Abstract
Multiple sclerosis is a complex autoimmune disease, and several therapies for multiple sclerosis have been developed and widely used. However, existing medications for multiple sclerosis were far from satisfactory due to their failure to suppress relapses and alleviate disease progression. Novel drug targets for multiple sclerosis prevention are still needed.
We performed Mendelian randomization to explore potential drug targets for multiple sclerosis using summary statistics from the International Multiple Sclerosis Genetics Consortium (nCase = 47 429, nControl = 68 374) and further replicated in UK Biobank (nCase = 1356, nControl = 395 209) and FinnGen cohorts (nCase = 1326, nControl = 359 815). Genetic instruments for 734 plasma and 154 CSF proteins were obtained from recently published genome-wide association studies. The reverse causality detection using bidirectional Mendelian randomization analysis and Steiger filtering, Bayesian co-localization, and phenotype scanning that searched previously reported genetic variant–trait associations were implemented to consolidate the Mendelian randomization findings further. In addition, the protein–protein interaction network was performed to reveal potential associations among proteins and/or present multiple sclerosis medications.
At Bonferroni significance (P < 5.63 × 10−5), Mendelian randomization analysis revealed six protein–multiple sclerosis pairs. In plasma, per standard deviation increase in FCRL3, TYMP and AHSG had a protective effect. Odds ratios for the proteins above were 0.83 (95% CI, 0.79–0.89), 0.59 (95% CI, 0.48–0.71) and 0.88 (95% CI, 0.83–0.94), respectively. In CSF, per 10-fold increase in MMEL1 (OR, 5.03; 95% CI, 3.42–7.41) increased the risk of multiple sclerosis, while SLAMF7 (OR, 0.42; 95% CI, 0.29–0.60) and CD5L (OR, 0.30; 95%CI, 0.18–0.52) decreased the risk. None of the six proteins had reverse causality. Bayesian co-localization suggested that FCRL3 [coloc.abf-posterior probability of hypothesis 4 (PPH4) = 0.889], TYMP (coloc.susie-PPH4 = 0.896), AHSG (coloc.abf-PPH4 = 0.957, coloc.susie-PPH4 = 0.973), MMEL1 (coloc.abf-PPH4 = 0.930) and SLAMF7 (coloc.abf-PPH4 = 0.947) shared the same variant with multiple sclerosis. FCRL3, TYMP and SLAMF7 interacted with target proteins of current multiple sclerosis medications. MMEL1 was replicated in both UK Biobank and FinnGen cohorts.
Our integrative analysis suggested that genetically determined levels of circulating FCRL3, TYMP, AHSG, CSF MMEL1 and SLAMF7 had causal effects on multiple sclerosis risk. These findings suggested those five proteins might be promising drug targets for multiple sclerosis and warrant further clinical investigation, especially FCRL3 and SLAMF7.
Keywords: multiple sclerosis, Mendelian randomization, drug target
Lin et al. use Mendelian randomization analysis to search for drug targets for multiple sclerosis, particularly progressive multiple sclerosis. They identify three plasma proteins and two CSF proteins whose genetically determined levels are associated with multiple sclerosis risk, and which may be promising drug targets.
Introduction
Multiple sclerosis (MS) is one of the most common chronic disabling neurological diseases that mainly affects young adults.1 Both the incidence and prevalence of MS are increasing worldwide. In most patients (∼85%), MS primarily manifests as recurrent attacks due to inflammation in the early stages (relapsing–remitting MS, RRMS) followed by progressive disability due to neurodegeneration (secondary progressive MS, SPMS).2 However, a minority of patients (10–15%) have a continuous non-relapsing progression from disease onset (primary progressive MS, PPMS). The pathological mechanisms of RRMS and progressive MS (PMS), including SPMS and PPMS, differ. Inflammation in RRMS is characterized by the influx of immune cells, while inflammation in PMS is chronic and compartmentalized behind a closed blood–brain barrier, with activation of microglia and continued involvement of T and B cells.3
Disease-modifying therapy is an integral part of MS management, which can reduce relapse occurrence and slow progression to permanent disability in patients with MS. A total of nine classes of disease-modifying therapy treatments with distinct mechanisms have become available, including interferons, glatiramer acetate, teriflunomide, sphingosine-1-phosphate receptor modulators, fumarates, cladribine, natalizumab, alemtuzumab and B-cell-depleting treatments (ocrelizumab and ofatumumab).4 However, most of these medications target the inflammatory component of the disease and mainly affect RRMS,5 while treatment of PMS remains unsatisfactory.
Human proteins play critical roles in a range of biological processes and are the predominant type of drug target.6 Nelson et al.7 demonstrated that a protein drug target whose link with the disease is supported by genetic association is twice as likely to reach market approval. Recently, Mendelian randomization (MR) analysis has been widely used for drug target development and drug repurposing.8 MR is a genetic instrumental variable analysis that usually utilizes single nucleotide polymorphisms (SNPs) from genome-wide association studies (GWAS) as genetic instruments to estimate the causal effect of an exposure on an outcome. In contrast to observational studies, MR can avoid the influence of confounders. Owing to advances in high-throughput genomic and proteomic techniques in both plasma and CSF, MR-based strategies have promoted the identification of potential therapeutic targets for many diseases, such as stroke and Alzheimer’s disease.9,10 However, to date, few MR studies integrating GWAS and protein quantitative trait loci (pQTL) data on MS have been reported.
