Abstract
Distal hereditary motor neuropathy represents a group of motor inherited neuropathies leading to distal weakness. We report a family of two brothers and a sister affected by distal hereditary motor neuropathy in whom a homozygous variant c.3G>T (p.1Met?) was identified in the COQ7 gene. This gene encodes a protein required for coenzyme Q10 biosynthesis, a component of the respiratory chain in mitochondria. Mutations of COQ7 were previously associated with severe multi-organ disorders characterized by early childhood onset and developmental delay.
Using patient blood samples and fibroblasts derived from a skin biopsy, we investigated the pathogenicity of the variant of unknown significance c.3G>T (p.1Met?) in the COQ7 gene and the effect of coenzyme Q10 supplementation in vitro.
We showed that this variation leads to a severe decrease in COQ7 protein levels in the patient’s fibroblasts, resulting in a decrease in coenzyme Q10 production and in the accumulation of 6-demethoxycoenzyme Q10, the COQ7 substrate. Interestingly, such accumulation was also found in the patient’s plasma. Normal coenzyme Q10 and 6-demethoxycoenzyme Q10 levels were restored in vitro by using the coenzyme Q10 precursor 2,4-dihydroxybenzoic acid, thus bypassing the COQ7 requirement. Coenzyme Q10 biosynthesis deficiency is known to impair the mitochondrial respiratory chain. Seahorse experiments showed that the patient’s cells mainly rely on glycolysis to maintain sufficient ATP production. Consistently, the replacement of glucose by galactose in the culture medium of these cells reduced their proliferation rate. Interestingly, normal proliferation was restored by coenzyme Q10 supplementation of the culture medium, suggesting a therapeutic avenue for these patients.
Altogether, we have identified the first example of recessive distal hereditary motor neuropathy caused by a homozygous variation in the COQ7 gene, which should thus be included in the gene panels used to diagnose peripheral inherited neuropathies. Furthermore, 6-demethoxycoenzyme Q10 accumulation in the blood can be used to confirm the pathogenic nature of the mutation. Finally, supplementation with coenzyme Q10 or derivatives should be considered to prevent the progression of COQ7-related peripheral inherited neuropathy in diagnosed patients.
Keywords: COQ7, distal hereditary motor neuropathy, Coenzyme Q10
Jacquier et al. describe a new homozygous variant in the COQ7 gene in a family with distal hereditary motor neuropathy. They show that the variant affects coenzyme Q10 production, leading to impaired mitochondrial metabolism. Coenzyme Q10 supplementation may represent a potential treatment for the disorder.
Introduction
Distal hereditary motor neuropathy (dHMN) is a motor inherited neuropathy, resulting in a distal weakness starting in the lower limbs, with a slowly progressive course.1 The prevalence of dHMN is around 2 per 100 000 individuals.2 To date, at least 15 genes have been associated with dHMN, according to the Neuromuscular Disease Center at Washington University.3 These genes are implicated in several crucial functions such as axonal transport, ionic homeostasis, RNA metabolism, and protein folding. However, in around 65% of dHMN cases, no causative mutation is identified, indicating that other genes and variations remain to be identified.2,4 Over recent decades, the development of next-generation sequencing has allowed the identification of novel genes associated with inherited neuropathies. Here, we report two brothers and a sister from a Portuguese family, presenting with an axonal, recessively inherited form of dHMN characterized by a motor deficit affecting distal muscles, associated with the identification of a homozygous mutation in the COQ7 gene located on chromosome 16. The COQ7 gene is transcribed in three main isoforms that provide alternative start codons in the mRNA. Isoform 1 (NM_016138, ENST00000321998, COQ7-201) is the longest and most abundant transcript. It encodes a protein (NP_057222) of 217 amino acids (AA), which is maturated in a 182 AA protein after cleavage of its signal peptide. The second isoform (NM_00190983, ENST00000544894, COQ7-202) is produced from an alternative start codon localized in-frame in exon 2 of isoform 1. It encodes a 179 AA protein (NP_001177912) homologous to isoform 1 except for three C-terminal AA. The third isoform (NM_001370489, ENST00000569127, COQ7-209) is produced from a start codon localized in the first intron of isoform 1 and encodes a 203 AA protein (NP_001357418) in which the first 10 AA are not homologous to isoform 1 (Fig. 1). COQ7 protein is mainly localized in the mitochondrial inner membrane and is directly involved in coenzyme Q10 (CoQ10) biosynthesis. Indeed, through the hydroxylation of 6-demethoxy-CoQ10 (6-DMQ), the COQ7 enzyme catalyses the last monooxygenase step in CoQ10 synthesis.5 CoQ10 is a component of the respiratory mitochondrial chain. It is involved in the transfer of electrons from the respiratory complex I and II to complex III and is therefore required for ATP production by mitochondria.5 COQ7 is also described to have a nuclear function to promote reactive oxygen species (ROS)-defensive gene expression and potentially regulate metabolic pathways that alter cellular ROS production independently of CoQ10.6
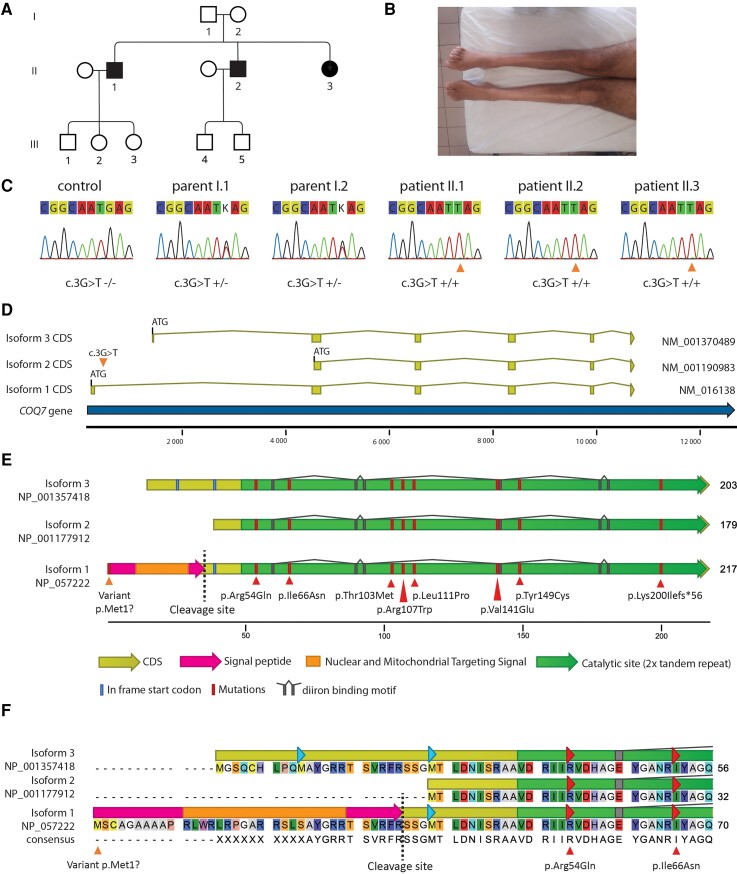
Figure 1.
Presentation of the family and COQ7 protein. (A) Pedigree of the family. Squares are males and circles are females. Filled symbols represent affected subjects and empty symbols unaffected subjects (B) Amyotrophy of Patient II-2 legs. (C) Sanger sequencing confirmation of the genotypes. (D) Schematic representations of the different isoforms showing the three possible ATG start codons. The orange triangle shows the point mutation c.3G>T on isoform 1 (NM_016138.5), which is the main isoform expressed in the spinal cord. (E) Schematic representation of the three COQ7 isoforms aligned on the major isoform 1 (NP_057222.2). Isoform 1 is composed of 217 AA, including a 35-AA signal peptide which will be cleaved, and a 182-AA mature protein. Isoforms 2 and 3 are respectively composed of 179 and 203 AA. The variant from the patients is localized in the isoform 1 start codon (orange triangle), leading to the disruption of this codon. Other pathogenic mutations previously described are localized within the catalytic site (red triangle). Translation of the coding DNA sequence (CDS) is in yellow. The nuclear and mitochondrial targeting signal in the isoform 1 signal peptide is in orange. The cleavage site of the mitochondrial processing peptidase is shown by a dashed line. The diiron binding site of the catalytic domain is in grey. In-frame start codons are in blue. (F) COQ7 isoform alignment showing the variable AA in the N-terminal region.
