 ms, for instance, by using an automated inkjet piezo dispensing and vitrification device, Spotiton (38),has been shown to ameliorate AWI effects (22) such as preferred orientation (39) and sample denaturation (40). However, since the sample is concentrated at the AWI, a side effect of reducing its impact is that a higher sample concentration is required to obtain an equivalent number of particles in each micrograph (25).
One interesting biological application of vitrification is the ability to trap intermediates at fixed time points, also called time-resolved cryo-EM. This approach has been demonstrated beginning with Nigel Unwin’s experiments in 1995 (41) and more recently by those of Joachim Frank’s group (reviewed in 42).Recently, Spotiton was upgraded to support time-resolved experiments by adding a second piezo dispense head, allowing for on-grid mixing by spraying two streams of droplets (~50-pL volume) of separate samples onto the same area of the grid. This allows for mixing on a timescale of 90–500 ms (43). All of these time-resolved instruments aim to achieve a similar goal of allowing biological reactions to be studied on a tens-of-milliseconds timescale, with potentially multiple assembly states in the reaction being visualized on-grid (44,45).Time-resolved Spotiton, for instance, has been applied to studying ribosome assembly, to studying conformation changes in calcium-gated potassium channels, and to trapping early RNA polymerase intermediates (43).
A major remaining challenge of cryo-EM imaging is to reduce beam-induced motion. During imaging with an electron beam, the sample and support foil can buckle, and this movement results in blurred images, limiting capture of high-resolution information (46, 47). The blurring can be corrected by applying motion-correction algorithms (46, 48) to the movie frames obtained from direct detectors. However, the early frames that contain the best high-resolution information also have the largest movements (49). It was demonstrated that beam-induced motion can be significantly reduced by replacing the traditional holey carbon support foil with one made of gold (50), which is also critical for tilted data collection that is sometimes used for samples with preferred orientation issues (51). UltrAuFoil grids (gold foil on gold grids, with 1.2-μm-diameter holes) were the sample support of choice for the highest resolution cryo-EM maps obtained thus far (apoferritin at ~1.2 Å) (3, 4). Nevertheless, it is worth noting that samples prepared over carbon foil also achieved very high resolution (apoferritin at ~1.4 Å) (5). Building on the gold substrate work, Naydenova et al. (52) showed recently that a thin layer of ice suspended across the holes of a grid—the sample itself—undergoes beam-induced motion via a mechanism independent of movement of the support foil. Naydenova et al. proposed that thin films of ice can store compressive stress during vitrification, which is released during electron irradiation, causing the ice to buckle. They went on to show that this stress in the ice could be reduced by using grids with sufficiently small holes (<0.3-μm diameter for most single-particle data sets) (52), which, when patterned into all-gold grids (called HexAuFoil grids), resulted in effectively motion-free imaging (Figure 3). Recently, Engstrom et al. (53) proposed plunging grids into boiling nitrogen instead of liquid ethane as an alternate method to reducing beam-induced motion. By reducing specimen movement at the sample preparation stage, better cryo-EM images can be taken, allowing for more efficient reconstructions at higher resolution.
Figure 3.
The development of all-gold HexAuFoil grids with small (<0.3 μm) holes dramatically reduced beam-induced motion during imaging. The gold foil and gold mesh reduce foil movement, while the small holes minimize ice movement during electron irradiation.
The method of plunge freezing to prepare samples for cryo-EM has remained remarkably unchanged since the 1980s, and a good theoretical knowledge of the process of creating thin ice is still lacking. In an effort to further our understanding of how blotting affects ice thickness, Armstrong et al. (54) analyzed in detail the behavior of water during blotting with filter paper in a Vitrobot. Future work on grid preparation should include efforts to better understand and control the forces that govern the formation of thin films of liquid on an EM grid and the behavior of macromolecules in these films and to capture biological reactions in a biologically relevant (millisecond) timescale. Improvements in the consistency and reliability of grid-making will be major advances toward improving speed, efficiency, and throughput of the entire cryo-EM pipeline.
INSTRUMENTATION
Acquiring images of a vitrified sample requires a cryogenic transmission electron microscope (cryo-TEM). Simply, these microscopes have a source that produces an electron beam, an ultrahigh vacuum system, a series of magnetic lenses that focus the beam, a cryogenic sample holder for translating the grid, and a detector that captures images. Most of these components are still undergoing significant development to improve images and speed up the imaging process.
Over the past several decades, electron sources have undergone major changes. The advent of the field emission gun (FEG) made it possible to generate a brighter electron beam with better spatial and temporal coherence than is possible for conventional thermionic electron sources such as tungsten filaments or LaB6 crystals. Most high-end electron microscopes used in cryo-EM, such as the Titan Krios, are equipped with a FEG operated at 1,700–1,800 K called XFEGs. In 2012, Ricolleau et al. (55) and JEOL developed the first TEM with a FEG operated at room temperature, called a cold field emission gun (CFEG). CFEGs generate an electron beam with an energy spread of around 0.3 eV without losing beam brightness compared to the 0.7-eV spread from the XFEG (3, 55). The smaller energy spread improves temporal coherence of the electron beam leading to a better signal-to-noise ratio in the images formed. This technology was applied in 2019 by Kato et al.(56) to obtain a 1.5-Å reconstruction of apoferritin using data collected from a JEOL CryoARM-300 equipped with a CFEG. More recently, Nakane et al. (3) used a prototype of a CFEG in conjunction with a new in-column energy filter, both developed by Thermo Fisher Scientific on a Titan Krios, and achieved a record-breaking 1.2-Å-resolution map of apoferritin where individual hydrogen atoms in the protein could be clearly visualized (Figure 4). The new energy filter, the Selectris, is expected to be a robust system capable of providing stable data collection for several days with minimum energy slit deviation while reducing higher-order distortions with its larger prism radius. As the CFEG and Selectris become more widely available, it will become more evident whether these advancements will have a general impact.
Figure 4.
Advances in cryo–electron microscopy instrumentation and processing recently produced an atomic-resolution map of apoferritin (3), shown here in blue, volume-rendered using PyMOL. The hydrogen difference map (green) shows ordered hydrogens, even on a water molecule (center). Modeled atoms are shown as spheres with bonds as sticks. Carbon is shown in light blue, oxygen in red, nitrogen in dark blue, and hydrogens in white. Image provided by Takanori Nakane.
Researchers now routinely achieve high-resolution structures using commonly available 300-keV cryo-TEMs. However, given the high operating costs of 300-keV microscopes and the complexity of support systems needed for sustained operation, efforts are being made to democratize TEM by setting up national facilities for general use access and to develop lower-cost microscopes capable of producing high-quality data (see the section titled Facilities). One way to reduce the cost of a cryo-TEM is by using instruments with lower voltage. Sub-2-Å-resolution structures have been obtained using a 200-keV TEM equipped with a FEG and direct detector (for example, in 57). Peet et al. (58) also advocate for developing a 100-keV instrument equipped with a FEG and direct detector as an affordable microscope, and they have shown that 100-keV electrons contribute 25% more elastic scattering per unit damage compared to 300-keV electrons, thereby making them better for imaging specimens up to 60 nm in thickness. As a proof of concept, Naydenova et al. (59) operated a commercial 200-keV FEG microscope at 100 keV and, using a hybrid pixel detector, obtained five reconstructions with resolutions ranging from 3.4 to 8.4 Å. Subsequently Thermo Fisher Scientific announced a new 100-keV microscope, the Tundra, to be released late in 2021, that is equipped with a FEG and a standard complementary metal oxide semiconductor (CMOS) detector. The instrument is anticipated to have a footprint that should allow it to fit into most standard labs and to be highly automated and easy to operate. These low-voltage instruments may be well suited as screening tools prior to sending the highest-quality grids to the national centers for high-resolution data collection or, if suitable detectors for 100-keV electrons are developed, as lower-cost data collection tools.
New developments in aberration correction have also been shown to increase the resolution of cryo-EM maps. While many of these aberrations, including beam tilt, coma, trefoil, and tetrafoil, can be corrected postacquisition through software (60), spherical (Cs) and chromatic (Cc) aberrations can be corrected with additional lenses added to the column of the microscope. Cs correctors use hexapole magnets to correct third-order aberrations, and one way to reduce Cc aberrations is by using a monochromator to reduce the energy spread of the electron beam (4). Recently, the power of the aberration correctors was demonstrated by Yip et al. (4) in reporting a 1.2-Å-resolution structure of apoferritin obtained by using a prototype Titan Krios equipped with a monochromator and a second-generation Cs corrector. Despite the advantages of Cs correctors and monochromators, these add considerable extra capital costs, and the additional components need to be tuned ahead of each data collection session, which can be time consuming and adds complexity. Nevertheless, aberration correction using either hardware or software was key to obtaining atomic-resolution data.
Recent improvements in direct detector device technology have provided significant speedups in data collection rates (61). A day of Krios data collection can now generate multiple terabytes of data, making data management an increasingly important consideration (62). One recent innovation for recording camera readouts has been to record images in the electron event representation (EER) format (63). Rather than recording and storing movies as a series of frames, the EER format records individual electron detection events (both position and time) at the detector’s physical frame rate, thereby preserving the spatial and temporal resolution in the data. Furthermore, the file sizes are small for most use cases, and motion correction is improved by preserving temporal resolution in the data and not requiring user-defined dose fractionation.
Interest in phase plates has been reignited by the recent development of the laser phase plate (64). Turnbaugh et al. (64) demonstrated the ability to generate sufficient intensity in a Fabry-Perot cavity to induce a 90° phase shift to electrons accelerated to 300 keV. While the Volta phase plate greatly improved contrast in cryo-EM images, it suffered from a continually developing phase shift and a reduction in high–spatial frequency information (65). The laser phase plate provides a defined phase shift, and high-resolution information is expected to be preserved: A 3.8-Å reconstruction of the 20S proteasome was presented to demonstrate the potential that laser phase plates have for single-particle cryo-EM (64).
It has now been demonstrated that microscope technology is no longer a limiting factor for achieving atomic resolution on well-behaved samples. Recent microscope innovations have contributed to obtaining significantly better data, thus improving the amount of information harvested from each sample and micrograph. Further improvements in data collection speed, for instance, with more stable microscope stages and faster cameras, will further reduce data collection time and costs. As data collection rates continue to speed up, new strategies will be critical to support robust data management and storage as well as on-the-fly data processing.