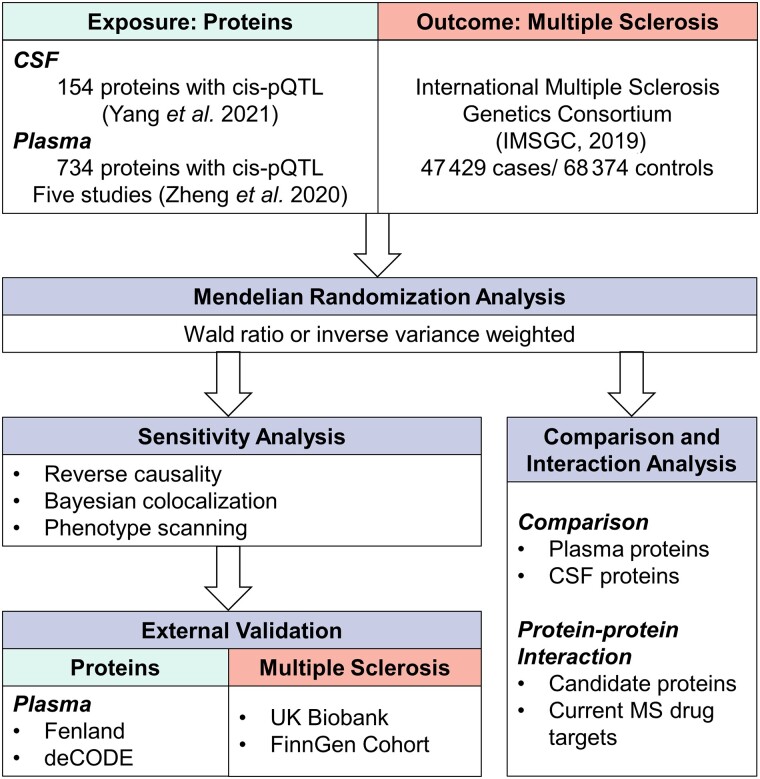
In this study, we aimed to identify both plasma and CSF proteins as potential therapeutic targets for MS. The study design is shown in Fig. 1. First, we used MR to identify potential causal plasma and CSF proteins for MS using GWAS data from the International Multiple Sclerosis Genetics Consortium (IMSGC),11 plasma pQTL data from Zheng’s study,6 and CSF pQTL data from Yang’s study.12 Second, the primary findings were further validated using reverse causality detection, Bayesian co-localization analysis and phenotype scanning. Third, we mapped the interaction network among the identified proteins, between the proteins based on plasma and CSF, and between the identified proteins and the targets of current MS medications. Finally, using the GWAS data from the UK Biobank13 and FinnGen14 cohorts and plasma pQTL data from two newly published studies,15,16 we replicated the analysis as an external validation to strengthen our conclusion.
Figure 1.
Study design for identification of plasma and CSF proteins causally associated with MS.
Materials and methods
CSF and plasma protein quantitative trait loci
CSF pQTL data were obtained from a study by Yang et al.,12 who reported 274 pQTLs of 184 CSF proteins. Only pQTLs satisfying the following criteria were included: (i) showed genome-wide significant association (P < 5 × 10−8); (ii) were located outside the major histocompatibility complex (MHC) region (chr6, 26–34 Mb); (iii) showed independent association [linkage disequilibrium (LD) clumping r2 < 0.001]; and (iv) was a cis-acting pQTL. Finally, 154 cis-pQTLs were identified for 154 proteins.
For the primary analysis, the plasma pQTL data were retrieved from the study by Zheng et al.,6 which integrated five previously published GWAS.17–21 Based on the screening criteria used above in the CSF pQTL dataset, 738 cis-acting SNPs for 734 proteins were included. Data were checked using the original documents as a reference to ensure reliability. In addition, the plasma pQTL data retrieved from two recently published studies by Pietzner et al.15 (4775 plasma proteins measured in 10 708 participants) and Ferkingstad et al.16 (4907 plasma proteins measured in 35 559 participants) were used for external validation.
For any missing information in the QTL GWAS summary statistics, such as effect allele frequency, we used the matched human genome build as a reference to complete the data (Supplementary Table 1).
GWAS summary statistics of multiple sclerosis
For the primary analysis, summary statistics were retrieved from the largest GWAS dataset of IMSGC,11 including 115 803 individuals (nCase = 47 429, nControl = 68 374) of European ancestry. For external validation, summary statistics were obtained from the UK Biobank (until 2017, nCase = 1356, nControl = 395 209)13 and the FinnGen study (nCase = 1326, nControl = 359 815, R6 release).14
Statistical analysis
Mendelian randomization analysis
In this study, we used the plasma and CSF proteins as the exposure and MS as the outcome to perform MR with ‘TwoSampleMR’ (https://github.com/MRCIEU/TwoSampleMR). The Wald ratio was used if only one pQTL was available for a given protein. When two or more genetic instruments were available, inverse variance weighted MR (MR-IVW) was applied and followed by heterogeneity analysis.22 Odds ratios (OR) for increased risk of MS were expressed as per standard deviation (SD) increase in plasma protein levels and per 10-fold increase in CSF protein levels.
For the primary analysis, Bonferroni correction was used to adjust for multiple testing, and a threshold P-value of 0.05/888 (P < 5.63 × 10−5) was used to prioritize the results for further analysis. MR was performed only on the preliminarily identified proteins for external validation and was set at a P-value threshold of 0.05. We implemented a same-variant strategy that used the same SNP used in the primary analysis as genetic instruments and a significant-variant strategy that used genome-wide significant SNP as a genetic instrument to validate preliminary findings.
Reverse causality detection
Following the same screening criteria for pQTLs, 134 genetic instruments for MS were selected from the GWAS of IMSGC11 for bidirectional MR analysis to detect potential reverse causality (Supplementary Table 2).23 Complete summary statistics for proteins were obtained from three previous studies.12,16,18 The effect was estimated using MR-IVW, MR-Egger, weighted median, simple mode and weighted mode. We also conducted Steiger filtering to ensure the directionality of the association between proteins and MS.24 The results were considered statistically significant at P < 0.05.
