In this review, Dever et al. discuss the nuances of eukaryotic translation initiation, particularly translation start site selection and the translation of upstream open reading frames (uORFs). Using select examples, they further expand on the mechanisms by which uORFs exert translational control, both repressive and stimulatory, on main ORFs.
Keywords: ATF4, GCN4, eIF2 phosphorylation, ribosome queuing, stringency, uORF
Abstract
In addition to the main, protein-coding, open reading frame (mORF), many eukaryotic mRNAs contain upstream ORFs (uORFs) initiated at AUG or near-cognate codons residing 5′ of the mORF start site. Whereas translation of uORFs generally represses translation of the mORFs, a subset of uORFs serves as a nexus for regulating translation of the mORF. In this review, we summarize the mechanisms by which uORFs can repress or stimulate mRNA translation, highlight uORF-mediated translational repression involving ribosome queuing, and critically evaluate recently described alternatives to the delayed reinitiation model for uORF-mediated regulation of the GCN4/ATF4 mRNAs.
While the basic mechanism of protein synthesis has been understood for >50 yr, new insights into the mechanism and regulation of translation continue to emerge. The rapid increase in genome sequences and the development of ribosomal profiling, which maps the positions of initiating and elongating ribosomes on mRNAs, have increased interest in the mechanism and regulation of translation start site selection and utilization of alternative translational start sites. Here, we describe the mechanism of translation start site selection in eukaryotes and the impact of alternative translation start sites on main open reading frame (mORF) translation. Using well-characterized examples, we highlight varied means by which upstream open reading frames (uORFs)—reading frames that initiate 5′ to the mORF—regulate mORF translation.
Mechanism of translation start site selection
Both mRNA features and the translational apparatus (the ribosome and translation factors) contribute to translation start site selection. While in bacteria ribosomes bind to mRNAs close to the mORF start codon, a distinguishing feature of eukaryotic translation initiation is ribosomal scanning. The small (40S) ribosomal subunit with associated translation factors binds an mRNA near the 5′ cap and then progresses down the mRNA while scanning the nucleotide sequence for a start codon. Recent genetic, biochemical, and structural studies have provided new insights into the mechanisms of scanning and start codon selection.
Overview of eukaryotic translation initiation
Here, we highlight the findings most relevant to uORF control of translation and direct the reader to other reviews and the primary literature for more detailed descriptions of the mechanisms of scanning and start site selection (Hinnebusch 2011, 2014) as well as the impact of uORF translation on nonsense-mediated mRNA decay (NMD) (Gaba et al. 2005; Arribere and Gilbert 2013).
Assembly of a translationally competent 80S ribosome on the start codon is a complex process requiring the assistance of multiple eukaryotic translation initiation factors (eIFs), many of which impact start codon selection (for reviews, see Hinnebusch 2011, 2014; Dever et al. 2016). The factor eIF2 bound to GTP is competent to bind initiator methionyl tRNA (Met-tRNAMeti) to form an eIF2–GTP–Met-tRNAMeti ternary complex (TC). Binding of eIF1A and eIF1 to the A site and near the P site, respectively, of the small (40S) ribosomal subunit opens the 40S mRNA binding channel and facilitates TC binding in the P site, with eIF1 contacting the eIF2β subunit. The multisubunit factor eIF3 binds across the back of the 40S subunit with contacts at both the mRNA entry and exit channels and with certain eIFs in the decoding center; the eIF3 also promotes TC binding to the 40S. Whereas an isolated 40S subunit is unable to bind an mRNA (with a few rare exceptions [CrPV IRES]) (Wilson et al. 2000), binding of the TC, eIF1, eIF1A, and eIF3 to the 40S subunit generates a 43S preinitiation complex (PIC) and licenses the complex to bind mRNA.
Binding of the 43S PIC to an mRNA is promoted by the eIF4F complex, which binds near the 5′ end of the mRNA owing to direct interaction of its cap-binding subunit eIF4E with the m7GTP mRNA cap. eIF4E also binds to the scaffolding subunit eIF4G, which additionally binds RNA, the mRNA poly(A) tail-binding protein (PABP or PABPC1), and the RNA helicase eIF4A. eIF4G also participates in recruiting the 43S PIC to mRNA via interactions with factors bound to the PIC (LeFebvre et al. 2006; Villa et al. 2013; Kumar et al. 2016; Brito Querido et al. 2020). eIF4A is thought to unwind cap-proximal secondary structures in mRNAs to prepare a binding site for the 43S PIC and also interacts with the PIC to enhance mRNA binding (Yourik et al. 2017). Following attachment near the cap, the PIC rapidly inspects the mRNA sequence (scans) for a start codon as it slides down the mRNA (Hinnebusch 2014; Wang et al. 2022).
The key feature directing selection of the translation start codon is the anticodon of the Met-tRNAMeti (Cigan et al. 1988). The Met-tRNAMeti in the PIC is thought to oscillate between Pout (not engaged with a codon) and Pin (the anticodon base-paired with an mRNA codon) conformations as the PIC scans the mRNA (Hinnebusch 2014). eIF1, bound adjacent to the P site, clashes with tRNAMeti in the Pin state to impede codon–anticodon pairing at non-AUG codons (Hussain et al. 2014; Thakur and Hinnebusch 2018). Movement of Met-tRNAMeti into the Pin conformation on pairing with an AUG codon triggers release of eIF1 (Maag et al. 2005). The N-terminal domain of eIF5, the GTPase-activating protein (GAP) for eIF2, binds the site vacated by eIF1 and stabilizes rather than clashes with tRNAMeti, with concurrent completion of GTP hydrolysis by eIF2 and release of the liberated Pi (Llácer et al. 2018) to finalize selection of the start site. Conformational changes in the PIC constrict the mRNA binding channel to arrest scanning, and additional eIFs are released. The eIF2-GDP is released, and guanine nucleotide exchange factor (GEF) eIF2B recycles it to functional eIF2–GTP for additional rounds of initiation (Pavitt 2018). The eIF1A remains bound in the A site and helps recruit the GTPase eIF5B, which reorients the position of the acceptor arm of the Met-tRNAMeti to enable joining of the large (60S) ribosomal subunit (Lapointe et al. 2022). Subsequent hydrolysis of GTP by eIF5B enables release of the factor, and the resulting 80S IC can proceed into the elongation phase of protein synthesis.
mRNA features contributing to translation start site selection
Given the vectorial nature of scanning, translation typically initiates at the AUG codon closest to the 5′ end of the mRNA (Kozak 2002). However, several mRNA features influence the efficiency of start site selection. First, AUG codons residing within ∼32 nt of the 5′ cap are inefficiently selected for initiation (Kozak 1991a). Second, as shown by Kozak (1999), the “context” nucleotides flanking a start codon influence the frequency of start codon selection. Sequence alignments of annotated start codons as well as mutational analyses of context nucleotides revealed the importance for purine nucleotides at the −3 position relative to the A of the AUG codon at position +1 and for G at position +4 (Kozak 1986b, 1987a,b; Nakagawa et al. 2008; Loughran et al. 2012; Noderer et al. 2014; Hernández et al. 2019). Mutational studies assessing translational output have established the optimum Kozak consensus sequence, 5′-GCCACCAUGG-3′ (Kozak 1986b, 1991b), with positions −3 and +4 playing the greatest roles in start site selection.
Whereas most ribosomes scanning an mRNA initiate translation at the first AUG codon they encounter, a fraction of ribosomes may continue scanning and initiate at a downstream start codon (see Kozak 2002). Such “leaky scanning” is more prominent when the first start codon is in poor context or under conditions of heightened global start codon selection stringency. As detailed below, elevated levels of eIF1 or reduced levels of eIF5 increase global stringency, while reduced eIF1 or elevated eIF5 levels decrease stringency and enhance initiation at start codons in poor context. In addition to eIF1 and eIF5, the factors eIF3 and eIF4G2 (DAP5) have been linked to start codon selection and uORF regulation. The studies on eIF4G2 have been recently reviewed (Shestakova et al. 2023), and it is unclear whether the impact of eIF3 and eIF4G2 on start codon selection is direct or indirect (She et al. 2023). Notably, lowered stringency was observed upon knockdown of ribosomal proteins, eIF3 subunits, or eIF4G2 and was correlated with the impact of the knockdowns on cell growth (She et al. 2023). Interestingly, reduced start codon selection stringency upon inhibition of general translation was recently correlated with altered relative levels of eIF1 and eIF5, possibly due to different half-lives of the two factors (Ivanov et al. 2022).
Near-cognate start codons that differ from AUG by a single nucleotide can also serve as initiation codons, albeit with lower efficiency than an AUG codon (Kozak 1989; Peabody 1989). Reporter assays established a hierarchy of near-cognate initiation codons in human cells with CUG (9.9% of AUG initiation efficiency) > ACG (4.1%) > GUG (2.5%) > AUU (1.3%) ≈ AUA (1.1%) > AUC (0.5%) ≈ UUG (0.4%) (Loughran et al. 2012). Similar preferences were seen in Saccharomyces cerevisiae and Neurospora crassa (Kolitz et al. 2009; Wei et al. 2013), and, notably, a purine at the +2 position has the greatest inhibitory effect on initiation. In human cells, initiation at a CUG codon in perfect context (9.9% of AUG) was more efficient than initiation at an AUG codon in poorest context (3.17%; with U at −6 to −1 and C at +4) (Loughran et al. 2012). Initiation at near-cognate start codons is more sensitive to mutations in the context nucleotides flanking the start codon (Diaz de Arce et al. 2018), such that near-cognate start codons in poor context are not generally used for initiation at significant levels.
