Abstract
Focal cortical dysplasia (FCD) type II is a highly epileptogenic developmental malformation and a common cause of surgically treated drug-resistant epilepsy. While clinical observations suggest frequent occurrence in the frontal lobe, mechanisms for such propensity remain unexplored. Here, we hypothesized that cortex-wide spatial associations of FCD distribution with cortical cytoarchitecture, gene expression and organizational axes may offer complementary insights into processes that predispose given cortical regions to harbour FCD.
We mapped the cortex-wide MRI distribution of FCDs in 337 patients collected from 13 sites worldwide. We then determined its associations with (i) cytoarchitectural features using histological atlases by Von Economo and Koskinas and BigBrain; (ii) whole-brain gene expression and spatiotemporal dynamics from prenatal to adulthood stages using the Allen Human Brain Atlas and PsychENCODE BrainSpan; and (iii) macroscale developmental axes of cortical organization.
FCD lesions were preferentially located in the prefrontal and fronto-limbic cortices typified by low neuron density, large soma and thick grey matter. Transcriptomic associations with FCD distribution uncovered a prenatal component related to neuroglial proliferation and differentiation, likely accounting for the dysplastic makeup, and a postnatal component related to synaptogenesis and circuit organization, possibly contributing to circuit-level hyperexcitability. FCD distribution showed a strong association with the anterior region of the antero-posterior axis derived from heritability analysis of interregional structural covariance of cortical thickness, but not with structural and functional hierarchical axes. Reliability of all results was confirmed through resampling techniques.
Multimodal associations with cytoarchitecture, gene expression and axes of cortical organization indicate that prenatal neurogenesis and postnatal synaptogenesis may be key points of developmental vulnerability of the frontal lobe to FCD. Concordant with a causal role of atypical neuroglial proliferation and growth, our results indicate that FCD-vulnerable cortices display properties indicative of earlier termination of neurogenesis and initiation of cell growth. They also suggest a potential contribution of aberrant postnatal synaptogenesis and circuit development to FCD epileptogenicity.
Keywords: focal cortical dysplasia, MRI, epilepsy, neurodevelopment, imaging-genetics
Focal cortical dysplasia (FCD) is particularly likely to affect the frontal lobe. By examining associations between FCD and cytoarchitecture, gene expression and axes of cortical organization, Lee et al. show that frontal lobe vulnerability may reflect relatively early termination of prenatal neurogenesis, as well as aberrant postnatal synaptogenesis.
Introduction
Focal cortical dysplasia (FCD) type II is the most prevalent epileptogenic developmental brain malformation and a common cause of surgically amenable epilepsy.1 This lesion is characterized by cortical dyslamination, cytomegaly and cortical thickening,2 likely due to atypical neuroglial proliferation, growth and migration.3 At a molecular scale, studies in resected FCD tissue have established a causal role of somatic mutations in genes implicated in the mechanistic target of the rapamycin (mTOR) pathway4-7; mTOR hyperactivity disrupts neuronal migration and cortical lamination.7 A recent multiomic study of somatic mutations in hemimegalencephaly and FCD also implicated genes related to calcium dynamics and synaptic function as potential contributors to epileptogenesis.8
Although FCD type II lesions may occur across the entire cortex, histopathological reports of surgically resected tissues in large cohorts1,9,10 as well as a recent atlas of lesion location11 suggest a propensity for frontal lobe involvement. However, mechanisms underpinning this regional vulnerability remain unexplored. Notably, the developing cortex undergoes area-specific, genetically regulated neurogenesis, synaptogenesis and circuit development that give rise to variations in cytoarchitecture.12 Given the strong genetic influence on regional cytoarchitecture,13 it is conceivable that architectural features of the putative FCD-prone cortices may inform on the morphopathogenic characteristics of this malformation.14 Likewise, given the substantial variability of gene expression profiles across the cortex,15 their relation to FCD topology may provide insights into the molecular pathways contributing to the pathogenesis of this brain malformation. Furthermore, cortical organization is thought to be governed by graded macroscale axes, emerging from gene expression,12,16,17 morphology and microstructure18-21 as well as functional and structural connectivity.22,23 Specifically, the antero-posterior axis related to the prenatal timetable of neuroglial proliferation and growth24-26 results in a gradient of neuronal density, size and cortical thickness that persists throughout adulthood.13,20,27 Another increasingly recognized axis marks the transition from sensory to transmodal association cortices.17,21,23,28,29 Recapitulating classic accounts formulated in non-human primates,30 this axis has been thought to mature during late prenatal and early postnatal stages31 and reflect the hierarchical organization of neural function. In sum, cortex-wide spatial associations of FCD distribution with cortical cytoarchitecture, gene expression and organizational axes may offer complementary insights into the neurogenic processes that predispose given cortical regions to harbour this developmental malformation.14,29
Whole-brain cross-modal associations are facilitated by the availability of human brain atlases based on histological features32-34 and spatiotemporal gene expression profiles.35,36 The overall purpose of this work was to investigate the intrinsic regional vulnerability of cortices harbouring FCD. To this end, we mapped the cortex-wide lesional distribution of a multicentric dataset collected from epilepsy centres worldwide, determined cellular and genetic factors based on post-mortem histology and transcriptomics and examined the embedding of FCD lesions within the axes of neurogenic patterning and structure–function hierarchy. Specifically, after creating a topographic map of FCD type II lesions on MRI-derived cortical surface models, we cross-referenced it against histological taxonomies32,33 and a 3D high-resolution human brain histological model.34 In parallel, we performed spatial correlation with whole-brain gene expression data from the Allen Human Brain Atlas (AHBA)35 and examined spatiotemporal gene expression dynamics from prenatal to adulthood stages using the PsychENCODE BrainSpan, an independent development-targeted genetic dataset.36,37 Targeted gene enrichment analysis probed transcriptomic associations for previously known pathogenic FCD variants,3,38,39 as well as non-FCD epilepsies40 and other neurological disorders. Finally, we contextualized the FCD distribution within the antero-posterior axis previously associated with genetic cortical patterning and timetable of neurogenesis,13,24-26 contrasting these findings with hierarchical cortical axes derived from myelin-sensitive MRI28 and resting-state MRI functional connectivity.23
Materials and methods
Study design and participants
We studied a consecutive retrospective cohort of 337 patients (153 females; mean ± SD age = 22.2 ± 12.7 years) with histologically verified FCD lesions collected from 13 tertiary epilepsy centres worldwide. All patients had been investigated for drug-resistant epilepsy with a standard presurgical workup including assessment of seizure history, routine MRI and video-EEG recordings. Histological examination of the surgical specimen2 determined FCD type II as disrupted cortical lamination with dysmorphic neurons in isolation (IIA, n = 134) or together with balloon cells (IIB, n = 203). Site-specific demographics are summarized in Table 1. The Ethics Committees and Institutional Review Boards at all participating sites approved the study, and written informed consent was obtained from all patients.
Table 1.
