Abstract
Psychiatric disorders and common epilepsies are heritable disorders with a high comorbidity and overlapping symptoms. However, the causative mechanisms underlying this relationship are poorly understood. Here we aimed to identify overlapping genetic loci between epilepsy and psychiatric disorders to gain a better understanding of their comorbidity and shared clinical features.
We analysed genome-wide association study data for all epilepsies (n = 44 889), genetic generalized epilepsy (n = 33 446), focal epilepsy (n = 39 348), schizophrenia (n = 77 096), bipolar disorder (n = 406 405), depression (n = 500 199), attention deficit hyperactivity disorder (n = 53 293) and autism spectrum disorder (n = 46 350). First, we applied the MiXeR tool to estimate the total number of causal variants influencing the disorders. Next, we used the conjunctional false discovery rate statistical framework to improve power to discover shared genomic loci. Additionally, we assessed the validity of the findings in independent cohorts, and functionally characterized the identified loci.
The epilepsy phenotypes were considerably less polygenic (1.0 K to 3.4 K causal variants) than the psychiatric disorders (5.6 K to 13.9 K causal variants), with focal epilepsy being the least polygenic (1.0 K variants), and depression having the highest polygenicity (13.9 K variants). We observed cross-trait genetic enrichment between genetic generalized epilepsy and all psychiatric disorders and between all epilepsies and schizophrenia and depression. Using conjunctional false discovery rate analysis, we identified 40 distinct loci jointly associated with epilepsies and psychiatric disorders at conjunctional false discovery rate <0.05, four of which were associated with all epilepsies and 39 with genetic generalized epilepsy. Most epilepsy risk loci were shared with schizophrenia (n = 31). Among the identified loci, 32 were novel for genetic generalized epilepsy, and two were novel for all epilepsies. There was a mixture of concordant and discordant allelic effects in the shared loci. The sign concordance of the identified variants was highly consistent between the discovery and independent datasets for all disorders, supporting the validity of the findings. Gene-set analysis for the shared loci between schizophrenia and genetic generalized epilepsy implicated biological processes related to cell cycle regulation, protein phosphatase activity, and membrane and vesicle function; the gene-set analyses for the other loci were underpowered.
The extensive genetic overlap with mixed effect directions between psychiatric disorders and common epilepsies demonstrates a complex genetic relationship between these disorders, in line with their bi-directional relationship, and indicates that overlapping genetic risk may contribute to shared pathophysiological and clinical features between epilepsy and psychiatric disorders.
Keywords: GWAS, conjunctional FDR, pleiotropy, MoBa, 23andMe, ILAE
Karadag et al. identify 40 distinct loci jointly associated with common epilepsies and psychiatric disorders in a genome-wide association study. The extensive genetic overlap with mixed effect directions points to a complex genetic relationship between these conditions.
Introduction
Brain disorders are major global causes of morbidity with high costs for society.1 Epilepsy is regarded as a heterogeneous neurological condition defined by recurrent seizures, affecting over 60 million people worldwide.1,2 Psychiatric comorbidity is frequent in people with epilepsy, including depression, anxiety, bipolar disorder, psychosis, attention deficit hyperactivity disorder (ADHD), and autism spectrum disorders (ASD).3,4 These comorbidities share some clinical features with epilepsy, may impede diagnostic accuracy and treatment approaches, and are associated with a lower quality of life for people with epilepsy.3,4 The relationship between epilepsy and psychiatric disorders seems to be bi-directional and the underlying aetiological mechanisms are poorly understood.3–5 Certain anti-seizure medications (valproate, lamotrigine and carbamazepine) are among the most effective drugs for treating bipolar disorder, suggesting shared biological mechanisms between epilepsy and bipolar disorder.6 Other anti-seizure medications are known to induce psychiatric adverse effects, while anti-psychotic drugs may alter seizure threshold.6,7 Potential shared neurobiological dysfunctions have been implicated across these disorders, such as perturbed calcium signalling, synaptic plasticity and neurotransmission.8–11 Uncovering shared genetic variants for epilepsy and psychiatric disorders may help identify people at risk and guide early treatment decisions.
Both psychiatric disorders and epilepsy are heritable.12–14 The heritability accounted for by common single nucleotide polymorphisms (SNPs) is estimated to range between 11% and 24% for depression, ADHD, ASD, bipolar disorder and schizophrenia.15–19 Epilepsy is broadly categorized under the two major subtypes, focal epilepsy and generalized epilepsy; the latter being primarily constituted by genetic generalized epilepsy (GGE). Focal epilepsy and GGE have substantial differences in their SNP heritability estimates, 9.2% and 32.1%, respectively, reflecting their different aetiologies.2 Recent large-scale genome-wide association studies (GWAS) have identified 287 risk loci for schizophrenia, 64 loci for bipolar disorder, 178 loci for depression, 27 loci for ADHD, five loci for ASD and 26 loci for common epilepsies.2,15–25 Further, two risk loci were recently identified as shared between ADHD and GGE.26 However, these loci only account for a small fraction of the SNP heritabilities. GWAS analyses have also indicated a high degree of shared genetic risk between psychiatric disorders, with substantial pairwise genetic correlations estimated between schizophrenia, bipolar disorder, depression, ADHD and ASD.27,28 A significant yet weak positive genome-wide correlation was recently reported between GGE and ADHD, indicating shared genetic risk, while weak negative genetic correlations between epilepsy phenotypes, schizophrenia and bipolar disorder did not survive correction for multiple comparisons.25,26 No significant genetic correlations were reported between epilepsies and depression or ASD. However, the estimates of genetic correlations do not provide a complete overview of genetic overlap between complex human phenotypes.29,30 First, genetic correlations are agnostic about the specific shared loci involved, and accumulating evidence has demonstrated substantial genetic overlap between complex human phenotypes despite weak or absent genetic correlations,29,31–33 including between psychiatric and neurological disorders.34–36
In the present study, we aimed to improve the understanding of the genetic relationship between common epilepsies and major psychiatric disorders using MiXeR,37 which quantifies the number of variants influencing a phenotype, and the conjunctional false discovery rate (conjFDR) approach, which boosts GWAS discovery by leveraging overlapping GWAS associations to identify shared genomic loci.30,38 This approach has improved discovery of shared genetic influences between several complex human phenotypes in recent years.30,32–36,39–42
Materials and methods
Sample description
GWAS data were obtained as summary statistics (P-values and effect sizes; Table 1). For each phenotype, available GWAS data with the largest sample size were chosen and overlapping samples were excluded, which might otherwise bias conjFDR results. In total, we analysed GWAS data on more than one million participants (258 230 cases and 773 053 controls).
Table 1.
Summary data from all GWAS used in the present study
| Phenotype | Sample size, n | Ancestry (n) | SNPs, n | Source |
|---|---|---|---|---|
| Discovery samples | ||||
| All epilepsy | 15 212 cases, 29 677 controls | 86% European (38 752), 8% Asian (3406), 6% African (2731) | 4 880 492 | ILAE2 |
| Focal epilepsy | 9671 cases, 29 677 controls | 84% European (33 313), 9% Asian (3365), 7% African (2670) | 4 862 782 | ILAE2 |
| GGE | 3769 cases, 29 677 controls | 83% European (27 926), 9% Asian (2875), 8% African (2645) | 4 867 068 | ILAE2 |
| SCZ | 45 313 cases, 67 472 controls | European | 7 634 648 | Trubetskoy et al.19 |
| BIP | 39 027 cases, 367 378 controls | European | 9 028 988 | Mullins et al.18 |
| DEP | 121 198 cases, 246 363 controls | European | 15 807 881 | Howard et al.17 |
| ADHD | 19 099 cases, 34 194 controls | European | 8 094 094 | Demontis et al.15 |
| ASD | 18 381 cases, 27 969 controls | European | 9 112 386 | Grove et al.16 |
| Independent samples | ||||
| All epilepsy | 2466 cases, 175 788 controls | European | 15 746 420 | https://r5.finngen.fi |
| SCZ | 22 778 cases, 35 362 controls | East Asian | 10 694 910 | Lam et al.22 |
| BIP | 4501 cases, 192 220 controls | European | 15 746 437 | https://r5.finngen.fi |
| DEP | 170 756 cases, 329 443 controls | European | 15 746 508 | https://r5.finngen.fi |
| ADHD | 4224 cases, 203 345 controls | European | 6 981 749 | MoBa; Magnus et al.43,44 |
| ASD | 925 cases, 206 644 controls | European | 6 981 749 | MoBa; Magnus et al.43,44 |
BIP = bipolar disorder; DEP = depression; SCZ = schizophrenia; ILAE = International League Against Epilepsy; MoBa = Norwegian Mother, Father and Child Cohort Study.