In addition to trans-acting eIFs, mRNA secondary structures can act in cis to control leaky scanning and start site selection. While secondary structures near the 5′ end of an mRNA impair ribosome loading, and secondary structures within the leader impair scanning and lower translational yields (for review, see Hinnebusch 2011), secondary structures downstream from start codons can enhance initiation. Kozak (1990) showed that a stem–loop structure (−19 kcal/mol) placed 14 nt downstream from a weak start codon enhanced initiation and nearly abolished leaky scanning in vitro. Altering the distance between the stem–loop and the upstream start codon to 8 or 32 nt dampened this stimulatory effect, indicating a need for precise positioning. As the length of the mRNA entry channel from the edge of the ribosome to the P site is 15 nt, the downstream stem–loop would position the scanning ribosome over the start codon and provide a greater chance for its selection. Similarly, a properly positioned downstream hairpin enhanced initiation at a near-cognate start codon in yeast (Wang et al. 2022) or at an AUG codon in poor context in mammalian cells (Clyde and Harris 2006). In accord with these findings, binding of the sex-lethal (SXL) protein downstream from the start codon of a uORF in the msl-2 mRNA impaired mORF translation in a manner dependent on the uORF start codon (Medenbach et al. 2011), suggesting that SXL binding enhanced uORF translation. Thus, properly positioned impediments to scanning can enhance selection of a weak start site by positioning a 40S PIC in the vicinity of the start codon.
uORF attributes that affect downstream translation
While uORFs generally impair translation of the mORF, a variety of attributes contribute to the impact of the uORF on mORF translation (Fig. 1). An AUG start codon in poor context or a near-cognate start codon will lead to higher rates of leaky scanning, diminishing any inhibitory impact of the uORF. If the uORF stop codon is 3′ of the mORF start codon such that the uORF overlaps the mORF, ribosomes translating the uORF are unlikely to translate the mORF. While ribosomes primarily scan in the 3′ direction, modest backward scanning in the 5′ direction has been reported (Matsuda and Dreher 2006; Li et al. 2022; Wang et al. 2022). An early study suggested efficient reinitiation at AUG codons located 13 nt upstream of an overlapping uORF stop codon (Peabody and Berg 1986); however, later studies showed that reinitiation was strongly impaired when the AUG start codon was 1, 7, or 13 nt upstream of the uORF stop codon (Kozak 1987c, 2001). Thus, it appears that following uORF translation, very few ribosomes will scan in the 5′ direction to reinitiate at an upstream start codon.
Figure 1.
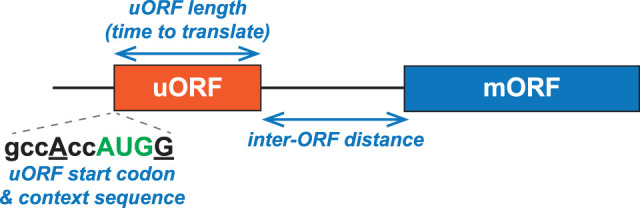
uORF attributes that affect mORF translation include the following: (1) The efficiency of initiation at the uORF start codon, impacted by the start codon context sequence, controls leaky scanning. (2) The length of the uORF and the time it takes to translate the uORF affect the ability of a ribosome to resume scanning and reinitiate at a downstream start site following translation termination at the uORF stop codon. (3) The distance between the uORF stop codon and downstream start codon impacts the ability of a ribosome sliding down the mRNA after termination at the uORF stop codon to reacquire the initiation factors and Met-tRNAMeti required to initiate at a downstream start site.
For uORFs that terminate upstream of the mORF start codon, ribosomes that translate the uORF must terminate, remain bound to the mRNA, reacquire Met-tRNAMeti and eIFs, and then scan to the mORF start codon to reinitiate translation and synthesize the mORF polypeptide. Each of these steps has the potential to impose control on mORF translation.
The efficiency of reinitiation increases as the distance between the uORF stop codon and the downstream start codon increases (Fig. 1). In mammalian cells, the efficiency of reinitiation ranged from <10% when the intercistronic distance was <11 nt to ∼70% when the distance between the uORF and mORF was ≥79 nt (Kozak 1987c). Similarly, in yeast, increasing the intercistronic distance between a short uORF and downstream reporter mORF from 50 to 100 nt increased reporter expression over twofold (Grant et al. 1994). This dependency of reinitiation on the spacing between the uORF and mORF is attributed to the time-dependent reacquisition of translation factors and especially Met-tRNAMeti by ribosomes that have translated the uORF (see GCN4 discussion below).
The length of a uORF is inversely correlated with the efficiency of reinitiation (Fig. 1). Studies in mammalian cells using mutated HIV tat mRNAs indicated that increasing the length of a uORF lead to stepwise decreases in reinitiation efficiency, with 40 codons eliminating reinitiation and 28 codons triggering ∼50% inhibition (Luukkonen et al. 1995). Similarly, increasing the length of a uORF in an altered hepatitis B virus mRNA from 13 codons (no inhibition of reinitiation) to 19 (∼50%) or 29 (∼80%) codons inhibited reinitiation (Hwang and Su 1998). In rabbit reticulocyte lysates, increasing the length of a uORF from three to 13 codons did not impact reinitiation; however, increasing the uORF length to 33 codons led to a threefold reduction in reinitiation efficiency (Kozak 2001). In yeast, mORF translation was inhibited by ∼65% to ∼95% when the length of a uORF increased from 13 to 35 codons (Rajkowitsch et al. 2004). While the precise impacts on reinitiation varied among the three studies, perhaps due to other attributes of the uORFs, uORF length was inversely correlated with reinitiation efficiency, and lengthening a uORF to >40 codons severely impaired reinitiation.
The inverse relationship between uORF length and reinitiation efficiency is commonly attributed to the time-dependent loss of eIFs from ribosomes elongating on the uORF. The notion is that reinitiation is enhanced if key initiation factors remain associated with the ribosome when it terminates uORF translation. Three lines of evidence support this model. First, slowing the rate of elongation either locally on the uORF or globally impaired reinitiation. Inserting a structured pseudoknot in a uORF to slow elongation increased the inhibitory impact of the uORF in rabbit reticulocyte lysates (Kozak 2001). In yeast, the inhibitory impact of a uORF containing rare codons was suppressed by overexpression of the complementary tRNA, consistent with the notion that slow elongation through the uORF inhibited reinitiation (Lin et al. 2019). Likewise, uORFs encoding stalling peptides that block elongation or termination on the uORF are strongly inhibitory (Dever et al. 2020). While these examples support the notion that slow elongation impairs reinitiation, the slowly elongating ribosomes could also be impairing mORF translation by blocking leaky scanning of the uORF. Consistent with slow elongation impairing reinitiation, globally impairing tRNA aminoacylation by mutating Cca1, the tRNA nucleotidyltransferase (CCA-adding enzyme) in yeast, exacerbated the inhibitory effect of a uORF on mORF translation (Rajkowitsch et al. 2004), though impaired reinitiation in this mutant might also reflect reduced levels of Met-tRNAMeti. Second, the mechanism of initiation on the uORF impacted reinitiation. uORFs translated by viral internal ribosome entry site (IRES) mechanisms that do not require the cap-binding complex eIF4F (classic swine fever virus IRES) or any factors (cricket paralysis virus IRES) were defective for reinitiation when compared with uORFs translated by the traditional scanning mechanism (Pöyry et al. 2004), as expected if eIFs used in initiating at the uORF must be retained and reused for reinitiation at the mORF. Third, eIF3 was found to remain associated with elongating ribosomes at the start of ORFs (Bohlen et al. 2020a; Lin et al. 2020; Wagner et al. 2020) and to dissociate from the elongating ribosomes with a half-life (length) of ∼12 translated codons (Bohlen et al. 2020a). Consistent with these findings, eIF3 association with uORFs decreased as the uORF length increased (Mohammad et al. 2017).
Following translation termination and peptide release, an 80S ribosome with deacylated tRNA in the P site is situated on the stop codon. The ribosome recycling factor ABCE1 (or Rli1 in yeast) dissociates the 60S subunit from this complex. The factor eIF2D (ligatin; Tma64 in yeast) or the combination of DENR and MCT-1 (Tma20 and Tma22, respectively, in yeast) releases the deacylated tRNA from the resulting 40S–tRNA–mRNA complex, enabling the 40S subunit to dissociate from the mRNA (for review, see Hellen 2018). Efficient reinitiation requires retention of the 40S subunit on the mRNA following termination and the first steps of recycling. How ribosome recycling factors contribute to translation reinitiation is not fully understood. In mammalian cells, MCT-1 and DENR promote reinitiation downstream from uORFs, most notably for uORFs encoding Met-stop (Schleich et al. 2014, 2017; Ahmed et al. 2018; Castelo-Szekely et al. 2019; Vasudevan et al. 2020). As discussed below, loss of these recycling factors in yeast increased translation in 3′ UTRs but did not significantly affect reinitiation following translation of the short uORFs on the GCN4 mRNA (Young et al. 2018; Gaikwad et al. 2021). Notably, mRNA sequences upstream of and downstream from GCN4 uORFs control permissiveness for reinitiation (Grant and Hinnebusch 1994; Grant et al. 1995; Munzarová et al. 2011; Gunišová and Valášek 2014; Gunišová et al. 2016). While the role of ribosome recycling in reinitiation requires more study, one possible model is that following uORF termination, the 60S subunit and deacylated tRNA are released from the 40S subunit. The persistence of eIF3 and/or eIF4G on the ribosome following translation of a short uORF enables retention of the posttermination 40S subunit on the mRNA. The 40S subunit then migrates down the mRNA, acquires an eIF2 TC, and then reinitiates at a downstream AUG codon.
Distinction between regulatory uORFs and inhibitory uORFs
It is important to differentiate between uORFs that simply impair ribosome access to the mORF (and thereby throttle down the translational capacity of an mRNA) and uORFs that enable translational regulation in response to changes in conditions. Not all uORFs are regulatory. A vast number of translated uORFs have been identified in ribosome profiling experiments (for example, see Ingolia et al. 2009, 2011; Chew et al. 2016; Johnstone et al. 2016; Zhang et al. 2018; Takahashi et al. 2020; Chothani et al. 2022; Liu et al. 2023); however, it is likely that many of these uORFs simply reduce the number of ribosomes that translate the mORF but do not alter translation of the mORF in response to changing conditions. Here, we focus on several well-characterized examples of regulatory uORFs and describe the varied ways by which they control mORF translation. Owing to space limitations, we cannot comprehensively discuss regulatory uORFs and we direct the reader to other reviews and primary literature for discussions of plant uORFs (von Arnim et al. 2014; van der Horst et al. 2020; Zhang et al. 2020) and other regulatory uORFs in metazoans (Zhang et al. 2019).
Examples of translational control by uORFs and stringency
The delayed reinitiation model of translational control of GCN4 and ATF4 mRNAs
Defining features of delayed reinitiation (REI)
One of the best-understood examples of translational regulation by uORFs applies to yeast GCN4 and mammalian ATF4 mRNAs, involving phosphorylation of the α subunit of eIF2 (eIF2α). Gcn4 and Atf4 are transcription factors that activate many genes enabling cells to adapt to starvation or stress, including genes promoting amino acid biosynthesis or tRNA aminoacylation. Gcn2, the sole eIF2α kinase in S. cerevisiae, is activated by amino acid starvation and mediates “general amino acid control” (GAAC) (for review, see Hinnebusch 2005). Mammalian cells contain multiple eIF2α kinases besides Gcn2 (each activated by different stresses but all regulating translation initiation by similar mechanisms) to comprise the “integrated stress response” (ISR) (Harding et al. 2000).