Mutations of COQ7 were previously associated with primary CoQ10 deficiencies (MIM 601683, COQ10D8) encompassing a large spectrum of severe phenotypes. Six families of patients with early childhood onsets and developmental delays were identified. The first patient was reported by Freyer et al.7 in 2015 with a c.422T>A homozygous substitution associated with encephalo-neuro-nephro-cardiopathy at birth. Then, Wang et al.8 and Hashemi et al.9 extended the phenotype of COQ7-associated diseases by reporting two patients affected by hereditary spastic paraplegia associated with developmental delay. These patients harboured a c.332T>C homozygous mutation, associated with a particular COQ7 polymorphism. Theunissen et al.10 reported two other patients with a paediatric onset affected by neuropathy and developmental delay, but without precise clinical description. These patients had compound heterozygous mutations of COQ7, c.197T>A, and c.446A>G. Kwong et al.11 described another patient presenting with multiorgan failure at birth harbouring a compound heterozygous mutation in COQ7. The last patient reported by Wang et al.12 was affected by a c.161G>A associated with neuromuscular defects, hypotonia, and developmental delay at birth. Here, we extend the spectrum of COQ7-associated disorders by reporting the first familial cases of patients affected by COQ7 mutation and presenting with a pure distal motor neuropathy. We show that the c.3G>T (p.1Met?) mutation in the COQ7 gene causes a decrease in COQ7 protein levels, leading to an accumulation of 6-DMQ, a decrease in CoQ10 levels in fibroblasts, and a reduction in the efficiency of the respiratory chain counterbalanced by an increase in glycolysis. These results thus provide a physiological basis for the pathogenicity of the COQ7 mutation that decreases the production of CoQ10 and alters glycolytic metabolism and mitochondrial ATP production, which could lead to motor neuron degeneration over time.
Material and methods
Patients and ethical considerations
Patients were all born in Portugal from Portuguese ascendants. All procedures performed in this study involving human participants were carried out in accordance with the ethical standards of the Hospices Civils de Lyon (ethical approval #22_923) and the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Patients displayed a clinical and electrical phenotype of length-dependent axonal motor neuropathy, with no mutation in known dHMN genes at that time. Two brothers and one sister from one family were included in the study.
Clinical assessment
Patients II-1, II-2, and II-3 were seen in a neuromuscular center and assessed by a senior neurologist (A.P.) and a resident (J.T.) specialized in neuromuscular disorders. The clinical assessment included medical history and neurological examination.
Neurophysiological study
Electroneuromyography (ENMG) included nerve conduction studies in the upper and lower limbs, and myography using concentric needle electrodes in at least three muscles. Conventional equipment and standard methods were used. Skin temperature was maintained between 32°C and 34°C.
Molecular analysis
Five whole-exome sequencings (WES) studies were performed for the two parents (Individuals I-1, I-2) and the three children (Individuals II-1, II-2, and III-3). Blood samples were collected with informed consent and in accordance with French ethical laws. Exome enrichment was done with the SeqCap EZ MedExome Target Enrichment Kit (Roche Diagnostics), and sequencing was done using the NextSeq 500 sequencing system (Illumina) following the manufacturers’ instructions. Sequences were aligned to the human reference genome hg19 using BWA-MEM. Variant calling was performed using GATK HaplotypeCaller, and the variants were then annotated using SnpEff. Variants were filtered as follows: allele depth superior or equal to 7, threshold for heterozygous variants was superior to 0.25. Maximal allele occurrence in ExAC database was 0 for homozygous variants and 125 for heterozygous. Maximal allele frequency in 1000Genome was under 1%. The confirmation of the putative deleterious COQ7 variants in patients was performed using Sanger sequencing.
Cell culture and transfection
Human neuroblastoma cell line SH-EP were grown in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher, Cat. No. 10569010) supplemented with 10% foetal bovine serum (FBS; Thermo Fisher, Cat. No. 10270-106) and 1% penicillin/streptomycin (Gibco) and transfected at 60% confluence with jetPRIME (Polyplus-transfection) according to the manufacturer’s protocol. Skin fibroblasts (AF400) were derived from a skin biopsy in Patient II-2 following standard procedures. Normal control fibroblasts (AB249 and V972F) were provided by the Centre de Biotechnologie Cellulaire (CBC) of the Hospices Civils de Lyon (Bron, France).
Fibroblasts were routinely cultured in high glucose DMEM (Thermo Fisher, Cat. No. 10569010) with 10% FBS (Thermo Fisher, Cat. No. 10270-106) and 1% penicillin/streptomycin (Gibco). Galactose medium was prepared with glucose-free DMEM (Thermo Fisher, Cat. No. 11966025) by adding galactose at the final concentration of 10 mM, 1 mM sodium pyruvate, 10% dialysed FBS (Thermo Fisher, Cat. No. 10270-106), and 1% penicillin/streptomycin (Gibco). Both routine culture medium and galactose medium contained 4 mM l-glutamine. For viability measurements in optimal or galactose media, cells were plated in high-glucose DMEM overnight and the medium was changed the following day. Cell viability was measured during the next 4 days.
Survival quantification
During the 4 days of incubation with the media of interest, cells were fixed daily with 4% paraformaldehyde (PFA) for 10 min, and nuclei were marked with DAPI. Ten images per condition were acquired randomly in the well with an Evos FL cell imaging system (Thermo Fisher), and cells were counted by MetaMorph Software version 7.8.4.0 (Molecular Devices, San Jose, CA, USA). Experiments were repeated at least three times. For treatment with CoQ10, the chemical was first solubilized in ethanol to a 1 mM concentration and then added into the galactose medium at a final concentration of 10 μM.
Reverse transcription quantitative PCR
RNA was extracted from the patient’s fibroblasts by use of an RNeasy Mini Kit (Qiagen) and reverse-transcribed into cDNA using a RevertAid H minus (Thermo Fisher), according to the manufacturers’ protocols. The coding sequence of COQ7 was then amplified by quantitative PCR using SsoAdvanced™ Universal SYBR Green Supermix (Bio-Rad) following the provider’s instructions, with two primer pairs: (i) forward: 5′-AATATGGAGCAAACCGCATC-3′, reverse: 5′- TCCACAAGGGCATCAGAACT-3′; and (ii) forward: 5′- AGCACATCACTACAACAACCA-3′, reverse: 5′- CAAGCTCTTCATCCCGAAA-3′. Housekeeping gene primers were the following: (i) TBP, forward: 5′-CACGAACCACGGCACTGATT-3′, reverse: 5′-TTTCTTGCTGCCAGTCTGGAC-3′; (ii) GAPDH: forward: 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse: 5′-GAAGATGGTGATGGGATTC 3′; and (iii) HPRT, forward: 5′-TGACAGTGGCAAAACAATGCA-3′, reverse: 5′-GGTCCTTTTCACCAGCAAGCT-3′.
Cellular extract for western blot analysis
Total protein extracts were obtained by adding 400 μl of Laemmli buffer (50 mM Tris-HCl pH 8, 10% glycerol, 100 mM DTT, 2% SDS, bromophenol blue) and 100 U of benzonase on 2 million cell pellets. Subcellular fractionation was performed according to Martini et al.13 Briefly, extracts were prepared from confluent fibroblast cells in 150 mm diameter dishes. After rinsing twice in PBS buffer, cells were allowed to swell for 10 min in 10 ml of low-salt (LS) buffer (HEPES 20 mM pH 7.8, potassium acetate 5 mM, MgCl2 0.5 mM, protease inhibitor 1×) per dish. Then, the extracts were collected in a 1.5 ml centrifugation tube with cell scrapers and disrupted with 25 strokes in a Dounce homogenizer using the loose pestle. The extract was centrifugated (3 min, 2100g, 4°C). After aspiration of the supernatant, the pellet was resuspended in LS buffer (typically 80 μl) with NaCl to reach a final concentration of 0.6 M and incubated for 90 min at 4°C. A new centrifugation (20 min, 21 000g, 4°C) allowed separation of the nuclear extract in the supernatant from the chromatin extract in the pellet. The pellet was resuspended in 80 μl of LS buffer containing 100 U of benzonase and incubated for 15 min at room temperature. After the last centrifugation (15 min, 20 000g, 4°C), solubilized chromatin was collected in the supernatant, while the final pellet containing non-solubilized chromatin and mitochondria was resuspended in 80 μl of Laemmli buffer.
Western blot analysis
Cellular extracts were resolved on 8–16% SDS-PAGE gels and transferred to nitrocellulose membranes (Bio-Rad). Western blot was performed with anti-α-tubulin (Sigma, Cat. No. T6074), anti-FLAG-tag (mouse; M2 clone; Sigma, Cat. No. F1804), anti-COQ7 (rabbit; Proteintech, Cat. No. 15083-1-AP), anti-TOM20 (rabbit mAB D8T4N; CST, Cat. No. 42406), and anti-histone H4 (Merk-Millipore, Cat. No. 04-858), followed by chemiluminescent detection using horseradish peroxidase-conjugated antibodies and the SuperSignal West Pico reagent (Thermo Scientific).