Bayesian co-localization analysis
Bayesian co-localization analyses were used to assess the probability that two traits share the same causal variant using the ‘coloc’ package (https://github.com/chr1swallace/coloc) with default arguments. As described previously,21 Bayesian co-localization provides the posterior probability for five hypotheses on whether a single variant is shared between two traits. In this study, we tested the posterior probability of hypothesis 3 (PPH3), in which both the protein and MS were associated with the region by different variants, and hypothesis 4 (PPH4), in which both the protein and MS were associated with the region by shared variants. Both the coloc.abf and coloc.susie algorithms were used, and we defined a gene as having evidence of co-localization based on gene-based PPH4 > 80%, determined by at least one algorithm.22,25
Phenotype scanning
We also performed phenotype scanning that searched previous GWAS to reveal associations of identified pQTLs with other traits. Phenotype scanning was conducted via both ‘phenoscanner’26 and looking up in the study of Ferkingstad et al.16 a plasma proteome GWAS. An SNP was considered pleiotropic when satisfying the following criteria: (i) the association was genome-wide significant (P < 5 × 10−8); (ii) the GWAS was conducted in a population of European ancestry; and (iii) the SNPs were associated with any known risk factors of MS, including metabolic traits, proteins or clinical traits. Besides, we calculated the LD r2 among pQTLs of prioritized proteins to reveal potential linkage.
Comparison analysis and protein–protein interaction network
We hypothesized that there would be little correlation between plasma- and CSF-identified pQTLs because of the blood–brain barrier. Therefore, the correlation between the shared pQTLs identified in the CSF and plasma using effect estimates from the MR analysis was investigated by Spearman correlation analysis, and the different P-value thresholds were set to explore whether the correlation changed as the significance level increased.
The protein–protein interaction (PPI) network of proteins suggestively associated with MS risk (primary MR analysis P < 0.05) in CSF or plasma analysis was explored. We aimed to investigate the interactions among the prioritized proteins and whether the proteins identified using plasma data could interact with those identified using CSF data. In addition, to explore the interactions between those MS-associated genes and the targets for medications already on the market, we obtained 13 disease-modifying drugs for MS from a recent review4 and corresponding drug targets based on the Drugbank database (https://www.drugbank.ca).27 We also searched current medications targeting the identified potential causal proteins. All PPI analyses were conducted using the Search Tool for the Retrieval of Interacting Genes (STRING) database version 11.5 (https://string-db.org/), with the minimum required interaction score at 0.4.28 In addition, we conducted MR by Wald ratio method and Bayesian co-localization by coloc.abf algorithm with prioritized proteins as both exposure and outcome. We regarded a P-value of MR smaller than 0.05 as a potential interaction and PPH4 >0.8 as potential co-localization.
Data availability
Genome-wide summary-level statistics for cis-pQTL were available from the original studies.12,15–21 GWAS summary statistics of the IMSGC were obtained from https://gwas.mrcieu.ac.uk/datasets/. GWAS summary statistics of the UK Biobank conducted by Zhou et al.29 were obtained from https://www.leelabsg.org/resources. Access to FinnGen (R6 release) can be obtained from https://www.finngen.fi/en/access_results.
Results
Screening the proteome for multiple sclerosis causal proteins
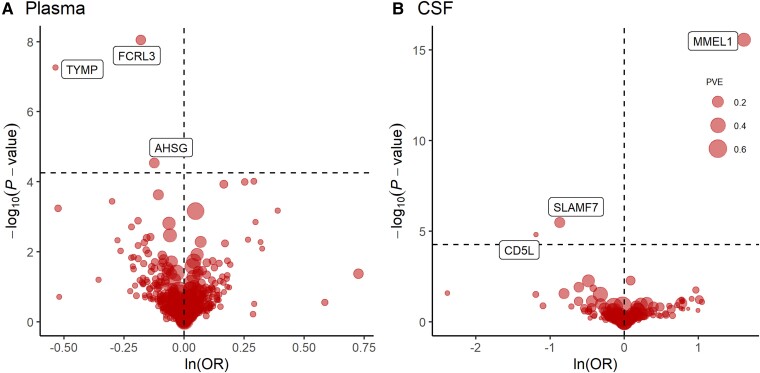
At Bonferroni significance (P < 5.63 × 10−5), MR analysis revealed six protein–MS pairs (Table 1 and Fig. 2A and B), including Fc receptor-like protein 3 (FCRL3), thymidine phosphorylase (TYMP) and alpha-2-HS-glycoprotein (AHSG) in the plasma, and membrane metallo-endopeptidase-like 1 (MMEL1), signalling lymphocytic activation molecule F7 (SLAMF7) and CD5 antigen-like (CD5L) in the CSF. Specifically, increased FCRL3 (OR = 0.83; 95% CI, 0.79–0.89; P = 8.93 × 10−9), TYMP (OR = 0.59; 95% CI, 0.48–0.71; P = 5.43 × 10−8), AHSG (OR = 0.88; 95% CI, 0.83–0.94; P = 2.91 × 10−5), SLAMF7 (OR = 0.42; 95% CI, 0.29–0.60; P = 3.28 × 10−6) and CD5L (OR = 0.30; 95% CI, 0.18–0.52; P = 1.52 × 10−5) decreased the risk of MS, whereas elevated MMEL1 (OR = 5.03; 95% CI, 3.42–7.41; P = 2.80 × 10−16) increased the risk of MS. No heterogeneity was detected for the proteins analysed in the primary analysis (Supplementary Table 3).
Table 1.
MR results for plasma and CSF proteins significantly associated with MS after Bonferroni correction
| Tissue | Protein | UniProt ID | SNPa | Effect allele | OR (95% CI)b | P value | PVE | F statistics | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Plasma | FCRL3 | Q96P31 | rs7528684 | G | 0.83 (0.79, 0.89) | 8.93 × 10−9 | 13.34% | 508.28 | Sun et al.18 |
| Plasma | TYMP | P19971 | rs131798 | G | 0.59 (0.48, 0.71) | 5.43 × 10−8 | 1.42% | 46.24 | Emilsson et al.20 |
| Plasma | AHSG | P02765 | rs35094235 | G | 0.88 (0.83, 0.94) | 2.91 × 10−5 | 15.40% | 601.12 | Sun et al.18 |
| CSF | MMEL1 | Q495T6 | rs10909839 | G | 5.03 (3.42, 7.41) | 2.80 × 10−16 | 29.27% | 345.53 | Yang et al.12 |
| CSF | SLAMF7 | Q9NQ25 | rs3766374 | G | 0.42 (0.29, 0.60) | 3.28 × 10−6 | 16.21% | 161.51 | Yang et al.12 |
| CSF | CD5L | O43866 | rs6427401 | G | 0.30 (0.18, 0.52) | 1.52 × 10−5 | 3.66% | 31.71 | Yang et al.12 |
PVE = proportion of variance explained.