The GWAS data on all epilepsies combined, focal epilepsy and GGE were obtained from the International League Against Epilepsy (ILAE) Consortium.2 GWAS data for schizophrenia19 and bipolar disorder18 were obtained from the Psychiatric Genomics Consortium (PGC). Depression data were obtained from a meta-analysis17 of data from PGC and 23andMe, Inc. Data on both ADHD15 and ASD16 were acquired from PGC and the iPSYCH cohort. The GWAS participants were predominantly of European ancestry.
All GWAS investigated in the present study were approved by the relevant ethics committees, and informed consent was obtained from all participants. The Norwegian Institutional Review Board for the South-East Norway Region has evaluated the current protocol and found that no additional institutional review board approval was needed because no individual data were used. See Supplementary material for more details.
Data analysis
Univariate causal mixture model: MiXeR
We applied the statistical tool, MiXeR v1.3, to estimate the number of causal variants explaining 90% of the SNP heritability of each phenotype, i.e. the polygenicity, using GWAS summary statistics.37 A ‘causal’ variant is here defined as a variant with non-zero additive genetic effects on a phenotype.37 Akaike information criterion (AIC) was used to evaluate the model fit. More information on the MiXeR method can be found in the Supplementary material.
Conjunctional false discovery rate analysis
We applied the conjFDR method to increase discovery of genomic loci jointly associated with epilepsies and psychiatric disorders. The conjFDR approach is an extension of the conditional FDR (condFDR), which leverages cross-trait enrichment between two phenotypes to improve genetic discovery30,38 CondFDR readjusts the test statistics in a primary phenotype (e.g. GGE) by conditioning on SNP associations with a secondary phenotype (e.g. schizophrenia). The conjFDR method performs two condFDR analyses (conditioning the first phenotype on the second phenotype and vice versa) and defines the conjFDR value as the maximum of the two condFDR values. The conjFDR threshold 0.05 was used in line with previous literature.30,38 The cross-trait enrichment is visualized using conditional Q-Q plots, which show the distribution of P-values for a primary phenotype for all SNPs, and for SNP strata defined by their association with the secondary phenotype. We excluded SNPs around the extended major histocompatibility complex (MHC) region, chromosome 8p23.1 and MAPT region (genome build 19 locations chr6:25119106-33854733; chr8:7200000-12500000; chr17:40000000-47000000, respectively) before fitting the FDR model to avoid bias in our cond/conjFDR analyses due to their complex regional linkage disequilibrium (LD)45 (Supplementary material).
Functional analyses
Genomic loci definition
Independent genomic loci were defined in line with the FUMA46 protocol. Independent significant SNPs were identified as r2 < 0.60 and conjFDR < 0.05. Of those, SNPs with r2 < 0.1 were defined as in approximate linkage equilibrium and chosen as lead SNPs. Candidate SNPs were defined as SNPs with a conjFDR value of <0.10 and an LD r2-value of >0.60 with an independent significant SNP. All loci < 250 kb apart were merged and the SNP with the most significant conjFDR value was chosen as the lead SNP of the merged locus. The borders of the loci were defined by identifying all candidate SNPs in LD (r2 ≥ 0.6) with one of the independent significant SNPs in the locus. All LD r2-values were obtained from the 1000 Genomes Project European-ancestry haplotype reference panel.47
We evaluated the directional effects of the shared loci by comparing their z-scores and odds ratios. Novel loci were defined as novel if they were not within 500 kb of the reported loci from the original GWAS or were not reported in the GWAS Catalogue48 or other post-GWAS analyses on epilepsy or psychiatric disorders.
Functional annotation
SNPs were functionally annotated with combined annotation dependent depletion scores (CADD), regulomeDB scores and chromatin states. These scores predict deleterious SNP effect on a protein, likelihood of regulatory functionality and transcriptional effects due to chromatin states, respectively. The candidate SNPs were mapped to putative causal genes using positional mapping, expression quantitative trait locus (eQTL) mapping and chromatin interaction mapping.46 Gene expression and gene-set analysis of the identified genes were performed using FUMA and Genotype-Tissue Expression data (GTEx)46,49 (Supplementary material).
Validation tests in independent samples
To validate our findings, we conducted sign concordance tests50 to compare the overall pattern of consistency in allelic effect directions of the lead SNPs between discovery and independent datasets on schizophrenia22 from PGC, bipolar disorder,51 depression,51 epilepsy combined51 from FinnGen, and ADHD and ASD43,44 from the Norwegian Mother, Father and Child Cohort Study (MoBa) conducted by the Norwegian Institute of Public Health (Table 1). To secure sufficient number of variants for valid analysis, we evaluated loci identified at a more relaxed significance threshold (conjFDR <0.10). We determined the number of lead SNPs in the shared loci that had the same allelic effect direction in the independent datasets by comparing the point-estimate of the beta coefficients. Under the null hypothesis that there is no genetic association with the trait of interest, observing sign concordance by chance has a probability of 50%. Using the two-tailed exact binomial test, we then evaluated whether the observed sign concordance rates were significantly higher than expected by chance.
Data availability and computational tools
Statistical analyses for the relevant methods were performed in MATLAB and Python, using existing tools available on GitHub, including MiXeR v1.3 (https://github.com/precimed/mixer) and condFDR/conjFDR (https://github.com/precimed/pleiofdr).
Results
MiXeR results
Using MiXeR,37 we estimated the number of ‘causal’ variants for each epilepsy phenotype and found that ∼3.0 K variants [standard deviation (SD) = 0.8 K] influence all epilepsy, ∼3.4 K variants (SD = 0.3 K) influence GGE and ∼1.0 K variants (SD = 1.1 K) influence focal epilepsy; reflecting their different genetic architectures (Table 2). The polygenicity estimates for psychiatric disorders were 9.6 K variants for schizophrenia (SD = 0.2 K), 8.6 K variants for bipolar disorder (SD = 0.2 K), 13.9 K variants for depression (SD = 0.6 K), 5.6 K variants for ADHD (SD = 0.4 K) and 12.3 K variants for ASD (SD = 1.5 K) (Table 2), in line with previous reports.13–15 The large standard deviations for the polygenicity estimates for focal epilepsy and ASD indicate that these estimates should be interpreted with caution, likely reflecting a combination of low SNP-heritability and insufficient GWAS power for these disorders. Moreover, we estimated the discoverability of each disorder and found that depression (7.43 × 10−6, SD = 2.65 × 10−7) and ASD (2.45 × 10−5, SD = 2.98 × 10−6) were the least discoverable disorders, while focal epilepsy (1.11 × 10−4, SD = 3.35 × 10−5) and GGE (2.57 × 10−4, SD = 1.89 × 10−5) were the most discoverable traits (Table 2).
Table 2.