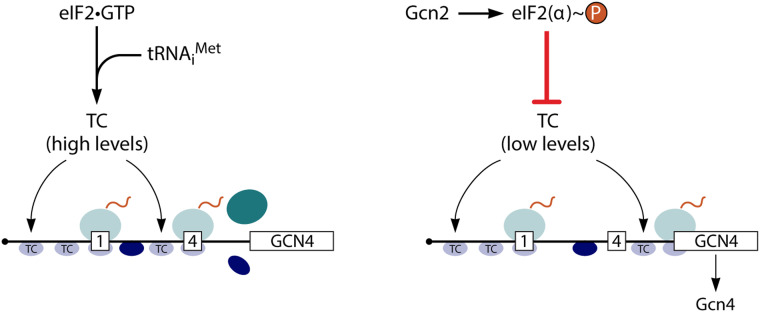
Phosphorylation of eIF2α on Ser51 by the eIF2α kinases converts eIF2–GDP from substrate to inhibitor of its GEF eIF2B, and the ensuing depletion of eIF2–GTP reduces TC assembly. This broadly reduces protein synthesis as one leg of the GAAC/ISR while activating translation of particular mRNAs (e.g., GCN4/ATF4) whose encoded products act to alleviate stress. The reduction in TC assembly preferentially impairs translation of mRNAs of lower translational efficiencies, presumably reflecting their inability to compete effectively for limiting 43S PICs (Gaikwad et al. 2021). GCN4 and ATF4 mRNAs are very inefficiently translated in nonstressed cells, owing to the presence of multiple uORFs, but their translation is paradoxically induced by eIF2α phosphorylation because, at low TC levels, ribosomes that have translated the first or second uORFs and resumed migrating downstream (as 40S subunits following termination) can bypass the start codons of the more inhibitory 5′-distal uORFs and reinitiate at the mORF AUG codons instead (Fig. 2).
Figure 2.
Schematic model of GCN4 translational control, simplified to show only uORF1 and uORF4, which are sufficient for nearly wild-type regulation. Following translation of uORF1 (boxed 1), posttermination 40S subunits remain attached to the GCN4 mRNA and resume scanning. (Left) Under nonstarvation conditions, they quickly rebind TC and reinitiate at uORF4 (boxed 4), and the 80S ribosome dissociates after terminating at uORF4. (Right) Under amino acid starvation conditions, the concentration of TC is reduced by eIF2α phosphorylation, such that many 40S ribosomes fail to rebind TC until after scanning past uORF4 and can thereby reinitiate at the GCN4 ORF instead. (Reproduced from Hinnebusch 2011 with permission from American Society for Microbiology).
The GCN4 mRNA leader is unusually long (∼600 nt) and contains four short uORFs of only two or three codons (uORF1 to uORF4, from 5′ to 3′) (Figs. 2, 3) whose AUG codons have optimal sequence contexts for yeast. The uORF1 of mouse ATF4 is also three codons long, whereas uORF2 is much longer and overlaps the mORF, and both uORF AUGs are in favorable Kozak context. According to the delayed REI model, uORF1 is translated by nearly all PICs scanning from the 5′ end, and a sizable fraction of the 40S subunits remaining at the uORF1 stop codon after termination and recycling of the 60S subunit resume migration downstream. Under nonstress conditions of abundant TC, the majority of these 40S subunits rebind TC and reinitiate translation at one of the distal uORFs (GCN4 uORF3 or uORF4 and ATF4 uORF2), after which essentially none of the resulting posttermination 40S subunits goes on to reinitiate at the GCN4 or ATF4 mORFs. When TC levels are reduced by eIF2α phosphorylation, rebinding of TC is delayed, and a fraction of migrating 40S subunits reacquires TC only after bypassing the inhibitory distal uORFs and initiates at the mORF instead (Fig. 2). Similar to uORF1, GCN4 uORF2 exhibits a high propensity for reinitiation and serves as a fail-safe for PICs scanning from the cap that leaky-scan uORF1 during the primary initiation event (Fig. 3). This confers stronger repression of GCN4 under nonstress conditions—but greater derepression in starved cells owing to a larger fraction of scanning PICs put into “reinitiation mode” that are thus able to reach GCN4—than occurs with uORF1 alone (Gunišová and Valášek 2014).
Figure 3.
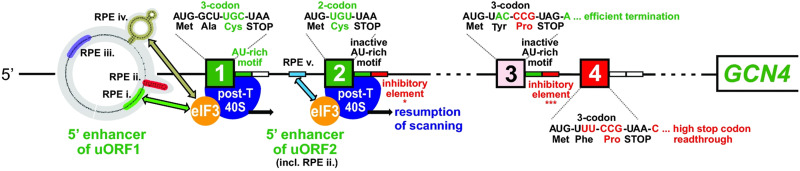
Schematic summary of the mRNA leader of GCN4 mRNA with its four short uORFs (uORF1 to uORF4), summarizing the positions and functions of elements surrounding uORF1 and uORF2 that promote reinitiation following their translation, including RPE(i) to RPE(iv) in the enhancer region upstream of uORF1 and the AU-rich motif following uORF1, as well as RPE(v) upstream of uORF2. RPE(i), RPE(iv), and RPE(v) interact with eIF3 (arrows) at the exit channel of the 40S subunit to enhance resumption of scanning by 40S posttermination complexes at the uORF stop codons. uORF3 and uORF4 allow much less reinitiation because they are closer to the GCN4 mORF, lack functional stimulatory elements found at uORF1 and uORF2, and contain the CCG Pro codon that impairs reinitiation. Additionally, the inefficient termination codon at uORF4 allows stop codon readthrough, placing posttermination 40S complexes even closer to the GCN4 mORF, thus rendering them less able to reinitiate there. (Reprinted with permission from Gunisova et al. 2016).
A defining feature of the delayed REI model is that the 5′-proximal uORFs promote, rather than inhibit, translation of the mORF under conditions of limiting TC. This positive effect is observed only in the presence of distal uORFs, as removing uORF1 from an otherwise uORF-less leader increases rather than decreases GCN4 translation. This finding indicated that uORF1 stimulates GCN4 translation indirectly by overcoming inhibition by the distal uORFs. In addition to GCN4 uORF1 and uORF2 (Mueller and Hinnebusch 1986; Gunišová and Valášek 2014), this behavior applies to uORF1 of mouse ATF4 (Lu et al. 2004; Vattem and Wek 2004) and the mRNAs encoding mammalian transcription factor Atf5 (Zhou et al. 2008) and the N. crassa Gcn4 ortholog cpc-1 (Ivanov et al. 2017)—all regulated by delayed REI. The positive function of uORF1 was attributed to its ability to allow retention of 40S posttermination complexes at its stop codon that resume migrating and bypass the distal inhibitory uORFs when TC levels are reduced (Abastado et al. 1991). In the absence of the 5′-proximal uORFs, 48S PICs harboring TC will scan directly from the cap to the distal uORFs, whose translation and termination eliminate virtually all scanning ribosomes from the mRNA.
A second defining feature of delayed REI, shown for GCN4, ATF4, ATF5, and cpc-1 in the studies cited above, is that removing the distal uORFs elevates mORF translation when TC is abundant, regardless of the presence of the proximal uORFs. This finding indicates that the distal uORFs are potent barriers to reinitiation (Figs. 2, 3). Strong evidence that the distal uORFs are bypassed by 40S subunits migrating from the proximal uORFs was that mutations that removed multiple in-frame stop codons of the distal GCN4 uORF4, rendering it a much longer uORF that overlaps the mORF, had no effect on GCN4 expression at limiting TC levels. As this extension would preclude reinitiation, the elongated uORF4 must be skipped by the 40S subunits migrating from the proximal uORFs en route to the GCN4 start codon (Abastado et al. 1991). Consistent with this, the native inhibitory uORF2s of both ATF4 and ATF5 extensively overlap the mORFs, and the inhibitory cpc-1 uORF2 is ∼10-fold longer than the three-codon inhibitory uORF3 and uORF4 of GCN4. When uORF1 was similarly elongated to overlap the GCN4 ORF in an allele lacking uORF2–uORF4, it became a strong barrier to GCN4 translation, demonstrating that most 40S subunits scanning from the 5′ end translate uORF1 and that its permissiveness to downstream translation does not arise from leaky scanning of its start codon but from frequent reinitiation (Grant et al. 1994).
A third defining feature of delayed REI is that mORF translation is highly sensitive to the distances separating the proximal and distal uORFs. Lengthening the uORF1–uORF4 interval in GCN4 mRNA by inserting spacer sequences of increasing length progressively reduced GCN4 translation under limiting TC, with derepression nearly eliminated when the expanded uORF1–uORF4 distance was close to that normally separating uORF1 from the mORF. This is the outcome expected if the expanded uORF1–uORF4 separation provides sufficient time for TC binding by all 40S subunits migrating from uORF1 before they encounter uORF4, such that none can bypass the uORF4 AUG codon. It also explains why (1) a heterologous short uORF was not bypassed under conditions of limiting TC when inserted just upstream of the GCN4 AUG, (2) GCN4 uORF1 as a solitary uORF becomes more inhibitory when moved closer to the GCN4 start codon (Grant et al. 1994), and (3) shortening the spacing between uORF1 and uORF4 in GCN4 (Grant et al. 1994) or between uORF1 and uORF2 of ATF4 (Lu et al. 2004) leads to greater bypass of the distal inhibitory uORFs in nonstress conditions of abundant TC. As noted, Kozak (1987c) first demonstrated that reinitiation declines as the separation between an uORF and the downstream mORF is reduced in mammalian cell-free extracts and further showed that an inhibitory uORF could be overcome by inserting a short uORF further upstream—a key principle of the delayed REI mechanism.
A fourth defining feature of delayed REI is that inhibiting scanning by insertions of stem–loops of strong predicted stability between uORF1 and the distal inhibitory uORFs blocks induction of mORF translation under conditions of reduced TCs, as shown for GCN4 (Abastado et al. 1991) and ATF4 (Vattem and Wek 2004). These experiments exploited knowledge about the inhibitory effects of stem–loops on PIC attachment or scanning (Pelletier and Sonenberg 1985; Kozak 1986a) to provide evidence that ribosomes do not bypass the inhibitory distal uORFs by hopping or internal initiation.