DATA
After a grid has been inserted into the microscope, a data collection strategy must be applied to target and image the regions of the grid that contain the sample of interest in optimal conditions. The goal of most single-particle data collections is to collect enough high-magnification images to obtain a reconstruction at a resolution required to answer a biological question. Commonly, a data set between 5,000 and 10,000 micrographs is collected with the goal of obtaining a reconstruction better than approximately 3.5 Å. A typical data collection workflow involves the following: After an initial low-magnification atlas of the grid is collected, squares of interest are selected either automatically or by an operator using visual inspection. Next, a higher-magnification image of each selected square is taken to allow more accurate targeting of the regions of interest. After identifying holes covered by ice of suitable thickness, drift and defocus are checked and adjusted for, then high-magnification images of the sample of interest are acquired at the desired dose and pixel size. This multiscale imaging workflow, if done manually, requires many user hours on the microscope and is a tedious, repetitive task. As a result, the workflow has been largely automated using data collection software, originating with Leginon (66–68), that is now also implemented in SerialEM (69), EPU (Thermo Fisher Scientific), JADAS (JEOL) (70), and Latitude S (Gatan). The automation of data collection since the 2000s has enabled major speedups in cryo-EM data collection rates and reduced the amount of operator input required during a data collection session.
Recently, a data collection strategy was introduced that greatly increased throughput by using beam-image shifts rather than stage movement to center exposure targets for data acquisition (Figure 5) (71). Because of this implementation, it is now possible to collect up to approximately 40 high-magnification exposure images per stage movement on a standard grid compared to a single high-magnification exposure image that was previously obtained using a stage shift data collection strategy. Using beam-image shift to move to a selected target minimizes the number of slower and less accurate stage movements, thereby increasing data collection rates. An undesirable consequence of using beam-image shift is the introduction of coma, which becomes more prevalent the larger the beam-image shift is. Cheng et al. (71) determined that the resolution of single-particle reconstructions is not as negatively impacted by this effect as expected, likely due to averaging of particles during single-particle analysis (72). Since then, corrections for imaging aberrations have been developed that have further improved reconstruction resolutions. Beam tilt and higher-order aberrations can be corrected either before data collection using hardware (68, 73) or after data collection with software (60,74) (see the section titled Processing).These corrections enable beam tilts capable of image shifts of up to 8–10 μm, increasing throughput without sacrificing resolution (68, 73).
Figure 5.
Data collection algorithms now allow for collection of up to approximately 40 exposures (white boxes) per stage movement by using large beam-image shifts. This reduces the number of slow stage movements and greatly increases data collection speeds.
A current bottleneck in data collection throughput is grid screening, which is the initial assessment of several grids to find ones that have ice of optimal thickness and particle composition. Starting with a previously uncharacterized sample, a microscope operator typically screens several grid squares with varying ice thicknesses then selects, by eye, the optimal squares for data collection. Even with on-the-fly feedback on incoming micrographs producing near-real-time information such as contrast transfer function (CTF) estimation, ice thickness, and 2D classes, the microscopist must still be able to integrate all this information and determine the best grid, squares, and holes to target for extended data collection. Optimizing this process manually can require many hours of data collection screening and experience with each specific sample. The obvious step to address these issues is to develop automated tools to switch grids, create an atlas and collect multiscale images, and assess them using automated on-the-fly processing. For the task of square selection, semisupervised deep learning is one obvious possible approach, and some promising results have been demonstrated (75). However, the performance of the classifier is sample dependent, so it is not yet a general solution for unattended data collection, as a new classifier must be trained for each novel sample (76). Work on automating and simplifying the data collection pipeline by invoking deep learning to assess micrographs and 2D classes is in development (77).For this automated workflow, Li et al.(77) created MicAssess, a convolutional neural network (CNN)-based micrograph evaluator, to automatically and accurately separate good micrographs from bad. Particle sizes were determined automatically with an edge detector, then particles were picked with crYOLO.Finally,2DAssess was developed by Li et al.to evaluate resulting 2D classes. 2DAssess was also trained on neural network–generated artificial good class averages to improve its applicability to protein classes the network had not yet seen (77). Fully automated grid screening and an operator-free preprocessing pipeline hold promise for significantly increasing microscope throughput and efficiency in the future.
It is rapidly becoming the norm to acquire data for multiple samples in a single microscope session. Fully automating the entire data collection pipeline will be required to make the most efficient use of the instrument and avoid burnout of the operators. This can begin with an automated grid screening protocol, which can automatically select useful regions of a grid for imaging. Based on desirable metrics (e.g., CTF fit, ice thickness, particle distribution, 2D classes), a neural network may then select optimal grid squares and ice thicknesses for an extended data collection or move on to screen the next grid, just as human operators now do (77).These approaches should increase microscope throughput and reduce the need for human intervention, therefore making the data collection process faster, more efficient, and more cost effective. This approach is intimately connected to the need to fully process the data and feed this information back to the data collection strategy. We discuss this further in the section titled Processing.
FACILITIES
A critical component of ensuring consistent and reliable cryo-EM experiments is the proper construction and operation of a cryo-EM facility. The performance of an electron microscope is affected by its environment, siting, installation, and maintenance (78,79). In addition, as cryo-EM is being applied to biomedical and translational fields that include experiments in model cells, tissues, or organisms, operations at appropriate biosafety levels are important to ensure that experiments are conducted safely (80). Given the high operating costs of cryo-EM laboratories and 300-keV microscopes ($4–10 million), there is a need to ensure efficient use of instrumentation (81). Facility monitoring to track the health and status of the systems is crucial, as well as the ability to handle high volume and throughput through the development of metrics and infrastructure specialized for the cryo-EM field (82, 83). Home source 100-keV screening microscopes are sufficient for initial sample characterization, but for high-resolution results,300-keV microscopes with direct detectors are required, which may necessitate funding by large research institutes. As technology advances, the ability to obtain protein structures using 100–200-keV microscopes ($1–2 million and $2–5 million, respectively) will encourage cryo-EM democratization by offering scientists access to instrumentation at lower costs (57,59).Resources such as COSMIC2 (84),a science gateway developed by Michael Cianfrocco that provides free access to computing resources at the San Diego Supercomputer Center, are also an important step toward lowering barriers imposed by expensive computational infrastructure, thereby making cryo-EM more accessible.
National facilities are another emerging pathway to serve the rapidly expanding cryo-EM community by increasing the access of biomedical scientists to instrumentation (85; https://commonfund.nih.gov/cryoem ).Ascryo-EM technology is adopted by more biomedical research fields, the need for training opportunities is growing. National centers have the ability to serve as platforms for standardization of cryo-EM curricula, offer workshops, and provide a resource for cross-training local core facilities (https://www.cryoemcenters.org/ ). As cryo-EM national centers emulate synchrotrons, which have been a successful model for lowering the barriers of access to X-ray crystallography, remote cryo-EM data collection will become routine and more accessible to the general scientist (86). These centralized facilities will allow biomedical researchers to focus on their biological questions knowing they are supported by the availability of excellent infrastructure and scientific staff knowledgeable on the best practices in the field.
PROCESSING
Once cryo-EM micrographs have been acquired, the next step in the pipeline is computational image processing. This typically includes motion correction of movie frames, CTF estimation, particle picking and extraction, 2D classification, initial model generation, 3D classification, and 3D refinement to obtain a cryo-EM map. This workflow, now streamlined and very fast, has been built on the development of many advances in data processing, including the development of reliable initial model generation algorithms (87,88) and graphics processing unit (GPU) acceleration (89, 90), to mention only two. While the standard processing workflow for single-particle analysis is now well established, recent developments have allowed more challenging samples to be processed and record-breaking resolutions to be obtained.
Particle picking for a single-particle data set is the process of locating the imaged molecules of interest in the cryo-EM micrographs. Particle picking can be challenging due to the inherently low signal-to-noise ratio of the micrographs. Further complications can arise from impurities or contaminants present in the micrographs (i.e., so-called dirty micrographs), as well as from various characteristics of the imaged specimens such as low molecular weight, nonglobular shape, uneven distribution of particles or of specific particle views, and a tendency to aggregate or to assemble into multiple oligomeric states. Initial particle picking software often relied on heuristic approaches. As such, substantial user intervention was required to optimize various mathematical parameters used by the software. In addition, the removal of incorrectly picked particles or selection of missed particles frequently involved time-consuming manual intervention. With the ever-increasing size of data sets and variety of imaged specimens, these shortcomings became increasingly cumbersome, sometimes to the point of representing the main bottleneck in a cryo-EM project. Consequently, the past few years have seen major efforts dedicated to the development of improved particle picking software, often incorporating CNNs.
CNN-based pickers consist of two steps: network training and automated picking. In the training step, the software learns the characteristics of the imaged specimen and of the acquired data. Training can be achieved using particles picked manually from a small subset of micrographs, using particles selected by an alternative automated picking algorithm [e.g., difference of Gaussians (91)] ideally assisted by manual curation, or based on similar data sets on which the CNN has already been trained. The software uses the information gained from training to select particle locations from entire data sets. As is the case for traditional particle picking algorithms, multiple rounds of training-picking-curating can be performed to improve results, provided that appropriate caution is taken to confirm that the particles picked are real. At least nine particle picking algorithms based on CNNs have been introduced in the last two years. Among these, crYOLO (92), Warp’s BoxNet (93), and Topaz (94) have been successfully employed in a broad range of single-particle cryo-EM projects. crYOLO is based on the “you only look once” framework introduced in 2015 (95). Notable issues that have been overcome using crYOLO include uneven particle distribution (96), elongated specimen shapes leading to substantially different top and side views (97), dirty micrographs (98), peculiar specimen shapes (99), and coexistence of multiple oligomeric states (100). Topaz employs positive-unlabeled CNNs and has been used to tackle extreme particle heterogeneity (101), uneven particle distribution (96), and elongated specimen shapes (102), as well as to capture more specimen views and a higher true positive to false positive ratio compared to alternative approaches (103, 104). It is expected that general CNN-based picking software will continue to improve as models are trained on increasingly diverse data sets. This could potentially lead to accommodating high-resolution studies of samples for which a clean purification is unachievable and of samples with intrinsic flexibility or heterogeneity. In order for neural network particle pickers to be used routinely in automated processing workflows, general pickers need to be further optimized to pick a broad range of sample shapes and sizes.