Plasmid construct and mutagenesis
The human COQ7 coding sequence of transcript NM_016138.5 was fused with a FLAG-tag in the c-terminal and cloned in the pcDNA3.1 plasmid by Genscript (Piscataway, NJ, USA). Then, the c.3G>T mutation was realized by Genscript in the same pcDNA3.1 backbone plasmid.
Immunohistochemistry
Fixed cells were blocked and permeabilized with PBS containing 4% bovine serum albumin, 100 mM glycine and 0.3% Triton X-100. The primary antibody was applied for 1 h at room temperature diluted in blocking solution, washed three times in PBS, incubated for 1 h in the secondary antibody at room temperature with DAPI, then washed three times in PBS, mounted in Fluoromount-G™ (Invitrogen) and imaged under a confocal microscope (Zeiss LSM800). The following antibody was used: mouse anti-FLAG (M2 clone; Cat. No. F1804 at 1/1000). MitoTracker Red (Thermo Fisher, Cat. No. M7512) at 50 nM for 10 min was used in living cells to mark mitochondria before fixation with 4% PFA.
CoQ10 level measurement by high-performance liquid chromatography
Plasma and fibroblast CoQ10 content were determined using high-performance liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) as previously described.14 For MS/MS detection, we followed the transition of m/z 863.4 → 197.1 (CoQ10), 872.9 → 206.3 (CoQ10-d9 as internal standard), and 833.4 → 167.1 (6-DMQ10). The protein content of fibroblast lysates was determined using the bicinchoninic acid assay.15
Seahorse analysis
Analysis of mitochondrial respiration and ATP production was performed by measuring the oxygen consumption rate (OCR) and extracellular acidification (ECAR) in real-time using the Seahorse Bioscience Extracellular Flux Analyzer Xfe96 (Seahorse Bioscience). The Seahorse XF Real-Time ATP Rate Assay Kit (Agilent Technologies, Cat. No. 103592-100) and the Seahorse XF Cell Mito Stress Test Kit (Agilent Technologies, Cat. No. 103015-100) were performed according to the manufacturer’s protocol. The day prior to the assay, V972F, AB249, and AF400 fibroblasts were plated at a density of 20 000 cells/well in a 96 well Xfe96 Seahorse microplate (Agilent, Cat. No. 101085-004) in 80 µl of growth media and incubated overnight at 37°C 5% CO2. On the day of the assay, 60 µl of growth media were removed, and cells were washed once with Seahorse Assay media (Agilent, Cat. No. 103575-100) supplemented with glucose 40 mM, pyruvate 1 mM, and glutamine 2 mM, then incubated with 200 µl of supplemented Seahorse Assay media at 37°C in a non-CO2 incubator for 45–60 min. Before measurement, media was then replaced by fresh supplemented Seahorse Assay media to a final volume of 180 µl per well. Using the real-time ATP rate assay template, the OCR and ECAR were measured in three cycles (mix: 3 min measure: 3 min), first at the basal level and then after sequential addition of oligomycin (1.5 µM/well) and rotenone/antimycin A (0.5 µM/well). Using the Cell Mito Stress Test template, OCR and ECAR were measured in three cycles, first at the basal level, then after sequential addition of oligomycin (1.5 µM/well), FCCP (1.5 µM/well), and rotenone/antimycin A (0.5 µM/well).
After measurement, cells were fixed with a solution of PFA 4%/Triton 0.2%/DAPI (1/500) and imaged on an EVOS microscope. Metamorph software was used to determine the number of cells per well according to DAPI staining. These values were then used to normalize all the OCR and ECAR recorded data. Glycolytic ATP production rate (glycoATP) and mitochondrial ATP production rate (mitoATP) were calculated using OCR and ECAR data based on Agilent validated equations present in the manufacturer’s protocol (Agilent, Cat. No. 103592-100).
For the rescue with CoQ10, the cell culture medium was supplemented with a final concentration of 10 μM every 2 days for 9 days prior to the respiration analysis.
Statistical analysis
Each experiment was repeated at least three times independently. Data were analysed with Excel (Microsoft®) or Prism (GraphPad). First, data distribution was analysed with the D’Agostino and Pearson omnibus normality test for Gaussian distribution and Bartlett’s test for equal variances. Data from two conditions following Gaussian distribution or equal variance were analysed by Student’s t-test. Data from several conditions each showing normality and equal variance were compared with one-way ANOVA, followed by Bonferroni’s multiple comparison test. Data that did not show a Gaussian distribution or an equal variance were analysed using non-parametric Kruskal–Wallis test, followed by Dunn’s multiple comparison test. Finally, a two-way ANOVA was used to estimate the difference in the mean of data according to two categorical variables.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Clinical results
The affected siblings were born from non-consanguineous healthy parents in Portugal (Table 1). The family displayed an autosomal recessive inheritance pattern. The three patients were two brothers and a sister (Fig. 1A).
Table 1.
Clinical features of patients with COQ7 mutations
| Patient/Sex/Age (years) | Age at onset (years) | Initial symptoms | Pattern of muscle weakness | Pes cavus | Sensory involvement | Pyramidal signs | Cognitive disorders | Ambulation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UL (prox.) | UL (dist.) | LL (prox.) | LL (dist.) | UL | LL | |||||||
| II-2/M/36 | 12 | Walking difficulties | − | ++ | − | +++ | + | − | − | +Brisk reflexes, Hoffman’s sign | − | Ambulant without help |
| II-3/F/25 | 9 | Walking difficulties | − | + | − | ++ | + | − | + | +Babinski’s sign on left foot | − | Ambulant without help |
| II-1/M/42 | 10 | Walking difficulties | − | ++ | + | +++ | + | − | − | − | − | Ambulant with ankle foot orthoses |
For column ‘pattern of muscle weakness’: − = absent; + = mild; ++ = moderate; +++ = severe; for other columns: − = absent; + = present; dist = distal; F = female; LL = lower limb; M = male; prox = proximal; UL = upper limb.
Patient II-2
The index case, Patient II-2, was a 36-year-old man (Fig. 1A). He had normal developmental milestones without walking difficulty in childhood and took part in sports. When he was 12 years old, he developed walking difficulties with a sensation of stiffness in the lower limbs and a difficulty walking on heels. Muscle weakness worsened gradually with a predominance in the distal part of the lower limbs (Fig. 1B). At the age of 32 years, he developed muscle weakness in the hands, leading to mild difficulties in activities of daily living. These symptoms also worsened progressively. He had never complained of sensory symptoms. When the patient was seen in the study unit at the age of 36 years, he could walk 45 min without aid and could still climb stairs. Clinical examination showed amyotrophy in the lower limbs and hands. Gower’s manoeuver was negative. Heel or tiptoe walking was impossible. Muscle strength examination revealed a severe deficit in the distal muscles of both upper and lower limbs, with a predominant involvement of foot dorsal flexion in the lower limbs [tibialis anterior 2/5 right side, 0/5 left side, extensor hallucis longus 0/5 both sides, gastrocnemius 3/5 both sides, abductor pollicis brevis 3/5 both sides, first interosseous muscle 3/5 both sides, according to the Medical Research Council (MRC) scale]. There was no fasciculation. He had pes cavus. Sensory clinical examination was normal. Deep tendon reflexes were brisk in the four limbs (except for Achilles reflexes which were abolished), and a Hoffman’s sign was found, without Babinski’s sign or spasticity. Cranial nerve examination was normal. There was no cognitive disorder.
Patient II-3
Patient II-3 was a 25-year-old woman. She had normal early development, without walking difficulty when she was a child. When she was 9 years old, she developed walking difficulties with fatigability and feet drop. She noticed a progressive degradation of her muscle weakness, with a distal predominance, and a rapid worsening between 12 and 15 years of age. She presented motor difficulties in her hands for precise movements when she was 13 years old, also with a progressive worsening course. Between 15 and 18 years of age, while she was treated with CoQ10 (30 mg per day), she reported symptom stabilization. A new progressive worsening started at the age of 18 years and the stop of CoQ10 supplementation. She complained of difficulties in feeling cold and warm temperatures on her feet. When she was seen at the age of 25 years, she estimated her walking distance to be 1 km without aid and could climb stairs. Gower’s manoeuver was positive. Heel or tiptoe walking was impossible. Muscle strength examination revealed a deficit in distal muscles primarily in the lower limbs, with a predominant involvement of foot dorsal flexion in the lower limbs (tibialis anterior 3/5 both sides, gastrocnemius 4/5 both sides, abductor pollicis brevis 4/5 both sides, interosseous muscles 4/5 both sides according to MRC scale). There was no fasciculation observed, but the patient reported some. She had pes cavus. Sensory clinical examination was normal. Deep tendon reflexes were found to be normal in the four limbs, and no Hoffman’s sign was found, but a Babinski’s sign was found on the left foot. Cranial nerve examination was normal. There was no cognitive disorder.
