Abstract
Live, attenuated viruses have been the most successful vaccines in monkey models of human immunodeficiency virus type 1 (HIV-1) infection. However, there are several safety concerns about using such an anti-HIV vaccine in humans, including reversion of the vaccine strain to virulence and recombination with endogenous retroviral sequences to produce new infectious and potentially pathogenic viruses. Because testing in humans would inevitably carry a substantial risk, we set out to test the genetic stability of multiply deleted HIV constructs in perpetuated tissue culture infections. The Δ3 candidate vaccine strain of HIV-1 contains deletions in the viral long terminal repeat (LTR) promoter and the vpr and nef genes. This virus replicates with delayed kinetics, but a profound enhancement of virus replication was observed after approximately 2 months of culturing. Analysis of the revertant viral genome indicated that the three introduced deletions were maintained but a 39-nucleotide sequence was inserted in the LTR promoter region. This insert was formed by duplication of the region encoding three binding sites for the Sp1 transcription factor. The duplicated Sp1 region was demonstrated to increase the LTR promoter activity, and a concomitant increase in the virus replication rate was measured. In fact, duplication of the Sp1 sites increased the fitness of the Δ3 virus (Vpr/Nef/U3) to levels higher than that of the singly deleted ΔVpr virus. These results indicate that deleted HIV-1 vaccine strains can evolve into fast-replicating variants by multiplication of remaining sequence motifs, and their safety is therefore not guaranteed. This insight may guide future efforts to develop more stable anti-HIV vaccines.
Relatively disappointing outcomes have been obtained thus far with a variety of anti-human immunodeficiency virus (HIV) vaccine candidates (41). However, the results of studies with live attenuated simian immunodeficiency viruses (SIVs) in monkey models are promising models for the possible use of live attenuated HIV as a protective vaccine. It has been repeatedly demonstrated that macaques or chimpanzees persistently infected with genetically attenuated, nonpathogenic isolates of SIV or HIV-1, respectively, strongly resist a subsequent challenge with pathogenic virus (2, 12, 36, 45, 47, 53, 57). In addition, there is some evidence that attenuated HIV-1 variants lacking the nef gene result in a benign course of infection in humans (16). These results warrant further investigation of this class of anti-HIV vaccines. However, the development of a live attenuated HIV-1 vaccine will face major safety issues, and the question has been raised of how much animal work remains to be done before human vaccine trials can proceed (7, 10, 17, 38). For instance, recent evidence suggests that SIV constructs with multiple gene deletions can be pathogenic in newborn monkeys (3, 58). Another major safety concerns is the fear that the vaccine strain can evolve from an attenuated form to a more virulent, pathogenic form. The latter process of virus evolution can occur either by spontaneous mutation of one of the remaining viral functions or by recombination-mediated repair with parts of the host cell genome, e.g., endogenous retroviral sequences.
In this study, we addressed the genetic stability of multiply deleted HIV-1 variants. To do so, we performed long-term tissue culture infections with the HIV-1 candidate vaccine strains to allow virus evolution to be studied on a laboratory timescale. Evolution of HIV-1 in a tissue culture setting has been reported repeatedly. A well-known example of tissue culture evolution occurs during culturing of primary isolates on T-cell lines, which results in the selection of “laboratory-adapted” HIV-1 variants with amino acid changes within the envelope glycoprotein that cause a shift in the host cell range and coreceptor use. Furthermore, many studies with replication-impaired HIV-1 mutants yielded revertant viruses after prolonged in vitro culturing. The analysis of revertant viruses has been used to study interactions within HIV-1 proteins, e.g., the Env glycoprotein (56) and the integrase enzyme (49). This genetic approach has been particularly useful in the dissection of complex RNA motifs, including the TAR hairpin (25, 32), the poly(A) hairpin (5, 14), and the dimerization initiation site DIS (6). Furthermore, mutations in the DIS RNA motif can be overcome by compensatory mutations within the viral Gag protein, which is most probably due to a direct interaction between the DIS RNA and the Gag protein (35). These combined results demonstrate the enormous genetic flexibility and repair capacity of the HIV-1 retroviral genome.
This genetic flexibility of HIV-1 is likely to be greater in in vivo infections, where a larger number of virion particles are produced. A prominent example is the appearance of drug-resistant HIV-1 variants in patients treated with potent antiviral drugs. For resistance to drugs against the protease enzyme, there is accumulating evidence that the primary resistance mutations within the protease protein cause a partial enzyme defect, which is subsequently restored by secondary mutations in protease and/or compensatory changes within the gag-encoded substrate sequences (11, 59). The in vivo capacity for repair of small attenuating gene deletions was also demonstrated by the evolution of an SIV variant with a 12-nucleotide (nt) deletion in the nef gene. A sequence duplication event was observed first, and multiple mutations were subsequently selected in the “insert” to create an amino acid sequence that is virtually indistinguishable from that of the wild-type virus (55). Despite these safety concerns, it seems unlikely that HIV-1 can repair large gene deletions. Therefore, whole genes have been deleted in the current live, attenuated vaccine candidates. Nevertheless, it has been reported recently that certain HIV-1 mutants can restore their fitness by second-site changes in unrelated functions encoded by the remaining sequences in the viral genome (15). We therefore tested the genetic stability of the current generation of HIV-1 vaccine candidates in long-term tissue culture infections.