Several approaches to increase the highest achievable resolution for a given data set have been introduced in the last few years. CTF refinement (74) estimates the defocus and astigmatism on a per-particle basis. Bayesian polishing (105) performs radiation dose weighting and corrects for beam-induced motion. Corrections for beam tilt (74), high-order aberrations (60), Ewald sphere curvature (106, 107), and anisotropic magnification (60) were also made available and were used to obtain one of the two 1.2-Å apoferritin maps (3). Currently there is no well-defined and widely accepted single sequence of performing these corrections that is guaranteed to achieve the highest possible resolution for any given data set. This is because resolution-limiting factors can vary across data sets, depending, for example, on the optical aberrations of the microscope, data collection scheme, and specimen characteristics. Users are usually advised to correct for the largest errors first and check the effects of each correction on both the resolution of the reconstruction and the performance of the subsequent corrections. An automated approach to this can be envisioned, whereby a user can submit a stack of particles from a 3D refinement into a job that tests the outcome of each correction to identify the optimal sequence of corrections.
One major advantage of cryo-EM is its ability to cope with high degrees of sample heterogeneity. Heterogeneity can be either discrete, in cases where the specimen conformations are readily discernible from one another, or continuous, in cases where the specimen conformations are either too many or too similar to be robustly distinguished. As previously established, discrete heterogeneity can be addressed by performing consecutive rounds of 3D classification, focused classification, and signal subtraction (108). Continuous heterogeneity, on the other hand, remained far more elusive until the recent development of a variety of novel approaches. Multibody refinement (109,110) seeks to tackle continuous heterogeneity by tracking user-defined bodies corresponding to specific subunits or components of the imaged specimen. Variability analysis based on principal component analysis fits 3D linear subspace models to cryo-EM data (111).Machine learning algorithms have also recently been employed to further advance analysis of continuous heterogeneity. For example, manifold embedding (112) is used to track the conformational motions across the energy landscape of the specimen, and EMAN2’s e2gmm (113) employs a 3D Gaussian mixture model to trace conformational spectrums. CryoDRGN (114) uses deep neural networks to identify continuous conformational changes while also providing tools for exploration of per-particle heterogeneity (Figure 6). As these new approaches for dealing with sample heterogeneity are still in their infancy, their effects on the analysis and interpretation of cryo-EM maps remain to be seen, but the prospects hold promise.
Figure 6.
CryoDRGN (114) proposes a deep learning framework for heterogeneous reconstruction that directly learns a continuous representation of 3D density maps without supervision from additional data sets or structures. Shown here are density maps reconstructed by cryoDRGN recapitulating a large continuous motion along a synthetic reaction coordinate. Image provided by Ellen Zhong and rendered with ChimeraX.
As more challenging and heterogeneous samples are imaged, better particle pickers and algorithms to manage heterogeneity will be critical for successfully interpreting these data. The development of general neural network particle pickers and 2D class selectors (77) will also be needed for better automation and throughput. When general neural network particle pickers and 2D class selectors are able to reliably select real particles, rare views, and challenging samples, the automation of the data collection process will be greatly simplified. Having these automated processes in place will enable high-throughput cryo-EM structure pipelines similar to those at X-ray crystallography beamlines, which will speed up both fundamental biological discoveries and drug discovery efforts.
MODELING
Once a cryo-EM map has been obtained, the next stage is to build and refine an appropriate model that accurately represents and interprets the experimental map. The accurate representation of cryo-EM maps with atomic models requires an extensive knowledge of chemistry and an understanding of the nature of the data and maps generated by cryo-EM. Thus far, cryo-EM has leaned heavily on the more mature field of X-ray crystallography for model building and validation. Some of the more commonly used tools for model building and refinement include the Collaborative Computational Project for Electron cryo-Microscopy (CCP-EM) (115), Phenix (116), Rosetta (117), UCSF Chimera (118), ISOLDE (119), and Coot (120). As with data processing, there is no one-size-fits-all approach to model building. Instead, the process depends on whether prior structures exist, the resolution and interpretability of the maps, and the user’s choice of refinement program. Model building into cryo-EM maps is a process with multiple challenges: Modeling large megadalton protein complexes is very labor intensive, conformational flexibility can limit map interpretability, and limited resolution can hinder accurate map interpretation. Furthermore, as cryo-EM has only attained true atomic resolution within the past year, technical details, including which electron scattering factor to use and how the hydrogens appear at high resolution, remain to be appropriately addressed (3, 4, 121, 122). Learning this information will enable better model building into cryo-EM maps at all resolutions.
Model building can incorporate a variety of supporting approaches. These approaches include generating homology models from existing nuclear magnetic resonance (NMR), X-ray, or cryo-EM structures; secondary structure prediction algorithms to aid sequencing and building; restraints from NMR or crosslinking mass spectrometry; and integrative structural modeling approaches (123). Recently developed automated approaches toward model building include Buccaneer (124), MAINMAST (125), and phenix.map_to_model (126). Deep learning is also being applied to automated backbone structure generation for high-resolution maps (127). Using a cascaded CNN, this approach leveraged GPU technology advances to process an entire 3D map with a fully connected network.
Neural networks can also be trained to predict protein structures from primary sequence information. DeepMind’s AlphaFold 2 (128) made headlines in 2020 by vastly outperforming competing software at the protein structure–prediction assessment CASP14 (129). Together with other neural network protein folding tools like RoseTTAFold (130), the ability to accurately predict protein structures will likely provide useful preliminary models for proteins with unknown structures that can be built into cryo-EM data, thereby speeding up the modeling process.
After a model is built and refined into a map, it must be validated. The EMDataResource (https://www.emdataresource.org) model challenge is held regularly to assess models that can be built using current state-of-the-art software and comprehensively evaluates existing model validation metrics. In the 2019 challenge (131), 63 models (51 of which were generated ab initio) built into 4 cryo-EM maps were submitted by 13 teams from the United States and Europe. The models were evaluated in four categories of validation tools: fit-to-map, coordinates-only, comparison-to reference, and comparison-among-models. From the challenge, recommendations were made for best practices in validating cryo-EM models, which provide a good standard in the field for better model building.
The model building field has recently leveraged the powers of neural networks to better predict and refine models for the interpretation of cryo-EM maps. The implementation of these tools in routine model building workflows will make the process faster and better. Community agreement on validation metrics and standards should continue to develop together with these tools so that structures being built manually or automatically, and the programs used to create these structures, will be more accurate and reliable.
CASE STUDY: ANTIBODIES TARGETING THE SARS-CoV-2 SPIKE
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread to more than 383 million people worldwide and killed more than 5 million as of February 2022 (132). Efforts to put an end to the pandemic are currently focused on the administration of multiple vaccines that have been developed, with a staggering 10 billion doses administered in only 13 months (133). However, in the absence of complete eradication, the need for effective treatment of the disease remains important. One possible treatment is the use of monoclonal antibodies as therapeutic agents.
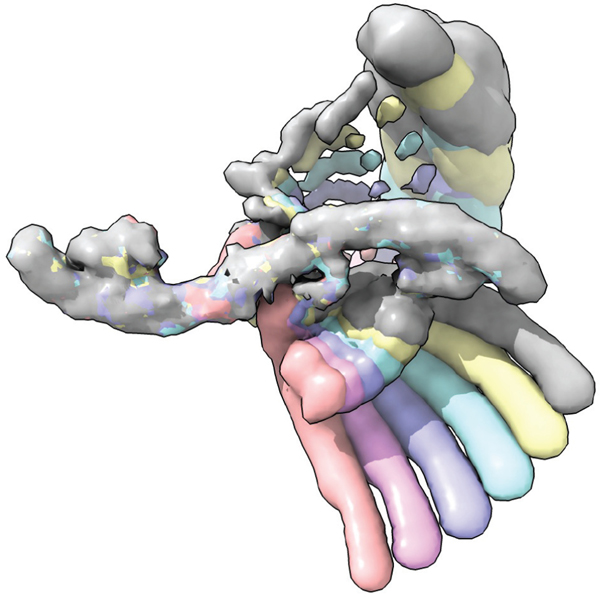
Structure determination of antibodies in complex with the spike (S) protein proceeded rapidly very soon after the pandemic was acknowledged. The S protein is a large oligomeric molecule with multiple conformations essential to the SARS-CoV-2 function, making it an ideal target for cryo-EM (Figure 7). Its primary role is to mediate fusion of the viral membrane with that of the host cell through binding to the human receptor ACE2 (134). Upon binding, the S1 subunit is shed, promoting a conformational change in the S2 subunit that initiates fusion. There are currently more than 80 structures of antibodies in complex with all or a part of the S protein that have been deposited to the PDB by many groups worldwide, most of which have been solved using cryo-EM. In this section, we expand on the collaborative efforts between the Shapiro lab at Columbia University, the Aaron Diamond AIDS Research Center, the National Center for CryoEM Access and Training (NCCAT), and the National Resource for Automated Molecular Microscopy (NRAMM) to combat COVID-19. While the work did not use all the recent cryoEM advances reviewed here, it is a useful example of how many of them were leveraged in a time of crisis when speed and accuracy were of global importance.
Figure 7.
Illustration of structures of antibodies (cyan, magenta, and green) targeting the SARS-CoV-2 spike (red) with interfaces highlighted. Advances across the entire cryo-EM pipeline were critical in enabling these structures to be determined in a very short time, thereby contributing to efforts to curb the COVID-19 pandemic.
Sample and Grid
The S protein purification protocol was standard, beginning with overexpression in suspension cell cultures and purification using affinity chromatography (135). One critical optimization step was lowering the pH of the sample from the standard 7.4 to a more acidic 5.5, a discovery that led to a fascinating project done in collaboration with the National Institutes of Health’s Vaccine Research Center (136). Work on other viral trimers, particularly HIV’s glycoprotein (gp) and influenza’s hemagglutinin (HA) protein, demonstrated the usefulness of detergents in vitrification. Even though these proteins are soluble since the transmembrane domains are not typically included for expression, the use of N-dodecyl-β-D-maltoside (DDM) was critical to obtaining high-quality micrographs. Without detergent, little protein could be observed in the holes of the support film, and what was there appeared to be denatured. Adding a small amount of DDM, about 0.005% w/v, improved image quality dramatically. As is routinely seen, the addition of detergent required an approximately tenfold increase in protein concentration compared to samples with no detergent. Additional challenges included the preference of protein for almost exclusively relatively thick ice, greater than 100 nm, despite the fact that the S protein’s longest dimension is only approximately 16 nm. Also, sample quality was poor on gold grids, at least in this work, although other groups used them successfully (137). This protocol, particularly the issues with gold grids and the benefit of a more acidic buffer, may be unique to this study, and the results produced were neither uniformly better nor worse than those of others. Why there is not currently a single, optimal protocol for even highly similar samples is unclear. This observation highlights the need for continued research into cryo-EM sample preparation theory and techniques and should remind researchers of the importance of extensive optimization (138, 139).