All SNPs used were cis-acting.
Odds ratios for increased risk of MS were expressed as per SD increase in plasma protein levels and per 10-fold increase in CSF protein levels.
Figure 2.
MR results for plasma and CSF proteins and the risk of MS.
Volcano plots of the MR results for (A) 734 plasma and (B) 154 CSF proteins on the risk of MS. A and B show MR analysis with Wald ratio or inverse variance weighted method on plasma and CSF proteins on the risk of MS, respectively. OR for increased risk of MS were expressed as per SD increase in plasma protein levels and per 10-fold increase in CSF protein levels. Dashed horizontal black line corresponded to P = 5.63 × 10−5 (0.05/888). ln = natural logarithm; PVE = proportion of variance explained.
Sensitivity analysis for multiple sclerosis causal proteins
Five of the six proteins revealed by the MR analysis were identified as potential drug targets for MS, including FCRL3, TYMP, AHSG, MMEL1 and SLAMF7. First, bidirectional MR analysis did not reveal any causal effect of MS on the level of six identified proteins and Steiger filtering further ensured directionality (Table 2 and Supplementary Fig. 1). Second, Bayesian co-localization strongly suggested that FCRL3 (coloc.abf-PPH4 = 0.889), TYMP (coloc.susie-PPH4 =0.896), AHSG (coloc.abf-PPH4 = 0.957, coloc.susie-PPH4 = 0.973), MMEL1 (coloc.abf-PPH4 = 0.930) and SLAMF7 (coloc.abf-PPH4 = 0.947) shared the same variant with MS (Table 2 and Supplementary Fig. 2). Finally, after phenotype scanning, FCRL3 (rs7528684) was found to be associated with type 1 diabetes, Fc receptor-like protein 4 (FCRL4), immunoglobulin superfamily member 11 (IGSF11) and killer cell immunoglobulin-like receptor 3DL3 (KIR3DL3); TYMP (rs131798) was found to be associated with blood cell traits and pyrimidine metabolism, such as mean corpuscular haemoglobin, mean corpuscular volume, uridine and 2-deoxyuridine; AHSG (rs35094235) was found to be associated with activated partial thromboplastin time (APTT); MMEL1 (rs10909839) was found to be a susceptibility gene for primary sclerosing cholangitis and indirectly influenced MS, rheumatoid arthritis and coeliac disease; and CD5L was found to be associated with Fc receptor-like protein 1 (FCRL1). In contrast, no significant association was observed for SLAMF7 (Table 2 and Supplementary Table 4). Meanwhile, FCRL3 and CD5L pQTLs showed a suggestive correlation (LD r2 = 0.65, Supplementary Table 5).
Table 2.
Summary of reverse causality detection, Bayesian co-localization analysis and phenotype scanning on six potential causal proteins
| Tissue | Protein | UniProt ID | SNP | Bidirectional MR (MR-IVW)a | Steiger filtering | Co-localization PPH4 (coloc.abf/coloc.susie) | Previously reported associations |
|---|---|---|---|---|---|---|---|
| Plasma | FCRL3 | Q96P31 | rs7528684 | 0.946 (0.860, 1.041) | Passed (1.637 × 10−95) | 0.889/2.14 × 10−2 | T1DMb,c, FCRL4c, IGSF11c, KIR3DL3c |
| Plasma | TYMP | P19971 | rs131798 | 1.007 (0.989, 1.026) | Passed (6.638 × 10−9) | 0.719/0.896 | Blood cellsb,c, pyrimidine metabolismc |
| Plasma | AHSG | P02765 | rs35094235 | 0.976 (0.929, 1.026) | Passed (4.925 × 10−115) | 0.957/0.973 | APTTc |
| CSF | MMEL1 | Q495T6 | rs10909839 | 0.970 (0.934, 1.008) | Passed (1.917 × 10−7) | 0.930/8.57 × 10−6 | MSc, PSCc, RAc |
| CSF | SLAMF7 | Q9NQ25 | rs3766374 | 1.011 (0.984, 1.039) | Passed (7.540 × 10−63) | 0.947/9.22 × 10−3 | N/A |
| CSF | CD5L | O43866 | rs6427401 | 1.021 (0.992, 1.052) | Passed (1.401 × 10−32) | 0.634/2.19 × 10−4 | FCRL1b |
APTT = activated partial thromboplastin time; MR-IVW = Mendelian randomization with inverse variance weighted method; N/A = not available; PP = posterior probability; PSC = primary sclerosing cholangitis; RA = rheumatoid arthritis; T1DM = type 1 diabetes.
Odds ratios per SD increase in plasma protein levels and per 10-fold increase in CSF protein levels as MS risk increased.
SNP associated with traits directly.
SNP associated with traits mediated by its proxy.
Comparison of analysed proteins in plasma and CSF
At the protein level, there was a non-significant negative correlation between the CSF and plasma MR results (Spearman correlation coefficient = −0.037, number of proteins = 66, no P-value threshold). Meanwhile, when restricting the number of proteins included in the analysis with different P-value thresholds, a negative correlation was still present and remained insignificant (Supplementary Fig. 3). Further PPI network analysis via text mining and coexpression suggested that plasma-based AHSG and CSF-based CD5L might be connected (Supplementary Fig. 4). Protein–protein MR also revealed that increasing levels of plasma FCRL3 led to increased CSF CD5L (β = 6.883, P = 1.140 × 10−66), whereas higher CSF CD5L inversely reduced plasma FCRL3 (β = −0.106, P = 1.296 × 10−7) (Supplementary Fig. 5). However, protein–protein co-localization did not support plasma–CSF proteins association (Supplementary Fig. 6).