MATERIALS AND METHODS
Plasmid constructs and PCR.
The HIV-1 molecular clone used in this study is the chimeric pNL4-3 plasmid (1), which contains the 5′ half of proviral NY5 and the 3′ half of proviral LAV sequences joined at a shared EcoRI site in the vpr gene. The different deletion mutants were described previously (21), and infectious virus was reconstructed from the 5′ and 3′ half genomes by EcoRI digestion and subsequent ligation.
Proviral DNA sequences were PCR amplified across the deletion points. To screen for the nef-U3 deletions, we used the primer pair NEF.SEQ and 3′TATA primer (positions 2990 to 3009 and 3756 to 3779 in plasmid p83-10 [21]). The vif-vpr region was analyzed by PCR with the primer pair Pol 5′FM and 6N (positions 4932 to 4964 in p83-2 and positions 213 to 237 in p83-10), and the vpu region was analyzed with the primer pair Tat-AUG and WS-3 (positions 88 to 107 and 801 to 820 in p83-10). The nef-U3 PCR fragment with an insert was introduced in a T/A cloning vector (Promega), and the sequence of multiple clones was analyzed by the Taq T7 Dyeprimer cycle-sequencing method (Applied Biosystems) on an Applied Biosystems 373 DNA sequencer.
Long terminal repeat (LTR)-chloramphenicol acetyltransferase (CAT) reporter constructs with either the wild-type, Δ3 mutant, or 6×Sp1 revertant sequences were constructed as follows. Viral DNA was PCR amplified with the upstream Xho-U3 primer (CCGCTCGAGTGGAAGGGCTAATTCACT), which places an XhoI restriction site (underlined) immediately upstream of the U3 region of the LTR, and the downstream U5 primer CN1. This DNA fragment was digested with XhoI and HindIII (position +77 in the R repeat region) and inserted into XhoI-HindIII-cleaved pBlue-3′LTR-CAT (33). The Tat expression vector pcDNA3-Tat and the CAT assay method were described previously (54).
The 6×Sp1 revertant sequence was introduced into the nef-U3 and Δ3 viruses as follows. First, the 6×Sp1 region was cloned into the 3′ LTR of the nef-U3-deleted 3′-half plasmid (p210-8 in reference 21) by replacement of a 216-bp 3×Sp1 fragment by a 255-bp 6×Sp1 fragment via EcoRV and BfrI restriction sites in the U3-R region. Infectious virus was obtained by ligation of this 3′-half plasmid (nef-U3-6×Sp1) with a 5′-half plasmid (either the wild type or the Vpr deletion variant) as described above. Virus stocks were produced in SupT1 cells.
Cells and viruses.
SupT1 cells were grown in RPMI medium containing 10% fetal calf serum and penicillin-streptomycin. Transfection of SupT1 cells by electroporation was carried out as described previously (13) with ligation mixtures that contain a total of 10 μg of DNA. Culture supernatants containing infectious virus were harvested at the peak of infection and stored in small aliquots at −70°C. These virus stocks were carefully quantitated by CA-p24 enzyme-linked immunosorbent assay (4) and used in infections of SupT1 cells. Infections were performed in 1.5 ml of RPMI medium containing 3 × 105 cells, and virus production was monitored by measuring CA-p24 antigen production. Donor peripheral blood mononuclear cells (PBMC) were prepared, cultured, and infected with HIV-1 as previously described (4).
Long-term culturing to select for revertant viruses was initiated by massive transfection of SupT1 cells (the ligation mixture containing 5 μg of both the 5′ and 3′ genome fragments was electroporated in 5 × 106 cells). In the first weeks, the cells were split when necessary. As soon as virus spread was apparent, as indicated by the presence of syncytia, the virus-containing culture supernatant was passaged onto uninfected SupT1 cells, initially with large samples of up to 1 ml and later with much smaller samples, as described previously (14).
Direct competition experiments with two virus variants were performed as described previously (34). We used equal amounts of the two viruses based on the CA-p24 measurements. The compositions of the 3×Sp1 and 6×Sp1 virus mixtures were analyzed by PCR amplification of the proviral genomes to monitor the LTR length polymorphism. These data were used to calculate the relative fitness as described previously (26). In brief, the relative fitness of the 6×Sp1 virus was approximated from the equation p/q = [p(0)/q(0)] × (fitness)T, where p is the proportion of 6×Sp1 virus, q is the proportion of 3×Sp1 virus, 0 indicates time zero, and T is the time in viral generations (2 days per generation).