Site-specific demographics
| Sample size (n) | FCD IIA/IIB | Age (years) | Sex (female/male) | Age at onset (years) | |
|---|---|---|---|---|---|
| All | 337 | 134/203 | 22.2 ± 12.7 | 153/184 | 7.6 ± 6.7 |
| S1 | 114 | 55/59 | 24.8 ± 10.5 | 56/58 | 9.1 ± 7.1 |
| S2 | 8 | 3/5 | 10.5 ± 6.4 | 2/6 | 5.5 ± 4.2 |
| S3 | 10 | 2/8 | 25.3 ± 14.2 | 5/5 | 7.2 ± 7.4 |
| S4 | 43 | 6/37 | 24.3 ± 14.4 | 20/23 | 7.3 ± 7.6 |
| S5 | 18 | 9/9 | 6.8 ± 5.6 | 8/10 | 5.6 ± 4.1 |
| S6 | 22 | 13/9 | 17.4 ± 13.5 | 8/14 | 5.0 ± 4.8 |
| S7 | 11 | 4/7 | 30.8 ± 14.0 | 7/4 | 4.1 ± 3.1 |
| S8 | 14 | 3/11 | 29.1 ± 11.8 | 5/9 | 7.5 ± 5.6 |
| S9 | 8 | 0/8 | 31.9 ± 15.3 | 3/5 | 8.9 ± 4.7 |
| S10 | 14 | 7/7 | 25.3 ± 7.5 | 6/8 | 9.9 ± 5.6 |
| S11 | 11 | 6/5 | 20.8 ± 6.8 | 7/4 | 6.8 ± 8.2 |
| S12 | 42 | 17/25 | 17.0 ± 10.7 | 17/25 | 6.6 ± 5.8 |
| S13 | 22 | 9/13 | 20.9 ± 15.5 | 9/13 | 7.1 ± 8.6 |
Data for age and age at onset are presented as mean ± standard deviation.
MRI acquisition and processing
All patients had high-resolution 3D T1-weighted MRI acquired as a part of the clinical presurgical investigation, consisting of images with isotropic 1 × 1 × 1 mm voxel resolution.41 Data underwent intensity non-uniformity correction and normalization and linear registration to the ICBM MNI152 symmetric template. To generate cortical surface models, we applied the Constrained Laplacian Anatomic Segmentation using Proximity algorithm, yielding GM-WM and GM-CSF surfaces with 41k surface points (or vertices) per hemisphere.42 Surface-based registration, which aligns individual participants based on cortical folding, was performed to optimize vertex-wise anatomical correspondence across participants.43
Cortex-wide MRI mapping of focal cortical dysplasia lesions
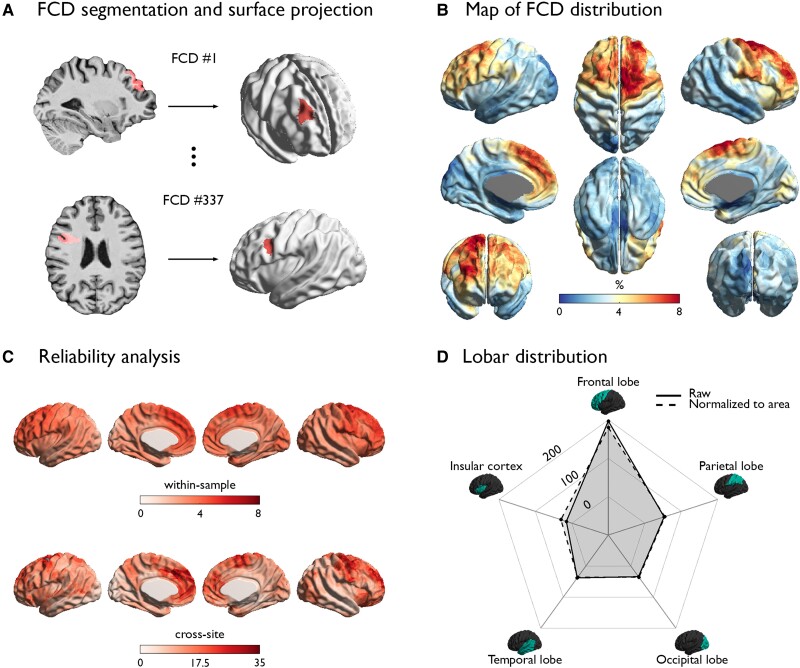
Two experts (A.B., N.B.) independently segmented each FCD lesion on the 3D MRI registered onto the ICBM MNI152 template. The consensus labels (the union of the two segmentations; inter-rate Dice index: 0.94 ± 0.13) was intersected with the cortical surfaces to generate surface-based FCD labels. To enhance regional sensitivity while retaining specificity, labels were minimally smoothed using a surface-based 4 mm full-width at half-maximum Gaussian kernel to maximize local specificity.44 We then calculated for each vertex the FCD probability, defined as the percentage of patients whose lesion label coincided with that vertex. To assess within-sample reliability, we calculated bootstrap certainty at each vertex, defined by mean of lesion probability from the bootstrap subsamples divided by their standard deviation. Similarly, we assessed cross-site reliability as defined by the mean divided by the standard deviation from leave-one-site-out subsamples. We assessed the lobar distribution by counting the number of FCD lesions located within each lobe; to account for lobar size, we divided the lesion counts by the relative surface areas of each lobe, defined based on automated anatomical labelling parcellation atlas.45
Association analyses
Histological atlases
To assess associations of regional FCD probability with histological markers, we used the von Economo–Koskinas MRI atlas (http://dutchconnectomelab.nl) indexed with quantitative histological information (cell size, cell density and cortical thickness) of 43 cortical regions per hemisphere.33 For independent validation, we leveraged the BigBrain atlas, a 3D reconstruction of a stained post-mortem human brain34; these histological data, mapped to intracortical surface models in standard space and to the Schaefer 400 parcellations,46 were obtained from https://github.com/MICA-MNI/micaopen/tree/master/bigbrain.
Cortex-wide gene expression
To investigate the molecular properties of cortical vulnerability, we related the FCD distribution with the anatomically comprehensive gene expression data from the AHBA (six post-mortem adult brains; one female; age = 42.5 ± 13.4 years; https://human.brain-map.org),35 which was mapped onto the 308 parcels of the Desikan–Killiany atlas (DKA).47 The microarray data of these donors were acquired using ∼500 samples per hemisphere, with each sample indexed with expression levels for ∼60 000 genes from at least two probes. Following an established procedure,48 the Maybrain package (https://github.com/rittman/maybrain) matched the closest AHBA sample in each donor to the centroids of 308 parcels of equal area (500 mm2) averaged across donors. Notably, data were averaged across probes corresponding to the same gene, excluding those not matched to gene symbols in the AHBA data. To reduce inter-donor variability, expression data for each probe were normalized through z-transformations across the 308 DKA parcels within each donor. The final output was a matrix of z-scored expression values for each of 20 737 genes mapped onto the 308 DKA parcels.
Spatiotemporal gene expression
We determined how genes associated with the FCD distribution are spatially and temporally regulated throughout pre- and postnatal development. To this end, we used PsychENCODE BrainSpan (http://development.psychencode.org),36 a dataset including tissue-level mRNA-sequencing of 607 samples across 16 anatomical brain regions of 41 post-mortem human brains ranging from eight postconceptional weeks to 40 postnatal years (18 females; post-mortem interval = 12.9 ± 10.4 h; tissue pH = 6.5 ± 0.3; RNA integrity number = 8.8 ± 1). After bulk tissue mRNA-sequencing, this dataset has yielded expression levels for 60 154 genes. The final output consisted of a matrix of reads per kilobase million transcript expression level for each of 17 584 genes overlapping with the 20 737 genes from the AHBA atlas.