Association of potential drug targets with current multiple sclerosis medications
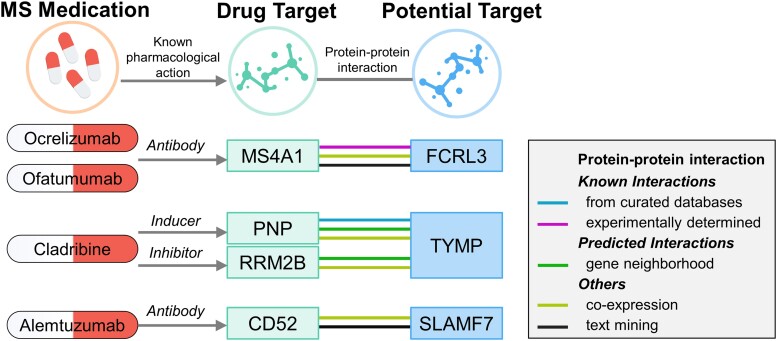
The PPI network revealed interactions between three prioritized proteins (FCRL3, TYMP and SLAMF7) and the targets of four current MS medications. Using STRING, MS4A1–FCRL3 and PNP–TYMP were determined to have the most reliable interactions (known interactions). Specifically, FCRL3 was associated with the B-lymphocyte antigen CD20 (MS4A1), the target of ocrelizumab and ofatumumab. STRING also revealed that FCRL3 and MS4A1 interacted physically, suggesting that the two proteins are in close proximity but not necessarily in direct contact. TYMP is associated with two targets of cladribine, purine-nucleoside phosphorylase (PNP) and ribonucleotide reductase regulatory TP53 inducible subunit M2B (RRM2B). Interestingly, PNP and TYMP were enriched in pyrimidine metabolism. SLAMF7 is associated with the target of alemtuzumab, CAMPATH-1 antigen (CD52) (Fig. 3, Supplementary Table 6 and Supplementary Fig. 4). We also searched for current medications targeting identified potential causal proteins in the Drugbank database. Two medications that might modify the disease were identified: floxuridine (an inducer of TYMP) and elotuzumab (a modulator of SLAMF7) (Supplementary Table 7).
Figure 3.
Interaction between current MS medications targets and identified potential drug targets.
External validation of potential drug targets for multiple sclerosis
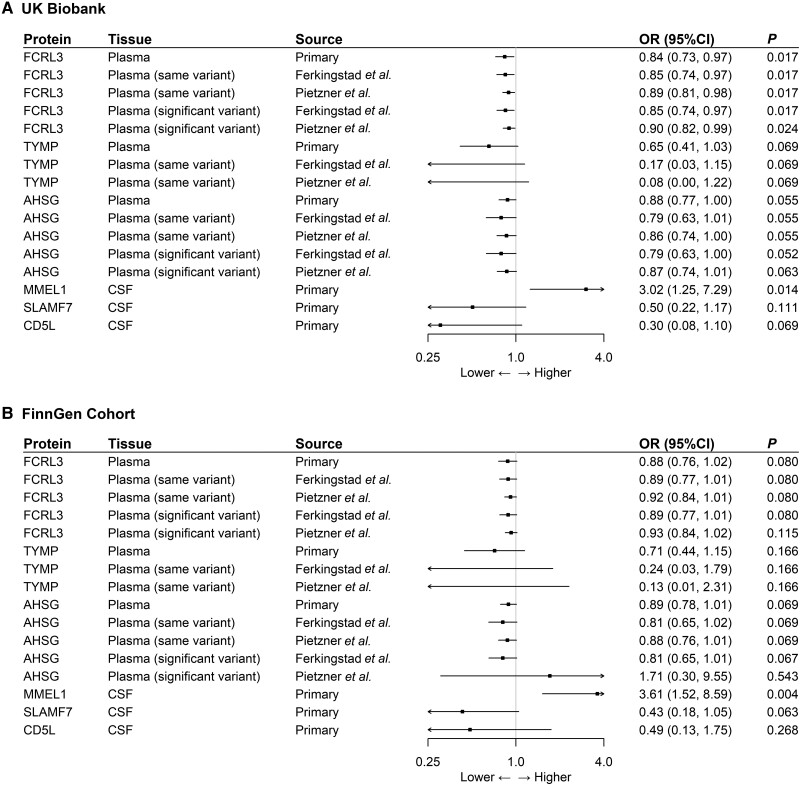
Using the same-variant and significant-variant strategies in different datasets to replicate the primary findings, MMEL1 was also found to be associated with MS in both the UK Biobank and FinnGen cohorts, whereas FCRL3 was associated with MS only in the UK Biobank. For example, using a genome-wide significant variant reported by Ferkingstad et al.16 as a genetic instrument, increasing FCRL3 increased the risk of MS (OR = 0.85; 95% CI, 0.74–0.97; P = 0.017). In addition, AHSG showed a marginally significant causal effect on MS in the UK Biobank (Fig. 4 and Supplementary Table 8).
Figure 4.
External validation of the causal relationship between six potential causal proteins and MS MR analysis on the causal relationship of six potential causal proteins on MS using data from (A) the UK Biobank and (B) the FinnGen cohort. OR for increased risk of MS were expressed as per SD increase in plasma protein levels and per 10-fold increase in CSF protein levels.
Discussion
To the best of our knowledge, this is the first study to combine plasma and CSF proteomic data to explore casual proteins for MS using two-sample MR and Bayesian co-localization. Finally, we identified five proteins as potential drug targets for MS, including circulating FCRL3, TYMP, AHSG, CSF MMEL1 and SLAMF7. Among these proteins, MMEL1 was also found to be associated with MS using a similar approach to analysis in both the UK Biobank and FinnGen cohorts, further suggesting the reliability of the potential drug targets identified in this study.