Patient II-1
Patient II-1 was a 42-year-old man. He had normal developmental milestones without walking difficulty in childhood, and took part in sports. When he was 10 years old, he noticed walking and biking difficulties, particularly when compared to his classmates, with foot drop. These symptoms progressively worsened and became more stable after the age of 20 years, when he required ankle-foot orthoses. At 22 years of age, he developed muscle weakness in his hands leading to difficulties in dressing. These symptoms also progressively worsened. He had never complained of sensory symptoms. When the patient was seen in the study unit at age 42 years, he could walk 500 m with ankle-foot orthoses and could still climb stairs. Heel or tiptoe walking was impossible. Gower’s manoeuver was positive. Clinical examination showed a severe amyotrophy in lower limbs, with a distal predominance, and an amyotrophy of hand muscles. Muscle strength examination revealed a severe deficit in distal muscles of both upper and lower limbs with a complete loss of foot plantar and dorsal flexions (tibialis anterior 0/5 both sides, gastrocnemius 0/5 both sides, abductor pollicis brevis 3/5 both sides, quadriceps and psoas 4/5 both sides, interosseous muscle 3/5 both sides, according to MRC scale). There was no fasciculation observed, but the patient reported some. He had pes cavus. Sensory clinical examination was normal. Achilles deep tendon reflexes were abolished. Hoffman and Babinski’s signs were not found. Cranial nerve examination was normal. There was no cognitive disorder.
Electrophysiological findings
ENMG data were only available for Patients 1 and 2 (Table 2). There was evidence of a pure motor axonal neuropathy predominantly affecting the distal part of the lower limbs. During needle examination, fibrillations were observed at rest, and a decrease in motor unit recruitment was recorded during contraction in the lower limb muscles in both patients and in the distal upper limb muscles in Patient 1.
Table 2.
Electrophysiological findings
| Patient (age at exam, years) | Motor nerve conduction | Sensory nerve conduction | Myography | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median nerve | Ulnar nerve | Common fibular nerve | Tibial nerve | Radial nerve | Sural nerve | Quadriceps | Tibialis anterior | Deltoid | 1st interosseous | |||||||||
| Amp (mV) (>4) | CV (m/s) (>45) | Amp (mV) (>4) | CV (m/s) (>45) | Amp (mV) (>2) | CV (m/s) (>40) | Amp (mV) (>4) | CV (m/s) (>40) | Amp (µV), (>18) | Amp (µV) (>10) | Rest | Contraction | Rest | Contraction | Rest | Contraction | Rest | Contraction | |
| II-2 (36) | 3.7 | 53.3 | 3.3 | 60.8 | NO | NO | NO | NO | 55 | 23 | Fib | Neurogenic | Fib | Neurogenic | N | N | Fib | Neurogenic |
| II-3 (22) | 1.6 | 50.0 | 2.3 | 53.2 | NO | NO | 1.2 | 30.5 | 72 | 15 | Fib | Neurogenic | Fib | Neurogenic | N | N | N | N |
In bold, abnormal result according to normal laboratory values (presented in parentheses). Amp = amplitude; CV = conduction velocity; Fib = fibrillations; N = normal; NO = not obtained.
Complementary exam
Patient 1 benefited from several complementary exams. Brain and spinal cord MRI were normal. Transthoracic echocardiography and ECG were normal. The ophthalmic exam was normal, expect for known myopia. Audiometry was normal. Biological testing showed a mild elevation of the creatine phosphokinase (CPK) level (215 U/l, normal <200 U/l).
Molecular analysis
In the family presented herein (Fig. 1A) only one variant, which was absent from the ExAC database but present once at the heterogenous state in gnomAD database (allele frequency 0.000004065) was shared by the three patients in a homozygous state and present in their unaffected parents in a heterozygous state, confirming the segregation (Fig. 1C). This variant is localized in the COQ7 gene (geneID:10229), with a pLI score equal to 0 that reflects the tolerance of a given gene to loss of function on the basis of the number of protein truncating variants weighted by the size of the gene. This pLI score suggests that COQ7 is tolerant to the loss of function in heterozygous carriers, as in our unaffected parents. The variant was a G>T substitution in the start codon of the longest transcript (isoform 1, NM_016138.5, c.3G>T; NP_057222.2, p.Met1?), which is the most common isoform of the COQ7 protein, and which is also the main isoform in the spinal cord as seen on the GTEX portal (Fig. 1D–F)16 and as confirmed in several cell types by RT-qPCR (Supplementary Fig. 1). This substitution leads to the disruption of the canonical ATG start codon into an ATT sequence in isoform 1, indicating that its translation should be affected (Fig. 1D–F). Interestingly, two alternative start codons are used by isoforms 2 and 3, indicating that these two alternative isoforms should not be affected by the variant. The mutations previously identified in the COQ7 gene in non-dHMN families7–12 were localized in the catalytic domain of the protein and caused severe phenotypes appearing at birth or during early childhood, characterized by developmental delay in all patients and often associated with multiorgan failure (Fig. 1E). The phenotype was never restricted to peripheral neuropathy. The precise genotypical and clinical description of these patients is available in Supplementary Table 1.
The c.3G>T mutation decreases the translation of COQ7 isoform 1
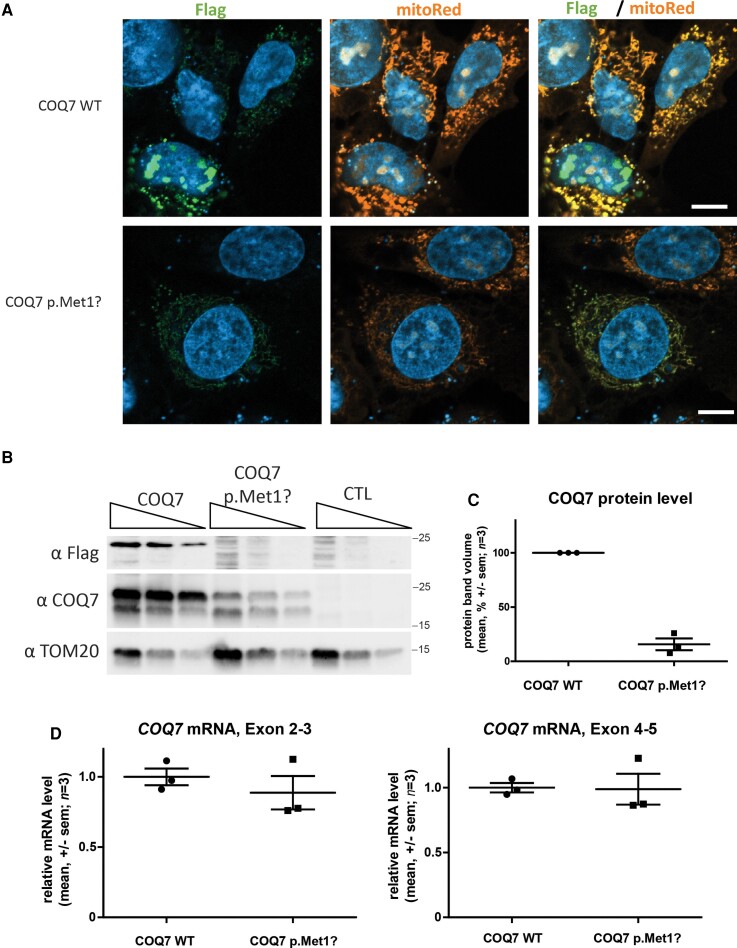
The c.3G>T mutation in COQ7 disrupts the start codon of the main COQ7 isoform 1 (NM_016138). The COQ7 transcript contains a second in-frame ATG codon at 111 nucleotides (37 AA) downstream of the first start codon. This second ATG is used to generate COQ7 isoform 2 (NM_001190983.2) (Fig. 1D–F) and deprives COQ7 of its mitochondrial targeting signal.6 Thus, we hypothesized that mutation of the first ATG could prevent mitochondrial translocation, thereby causing mislocalization of the protein. To evaluate COQ7 subcellular localization, plasmids encoding wild-type and mutant COQ7 were transfected into the human neuroblastoma cell line SH-EP. A FLAG octapeptide was fused to the C-terminal end of COQ7 to allow detection with an anti-FLAG antibody. MitoTracker Red and DAPI were used to visualize mitochondria and nuclei, respectively. FLAG-tag, mitochondria, and nuclei staining indicated that the localization of wild-type and mutant COQ7 were similar (Fig. 2A and Supplementary Fig. 2). Consistent with the fact that the mutation is localized in the translation initiation codon, the FLAG labelling was significantly lower in intensity and in number of cells in SH-EP cells expressing the mutant COQ7 than in those expressing wild-type COQ7 (Fig. 2A and Supplementary Fig. 2A and B). This was confirmed by Western blot using a polyclonal anti-COQ7 antibody: COQ7 levels were reduced by 85% in SH-EP cells expressing the mutant COQ7 compared to SH-EP cells expressing the wild-type COQ7 (Fig. 2B and C and Supplementary Fig. 3A and B). This decrease could not be attributed to a transcriptional effect since COQ7 mRNA levels evaluated by qRT-PCR did not differ significantly between cells transfected with the wild-type or mutant COQ7 (Fig. 2D). Altogether, these results show that the c.3G>T mutation severely reduces cellular COQ7 protein isoform 1 levels, most likely by reducing translation, since transcription is unaffected.