The key tenet of delayed REI is that migrating 40S subunits generated by the proximal uORF bypass the start codons of the inhibitory distal uORFs when TC becomes limiting and reinitiate further downstream at the mORF instead. This tenet was supported by observing reductions in rates of GCN4 uORF4 or ATF4 uORF2 translation on limiting TC and quantified by immunoprecipitation of pulse-labeled reporter polypeptides fused to these uORFs (Abastado et al. 1991; Lu et al. 2004). A moderate reduction in initiation at these inhibitory uORFs should be sufficient to account for a large increase in initiation at the downstream mORFs because almost none of the migrating 40S subunits generated by uORF1 translation bypasses the distal uORFs when TC is abundant. The increased fraction that bypasses when TC is limiting can be predicted as ∼20%–40% by comparing GCN4 translation with or without the distal uORFs (Mueller and Hinnebusch 1986), which is consistent with the observed approximately twofold reduction in synthesis of an uORF4-β-galactosidase fusion (Abastado et al. 1991). The inferred reduction in ATF4 uORF2 translation on eIF2α phosphorylation by PERK corresponds to only ∼18% (Lu et al. 2004; Vattem and Wek 2004) and to only 8% for ATF5 (Zhou et al. 2008). Indeed, a small reduction in ATF4 uORF2 expression in response to eIF2α phosphorylation induced by arsenite was observed by quantifying a tracer peptide encoded by a modified version of uORF2 (Starck et al. 2016), though it was unjustifiably interpreted as being contradictory to the delayed REI model because of its small magnitude. Because the steady-state level rather than rate of synthesis of the uORF2 tracer was measured, an even larger decrease in synthesis could have been obscured by the pre-existing tracer synthesized prior to arsenite treatment. Increased ATF4 mRNA expression on eIF2α phosphorylation (Dey et al. 2010; Zhou et al. 2018) could also have obscured a larger decrease in uORF2 translation at limiting TC levels.
Consistent with delayed REI, ribosome profiling shows modest reductions in ribosome footprint (RFP) density in uORF2 of native ATF4 and ATF5 mRNAs in arsenite-treated cells (Andreev et al. 2015). The opposite conclusion was reached for ATF4 uORF2 translation during amino acid starvation by calculating the uORF2/uORF1 ratio of RPFs (Zhou et al. 2018), which may be problematic because RPF densities in short uORFs (like three-codon uORF1) cannot be measured reliably (Gerashchenko and Gladyshev 2014). In this last study, an uORF2-FLUC reporter showed no change in steady-state expression in starved cells; however, an increase in reporter mRNA abundance that parallels native ATF4—or the complication of pre-existing luciferase made prior to starvation—could have obscured the predicted small decrease in uORF2-FLUC translation (Zhou et al. 2018). In fact, quantification of 80S ICs by ribosome profiling in the presence of lactimidomycin (QTI-seq) showed reduced initiation at the uORF2 start codon during starvation (Zhou et al. 2018), in agreement with the delayed REI model.
Surprisingly, the QTI-seq results just mentioned revealed no 80S ICs at the ATF4 mORF AUG, leading to the conclusion that reinitiation following uORF1 translation involves scanning 80S versus 40S posttermination ribosomes. However, this conclusion was based on the ad hoc assumption that lactimidomycin cannot bind to an 80S IC formed by a scanning 80S ribosome (Zhou et al. 2018). This proposal is at odds with findings that reinitiation by 80S ribosomes in 3′ UTRs generally does not occur at AUG codons in yeast cells (Young et al. 2015a) or in a mammalian reconstituted system where it is dictated by complementarity to the elongator tRNA decoding the last codon of the uORF (Skabkin et al. 2013). It is also difficult to envision how a delay in reinitiation by 80S ribosomes could arise from reduced TC levels, as TC binding to the 40S subunit (Hussain et al. 2014) is incompatible with the 40S:60S interface of an 80S ribosome (Ben-Shem et al. 2011). Another line of evidence advanced to support reinitiation by scanning 80S ribosomes, of no increase in the 40S:60S ratio bound to ATF4 mRNA on TC limitation (Zhou et al. 2018), is not compelling because delayed REI predicts that the number of 40S subunits scanning from the cap to uORF1 or migrating downstream from uORF1 is constitutive, and only ∼20% of the latter bypass uORF2 and continue downstream to the mORF in starved cells rather than dissociating after uORF2 translation. The small increase in 40S subunits between uORF2 and the ATF4 AUG on TC limitation predicted by delayed REI would be challenging to detect biochemically.
Other evidence from Zhou et al. (2018) led to the proposal that m6A methylation of nucleotide A225 in uORF2 acts as a fail-safe to prevent translation of the ATF4 mORF in nonstarved cells by promoting initiation at the ORF2 start codon such that demethylation of A225 is required to induce mORF translation in starved cells. This model is supported by the findings (1) of reduced methylation at uORF2 during starvation, (2) that knocking down m6A demethylase ALKBH5 or FTO impaired induction of both Atf4 and ATF4-FLUC expression in starved cells, (3) of cross-linking of ALKBH5 to ATF4 mRNA, and (4) that overexpressing FTO in the livers of transgenic mice derepressed Atf4 expression in nonstarvation conditions. However, the fail-safe model seems inconsistent with the findings that either overexpressing FTO, knocking down methylase METTL3 or mutating A225 in the ATF4-LUC reporter produced no increase in Atf4 protein or ATF4-FLUC reporter expression, respectively, in nonstarved cells. Additionally, A225G mutation did not increase 80S ICs at the ATF4 AUG codon, even though it reduced ICs at the uORF2 AUG, when assayed in lysates by toeprint analysis. Overexpressing FTO, depleting METTL3, or mutating A225, however, did enhance induction of Atf4 or ATF4-FLUC, respectively, in starved cells, and the A225G mutation both eliminated the enhanced induction conferred by depleting METTL3 and restored induction of ATF4-LUC in cells depleted of demethylase ALKBH5 (Zhou et al. 2018). These last findings could indicate that A225 methylation dampens induction of the mORF in response to eIF2α phosphorylation rather than preventing its induction in nonstarved cells. The proposal that A225 methylation increases initiation at the uORF2 AUG predicts a reduction in ORF2 translation during starvation (when methylation is reduced) that will be diminished by the A225G mutation, which should be tested in the future.
Determinants of the differing reinitiation potentials of 5′-proximal vs. 5′-distal uORFs.
The delayed REI mechanism depends on a high frequency of reinitiation following translation of the 5′-proximal uORFs. For GCN4 uORF1, this depends partly on its short three-codon length (Stiles et al. 1981; Williams et al. 1988; Miller and Hinnebusch 1989) and its location far upstream of the mORF (350 nt) (Grant et al. 1994), both in line with determinants of reinitiation mentioned above. It also requires cis-acting sequences upstream of uORF1 (Grant et al. 1995), including four reinitiation-promoting elements (RPEs), of which RPE(ii) and RPE(iv) form stem–loop structures. The relatively lower reinitiation potential of uORF2 also depends on RPE(ii) plus an additional element, RPE(v), which is similar in sequence to uORF1 RPE(i) (Fig. 3; Munzarová et al. 2011; Gunišová and Valášek 2014). uORF1 contains an AU-rich element just downstream from its stop codon that also promotes reinitiation (Miller and Hinnebusch 1989; Grant and Hinnebusch 1994; Gunišová et al. 2016). uORF2 and uORF3 contain similar sequences of consensus AU1-2A/UUAU2 that can functionally replace the uORF1 sequence but are nullified by other sequences at these two uORFs (Fig. 3; Gunišová et al. 2016). Combining the functions of these various positive and negative elements helps to explain why the reinitiation potential varies as uORF1 > uORF2 > uORF3 (Gunišová et al. 2016).
Genetic evidence indicates that RPE(i) and RPE(iv) of uORF1 and RPE(v) of uORF2 (Fig. 3) functionally cooperate with two segments in the N-terminal domain (NTD) of the largest subunit of eIF3 (a/Tif32), shown to be required in trans for efficient reinitiation at GCN4 following translation of uORF1 and uORF2 (Szamecz et al. 2008; Munzarová et al. 2011; Gunišová and Valášek 2014). Located upstream of the uORFs, these RPEs will lie just outside of the exit channel of 40S posttermination complexes positioned at the uORF stop codons, in proximity to the eIF3a-NTD in the yeast 48S PIC (Llácer et al. 2015). It was proposed that eIF3 remains associated with the 40S subunit during elongation and termination at uORF1 and uORF2 and that the eIF3a-NTD interaction with RPE elements impedes dissociation of the 40S posttermination complex to enable reinitiation (Munzarová et al. 2011). Biochemical evidence supports the predicted association of eIF3 with GCN4 mRNA sequence intervals containing uORF1 or uORF2 (with RPEs) versus uORF3 or uORF4 (lacking RPEs), dependent on the RPEs and key segments of the eIF3a-NTD (Mohammad et al. 2017). Importantly, the eIF3/uORF1 association was impaired by lengthening uORF1 by two or five alanine codons, which were shown previously to impair reinitiation by placing RPEs beyond the reach of eIF3a-NTD at the exit channel (two-codon insertion) or by increasing eIF3 dissociation during translation of a longer version of uORF1 (five-codon insertion) (Mohammad et al. 2017). Consistent with these last findings, selective ribosome profiling revealed high occupancies of 40S subunits bound to eIF3 and eIF2 selectively at GCN4 uORF1 and uORF2 that were attributed to posttermination complexes at the proximal uORFs that retained eIF3, reacquired TC, and became competent to reinitiate downstream, whereas posttermination complexes at uORF3 and uORF4 are recycled from the mRNA. Even larger occupancies of 40S:eIF3 and 40S:eIF2 complexes were observed upstream of uORF1, which might serve as a reservoir of mRNA-bound PICs that can be mobilized for GCN4 translation in response to stress (Wagner et al. 2020). There is evidence that sequences upstream of or downstream from the ATF4 uORF1 similarly promote reinitiation, possibly via the h subunit of mammalian eIF3 (Hronová et al. 2017).
The low reinitiation potential of the ATF4 uORF2 can probably be explained by the combination of its extended length and extensive overlap with the mORF; however, other explanations are required for the three-codon inhibitory uORF3 and uORF4 at GCN4. Being closer to the GCN4 AUG codon and lacking both REI elements that retain eIF3 and stimulatory AU-rich sequences downstream from uORF3 and uORF4 stop codons (Fig. 3) helps to explain their lower reinitiation potential versus uORF1 and uORF2. The latter also depends on the third CCG Pro codons at both uORF3 and uORF4 (Fig. 3), conserved among related yeasts (Gunišová et al. 2016), which can be partially mimicked by the other three Pro codons (Miller and Hinnebusch 1989; Grant and Hinnebusch 1994; Gunišová et al. 2016). This effect could involve the known slow decoding of Pro codons or inefficient peptide release at stop codons following a Pro codon (Doerfel et al. 2015), which would mimic the time required to translate a longer uORF.