Instrumentation and Data
After obtaining samples, it was crucial to obtain data as quickly as possible. In this, national cryo-EM centers, particularly NCCAT, were key. Having a center staffed with experts in sample preparation, data collection, and processing allowed a moderately sized lab to increase data production by orders of magnitude almost overnight. Part of this was made possible by the advancements in data collection throughput, with improvements in camera speeds, data acquisition strategies, and data processing algorithms discussed in this review. All of these improvements were important because large amounts of data were required to solve structures of the highly heterogeneous protein complexes of spike proteins bound to receptors or antibodies. Each compositional and conformational class, of which there were many, needed enough particles to be accurately classified and reconstructed. The SARS-CoV-2 spike had multiple conformations, with each of the three receptor binding domains (RBDs) being in either the up or down state. When studying complexes bound to antibody fragment antigen-binding (Fab) regions, each RBD could have either zero or one Fab bound. The combinatorial nature of composition and conformation drastically increased the number of possible states. Sorting these states out required between 4,000 and 10,000 micrographs to solve a single structure. Utilizing current advancements in microscope speed and quality, together with an optimized sample and microscope setup, this process typically required no more than 24 h of data collection time.
Processing
As with data collection, improvements to the speed of processing algorithms greatly aided this work. GPU acceleration is now commonplace in cryo-EM data processing. Combined with advanced optimization methods like stochastic gradient descent (88), the large data sets collected could be processed very rapidly. The use of CNN particle pickers, namely Topaz (94), also greatly improved the quality of reconstructions by picking particles with a wider range of orientations compared to blob or manual picking. Like many other flexible complexes, the use of nonuniform refinement and local refinement in cryoSPARC (140) greatly improved the quality of the maps, particularly at the spike:antibody interface. Post-processing techniques such as density modification (141) also aided in interpretation of the density.
Modeling
The final stage, model building, benefited greatly from prior knowledge. Very little manual building was required, as most structures were of the previously solved spike protein (142) and antibodies that can be built using homology modeling programs such as Schrödinger’s BioLuminate (https://www.schrodinger.com/products/bioluminate) and ABodyBuilder (143). The largest hurdle was refining these structures into densities with different conformations; for example, not all RBDs in the up conformation are identical. We used a combination of programs such as Coot (120), Phenix (116), REFMAC (144), and ISOLDE (119) to interactively build and refine these structures in a relatively short amount of time.
IN SITU
Cryo-ET provides a method for determining the structure of macromolecules in their native environment, or in situ (2).Just as for single-particle cryo-EM, the resolution of the structures determined this way depends on the ability to align and average many copies of the particles, which for cryo-ET are individual 3D volumes (subtomograms) or coherent 2D slices of the subtomograms segmented out of the 3D tomogram (145, 146). A unique challenge for cryo-ET is that samples must be thin enough (<0.5 μm) to allow for the electron beam to pass through the sample coherently. Most cells and tissues exceed the thickness limit for cryo-EM/ET and must therefore be thinned. Two methods for thinning vitrified biological specimens have been developed and implemented: cryo-sectioning and cryo–focused ion beam scanning electron microscopy (cryoFIB/SEM) (reviewed in 147). For cryo-sectioning, thin slices of the specimen are cut using a diamond knife and transferred onto the surface of a grid (148, 149). In cryo-FIB/SEM, the bulk of a vitrified specimen is ablated away by a focused beam of gallium ions, leaving behind a thin area, a lamella typically 100–250 nm thick (150). An advantage of cryo-sectioning is that it allows for an entire specimen to be imaged by collecting and imaging consecutive slices, but there are many technical challenges, and the method has not been widely adopted. Cryo-FIB/SEM provides only a small window into the sample but results in higher-fidelity specimen preservation. Cryo-FIB/SEM can also provide sequential SEM imaging between each FIB-milling ablation step, called cryo-auto-slice-and-view (cryo-ASV) (151), but resolution is limited (~10 nm). Both cryo-FIB/SEM and cryo-sectioning may be coupled with fluorescent object identification using cryo–fluorescent light microscopy (cryo-FLM), also referred to as cryo–correlative light and electron microscopy, which can be performed before, during, and after vitrification and thinning (152–154). Fluorescent imaging can be achieved using a standalone cryo-FLM or by incorporating a cryo-FLM inside the chamber of a cryo-FIB/SEM for object identification during imaging (155). Cryo-ASV and/or cryo-FLM may be combined with cryo-FIB/SEM to more accurately locate regions of interest (156).
Methods for more routinely studying complex biological specimens by cryo-ET are beginning to be developed and put into practice. For example, cryo-FIB lift-out (157) removes a segment (typically tens of microns in each dimension) from a bulk specimen, moves it, attaches it to a TEM grid, and then FIB-mills the large segment to reduce it to a thin lamella suitable for cryo-ET. Cryo-FIB lift-out makes it possible to obtain high-resolution images from specimens with dimensions on the order of tens of microns if the bulk sample, for example, tissue, can be vitrified using a high-pressure freezer (HPF) (158, 159). Another recently developed method, called the waffle method (160), also uses HPF to vitrify a specimen onto a cryo-EM grid, but instead of lifting out a large piece of specimen, the bulk sample is FIB-milled on the original grid (Figure 8). This potentially increases throughput of lamellae-making by producing more lamellae than cryo-FIB lift-out and larger lamellae than conventional cryo-FIB/SEM.
Figure 8.
Schematic depicting lamellae cryo–focused ion beam (cryo-FIB)-milled using the waffle method. By high-pressure freezing then FIB-milling bulk sample on a grid, more and larger lamellae can be made, potentially increasing throughput. Several large (>10 × 10 μm) lamellae are depicted, and the inset shows the relative scale of the lamellae on the grid. Figure adapted from Reference 160 (CC BY-NC-ND 4.0).
In situ 3D visualization of complex biological environments such as cells and tissues at nanometer resolutions comes with many challenges. Cells are extremely crowded, heterogeneous, and complex environments involving numerous interacting protein complexes, organelles, membranes, transmembrane proteins, filaments and tubules, DNA, and RNA. Integrative modeling of existing structures and fragments into tomograms and subtomograms allows for medium-resolution localizations of higher-resolution complexes within their cellular contexts (161). However, to determine high-resolution structures of macromolecules or complexes within the cell, individual copies must be identified, extracted from the volume, and subsequently aligned and averaged. Cryo-ET provides high-resolution 3D volumes (6), but there are no labels available to identify individual molecules in the very noisy tomography volumes. Overlaying cryo-FLM data onto cryo-ET tomograms identifies areas of interest but only provides low-resolution localization (on the order of 100 nm). A developing method, super-res-cryo–fluorescent light microscopy, has the potential to provide localization on the order of 10 nm, but it is currently hampered in practice by the need for high beam intensities and concomitant devitrification issues (162–164).
Currently, in situ tomography is undergoing significant enhancements in data processing in three key areas: tomogram visualization, object identification in tomograms, and high-resolution subtomogram processing pipelines. Emerging tomogram reconstruction methods and deep learning denoising models are enabling higher-confidence visualization of molecules and interactions in 3D (165–168). While these improvements assist human interpretation of tomography data, semiautomated deep learning methods for object identification are also being developed in parallel. These deep learning methods build on the successes of the revolutions in computer vision in the past decade with CNNs and transformers. Advances in algorithms and computer hardware have significantly improved the capacity and accuracy of deep learning models. Self-supervised and unsupervised object detection requiring little or no human intervention are becoming a reality (169–173). As such, tomogram annotation tasks now take orders of magnitude less time and effort. An intriguing approach for localizing proteins of interest or their fragments uses high-resolution templates of known structures to accurately pinpoint protein locations and orientations from individual 2D projections of a vitrified cell (174). Lastly, algorithms for high-resolution processing of repeated objects in tomograms that successfully take into account local movements of objects in the ice are being developed (6, 175–177). Most notably, it was recently shown that ribosome particles extracted from thin cell-edge lamellae can be reconstructed to better than 4-Å resolution, and a bound ligand was identified (6). Given the highly heterogeneous nature of molecules in the cell, future advances in flexibility analysis of structures from in situ tomography, possibly through the use of deep learning methods (111, 114, 178), may increase the number of heterogeneous structures that may be analyzed at moderate to high resolutions.
As each bottleneck in the in situ tomography workflow is addressed, higher-throughput and more complete studies of native cellular and tissue specimens will be performed with less human intervention and bias. As in situ studies become routine, cryo-FIB/SEM may become part of the drug discovery and drug delivery analysis platform. It is even conceivable that in situ tomography may extend to patient tissue studies, opening the door to personalized medicine at molecular scales.
MicroED
MicroED (179), also known as continuous rotation electron diffraction (180), uses a TEM to record data in diffraction mode from 3D crystals. The strong interactions between electrons and matter make the method suitable for much smaller crystals, typically 0.2–1 μm in length, than can be used for X-ray crystallography (as small as ~5–10 μm at microfocus beamlines). The requirement for diffraction with minimal interference from dynamical scattering effects necessitates that crystals be smaller than ~1 μm thick. The method can be applied to biological macromolecules and small molecules (<~1,000 Da) of interest for medicinal chemistry, chemical biology, natural product research, organic semiconductor research, and synthetic organic chemistry. Along with MicroED methods that make use of continuous rotation of crystals in the microscope, additional 3D crystal electron (3D-ED) diffraction methods include SerialED (181) and nanobeam precession-assisted 3D diffraction (182).
While MicroED has been established for several years, contributions to protein structure determination have been limited to a small subset of proteins. Aside from proof-of-concept studies of proteins with previously solved structures, novel protein structures solved using MicroED have been limited to amyloid-forming oligopeptides (up to 12 amino acids) (183) and one protein (184).
