Abstract
Objective
To study the association of serum IFNα2 levels measured by ultrasensitive single-molecule array (Simoa) and the IFN-I gene signature (IGS) with disease activity and determine whether these assays can mark disease activity states in a longitudinal cohort of childhood-onset SLE (cSLE) patients.
Methods
Serum IFNα2 levels were measured in 338 samples from 48 cSLE patients and 67 healthy controls using an IFNα Simoa assay. Five-gene IGS was measured by RT-PCR in paired whole blood samples. Disease activity was measured by clinical SELENA-SLEDAI and BILAG-2004. Low disease activity was defined by Low Lupus Disease Activity State (LLDAS) and flares were characterized by SELENA-SLEDAI flare index. Analysis was performed using linear mixed models.
Results
A clear positive correlation was present between serum IFNα2 levels and the IGS (r = 0.78, P < 0.0001). Serum IFNα2 levels and IGS showed the same significant negative trend in the first 3 years after diagnosis. In this timeframe, mean baseline serum IFNα2 levels decreased by 55.1% (Δ 201 fg/ml, P < 0.001) to a mean value of 164 fg/ml, which was below the calculated threshold of 219.4 fg/ml that discriminated between patients and healthy controls. In the linear mixed model, serum IFNα2 levels were significantly associated with both cSELENA-SLEDAI and BILAG-2004, while the IGS did not show this association. Both IFN-I assays were able to characterize LLDAS and disease flare in receiver operating characteristic analysis.
Conclusions
Serum IFNα2 levels measured by Simoa technology are associated with disease activity scores and characterize disease activity states in cSLE.
Keywords: type I IFN, SLE, Simoa, IFN signature, disease activity
Rheumatology key messages.
IFNα detected by Simoa, but not the IGS, is associated with disease activity in cSLE.
Both IFN-I assays can characterize patients with a disease flare or in LLDAS.
Simoa technology could help to select patients for targeted treatment.
Introduction
An accepted paradigm in the pathogenesis of SLE is the role of IFN type I (IFN-I) [1]. This notion is supported by the finding that increased serum IFN-I activity precedes SLE diagnosis in patients at risk and the upregulation of IFN-stimulated genes (ISGs) in the vast majority of SLE patients. The line of evidence is further strengthened by the promising results of anifrolumab (a blocking antibody against the IFN-I receptor (IFNAR)) and tofacitinib (a Janus kinase inhibitor) for the treatment of SLE [2–6]. Measuring IFN-I pathway activation could be considered as a tool to select candidates for these novel treatments. This highlights the need for a validated assay to measure IFN-I for daily clinical use.
So far, there has been great heterogeneity in assays measuring IFN-I pathway activation, as the assays measure different elements of the IFN-I pathway. The most frequently used IFN-I assay describes the downstream cellular response of IFN-I by quantifying the expression of ISGs, followed by calculation of a score known as the IFN-I gene signature (IGS) [7]. The IFN-I family consists of many different subtypes, which generally are expressed at low levels, even under inflammatory conditions. This complicates their measurement by conventional antibody-based techniques [8]. The fairly new ultrasensitive single-molecule array (Simoa) digital ELISA compensates for this caveat by measuring IFN-I proteins at subfemtomolar concentrations [9, 10]. There is substantial evidence, but only in cross-sectional studies, that measurement of IFN-I pathway activation with both IFN-I assays is associated with disease activity in SLE [11–13]. However, longitudinal studies in SLE, which compare the different IFN-I assays in paired samples and study their relation with disease activity over time, are needed to unravel the added value of the Simoa assay to clinical management. Here, we study for the first time the association of IGS and serum IFNα2 levels measured by Simoa with disease activity in a longitudinal cohort of childhood-onset SLE (cSLE) patients and determine their ability to mark specific disease activity states.
Methods
Study design
In this longitudinal cohort study, all patients fulfilled the Systemic Lupus International Collaborating Clinics (SLICC) classification criteria and were diagnosed with SLE before 18 years of age [14]. Demographics, clinical characteristics and history of medication were prospectively collected from the date of inclusion and during each follow-up visit using an electronic case record file. Disease activity was assessed by the clinical Safety of Estrogen in Lupus Erythematosus National Assessment (cSELENA)–SLEDAI and BILAG-2004 [15, 16]. The following numerical scoring scheme was used for the BILAG-2004: A = 12, B = 5, C = 1, D = 0 and E = 0 [17]. Disease flares were indicated by the SELENA-SLEDAI Flare Index (SFI) and low disease activity was assessed by the Low Lupus Disease Activity State (LLDAS) as described before [18]. Written informed consent was obtained from all participants and/or their parents. The study was approved by the Medical Ethics Review Committee of the Erasmus Medical Center (MEC2019-0412).