The lower reinitiation permitted by uORF4 versus uORF3 was attributed partly to (1) their different second codons, but in a manner that likely involves their different sequences rather than differing cognate tRNAs, and (2) their different stop codons and the adjacent nucleotide. The latter tetranucleotide sequence at uORF4 (TAA-C), but not that at uORF3 (TAG-A), confers a high frequency (>30%) of stop codon readthrough, dependent on the +4 C residue (Fig. 3; Gunišová et al. 2016). Readthrough might be associated with slow decoding of the stop codon by release factor, and when readthrough occurs, it will effectively increase the length of uORF4. Either effect would increase the time required to complete uORF4 translation and reduce reinitiation frequency accordingly. The important cis-acting elements identified at the four GCN4 uORFs that collaborate to establish the reinitiation hierarchy uORF1 > uORF2 > uORF3 > uORF4 are summarized in Figure 3.
Role of 40S recycling factors in reinitiation on human ATF4 mRNA
In contrast to the third (penultimate) CCG Pro codon at GCN4 uORF3 and uORF4, which contributes to the low reinitiation potential of these uORFs, the penultimate UGC and UGU Cys codons of uORF1 and uORF2 (Fig. 3) appear to be dispensable, as substitutions with many other triplets was compatible with high reinitiation downstream (Grant et al. 1995; Gunišová et al. 2016). Analysis of human ATF4 uORF1 and short uORFs in other human mRNAs indicated that certain codons at either the penultimate or antepenultimate codons preceding the uORF stop codons, including the antepenultimate GCG Ala codon in ATF4 uORF1, confer a requirement for 40S recycling factors (the DENR/MCT-1 heterodimer and related eIF2D protein) for efficient reinitiation at the mORFs in response to eIF2α phosphorylation. As penultimate codons, these triplets also generally increase dependence on these factors for recycling of posttermination 40S subunits at uORFs and main CDSs. It was proposed that efficient recycling is required to dissociate the tRNAs decoding the sensitive triplets in 40S posttermination complexes to allow the vacant 40S subunits to resume migrating and reinitiate downstream (Bohlen et al. 2020b). It seems difficult to explain by this model the observed effects in reinitiation of antepenultimate codons, which should occupy the empty E site in 40S posttermination complexes. Translation of GCN4 is unaffected by simultaneously eliminating the yeast homologs of the DENR/MCT-1 heterodimer and eIF2D (Gaikwad et al. 2021). It remains to be seen whether replacing the WT triplets of uORF1 with others that confer a greater dependence on these factors for 40S recycling at stop codons (Young et al. 2018) confers a similar dependence on these proteins for efficient reinitiation and induction of GCN4 translation.
Single uORFs that regulate translation in response to changes in TC levels
A key hallmark of the delayed REI model is the presence of multiple uORFs in the mRNA. However, eIF2α phosphorylation also regulates translation of select mRNAs containing a single regulatory uORF, though the mechanism is less well understood. Treating mammalian cells with arsenite triggers eIF2α phosphorylation and translationally derepresses numerous mRNAs containing a single uORF (Andreev et al. 2015), including GADD34 (PPP1R15A), which together with the protein phosphatase 1 (PP1c) directs dephosphorylation of eIF2α-P to restore protein synthesis. Although the GADD34 mRNA leader contains two uORFs, the second uORF is sufficient to confer derepression under conditions of high eIF2α phosphorylation (Lee et al. 2009; Young et al. 2015b). The regulatory uORF encodes a peptide that is under purifying selection near its C terminus, which ends with the sequence PPG (Young et al. 2015b). Translation of the uORF peptide directs ribosomes to stall during decoding of its termination codon (Fig. 4A, panel ii; Young et al. 2015b). The C-terminal PPG motif is critical for the regulatory properties of the uORF and is thought to impede termination. The slow termination could lead to greater loss of initiation factors from the ribosomes translating the uORF, leading to ribosome dissociation from the mRNA following uORF translation and thus preventing reinitiation. Ribosomes are thought to leaky-scan the regulatory uORF when eIF2α is phosphorylated, facilitated by the imperfect context of its AUG codon (Fig. 4A, panel i); however, the imperfect context is not essential for the regulation. Mutating the start codon of the regulatory uORF of GADD34 to a noninitiating codon or mutating the sequence of the encoded peptide derepresses mORF translation even under conditions of high TC (Young et al. 2015b).
Figure 4.
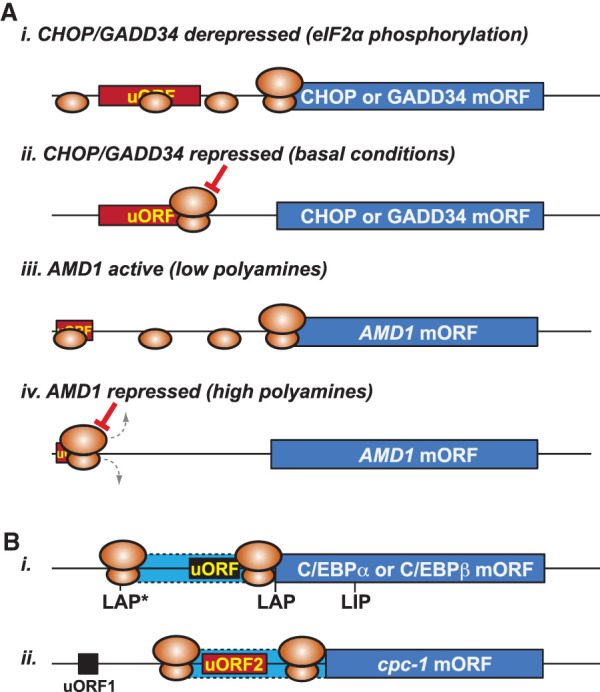
uORF and stringency regulation of translation. (A) CHOP, GADD34, and AMD1 regulation. (Panels i,ii) Elongation/termination pause impairs leaky scanning over the uORF and represses CHOP and GADD34 translation, and eIF2α phosphorylation enhances leaky scanning to derepress mORF translation. (Panels iii,iv) Ribosomes access the vertebrate AMD1 mORF by leaky scanning over the cap-proximal uORF. Polyamine-triggered pausing of a ribosome on the uORF precludes additional ribosomes from loading, repressing mORF translation. (B) Alternative initiation enables bypass of a regulatory uORF. (Panel i) Initiation at upstream weak start sites in C/EBP mRNAs, generating LAP*, leads to ribosomes bypassing the short regulatory uORF that controls synthesis of LAP or LIP isoforms in response to eIF2α phosphorylation. (Panel ii) Reinitiation at the near-cognate start codon upstream of inhibitory uORF2 generates N-terminally extended cpc-1.
CHOP (DDIT3), like GCN4/ATF4, is a bZIP transcription factor that modulates gene expression programs during cellular stress. The leader of the CHOP mRNA contains a single uORF that confers derepression under conditions of high eIF2α phosphorylation. Like GADD34, the uORF of the CHOP mRNA inhibits mORF translation under conditions of high TC. Also, like GADD34, the C terminus of the encoded uORF peptide is under purifying selection, though the conserved sequences are dissimilar to each other. Unlike GADD34, the CHOP uORF peptide sequence appears to pause ribosomes during elongation rather than termination (Fig. 4A, panel ii). Mutations that alter the uORF peptide sequence affect the level of repression of the mORF and impair translational regulation. Interestingly, the uORF is initiated by two in-frame AUG codons, though both are in suboptimal contexts. Introducing the optimal context at the uORF start codons increases repression of the downstream mORF and dampens, but does not eliminate, the eIF2α phosphorylation-mediated regulation (Young et al. 2016). Current models suggest a common mechanism for GADD34 and CHOP regulation based on peptide sequence-dependent impairment of reinitiation that is relieved by increased leaky scanning of the uORF following eIF2α phosphorylation (Fig. 4A, panel i); however, the mechanism whereby eIF2α phosphorylation increases leaky scanning remains unclear.
The GCN4 homolog in Candida albicans contains multiple uORFs; however, the single uORF3 is sufficient for translational regulation in response to eIF2α phosphorylation (Sundaram and Grant 2014), raising the possibility that it is regulated in a manner similar to CHOP and GADD34. While Schizosaccharomyces pombe lacks a GCN4 homolog, the Fil1 protein performs a similar function, regulating expression of amino acid biosynthetic enzyme genes (Duncan et al. 2018). The fission yeast fil1 mRNA contains multiple uORFs that enable eIF2α phosphorylation control, but it is unclear whether the regulation is via delayed REI or a distinct mechanism (Duncan and Mata 2022).
uORFs responding to small metabolites
A rate-limiting step in the synthesis of spermidine and spermine is decarboxylation of S-adenosyl-L-methionine, a reaction catalyzed by the enzyme S-adenosylmethionine decarboxylase (AdoMetDC) encoded by the gene AMD1. The vertebrate AMD1 mRNA contains a short conserved uORF that starts 13–14 nt downstream from the 5′ cap and encodes the peptide sequence MAGDIS (Hill and Morris 1993; Ruan et al. 1994, 1996; Ivanov et al. 2010a). The uORF in the AMD1 mRNA confers polyamine-dependent translational regulation of AdoMetDC synthesis with high-level synthesis in low-polyamine conditions (Fig. 4A, panel iii) and repression of AdoMetDC synthesis in high-polyamine conditions (Fig. 4A, panel iv). Mutagenesis of the uORF codons showed the importance of Asp in the fourth position and Ile in the penultimate position for polyamine regulation (Mize et al. 1998). Increased polyamine concentrations cause ribosomal stalling during termination on the uORF stop codon (Raney et al. 2000; Law et al. 2001), dependent on the DI codons. However, elevated polyamine levels also increase stalling at the termination codon of a mutant peptide that is less inhibitory on downstream translation, indicating that polyamines might have a general inhibitory effect on translation with exacerbating impacts on inefficient terminators (Law et al. 2001). The inefficient termination of the uORF might impair reinitiation. Alternatively, a ribosome stalled at the stop codon of the MAGDIS uORF could, by virtue of its size and distance from the 5′ end, interfere with loading of 40S PICs at the cap (Ivanov et al. 2010a). Together, these two effects of polyamines on uORF translation termination—reduced reinitiation and impaired ribosome loading—impair AdoMetDC synthesis (Fig. 4A, panel iv).