Developmental axes of cortical network organization
Gradient axes of cortical structural and functional network organization are shaped by gene expression and cytoarchitecture during the pre- and postnatal development. The antero-posterior axis relates to the prenatal timetable of neurogenesis and growth24-26; we derived this axis from a heritability analysis of structural covariance networks13 mapped on the Schaefer 400 parcellations.46 Structural and functional hierarchical axes are thought to mature during late prenatal and early postnatal circuit development31; we derived these axes from MRI-based covariance of microstructural profiles28 and resting-state functional connectivity,23 which we mapped to the Schaefer 400 parcellations using the BrainSpace toolbox (https://github.com/MICA-MNI/BrainSpace).49 The FCD distribution and developmental axes were mapped to Schaefer 400 parcellations prior to correlation analysis to achieve anatomical correspondence between them.
Statistical analysis
Multivariate analysis
Cortex-wide linear models assessed associations of regional FCD probability with histological markers and neurodevelopmental axes. For the gene expression analysis, given the high dimensionality of AHBA data, we used partial least squares (PLS) regression, a multivariate linear model, to uncover weighted combinations of genes (or PLS components) that best explained the regional variance in FCD probability. The statistical significance of the variance explained by the PLS components was tested based on 10 000 spin permutations of the FCD distribution, accounting for spatial autocorrelations.50 The regional expression profile of each PLS component was defined as the average of the spatial expression profile of 20 757 genes, adjusted by their PLS weight; weight stability was estimated by dividing the PLS weight by the bootstrap SD.
Enrichment analysis
A web-based gene set analysis toolkit (https://webgestalt.org)51 was utilized to uncover biological processes enriched in the list of genes whose bootstrap weights (absolute value) were ranked within the top 10 percentile of 20 757 genes. In other words, this analysis quantified the significance and enrichment ratio, namely the number of PLS-derived genes overlapping with each biological process divided by the number of genes expected to overlap by random permutations.
Spatiotemporal gene expression profiles
Using the PsychENCODE BrainSpan dataset, we calculated the spatiotemporal profile for each PLS component obtained in the gene expression analysis. This profile, defined as the regional average of each gene’s expression level weighted by its bootstrap weight, was obtained across 16 cortical regions and time points based on major neurodevelopmental milestones derived from whole-brain transcriptomic signatures.52 Student’s t-tests compared the expression levels between time windows, and between different regions within time windows.
Specificity analysis
We assessed whether known genes of the pathways causing FCD via somatic mutations were enriched in the PLS components, including the PI3K–AKT–mTOR pathway,5,6,38,53,54 the PI3K–PTEN–AKT–TSC–RHEB pathway,6,53,55-57 the TSC1–TSC2 complex,58-61 the GATOR1 complex6,55,57,59,62-65 and other reported variants (IRS1, RAB6B, ZNF337, RALA and HTR6).61 These genes are listed in Supplementary Table 1. We also assessed associations with risk genes of focal epilepsy with hippocampal sclerosis, generalized epilepsy and all epilepsies as determined by a recent genome-wide association study (GWAS),40 neurodevelopmental conditions, namely autism66 and bipolar spectrum.67 Finally, our specificity analysis included frontotemporal dementia68 due to the preferential involvement of the frontal lobe.
For each PLS component, we quantified the enrichment ratio (ER; defined as the difference between the mean bootstrap weight of the candidate genes and the mean bootstrap weight of the same number of randomly permuted genes), which was then divided by the standard deviation weight of the permutated genes. Significance was determined by percentile of the bootstrap weight of the candidate genes relative to the bootstrap weights of randomly selected genes from 10 000 permutations. Positive/negative ER of a given condition indicates that the risk genes are expressed to a higher/lower degree relative to the baseline expression level. In addition, the function of the risk genes needs to be considered when interpreting ER. For example, the FCD candidate genes are inhibitory regulators of mTOR pathway; thus, negative ER for these genes indicates activation of mTOR pathway.
Corrections for multiple comparisons
For all spatial correlation analyses, findings were corrected using spin permutation tests at Pspin = 0.05.50 Remaining results were corrected for multiple comparisons using false discovery rate (FDR) at 0.05.69
Data availability
The data supporting findings of this study are available from the corresponding author upon request. The datasets are not publicly available as they contain information that could compromise the privacy of the research participants.
Results
Cortex-wide MRI distribution of focal cortical dysplasia
The vertex-wise MRI mapping of FCD lesions across the cortex (Fig. 1) showed aggregation within the frontal lobe, particularly in prefrontal (dorsolateral, ventrolateral, dorsomedial and medial frontopolar; Brodmann areas 4, 9, 10, 44, 45, 46, 57) and cingulate (anterior-mid and pre-genual; Brodmann areas 24, 32, 33) cortices. The reliability of these areas was supported by higher within-sample and cross-site certainty as compared to the other regions. Lobar mapping also confirmed higher occurrence in the frontal lobe compared to other areas, even after normalizing for lobar surface area.
Figure 1.
Cortex-wide FCD distribution. (A) For each patient, the FCD lesion was manually segmented on MRI and mapped onto its cortical surface. (B) Map of FCD distribution. (C) Reliability analysis. Within-sample and cross-site robustness of regional FCD probability is high where the FCD probability is high. (D) Lobar distribution. The spider plot of the FCD distribution across lobes demonstrates remarkable preference towards the frontal lobe, which holds after normalizing for the surface area of each lobe (dotted line).
Association between focal cortical dysplasia distribution and cytoarchitecture
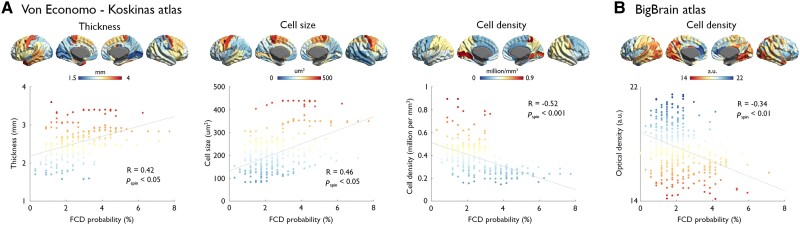
With respect to the von Economo and Koskinas data (Fig. 2), mapping 43 regions per hemisphere, we found a positive correlation between FCD distribution and cortical thickness (R = 0.35, Pspin < 0.05) and cell size (R = 0.46, Pspin < 0.05) and a negative correlation with cell density (R = −0.52, Pspin < 0.001). We also found a negative correlation with cell density obtained from the BigBrain atlas (R = −0.34, Pspin < 0.01). In other words, frontal lobe areas with the highest probability of lesions were those displaying lower neuronal density, larger neurons and higher cortical thickness.
Figure 2.
Associations between FCD distribution and histological measures. Plots show correlations between FCD probability and cortical thickness, cell size and cell density derived from the von Economo–Koskinas atlas (A), as well as cell density (in arbitrary units, a.u.) indexed by optical density of silver-stained cells in the BigBrain atlas (B). In the scatterplots, x- and y-axes represent FCD probability (in %) and histological quantities, respectively; dots indicate 308 parcels of the DKA. Colour-coding is identical for brain maps and dots; Pspin indicates P-value after adjusting for spatial autocorrelation.