Univariate MiXeR estimates for the epilepsies and psychiatric disorders
| Phenotype | ADHD | ASD | BIP | DEP | SCZ | EP | FEP | GGE |
|---|---|---|---|---|---|---|---|---|
| pi (mean) | 1.76 × 10−3 | 3.88 × 10−3 | 2.71 × 10−3 | 4.38 × 10−3 | 3.00 × 10−3 | 9.48 × 10−4 | 3.04 × 10−4 | 1.07 × 10−3 |
| pi (SD) | 1.37 × 10−4 | 4.75 × 10−4 | 7.43 × 10−5 | 1.79 × 10−4 | 7.56 × 10−5 | 2.59 × 10−4 | 3.33 × 10−4 | 9.47 × 10−5 |
| sig2_beta (mean) | 6.31 × 10−5 | 2.45 × 10−5 | 3.37 × 10−5 | 7.43 × 10−6 | 6.13 × 10−5 | 6.15 × 10−5 | 1.11 × 10−4 | 2.57 × 10−4 |
| sig2_beta (SD) | 4.59 × 10−6 | 2.98 × 10−6 | 8.52 × 10−7 | 2.65 × 10−7 | 1.43 × 10−6 | 1.58 × 10−5 | 3.35 × 10−5 | 1.89 × 10−5 |
| sig2_zero (mean) | 1.09 × 100 | 1.02 × 100 | 1.12 × 100 | 1.05 × 100 | 1.22 × 100 | 1.19 × 100 | 1.17 × 100 | 1.13 × 100 |
| sig2_zero (SD) | 2.98 × 10−3 | 5.89 × 10−4 | 3.01 × 10−3 | 5.95 × 10−4 | 4.36 × 10−3 | 1.57 × 10−3 | 9.32 × 10−4 | 1.64 × 10−3 |
| h2 (mean) | 2.29 × 10−1 | 1.95 × 10−1 | 1.89 × 10−1 | 6.74 × 10−2 | 3.82 × 10−1 | 1.13 × 10−1 | 5.23 × 10−2 | 5.64 × 10−1 |
| h2 (SD) | 3.37 × 10−3 | 4.20 × 10−3 | 2.12 × 10−3 | 1.14 × 10−3 | 4.14 × 10−3 | 5.94 × 10−3 | 4.63 × 10−3 | 2.01 × 10−2 |
| nc@p9 (mean) | 5.60 × 103 | 1.24 × 104 | 8.60 × 103 | 1.40 × 104 | 9.60 × 103 | 3.02 × 103 | 9.69 × 102 | 3.40 × 103 |
| nc@p9 (SD) | 4.00 × 102 | 1.52 × 103 | 2.00 × 102 | 5.71 × 102 | 2.00 × 102 | 8.27 × 102 | 1.06 × 103 | 3.02 × 102 |
| AIC | 3.08 × 101 | 1.69 × 100 | 1.68 × 102 | 1.97 × 101 | 4.10 × 102 | 3.34 × 100 | 1.72 × 100 | 2.42 × 101 |
BIP = bipolar disorder; DEP = depression; EP = all epilepsy; FEP = focal epilepsy; h2 = heritability; nc@p9 = number of causal variants with strongest effects required to explain 90% variance at genome-wide significance; pi = polygenicity; SCZ = schizophrenia; sig2_beta = discoverability.
Cross-trait enrichment
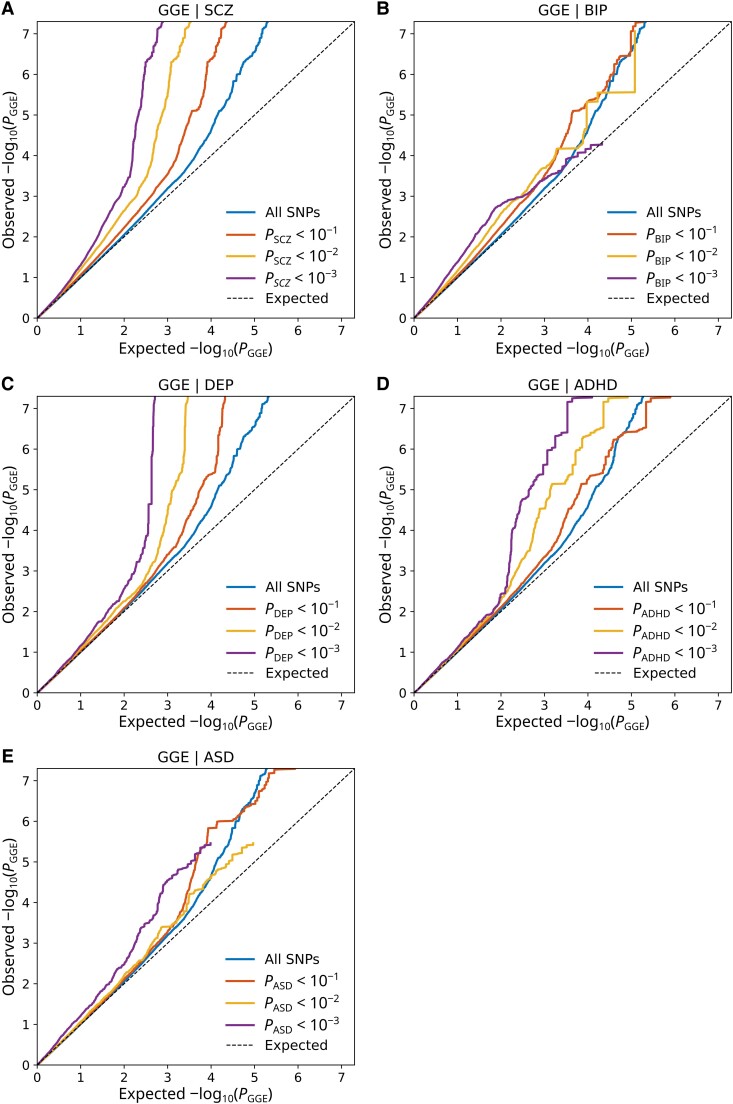
The conditional Q-Q plots demonstrated substantial enrichment of SNP associations with GGE as a function of increasing levels of SNP associations with all psychiatric disorders, indicating polygenic overlap (Fig. 1). This enrichment was also consistent in the reverse conditional Q-Q plots (Supplementary Fig. 1). We also observed bi-directional cross-trait enrichment between all epilepsies and schizophrenia and depression, but not with the other disorders (Supplementary Figs 2 and 3). We observed enrichment of SNP associations with bipolar disorder conditional on focal epilepsy, but not in the other direction (Supplementary Figs 4 and 5). There was no evident cross-trait enrichment between focal epilepsy and the other psychiatric disorders.
Figure 1.
Cross-trait enrichment between GGE and psychiatric disorders. Quantile-quantile (Q-Q) plots show SNP enrichment for GGE conditional on SNP associations with (A) schizophrenia (SCZ), (B) bipolar disorder (BIP), (C) depression (DEP), (D) ADHD and (E) ASD. Conditional Q-Q plots of nominal versus empirical −log10 P-values (corrected for inflation) in GGE below the standard GWAS threshold of P < 5 × 10−8 as a function of significance of association with the psychiatric disorders, at the level of P < 0.10, P < 0.01 and P < 0.001. The blue lines indicate all SNPs. The dashed lines indicate the null hypothesis.