Additional examples of uORFs conferring translational control in response to metabolites include the N. crassa arg-2 and mammalian PTP4A2 (PRL-2) mRNAs. Arginine-triggered stalling of a translating ribosome on a conserved uORF in the arg-2 mRNA and its S. cerevisiae homolog, CPA1 mRNA, represses mORF translation, and, likewise, apparent magnesium-induced ribosome stalling on a conserved uORF in the PTP4A2 mRNA represses mORF translation (for review, see Dever et al. 2020).
uORFs regulated in response to changes in global stringency of start codon selection
Although the stringency of start codon selection in cells was originally thought to be static, with perhaps variations in stringency existing only between different organisms or different cell types, the levels of eIF1 and eIF5 modulate the stringency of start codon selection. Whereas eIF1 binding to the 43S PIC favors scanning, eIF1 dissociation and eIF5 binding favor initiation (Hinnebusch 2011). The genes encoding eIF1 and eIF5 in eukaryotes have evolved features that mediate cybernetic feedback regulation and cross-regulation to maintain the stringency of start codon selection and homeostasis of eIF1 and eIF5 expression (Fig. 5; Ivanov et al. 2010c; Martin-Marcos et al. 2011; Loughran et al. 2012). The eIF1 CDS is initiated by an AUG codon in conserved poor context (Miyasaka et al. 2010; Martin-Marcos et al. 2011). High levels of eIF1 confer high stringency, leading to repression of eIF1 mRNA translation for a negative autoregulatory loop (Fig. 5B; Ivanov et al. 2010c; Martin-Marcos et al. 2011). The leader of the eIF5 mRNA contains inhibitory uORFs initiated by AUG codons in conserved poor context (Loughran et al. 2012). High levels of eIF5 lower stringency, which enhances initiation at the inhibitory uORFs in the eIF5 mRNA and impairs eIF5 synthesis—a second negative autoregulatory loop (Fig. 5C; Loughran et al. 2012). In parallel, high-level eIF5 increases initiation at the poor context AUG codon of eIF1 mRNA to increase eIF1 synthesis (Fig. 5A), whereas high-level eIF1 decreases initiation at the poor context AUG of the eIF5 mRNA uORF to elevate eIF5 synthesis (Fig. 5D). These cross-regulatory effects combine with the autoregulatory controls to maintain a constant ratio of eIF1:eIF5 and level of global stringency. The cellular levels of BZW (or eIF5-mimic protein) also modulate global stringency of start codon selection in cells by apparently counteracting the activity of eIF5 (Tang et al. 2017; Loughran et al. 2018).
Figure 5.
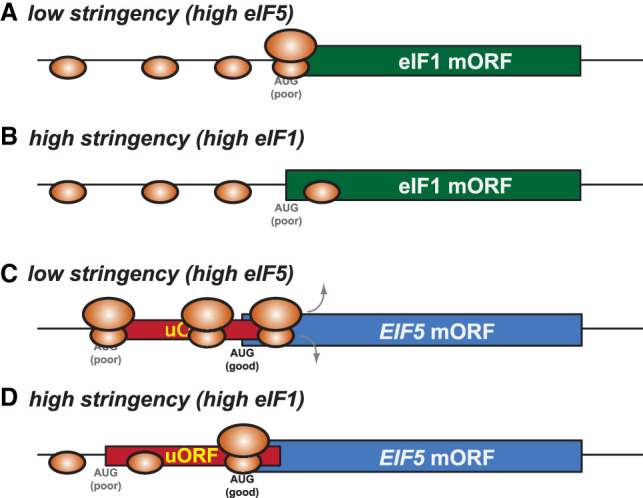
Stringency control of eIF1 and eIF5 expression. Suboptimal AUG start codons of EIF1 mORF and inhibitory uORF in EIF5 mRNA sense global stringency of start codon selection: Low stringency enhances EIF1 mORF and EIF5 uORF translation, thereby increasing eIF1 and repressing eIF5 synthesis. High stringency increases leaky scanning over poor AUG codons, repressing EIF1 mORF and EIF5 uORF translation, thereby repressing eIF1 and increasing eIF5 synthesis.
The existence of the eIF1/eIF5 homeostatic autoregulatory and cross-regulatory system and its conservation over a vast evolutionary time scale strongly suggest that perturbations of global stringency occur under some physiological conditions (see discussion on cpc-1 and C/EBP below) and that conserved uORFs in the leaders of some mRNAs have evolved to sense changes in global stringency. Such uORFs, like the one in eIF5 mRNA, should inhibit expression of the mORF (i.e., either be long or overlap the mORF) and should be initiated by inefficient start codons (i.e., poor context AUGs or near-cognate start codons). Likely examples are the conserved uORFs in the leaders of the mouse Hoxa1, Hoxa9, and Hoxa11 mRNAs that have been shown to mediate differential mORF expression in response to global changes in the stringency of start codon selection (Ivanov et al. 2022). The total number of mammalian genes with similar characteristics is unknown, as is the number of uORFs that fit the eIF5 paradigm and mediate translational control in response to changes in global stringency.
uORF-mediated ribosomal queuing controls local stringency of start codon selection
Human antizyme inhibitor (AZIN1) is a key component of another cybernetic mechanism that maintains intracellular polyamine homeostasis. AZIN1 binds to and inhibits the protein antizyme, which in turn inhibits the enzyme ornithine decarboxylase (ODC) catalyzing the first step in the biosynthesis of polyamines. The mRNAs encoding all three proteins are translationally regulated by polyamines in mammals (Dever and Ivanov 2018). AZIN1 and ODC are homologs that started diverging from each other in early vertebrate evolution (Ivanov et al. 2010b). The leader of the AZIN1 mRNA contains three conserved AUG-initiated short uORFs that do not have known regulatory functions. However, the mRNA also contains an upstream conserved coding region (uCC) that has two defining features: a conserved AUU near-cognate start codon and a conserved C terminus that often encodes the amino acid sequence PPW (Fig. 6; Ivanov et al. 2008). Both features are ancient and predate the divergence of AZIN1 and ODC but have been lost in mammalian ODC. Similar sequences have also evolved apparently independently in fungi. The uCC of AZIN1 directs polyamine-dependent regulation, with high polyamine levels inhibiting mORF translation and low polyamine levels stimulating translation (Ivanov et al. 2008). Both the near-cognate AUU start codon of the uCC and the conserved C-terminal sequence of the encoded peptide are essential for polyamine regulation. Polyamines were found to enhance the efficiency of initiation on the near-cognate AUU start codon of the uCC. While high polyamine levels were initially proposed to reduce the global stringency of start codon selection (Ivanov et al. 2008) and by this mechanism account for enhanced AZIN1 translation, subsequent studies showed that the enhanced translation of the AZIN1 uCC in the presence of high polyamine levels is strictly dependent on the conserved uCC peptide sequence (Ivanov et al. 2018). High polyamine levels were also shown to inhibit the activity of the translation factor eIF5A, which is required for translating the PPW motif in the uCC, resulting in ribosomal stalling. Interestingly, substitution of the uCC PPW motif with the arginine-regulated stalling peptide from the N. crassa arg-2 mRNA (Wang and Sachs 1997; Fang et al. 2004) conferred enhanced initiation at the AUU near-cognate start codon in the presence of high arginine levels (Ivanov et al. 2018).
Figure 6.
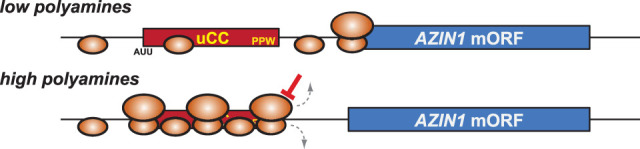
Schematic model of AZIN1 translational control. (Top) Under low-polyamine conditions, most ribosomes leaky-scan over the near-cognate AUU start codon of the uCC uORF and translate the mORF. (Bottom) Polyamine-triggered pausing of a ribosome on the PPW motif in the uCC causes ribosomes to queue and enhances initiation at the near-cognate uCC start codon, repressing AZIN1 synthesis.
Taken together, these results suggested a model in which a ribosome elongating on the uCC pauses in response to high concentration of polyamines. The paused ribosome triggers ribosomal queuing on the uCC that causes a trailing 40S ribosome to dwell in the vicinity of the near-cognate AUU start codon, enhancing the efficiency of initiation at this site (Fig. 6). At low polyamine levels, ribosome pausing in the uCC is diminished, dissipating the queue and allowing trailing 40S subunits to leaky-scan the uCC and initiate at the mORF instead (Fig. 6). Thus, the stringency of selecting the poor uCC start codon is modulated locally by expanding or contracting the queue of scanning 40S PICs (and 80S elongating ribosomes) to make it either include or exclude the uCC start codon. This model is strengthened by the observation that lowering PIC loading on mRNA, either by phosphorylation of eIF2α or by disrupting the interaction between eIF4G and eIF4E, reduces uCC translation and derepresses AZIN1 synthesis in the presence of high polyamine levels (Ivanov et al. 2018). The decreased PIC loading will diminish the queue despite persistence of polyamine-induced ribosomal pausing within the uCC. It is currently unknown whether uCC-like elements in other mRNAs confer similar regulation involving ribosomal queuing in response to different metabolites. However, the uORFs described above controlling GADD34 and CHOP mRNA translation and the ascorbate-mediated translational regulation of the GGP mRNA in plants (Laing et al. 2015) are good candidates in containing uORFs initiated at a suboptimal or near-cognate start codon.
Upstream initiation enables bypass of regulatory uORFs
As noted earlier, the uORF architecture of N. crassa cpc-1 and its orthologs in other filamentous fungi is similar to the uORF architecture in mammalian ATF4 with a short uORF1 (three to six codons long) and a longer uORF2 (35–70 codons), except that cpc-1 uORF2 does not overlap the mORF. cpc-1 and all its >100 known orthologs in Pezizomycotina share a feature that distinguishes them from GCN4/ATF4 and suggests an additional mode of regulation. No in-frame stop codons are present in the >500 nt preceding the cpc-1 mORF and extending beyond the beginning of uORF2. Thus, translation initiating upstream of uORF2 and in-frame with the mORF will generate an N-terminally extended CPC1 protein and bypass the inhibitory consequences of translating uORF2 (Fig. 4B, panel ii; Ivanov et al. 2017). The cpc-1 mRNA sequence between uORF2 and the mORF shows evidence of purifying selection in the mORF, providing evidence that the region is translated, which is supported by ribosome profiling data. Moreover, phylogenetic analysis identified conserved near-cognate start codons in-frame with the mORF and upstream of uORF2 that could initiate N-terminal extensions of CPC1. The cpc-1 homologs in Basidiomycota appear to have independently acquired an mRNA architecture analogous to cpc-1 (Ivanov et al. 2017). These findings suggest that, in addition to translational control by delayed REI, the Pezizomycotina and Basidiomycota cpc-1 homologs are also translationally regulated in response to altered global or local stringency of start codon selection.