Transcriptomic associations and relation to spatiotemporal gene expression
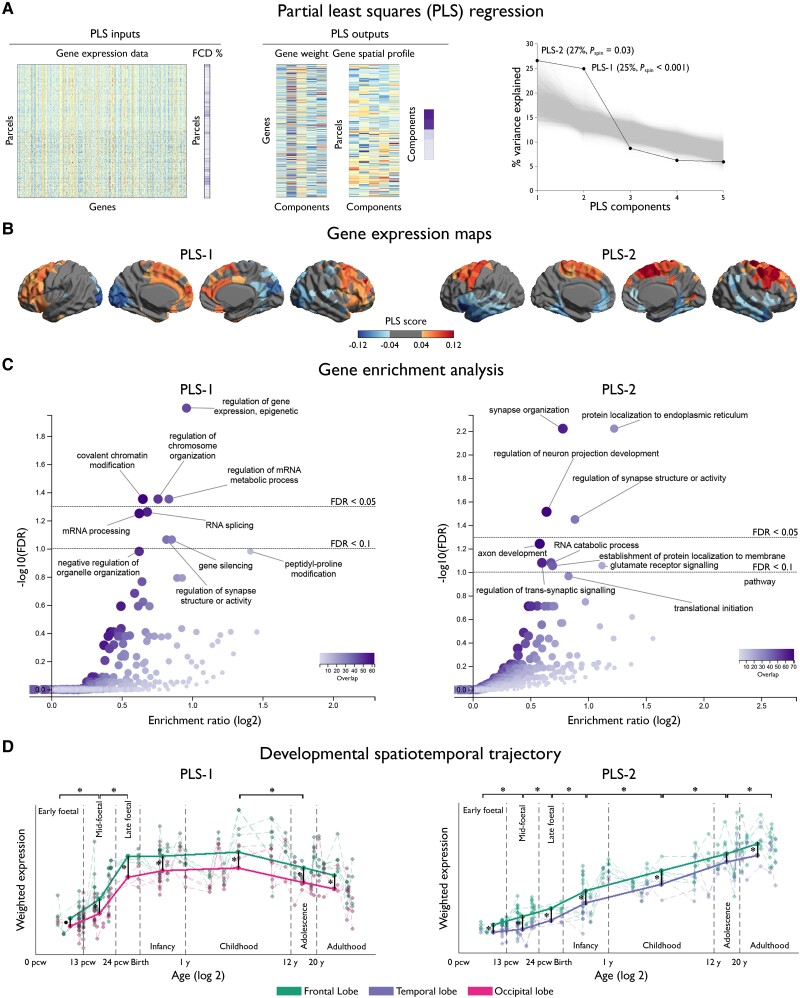
Two PLS components explained 25% (PLS-1: Pspin < 0.001) and 27% (PLS-2: Pspin = 0.03) of the covariance between the FCD probability and AHBA gene expression (Fig. 3). As shown by the gene enrichment analysis, PLS-1 reflected regulation at epigenetic, RNA and post-translational levels, as well as covalent chromatin modification and chromosome organization (FDR < 0.05), both critical for mitotic cell division and differentiation. Conversely, PLS-2 was mainly characterized by general synaptic organization and activity (FDR < 0.05) and marginally by glutamate receptor signalling (FDR < 0.1).
Figure 3.
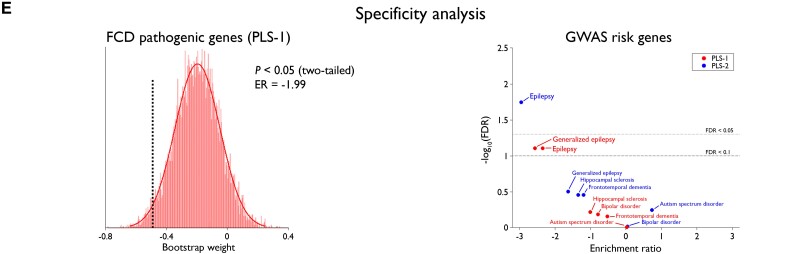
Cortex-wide association between FCD topography and gene expression. (A) PLS regression identified weighted combinations of genes, or PLS components, and their spatial expression profiles that best explained the regional variance in FCD distribution, or per cent variance explained; Pspin indicates P-value after adjusting for spatial autocorrelation). Inputs to PLS include the whole-brain gene expression data matrix (parcels by genes) and FCD distribution across parcels (in %). Outputs include gene weights (genes by components), gene spatial profiles (parcels by components) and per cent variance explained by PLS components. (B) Maps of gene expression. The colour scale indicates the score for PLS-1 and -2, namely the weighted average expression level of 20 737. (C) Gene enrichment analysis. Genes associated with PLS-1 were enriched for epigenetic, RNA and post-translational levels as well as covalent chromatin modification and chromosome organization; and PLS-2 for general synapse organization and activity. In the volcano plots, the x-axis indicates log2 of enrichment ratio and the y-axis indicates -log10 of FDR. Colour codes indicate the number of genes related to the biological processes that overlap with the input list of top 10 percentile genes; upper/lower dotted lines indicate FDR = 0.05/0.1. (D) Developmental spatiotemporal trajectory. The expression of genes associated with PLS-1 sharply increased from early to late foetal stages, plateaued during infancy and childhood, and decreased thereafter. Conversely, PLS-2 showed monotonic increase from early foetal stage to adulthood. In both instances, expressions were more marked in the frontal lobe. Dots represent cortical samples at a given time point colour-coded by lobes; dotted lines connecting dots correspond to the same region of interest. Thick coloured lines connect the average of samples within each time window, thereby showing the overall trajectory. Asterisks indicate FDR < 0.05. (E) Specificity analysis. PLS-1 was significantly enriched for FCD pathogenic genes; the histogram shows bootstrap weights of 10 000 permutations; the dotted line indicates the bootstrap weight of the candidate genes. In relation to GWAS-risk genes, PLS-2 (blue) was enriched for genes associated with all epilepsies, while PLS-1 (red) was marginally enriched for those associated with all and generalized epilepsies. Top dotted line indicates FDR = 0.05; bottom dotted line indicates FDR = 0.1.
Evaluating the developmental spatiotemporal trajectories, the expression of genes associated with PLS-1 sharply increased from early to late foetal stages (FDR < 0.05), plateaued during infancy and childhood and decreased thereafter (FDR < 0.05). Conversely, the expression of genes associated with PLS-2 showed a monotonic increase from early foetal stage to adulthood (FDR < 0.05). Expressions were more marked in the frontal lobe, with a fronto-occipital gradient for PLS-1 and a fronto-temporal gradient for PLS-2. We did not find differential associations between early and late-onset lesional distribution and the PLS components.
Supplementary Table 1 lists the risk genes used for each condition. Specificity analysis revealed that PLS-1 and PLS-2 were enriched for the risk genes of all epilepsies (PLS-1: FDR = 0.08; ER = −2.60; PLS-2: FDR < 0.001, ER = −3.01), with PLS-1 additionally enriched for genes causing FCD via somatic mutations (P < 0.05, ER = −1.99) and risk genes of generalized epilepsy (FDR = 0.08, ER = −2.6). Neither PLS showed associations to genes for focal epilepsy with hippocampal sclerosis, frontotemporal dementia, bipolar or autism spectrum disorders; Supplementary Table 2 provides uncorrected P-values for the enrichment of the GWAS risk genes.