RESULTS
We analyzed the in vitro replication capacity of a set of 20 HIV-1 variants with single or multiple deletions of accessory genes (vif, vpr, vpu, and nef) and of the upstream part of the U3 promoter region (21). Most variants with multiple deletions were severely replication impaired in primary human lymphocytes, e.g., vpr-nef-U3 (Δ3), vpr-vpu-nef, and all viruses with the vif deletion (data not shown; see also reference 21). The evolutionary capacity of such crippled viruses will be severely limited because the generation of new variants depends on random mutations introduced during viral replication by the error-prone reverse transcriptase. Therefore, we tested the deleted HIV-1 strains in a human T-cell line in which the defects caused by inactivation of some of the HIV-1 accessory genes are less pronounced (51). For instance, the Δ3 vaccine strain replicated with delayed kinetics in the SupT1 T-cell line but was eventually able to produce massively infected cultures as measured by the CA-p24 levels in the culture supernatant and the appearance of syncytia (data not shown). This optimized culture system was used for the long-term evolution studies of several HIV-1 constructs with multiple deletions. Virus was passaged onto uninfected cells for 4 months to select for faster-replicating revertants, and increased virus replication was noticed in several cultures. To accurately compare the replication kinetics of viruses sampled over time, we used identical amounts of these virus samples to infect SupT1 cells. The results obtained with the samples of the Δ3 virus are shown in Fig. 1 and indicate that replication improved dramatically after approximately 2 months of culturing. A similar gain of replication capacity was observed with other HIV-1 deletion constructs (e.g., vif-vpu and vif-nef-U3) but not with all variants.
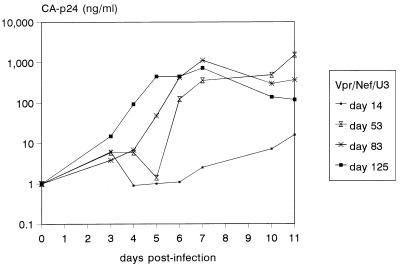
FIG. 1.
Improved growth kinetics of Δ3 revertant viruses. The HIV-1 samples represent the Δ3 virus after culturing on SupT1 cells for increasing periods. The perpetuated SupT1 infection was started by electroporation of 10 μg of Δ3 construct, and at the peak of infection, virus was passaged onto fresh, uninfected SupT1 cells. Supernatant samples were taken at several days posttransfection, frozen, and subsequently used to infect SupT1 cells with the same amount of input virus (1 ng CA-p24). Cell samples were also stored for proviral DNA analysis (see Fig. 2).
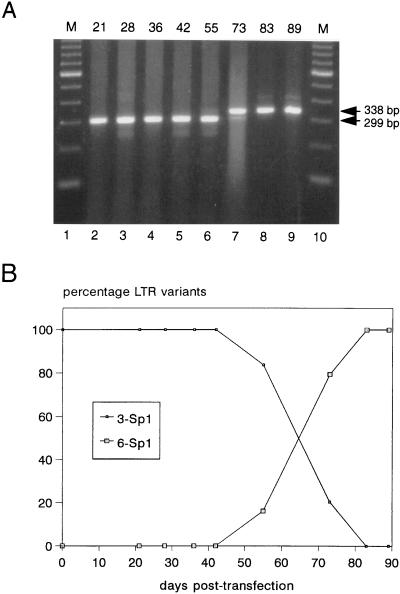
One possibility for repair of deleted gene functions is the insertion of a cellular gene with a similar function through recombination. To check whether the deletions in the HIV-1 genome were maintained, we performed PCR analyses across the deletion sites for all viruses that were cultured for 4 months. No insertions were observed for any of the virus samples, except for the Δ3 virus, in which a large insert appeared over time in the nef-LTR region. To analyze this process in more detail, SupT1 cell samples taken at different times were used to extract genomic DNA and to amplify the nef-LTR region of integrated proviruses (Fig. 2A). The 299-bp fragment predicted for the Δ3 virus was observed in the first 2 months of culturing, but a larger fragment appeared around day 55. This new fragment became the most prominent band at day 73 and completely replaced the original fragment at later times. Quantitation of the data indicated that the size variant was able to efficiently outgrow the Δ3 virus, with an increase in relative concentration from 16 to 80% in only 18 days (Fig. 2B).
FIG. 2.
The Δ3 mutant creates an LTR promoter with six Sp1 sites. (A) Cell samples taken on days 21, 28, 36, 42, 55, 73, 83, and 89 of the perpetuated SupT1 infection were used to extract cellular DNA as described previously (14), and the nef LTR region of the HIV-1 genome was PCR amplified. A 299-bp fragment is produced with the Δ3 mutant template (three Sp1 sites), and a revertant fragment of 338 bp is observed (six Sp1 sites). The day of cell harvest is indicated at the top of the gel. A 100-bp DNA ladder is provided in lanes 1 and 10 (lanes labeled M). (B) Quantitation of the ethidium bromide-stained gel was performed with the Kodak digital science 1D system and used to calculate the fractions of Δ3 mutant (three Sp1 sites) and Δ3 revertant (six Sp1 sites).