Type I interferon gene signature
Blood was collected in PAXgene RNA tubes (PreAnalytix GmbH, Becton Dickinson, Vianen, the Netherlands) and was stored in −80°C until use for whole blood RNA purification. RNA isolation (RNA easy mini kit, Qiagen, Hilden, Germany), cDNA preparation (High-Capacity Reverse Transcription kit, Applied Biosystems, Foster City, USA) and real-time PCR (QuantstudioTM 5 Real Time PCR System, Applied Biosystems) were performed according to the manufacturer’s protocol. In short, total RNA was isolated from PAXgene tubes and reverse-transcribed to cDNA. For calculation of relative gene expressions, samples were normalized to expression of the household gene Abl. Whole blood expression of ISGs MxA, IFIT1, IFIT3, IF144 and Ly6e was used to calculate the IGS, according to the following formula:
Relative expression values were determined from normalized CT values using the 2−ΔCT method. Threshold of the IGS was identified by Youden’s index calculated from a receiver operating characteristic (ROC) analysis between healthy controls (HC) and cSLE patients [19].
Ultrasensitive IFNα single-molecule array
IFNα2 was measured in duplicate from serum samples (diluted 2-fold in sample diluent) using the Simoa IFNα Advantage Kit (no. 100860, Quanterix, Billerica, MA, USA) following the manufacturer’s instructions. Sample processing and analysis were done using an HD-X analyser (software version 1.6.1905.300; Quanterix). The lower limit of quantification was 66 fg/ml. In cases in which a sample was below the lower limit of quantification, its value was adjusted to 66 fg/ml for further analysis. The threshold for normal serum IFNα2 levels was identified by Youden’s index calculated from a ROC analysis between HC and cSLE patients [19].
Fluorescent enzyme immune assay for anti-dsDNA levels
Serum samples were diluted 1:10 and added to antigen-coated wells. Recombinant circular plasmid dsDNA was used as antigen. Monoclonal γ-chain-specific anti-human IgG conjugated with β-galactosidase was added to each well after incubation and wash steps. 4-Methylumbelliferyl-β-D-galactoside (0.01%) was then applied as development solution and the reaction was terminated by adding a 4% sodium carbonate. Antibody concentrations ≥20 IU/ml were considered as positive. All samples were run on the Phadia250, a random access immunoassay system (Thermo Fisher Scientific, Freiburg, Germany).
Statistical analysis
All data were analysed using R (version 4.1.0; R Foundation for Statistical Computing, Vienna, Austria). We used package nlme for the mixed models. Non-parametrically distributed data were log-transformed before analysis. In our models the cSELENA-SLEDAI or BILAG-2004 was the outcome (dependent variable) while the assays were included as predictors (independent variables). To correct for confounders, variables such as gender, ethnicity, age, follow-up time and medication use were also included in the model. Spearman’s rho coefficient was calculated to assess correlation. The pROC package (version 1.16.2) was used for ROC analysis [20]. Values of P < 0.05 were considered statistically significant.
Results
Study population
Forty-eight patients with cSLE were included within a total of 338 visits. The median age at enrolment was 15.2 (4.6–18.2) years (median range). The median number of visits was 5 (1–23) with a median follow-up time of 401.5 (0–2518.5) days. The median disease duration at enrolment was 54 (0–2498) days and the median cSELENA-SLEDAI at this time point was 3 (0–14). Fourteen (29.2%) patients ever flared during the study period, which occurred in a median time of 276.5 (111–2591) days. Patients were in LLDAS in the majority (62.1%) of the visits. Ten patients were treatment-naïve at enrolment. All children ≤10 years at diagnosis were tested on monogenetic SLE- and interferonopathy-related genes, which were all found to be negative. Additionally, CH50 and AP50 were determined in all cSLE patients; none had complement deficiency.
Sixty-seven healthy controls were included in the study. The median age was 50 (22–72) years. None of these individuals had symptoms of underlying viral infections or used any medication. More details can be found in Table 1.
Table 1.