The first step in any 3D-ED pipeline is to grow crystals of suitable size. However, the requirement for 0.2–1-μm crystals means they cannot be readily seen under a light microscope, thus requiring alternative methods to detect crystal growth. High-throughput methods for identifying microcrystals in crystallization trials are not yet routine, and microcrystal detection is currently the rate-limiting step in the pipeline. Cryogenic and negative-stain TEM have been used as screening methods for microcrystal formation, but higher-throughput approaches, including second-order nonlinear imaging of chiral crystals (185), dynamic light scattering (186), UV fluorescence imaging, and X-ray powder diffraction, have also been employed. These methods also require specialized equipment but can achieve higher throughput than TEM. Better and higher-throughput detection for microcrystals is an important requirement for making 3D-ED faster and more viable.
After microcrystals are obtained, they must be vitrified on a grid for imaging on a TEM. Dry powders of organic small-molecule crystals can simply be dusted onto a TEM grid (187). Preparing protein crystals typically follows the same workflow as for single-particle analysis: A solution of crystals is pipetted onto a TEM grid, excess liquid is blotted away with filter paper, then the crystals are plunge frozen in liquid ethane. Obtaining suitable ice thickness is nontrivial: Protein crystals must remain hydrated, but if the surrounding solvent on the frozen grid is too thick, it will negatively impact the signal-to-noise ratio. However, many crystallization solutions are composed of viscous precipitants that are difficult to blot away with conventional plunge freezing techniques, so novel blotting methods have been proposed (188). The conversion of highly viscous lipid matrix used for some membrane proteins to a more blottable phase has also been reported (189). Another approach makes use of large microcrystals that are opaque to the electron beam but can be thinned after vitrification by using FIB-milling, producing crystalline lamellae that can diffract (190).Crystalline lamellae have been shown to provide high-quality data (190),but the addition of another low-throughput step of FIB-milling into the pipeline currently impedes the widespread uptake of this method.
The collection of 3D-ED data can be done on a wide variety of instruments already used for cryo-EM imaging, although the optimal data collection setup is unique. ED does not require as high an optical performance as traditional cryo-EM imaging to achieve the same resolution. Detector requirements are also unique. Diffraction imaging concentrates the scattering power into a small number of diffraction spots. A detector with large dynamic range is therefore important to faithfully record both strong reflections and background. On the other hand, high detector quantum efficiency and the large number of pixels in the image that are important in cryo-EM imaging are not essential for diffraction data collection. A smooth rotation of the crystal within the fixed electron beam is crucial for continuous rotation diffraction, and it is even more important than in cryo-ET because beam-image shift, typically used to compensate feature movement during stage tilting in tomography, causes beam tilt, which complicates the phase recovery in diffraction experiments. Existing TEM hardware meets these requirements for stage tilt and low-dose diffraction procedures, allowing for collection of almost complete data from one crystal of high symmetry on many existing TEMs. Specialized electron diffraction setups are also being developed (191). The rate-limiting step once the crystals are inthe TEM is screening for diffraction-quality crystals. Not all crystals frozen on a grid will diffract to the required resolution, if at all. Thus, multiple crystals need to be surveyed, making this process time consuming. Automated data collection software such as Leginon (68), Instamatic (192), yoneoLocr (193), and EPU-D (ThermoFisher Scientific) can be used to streamline the screening and collection of diffraction data from crystals scattered across a grid, greatly enhancing the speed and efficiency of screening and data collection.
The final step in the 3D-ED pipeline is the solving of the structure from diffraction data. As with X-ray crystallography, phase information is not measured directly and must be independently determined. Small-molecule and short-peptide crystals that diffract to sufficient resolution (better than 1.2 Å) can be phased ab initio (187, 194). For crystals of larger biomacromolecules, molecular replacement (MR) is the only routine phasing option, although ab initio methods were recently used to solve structures of test proteins where data were collected to 1.5-Å resolution; or better (195). MR is an established method, and recent advancements in structure prediction such as with AlphaFold may provide successful MR models for targets with no known structural homologs (129). Continued development of electron diffraction data processing and analysis software that incorporates specific electron scattering factors is also advancing the utility of MicroED (196). For example, MicroED has been shown to allow for refinement of the charge state of bound metals (197–199). Another recent application is ligand soaking of microcrystals (200), a technique that could be useful in drug design pipelines when soaking into larger crystals is inefficient due to slow diffusion.
The utility of MicroED is clear, from solving structures of molecules that form only microcrystals to providing complementary data for systems that can also form larger crystals. Whereas MicroED for small-molecule structure determination is steadily growing among structural chemists, the technique remains underutilized for protein structure determination, possibly due to the low throughput of crystal screening and difficulties in grid preparation. However, in cases where atomic resolution information is needed and the molecules are too small for single-particle cryo-EM, the unique capabilities of electron diffraction will drive developments to make MicroED better, faster, and cheaper and therefore more accessible to the structural biology community.
DISSEMINATION
The final step in the cryo-EM pipeline is data deposition, where the maps, models, and raw data are made available to the general public. This critically important step in the pipeline has been a strong driving force for rapid developments in the field by making data available for other scientists and software developers to analyze and test new algorithms and software (201). Cryo-EM maps can be deposited into the EMDB (https://www.ebi.ac.uk/pdbe/emdb) (202) and their corresponding atomic models into the PDB (https://www.rcsb.org) (reviewed in 203). Primary data in the form of cryo-EM movies, micrographs, and particle stacks can be deposited into the Electron Microscopy Public Image Archive (EMPIAR; https://www.ebi.ac.uk/pdbe/emdb/empiar/) (204), and primary MicroED data into Zenodo (https://zenodo.org/communities/microed/). With ever-increasing amounts of data being generated at higher rates, in terms of both primary data (e.g., due to movie frames and higher microscope throughput) and processed data (e.g., flexibility data, multiple structures, movies of structures), data archiving platforms will need to continue to innovate and provide robust data archiving, annotation, and retrieval.
The data and structures generated by cryo-EM methods can be situated within their larger biological contexts. To this end, the BioImage Archive (https://www.ebi.ac.uk/bioimagearchive/about-us/) (205) was recently developed as a value-added database with the goal of providing careful curation and integration of data from different imaging modalities: for example, cryo-EM, super-resolution light microscopy, and light-sheet microscopy. In doing so, the archive aims to integrate knowledge across different imaging methods. By bringing together complementary information on the same system, new insights can be gained (206).
Open access to cryo-EM data and programs will be important for continuing to drive discovery and innovation in the field. Integrating information across databases from different imaging methods and scales has the potential to provide truly multiscale imaging for understanding the biology of a system, from atom to animal. The global cryo-EM community has been free flowing, with scholars depositing data on public repositories, distributing software developments as open source, prepublishing papers on bioRxiv, and having open and constructive discussions on public forums such as the mailing list for the CCP-EM. The openness with which the cryo-EM community has engaged in disseminating ideas, data, and research has and will continue to enable rapid advances in the field.
THE FUTURE
Cryo-EM as a field is continuing its trajectory of exponential growth, driven by developments along the entire pipeline from sample preparation to data dissemination. Here we have attempted to describe what we consider to be significant recent developments in each step of the pipeline as well as some outlook for the future. In the past decade, significant advances such as direct electron detectors and the development of motion correction software overcame the resolution limitations of cryo-EM. While innovative sample preparation devices have been developed to overcome issues with the AWI and to achieve biologically relevant timescales for imaging, these methods need much more development in the coming years for better throughput and widespread applicability. In addition, we anticipate that machine learning in cryo-EM will continue to make significant advances in automation for data collection and processing. As methods continue to develop for in situ imaging and reconstructions, the ability to image biological objects in their native environments at high resolutions promises to shed significant light on native biological processes within the next decade. Integrating biological information from single-particle and in situ tomography data and adding further context by combining these data with information from other imaging modalities will provide more holistic insight into the biology of a system. Altogether, cryo-EM continues to contribute to our understanding of the function of molecular machines both in vitro and in vivo. With open input from the community, structures are being determined at ever-improving resolutions, with information on the dynamics of the systems, and all this at higher throughputs and with greater accessibility to the wider biological research community. The future of cryo-EM remains very bright.