Shared loci between common epilepsies and psychiatric disorders
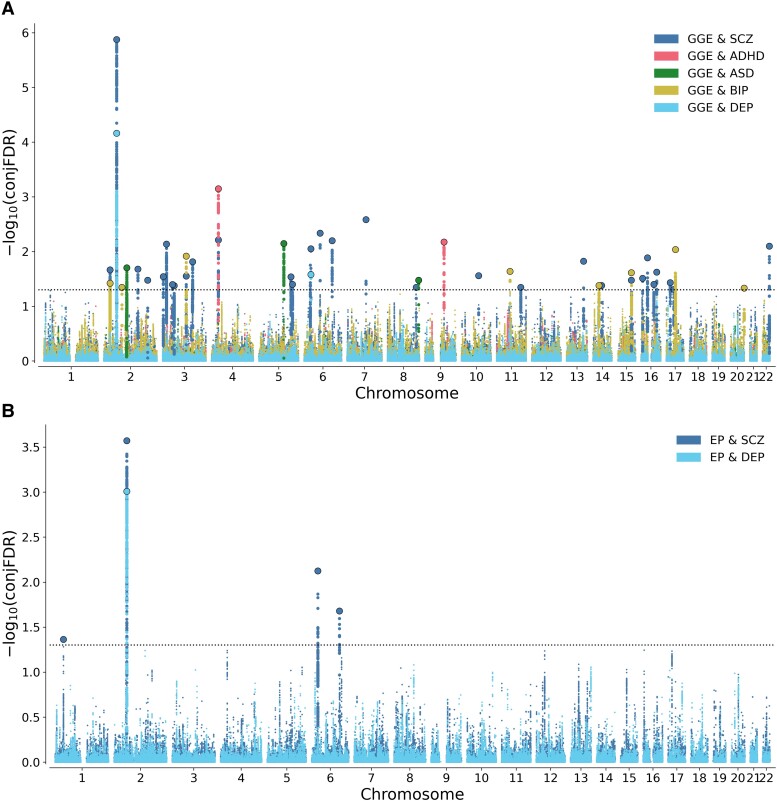
Next, we leveraged the bi-directional cross-trait enrichment to increase statistical power for discovery of shared loci using conjFDR analysis. At conjFDR <0.05, we identified 30 loci significantly associated with both GGE and schizophrenia, eight loci shared between GGE and bipolar disorder, two loci shared between GGE and depression, two loci shared between GGE and ADHD, and three loci shared between GGE and ASD (Fig. 2A and Supplementary Tables 1–6). Taken together, we identified a total of 39 distinct loci associated with GGE of which 32 are novel risk loci for GGE (Table 3 and Supplementary Table 6). Four of the loci were novel for psychiatric disorders; one locus on chromosome 11 at HNRNPA1P60 for schizophrenia, two loci at chromosomes 2 (at RN7SL201P) and 20 (at ZNF512B:LINC00176) for bipolar disorder and one locus at chromosome 2 (at AC018880.2) for ASD (Supplementary Tables 1–5). Further, we identified four loci jointly associated with all epilepsies and schizophrenia, and one locus shared between all epilepsies and depression, the same that was linked to GGE (Fig. 2B and Table 4). Among these loci, the risk locus at chromosome 1 at MACF1 and at chromosome 6 at ZSCAN23 are novel findings for epilepsy (Supplementary Tables 7 and 8).
Figure 2.
Shared loci between epilepsy and psychiatric disorders at conjFDR < 0.05. (A) Common genetic variants jointly associated with GGE and psychiatric disorders at conjFDR < 0.05. SCZ = schizophrenia; BIP = bipolar disorder; DEP = depression; Manhattan plots showing the −log10 transformed conjFDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal lines represent the threshold for significant shared associations [conjFDR <0.05, i.e. −log10(conjFDR) > 1.3]. Independent lead SNPs are circled in black. The significant shared signals in the MHC region are represented by one lead SNP only. For further information about the identified variants and loci, see Supplementary Tables 1–5. (B) Common genetic variants jointly associated with all epilepsy and schizophrenia and depression at conjFDR < 0.05. EP = all epilepsy. Manhattan plots showing the −log10 transformed conjFDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal lines represent the threshold for significant shared associations [conjFDR <0.05, i.e. −log10(conjFDR) > 1.3]. Independent lead SNPs are circled in black. The significant shared signals in the MHC region are represented by one lead SNP only. For further information about the identified variants and loci, see Supplementary Tables 7 and 8.
Table 3.
All distinct loci associated with GGE at conjFDR < 0.05
| CHR | LEAD_SNP | Nearest gene | A1/A2 | P-value | Novel in epilepsy | Psychiatric disorder shared with | Concord effect |
|---|---|---|---|---|---|---|---|
| 2 | rs6708889 | MRPL33:RBKS | T/C | 6.22 × 10−5 | Novel | BIP, SCZ | Yes |
| 2 | rs1040225 | VRK2 | G/A | 1.53 × 10−10 | ILAE 2014 | SCZ, DEP | No |
| 2 | rs6715448 | RN7SL201P | C/T | 8.24 × 10−5 | Novel | BIP | No |
| 2 | rs1673468 | AC018880.2 | T/C | 4.30 × 10−5 | ILAE25 | ASD | No |
| 2 | rs249697 | AC009227.3 | A/G | 7.90 × 10−5 | Novel | SCZ | No |
| 2 | rs6714133 | SATB2:SATB2-AS1 | G/T | 1.60 × 10−4 | Novel | SCZ | Yes |
| 3 | rs17194427 | CNTN4 | C/A | 1.25 × 10−4 | Novel | SCZ | Yes |
| 3 | rs10428260 | TBC1D5 | A/G | 1.85 × 10−5 | Novel | SCZ | Yes |
| 3 | rs75298156 | RP11-944L7.4:ZNF197 | A/G | 2.14 × 10−4 | Novel | SCZ | Yes |
| 3 | rs62256903 | SMIM4 | G/A | 2.30 × 10−4 | Novel | SCZ | No |
| 3 | rs3804640 | CD47 | A/G | 1.20 × 10−5 | Novel | BIP, SCZ | Yes |
| 3 | rs4678442 | RP11-731C17.1 | A/G | 5.06 × 10−5 | Novel | SCZ | No |
| 4 | rs6448744 | PCDH7 | T/G | 4.20 × 10−8 | ILAE 2014 | ADHD, SCZ | No |
| 5 | rs434517 | RP11-492A10.1 | C/A | 7.25 × 10−7 | ILAE2 | ASD | No |
| 5 | rs4515335 | PPP2R2B | G/A | 1.27 × 10−4 | Novel | SCZ | No |
| 5 | rs815624 | MFAP3 | T/C | 2.10 × 10−4 | Novel | SCZ | No |
| 6 | rs2260000 | PRRC2A | C/T | 2.40 × 10−5 | Novel | SCZ, DEP | Yes |
| 6 | rs1572208 | RIMS1 | T/C | 1.03 × 10−5 | Novel | SCZ | Yes |
| 6 | rs7742212 | PTPRK | A/G | 9.10 × 10−6 | ILAE2 | SCZ | Yes |
| 7 | rs12704290 | GRM3 | G/A | 5.03 × 10−6 | Song et al.52 | SCZ | No |
| 8 | rs7016267 | FAM49B | C/T | 2.60 × 10−4 | Novel | SCZ | Yes |
| 8 | rs11782665 | AC138647.1 | A/C | 8.40 × 10−5 | Novel | ASD | Yes |
| 9 | rs13290882 | KIF27 | G/A | 1.16 × 10−5 | ILAE25 | ADHD | Yes |
| 10 | rs1873691 | KCNMA1 | G/A | 1.19 × 10−4 | Novel | SCZ | No |
| 11 | rs174605 | FADS2 | G/T | 2.87 × 10−5 | Novel | BIP | No |
| 11 | rs56186611 | HNRNPA1P60 | C/T | 1.53 × 10−4 | Novel | SCZ | No |
| 13 | rs73550679 | HS6ST3 | C/T | 4.17 × 10−5 | Novel | SCZ | No |
| 14 | rs12885033 | MDGA2:MDGA2 | A/C | 7.20 × 10−5 | Novel | BIP | Yes |
| 14 | rs55643369 | CTD-2315A10.2 | T/C | 7.56 × 10−5 | Novel | SCZ | Yes |
| 15 | rs2055891 | HOMER2 | A/G | 3.10 × 10−5 | Novel | BIP, SCZ | No |
| 16 | rs4350587 | RBFOX1 | A/G | 1.40 × 10−4 | ILAE25 | SCZ | Yes |
| 16 | rs12325539 | DOC2A | T/C | 4.00 × 10−5 | Novel | SCZ | No |
| 16 | rs72790284 | CNOT1 | C/T | 2.11 × 10−4 | Novel | SCZ | Yes |
| 16 | rs13333786 | AC010547.9:ZNF19 | T/C | 9.40 × 10−5 | Novel | SCZ | Yes |
| 17 | rs8071147 | PRPSAP2 | G/A | 1.44 × 10−4 | Novel | SCZ | Yes |
| 17 | rs2306593 | MYO19 | T/C | 3.00 × 10−4 | Novel | SCZ | Yes |
| 17 | rs4473241 | CTB-175E5.7 | G/T | 8.40 × 10−6 | Novel | BIP | No |
| 20 | rs3829704 | ZNF512B:LINC00176 | C/T | 8.70 × 10−5 | Novel | BIP | No |
| 22 | rs133568 | MIR3201 | A/G | 2.50 × 10−6 | Novel | SCZ | No |
P-values are reported for the epilepsy phenotype. Detailed information about the reported loci can be found in Supplementary Tables 1–8. BIP = bipolar disorder; DEP = depression; SCZ = schizophrenia.