The human proteome is a major therapeutic target. Therefore, to find the novel drug targets for MS, we employed an integrative analysis that combined MR with co-localization to assess causal proteins for MS as a clinical translation of previous GWAS findings.30 A ‘causality’ identified by MR might be reverse causality, horizontal pleiotropy or genetic confounding due to LD.6 Hence, bidirectional MR was conducted in the study and no proteins identified by the primary MR analysis showed reverse causality, which was further supported by Steiger filtering.31 To limit the bias from horizontal pleiotropy, we only used cis-pQTLs as the instruments, given their direct role in the transcription and/or translation of related genes.32 In addition, Bayesian co-localization was also used to exclude the bias introduced by LD. With 0.8 as the critical threshold for posterior probability, five proteins identified by MR (FCRL3, TYMP, AHSG, MMEL1 and SLAMF7) were likely to share the same variant of MS.33 Four of the five identified proteins (FCRL3, TYMP, AHSG and MMEL1) were found to be associated with other traits via phenotype scanning, but none of the associations could fullyexplain the relationship between identified proteins and MS. For example, we investigated the role of FCRL4 (Fig. 2 and Supplementary Table 1), which was associated with FCRL3-related SNP (rs7528684), but did not find a significant effect. It was also found that IGSF11 and KIR3DL3 are associated with rs7528684 but in a trans-acting manner similar to FCRL4, rendering them less likely to bias the FCRL3–MS association. Phenoscanner also revealed associations between SNP and several autoimmune diseases, while co-occurrence of MS and type 1 diabetes,34 primary sclerosing cholangitis35 and rheumatoid arthritis36 has been reported previously, suggesting a common aetiology of the aforementioned diseases. However, we cannot fully exclude the possibility that APTT might bias the AHSG–MS relationship, as statistically higher APTT levels were reported in patients with MS compared with those in the control group.37 Therefore, FCRL3, TYMP, AHSG, MMEL1 and SLAMF7 might be potential drug targets for MS, but the role of AHSG should be carefully interpreted.
Despite the development of new therapies in recent years, current therapeutic options for PMS remain comparatively disappointing and challenging.38 Considering the pathogenesis of PMS, which is characterized by chronic inflammation compartmentalized behind a closed blood–brain barrier,3 we explored the causal proteins for MS not only in the plasma but also in the CSF and compared their effects thereafter. Notably, in our study, the identified proteins in the plasma and CSF were different, and no correlation of these proteins was found between the plasma and CSF. We also investigated the bidirectional causal relationship among the five prioritized proteins and failed to identify any significant associations. The effect of the blood–brain barrier might explain the absence of a correlation. Although the evidence is still preliminary, these results suggest that both the CSF and plasma might be worthwhile avenues for detecting proteins associated with MS and that the proteins identified in the CSF might be promising drug targets for PMS.
Among the five proteins identified in this study, three (FCRL3, TYMP and MMEL1) have been previously implicated in a recent summary-data–based MR study by Jacob et al.39 Unlike our study, which used the pQTL in the plasma and CSF, this study used the blood expression and methylation QTL. Owing to the importance of protein drug targets for the success of market approval,6,7 our study is a key addition to the research by Jacobs et al.39
Human FCRL3 is a transmembrane glycoprotein encoded by FCRL3.40 As a member of the immunoglobulin receptor superfamily, FCRL3 was preferentially expressed by B lymphocytes and had dual signalling capacity in B cells.40,41 Consistent with our results, previous genetic studies have reported that the high producer variant of the FCRL3 gene protected against MS in Spanish and Chinese populations.42,43 Besides, we found a suggestive linkage between the pQTLs of FCRL3 (rs7528684) and CD5L (rs6427401) (LD r2 = 0.65). Given the failure of co-localization of CD5L and the significant association of FCRL3 and CD5L revealed by protein–protein MR, we speculated that the effect of CD5L in CSF might be a proxy of FCRL3’s effect. In other words, FCRL3 might be the only potential target that acts in both plasma and CSF. In addition, our PPI analysis showed that FCRL3 interacted with MS4A1, the treatment target for ocrelizumab and ofatumumab, two newly licensed medications for PPMS and RMS, respectively. Therefore, FCRL3 may be a promising new druggable target for both RMS and PMS. We also noticed that pQTLs for FCRL3 and CD5L were also reported to be associated with FCRL4 and FCRL1, which might suggest FCRL family proteins might play a crucial role in the pathogenesis of MS and deserve further study. MMEL1 is a member of the M13 family of metalloendopeptidases that contribute to neuropeptide degradation. Similar to our results, a previous GWAS study suggested that MMEL1 is a susceptibility gene for MS.44 Although we did not find that MMEL1 interacted with the target for current MS medications by PPI analysis, it was the only biomarker that was externally validated in both the UK Biobank and FinnGen cohorts in this study. Because MMEL1 is highly expressed in the CNS,44 we hypothesized that it might become a drug target for PMS.