ACKNOWLEDGMENTS
We would like to thank the staff at the Simons Electron Microscopy Center for enabling the cryo-EM work at the facility, Sjors Scheres for providing valuable feedback, and Takanori Nakane and Ellen Zhong for providing figures of their beautiful work. Some of this work was supported by the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247) and the National Institutes of Health National Institute of General Medical Sciences (GM103310). Some of this work was also supported by the National University of Singapore Presidential Young Professorship Award (R-154-000-C62-133 to Y.Z.T.), Ministry of Education Singapore (to Y.Z.T.), and Agency for Science Technology and Research Singapore (to Y.Z.T.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Cheng Y. 2018. Single-particle cryo-EM—How did it get here and where will it go. Science 361:876–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turk M, Baumeister W. 2020. The promise and the challenges of cryo-electron tomography. FEBS Lett. 594:3243–61 [DOI] [PubMed] [Google Scholar]
- 3.Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, et al. 2020. Single-particle cryo-EM at atomic-resolution. Nature 587:152–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yip KM, Fischer N, Paknia E, Chari A, Stark H. 2020. Atomic-resolution protein structure determination by cryo-EM. Nature 587:157–61 [DOI] [PubMed] [Google Scholar]
- 5.Zhang K, Pintilie GD, Li S, Schmid MF, Chiu W. 2020. Resolving individual atoms of protein complexby cryo-electron microscopy. Cell Res. 30:1136–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J. 2021. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5A in cells.˚ Nat. Methods 18:186–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandey VP, Kahn P, Wei H, Carragher B, Potter CS. 2019. Scorpion: facilitating high throughput electron microscopy. Microsc. Microanal 25:1002–3 [Google Scholar]
- 8.Young G, Hundt N, Cole D, Fineberg A, Andrecka J, et al. 2018. Quantitative mass imaging of single biological macromolecules. Science 360:423–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soltermann F, Foley EDB, Pagnoni V, Galpin M, Benesch JLP, et al.2020.Quantifying protein-protein interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed 59:10774–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olerinyova A, Sonn-Segev A, Gault J, Eichmann C, Schimpf J, et al. 2021. Mass photometry of membrane proteins. Chem 7:224–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Struwe WB, Kukura P. 2020. Single molecule mass photometry of nucleic acids. Nucleic Acids Res. 48:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choy BC, Cater RJ, Mancia F, Pryor EE Jr. 2021. A 10-year meta-analysis of membrane protein structural biology: detergents, membrane mimetics, and structure determination techniques. Biochim.Biophys. Acta Biomembranes 1863:183533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Autzen HE, Julius D, Cheng Y. 2019. Membrane mimetic systems in CryoEM: keeping membrane proteins in their native environment. Curr. Opin. Struct. Biol 58:259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho C-M, Beck JR, Lai M, Cui Y, Goldberg DE, et al. 2018. Malaria parasite translocon structure and mechanism of effector export. Nature 561:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Bon C, Michon B, Popot JL, Zoonens M. 2021. Amphipathic environments for determining the structure of membrane proteins by single-particle electron cryo-microscopy. Q. Rev. Biophys 54:e6. [DOI] [PubMed] [Google Scholar]
- 16.Yao X, Fan X, Yan N. 2020. Cryo-EM analysis of a membrane protein embedded in the liposome. PNAS 117:18497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonggu L, Wang L. 2020. Cryo-EM sample preparation method for extremely low concentration liposomes. Ultramicroscopy 208:112849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Drie JH, Tong L. 2020. Cryo-EM as a powerful tool for drug discovery. Bioorg. Med. Chem. Lett 30:127524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan YZ,Zhang L,Rodrigues J,Zheng RB,Giacometti SI,et al.2020.Cryo-EM structures and regulation of arabinofuranosyltransferase AftD from mycobacteria. Mol. Cell 78(4):683–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaeser RM, Han BG. 2017. Opinion: hazards faced by macromolecules when confined to thin aqueousfilms. Biophys. Rep 3:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor KA, Glaeser RM. 2008. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol 163:214–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble AJ, Dandey VP, Wei H, Brasch J, Chase J, et al. 2018. Routine single particle CryoEM sample and grid characterization by tomography. eLife 7:e34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Imprima E, Floris D, Joppe M, Sánchez R, Grininger M, Kühlbrandt W. 2019. Protein denaturation at the air-water interface and how to prevent it. eLife 8:e42747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinothkumar KR, Henderson R.2016.Single particle electron cryomicroscopy: trends, issues and future perspective. Q. Rev. Biophys 49:e13. [DOI] [PubMed] [Google Scholar]
- 25.Klebl DP, Gravett MSC, Kontziampasis D, Wright DJ, Bon RS, et al. 2020. Need for speed: examining protein behavior during cryoEM grid preparation at different timescales. Structure 28:1238–48.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Fan X, Wang H, Zhao F, Tully CG, et al. 2020. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. PNAS 117:1009–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naydenova K, Peet MJ, Russo CJ. 2019. Multifunctional graphene supports for electron cryomicroscopy. PNAS 116:11718–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han BG, Watson Z, Kang H, Pulk A, Downing KH, et al. 2016. Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J. Struct. Biol 195:238–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llaguno MC, Xu H, Shi L, Huang N, Zhang H, et al. 2014. Chemically functionalized carbon films for single molecule imaging. J. Struct. Biol 185:405–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Li K, Jiang W. 2016. Antibody-based affinity cryo-EM grid. Methods 100:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastner B, Fischer N, Golas MM, Sander B, Dube P, et al. 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5:53–55 [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Fu Z, Kaledhonkar S, Jia Y, Shah B, et al. 2017. A fast and effective microfluidic spraying-plunging method for high-resolution single-particle cryo-EM. Structure 25:663–70.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashtiani D, Venugopal H, Belousoff M, Spicer B, Mak J, et al.2018.Delivery of femtolitre droplets using surface acoustic wave based atomisation for cryo-EM grid preparation. J. Struct. Biol 203:94–101 [DOI] [PubMed] [Google Scholar]
- 34.Arnold SA, Albiez S, Bieri A, Syntychaki A, Adaixo R, et al.2017.Blotting-free and lossless cryo-electronmicroscopy grid preparation from nanoliter-sized protein samples and single-cell extracts. J. Struct. Biol 197:220–26 [DOI] [PubMed] [Google Scholar]
- 35.Ravelli RBG, Nijpels FJ, Henderikx RJ, Weissenberger G, Thewessem S, et al. 2020. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun 11:2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinstein JL, Guo H, Ripstein ZA, Haydaroglu A, Au A, et al. 2019. Shake-it-off: a simple ultrasonic cryo-EM specimen-preparation device. Acta Crystallogr. D Struct. Biol 75:1063–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan YZ, Rubinstein JL.2020.Through-grid wicking enables high-speed cryoEM specimen preparation. Acta Crystallogr. D Struct. Biol 76:1092–1103 [DOI] [PubMed] [Google Scholar]
- 38.Jain T, Sheehan P, Crum J, Carragher B, Potter CS. 2012. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J. Struct. Biol 179:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han H, Fulcher JM, Dandey VP, Iwasa JH, Sundquist WI, et al. 2019. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. eLife 8:e44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, et al. 2020. FACT caught in the act of manipulating the nucleosome. Nature 577:426–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unwin N. 1995. Acetylcholine receptor channel imaged in the open state. Nature 373:37–43 [DOI] [PubMed] [Google Scholar]
- 42.Frank J. 2017. Time-resolved cryo-electron microscopy: recent progress. J. Struct. Biol 200:303–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dandey VP, Budell WC, Wei H, Bobe D, Maruthi K, et al. 2020. Time-resolved cryo-EM using Spotiton. Nat. Methods 17:897–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Z, Shaikh TR, Barnard D, Meng X, Mohamed H, et al. 2009. Monolithic microfluidic mixing–spraying devices for time-resolved cryo-electron microscopy. J. Struct. Biol 168:388–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, et al. 2015. Structural dynamics of ribosome subunitassociation studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure 23:1097–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC,et al. 2012. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol 177:630–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, et al.2012.Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20:1823–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, et al. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10:584–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres SH. 2014. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3:e03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo CJ, Passmore LA. 2014. Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346:1377–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, et al. 2017. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14:793–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naydenova K, Jia P, Russo CJ. 2020. Cryo-EM with sub–1 Å specimen movement. Science 370:223–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engstrom T, Clinger JA, Spoth KA, Clarke OB, Closs DS, et al. 2021. High-resolution single-particle cryo-EM of samples vitrified in boiling nitrogen. IUCrJ 8:867–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armstrong M,Han B-G, Gomez S, Turner J, Fletcher DA, Glaeser RM.2020.Microscale fluid behavior during cryo-EM sample blotting. Biophys. J 118:708–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ricolleau C, Nelayah J, Oikawa T, Kohno Y, Braidy N, et al. 2012. High resolution imaging and spectroscopy using CS-corrected TEM with cold FEG JEM-ARM200F. JEOL News 47:2–8 [Google Scholar]
- 56.Kato T, Makino F, Nakane T, Terahara N, Kaneko T, et al. 2019. CryoTEM with a cold field emission gun that moves structural biology into a new stage. Microsc. Microanal 25:998–9931232262 [Google Scholar]
- 57.Wu M, Lander GC, Herzik MA Jr. 2020. Sub-2 Angstrom resolution structure determination using single-particle cryo-EM at 200 keV. J. Struct. Biol. X 4:100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peet MJ, Henderson R, Russo CJ. 2019. The energy dependence of contrast and damage in electron cryomicroscopy of biological molecules. Ultramicroscopy 203:125–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naydenova K, McMullan G, Peet M, Lee Y, Edwards P, et al. 2019. CryoEM at 100keV: ademonstration and prospects. IUCrJ 6:1086–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zivanov J, Nakane T, Scheres SH. 2020. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7:253–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun M, Azumaya CM, Tse E, Bulkley DP, Harrington MB, et al.2021.Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM. J. Struct. Biol 213:107745. [DOI] [PubMed] [Google Scholar]
- 62.Eng ET, Kopylov M, Negro CJ, Dallaykan S, Rice WJ, et al. 2019. Reducing cryoEM file storage using lossy image formats. J. Struct. Biol 207:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo H, Franken E, Deng Y, Benlekbir S, Singla Lezcano G, et al. 2020. Electron-event representation data enable efficient cryoEM file storage with full preservation of spatial and temporal resolution. IUCrJ 7:860–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh C, Axelrod JJ, Campbell SL, Dioquino JY, Petrov PN, et al. 2021. High-power near-concentric Fabry-Perot cavity for phase contrast electron microscopy. Rev. Sci. Instrum 92:053005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buijsse B, Trompenaars P, Altin V, Danev R, Glaeser RM. 2020. Spectral DQE of the Volta phase plate. Ultramicroscopy 218:113079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, et al. 2000. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol 132:33–45 [DOI] [PubMed] [Google Scholar]
- 67.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, et al. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151:41–60 [DOI] [PubMed] [Google Scholar]
- 68.Cheng A, Negro C, Bruhn JF, Rice WJ, Dallakyan S, et al.2021.Leginon: new features and applications. Protein Sci. 30:136–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152:36–51 [DOI] [PubMed] [Google Scholar]