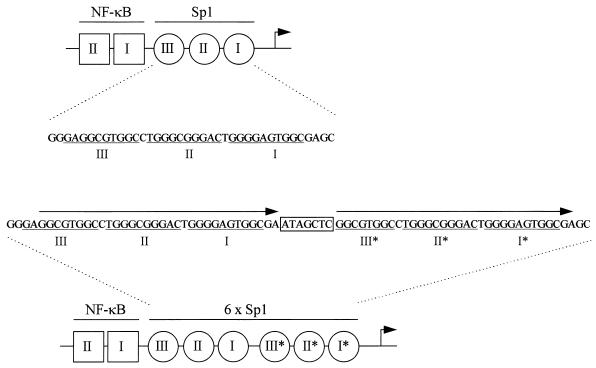
The complete Nef-LTR region of the Δ3 revertant was sequenced (Fig. 3). Interestingly, both deletions in the Nef and U3 region were still present, but a 39-nt fragment was inserted in the promoter region containing the Sp1 sites that bind the constitutively expressed Sp1 transcription factor (31). The insert consists of a 32-nt duplication and a 7-nt sequence of unknown identity. It seems likely that the Sp1 region was duplicated during reverse transcription of the viral genome. Tandem repeat sequences such as the Sp1 sites are known to be subject to deletion or duplication by a slippage-realignment mechanism (42). Inspection of the nucleotide sequence of the insert clearly indicates that the whole Sp1 region was copied in a single step. The alternative, i.e., multiple rounds of duplication of a single Sp1 site, can be excluded also because no PCR products of intermediate length were observed in the evolution experiment (Fig. 2A). Thus, a novel LTR promoter configuration consisting of two NF-κB and six Sp1 binding sites was created during replication of the Δ3 virus.
FIG. 3.
Duplication of the three Sp1 sites in the LTR promoter. The wild-type LTR promoter contains two NF-κB sites (squares) at positions −105 to −96 and −91 to −82 relative to the RNA start site at +1 (arrow) and three Sp1 sites (circles) at positions −78 to −69, −67 to −58, and −56 to −47. The Δ3 LTR carries a deletion of the upstream part of the U3 promoter region (starting at position −150). The nucleotide sequence of the three Sp1 sites is shown, with the 10-mer binding sites underlined. The lower panel shows the Δ3 revertant with the 39-nt insert. The insert consists of a 32-nt duplication (arrows) and a 7-nt sequence of unknown origin (boxed). Of the three new Sp1 motifs, the upstream site III* is a partial copy of site III, and it is therefore unknown whether site III* can bind the Sp1 factor.
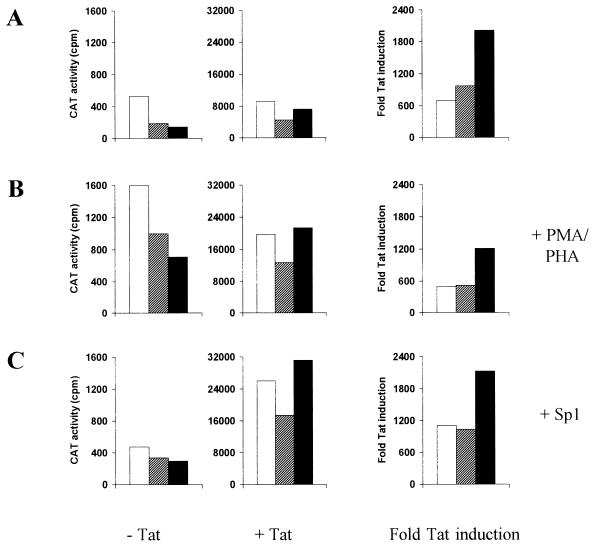
To test whether duplication of the three Sp1 motifs improved the transcriptional activity of the Δ3 promoter, we constructed a set of LTR-CAT reporter plasmids, including the wild-type LTR promoter, the Δ3 mutant lacking the upstream part of the U3 region, and the Δ3 revertant with six instead of three Sp1 sites. These plasmids were transfected into SupT1 cells in the presence or absence of a second plasmid encoding the viral Tat trans-activator protein. The results of a representative experiment are shown in Fig. 4A (left and middle), and the fold transcriptional activation was calculated (right). To boost the low level of basal LTR transcription, similar transfections were performed in cell cultures that were activated with phorbol myristate acetate and phyrohemagglutinin (PMA-PHA) on day 1 posttransfection (Fig. 4B) and in the presence of additional Sp1 encoded by an expression plasmid (Fig. 4C). Comparison of the wild-type and Δ3 promoters indicated an approximately twofold reduction of transcriptional activity upon deletion of the upstream U3 sequences, in both the absence and presence of Tat. A further reduction of the LTR activity in the absence of Tat was measured for the Δ3 revertant with six Sp1 sites. However, Tat-activated transcription of the Δ3 revertant was improved relative to that of the Δ3 mutant, and an expression level comparable to that of the wild-type LTR was reached. Similar results were obtained in cells activated with PMA-PHA and upon overexpression of Sp1. Our finding that the six Sp1 sites are beneficial only in the presence of Tat is consistent with the proposed functional interaction between the Tat and Sp1 proteins during LTR-mediated transcription (30, 39, 48).
FIG. 4.