Relation to developmental axes of cortical organization
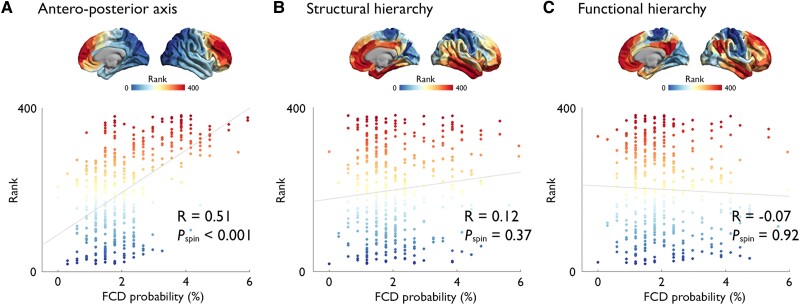
The multisite-derived FCD distribution showed a strong positive association with the anterior region of the antero-posterior axis derived from heritability analysis of interregional structural covariance of cortical thickness (R = 0.51, Pspin < 0.001), but not with structural (R = 0.12, P = 0.37) and functional (R = −0.07, P = 0.92) hierarchical axes (Fig. 4).
Figure 4.
Relation to developmental axes of cortical organization. FCD distribution showed a strong association with the anterior region of the antero-posterior axis derived from heritability analysis of interregional structural covariance of cortical thickness (A), but not with structural (B) and functional (C) hierarchical axes; x- and y-axes represent the FCD probability (in %) and the rank along the gradient axes, also represented as maps. The colour scale represents the percentage of patients in whom the FCD is located at a given vertex.
Discussion
We systematically investigated the cellular, genetic and organizational features of cortices harbouring FCD. Mapping the cortex-wide MRI distribution of 337 histologically verified lesions collected from 13 sites worldwide, we found a propensity for the frontal lobe. Associations with histological markers derived from Von Economo and Koskinas and BigBrain atlases showed that in the healthy brain these areas display lower neuronal density, larger neurons and thicker cortices. Using whole-brain and spatiotemporal gene expression datasets, we identified two genetic factors related to FCD distribution: one defined by prenatal regulation of gene expression and chromosome organization and another related to postnatal synapse organization and activity driving neural circuits.70 At macroscale, FCD distribution was associated with the antero-posterior organizational axis reflective of the timetable of neurogenesis. Concordant with a causal role of atypical neuroglial proliferation and growth, our results indicate that FCD-vulnerable cortices display cytoarchitectural, molecular and organizational properties indicative of earlier termination of neurogenesis and initiation of cell growth. Our findings also suggest a potential contribution of postnatal synaptogenesis and circuit development to FCD epileptogenicity.
While propensity for frontal lobe involvement is in keeping with previous observations,1,9-11 our multisite dataset refined this knowledge by demonstrating locoregional vulnerability of prefrontal and fronto-limbic cortices, the consistency of which was supported by high within-sample and cross-site reliability. Notably, normalizing for lobar surface did not modify results, attesting that such susceptibility is not merely due to the frontal lobe’s larger size, but rather linked to intrinsic developmental, likely multifactorial vulnerability. With respect to cytoarchitectural markers, frontal cortices are typified by lower neuronal density, larger cell soma and thicker grey matter. Given that these are also key histopathological traits of FCD,2,71 the association we found may hint at potential pathophysiological developmental processes linked to intrinsic anatomical characteristics of the prefrontal and fronto-limbic cortices. In this context, the timetables of neurogenesis and synaptogenesis of the prefrontal cortices are distinct from other cortices,72 as they undergo earlier initiation of proliferation, transition from symmetric (cloning) to asymmetric (differentiation) division, reduction of cell cycle rates and termination of neurogenesis, resulting in lower neuronal density. This is followed by early initiation of neuronal growth leading to larger soma and more complex dendritic arborization of frontal relative to occipital cortices.24-26 Hence, although subtle somatic mutations can occur randomly throughout the developing cortex,73 this tighter regulation of neurogenesis in the frontal cortex may explain its heightened susceptibility to harbouring FCD. This longer period of cell growth sets the basis for the frontal neurons to undergo a longer period of synaptogenesis,72,74-76 resulting in the overproduction of synapses and a protracted period of pruning.74,75,77,78 Similarly, limbic cortices, marked by agranular or dysgranular laminar patterns, develop earlier and undergo longer period of synaptic plasticity through adulthood relative to the isocortex.79,80 Fronto-limbic cortices have shown vulnerability for other developmental disorders, such as schizophrenia81,82 and autism,83-86 while temporo-limbic cortices preferentially harbour neurodegenerative disorders, namely Alzheimer’s and Parkinson’s diseases.87-90 Interestingly, tau pathology has been suggested to mediate premature neurodegeneration and cell injury in FCD.91,92 The frontal and limbic regions have been shown to become central hubs in the mature cortical network architecture, which also render themselves vulnerable to structural pathology in numerous lesional and degenerative conditions.93,94
Contextualizing lesional distribution within axes of developmental cortical organization revealed that FCD preferentially occurs in the rostral portion of the anterior–posterior axis defined by genetically determined interregional synchrony of cortical development.13,95,96 Given that this axis reflects the prenatal timetable of neurogenesis and cell growth, the rostral concentration of FCD supports the predisposing roles of aberrant neurogenesis and cell growth as contributors to the histopathological make up of FCD. In contrast, FCD distribution was disassociated from the sensory-association axis established during late prenatal and postnatal neural circuit development,31 a finding consistent with the prenatal occurrence of this malformation.3 A potential genetic underpinning of FCD distribution was also suggested assessing associations to whole-brain gene expression. Indeed, transcriptomic associations based on data-driven PLS regression uncovered a component (PLS-1) reflecting regulation of gene expression at epigenetic, RNA and post-translational levels, as well as covalent chromatin modification and chromosome organization. Chromatin architecture is tightly coupled to mitotic cell cycle and fate. As such, its modification regulated by epigenetic, transcriptional and post-transcriptional mechanisms plays a key role in cell division97 and differentiation.98 Chromosome organization, which involves assembly, arrangement or disassembly of chromosomes, is the process that allows the parent cell to replicate its DNA such that each daughter cell receives a copy during mitosis.99 Therefore, within the cortex, PLS-1 likely represents molecular mechanisms underpinning neuroglial proliferation and differentiation. On the other hand, PLS-2 was related to general synaptic organization and activity, circuit organization,37 as well as glutamate receptor signalling. Evaluating the developmental spatiotemporal trajectories, PLS-1 expression sharply increased from the early foetal stage to late foetal stage, while PLS-2 expression showed steady increase from foetal stages to adulthood. The relevance of these PLS components was supported by the disease specificity analysis. Indeed, while PLS-1 and -2 were both associated with risk genes for all epilepsies, PLS-1 was additionally associated with genes causing FCD via somatic mutations and risk genes of generalized seizures. Therefore, on one hand, it is conceivable that PLS-1 may indicate early cortical vulnerability to aberrant neurogenesis and cell growth, ultimately resulting in a dysplastic lesion. On the other hand, PLS-2 may account for the susceptibility to aberrant synaptogenesis and neurotransmitter systems for hyperexcitable circuits during a latent period following the precipitating lesion,100 thereby promoting epileptogenesis. Although synaptic and white matter maturation have been postulated to contribute to FCD occurrence,101 the presented work is the first to provide evidence for the role of postnatal synaptogenesis and circuit development for FCD epileptogenesis.