Patient characteristics
| Characteristic | cSLE | HC |
|---|---|---|
| Number of individuals | 48 | 67 |
| Total number of visits | 338 | |
| Gender, n (%) | ||
| Male | 7 (14.6) | 6 (9.0) |
| Female | 41 (85.4) | 61 (91) |
| Ethnicity, n (%) | ||
| White | 27 (56.2) | 67 (70) |
| Non-white | 21 (43.8) | 20 (30) |
| Age at enrolment, median (range), years | 15.2 (4.6–18.2) | 50 (22–72) |
| Number of visits, median (range) | 5 (1–23) | |
| Follow-up time, median (range), days | 401.5 (0–2518.5) | |
| Disease duration at enrolment, median (range) | 54 (0–2498) | |
| cSELENA-SLEDAI at enrolment, median (range) | 3 (0–14) | |
| BILAG-2004 at enrolment, median (range) | 4 (0–42) | |
| Patients ever flared, n (%) | 14 (29.2) | |
| Time to first flare, median (range) | 276.5 (111–2591) | |
| Visits ever in LLDAS, n (%) | 210 (62.1) | |
| Positive anti-dsDNA at enrolment, n (%) | 16 (33.3) | |
| Positive auto-antibodies, median (range) | 1 (0–5) | |
| Treatment-naïve at enrolment, n (%) | 10 (20.8) | 67 (100) |
| Treatment ever, n (%) | ||
| Hydroxychloroquine | 48 (100) | |
| Prednisone | 37 (77.1) | |
| Mycophenolate mofetil | 29 (60.4) | |
| NSAIDs | 22 (45.8) | |
| Azathioprine | 3 (6.3) | |
| Cyclophosphamide | 1 (2.1) | |
| Methotrexate | 4 (8.3) | |
| Rituximab | 5 (10.4) | |
| Belumimab | 2 (4.2) |
cSLE: childhood-onset SLE; cSELENA: clinical Safety of Estrogen in Lupus Erythematosus National Assessment; HC: healthy controls; LLDAS: Low Lupus Disease Activity State.
IGS is more accurate in discriminating between HC and cSLE patients than serum IFNα2 levels
Serum IFNα2 levels and IGS were measured in 338 paired samples from the cSLE patients. A positive significant association (r = 0.77; P < 0.0001) was observed between these assays (Supplementary Fig. S1A, available at Rheumatology online). Next, using the first sample from each cSLE patient, a ROC analysis was performed to assess the accuracy in which both assays were able to discriminate between HC and patients. The area under curve (AUC) was significantly higher for the IGS (0.894; P = 0.0004) when compared with the serum IFNα2 levels (AUC = 0.755) (Supplementary Fig. S1B, available at Rheumatology online). In the same analysis, the maximum Youden’s J index was obtained for serum IFNα2 at a threshold of 219.4 fg/ml and 11.7 for the IGS, respectively (Supplementary Fig. S1C, available at Rheumatology online). Using these thresholds, there was a comparable specificity, positive predictive value (PPV) and negative predictive value (NPV) between both assays (Supplementary Fig. S1C, available at Rheumatology online). The sensitivity was yet lower (49%) for the serum IFNα2 levels compared with the IGS (76%) (Supplementary Fig. S1C, available at Rheumatology online). When these thresholds were applied to all visits, the majority of patients had a positive IGS (67.5%) while a smaller proportion had serum IFNα2 levels (37.9%) above the calculated threshold (Supplementary Fig. S1D, available at Rheumatology online). Almost all HC had a serum IFNα2 level and IGS below the calculated threshold. These data support the theoretical overlap between serum IFNα2 levels and IGS, yet indicate IGS as a better IFN-I assay to discriminate between HC and patients.
Both IFN-I assays show the same downward trend as disease activity in the first 1000 days after diagnosis
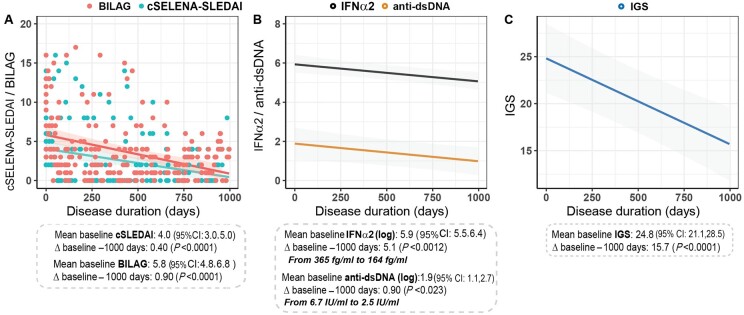
Next, linear mixed models were performed to study the trajectory of disease activity measures and two IFN-I assays in the first 1000 days after diagnosis. In addition, anti-dsDNA autoantibodies were measured due to their known association with disease activity and the fact they are often considered as gold standard to assess disease activity in clinical practice [21]. Patients had a mean baseline cSELENA-SLEDAI of 4.0 (95% CI: 3.0, 5.0). Within 1000 days after diagnosis, cSELENA-SLEDAI decreased significantly by 90% (Δ −3.6, P < 0.0001) to 0.4 (Fig. 1A, blue; Supplementary Table S1, available at Rheumatology online). BILAG-2004 likewise decreased significantly by 84.5% (Δ −4.9, P < 0.0001) to 0.9 (Fig. 1A, red; Supplementary Table S1, available at Rheumatology online). Mean baseline of the log-transformed serum IFNα2 levels was 5.9 (95% CI: 5.5, 6.4). These levels significantly decreased to 5.1 (Δ −0.8, P < 0.0012) within 1000 days. When these log values were converted to numbers, the serum IFNα2 levels decreased by 55.1% to 164 fg/ml (Fig. 1B, black; Supplementary Table S1, available at Rheumatology online). Next, the log-transformed anti-dsDNA levels had a mean baseline of 1.9 (95% CI: 1.1, 2.7), which significantly decreased to 0.9 (Δ −1.0, P < 0.023). Converting the log data to numbers, showed a decrease of 63.2%, which matches with a mean anti-dsDNA level of 2.5 IU/ml after 1000 days (Fig. 1B, orange; Supplementary Table S1, available at Rheumatology online). Lastly, the IGS decreased by 36.7% from 24.8 to 15.7 (Δ −9.1, P < 0.0001; Fig. 1C, Supplementary Table S1, available at Rheumatology online). Although both IFN-I assays showed a significant downward trend in agreement with disease activity, only serum IFNα2 levels reached a value below its threshold within 1000 days after diagnosis indicating that this assay could be more related to disease activity than the IGS.