The Δ3 LTR promoter gains activity by duplication of the Sp1 sites. (A) SupT1 T cells (5 × 106) were electroporated with 40 μg of LTR-CAT reporter construct (wild type [open bars], Δ3 mutant [hatched bars], and Δ3 revertant [solid bars]) in the absence of Tat (left) or with 1 μg of LTR-CAT plasmid in combination with 2.5 μg of pcDNA3-Tat (middle). The cultures were harvested on day 3 for CAT assays. The fold Tat-mediated activation of LTR-transcription was calculated and is plotted (right). (B and C) Parallel transfections were performed on cells that were treated with PMA-PHA (final concentrations 25 ng/ml and 1 μg/ml, respectively) on day 1 posttransfection (B), and transfections were repeated in the presence of Sp1 expression plasmid pSVSp-1 (44a) (C). A representative experiment is shown, and similar results were obtained in four independent transfections. Furthermore, similar results were obtained in transfections with other cell types, including non-T cells. The basal and Tat-induced promoter activities cannot be compared directly because different amounts of LTR-CAT plasmid were used. When the results were corrected for this difference, an approximately 200-fold induction of LTR activity was measured.
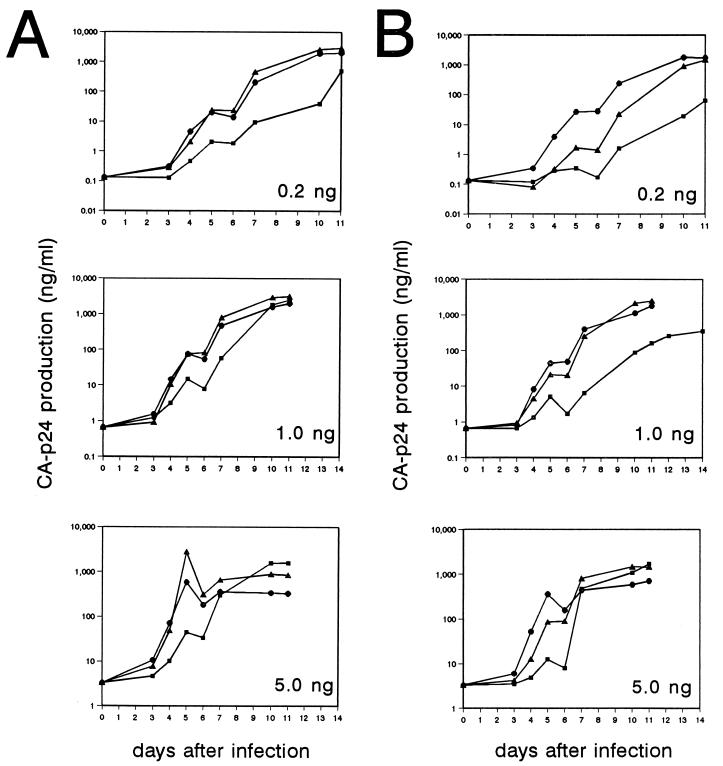
Enhanced LTR promoter activity of the 6×Sp1 variant is consistent with the improved replication of the Δ3 revertant virus, but it cannot be excluded that other genomic changes contribute to the reversion phenotype. To unequivocally prove that the duplication of the Sp1 region is responsible for the observed fitness gain, we introduced the 6×Sp1 sites in the 3′ LTR region of two molecular clones, the nef-U3 plasmid and the vpr-nef-U3 (Δ3) variant. Virus stocks were produced and used to infect SupT1 cells. We used equal amounts (based on CA-p24) of the 6×Sp1 viruses and the appropriate control viruses (Fig. 5). The contribution of the six Sp1 sites in the nef-U3 background is shown in Fig. 5A for three infections with a variable amount of input virus (top, 0.2 ng; middle, 1.0 ng; bottom, 5.0 ng). Introduction of the six Sp1 sites improved the replication of the nef-U3 virus to a level very similar to that of the wild-type control. The increase in virus replication capacity is even more prominent in the Δ3 background (Fig. 5B). In fact, the Δ3 virus with six Sp1 sites replicated faster than did the singly deleted vpr virus, which was included as a control. We also performed mixed-infection experiments with pairs of viruses to demonstrate the gain of fitness by duplication of the Sp1 region. Infections were initiated with equal amounts of the 6×Sp1 virus and the 3×Sp1 control, proviral samples were analyzed over time by LTR PCR amplification, and the composition of the viral mixture was determined by size separation of the LTR fragments on a gel as in Fig. 2 (data not shown). Rapid outgrowth of the 6×Sp1 variant was observed in both the nef-U3 and vpr-nef-U3 (Δ3) contexts. Based on the results of this internally controlled competition experiment, we calculated a relative gain of virus fitness of 30 and 60%, respectively.
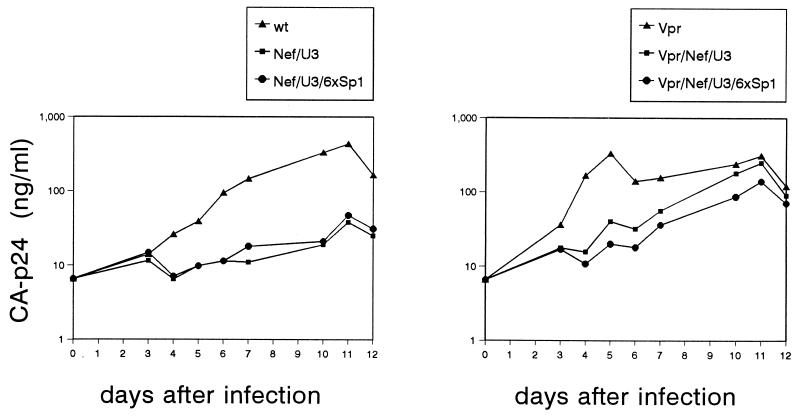
FIG. 5.