Figure 2.
COQ7 c.3G>T mutation reduces COQ7 protein expression in SH-EP cells. (A) Confocal images of SH-EP cells transfected with expression vectors for wild-type (WT) or mutant COQ7 fused to c-terminal FLAG-tag and counterstained for mitochondria in orange (MitoTracker), COQ7 in green (anti-FLAG), and nucleus (DAPI) in blue. Scale bar = 10 µm. (B) Western blot analysis of SH-EP transfected with COQ7 expression vectors. COQ7 was detected with anti-FLAG and anti-COQ7 antibodies. The mitochondrial protein TOM20 is detected with an anti-TOM20 antibody. Protein sample dilution ratio: 1, 1:2, and 1:4 for each condition. The two bands observed with the anti-COQ7 antibody and anti-Flag (Supplementary Fig. 3A and B) probably correspond to mature and immature isoform 1 with 36 additional AA before cleavage of the signal peptide. (C) Quantification of COQ7 protein by western blot (mean ± standard error of the mean). (D) Quantification of COQ7 mRNA in SH-EP cells transfected with expression plasmids for wild-type or mutant COQ7, using two primer pairs localized in exons 2–3 and exons 4–5 respectively. Values represent mean ± standard error of the mean.
To investigate whether the isoforms of COQ7 that use alternative start codons could provide some COQ7 activity to the mitochondria, a Flag-tagged COQ7 isoform 2 (NM_001190983) expression vector was transfected in SH-EP cells. Immunostaining of the Flag and mitochondrial protein TOM20 showed that COQ7 isoform 2 was expressed and co-localized with mitochondria (Supplementary Fig. 2C). Altogether, these results suggest that the translation of COQ7 isoform 1 is impacted by the variant and that other isoforms of the protein can provide mitochondria with COQ7, suggesting that they could partially compensate for the loss of isoform 1.
The COQ7 mutation decreases the protein level in patient’s fibroblasts
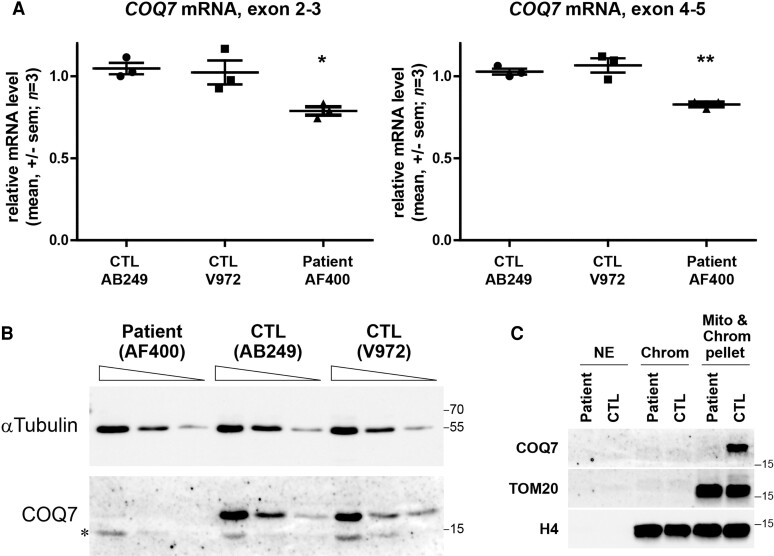
Similar to motor neurons, fibroblasts mainly express COQ7 isoform 1, which carries the mutation (Supplementary Fig. 1).16 To investigate whether the drastic reduction in the mutant COQ7 protein level was also present in the cells of the patient, a fibroblast culture was derived from a skin biopsy from Patient II-2. After a day in culture, total RNAs were extracted and COQ7 mRNA levels were measured by RT-qPCR in control fibroblasts and in those of the patient. PCR amplification between exons 2–3 or exons 4–5 showed a 21 and 17% decrease, respectively, in COQ7 mRNA levels in the patient’s fibroblasts compared with the controls (Fig. 3A), indicating that the c.3G>T mutation had a low impact on COQ7 mRNA levels.
Figure 3.
COQ7 c.3G>T mutation decreases COQ7 protein level in the patient’s fibroblasts. (A) Quantification of COQ7 mRNA in the control (CTL) and patient’s fibroblasts using two primer pairs targeting exon 2–3 and exon 4–5. Values represent mean ± standard error of the mean. *P < 0.05; **P < 0.01; n = 3; one-way ANOVA followed by Bonferroni’s multiple comparison test. (B) Western blot analysis of control and patient’s fibroblasts using anti-COQ7 antibodies and α-tubulin gene to normalize. Protein loading 4×, 2×, and 1× for each condition. Stars show the non-specific histone band (Supplementary Fig. 3C and D). (C) Nuclear extract (NE), benzonase soluble (Chrom), and insoluble (Chrom & Mito Pellet) fractionation revealed by western blotting using anti-COQ7 antibodies to stain COQ7 protein, anti-TOM20 antibodies to stain mitochondria, and anti-H4 antibodies to stain H4 histones in the control and patient’s fibroblasts.
Total COQ7 protein levels were then analysed by western blot. COQ7 protein was barely detected in the patient’s fibroblasts, whereas it was easily detected in control cells (Fig. 3B), suggesting a drastic reduction in the COQ7 protein level. Western blot analysis was then performed after subcellular fractionation. As expected, in control cells, COQ7 was present exclusively in the insoluble chromatin fraction that also contained the mitochondria as evidenced by the TOM20 labelling. Again, the COQ7 level was barely detected in the fractions from the patient’s fibroblasts (Fig. 3C). The TOM20 labelling also indicated that the amount of mitochondria was similar in the patient’s and control fibroblasts (Fig. 3C). Altogether, these results show that the c.3G>T mutation affecting the start codon of COQ7 isoform 1 causes a strong reduction in the level of COQ7, probably consecutive to poor translation of COQ7 isoform 1, which accounts for most cellular COQ7.
COQ7 protein deficit decreases CoQ10 levels and disrupts mitochondrial metabolism
CoQ10 is essential for respiratory chain activity. By catalysing the hydroxylation of 6-DMQ in mitochondria, COQ7 is one of the main enzymes involved in CoQ10 synthesis.5 To study the impact of the COQ7 mutation on CoQ10 levels, CoQ10 and 6-DMQ levels were measured by HPLC-MS/MS in the fibroblasts and blood plasma of Patient II-2. We observed a major decrease in the CoQ10 level from 71.5 pmol/mg of protein in control fibroblasts to 12.5 pmol/mg of protein in patient fibroblasts together with the appearance of 6-DMQ in the fibroblasts of the patient compared with the controls (Fig. 4A, B and D). The appearance of 6-DMQ was also found in the patient’s plasma (Fig. 4B). These results are consistent with the drastic reduction of COQ7 protein in the patient’s fibroblasts (Fig. 3B and C). These results also indicate that fibroblasts provide a convenient model to study the metabolic changes in patients with COQ7 mutations.