- 70.Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, et al. 2009. JADAS: a customizable automated dataacquisition system and its application to ice-embedded single particles. J. Struct. Biol 165:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng A, Eng ET, Alink L, Rice WJ, Jordan KD, et al. 2018. High resolution single particle cryoelectron microscopy using beam-image shift. J. Struct. Biol 204:270–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glaeser RM, Typke D, Tiemeijer PC, Pulokas J, Cheng A. 2011. Precise beam-tilt alignment and collimation are required to minimize the phase error associated with coma in high-resolution cryo-EM. J. Struct. Biol 174:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C, Huang X, Cheng J, Zhu D, Zhang X. 2019. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol 208:107396. [DOI] [PubMed] [Google Scholar]
- 74.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, et al. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Timm DE, Elhabian SY. 2020. Attention-guided quality assessment for automated cryo-EM gridscreening. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020, ed.Martel AL, pp. 56–65. Cham, Switz.: Springer [Google Scholar]
- 76.Yokoyama Y, Terada T, Shimizu K, Nishikawa K, Kozai D, et al. 2020. Development of a deep learning-based method to identify “good” regions of a cryo-electron microscopy grid. Biophys. Rev 12:349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Cash JN, Tesmer JJG, Cianfrocco MA. 2020. High-throughput cryo-EM enabled by user-free preprocessing routines. Structure 28:858–69.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mills DJ. 2021. Setting up and operating a cryo-EM laboratory. Q. Rev. Biophys 54:e2. [DOI] [PubMed] [Google Scholar]
- 79.Sader K, Matadeen R, Castro Hartmann P, Halsan T, Schlichten C. 2020. Industrial cryo-EM facility setup and management. Acta Crystallogr. D Struct. Biol 76:313–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherman MB, Trujillo J, Leahy I, Razmus D, Dehate R, et al. 2013. Construction and organization of aBSL-3 cryo-electron microscopy laboratory at UTMB. J. Struct. Biol 181:223–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alewijnse B, Ashton AW, Chambers MG, Chen S, Cheng A, et al. 2017. Best practices for managing large cryoEM facilities. J. Struct. Biol 199:225–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alink LM, Eng ET, Gheorghita R, Rice W, Cheng A, et al.2021. System environmental metrics collector for EM facilities. bioRxiv 2021.11.04.467268. 10.1101/2021.11.04.467268 [DOI] [Google Scholar]
- 83.Scapin G, Prosise WW, Wismer MK, Strickland C. 2017. A novel storage system for cryoEM samples. J. Struct. Biol 199:84–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cianfrocco MA, Wong-Barmun M, Youn C, Wagner R, Leschziner A. 2017. COSMIC2: a science gateway for cryo-electron microscopy structure determination. In PEARC17: Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact, ed. Hart D, No. 22. New York: Assoc. Comput. Mach. [Google Scholar]
- 85.Bhella D. 2019. Cryo-electron microscopy: an introduction to the technique, and considerations when working to establish a national facility. Biophys. Rev 11:515–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kandiah E, Giraud T, de Maria Antolinos A, Dobias F, Effantin G, et al. 2019. CM01: a facility for cryo-electron microscopy at the European Synchrotron. Acta Crystallogr. D Struct. Biol 75:528–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elmlund H, Elmlund D, Bengio S. 2013. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure 21:1299–306 [DOI] [PubMed] [Google Scholar]
- 88.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA.2017.cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14:290–96 [DOI] [PubMed] [Google Scholar]
- 89.Li X, Grigorieff N, Cheng Y. 2010. GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors. J. Struct. Biol 172:407–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tagare HD, Barthel A, Sigworth FJ. 2010. An adaptive Expectation-Maximization algorithm with GPU implementation for electron cryomicroscopy. J. Struct. Biol 171:256–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. 2009. DoG Picker and TiltPicker: software tools to facilitate particle selection insingle particle electron microscopy. J.Struct.Biol 166:205–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, et al. 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol 2:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tegunov D, Cramer P. 2019. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16:1146–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, et al. 2019. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16:1153–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Redmon J, Divvala S, Girshick R, Farhadi A. 2016. You only look once: unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 779–88. Piscataway, NJ: IEEE [Google Scholar]
- 96.Rogala KB, Gu X, Kedir JF, Abu-Remaileh M, Bianchi LF, et al. 2019. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366:468–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen K, Rogala KB, Chou HT, Huang RK, Yu Z, Sabatini DM. 2019. Cryo-EM structure of the humanFLCN-FNIP2-Rag-Ragulator complex. Cell 179:1319–29.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, et al. 2020. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol 27:1185–93 [DOI] [PubMed] [Google Scholar]
- 99.Consolati T, Locke J, Roostalu J, Chen ZA, Gannon J, et al. 2020. Microtubule nucleation properties of single human γ TuRCs explained by their cryo-EM structure. Dev. Cell 53:603–17.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bilokapic S, Suskiewicz MJ, Ahel I, Halic M. 2020. Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin. Nature 585:609–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sears AE, Albiez S, Gulati S, Wang B, Kiser P, et al. 2020. Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody. FASEB J. 34:13918–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stanisich JJ, Zyla DS, Afanasyev P, Xu J, Kipp A, et al. 2020. The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism. eLife 9:e60265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, et al. 2021. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371:abc3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kern DM, Sorum B, Mali SS, Hoel CM, Sridharan S, et al. 2021. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol 28:573–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zivanov J, Nakane T, Scheres SH. 2019. A Bayesian approach to beam-induced motion correction incryo-EM single-particle analysis. IUCrJ 6:5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Russo CJ, Henderson R. 2018. Ewald sphere correction using a single side-band image processing algorithm. Ultramicroscopy 187:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.DeRosier DJ. 2000. Correction of high-resolution data for curvature of the Ewald sphere. Ultramicroscopy 81:83–98 [DOI] [PubMed] [Google Scholar]
- 108.Bai XC,Rajendra E,Yang G,Shi Y,Scheres SH.2015.Sampling the conformational space of the catalyticsubunit of human γ-secretase. eLife 4:e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakane T, Kimanius D, Lindahl E, Scheres SH. 2018. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7:e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakane T, Scheres SH. 2021. Multi-body refinement of cryo-EM images in RELION. In CryoEM: Methods and Protocols, ed. Gonen T, Nannenga BL, pp. 145–60. New York: Humana Press; [DOI] [PubMed] [Google Scholar]
- 111.Punjani A, Fleet DJ. 2021. 3D Variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol 213:107702. [DOI] [PubMed] [Google Scholar]
- 112.Maji S, Liao H, Dashti A, Mashayekhi G, Ourmazd A, Frank J. 2020. Propagation of conformational coordinates across angular space in mapping the continuum of states from cryo-EM data by manifold embedding. J. Chem. Inf. Model 60:2484–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen M, Ludtke SJ. 2021. Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM. Nat. Methods 18:930–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong ED, Bepler T, Berger B, Davis JH. 2021. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 18:176–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burnley T, Palmer CM, Winn M. 2017. Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol 73:469–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, et al. 2018. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol 74:531–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang RY- R, Song Y, Barad BA, Cheng Y, Fraser JS, DiMaio F. 2016. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 5:e17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. 2004. UCSF Chimera—avisualization system for exploratory research and analysis. J. Comput. Chem 25:1605–12 [DOI] [PubMed] [Google Scholar]
- 119.Croll TI. 2018. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol 74:519–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Casañal A,Lohkamp B,Emsley P.2020.Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29:1055–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marques MA, Purdy MD, Yeager M. 2019. CryoEM maps are full of potential. Curr. Opin. Struct. Biol 58:214–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan YZ, Aiyer S, Mietzsch M, Hull JA, McKenna R, et al. 2018. Sub-2 Å Ewald curvature corrected structure of an AAV2 capsid variant. Nat. Commun 9:3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rout MP, Sali A. 2019. Principles for integrative structural biology studies. Cell 177:1384–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoh SW,Burnley T,Cowtan K.2020.Current approaches for automated model building into cryo-EM maps using Buccaneer with CCP-EM. Acta Crystallogr. D Struct. Biol 76:531–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Terashi G, Kihara D. 2018. De novo main-chain modeling for EM maps using MAINMAST. Nat. Commun 9:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Terwilliger TC, Adams PD, Afonine PV, Sobolev OV. 2018. A fully automatic method yielding initial models from high-resolution cryo-electron microscopy maps. Nat. Methods 15:905–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Si D, Moritz SA, Pfab J, Hou J, Cao R, et al. 2020. Deep learning to predict protein backbone structure from high-resolution cryo-EM density maps. Sci. Rep 10:4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, et al. 2020. Improved protein structure prediction using potentials from deep learning. Nature 577:706–10 [DOI] [PubMed] [Google Scholar]
- 129.Callaway E. 2020. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving proteinstructures. Nature 588:203–4 [DOI] [PubMed] [Google Scholar]
- 130.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, et al.2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373:871–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lawson CL, Kryshtafovych A, Adams PD, Afonine PV, Baker ML, et al. 2021. Cryo-EM model validation recommendations based on outcomes of the 2019 EMDataResource challenge. Nat. Methods 18:156–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.WHO (World Health Organ.).2022.WHO coronavirus (COVID-19) dashboard. Internet Resour.,WHO, Geneva, Switz. https://covid19.who.int/ [Google Scholar]
- 133.Our World in Data. 2022. Coronavirus (COVID-19) vaccinations. Internet Resour., Our World in Data, Oxford, UK. https://ourworldindata.org/covid-vaccinations#citation [Google Scholar]
- 134.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu L, Wang P, Nair MS, Yu J, Rapp M, et al. 2020. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584:450–56 [DOI] [PubMed] [Google Scholar]
- 136.Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, et al. 2020. Cryo-EM structures of SARS-CoV2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28:867–79.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, et al. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588:682–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.White JBR, Maskell DP, Howe A, Harrow M, Clare DK, et al. 2021. Single particle cryo-electron microscopy: from sample to structure. J. Vis. Exp 171:e62415. [DOI] [PubMed] [Google Scholar]
- 139.Weissenberger G, Henderikx RJM, Peters PJ. 2021.Understanding the invisible hands of sample preparation for cryo-EM. Nat. Methods 18:463–71 [DOI] [PubMed] [Google Scholar]
- 140.Punjani A, Zhang H, Fleet DJ. 2020. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17:1214–21 [DOI] [PubMed] [Google Scholar]