AHSG, TYMP and SLAMF7 were protective proteins against MS in our study. AHSG, also known as fetuin-A, is a multifunctional glycoprotein that plays a role in the secretion levels of some inflammatory cytokines and exosomes.45 The relationship between circulating AHSG levels and MS risk remains uncertain, as previous studies on CSF biomarkers have reported conflicting direction of effects.46–48 Although the different expression patterns of AHSG due to tissue specificity might account for this discrepancy, AHSG was the only protein that was validated by both co-localization methods, indicating a higher probability of AHSG being a causal protein for MS. Future studies are warranted to clarify the directionality of these associations. TYMP is a thymidine phosphorylase that catalyses the reversible phosphorolysis of thymidine. TYMP interacts with PNP and RRM2B, the targets of cladribine, suggesting that TYMP might act by inhibiting DNA synthesis and repair, mainly in lymphocytes, to influence MS.49,50 Notably, TYMP is also an astrocyte-derived blood–brain barrier permeability modulator that interacts with VEGFA, another blood–brain barrier disruption inducer.51,52 Together, TYMP might be a potential therapeutic target for MS, and current floxuridine targeting might affect MS. SLAMF7 is a cell-surface receptor protein selectively expressed on natural killer cells leading to antibody-dependent cellular cytotoxicity and direct natural killer cell activation.53 Consistent with our results, a recent genetic study also observed that the methylation of SLAMF7 was upregulated in B cells of patients with MS, suggesting the association of SLAMF7 with MS.54 Different from the three identified MS targets in this study, the therapeutic agent targeting SLAMF7 has been well developed and evaluated in phase III clinical trials. Elotuzumab, a humanized IgG1 monoclonal antibody that binds to SLAMF7, has been approved for the treatment of relapsed/refractory multiple myeloma. Based on our results, we hypothesized that elotuzumab might have an effect on MS53 and future clinical studies should be conducted.
Limitations
Our study has several limitations. First, we tested the effect of proteins from different studies, and measurement inconsistency across studies might have led to biased results. However, circulating protein data from the GWAS conducted by Sun et al.,18 Emilsson et al.,20 Pietzner et al.15 and Ferkingstad et al.16 were all aptamer-based. Second, all prioritized proteins had only one cis-acting SNP and lacked trans-pQTLs, limiting the application of analyses, including alternative MR algorithms, heterogeneity tests and pleiotropy tests. However, our investigation of the SNPs for the top findings showed that most SNPs were strong instruments with F statistics larger than 10, and the proportion of variance explained more than 10%, except for plasma TYMP and CSF CD5L. Besides, the effect allele frequency of CSF pQTLs was retrieved from matched human genome build, and the corresponding data for CD5L were close to 0.5 (Supplementary Table 1), rendering less reliability of its effect direction. Therefore, the roles of TYMP and CD5L should be carefully interpreted. Third, we performed our analysis on populations of European ancestry and it was difficult to generalize the results to other ancestries. However, we validated the causal role of FCRL3 in the UK Biobank and MMEL1 in both the UK and Finnish populations. More studies on non-European ancestry are required to translate these findings into clinical applications further. Finally, although we found some interactions between the causal proteins and drug targets of current MS medications, the PPI analysis result was suggestive rather than conclusive.
Conclusions
In summary, our integrative analysis suggests that genetically determined levels of circulating FCRL3, TYMP, AHSG, CSF MMEL1 and SLAMF7 are causally associated with MS risk. The identified proteins may be appealing drug targets for MS, especially FCRL3 and SLAMF7. Further studies are needed to explore the roles of these candidate proteins in MS.
Supplementary Material
Acknowledgements
We acknowledge the participants and investigators of the IMSGC, UK Biobank and FinnGen studies.
Contributor Information
Jianfeng Lin, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100730, PR China.
Jiawei Zhou, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100730, PR China.
Yan Xu, Center of Multiple Sclerosis and Related Disorders, Peking Union Medical College Hospital, Beijing, 100730, PR China; Department of Neurology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100730, PR China.
Funding
This study was supported by the Peking Union Medical College Deposition Research Fund (grant number: ZC201904188).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Zhang Y, Xu Y, Xu T, et al. Prediction of long-term disability in Chinese patients with multiple sclerosis: A prospective cohort study. Mult Scler Relat Disord. 2020;46:102461. [DOI] [PubMed] [Google Scholar]
- 2. Dobson R, Giovannoni G. Multiple sclerosis—A review. Eur J Neurol. 2019;26:27–40. [DOI] [PubMed] [Google Scholar]
- 3. Faissner S, Plemel JR, Gold R, Yong VW. Progressive multiple sclerosis: From pathophysiology to therapeutic strategies. Nat Rev Drug Discov. 2019;18:905–922. [DOI] [PubMed] [Google Scholar]
- 4. McGinley MP, Goldschmidt CH, Rae-Grant AD. Diagnosis and treatment of multiple sclerosis: A review. JAMA. 2021;325:765–779. [DOI] [PubMed] [Google Scholar]
- 5. Macaron G, Ontaneda D. Diagnosis and management of progressive multiple sclerosis. Biomedicines. 2019;7(3):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng J, Haberland V, Baird D, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. 2020;52:1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. [DOI] [PubMed] [Google Scholar]
- 8. Reay WR, Cairns MJ. Advancing the use of genome-wide association studies for drug repurposing. Nat Rev Genet. 2021;22:658–671. [DOI] [PubMed] [Google Scholar]
- 9. Chong M, Sjaarda J, Pigeyre M, et al. Novel drug targets for ischemic stroke identified through Mendelian randomization analysis of the blood proteome. Circulation. 2019;140:819–830. [DOI] [PubMed] [Google Scholar]
- 10. Wingo AP, Liu Y, Gerasimov ES, et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer's disease pathogenesis. Nat Genet. 2021;53:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365:eeav7188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang C, Farias FHG, Ibanez L, et al. Genomic atlas of the proteome from brain, CSF and plasma prioritizes proteins implicated in neurological disorders. Nat Neurosci. 2021;24:1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FinnGen. FinnGen Documentation of R6 release. 2022.https://finngen.gitbook.io/documentation/