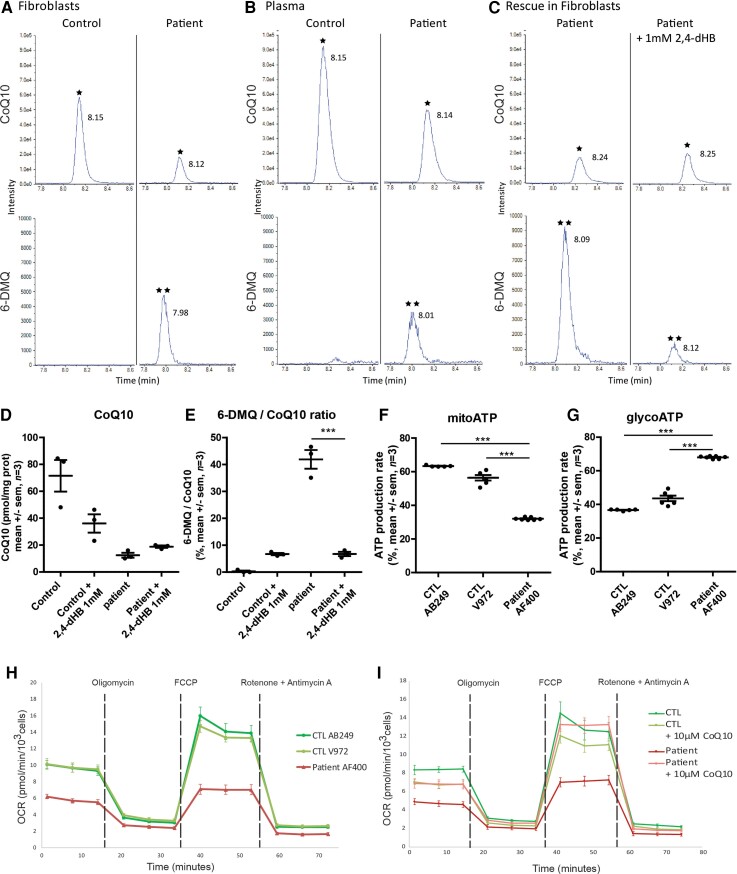
Figure 4.
Metabolic changes in the patient’s plasma and fibroblasts. Representative HPLC-MS/MS chromatograms for the detection of CoQ10 and 6-DMQ (A) in fibroblasts, (B) in the plasma of healthy controls and Patient II-2, and (C) in the patient’s fibroblasts treated with 1 mM 2,4-dHB. The top chromatogram represents the detected signal for CoQ10 (indicated by asterisk) and the bottom chromatogram represents the detected signal for 6-DMQ (indicated by double asterisk). (D and E) Graphical representation of the rescue of CoQ10 and 6-DMQ levels by 1 mM 2,4-dHB in the patient’s fibroblasts (one-way ANOVA followed by Bonferroni tests ***P < 0.001; n = 3). (F and G) Graphical representation of ATP production by mitochondrial respiration (mitoATP) and glycolysis (glycoATP) measured in real-time using the Seahorse Bioscience Extracellular Flux Analyzer on control (#AB249 and #V972) or patient’s (#AF400) fibroblasts (one-way ANOVA followed by Bonferroni tests ***P < 0.001; n = 3). (H) Seahorse Mitostress analysis. OCR traces, expressed as pmol O2/min/103 cells of fibroblast cell lines from the patient and controls. The dashed lines indicate the time of addition of oligomycin, FCCP, and antimycin A/rotenone. The OCR profile is representative of three independent experiments. (I) Effect of 10 μM CoQ10 supplementation on OCR traces of fibroblast cell lines from the patient and control. The OCR profile is representative of three independent experiments.
A synthetic precursor for CoQ10 biosynthesis, 2,4-dihydroxybenzoic acid (2,4-dHB), allows the need for hydroxylation by COQ7 to be bypassed.17 To investigate whether CoQ10 biosynthesis could be rescued in the absence of COQ7, 2,4-dHB was added to the fibroblast culture medium. After 14 days of supplementation, cells were harvested, and CoQ10 and 6-DMQ levels were measured by HPLC-MS/MS (Fig. 4C). The results indicated that 2,4-dHB slightly increased CoQ10 levels from 12.5 to 18.8 pmol/mg and normalized 6-DMQ accumulation in the patient’s fibroblasts (Fig. 4D and E). These results confirm that the decrease in CoQ10 levels was due to a lack of COQ7 activity (Fig. 4C and D). Altogether, these experiments demonstrate that decreased levels of COQ7 in patient cells result in lower production of CoQ10 and accumulation of 6-DMQ.
Low levels of CoQ10 are expected to affect mitochondrial respiration. A Seahorse analysis (ATP rate assay) showed that mitochondrial ATP production was decreased from 63.3% or 56.4% in control fibroblasts to 42% in the patient’s fibroblasts (Fig. 4F), while ATP production by glycolysis was increased from 36.7% or 43.6% in control fibroblasts to 68% in the patient’s fibroblasts (Fig. 4G). To confirm that the activity of the respiratory chain was decreased in COQ7 mutated cells, a Seahorse analysis was used to evaluate oxygen consumption by the cells (Cell Mito Stress test). Both basal and maximal respirations were significantly reduced in fibroblasts carrying the c.3G>T mutation (Fig. 4H). Interestingly, these basal and maximal respiration rates could be rescued by CoQ10 supplementation in the culture medium prior to the Seahorse analysis (Fig. 4I). Altogether, these results show that the c.3G>T mutation in COQ7 causes a deficit in CoQ10 levels, leading to reduced mitochondrial ATP production, which is compensated for by an increase in the glycolytic activity of the cells to maintain normal ATP levels. This reduced mitochondrial ATP production is explained by a decrease in the activity of the respiratory chain, which can be rescued by CoQ10 supplementation.
COQ7 protein deficit lowers cell proliferation but can be rescued by CoQ10
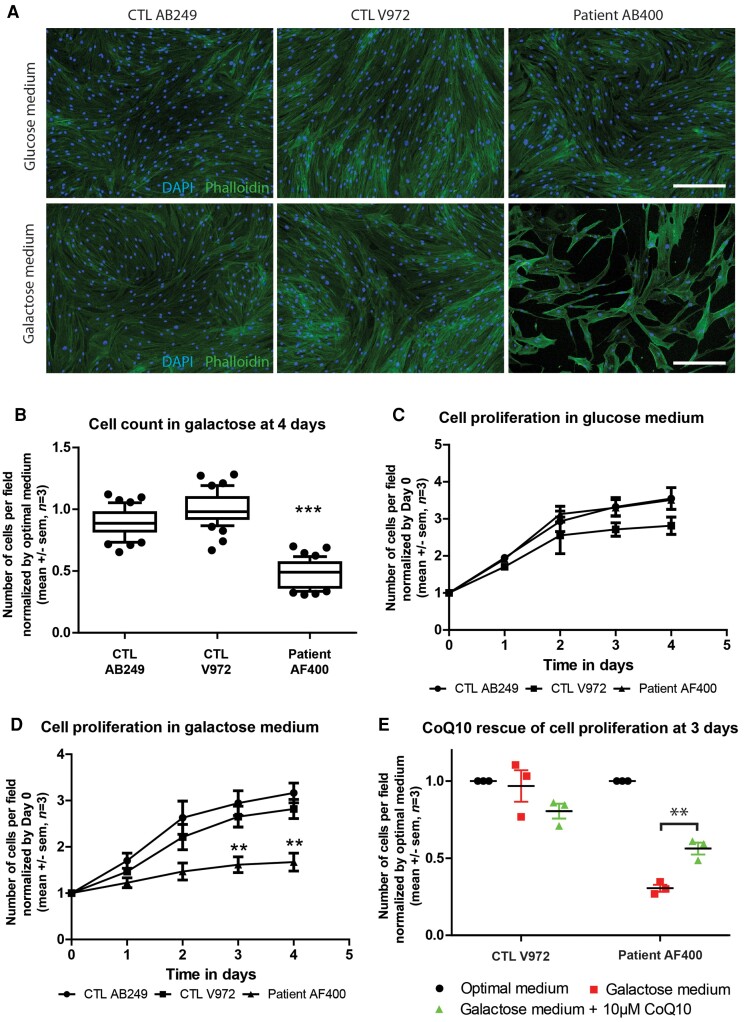
Galactose enters glycolysis by being converted to glucose-1-phosphate, which considerably slows ATP production. Therefore, replacing glucose by galactose in the culture medium reduces the efficiency of glycolysis and causes cultured cells to rely more on mitochondrial metabolism.13 To study the functional consequences of the COQ7 deficiency, fibroblasts were grown with either glucose or galactose as the source of carbohydrate. After 4 days, the cells were counted and their number in galactose medium was normalized to their number in glucose medium. Cell counting revealed a 50% decrease in the number of fibroblasts from the patient compared with the controls (Fig. 5A and B). To decipher whether this reduction was due to reduced proliferation or increased apoptosis, immunohistochemistry was performed to detect activated caspase 3. No active caspase 3 was detected in the control and COQ7 mutated cells (data not shown). In addition, no pyknotic nuclei were observed, suggesting that the COQ7 mutation affected cell proliferation rather than survival. The cell proliferation curve was thus determined by counting the cells every day and normalizing to the number of cells at Day 0. In glucose medium, fibroblasts from the patient had the same growth rate as the controls (Fig. 5C). Conversely, in galactose medium, the growth rate of the patient’s fibroblasts was significantly decreased compared with that of the controls (Fig. 5D).