Table 4.
All distinct loci associated with all epilepsy at conjFDR < 0.05
| CHR | LEAD_SNP | Nearest gene | A1/A2 | P-value | Novel in epilepsy | Psychiatric disorder shared with | Concord effect |
|---|---|---|---|---|---|---|---|
| 1 | rs13374459 | MACF1 | T/C | 3.86 × 10−5 | Novel | SCZ | No |
| 2 | rs2717055 | CTD-2026C7.1 | A/G | 5.53 × 10−7 | ILAE 2014 | DEP, SCZ | No |
| 6 | rs7766356 | ZSCAN23 | T/C | 5.76 × 10−6 | Novel | SCZ | Yes |
| 6 | rs13219424 | PTPRK | C/T | 2.40 × 10−5 | ILAE2 | SCZ | Yes |
P-values are reported for the epilepsy phenotype. Detailed information about the reported loci can be found in Supplementary Tables 1–8. BIP = bipolar disorder; DEP = depression; SCZ = schizophrenia.
Next, we evaluated the effect directions of the lead variants for each shared locus (Supplementary Tables 1–8). In the schizophrenia loci shared with GGE, 15 of the 29 lead SNPs had the same allelic effect directions. In the schizophrenia loci shared with all epilepsies, one of four lead SNPs had the same allelic effect directions. In the locus shared between depression, GGE and all epilepsies, the risk for depression was linked to lower risk of epilepsy. In the bipolar disorder loci, three of the eight lead SNPs had the same allelic effect directions in GGE. In the ADHD loci, one of the two lead SNPs, and in the ASD loci, one of the three lead SNPs had the same allelic effect directions in GGE.
Functional annotation
Functional annotation of the candidate SNPs showed that the majority of the SNPs were located in intergenic and intronic regions (Supplementary Tables 9–13, 15 and 16). There was a total of 11 non-synonymous exonic variants, which were detected within the seven loci implicating the genes VRK2, GNL3, FAM114A2, PRPSAP2, C15orf40, C17orf53, ASB16, KIF27 and RMI1 (Supplementary Table 14). Among the shared loci, 84 candidate SNPs had a CADD-score higher than 12.37, which is suggested to reflect deleteriousness.53 Using the three-way gene mapping strategy, the 40 distinct loci were linked to 560 genes (Supplementary Tables 17–23). We identified 19 gene-sets significantly enriched with the genes mapped to the loci shared between schizophrenia and GGE after correcting for multiple comparisons (Supplementary Table 24). The most strongly associated gene-sets were linked to cell cycle regulation and protein serine threonine phosphatase activity, as well as membrane and vesicle function (Supplementary Table 24). The gene-set analyses of the other groups of shared loci were underpowered. Additionally, we determined the differential gene expression of the mapped genes across human tissues (Supplementary Figs 6–19).
Sign concordance test
For GGE, 64 of 91 lead SNPs with conjFDR < 0.10 were sign concordant in the discovery and the independent samples (binomial test P-value = 6.6 × 10−5) (Supplementary Tables 1–8). For schizophrenia, 41 of 55 lead SNPs with conjFDR < 0.10 were sign concordant (P-value = 0.001). For bipolar disorder, 17 of 22 lead SNPs with conjFDR < 0.10 were sign concordant (P-value = 0.0084). All eight lead SNPs associated with depression at conjFDR <0.10 were sign concordant (P-value = 0.0039). For ADHD, three of five lead SNPs with conjFDR < 0.10 were sign concordant (P-value = 0.5). Finally, all four lead SNPs associated with ASD at conjFDR < 0.10 had concordant effects (P-value = 0.0625).
Discussion
In the current study, we demonstrate substantial overlap in common genetic variants influencing common epilepsies and major psychiatric disorders, along with differences in their genetic architectures. First, we demonstrate that the epilepsy phenotypes were considerably less polygenic (1.0 K–3.4 K causal variants) than the psychiatric disorders (5.6 K–13.9 K causal variants; Table 2). Hence, the overlapping genomic loci represent a larger fraction of the genetic architecture underlying the epilepsies than the psychiatric disorders. Then, using the conjFDR method, we leveraged the substantial cross-trait enrichment between epilepsies and psychiatric disorders to boost statistical power. We identified a total of 39 loci shared between GGE and psychiatric disorders and four loci shared between all epilepsy and psychiatric disorders (Fig. 2A, B and Tables 3 and 4). Among the 40 distinct loci identified in total, 32 were novel GGE risk loci and two were novel for all epilepsy (Tables 3 and 4). For schizophrenia, bipolar disorder, depression and GGE, we observed a high degree of sign concordance of the identified lead variants between the discovery and independent samples, supporting the reliability of the findings, while the sign tests for the remaining phenotypes were underpowered. Altogether, the study aligns with recent GWAS analyses suggesting shared genetic risk between neurological and psychiatric disorders,34–36 indicating that common genetic variants may jointly influence the risk of common epilepsies and psychiatric disorders and providing new insights into their shared genetic aetiology.