Associations with cytoarchitecture, whole-brain and spatiotemporal gene expression, as well as macroscale organizational axes, collectively suggest a vulnerability continuum spanning from prenatal neurogenesis and cell growth to postnatal synaptogenesis. Although age at epilepsy onset has been postulated to account at least partly to variability in FCD histological features,102 the link to molecular or cellular pathogenic processes remains still unclear. In our study, while we did not find differential associations between early and late disease onset lesional distribution with the PLS components, our findings clearly establish developmental underpinnings of FCD occurrence. To date, a plethora of molecular studies of resected FCD tissues have established a causal role of somatic variants that lead to hyperactivity of the mTOR pathway.5,38,39,57,59,61,103-105 A recent large-scale multiomic study of somatic mutations suggested genes implicated in calcium dynamics and synaptic function as potential causes for epileptogenesis.8 Nevertheless, given that the variant allelic frequency is typically below 5% in FCD, uncovering variants distinct from mTOR pathway may be difficult, even with a large sample of resected lesions.59 Notably, the present study circumvents this logistical and statistical burdens by identifying the genetic fingerprints of the FCD-prone cortices based on non-invasive imaging and offers novel insights that may be difficult to obtain otherwise. It has been shown that somatic activating mutations in the mTOR pathway cause a continuum of malformations, spanning from hemimegaloencephaly to posterior quadrantic dysplasia. Although these malformations share some of the genetic determinants with FCD, the time of molecular insult, as well as additional genetic mutations, may lead to varying phenotypes, as suggested by the two-hit germline and somatic mechanisms in hemimegaloencephaly.57 As for the posterior quadrantic dysplasia, prolonged neurogenesis in the posterior isocortex involving higher number and rate of proliferation cycles translates to a greater amplification of abnormal founder cells lesion.106 Subtle structural, possibly neurodevelopmental anomalies have been reported in generalized genetic epilepsy (GGE) and have been described as microdysgenesis in neuropathological studies107,108 that share histological similarity with FCD type IA.109 However, such reports have been sparse, as GGE patients generally do not undergo surgery. Furthermore, the replicability of identifying microdysgenesis in GGE has been limited, thereby not establishing it as a common feature of this condition.110 In terms of genotype–phenotype associations, while the cellular mechanisms that drive the histopathological features of dysplasia are being elucidated,7 those underlying circuit-level alterations that drive recurrent seizures in this condition remain elusive. Conceivably, mitigating the circuit-level alterations precipitated by FCD may reduce seizures.100 Hence, future work should elucidate the molecular and cellular mechanisms of aberrant postnatal synaptogenesis that drive circuit hyperexcitability and identify novel therapeutic targets, possibly combined with mTOR inhibitors, for improved seizure control.
Supplementary Material
Contributor Information
Hyo M Lee, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada.
Seok-Jun Hong, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada; Center for Neuroscience Imaging, Research Institute for Basic Science, Department of Global Biomedical Engineering, SungKyunKwan University, Suwon, KoreaSuwon, Korea.
Ravnoor Gill, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada.
Benoit Caldairou, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada.
Irene Wang, Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH, USA.
Jian-guo Zhang, Department of Functional Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China.
Francesco Deleo, Epilepsy Unit, Fondazione IRCCS Istituto Neurologico C. Besta, Milano, Italy.
Dewi Schrader, Department of Pediatrics, British Columbia Children’s Hospital, Vancouver, Canada.
Fabrice Bartolomei, Aix Marseille Univ, INSERM, INS, Institut de Neurosciences des Systèmes, Marseille, 13005, France.
Maxime Guye, Aix Marseille University, CNRS, CRMBM UMR 7339, Marseille, France.
Kyoo Ho Cho, Department of Neurology, Yonsei University College of Medicine, Seoul, Korea.
Carmen Barba, Meyer Children's Hospital IRCCS, Florence, Italy; University of Florence, 50121 Florence, Italy.
Sanjay Sisodiya, Department of Clinical and Experimental Epilepsy, UCL Queen Square Institute of Neurology, London, UK.
Graeme Jackson, The Florey Institute of Neuroscience and Mental Health and The University of Melbourne, Victoria, Australia.
R Edward Hogan, Department of Neurology, Washington University School of Medicine, St Louis, MO, USA.
Lily Wong-Kisiel, Mayo Clinic, Department of Neurology, Rochester, MN, USA.
Gregory D Cascino, Mayo Clinic, Department of Neurology, Rochester, MN, USA.
Andreas Schulze-Bonhage, Epilepsy Center, University Medical Center-University of Freiburg, Freiburg, Germany.
Iscia Lopes-Cendes, Department of Translational Medicine, School of Medical Sciences, University of Campinas (UNICAMP) and the Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), Campinas SP, Brazil.
Fernando Cendes, Department of Neurology, School of Medical Sciences, University of Campinas (UNICAMP), and the Brazilian Institute of Neuroscience and Neurotechnology (BRAINN), Campinas SP, Brazil.
Renzo Guerrini, Meyer Children's Hospital IRCCS, Florence, Italy; University of Florence, 50121 Florence, Italy.
Boris Bernhardt, Multimodal Imaging and Connectome Analysis Lab, McConnell Brain Imaging Centre, Montreal Neurological Institute and Hospital, McGill University, Montreal, QC, Canada.
Neda Bernasconi, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada.
Andrea Bernasconi, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute, McGill University, Montreal, Canada.
Funding
This work was supported by the Canadian Institutes of Health Research to A.B. and N.B. (MOP-57840 and 123520), Natural Sciences and Engineering Research Council of Canada (Discovery-243141 to AB and 24779 to N.B.), Epilepsy Canada (Jay & Aiden Barker Breakthrough Grant in Clinical & Basic Sciences to A.B.) and Brain Canada. Salary supports were provided by Fonds de Recherche Sante—Quebec, Savoy Foundation for Epilepsy (H.-M.L.), and Lloyd Carr-Harris Foundation (B.C.).