Figure 5.
The cell growth of the patient’s fibroblasts is lowered in galactose medium and is partially rescued by CoQ10 supplementation. (A) Fibroblast cultures from two controls and Patient II-2 after 4 days of cell growth, stained for nucleus (DAPI) in blue and actin (Phalloidin) in green. Scale bar = 300 μm. (B) Number of cells per field in the galactose medium after 4 days of growth, normalized to the number of cells in glucose medium, in the fibroblasts of the patient (#AF400) and controls (#AB249 and #V972) (Kruskal–Wallis test followed by Dunn’s multiple comparison test; ***P < 0.001; n = 3). (C) Number of cells per field in the optimal medium during 4 days of growth, normalized to the number of cells at Day 0 in the optimal medium, in the fibroblasts of the patient (#AF400) and controls (#AB249 and #V972). (D) Change in the number of cells per field in galactose medium over 4 days, normalized to the number of cells at Day 0 in glucose medium, in the fibroblasts of the patient (#AF400) and controls (#AB249 and #V972) (two-way ANOVA followed by Bonferroni tests **P < 0.01; n = 3). (E) Number of cells per field in glucose, galactose, and galactose with 10 μM CoQ10 medium after 3 days in culture, normalized to the number of cells at Day 0 in glucose medium, in the fibroblasts of the patient (#AF400) and controls (#V972) (two-way ANOVA followed by Bonferroni tests **P < 0.01; n = 3).
We next investigated whether CoQ10 supplementation could restore normal growth of the patient’s fibroblasts in the galactose medium. CoQ10 (10 μM) was added to the galactose medium, and 3 days later the number of cells was compared to the number of cells in the glucose medium. Cell counting revealed that CoQ10 supplementation induced a 100% increase in the number of COQ7 mutated fibroblasts after 3 days (Fig. 5E). Altogether, these results show that the cell proliferation of COQ7 mutated fibroblasts is diminished when the rate of glycolysis is reduced and that CoQ10 supplementation partially restores a normal growth rate.
Discussion
We reported a family of two brothers and a sister affected by a recessive dHMN, and for which molecular analysis revealed a homozygous c.3T>G substitution in the start codon of the COQ7 gene. All of the patients presented with length-dependent pure motor axonal neuropathy, leading to progressive walking difficulties and disability, with childhood onset. The results showed that this mutation leads to a decreased level of COQ7 protein, while mRNA levels are not strongly affected, suggesting a translation deficiency secondary to a translation initiation defect. Interestingly, transfection with COQ7 expression vectors indicated that translation of the mutated RNA was only partially prevented, suggesting the possibility that non-ATG-initiation of translation could occur.18 The decrease in COQ7 protein level reduces the production of CoQ10, resulting in a decrease in the efficiency of the mitochondrial respiratory chain and a higher dependence on glycolysis for ATP production. As a consequence, a reduction in the growth rate is observed when cells are placed in a medium where glycolysis is impeded. Treatment with exogenous CoQ10 can rescue these deficits of mitochondrial respiration and cell proliferation in the cellular model.
Compared to previously reported patients with COQ7 mutations, the family studied here presented with a less severe phenotype characterized by isolated neurological damage of motor neurons, despite comprehensive systemic explorations. Conversely, two of the seven patients previously described presented with multiorgan failure in early childhood, while the other five experienced severe locomotor difficulties with motor developmental delay.7–12 However, only two of the seven patients were investigated using ENMG. One showed PNS involvement, with an axonal and demyelinating sensorimotor neuropathy, while the other had a normal ENMG.7–9,11 Interestingly, the patients reported by Theunissen et al.10 and Wang et al.12 were described as having neuropathy. However, no ENMG was available to confirm this clinical hypothesis. Moreover, they also presented with developmental delay and had an early childhood onset, which did not correspond to the pure dHMN phenotype of the patients we presented herein.
A question that remains is how to explain the difference in severity between our patient with dHMN manifestation and those previously reported, when all these mutations were recessive and should affect the same molecular CoQ10 synthesis pathway. We speculate that in our patient the mutation affects only the translation of the main COQ7 isoform, and does not affect the two other COQ7 isoforms, and as a consequence does not completely prevent CoQ10 synthesis.
Interestingly, we have observed that the isoform of COQ7 produced from the second start codon (isoform 2) can reach the mitochondria. Although alternative isoforms are much less expressed than isoform 1, they may allow a basal level of COQ7 to be maintained in the mitochondria, thus explaining the moderate severity of the disease compared to the syndromes caused by the previously described mutations. Consistently, all previously described COQ7 mutations were localized in the catalytic site and therefore affected all COQ7 isoforms. These mutation generating mutated COQ7 proteins could cause protein misfolding or degradation, destabilization of the COQ7:COQ9 heterodimer or tetramer,19 modification of the substrate binding capacity or the hydrophobic access channel, or diminished activity of the iron-dependent hydroxylase catalytic site. Finally, we cannot exclude that other genes implicated in mitochondrial metabolism and differentially expressed among patients could influence the clinical manifestation of the mutations.
The present experiments were conducted on patient fibroblasts, a non-affected tissue in the studied family, which is easily accessible. This allowed us to show the functional consequences of the mutation under stress conditions. In other peripheral inherited neuropathies such as spinal muscular amyotrophy (SMA) caused by inactivation of the SMN gene or peripheral inherited neuropathies secondary to SORD mutations, fibroblasts were also used to show the functional impairment under stress conditions, even if the patients had no cutaneous symptoms.20,21 The presence of isolated neurological symptoms in such inherited neuropathies associated with mutations in ubiquitously expressed genes is classically explained by a higher sensitivity of neurons to stress, but in most cases the cellular specificity of the deficits remains to be fully elucidated.21–23 In the case of COQ7 mutations, fibroblasts provide the advantage of expressing the same major COQ7 transcript as motor neurons at comparable levels. Nevertheless, it will be interesting to further evaluate the consequences of the c.3G>T substitution in human motor neurons derived from human pluripotent stem cells either from patients or modified by CRISPR-CAS9.
The accumulation of 6-DMQ (the COQ7 substrate) in the patient’s fibroblasts was rescued by 2,4-dHB supplementation, indicating a loss of COQ7 enzymatic activity. Whether 6-DMQ can be toxic or beneficial for cells is a matter of controversy. While some authors report that it could replace CoQ10 as a mitochondrial electron carrier in the respiratory chain, others argue that 6-DMQ competes with CoQ10 in the respiratory chain.24,25 The present results do not allow us to conclude whether one or the other is true. Nevertheless, they suggest that if 6-DMQ can replace CoQ10, it does so with low efficiency, since the respiratory activity in the patient’s cells is low. Conversely, if 6-DMQ competes with CoQ10 in the respiratory chain, it also probably does so with low efficiency, since CoQ10 supplementation efficiently rescues cell growth despite the presence of 6-DMQ.
We reported herein the first family of patients affected by pure peripheral inherited neuropathy associated with COQ7 mutation. The clinical phenotype of dHMN with pyramidal features reported here is suggestive of upper motor neuron involvement. Such a phenotype has already been described for other dHMN genes such as BSCL2.26 The COQ7 mutation prevalence in dHMN, and also in Charcot-Marie-Tooth (CMT) disease, needs to be elucidated, and this gene should be included in the peripheral neuropathy gene panels for molecular analysis. Indeed, the same mutation can lead to different peripheral phenotypes with potential overlaps.27 When COQ7 variants are identified, the accumulation of 6-DMQ levels in the plasma observed herein will be useful to confirm the pathogenicity of these variants.
The search for COQ7 mutations in CMT and dHMN patients is particularly relevant, since it could indicate potential treatments. Indeed, we showed that CoQ10 can rescue cells from patients under stress conditions. This supplementation should be considered as a potential therapy. In the four patients previously reported with COQ7 mutations, this treatment was used and the follow-up was available for three of them: while the patient who presented with the most severe phenotype did not improve, the others reported clinical stabilization or improvement after CoQ10 supplementation.7,8,11 Moreover, this treatment has also been used with success in other neurological conditions such as hereditary ataxia, where CoQ10 needs to cross the blood–brain barrier, contrary to peripheral neuropathy in which a peripheral uptake is possible.28 Finally, such supplementation has been proven to be safe.29 Previous studies suggest potential interest in 2,4-dHB supplementation for patients affected by COQ7 mutations, especially in those with severe CoQ10 deficiency.7,8,12 In this study, such a supplementation led to a slight increase in CoQ10 synthesis by the patient’s fibroblasts, although the gain was far from rescuing normal levels. Moreover, in a previous study, mice benefited from such a supplementation, but with a minimal concentration of 0.0125 g/kg/day, which seems difficult to achieve in humans.8 Taken together, these findings are not in favour of a practical and clinical use of this treatment.