The reconstituted 6×Sp1 virus replicates with wild-type kinetics. Virus production in SupT1 cultures after infection with wild-type virus (▴) and the nef-U3 (■) and nef-U3-6×Sp1 (•) variants (A) and the vpr single-deletion mutant (▴), the vpr-nef-U3 (Δ3) mutant (■), and the vpr-nef-U3-6×Sp1 revertant (•) (B). The infections were performed in triplicate with different amounts of input virus: 0.2 ng of CA-p24 (top), 1.0 ng of CA-p24 (middle), and 5.0 ng of CA-p24 (bottom). Virus replication was monitored by measuring CA-p24 production in the culture supernatant. The cultures were split 1:5 at several times postinfection to sustain cell growth and virus replication; this resulted in small decreases in CA-p24 values.
There is abundant evidence that certain LTR promoter-enhancer motifs can affect virus replication in a cell-type-specific manner (8, 9, 29, 40, 43). Although we showed a gain of fitness of the 6×Sp1 variant virus in the SupT1 cell line that was used for the evolution experiment, we also wanted to know whether this promoter adaptation is beneficial in primary cells. PBMC were infected with equal amounts (10 ng of CA-p24) of the 6×Sp1 viruses (in both the nef-U3 and Δ3 backgrounds) and the appropriate control viruses (Fig. 6). Interestingly, the 6×Sp1 sites did not significantly improve replication in the nef-U3 background (Fig. 6, left), and a small negative effect was measured in the Δ3 context. This effect was verified in more sensitive competition experiments (data not shown).
FIG. 6.
The 6×Sp1 variant does not improve replication in primary cells. PBMC cultures were infected with the wild-type virus and the nef-U3 and nef-U3-6×Sp1 variants (left) and the vpr single deletion mutant, the vpr-nef-U3 (Δ3) mutant, and the vpr-nef-U3-6×Sp1 revertant (right). Equal amounts of input virus was used (10 ng of CA-p24). Virus replication was monitored by measuring CA-p24 production in the culture supernatant.
DISCUSSION
We described a dramatic gain of fitness by the Δ3 candidate vaccine strain (vpr-nef-U3) in prolonged tissue culture infections. In particular, this Δ3 virus restored LTR-mediated transcription and virus replication by multiplication of the Sp1 binding sites in the core promoter. Similar replication gains were observed for other multiply deleted HIV-1 variants (e.g., vif-vpu and vif-nef-U3 [data not shown]). Although we did not analyze the latter revertant viruses in detail, they do not have the characteristic duplication of Sp1 sites that we observed for the Δ3 revertant. Thus, HIV-1 exhibits an enormous evolutionary potential to restore replication. This may not come as a surprise, because there is ample evidence that HIV-1, which replicates as a quasispecies, is capable of overcoming a variety of selective pressures that are intended to limit its replication, including potent antiviral drugs (18). We demonstrate that replication-impaired HIV-1 variants with multiple gene deletions can improve their fitness within a relatively short culture period in an optimized in vitro system. We are obviously unable to directly translate the evolutionary potential of the Δ3 virus as measured in tissue culture to HIV-1 infections in humans. In fact, we found that this particular LTR modification does not improve virus replication in primary cells, suggesting that we may have selected for a SupT1-specific promoter change. Other LTR promoter motifs have also been demonstrated to function in a cell-type-specific manner (9, 40, 43). Nevertheless, because many more viruses are usually replicating in the in vivo situation, it seems unavoidable that other escape routes will be found in vivo.
Although the precise correlation between replication and pathogenicity is unknown, it is likely that a virus revertant with improved fitness will also regain pathogenic potential. For instance, the vpr-nef-U3-6×Sp1 revertant replicates more efficiently than does the singly deleted vpr virus in SupT1 cells, and an SIV variant with a single Vpr gene deletion induces AIDS in rhesus monkeys (20, 28). These combined results cast serious doubts on the safety of the current generation of multiply deleted HIV-1 vaccine strains. Most importantly, our results indicate that virus strains with multiple gene deletions can apparently restore their replication capacity without repairing the deleted gene functions. Can we mechanistically explain the reversion of a virus with deletions of the vpr-nef-U3 functions by acquisition of additional Sp1 sites in the core LTR promoter? First, the LTR-CAT transcription assays (Fig. 4) indicate that the twofold inhibition of LTR activity caused by the deletion of the upstream U3 region is compensated for by the six Sp1 sites. Multiplication of the Sp1 motifs may be particularly beneficial in SupT1 cells because these cells contain extremely low levels of the NF-κB transcription factor (9). Second, because one role of vpr is to maintain the host cell in a stage of the cell cycle where viral gene expression is optimal (22), changes in the LTR motifs may also indirectly compensate for a vpr defect. Consistent with this idea, expansion of the Sp1 region caused a more dramatic gain of fitness in the vpr-nef-U3 mutant than in the nef-U3 mutant in SupT1 cells (Fig. 5). Thus, part of the reversion is likely to occur at the level of viral gene expression. The Δ3 revertant with six Sp1 sites does not fully regain the wild-type fitness, which may be due in part to the inability to rescue the deleted Nef function.