CCAAT enhancer-binding proteins (C/EBPs) are a family of bZIP transcription factors in vertebrates that regulate gene expression in response to developmental or environmental cues. C/EBPα and C/EBPβ, two members of this paralogous group, are important for adipogenesis and macrophage functioning, respectively, among other documented roles. The mRNAs of both C/EBPα and C/EBPβ contain a short uORF that generally inhibits downstream translation (Fig. 4B, panel i; Calkhoven et al. 1994; Lincoln et al. 1998). This uORF controls the production of two alternative isoforms of the proteins in response to eIF2α phosphorylation, though in a manner distinct from the delayed REI model for GCN4 and ATF4 (Calkhoven et al. 2000). In addition, the leaders of both C/EBPα and C/EBPβ mRNAs have conserved suboptimal initiation codons, a near-cognate CUG codon in C/EBPα and an AUG in poor context in C/EBPβ, located upstream of the short regulatory uORF and in-frame with the mORF. Like cpc-1 in N. crassa, translation initiation at these suboptimal start codons will enable synthesis of N-terminally extended isoforms and will bypass the regulatory effects of the short uORF (Fig. 4B, panel ii; Calkhoven et al. 2000). Also, like cpc-1, this mRNA architecture suggests that the C/EBPα and C/EBPβ mRNAs have evolved features to sense changes in the global stringency of start codon selection, perhaps in response to specific physiological conditions.
In conclusion, uORFs are powerful elements of translational control that generally act to repress mORF translation. If the uORF contains the features required to allow reinitiation and is located 5′-proximal of another uORF, it can stimulate mORF translation indirectly by suppressing initiation at the 5′-distal uORF. This stimulation can be enhanced under conditions that reduce TC abundance. Repression by solitary uORFs frequently involves uORF coding sequences that stall ribosomes during elongation or termination, which can be modulated by metabolites to establish a negative regulatory loop. The negative impact of the elongation/termination pause can be amplified by formation of a ribosome queue in the uORF that delays migration of scanning PICs to elevate translation of the uORF, creating a positive feedback loop, which is crucial when the uORF has a poor context AUG or near-cognate start codon. Formation of the queue and hence translational repression by the uORF can be diminished by reducing the rate of PIC attachment to the mRNA, providing another layer of control over uORF repression. Alteration of global stringency can also modulate translational repression by altering the rate of uORF initiation. Translated uORFs are very frequent in eukaryotic mRNAs, making it likely that many new instances of translational control involving delayed REI, elongation/termination pausing with or without ribosome queuing, altered global stringency, or other yet to be discovered mechanisms will be uncovered in the future.
Acknowledgments
Research in our laboratories is supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350752.123.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed YL, Schleich S, Bohlen J, Mandel N, Simon B, Sinning I, Teleman AA. 2018. DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation. PLoS Biol 16: e2005160. 10.1371/journal.pbio.2005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 4: e03971. 10.7554/eLife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23: 977–987. 10.1101/gr.150342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bohlen J, Fenzl K, Kramer G, Bukau B, Teleman AA. 2020a. Selective 40S footprinting reveals Cap-tethered ribosome scanning in human cells. Mol Cell 79: 561–574.e5. 10.1016/j.molcel.2020.06.005 [DOI] [PubMed] [Google Scholar]
- Bohlen J, Harbrecht L, Blanco S, Clemm von Hohenberg K, Fenzl K, Kramer G, Bukau B, Teleman AA. 2020b. DENR promotes translation reinitiation via ribosome recycling to drive expression of oncogenes including ATF4. Nat Commun 11: 4676. 10.1038/s41467-020-18452-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito Querido J, Sokabe M, Kraatz S, Gordiyenko Y, Skehel JM, Fraser CS, Ramakrishnan V. 2020. Structure of a human 48S translational initiation complex. Science 369: 1220–1227. 10.1126/science.aba4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Bouwman PRJ, Snippe L, Geert AB. 1994. Translationa start site multiplicity of the CCAAT/enhancer binding protein α mRNA is dictated by a small 5′ open reading frame. Nucl Acids Res 22: 5540–5547. 10.1093/nar/22.25.5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CR, Müller C, Leutz A. 2000. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev 14: 1920–1932. 10.1101/gad.14.15.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Szekely V, De Matos M, Tusup M, Pascolo S, Ule J, Gatfield D. 2019. Charting DENR-dependent translation reinitiation uncovers predictive uORF features and links to circadian timekeeping via Clock. Nucleic Acids Res 47: 5193–5209. 10.1093/nar/gkz261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7: 11663. 10.1038/ncomms11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothani SP, Adami E, Widjaja AA, Langley SR, Viswanathan S, Pua CJ, Zhihao NT, Harmston N, D'Agostino G, Whiffin N, et al. 2022. A high-resolution map of human RNA translation. Mol Cell 82: 2885–2899.e8. 10.1016/j.molcel.2022.06.023 [DOI] [PubMed] [Google Scholar]
- Cigan AM, Feng L, Donahue TF. 1988. tRNAiMet functions in directing the scanning ribosome to the start site of translation. Science 242: 93–97. 10.1126/science.3051379 [DOI] [PubMed] [Google Scholar]
- Clyde K, Harris E. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 80: 2170–2182. 10.1128/JVI.80.5.2170-2182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Ivanov IP. 2018. Roles of polyamines in translation. J Biol Chem 293: 18719–18729. 10.1074/jbc.TM118.003338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Kinzy TG, Pavitt GD. 2016. Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics 203: 65–107. 10.1534/genetics.115.186221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Ivanov IP, Sachs MS. 2020. Conserved upstream open reading frame nascent peptides that control translation. Annu Rev Genet 54: 237–264. 10.1146/annurev-genet-112618-043822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. 2010. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem 285: 33165–33174. 10.1074/jbc.M110.167213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz de Arce AJ, Noderer WL, Wang CL. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46: 985–994. 10.1093/nar/gkx1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kubyshkin V, Starosta AL, Wilson DN, Budisa N, Rodnina MV. 2015. Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J Am Chem Soc 137: 12997–13006. 10.1021/jacs.5b07427 [DOI] [PubMed] [Google Scholar]
- Duncan CDS, Mata J. 2022. Translation-complex profiling of fission yeast cells reveals dynamic rearrangements of scanning ribosomal subunits upon nutritional stress. Nucleic Acids Res 50: 13011–13025. 10.1093/nar/gkac1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Rodriguez-Lopez M, Ruis P, Bahler J, Mata J. 2018. General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc Natl Acad Sci 115: E1829–E1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Spevak CC, Wu C, Sachs MS. 2004. A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci 101: 4059–4064. 10.1073/pnas.0400554101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Jacobson A, Sachs MS. 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell 20: 449–460. 10.1016/j.molcel.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Gaikwad S, Ghobakhlou F, Young DJ, Visweswaraiah J, Zhang H, Hinnebusch AG. 2021. Reprogramming of translation in yeast cells impaired for ribosome recycling favors short, efficiently translated mRNAs. Elife 10: e64283. 10.7554/eLife.64283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42: e134. 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Hinnebusch AG. 1994. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol 14: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Miller PF, Hinnebusch AG. 1994. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol Cell Biol 14: 2616–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Miller PF, Hinnebusch AG. 1995. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nuc Acids Res 23: 3980–3988. 10.1093/nar/23.19.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunišová S, Valášek LS. 2014. Fail-safe mechanism of GCN4 translational control—uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res 42: 5880–5893. 10.1093/nar/gku204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunišová S, Beznosková P, Mohammad MP, Vlčková V, Valášek LS. 2016. In-depth analysis of cis-determinants that either promote or inhibit reinitiation on GCN4 mRNA after translation of its four short uORFs. RNA 22: 542–558. 10.1261/rna.055046.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10: a032656. 10.1101/cshperspect.a032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Osnaya VG, Pérez-Martínez X. 2019. Conservation and variability of the AUG initiation codon context in eukaryotes. Trends Biochem Sci 44: 1009–1021. 10.1016/j.tibs.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Hill JR, Morris DR. 1993. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem 268: 726–731. 10.1016/S0021-9258(18)54212-5 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467. 10.1128/MMBR.00008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hronová V, Mohammad MP, Wagner S, Pánek J, Gunišová S, Zeman J, Poncová K, Valášek LS. 2017. Does eIF3 promote reinitiation after translation of short upstream ORFs also in mammalian cells? RNA Biol 14: 1660–1667. 10.1080/15476286.2017.1353863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. 10.1016/j.cell.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Su TS. 1998. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J Gen Virol 79(Pt 9): 2181–2189. 10.1099/0022-1317-79-9-2181 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Atkins JF. 2008. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci 105: 10079–10084. 10.1073/pnas.0801590105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ. 2010a. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38: 353–359. 10.1093/nar/gkp1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Atkins JF. 2010b. Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J Mol Evol 70: 289–302. 10.1007/s00239-010-9331-5 [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010c. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. 10.1073/pnas.1009269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Wei J, Caster SZ, Smith KM, Michel AM, Zhang Y, Firth AE, Freitag M, Dunlap JC, Bell-Pedersen D, et al. 2017. Translation initiation from conserved non-AUG codons provides additional layers of regulation and coding capacity. MBio 8: e00844-00817. 10.1128/mBio.00844-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, Dever TE. 2018. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70: 254–264.e6. 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Saba JA, Fan CM, Wang J, Firth AE, Cao C, Green R, Dever TE. 2022. Evolutionarily conserved inhibitory uORFs sensitize Hox mRNA translation to start codon selection stringency. Proc Natl Acad Sci 119: e2117226119. 10.1073/pnas.2117226119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35: 706–723. 10.15252/embj.201592759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Takacs JE, Lorsch JR. 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15: 138–152. 10.1261/rna.1318509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986a. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci 83: 2850–2854. 10.1073/pnas.83.9.2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986b. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. 10.1016/0092-8674(86)90762-2 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987a. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1987b. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196: 947–950. 10.1016/0022-2836(87)90418-9 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987c. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol 7: 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1989. Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol 9: 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci 87: 8301–8305. 10.1073/pnas.87.21.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1991a. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1: 111–115. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1991b. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870. 10.1016/S0021-9258(18)54860-2 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. 10.1016/S0378-1119(99)00210-3 [DOI] [PubMed] [Google Scholar]