Most of the identified epilepsy loci (both all epilepsy and GGE) were shared with schizophrenia. The different number of loci shared with each psychiatric disorder may partly reflect the differences in GWAS sample sizes and power, with schizophrenia GWAS being considerably well powered.19 Despite a considerable comorbidity between epilepsy and ASD54 and rare protein-disrupting genetic variants jointly linked to these disorders,55 we only identified three loci shared between ASD and GGE. However, this is not unexpected given the low power of the ASD GWAS.16 Further, ASD is also one of the least discoverable traits; in comparison schizophrenia is estimated to be 2.5 times more discoverable than ASD (Table 2). Recent cross-disorder analyses of GWAS data56 suggest substantial polygenic overlap between psychiatric disorders, and it is likely that many of the shared loci identified here will be linked to several psychiatric disorders as GWAS increase in size. The smaller genetic enrichment observed for focal epilepsy and all epilepsy is likely due to the lower SNP heritabilities (h2 = 5% and 11%, respectively) of these epilepsy phenotypes compared to GGE (h2 = 56%; Table 2). In line with this, MiXeR estimates that the genetic variance underlying GGE is twice more discoverable than focal epilepsy (Table 2). Taken together with polygenicity and heritability estimates, these differences in the genetic architectures of epilepsies may explain why we discovered more genetic loci associated with GGE than with focal epilepsy despite similar sample sizes. There was a mixed pattern of allelic effect directions among the shared loci, suggesting a complex genetic relationship between the disorders, in line with the bi-directional relationship between epilepsies and psychiatric disorders. This mirrors findings from other cross-trait investigations of genetic overlap in the recent years, demonstrating extensive pleiotropy of common variants with mixed effect directions among brain-related traits and disorders.34–36 While the weak or absent genetic correlations2,25 between epilepsy and psychiatric disorders cannot explain the comorbidity between these disorders, the present findings of mixed effects may indicate that in subgroups of patients, common genetic variants may increase susceptibility of both epileptic seizures and mental illness. Further research is needed to evaluate whether genomic prediction tools may help pinpoint individuals at higher risk of such comorbidity. Despite the discovery of many novel loci, these loci only represent a minor fraction of the total genetic risk architectures underlying these disorders, which involve thousands of common genetic variants (Table 2). To achieve clinically meaningful prediction of genomic prediction tools it is necessary to identify a considerably larger proportion of the common variants explaining variation in risk of these disorders.57,58 Hence, it is important to continue the efforts to assemble large-scale GWAS on diverse, well phenotyped populations to enable clinical translation of the emerging genomic findings.
Among the identified loci, five (at genes MRPL33:RBKS, VRK2, CD47, PRRC2A, PCDH7 and HOMER2) were shared between GGE and more than one psychiatric disorder, and three (at genes VRK2, ZSCAN23 and PTPRK) were associated with both GGE and all epilepsy phenotypes (Tables 3 and 4), indicating considerable cross-disorder effects of these loci. In total, we identified 33 novel epilepsy risk loci. Three of the identified loci (near AC018880.2, KIF27 and RBFOX1) were not reported in the original epilepsy GWAS,2 but reached genome-wide significance in the most recent ILAE GWAS,25 which has not yet been peer-reviewed. Similarly, two of the loci (near PCDH7 and KIF27) jointly associated with ADHD and GGE were recently reported in another study.26 We also detected a GGE risk locus at the metabotropic glutamate receptor GRM3 shared with schizophrenia, which was not identified in the original ILAE epilepsy GWAS2 but was detected in another GWAS on epilepsy,52 supporting the validity of this finding. Moreover, the novel risk locus for all epilepsy cases at MACF1 was previously found to be enriched for rare exonic variants in patients with epileptic encephalopathies.59 We also detected a novel locus for all epilepsy and GGE within the extended MHC region (Tables 3 and 4). In our analysis, this locus was shared between GGE, schizophrenia and depression, as well as between all epilepsy and schizophrenia; and have previously been associated with both schizophrenia and depression.20,21,60,61 Genetic variants within the MHC region are linked to both the innate immune system and synaptic maturation during brain development.62 Given the long-range complex LD in the MHC region, which spans numerous genes,63 we consider this finding to reflect the joint involvement of MHC region in these disorders, rather than any specific locus or gene, warranting further studies to disentangle the underlying genetic signal.
We linked all candidate SNPs in the loci to genes using the three-way gene-mapping procedure implemented in FUMA.46 However, functional validation is required to determine if the implicated genes play a role in the aetiology of epilepsy and psychiatric disorders. The genes should instead be considered as starting points for generating hypotheses that can be functionally tested. For instance, among the epilepsy risk loci, there were several potassium channel genes (e.g. KCNMA1, KCNJ3, KCNH8, KCNQ5, KCNN2), which could be prioritized for functional follow-up experiments. Potassium channels play key roles in neuronal excitability and have previously been linked to epilepsy pathogenesis,64–66 possibly representing novel treatment targets. Within the shared loci, we detected 11 non-synonymous exonic variants (Supplementary Table 14), which provide more direct mechanistic hypotheses since they may impact the phenotype directly by disrupting protein function or structure. The implicated genes were VRK2, GNL3, FAM114A2, PRPSAP2, C15orf40, C17orf53, ASB16, KIF27 and RMI1; all of which seem to be involved in various metabolic processes and many are linked to signal transduction.67–76 Nine of the non-synonymous exonic variant associations were novel for GGE, except the variants within KIF27 and RMI1.25 However, like in other GWAS,77 most of the detected genetic variants were located in non-coding regions, indicating regulatory effects on gene expression, complicating biological interpretation. Nineteen gene-sets were significantly enriched with the genes mapped to the loci shared between schizophrenia and GGE (Supplementary Table 24). Many of these gene-sets implicated pathways related to cell cycle and phosphatase activity, particularly serine/threonine-specific phosphatases, as well as membrane and vesicle function, which play important roles in neurotransmitter support and synaptic transmission.78,79 Of note, we did not perform gene mapping of candidate SNPs in the regions with complex long-range LD (the MHC region, 8p23.1 region and the MAPT region). These broad regions involve hundreds of candidate SNPs and a vast number of genes, which could be involved. However, their inclusion in the functional analysis would considerably bias these analyses.
There are some limitations to our study. Like standard GWAS, the conjFDR method detects SNP associations, but it is agnostic about the causal variant underlying the genetic signal, since multiple SNPs may be in LD with the lead SNP. Hence, we cannot exclude the possibility that the overlapping loci reflect separate causal variants.80 Within each locus, we nevertheless identified and functionally characterized the candidate SNPs with the highest probability of being a causal variant, to allow further inspection of plausible causal variants that can be selected for experimental follow-up studies. It is likely that some of the cases in the investigated GWAS on epilepsy and psychiatric disorders had or may develop comorbid mental illness or epilepsy, which might bias our investigation of genetic overlap. However, this potential bias cannot explain the presence of mixed effect directions among the shared loci. Another limitation was the focus on individuals of European ancestry. Participants in the discovery samples were predominantly of European ancestry to ensure compatibility in LD patterns, which might otherwise bias the conjFDR analyses. However, the high degree of sign consistency observed in the validation schizophrenia GWAS in East Asian individuals19 suggests that the findings are also generalizable to this population. Improving the diversity in GWAS populations remains a key issue for the genomics research.81
In conclusion, we demonstrate extensive polygenic overlap between common epilepsies and major psychiatric disorders and identified 40 shared risk loci with mixed effect directions, 32 of which are novel loci for GGE, two novel loci associated with all epilepsy phenotypes and four novel loci for psychiatric disorders. As GWAS get larger, we expect that many more loci will be found to jointly influence epilepsy and psychiatric disorders, which may eventually inform clinical practice.
Supplementary Material
Acknowledgements
All GWAS investigated in the present study were approved by the local ethics committees, and informed consent was obtained from all participants. We thank the ILAE, PGC, iPSYCH and 23andMe consortia as well as the FinnGen biobank and UK biobank for access to data, and the many participants who provided DNA samples. We also thank the Norwegian Mother, Father and Child Cohort Study (MoBa) for access to data and the Norwegian Institute of Public Health (NIPH) for generating high-quality genomic data. The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are grateful to all the participating families in Norway who take part in this on-going cohort study. This work was performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated, and developed by the TSD service group at the University of Oslo, IT-Department (USIT). (tsd-drift@usit.uio.no).
Contributor Information
Naz Karadag, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Alexey A Shadrin, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; K.G. Jebsen Centre for Neurodevelopmental disorders, University of Oslo and Oslo University Hospital, 0424, Oslo, Norway.
Kevin S O’Connell, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Guy F L Hindley, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London SE5 8AB, UK.