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Blumcke I, Spreafico R, Haaker G, et al. Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377:1648–1656. [DOI] [PubMed] [Google Scholar]
- 2. Blumcke I, Thom M, Aronica E, et al. The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc task force of the ILAE diagnostic methods commission. Epilepsia. 2011;52:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iffland PH, Crino PB. Focal cortical dysplasia: Gene mutations, cell signaling, and therapeutic implications. Annu Rev Pathol. 2017;12:547–571. [DOI] [PubMed] [Google Scholar]
- 4. Jamuar SS, Lam A-TN, Kircher M, et al. Somatic mutations in cerebral cortical malformations. N Engl J Med. 2014;371:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lim JS, Kim WI, Kang HCet al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21:395–400. [DOI] [PubMed] [Google Scholar]
- 6. D'Gama AM, Geng Y, Couto JA, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol. 2015;77:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park SM, Lim JS, Ramakrishina S, et al. Brain somatic mutations in MTOR disrupt neuronal ciliogenesis, leading to focal cortical dyslamination. Neuron. 2018;99:83–97.e7. [DOI] [PubMed] [Google Scholar]
- 8. Chung C, Yang X, Bae T, et al. Comprehensive multiomic profiling of somatic mutations in malformations of cortical development. bioRxiv. [Preprint] 10.1101/2022.04.07.487401 [DOI] [PMC free article] [PubMed]
- 9. Lamberink HJ, Otte WM, Blümcke I, et al. Seizure outcome and use of antiepileptic drugs after epilepsy surgery according to histopathological diagnosis: A retrospective multicentre cohort study. Lancet Neurol. 2020;19:748–757. [DOI] [PubMed] [Google Scholar]
- 10. Gill RS, Lee H-M, Caldairou B, et al. Multicenter validation of a deep learning detection algorithm for focal cortical dysplasia. Neurology. 2021;97:e1571–e1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagstyl K, Whitaker K, Raznahan A, et al. Atlas of lesion locations and postsurgical seizure freedom in focal cortical dysplasia: A MELD study. Epilepsia. 2021;63:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG, Nowakowski TJ. Development and arealization of the cerebral cortex. Neuron. 2019;103:980–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valk SL, Xu T, Margulies DS, et al. Shaping brain structure: Genetic and phylogenetic axes of macroscale organization of cortical thickness. Sci Adv. 2020;6:eabb3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klingler E, Francis F, Jabaudon D, Cappello S. Mapping the molecular and cellular complexity of cortical malformations. Science. 2021;371:eaba4517. [DOI] [PubMed] [Google Scholar]
- 15. Hawrylycz M, Miller JA, Menon V, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89:248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burt JB, Demirtaş M, Eckner WJ, et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat Neurosci. 2018;21:1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paquola C, Benkarim O, DeKraker J, et al. Convergence of cortical types and functional motifs in the human mesiotemporal lobe. Elife. 2020;9:e60673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demirtaş M, Burt JB, Helmer M, et al. Hierarchical heterogeneity across human cortex shapes large-scale neural dynamics. Neuron. 2019;101:1181–1194.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wagstyl K, Ronan L, Goodyer IM, Fletcher PC. Cortical thickness gradients in structural hierarchies. Neuroimage. 2015;111:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huntenburg JM, Bazin P-L, Margulies DS. Large-scale gradients in human cortical organization. Trends Cogn Sci (Regul Ed). 2018;22:21–31. [DOI] [PubMed] [Google Scholar]
- 22. Park B-Y, de Wael RV, Paquola C, et al. Signal diffusion along connectome gradients and inter-hub routing differentially contribute to dynamic human brain function. NeuroImage. 2021;224:117429. [DOI] [PubMed] [Google Scholar]
- 23. Margulies DS, Ghosh SS, Goulas A, et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 2016;113:12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen C-H, Panizzon MS, Eyler LT, et al. Genetic influences on cortical regionalization in the human brain. Neuron. 2011;72:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cahalane DJ, Charvet CJ, Finlay BL. Systematic, balancing gradients in neuron density and number across the primate isocortex. Front Neuroanat. 2012;6:28–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charvet CJ, Finlay BL. Evo-devo and the primate isocortex: The central organizing role of intrinsic gradients of neurogenesis. Brain Behav Evol. 2014;84:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paquola C, Royer J, Lewis LB, et al. The BigBrainWarp toolbox for integration of BigBrain 3D histology with multimodal neuroimaging. eLife. 2021;10:e70119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paquola C, De Wael R V, Wagstyl K, et al. Microstructural and functional gradients are increasingly dissociated in transmodal cortices. PLoS Biol. 2019;17:e3000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sydnor VJ, Larsen B, Bassett DS, et al. Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology. Neuron. 2021;109:2820–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mesulam MM. From sensation to cognition. Brain. 1998;121:1013–1052. [DOI] [PubMed] [Google Scholar]
- 31. Price DJ, Kennedy H, Dehay C, et al. The development of cortical connections. Eur J Neurosci. 2006;23:910–920. [DOI] [PubMed] [Google Scholar]
- 32. von Economo CF, Koskinas GN. Die Cytoarchitektonik der Hirnrinde des erwachsenen Menschen. Springer; 1925. [Google Scholar]
- 33. Scholtens LH, de Reus MA, de Lange SC, Schmidt R, van den Heuvel MP. An MRI von Economo–Koskinas atlas. Neuroimage. 2018;170:249–256. [DOI] [PubMed] [Google Scholar]
- 34. Amunts K, Lepage C, Borgeat L, et al. Bigbrain: An ultrahigh-resolution 3D human brain model. Science. 2013;340:1472–1475. [DOI] [PubMed] [Google Scholar]
- 35. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M, Santpere G, Imamura Kawasawa Y, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Sousa AMM, Gao T, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018;362:eaat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marsan E, Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44:6–17. [DOI] [PubMed] [Google Scholar]
- 39. Avansini SH, Torres FR, Vieira AS, et al. Dysregulation of NEUROG2 plays a key role in focal cortical dysplasia. Ann Neurol. 2018;83:623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Consortium TILAE. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernasconi A, Cendes F, Theodore WH, et al. Recommendations for the use of structural magnetic resonance imaging in the care of patients with epilepsy: A consensus report from the International League Against Epilepsy neuroimaging task force. Epilepsia. 2019;60:1054–1068. [DOI] [PubMed] [Google Scholar]
- 42. Kim JS, Singh V, Lee JK, et al. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. NeuroImage. 2005;27:210–221. [DOI] [PubMed] [Google Scholar]
- 43. Lyttelton O, Boucher M, Robbins S, Evans A. An unbiased iterative group registration template for cortical surface analysis. Neuroimage. 2007;34:1535–1544. [DOI] [PubMed] [Google Scholar]
- 44. Hong S-J, Bernhardt BC, Caldairou B, et al. Multimodal MRI profiling of focal cortical dysplasia type II. Neurology. 2017;88:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. [DOI] [PubMed] [Google Scholar]
- 46. Schaefer A, Kong R, Gordon EM, et al. Local–global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28:3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. [DOI] [PubMed] [Google Scholar]
- 48. Whitaker KJ, Vértes PE, Romero-Garcia R, et al. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci USA. 2016;113:9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Wael RV, Benkarim O, Paquola C, et al. BrainSpace: A toolbox for the analysis of macroscale gradients in neuroimaging and connectomics datasets. Commun Biol. 2020;3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alexander-Bloch AF, Shou H, Liu S, et al. On testing for spatial correspondence between maps of human brain structure and function. NeuroImage. 2018;178:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138:1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Terrone G, Voisin N, Alfaiz AA, et al. De novo PIK3R2 variant causes polymicrogyria, corpus callosum hyperplasia and focal cortical dysplasia. Eur J Hum Genet. 2016;24:1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kobow K, Ziemann M, Kaipananickal H, et al. Genomic DNA methylation distinguishes subtypes of human focal cortical dysplasia. Epilepsia. 2019;60:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schick V, Majores M, Engels G, et al. Activation of Akt independent of PTEN and CTMP tumor-suppressor gene mutations in epilepsy-associated Taylor-type focal cortical dysplasias. Acta Neuropathol. 2006;112:715–725. [DOI] [PubMed] [Google Scholar]