- Kozak M. 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232. 10.1093/nar/29.24.5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34. 10.1016/S0378-1119(02)01056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Hellen CU, Pestova TV. 2016. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev 30: 1573–1588. 10.1101/gad.282418.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. 2015. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786. 10.1105/tpc.114.133777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Grosely R, Sokabe M, Alvarado C, Wang J, Montabana E, Villa N, Shin BS, Dever TE, Fraser CS, et al. 2022. eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining. Nature 607: 185–190. 10.1038/s41586-022-04858-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner C, Morris DR. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276: 38036–38043. 10.1074/jbc.M105944200 [DOI] [PubMed] [Google Scholar]
- Lee YY, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J Biol Chem 284: 6661–6673. 10.1074/jbc.M806735200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932. 10.1074/jbc.M605418200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Kong J, Zhang S, Zhao T, Qian W. 2022. Distance-dependent inhibition of translation initiation by downstream out-of-frame AUGs is consistent with a Brownian ratchet process of ribosome scanning. Genome Biol 23: 254. 10.1186/s13059-022-02829-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, May GE, Kready H, Nazzaro L, Mao M, Spealman P, Creeger Y, McManus CJ. 2019. Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Res 47: 9358–9367. 10.1093/nar/gkz681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li F, Huang L, Polte C, Duan H, Fang J, Sun L, Xing X, Tian G, Cheng Y, et al. 2020. eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol Cell 79: 575–587.e7. 10.1016/j.molcel.2020.06.003 [DOI] [PubMed] [Google Scholar]
- Lincoln AJ, Monczak Y, Williams SC, Johnson PF. 1998. Inhibition of CCAAT/enhancer-binding protein α and β translation by upstream open reading frames. J Biol Chem 273: 9552–9560. 10.1074/jbc.273.16.9552 [DOI] [PubMed] [Google Scholar]
- Liu Q, Peng X, Shen M, Qian Q, Xing J, Li C, Gregory RI. 2023. Ribo-uORF: a comprehensive data resource of upstream open reading frames (uORFs) based on ribosome profiling. Nucleic Acids Res 51: D248–D261. 10.1093/nar/gkac1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. 10.1016/j.molcel.2015.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Hussain T, Saini AK, Nanda JS, Kaur S, Gordiyenko Y, Kumar R, Hinnebusch AG, Lorsch JR, Ramakrishnan V. 2018. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. Elife 7: e39273. 10.7554/eLife.39273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. 10.1093/nar/gkr1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Firth AE, Atkins JF, Ivanov IP. 2018. Translational autoregulation of BZW1 and BZW2 expression by modulating the stringency of start codon selection. PLoS One 13: e0192648. 10.1371/journal.pone.0192648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33. 10.1083/jcb.200408003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, Schwartz S. 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol 69: 4086–4094. 10.1128/jvi.69.7.4086-4094.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275. 10.1016/j.molcel.2004.11.051 [DOI] [PubMed] [Google Scholar]
- Martin-Marcos P, Cheung YN, Hinnebusch AG. 2011. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 31: 4814–4831. 10.1128/MCB.05819-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. 2006. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 12: 1338–1349. 10.1261/rna.67906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW. 2011. Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913. 10.1016/j.cell.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Miller PF, Hinnebusch AG. 1989. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev 3: 1217–1225. 10.1101/gad.3.8.1217 [DOI] [PubMed] [Google Scholar]
- Miyasaka H, Endo S, Shimizu H. 2010. Eukaryotic translation initiation factor 1 (eIF1), the inspector of good AUG context for translation initiation, has an extremely bad AUG context. J Biosci Bioeng 109: 635–637. 10.1016/j.jbiosc.2009.11.022 [DOI] [PubMed] [Google Scholar]
- Mize GJ, Ruan H, Low JJ, Morris DR. 1998. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem 273: 32500–32505. 10.1074/jbc.273.49.32500 [DOI] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS. 2017. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45: 201–207. 10.1016/0092-8674(86)90384-3 [DOI] [PubMed] [Google Scholar]
- Munzarová V, Panek J, Gunišová S, Dányi I, Szamecz B, Valášek LS. 2011. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet 7: e1002137. 10.1371/journal.pgen.1002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. 2008. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36: 861–871. 10.1093/nar/gkm1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, Diaz de Arce AJ, Zhang J, Khavari PA, Wang CL. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10: 748. 10.15252/msb.20145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD. 2018. Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wiley Interdiscip Rev RNA 9: e1491. 10.1002/wrna.1491 [DOI] [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem 264: 5031–5035. 10.1016/S0021-9258(18)83694-8 [DOI] [PubMed] [Google Scholar]
- Peabody DS, Berg P. 1986. Termination–reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol 6: 2695–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40: 515–526. 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75. 10.1101/gad.276504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol 335: 71–85. 10.1016/j.jmb.2003.10.049 [DOI] [PubMed] [Google Scholar]
- Raney A, Baron AC, Mize GJ, Law GL, Morris DR. 2000. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J Biol Chem 275: 24444–24450. 10.1074/jbc.M003364200 [DOI] [PubMed] [Google Scholar]
- Ruan H, Hill JR, Fatemie-Nainie S, Morris DR. 1994. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J Biol Chem 269: 17905–17910. 10.1016/S0021-9258(17)32395-5 [DOI] [PubMed] [Google Scholar]
- Ruan H, Shantz LM, Pegg AE, Morris DR. 1996. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271: 29576–29582. 10.1074/jbc.271.47.29576 [DOI] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Küchler K, Stoecklin G, Duncan KE, et al. 2014. DENR–MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. 10.1038/nature13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Acevedo JM, Clemm von Hohenberg K, Teleman AA. 2017. Identification of transcripts with short stuORFs as targets for DENR • MCTS1-dependent translation in human cells. Sci Rep 7: 3722. 10.1038/s41598-017-03949-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R, Luo J, Weissman JS. 2023. Translational fidelity screens in mammalian cells reveal eIF3 and eIF4G2 as regulators of start codon selectivity. Nucleic Acids Res 51: gkad329. 10.1093/nar/gkad329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova ED, Smirnova VV, Shatsky IN, Terenin IM. 2023. Specific mechanisms of translation initiation in higher eukaryotes: the eIF4G2 story. RNA 29: 282–299. 10.1261/rna.079462.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. 2013. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell 51: 249–264. 10.1016/j.molcel.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles JI, Szostak JW, Young AT, Wu R, Consaul S, Sherman F. 1981. DNA sequence of a mutation in the leader region of the yeast iso-1-cytochrome c mRNA. Cell 25: 277–284. 10.1016/0092-8674(81)90253-1 [DOI] [PubMed] [Google Scholar]
- Sundaram A, Grant CM. 2014. A single inhibitory upstream open reading frame (uORF) is sufficient to regulate Candida albicans GCN4 translation in response to amino acid starvation conditions. RNA 20: 559–567. 10.1261/rna.042267.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG, Valášek L. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425. 10.1101/gad.480508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyaki S, Onouchi H, Motomura T, Idesako N, Takahashi A, Murase M, Fukuyoshi S, Endo T, Satou K, et al. 2020. Exhaustive identification of conserved upstream open reading frames with potential translational regulatory functions from animal genomes. Sci Rep 10: 16289. 10.1038/s41598-020-73307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Morris J, Wan J, Moore C, Fujita Y, Gillaspie S, Aube E, Nanda J, Marques M, Jangal M, et al. 2017. Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide. Nucleic Acids Res 45: 11941–11953. 10.1093/nar/gkx808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A, Hinnebusch AG. 2018. eIF1 loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex. Proc Natl Acad Sci 115: E4159–E4168. 10.1073/pnas.1800938115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst S, Filipovska T, Hanson J, Smeekens S. 2020. Metabolite control of translation by conserved peptide uORFs: the ribosome as a metabolite multisensor. Plant Physiol 182: 110–122. 10.1104/pp.19.00940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D, Neuman SD, Yang A, Lough L, Brown B, Bashirullah A, Cardozo T, Ryoo HD. 2020. Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat Commun 11: 4677. 10.1038/s41467-020-18453-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274. 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JW, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288: 32932–32940. 10.1074/jbc.M113.517011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia Q, Vaughn JN. 2014. Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12. 10.1016/j.plantsci.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Wagner S, Herrmannová A, Hronová V, Gunišová S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, Valášek LS. 2020. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol Cell 79: 546–560.e7. 10.1016/j.molcel.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sachs MS. 1997. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol 17: 4904–4913. 10.1128/MCB.17.9.4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shin BS, Alvarado C, Kim JR, Bohlen J, Dever TE, Puglisi JD. 2022. Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation. Cell 185: 4474–4487.e17. 10.1016/j.cell.2022.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zhang Y, Ivanov IP, Sachs MS. 2013. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J Biol Chem 288: 9549–9562. 10.1074/jbc.M112.447177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NP, Mueller PP, Hinnebusch AG. 1988. The positive regulatory function of the 5′-proximal open reading frames in GCN4 mRNA can be mimicked by heterologous, short coding sequences. Mol Cell Biol 8: 3827–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. 10.1016/S0092-8674(00)00055-6 [DOI] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. 2015a. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. 10.1016/j.cell.2015.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Willy JA, Wu C, Sachs MS, Wek RC. 2015b. Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J Biol Chem 290: 28257–28271. 10.1074/jbc.M115.693184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Palam LR, Wu C, Sachs MS, Wek RC. 2016. Ribosome elongation stall directs gene-specific translation in the integrated stress response. J Biol Chem 291: 6546–6558. 10.1074/jbc.M115.705640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Makeeva DS, Zhang F, Anisimova AS, Stolboushkina EA, Ghobakhlou F, Shatsky IN, Dmitriev SE, Hinnebusch AG, Guydosh NR. 2018. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR recycle post-termination 40S subunits in vivo. Mol Cell 71: 761–774.e5. 10.1016/j.molcel.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yourik P, Aitken CE, Zhou F, Gupta N, Hinnebusch AG, Lorsch JR. 2017. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. Elife 6: e31476. 10.7554/eLife.31476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dou S, He F, Luo J, Wei L, Lu J. 2018. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol 16: e2003903. 10.1371/journal.pbio.2003903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Lu J. 2019. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem Sci 44: 782–794. 10.1016/j.tibs.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Zhang T, Wu A, Yue Y, Zhao Y. 2020. uORFs: important cis-regulatory elements in plants. Int J Mol Sci 21: 6238. 10.3390/ijms21176238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem 283: 7064–7073. 10.1074/jbc.M708530200 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Brüning JC, Qian SB. 2018. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell 69: 636–647.e7. 10.1016/j.molcel.2018.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]