Altogether, this work opens new perspectives in the field of inherited neuropathy, especially dHMN and possibly CMT. The COQ7 gene represents a new potential target for molecular analysis in these patients. The identification of mutations in this gene should lead clinicians to consider oral CoQ10 supplementation as a potentially useful therapy.
Supplementary Material
Acknowledgements
J.T. thanks the Fondation pour la Recherche Médicale and the Société Francophone du Nerf Périphérique for their financial support. We thank Dr Cécile Acquaviva for her help in establishing helpful collaboration. We thank Véréna Landel (Direction de la Recherche en Santé, Hospices Civils de Lyon) for her help in manuscript preparation. Original thumbnail image has been created with BioRender.
Contributor Information
Arnaud Jacquier, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Centre de Biotechnologie Cellulaire, CBC Biotec, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Julian Theuriet, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Hôpital Neurologique Pierre Wertheimer, Service d’électroneuromyographie et de pathologies neuromusculaires, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Fanny Fontaine, Service de Biochimie, CHU de Caen, UMRS 1237 PhIND, Université de Caen, Caen, France.
Valentine Mosbach, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Nicolas Lacoste, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Shams Ribault, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Hôpital Henry Gabrielle, Service de Médecine Physique et de Réadaptation, CHU de Lyon—Hospices Civils de Lyon (HCL), Saint-Genis-Laval, France.
Valérie Risson, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Julien Carras, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Laurent Coudert, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Thomas Simonet, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Philippe Latour, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Unité fonctionnelle de neurogénétique moléculaire, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Tanya Stojkovic, Institut de Myologie, Hôpital Pitié-Salpêtrière, Assistance Publique des Hôpitaux de Paris (APHP), Paris, France.
Juliette Piard, Centre de Génétique Humaine, CHU, Besançon, France; UMR-Inserm 1231 GAD, Génétique des Anomalies du Développement, Université de Bourgogne Franche-Comté, Dijon, France.
Anne Cosson, Neurologie Électrophysiologie Clinique, CHU Jean-Minjoz, Besançon, France.
Gaëtan Lesca, Service de génétique, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Françoise Bouhour, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Hôpital Neurologique Pierre Wertheimer, Service d’électroneuromyographie et de pathologies neuromusculaires, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Stéphane Allouche, Service de Biochimie, CHU de Caen, UMRS 1237 PhIND, Université de Caen, Caen, France.
Hélène Puccio, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France.
Antoine Pegat, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Hôpital Neurologique Pierre Wertheimer, Service d’électroneuromyographie et de pathologies neuromusculaires, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Laurent Schaeffer, Pathophysiology and Genetics of Neuron and Muscle, CNRS UMR 5261, INSERM U1315, Université Lyon1, Faculté de Médecine Lyon Est, Lyon, France; Centre de Biotechnologie Cellulaire, CBC Biotec, CHU de Lyon—Hospices Civils de Lyon (HCL) groupement Est, Bron, France.
Funding
A.J. was funded by the Association pour le Développement de la Neurogénétique (ADN). J.T. was funded by the Fondation pour la Recherche Médicale (FRM, number M2R202106013343) and the Société Francophone du Nerf Périphérique (SFNP). The project was supported by AFM Téléthon through the MyoNeurALP strategic grant and by the FRM through the equipe FRM grant to L.S., A.J. and J.C. were supported by AFM Téléthon through a MyoPharm grant.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Carroll AS, Burns J, Nicholson G, Kiernan MC, Vucic S. Inherited neuropathies. Semin Neurol. 2019;39:620–639. [DOI] [PubMed] [Google Scholar]
- 2. Bansagi B, Griffin H, Whittaker RG, et al. . Genetic heterogeneity of motor neuropathies. Neurology. 2017;88:1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neuromuscular Home Page. Accessed 21 June 2022. https://neuromuscular.wustl.edu/
- 4. Frasquet M, Rojas-García R, Argente-Escrig H, et al. . Distal hereditary motor neuropathies: Mutation spectrum and genotype–phenotype correlation. Eur J Neurol. 2021;28:1334–1343. [DOI] [PubMed] [Google Scholar]
- 5. Acosta MJ, Vazquez Fonseca L, Desbats MA, et al. . Coenzyme Q biosynthesis in health and disease. Biochim Biophys Acta. 2016;185:1079–1085. [DOI] [PubMed] [Google Scholar]
- 6. Monaghan RM, Barnes RG, Fisher K, et al. . A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nat Cell Biol. 2015;17:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freyer C, Stranneheim H, Naess K, et al. . Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4–dihydroxybensoic acid. J Med Genet. 2015;52:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S. Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J Cell Mol Med. 2017;21:2329–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashemi SS, Zare-Abdollahi D, Bakhshandeh MK, et al. . Clinical spectrum in multiple families with primary COQ 10 deficiency. Am J Med Genet A. 2021;185:440–452. [DOI] [PubMed] [Google Scholar]
- 10. Theunissen TEJ, Nguyen M, Kamps R, et al. . Whole exome sequencing is the preferred strategy to identify the genetic defect in patients with a probable or possible mitochondrial cause. Front Genet. 2018;9:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwong AKY, Chiu ATG, Tsang MHY, et al. . A fatal case of COQ7-associated primary coenzyme Q 10 deficiency. JIMD Rep. 2019;47:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Gumus E, Hekimi S. A novel COQ7 mutation causing primarily neuromuscular pathology and its treatment options. Mol Genet Metabol Rep. 2022;31:100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martini E, Roche DMJ, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontaine F, Legallois D, Créveuil C, et al. . Is plasma concentration of coenzyme Q10 a predictive marker for left ventricular remodelling after revascularization for ST-segment elevation myocardial infarction? Ann Clin Biochem. 2021;58:327–334. [DOI] [PubMed] [Google Scholar]
- 15. Smith PK, Krohn RI, Hermanson GT, et al. . Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. [DOI] [PubMed] [Google Scholar]
- 16. GTEx portal. Accessed 21 June 2022. https://gtexportal.org/home/gene/COQ7
- 17. Xie LX, Ozeir M, Tang JY, et al. . Overexpression of the Coq8 kinase in saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J Biol Chem. 2012;287:23571–23581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kearse MG, Wilusz JE. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017;31:1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manicki M, Aydin H, Abriata LA, et al. . Structure and functionality of a multimeric human COQ7:COQ9 complex. Mol Cell. 2022;82:4307–4323.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wadman RI, Stam M, Jansen MD, et al. . A comparative study of SMN protein and mRNA in blood and fibroblasts in patients with spinal muscular atrophy and healthy controls. PLoS One. 2016;11:e0167087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cortese A, Zhu Y, Rebelo AP, et al. . Biallelic mutations in SORD cause a common and potentially treatable hereditary neuropathy with implications for diabetes. Nat Genet. 2020;52:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prior TW, Nagan N. Spinal muscular atrophy: Overview of molecular diagnostic approaches. Curr Protoc Hum Genet. 2016;88:9.27.1–9.27.13. [DOI] [PubMed] [Google Scholar]
- 23. Motley WW, Talbot K, Fischbeck KH. GARS Axonopathy: Not every neuron’s cup of tRNA. Trends Neurosci. 2010;33:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Hekimi S. Minimal mitochondrial respiration is required to prevent cell death by inhibition of mTOR signaling in CoQ-deficient cells. Cell Death Discov. 2021;7:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang YY, Vasta V, Hahn S, et al. . The role of DMQ9 in the long-lived mutant clk-1. Mech Ageing Dev. 2011;132(6-7):331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossor AM, Kalmar B, Greensmith L, Reilly MM. The distal hereditary motor neuropathies. J Neurol Neurosurg Psychiatry. 2012;83:6–14. [DOI] [PubMed] [Google Scholar]
- 27. Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot–Marie–Tooth disease. Lancet Neurol. 2009;8:654–667. [DOI] [PubMed] [Google Scholar]
- 28. Salviati L, Trevisson E, Doimo M, Navas P. Primary Coenzyme Q10 Deficiency. 28.
- 29. Hathcock JN, Shao A. Risk assessment for coenzyme Q10 (ubiquinone). Regulat Toxicol Pharmacol. 2006;45:282–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.