There is some precedent for variation in the number of Sp1 binding sites in the LTR promoter of HIV-1. Several HIV-infected individuals were found to contain isolates with four Sp1 sites (34), and one natural isolate with five Sp1 sites was recently identified (44). It was shown that introduction of a fourth Sp1 site has a small but significant effect on LTR transcription and virus replication in SupT1 cells (34). Multiple Sp1 binding sites are found in various viral and cellular promoters, but the number of motifs varies widely, with up to eight sites in the cardiac α-actin gene (24). There is ample evidence for changes in host cell tropism or modulation of the viral oncogenic or pathogenic properties by variation in LTR promoter motifs in animal retroviruses (reviewed in reference 52). For instance, a point mutation in the Moloney murine leukemia virus LTR was shown to increase transcription and enable replication in embryonal cells because of the generation of an Sp1 binding site (23). There are also numerous examples of more blatant LTR rearrangements, including deletion and duplication of motifs in different clades of HIV-1 (37). Finally, we should also mention the reversion analysis performed with enhancer mutants of the DNA virus simian virus 40. Similar to our results, duplication of existing elements was the predominant mechanism for regaining promoter function (27). This apparent evolutionary flexibility of eukaryotic promoters is largely due to the modular architecture of these elements (19).
These in vitro studies demonstrate that multiply deleted HIV-1 strains are genetically unstable and therefore potentially unsafe. At the same time, these in vitro observations may also guide us toward the construction of improved HIV-1 vaccine candidates. Removal of all accessory genes from HIV-1 creates a replication-incompetent virus that will be useless as a vaccine. However, the Δ3 revertant virus described in this study may allow the removal of two additional genes (vpu and vif) without leading to complete loss of replication capacity. Subsequently, another round of in vitro evolution can be used to optimize the replication capacity of this Δ5 virus. Thus, repeated cycles of gene deletion and optimization of replication by means of forced evolution in tissue culture are proposed to generate a replication-competent version of HIV-1 that encodes only the basic set of retroviral proteins (Gag, Pol, Env, and perhaps Tat and Rev). This strategy may allow one to convert the complex HIV-1 genome into the form of a simple retrovirus, a vaccine approach that was originally proposed by Temin (50). This study indicates that such an evolutionary strategy should ideally be performed with primary cells, otherwise the selected variants may have an unpredictable in vivo replication phenotype. Other safety features can be added to such a mini-HIV backbone. For instance, insertion of the herpes simplex virus thymidine kinase gene will allow the elimination of cells carrying proviruses by treatment with ganciclovir (46). It remains to be tested whether such HIV-1 variants have lost their pathogenicity and whether humans can mount a response that protects against wild-type HIV-1 infection. Besides its use as a live, attenuated virus vaccine, the mini-HIV construct could be used as inactivated virus vaccine.
ACKNOWLEDGMENTS
We thank R. Desrosiers for providing the set of HIV-1 deletion mutants that was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The Sp1 plasmid was kindly donated by Jeff Saffer. We thank Rogier Sanders for technical assistance with LTR-CAT transfections and Wim van Est for photography and artwork.
This research was supported in part by the European Community and the Dutch AIDS Fund.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 3.Baba T W, Jeong Y S, Penninck D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 4.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, Van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout B, Klaver B, Das A T. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–947. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birmigham K. AIDS vaccine trial:publicity stunt or genuine attempt at progress? Nat Med. 1997;3:1055–1056. doi: 10.1038/nm1097-1055. [DOI] [PubMed] [Google Scholar]
- 8.Chang L J, McNulty E, Martin M. Human immunodeficiency viruses containing heterologous enhancer/promoters are replication competent and exhibit different lymphocyte tropisms. J Virol. 1993;67:743–752. doi: 10.1128/jvi.67.2.743-752.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B K, Feinberg M B, Baltimore D. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J Virol. 1997;71:5495–5504. doi: 10.1128/jvi.71.7.5495-5504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J. Novel campaign to test live HIV vaccine. Science. 1997;277:1035. doi: 10.1126/science.277.5329.1035. [DOI] [PubMed] [Google Scholar]
- 11.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 13.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys) J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das A T, Klaver B, Klasens B I F, van Wamel J L B, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A T, van Dam A P, Klaver B, Berkhout B. Improved envelope function selected by long-term cultivation of a translation-impaired HIV-1 mutant. Virology. 1998;244:552–562. doi: 10.1006/viro.1998.9124. [DOI] [PubMed] [Google Scholar]
- 16.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasispecies of HIV-1 from blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 17.Desrosiers R C. HIV with multiple gene deletions as a live attenuated vaccine for AIDS. AIDS Res Hum Retroviruses. 1992;8:411–421. doi: 10.1089/aid.1992.8.411. [DOI] [PubMed] [Google Scholar]