Zillur Rahman, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Nadine Parker, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Shahram Bahrami, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Vera Fominykh, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Weiqiu Cheng, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Børge Holen, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; Department of Neurology, Oslo University Hospital, 0372, Oslo, Norway.
Silje Alvestad, Department of Clinical Medicine, University of Bergen, 5020, Bergen, Norway; National Center for Epilepsy, Oslo University Hospital, 1337, Sandvika, Norway.
Erik Taubøll, Department of Neurology, Oslo University Hospital, 0372, Oslo, Norway; Faculty of Medicine, University of Oslo, 0372, Oslo, Norway.
Nils Eiel Steen, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; Division of Mental Health and Addiction, Oslo University Hospital, 0407, Oslo, Norway.
Srdjan Djurovic, Department of Medical Genetics, Oslo University Hospital, 0450, Oslo, Norway; NORMENT Centre, Department of Clinical Science, University of Bergen, 5020, Bergen, Norway.
Anders M Dale, Department of Cognitive Science, University of California, San Diego, La Jolla, CA 92093, USA; Multimodal Imaging Laboratory, University of California, San Diego, La Jolla, CA 92093, USA; Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA; Department of Neurosciences, University of California, San Diego, La Jolla, CA 92093, USA.
Oleksandr Frei, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway.
Ole A Andreassen, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; K.G. Jebsen Centre for Neurodevelopmental disorders, University of Oslo and Oslo University Hospital, 0424, Oslo, Norway; Division of Mental Health and Addiction, Oslo University Hospital, 0407, Oslo, Norway.
Olav B Smeland, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, 0407, Oslo, Norway; Division of Mental Health and Addiction, Oslo University Hospital, 0407, Oslo, Norway.
Funding
This research is part of the HARVEST collaboration, supported by the Research Council of Norway (#229624). We also thank the NORMENT Centre for providing genotype data, funded by the Research Council of Norway (#223273), South-East Norway Health Authorities and Stiftelsen Kristian Gerhard Jebsen. We further thank the Center for Diabetes Research, the University of Bergen for providing genotype data and performing quality control and imputation of the data funded by the ERC AdG project SELECTionPREDISPOSED, Stiftelsen Kristian Gerhard Jebsen, Trond Mohn Foundation, the Research Council of Norway, the Novo Nordisk Foundation, the University of Bergen, and the Western Norway Health Authorities. The project is funded by ERA-NET-NEURON (Application no: ES609126, Project NMDAR-PSY), which is funded by participating Research Councils across Europe. The work was supported by European Union Horizon 2020 Research and Innovation Action Grant (847776, CoMorMent), the Research Council of Norway (262656, 249711, 248980, 248778, 226971 and 223273), South-East Norway Regional Health Authority (2016–064) and KG Jebsen Stiftelsen (SKGJ-MED-008). A.M.D. was supported by NIH (U24DA041123). S.A. was supported by NordForsk (project no. 83796).
Competing interests
O.A.A. has received speaker’s honorarium from Lundbeck and is a consultant for Healthlytix. A.M.D. is a founder of and holds equity interest in CorTechs Labs and serves on its scientific advisory board. He is also a member of the Scientific Advisory Board of Healthlytix and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. Remaining authors have no conflicts of interest to declare.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. GBD 2019 Diseases and Injuries Collaborators . Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the global burden of disease study 2019. Lancet. 2020;396:1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. International League Against Epilepsy Consortium on Complex Epilepsies . Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brooks-Kayal AR, Bath KG, Berg AT, et al. . Issues related to symptomatic and disease-modifying treatments affecting cognitive and neuropsychiatric comorbidities of epilepsy. Epilepsia. 2013;54(Suppl 4):44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu E, Pyatka N, Burant CJ, Sajatovic M. Systematic literature review of psychiatric comorbidities in adults with epilepsy. J Clin Neurol. 2021;17:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hesdorffer DC, Ishihara L, Mynepalli L, Webb DJ, Weil J, Hauser WA. Epilepsy, suicidality, and psychiatric disorders: A bidirectional association. Ann Neurol. 2012;72:184–191. [DOI] [PubMed] [Google Scholar]
- 6. Devinsky O. Psychiatric comorbidity in patients with epilepsy: Implications for diagnosis and treatment. Epilepsy Behav. 2003;4(Suppl 4):S2–10. [DOI] [PubMed] [Google Scholar]
- 7. Kanner AM. Management of psychiatric and neurological comorbidities in epilepsy. Nat Rev Neurol. 2016;12:106–116. [DOI] [PubMed] [Google Scholar]
- 8. Nanou E, Catterall WA. Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron. 2018;98:466–481. [DOI] [PubMed] [Google Scholar]
- 9. Sullivan PF, Geschwind DH. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177:162–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devinsky O, Vezzani A, O’Brien TJ, et al. . Epilepsy. Nat Rev Dis Primers. 2018;4:18024. [DOI] [PubMed] [Google Scholar]
- 11. Egervari G, Kozlenkov A, Dracheva S, Hurd YL. Molecular windows into the human brain for psychiatric disorders. Mol Psychiatry. 2019;24:653–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kjeldsen MJ, Kyvik KO, Christensen K, Friis ML. Genetic and environmental factors in epilepsy: A population-based study of 11900 Danish twin pairs. Epilepsy Res. 2001;44(2–3):167–178. [DOI] [PubMed] [Google Scholar]
- 13. Peljto AL, Barker-Cummings C, Vasoli VM, et al. . Familial risk of epilepsy: A population-based study. Brain. 2014;137(Pt ):795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polderman TJ, Benyamin B, de Leeuw CA, et al. . Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. [DOI] [PubMed] [Google Scholar]
- 15. Demontis D, Walters RK, Martin J, et al. . Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grove J, Ripke S, Als TD, et al. . Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howard DM, Adams MJ, Shirali M, et al. . Genome-wide association study of depression phenotypes in UK biobank identifies variants in excitatory synaptic pathways. Nat Commun. 2018;9:1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mullins N, Forstner AJ, O’Connell KS, et al. . Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trubetskoy V, Pardinas AF, Qi T, et al. . Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hyde CL, Nagle MW, Tian C, et al. . Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wray NR, Ripke S, Mattheisen M, et al. . Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lam M, Chen CY, Li Z, et al. . Comparative genetic architectures of schizophrenia in east Asian and European populations. Nat Genet. 2019;51:1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levey DF, Stein MB, Wendt FR, et al. . Bi-ancestral depression GWAS in the million veteran program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. 2021;24:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demontis D, Walters GB, Athanasiadis G,. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet. 2023;55:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International League Against Epilepsy Consortium on Complex Epilepsies, Berkovic SF, Cavalleri GL, Koeleman BPC . Genome-wide meta-analysis of over 29,000 people with epilepsy reveals 26 loci and subtype-specific genetic architecture. medRxiv. [Preprint] 10.1101/2022.06.08.22276120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Y, Li Y, Zhu J, Long J. Shared genetics and causality underlying epilepsy and attention-deficit hyperactivity disorder. Psychiatry Res. 2022;316:114794. [DOI] [PubMed] [Google Scholar]
- 27. Brainstorm Consortium, Anttila V, Bulik-Sullivan B, et al. . Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cross-Disorder Group of the Psychiatric Genomics Consortium . Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smeland OB, Frei O, Dale AM, Andreassen OA. The polygenic architecture of schizophrenia—Rethinking pathogenesis and nosology. Nat Rev Neurol. 2020;16:366–379. [DOI] [PubMed] [Google Scholar]
- 30. Smeland OB, Frei O, Shadrin A, et al. . Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139:85–94. [DOI] [PubMed] [Google Scholar]