- 141.Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, Afonine PV. 2020. Improvement of cryo-EM maps by density modification. Nat. Methods 17:923–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, et al. 2020. Cryo-EM structure of the 2019nCoV spike in the prefusion conformation. Science 367:1260–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leem J, Dunbar J, Georges G, Shi J, Deane CM. 2016. ABodyBuilder: automated antibody structure prediction with data-driven accuracy estimation. mAbs 8:1259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, et al. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr 67:355–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Briggs JA. 2013. Structural biology in situ—the potential of subtomogram averaging. Curr. Opin. Struct. Biol 23:261–67 [DOI] [PubMed] [Google Scholar]
- 146.Pyle E, Zanetti G. 2021. Current data processing strategies for cryo-electron tomography and subtomogram averaging. Biochem. J 478:1827–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaplan M, Nicolas WJ, Zhao W, Carter SD, Metskas LA, et al. 2021. In situ imaging and structure determination of biomolecular complexes using electron cryo-tomography. In CryoEM: Methods and Protocols, ed. Gonen T, Nannenga BL, pp. 83–111. New York: Humana Press; [DOI] [PubMed] [Google Scholar]
- 148.Tan ZY, Cai S, Noble AJ, Chen JK, Shi J, Gan L. 2021. Heterogeneous non-canonical nucleosomes predominate in yeast cells in situ. bioRxiv 2021.04.04.438362. 10.1101/2021.04.04.438362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bharat TA, Hoffmann PC, Kukulski W. 2018. Correlative microscopy of vitreous sections provides insights into BAR-domain organization in situ. Structure 26:879–86.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wagner FR, Watanabe R, Schampers R, Singh D, Persoon H, et al.2020.Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat. Protoc 15:2041–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuba J, Mitchels J, Hovorka M, Erdmann P, Berka L, et al. 2021. Advanced cryo-tomography workflow developments–correlative microscopy, milling automation and cryo-lift-out. J. Microsc 281:112–24 [DOI] [PubMed] [Google Scholar]
- 152.Jun S, Ro H-J, Bharda A, Kim SI, Jeoung D, Jung HS. 2019. Advances in cryo-correlative light and electron microscopy: applications for studying molecular and cellular events. Protein J. 38:609–15 [DOI] [PubMed] [Google Scholar]
- 153.Carter SD, Mamede JI, Hope TJ, Jensen GJ. 2020. Correlated cryogenic fluorescence microscopy and electron cryo-tomography shows that exogenous TRIM5α can form hexagonal lattices or autophagy aggregates in vivo. PNAS 117:29702–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang JE, Larson MR, Sibert BS, Shrum S, Wright ER. 2021. CorRelator: interactive software for real-time high precision cryo-correlative light and electron microscopy. J. Struct. Biol 213:107709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gorelick S, Buckley G, Gervinskas G, Johnson TK, Handley A, et al. 2019. PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. eLife 8:e45919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klumpe S, Fung HKH, Goetz SK, Zagoriy I, Hampoelz B, et al. 2021. A modular platform for streamlining automated cryo-FIB workflows. bioRxiv 2021.05.19.444745. 10.1101/2021.05.19.444745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, et al. 2019. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods 16:757–62 [DOI] [PubMed] [Google Scholar]
- 158.Moor H. 1987. Theory and practice of high pressure freezing. In Cryotechniques in Biological Electron Microscopy, ed. Steinbrecht RA, Zierold K, pp. 175–91. Berlin: Springer [Google Scholar]
- 159.Dahl R, Staehelin LA. 1989. High-pressure freezing for the preservation of biological structure: theory and practice. J. Electron Microsc. Tech 13:165–74 [DOI] [PubMed] [Google Scholar]
- 160.Kelley K, Raczkowski AM, Klykov O, Jaroenlak P, Bobe D, et al. 2021. Waffle method: a general and flexible approach for improving throughput in FIB-milling. bioRxiv 2020.10.28.359372. 10.1101/2020.10.28.359372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Watanabe R, Buschauer R, Böhning J, Audagnotto M, Lasker K, et al. 2020. The in situ structure of Parkinson’s disease-linked LRRK2. Cell 182:1508–18.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tuijtel MW, Koster AJ, Jakobs S, Faas FG, Sharp TH. 2019. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep 9:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Moser F, Pražák V, Mordhorst V, Andrade DM, Baker LA, et al. 2019. Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. PNAS 116:4804–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoffman DP, Shtengel G,Xu CS, Campbell KR, Freeman M, et al.2020.Correlative three-dimensionalsuper-resolutionandblock-faceelectronmicroscopyofwholevitreouslyfrozencells.Science 367:aaz5357 [Google Scholar]
- 165.Yan R, Venkatakrishnan SV, Liu J, Bouman CA, Jiang W. 2019. MBIR: a cryo-ET 3D reconstruction method that effectively minimizes missing wedge artifacts and restores missing information. J. Struct. Biol 206:183–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen Y, Wang Z, Zhang J, Li L, Wan X, et al. 2017. Accelerating electron tomography reconstruction algorithm ICON with GPU. Biophys. Rep 3:36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Buchholz T-O, Jordan M, Pigino G, Jug F. 2019. Cryo-care: content-aware image restoration for cryo-transmission electron microscopy data. Presented at 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, April 8–11 [Google Scholar]
- 168.Bepler T, Kelley K, Noble AJ, Berger B. 2020. Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun 11:5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen M, Dai W, Sun SY, Jonasch D, He CY, et al. 2017. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zeng X, Leung MR, Zeev-Ben-Mordehai T, Xu M. 2018. A convolutional autoencoder approach for mining features in cellular electron cryo-tomograms and weakly supervised coarse segmentation. J. Struct. Biol 202:150–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu M, Singla J, Tocheva EI, Chang YW, Stevens RC, et al. 2019. De novo structural pattern mining in cellular electron cryotomograms. Structure 27:679–91.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martinez-Sanchez A, Kochovski Z, Laugks U, Meyer zum Alten Borgloh J, Chakraborty S, et al. 2020. Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms. Nat. Methods 17:209–16 [DOI] [PubMed] [Google Scholar]
- 173.GubinsI Chaillet ML, vander Schot G Veltkamp RC, Förster F, et al.2020.SHREC2020:classificationin cryo-electron tomograms. Comput. Graph 91:279–89 [Google Scholar]
- 174.Lucas BA, Himes BA, Xue L, Grant T, Mahamid J, Grigorieff N. 2021. Locating macromolecular assemblies in cells by 2D template matching with cisTEM. eLife 10:e68946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Himes BA, Zhang P. 2018. emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat. Methods 15:955–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen M, Bell JM, Shi X, Sun SY, Wang Z, Ludtke SJ. 2019. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16:1161–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bouvette J, Liu H-F, Du X, Zhou Y, Sikkema AP, et al. 2021. Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography. Nat. Commun 12:1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen M, Ludtke S. 2021. Deep learning based mixed-dimensional GMM for characterizing variability in CryoEM. arXiv:2101.10356 [q-bio.BM] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nguyen C, Gonen T. 2020. Beyond protein structure determination with MicroED. Curr. Opin. Struct. Biol 64:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Y, Yang T, Xu H, Zou X, Wan W. 2018. On the quality of the continuous rotation electron diffraction data for accurate atomic structure determination of inorganic compounds. J.Appl. Crystallogr 51:1094–101 [Google Scholar]
- 181.Bucker R, Hogan-Lamarre P, Mehrabi P, Schulz EC, Bultema LA, et al. 2020. Serial protein crystallography in an electron microscope. Nat. Commun 11:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lanza A, Margheritis E, Mugnaioli E, Cappello V, Garau G, Gemmi M. 2019. Nanobeam precession assisted 3D electron diffraction reveals a new polymorph of hen egg-white lysozyme. IUCrJ 6:178–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nannenga BL, Gonen T.2019.The cryo-EM method microcrystal electron diffraction (MicroED). Nat. Methods 16:369–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Xu H, Lebrette H, Clabbers MTB, Zhao J, Griese JJ, et al. 2019. Solving a new R2lox protein structure by microcrystal electron diffraction. Sci. Adv 5:eaax4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wampler RD, Begue NJ, Simpson GJ. 2008. Molecular design strategies for optimizing the nonlinear optical properties of chiral crystals. Cryst. Growth Des 8:2589–94 [Google Scholar]
- 186.de Wijn R, Rollet K, Engilberge S, McEwen AG, Hennig O, et al. 2020. Monitoring the production of high diffraction-quality crystals of two enzymes in real time using in situ dynamic light scattering. Crystals 10:65 [Google Scholar]
- 187.Gruene T, Wennmacher JTC, Zaubitzer C, Holstein JJ, Heidler J, et al. 2018. Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angew. Chem. Int. Ed 57:16313–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao J, Xu H, Lebrette H, Carroni M, Taberman H, et al. 2021. A simple pressure-assisted method for MicroED specimen preparation. bioRxiv 665448. 10.1101/665448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Martynowycz MW, Shiriaeva A, Ge X, Hattne J, Nannenga BL, et al. 2020. MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP. bioRxiv 2020.09.27.316109. 10.1101/2020.09.27.316109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Duyvesteyn HME, Kotecha A, Ginn HM, Hecksel CW, Beale EV, et al. 2018. Machining protein microcrystals for structure determination by electron diffraction. PNAS 115:9569–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yonekura K, Ishikawa T, Maki-Yonekura S. 2019. A new cryo-EM system for electron 3D crystallography by eEFD. J. Struct. Biol 206:243–53 [DOI] [PubMed] [Google Scholar]
- 192.Cichocka MO, Angstrom J, Wang B, Zou X, Smeets S. 2018. High-throughput continuous rotation electron diffraction data acquisition via software automation. J. Appl. Crystallogr 51:1652–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yonekura K,Maki-Yonekura S,Naitow H,Hamaguchi T,Takaba K.2021.Machine learning-based real-time object locator/evaluator for cryo-EM data collection. Commun. Biol 4:1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jones CG, Martynowycz MW, Hattne J, Fulton TJ, Stoltz BM, et al. 2018. The cryoEM method microED as a powerful tool for small molecule structure determination. ACS Cent. Sci 4:1587–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Martynowycz MW, Clabbers MTB, Hattne J, Gonen T. 2021. Ab initio phasing macromolecular structures using electron-counted MicroED data. bioRxiv 2021.10.16.464672. 10.1101/2021.10.16.464672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wolff AM, Young ID, Sierra RG, Brewster AS, Martynowycz MW, et al. 2020. Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals. IUCrJ 7:306–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yonekura K, Maki-Yonekura S. 2016. Refinement of cryo-EM structures using scattering factors of charged atoms. J. Appl. Crystallogr 49:1517–23 [Google Scholar]
- 198.Blum TB, Housset D, Clabbers MTB, van Genderen E, Bacia-Verloop M, et al. 2021. Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals. Acta Crystallogr. D Struct. Biol 77:75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yonekura K, Kato K, Ogasawara M, Tomita M, Toyoshima C.2015.Electron crystallography of ultrathin3D protein crystals: atomic model with charges. PNAS 112:3368–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martynowycz MW, Gonen T. 2021. Ligand incorporation into protein microcrystals for microED byon-grid soaking. Structure 29:88–95.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lawson CL, Berman HM, Chiu W. 2020. Evolving data standards for cryo-EM structures. Struct. Dyn 7:014701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Patwardhan A. 2017. Trends in the electron microscopy data bank (EMDB). Acta Crystallogr. D Struct. Biol 73:503–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chiu W, Schmid MF, Pintilie G, Lawson CL. 2021. Evolution of standardization and dissemination of cryo-EM structures and data jointly by the community, PDB and EMDB. J. Biol. Chem 296:100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. 2016. EMPIAR: a public archive for raw electron microscopy image data. Nat. Methods 13:387–88 [DOI] [PubMed] [Google Scholar]
- 205.Ellenberg J, Swedlow JR, Barlow M, Cook CE, Sarkans U, et al. 2018. A call for public archives for biological image data. Nat. Methods 15:849–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Berman HM, Adams PD, Bonvin AA, Burley SK, Carragher B, et al. 2019. Federating structural models and data: outcomes from a workshop on archiving integrative structures. Structure 27:1745–59 [DOI] [PMC free article] [PubMed] [Google Scholar]