- 57. D’Gama AM, Woodworth MB, Hossain AA, et al. Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21:3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lim JS, Gopalappa R, Kim SH, et al. Somatic mutations in TSC1 and TSC2 cause focal cortical dysplasia. Am J Hum Genet. 2017;100:454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baldassari S, Ribierre T, Marsan E, et al. Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138:885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sim JC, Scerri T, Fanjul-Fernández M, et al. Familial cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann Neurol. 2016;79:132–137. [DOI] [PubMed] [Google Scholar]
- 61. Zhang Z, Gao K, Liu Q, et al. Somatic variants in new candidate genes identified in focal cortical dysplasia type II. Epilepsia. 2020;61:667–678. [DOI] [PubMed] [Google Scholar]
- 62. Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015;77:675–683. [DOI] [PubMed] [Google Scholar]
- 63. Carvill GL, Crompton DE, Regan BM, et al. Epileptic spasms are a feature of DEPDC5 mTORopathy. Neurol Genet. 2015;1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Scerri T, Riseley JR, Gillies G, et al. Familial cortical dysplasia type IIA caused by a germline mutation in DEPDC5. Ann Clin Transl Neurol. 2015;2:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ying Z, Wang I, Blümcke I, et al. A comprehensive clinico-pathological and genetic evaluation of bottom-of-sulcus focal cortical dysplasia in patients with difficult-to-localize focal epilepsy. Epileptic Disord. 2019;21:65–77. [DOI] [PubMed] [Google Scholar]
- 66. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stahl EA, Breen G, Forstner AJ, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ferrari R, Hernandez DG, Nalls MA, et al. Frontotemporal dementia and its subtypes: A genome-wide association study. Lancet Neurol. 2014;13:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 70. Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. [DOI] [PubMed] [Google Scholar]
- 71. Thom M, Martinian L, Sen A, Cross JH, Harding BN, Sisodiya SM. Cortical neuronal densities and lamination in focal cortical dysplasia. Acta Neuropathol. 2005;110:383–392. [DOI] [PubMed] [Google Scholar]
- 72. Kolk SM, Rakic P. Development of prefrontal cortex. Neuropsychopharmacology. 2022;47:41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Y, Bae T, Thorpe J, et al. Comprehensive identification of somatic nucleotide variants in human brain tissue. Genome Biol. 2021;22:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kroon T, van Hugte E, van Linge L, Mansvelder HD, Meredith RM. Early postnatal development of pyramidal neurons across layers of the mouse medial prefrontal cortex. Sci Rep. 2019;9:5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. [DOI] [PubMed] [Google Scholar]
- 76. Lenroot RK, Giedd JN. Brain development in children and adolescents: Insights from anatomical magnetic resonance imaging. Neurosci & biobehav Rev. 2006;30:718–729. [DOI] [PubMed] [Google Scholar]
- 77. Petanjek Z, Judaš M, Šimić G, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 2011;108:13281–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mrzljak L, Uylings HB, Kostovic I, van Eden CG. Prenatal development of neurons in the human prefrontal cortex: I. A qualitative Golgi study. J Comp Neurol. 1988;271:355–386. [DOI] [PubMed] [Google Scholar]
- 79. García-Cabezas MÁ, Zikopoulos B, Barbas H. The structural model: A theory linking connections, plasticity, pathology, development and evolution of the cerebral cortex. Brain Struct Funct. 2019;224:985–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Barbas H. General cortical and special prefrontal connections: Principles from structure to function. Annu Rev Neurosci. 2015;38:269–289. [DOI] [PubMed] [Google Scholar]
- 81. Benes FM, Vincent SL, Todtenkopf M. The density of pyramidal and nonpyramidal neurons in anterior cingulate cortex of schizophrenic and bipolar subjects. Biol Psychiatry. 2001;50:395–406. [DOI] [PubMed] [Google Scholar]
- 82. Park B-Y, Kebets V, Larivière S, et al. Multilevel neural gradients reflect transdiagnostic effects of major psychiatric conditions on cortical morphology. bioRxiv. 2021:2021.10.29.466434. [DOI] [PMC free article] [PubMed]
- 83. Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. Journal of Neuroscience. 2010;30:14595–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wegiel J, Kuchna I, Nowicki K, et al. The neuropathology of autism: Defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Casanova MF, El-Baz AS, Kamat SS, et al. Focal cortical dysplasias in autism spectrum disorders. Acta Neuropathol Commun. 2013;1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Valk SL, Di Martino A, Milham MP, Bernhardt BC. Multicenter mapping of structural network alterations in autism. Hum Brain Mapp. 2015;36:2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer's disease. Cereb Cortex. 1991;1:103–116. [DOI] [PubMed] [Google Scholar]
- 88. Duyckaerts C, CoLLE M-A, Dessi F, Piette F, Hauw J. Progression of Alzheimer histopathological changes. Acta Neurol Belg. 1998;98:180–185. [PubMed] [Google Scholar]
- 89. Braak H, Del Tredici K. Spreading of tau pathology in sporadic Alzheimer’s disease along cortico-cortical top-down connections. Cereb Cortex. 2018;28:3372–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Brettschneider J, Tredici KD, Lee VM-Y, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat Rev Neurosci. 2015;16:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sen A, Thom M, Martinian L, et al. Pathological tau tangles localize to focal cortical dysplasia in older patients. Epilepsia. 2007;48:1447–1454. [DOI] [PubMed] [Google Scholar]
- 92. Iyer A, Prabowo A, Anink J, Spliet WGM, van Rijen PC, Aronica E. Cell injury and premature neurodegeneration in focal malformations of cortical development. Brain Pathology. 2014;24:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Crossley NA, Mechelli A, Scott J, et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sporns O. Towards network substrates of brain disorders. Brain. 2014;137:2117–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Raznahan A, Lerch Jason P, Lee N, et al. Patterns of coordinated anatomical change in human cortical development: A longitudinal neuroimaging study of maturational coupling. Neuron. 2011;72:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 2013;33:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ma Y, Kanakousaki K, Buttitta L. How the cell cycle impacts chromatin architecture and influences cell fate. Review. Front Genet. 2015;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. [DOI] [PubMed] [Google Scholar]
- 99. Batty P, Gerlich DW. Mitotic chromosome mechanics: How cells segregate their genome. Trends Cell Biol. 2019;29:717–726. [DOI] [PubMed] [Google Scholar]
- 100. Goldberg EM, Coulter DA. Mechanisms of epileptogenesis: A convergence on neural circuit dysfunction. Nature Reviews Neuroscience. 2013;14:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cohen NT, You X, Krishnamurthy M, et al. Networks underlie temporal onset of dysplasia-related epilepsy: A MELD study. Ann Neurol. 2022;92:503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fauser S, Huppertz H-J, Bast T, et al. Clinical characteristics in focal cortical dysplasia: A retrospective evaluation in a series of 120 patients. Brain. 2006;129:1907–1916. [DOI] [PubMed] [Google Scholar]
- 103. Møller RS, Weckhuysen S, Chipaux M, et al. Germline and somatic mutations in the MTOR gene in focal cortical dysplasia and epilepsy. Neurology Genetics. 2016;2:e118–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nakashima M, Saitsu H, Takei N, et al. Somatic mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol. 2015;78:375–386. [DOI] [PubMed] [Google Scholar]
- 105. Zhao S, Li Z, Zhang M, et al. A brain somatic RHEB doublet mutation causes focal cortical dysplasia type II. Exp Mol Med. 2019;51:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. D’Agostino MD, Bastos A, Piras C, et al. Posterior quadrantic dysplasia or hemi-hemimegalencephaly. A characteristic brain malformation. Neurology. 2004;62:2214–2220. [DOI] [PubMed] [Google Scholar]
- 107. Kasper BS, Chang BS, Kasper EM. Microdysgenesis: Historical roots of an important concept in epilepsy. Epilepsy Behav. 2009;15:146–153. [DOI] [PubMed] [Google Scholar]
- 108. Najm I, Lal D, Alonso Vanegas M, et al. The ILAE consensus classification of focal cortical dysplasia: An update proposed by an ad hoc task force of the ILAE diagnostic methods commission. Epilepsia. 2022;63:1899–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Palmini A, Najm I, Avanzini G, et al. Terminology and classification of the cortical dysplasias. Neurology. 2004;62(6 suppl 3):S2–S8. [DOI] [PubMed] [Google Scholar]
- 110. Opeskin K, Kalnins RM, Halliday G, Cartwright H, Berkovic SF. Idiopathic generalized epilepsy: Lack of significant microdysgenesis. Neurology. 2000;55:1101–1106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting findings of this study are available from the corresponding author upon request. The datasets are not publicly available as they contain information that could compromise the privacy of the research participants.