- 31. Le Hellard S, Wang Y, Witoelar A, et al. . Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophr Bull. 2017;43:654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng W, Frei O, van der Meer D, et al. . Genetic association between schizophrenia and cortical brain surface area and thickness. JAMA Psychiatry. 2021;78:1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smeland OB, Bahrami S, Frei O, et al. . Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2020;25:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drange OK, Smeland OB, Shadrin AA, et al. . Genetic overlap between Alzheimer’s disease and bipolar disorder implicates the MARK2 and VAC14 genes. Front Neurosci. 2019;13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smeland OB, Shadrin A, Bahrami S, et al. . Genome-wide association analysis of Parkinson’s disease and schizophrenia reveals shared genetic architecture and identifies novel risk loci. Biol Psychiatry. 2021;89:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bahrami S, Hindley G, Winsvold BS, et al. . Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain. 2022;145:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holland D, Frei O, Desikan R, et al. . Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet. 2020;16:e1008612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Andreassen OA, Thompson WK, Schork AJ, et al. . Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karch CM, Wen N, Fan CC, et al. . Selective genetic overlap between amyotrophic lateral sclerosis and diseases of the frontotemporal dementia spectrum. JAMA Neurol. 2018;75:860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wistrom ED, O’Connell KS, Karadag N, et al. . Genome-wide analysis reveals genetic overlap between alcohol use behaviours, schizophrenia and bipolar disorder and identifies novel shared risk loci. Addiction. 2022;117:600–610. [DOI] [PubMed] [Google Scholar]
- 41. Yokoyama JS, Wang Y, Schork AJ, et al. . Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol. 2016;73:691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smeland OB, Frei O, Kauppi K, et al. . Identification of genetic loci jointly influencing schizophrenia risk and the cognitive traits of verbal-numerical reasoning, reaction time, and general cognitive function. JAMA Psychiatry. 2017;74:1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Magnus P, Irgens LM, Haug K, et al. . Cohort profile: The Norwegian mother and child cohort study (MoBa). Int J Epidemiol. 2006;35:1146–1150. [DOI] [PubMed] [Google Scholar]
- 44. Magnus P, Birke C, Vejrup K, et al. . Cohort profile update: The Norwegian mother and child cohort study (MoBa). Int J Epidemiol. 2016;45:382–388. [DOI] [PubMed] [Google Scholar]
- 45. Schwartzman A, Lin X. The effect of correlation in false discovery rate estimation. Biometrika. 2011;98:199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Genomes Project C, Auton A, Brooks LD, et al. . A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buniello A, MacArthur JAL, Cerezo M, et al. . The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lonsdale J, Thomas J, Salvatore Met al. . The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Savage JE, Jansen PR, Stringer S, et al. . Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurki MI, Karjalainen J, Palta Pet al. . Finngen: Unique genetic insights from combining isolated population and national health register data. medRxiv. [Preprint] 10.1101/2022.03.03.22271360 [DOI] [Google Scholar]
- 52. Song M, Liu J, Yang Y, Lv L, Li W, Luo XJ. Genome-wide meta-analysis identifies two novel risk loci for epilepsy. Front Neurosci. 2021;15:722592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sundelin HEK, Larsson H, Lichtenstein Pet al. . Autism and epilepsy: A population-based nationwide cohort study. Neurology. 2016;87(2):192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Willsey HR, Willsey AJ, Wang B, State MW. Genomics, convergent neuroscience and progress in understanding autism spectrum disorder. Nat Rev Neurosci. 2022;23:323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O’Connell KS, Hindley G, Smeland OB, Shadrin Aet al. . Shared heritability among psychiatric disorders and traits. Psychiatric Genomics. 2022:341–360. [Google Scholar]
- 57. Murray GK, Lin T, Austin J, McGrath JJ, Hickie IB, Wray NR. Could polygenic risk scores be useful in psychiatry? : A review. JAMA Psychiatry. 2021;78:210–219. [DOI] [PubMed] [Google Scholar]
- 58. Smeland OB, Andreassen OA. Polygenic risk scores in psychiatry—Large potential but still limited clinical utility. Eur Neuropsychopharmacol. 2021;51:68–70. [DOI] [PubMed] [Google Scholar]
- 59. Cox A, Grady F, Velez G, et al. . In trans variant calling reveals enrichment for compound heterozygous variants in genes involved in neuronal development and growth. Genet Res (Camb). 2019;101:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cross-Disorder Group of the Psychiatric Genomics C . Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. International Schizophrenia C, Purcell SM, Wray NR, et al. . Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sekar A, Bialas AR, de Rivera H, et al. . Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Horton R, Wilming L, Rand V, et al. . Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. [DOI] [PubMed] [Google Scholar]
- 64. Kohling R, Wolfart J. Potassium channels in epilepsy. Cold Spring Harb Perspect Med. 2016;6:a022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Allen NM, Weckhuysen S, Gorman K, King MD, Lerche H. Genetic potassium channel-associated epilepsies: Clinical review of the Kv family. Eur J Paediatr Neurol. 2020;24:105–116. [DOI] [PubMed] [Google Scholar]
- 66. Jentsch TJ. Neuronal KCNQ potassium channels: Physiology and role in disease. Nat Rev Neurosci. 2000;1:21–30. [DOI] [PubMed] [Google Scholar]
- 67. Bizard AH, Hickson ID. The dissolution of double holliday junctions. Cold Spring Harb Perspect Biol. 2014;6:a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boultwood J, Fidler C, Strickson AJ, et al. . Transcription mapping of the 5q- syndrome critical region: Cloning of two novel genes and sequencing, expression, and mapping of a further six novel cDNAs. Genomics. 2000;66:26–34. [DOI] [PubMed] [Google Scholar]
- 69. Fernandez IF, Perez-Rivas LG, Blanco S, Castillo-Dominguez AA, Lozano J, Lazo PA. VRK2 Anchors KSR1-MEK1 to endoplasmic reticulum forming a macromolecular complex that compartmentalizes MAPK signaling. Cell Mol Life Sci. 2012;69:3881–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform. 2011;12:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hustedt N, Saito Y, Zimmermann M, et al. . Control of homologous recombination by the HROB-MCM8-MCM9 pathway. Genes Dev. 2019;33(19–20):1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Katashima R, Iwahana H, Fujimura M, et al. . Molecular cloning of a human cDNA for the 41-kDa phosphoribosylpyrophosphate synthetase-associated protein. Biochim Biophys Acta. 1998;1396:245–250. [DOI] [PubMed] [Google Scholar]
- 73. Katoh Y, Katoh M. KIF27 Is one of orthologs for Drosophila costal-2. Int J Oncol. 2004;25:1875–1880. [PubMed] [Google Scholar]
- 74. Liu P, Verhaar AP, Peppelenbosch MP. Signaling size: Ankyrin and SOCS box-containing ASB E3 ligases in action. Trends Biochem Sci. 2019;44:64–74. [DOI] [PubMed] [Google Scholar]
- 75. Luck K, Kim DK, Lambourne L, et al. . A reference map of the human binary protein interactome. Nature. 2020;580:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Visscher PM, Yengo L, Cox NJ, Wray NR. Discovery and implications of polygenicity of common diseases. Science. 2021;373:1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. van Spronsen M, Hoogenraad CC. Synapse pathology in psychiatric and neurologic disease. Curr Neurol Neurosci Rep. 2010;10:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Appelbaum LG, Shenasa MA, Stolz L, Daskalakis Z. Synaptic plasticity and mental health: Methods, challenges and opportunities. Neuropsychopharmacology. 2022;48:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: Challenges and strategies. Nat Rev Genet. 2013;14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martin AR, Gignoux CR, Walters RK, et al. . Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistical analyses for the relevant methods were performed in MATLAB and Python, using existing tools available on GitHub, including MiXeR v1.3 (https://github.com/precimed/mixer) and condFDR/conjFDR (https://github.com/precimed/pleiofdr).