- 18.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 19.Dynan W S. Modularity in promoters and enhancers. Cell. 1989;58:1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of a gene for Vpr or Vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 22.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 23.Grez M, Zörnig M, Nowock J, Ziegler M. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J Virol. 1991;65:4691–4698. doi: 10.1128/jvi.65.9.4691-4698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafson T A, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac α-actin promoter. Mol Cell Biol. 1989;9:3269–3283. doi: 10.1128/mcb.9.8.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrich D, Mavankal G, Mette-Snider A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrigan P R, Bloor S, Larder B A. Relative replication fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herr W, Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986;45:461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- 28.Hoch J, Lang S M, Weeger M, Stahl-Hennig C, Coulibaly C, Dittmer U, Hunsmann G, Fuchs D, Muller J, Sopper S, Fleckenstein B, Uberla K T. Vpr deletion mutant of simian immunodeficiency virus induces AIDS in rhesus monkeys. J Virol. 1995;69:4807–4813. doi: 10.1128/jvi.69.8.4807-4813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilyinskii P O, Desrosiers R C. Efficient transcription and replication of simian immunodeficiency virus in the absence of NF-κB and Sp1 binding elements. J Virol. 1996;70:3118–3126. doi: 10.1128/jvi.70.5.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Activation of the AIDS retrovirus by the cellular transcription factor Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 32.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koken S E C, van Wamel J L, Goudsmit J, Berkhout B, Geelen J L. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology. 1992;191:968–972. doi: 10.1016/0042-6822(92)90274-s. [DOI] [PubMed] [Google Scholar]
- 35.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, van Rompay K K, Reay E, Antipa L, Pedersen N C, Marthas M L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montano M A, Novitsky V A, Blackard J T, Cho N L, Katzenstein D A, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphey-Corb M. Live-attenuated HIV vaccines: how safe is safe enough. Nat Med. 1997;3:17–18. doi: 10.1038/nm0197-17. [DOI] [PubMed] [Google Scholar]
- 39.Pagtakhan A S, Tong-Starksen S E. Interactions between Tat of HIV-2 and transcription factor Sp1. Virology. 1997;238:221–230. doi: 10.1006/viro.1997.8847. [DOI] [PubMed] [Google Scholar]
- 40.Parrott C, Seidner T, Duh E, Leonard J, Theodore T S, Buckler-White A J, Martin M A, Rabson A B. Variable role of the long terminal repeat Sp1 binding sites in human immunodeficiency virus replication in T lymphocytes. J Virol. 1991;65:1414–1419. doi: 10.1128/jvi.65.3.1414-1419.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul W E. Can the immune response control HIV infection? Cell. 1995;82:177–182. doi: 10.1016/0092-8674(95)90304-6. [DOI] [PubMed] [Google Scholar]
- 42.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross E K, Buckler White A J, Rabson A B, Englund G, Martin M A. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol. 1991;65:4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rousseau C, Abrams E, Lee M, Urbano R, King M-C. Long terminal repeat and nef gene variants of human immunodeficiency virus type 1 in perinatally infected long-term survivors and rapid progressors. AIDS Res Hum Retroviruses. 1997;13:1611–1623. doi: 10.1089/aid.1997.13.1611. [DOI] [PubMed] [Google Scholar]
- 44a.Saffer J D, Jackson S P, Thurston S J. SV40 stimulates expression of the trans-acting factor Sp1 at the mRNA level. Genes Dev. 1990;4:659–666. doi: 10.1101/gad.4.4.659. [DOI] [PubMed] [Google Scholar]
- 45.Shibata R, Siemon C, Czajak S C, Desrosiers R C, Martin M A. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith S M, Markham R B, Jeang K-T. Conditional reduction of human immunodeficiency virus type 1 replication by a gain-of-herpes simplex virus 1 thymidine kinase function. Proc Natl Acad Sci USA. 1996;93:7955–7960. doi: 10.1073/pnas.93.15.7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stahl-Hennig C, Dittmer U, Nisslein T, Petry H, Jurkiewicz, Fuchs D, Wachter H, Matz-Rensing K, Kuhn E M, Kaup F J, Rud E W, Hunsmann G. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J Gen Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 48.Sune C, Garcia-Blanco M A. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6576. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taddeo B, Carlini F, Verani P, Engelman A. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J Virol. 1996;70:8277–8284. doi: 10.1128/jvi.70.12.8277-8284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Temin H M. A proposal for a new approach to a preventive vaccine against human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:4419–4420. doi: 10.1073/pnas.90.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 52.Tsichlis P N, Lazo P A. Virus-host interactions and the pathogenesis of murine and human oncogenic retroviruses. In: Kung H J, Vogt P K, editors. Retroviral insertion and oncogene activation. Berlin, Germany: Springer-Verlag KG; 1991. pp. 95–171. [DOI] [PubMed] [Google Scholar]
- 53.van Rompay K K A, Spinner A, Otsyula M, McChesney M B, Marthas M L. Attenuated retrovirus vaccines and AIDS. Science. 1995;270:1218. [PubMed] [Google Scholar]
- 54.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 55.Whatmore A M, Cook N, Hall G A, Sharpe S, Rud E W, Cranage M P. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willey R L, Ross E K, Buckler-White A J, Theodore T S, Martin M A. Functional interaction of constant and variable domains of human immunodeficiency virus type gp120. J Virol. 1989;63:3595–3600. doi: 10.1128/jvi.63.9.3595-3600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 59.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]