- 15. Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374:eabj1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53:1712–1721. [DOI] [PubMed] [Google Scholar]
- 17. Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao C, Chen G, Song C, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9:3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Folkersen L, Fauman E, Sabater-Lleal M, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng YT, Ou YN, Wu BS, et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol Psychiatry. 2022;27:2849–2857. [DOI] [PubMed] [Google Scholar]
- 23. Davey Smith G, Hemani G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgess S, Smith GD, Davies NM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamat MA, Blackshaw JA, Young R, et al. Phenoscanner V2: An expanded tool for searching human genotype–phenotype associations. Bioinformatics. 2019;35:4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wishart DS, Feunang YD, Guo AC, et al. Drugbank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szklarczyk D, Gable AL, Lyon D, et al. STRING V11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou W, Nielsen JB, Fritsche LG, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGowan LM, Davey Smith G, Gaunt TR, Richardson TG. Integrating Mendelian randomization and multiple-trait colocalization to uncover cell-specific inflammatory drivers of autoimmune and atopic disease. Hum Mol Genet. 2019;28:3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vimaleswaran KS, Berry DJ, Lu C, et al. Causal relationship between obesity and vitamin D status: Bi-directional Mendelian randomization analysis of multiple cohorts. PLoS Med. 2013;10:e1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–282. [DOI] [PubMed] [Google Scholar]
- 33. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Handel AE, Handunnetthi L, Ebers GC, Ramagopalan SV. Type 1 diabetes mellitus and multiple sclerosis: Common etiological features. Nat Rev Endocrinol. 2009;5:655–664. [DOI] [PubMed] [Google Scholar]
- 35. Sattar M, Poursadeghfard M. Concurrence of multiple sclerosis and primary biliary cholangitis: Report of 3 cases. Caspian J Intern Med. 2020;11:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toussirot E, Pertuiset E, Martin A, et al. Association of rheumatoid arthritis with multiple sclerosis: Report of 14 cases and discussion of its significance. J Rheumatol. 2006;33:1027–1028. [PubMed] [Google Scholar]
- 37. Zhang Y, Zhang X, Liu D, et al. Elevated fibrinogen levels in neuromyelitis optica is associated with severity of disease. Neurol Sci. 2016;37:1823–1829. [DOI] [PubMed] [Google Scholar]
- 38. Correale J, Gaitán MI, Ysrraelit MC, Fiol MP. Progressive multiple sclerosis: From pathogenic mechanisms to treatment. Brain. 2016;140:527–546. [DOI] [PubMed] [Google Scholar]
- 39. Jacobs BM, Taylor T, Awad A, et al. Summary-data-based Mendelian randomization prioritizes potential druggable targets for multiple sclerosis. Brain Commun. 2020;2:fcaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li FJ, Schreeder DM, Li R, Wu J, Davis RS. FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation. Eur J Immunol. 2013;43:2980–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agarwal S, Kraus Z, Dement-Brown J, Alabi O, Starost K, Tolnay M. Human Fc receptor-like 3 inhibits regulatory T cell function and binds secretory IgA. Cell Rep. 2020;30:1292–1299.e3. [DOI] [PubMed] [Google Scholar]
- 42. Matesanz F, Fernández O, Milne RL, et al. The high producer variant of the Fc-receptor like-3 (FCRL3) gene is involved in protection against multiple sclerosis. J Neuroimmunol. 2008;195(1–2):146–150. [DOI] [PubMed] [Google Scholar]
- 43. Yuan M, Wei L, Zhou R, et al. Four FCRL3 gene polymorphisms (FCRL3_3, _5, _6, _8) confer susceptibility to multiple sclerosis: Results from a case-control study. Mol Neurobiol. 2016;53:2029–2035. [DOI] [PubMed] [Google Scholar]
- 44. Ban M, McCauley JL, Zuvich R, et al. A non-synonymous SNP within membrane metalloendopeptidase-like 1 (MMEL1) is associated with multiple sclerosis. Genes Immun. 2010;11:660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Icer MA, Yıldıran H. Effects of fetuin-A with diverse functions and multiple mechanisms on human health. Clin Biochem. 2021;88:1–10. [DOI] [PubMed] [Google Scholar]
- 46. Harris VK, Diamanduros A, Good P, Zakin E, Chalivendra V, Sadiq SA. Bri2-23 is a potential cerebrospinal fluid biomarker in multiple sclerosis. Neurobiol Dis. 2010;40:331–339. [DOI] [PubMed] [Google Scholar]
- 47. Harris VK, Donelan N, Yan QJ, et al. Cerebrospinal fluid fetuin-A is a biomarker of active multiple sclerosis. Mult Scler. 2013;19:1462–1472. [DOI] [PubMed] [Google Scholar]
- 48. Ottervald J, Franzén B, Nilsson K, et al. Multiple sclerosis: Identification and clinical evaluation of novel CSF biomarkers. J Proteomics. 2010;73:1117–1132. [DOI] [PubMed] [Google Scholar]
- 49. Li W, Yue H. Thymidine phosphorylase: A potential new target for treating cardiovascular disease. Trends Cardiovasc Med. 2018;28:157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jacobs BM, Ammoscato F, Giovannoni G, Baker D, Schmierer K. Cladribine: Mechanisms and mysteries in multiple sclerosis. J Neurol Neurosurg Psychiatry. 2018;89:1266–1271. [DOI] [PubMed] [Google Scholar]
- 51. Argaw AT, Gurfein BT, Zhang Y, Zameer A, John GR. VEGF-mediated disruption of endothelial CLN-5 promotes blood–brain barrier breakdown. Proc Natl Acad Sci U S A. 2009;106:1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chapouly C, Tadesse Argaw A, Horng S, et al. Astrocytic TYMP and VEGFA drive blood–brain barrier opening in inflammatory central nervous system lesions. Brain. 2015;138(Pt 6):1548–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Markham A. Elotuzumab: First global approval. Drugs. 2016;76:397–403. [DOI] [PubMed] [Google Scholar]
- 54. Ma Q, Caillier SJ, Muzic S, et al. Specific hypomethylation programs underpin B cell activation in early multiple sclerosis. Proc Natl Acad Sci U S A. 2021;118:e21111920118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome-wide summary-level statistics for cis-pQTL were available from the original studies.12,15–21 GWAS summary statistics of the IMSGC were obtained from https://gwas.mrcieu.ac.uk/datasets/. GWAS summary statistics of the UK Biobank conducted by Zhou et al.29 were obtained from https://www.leelabsg.org/resources. Access to FinnGen (R6 release) can be obtained from https://www.finngen.fi/en/access_results.