Figure 1.
IFN-I assays show the same downward trend as disease activity in the first 1000 days after diagnosis. Bold lines indicate the modelled trajectory based on the mixed model results, including the 95% CI (shaded), mean at baseline, Δ and outcome after 1000 days for the cSELENA-SLEDAI (A), BILAG-2004, IFNα2 and anti-dsDNA (B), and IGS (C). The IFNα2 and anti-dsDNA values were log-transformed. cSLEDAI: clinical Safety of Estrogen in Lupus Erythematosus National Assessment–SLEDAI; IGS: IFN type I gene signature
Serum IFNα2 levels but not the IGS are significantly associated with changes in disease activity over time
To analyse the relationship between disease activity and IFN-I assays, multiple linear mixed effects models were built. In each model disease activity assessed by the cSELENA-SLEDAI or BILAG-2004 was used as outcome. In the first model there was a statistically significant association between serum IFNα2 levels and the cSELENA-SLEDAI (Table 2). Here, 1 unit change of log-transformed serum IFNα2 levels was associated with a 71.53% (27.11%, 131.49%; P < 0.001) increase in the cSELENA-SLEDAI (Table 2). In the same model, a 1 unit change of log-transformed anti-dsDNA level was associated with a 47.06% (18.77%, 82.08%; P < 0.001) increase in the cSELENA-SLEDAI. Moreover, 1 unit change of log-transformed disease duration was associated with a 48.66% (62.32%, 30.04%; P < 0.001) decrease in the cSELENA-SLEDAI (Table 2). The second model used the BILAG-2004 as the outcome and showed similar results (Supplementary Table S2, available at Rheumatology online). As there is a high association between serum IFNα2 levels and IGS, the IGS could not be added to the same model. Therefore, serum IFNα2 levels were replaced with the IGS leading to two new models assessing the association between IGS and disease activity. Here, no statistically significant association was found between changes in the IGS and the cSELENA-SLEDAI (P = 0.35) or the BILAG-2004 (P = 0.23) (Supplementary Tables S3 and S4, available at Rheumatology online). Overall, these data highlight serum IFNα2 levels as a good marker for disease activity.
Table 2.
Linear mixed effects model for the cSELENA-SLEDAI and IFNα2, with cSELENA-SLEDAI as the outcome
| Covariate | Estimate | Standard error | P-value | Percentage change (95% CI) |
|---|---|---|---|---|
| (Intercept) | 3.35 | 1.74 | 0.06 | NA |
| Log(IFNα2) | 0.54 | 0.15 | 0.001a | 71.53 (27.11, 131.49) |
| log(anti-dsDNA) | 0.39 | 0.11 | 0.001b | 47.06 (18.77, 82.08) |
| Gender | −0.18 | 0.78 | 0.82 | −16.59 (−81.81, 282.35) |
| Ethnicity | −1.43 | 0.58 | 0.02c | −75.95 (−92.36, −24.33) |
| Baseline age | −0.03 | 0.08 | 0.68 | −3.39 (−17.72, 13.44) |
| Log(disease duration) | −0.67 | 0.16 | 0.001d | −48.66 (−62.32, −30.04) |
| Hydroxychloroquine | −0.1 | 1.04 | 0.92 | −9.95 (−88.22, 588.58) |
| Mycophenolate mofetil | 0.56 | 0.5 | 0.26 | 75.32 (−33.72, 363.76) |
| Prednisone | 0.74 | 0.45 | 0.1 | 109.53 (−12.61, 402.37) |
| NSAID | −0.6 | 0.55 | 0.27 | −45.07 (−81.16, 60.11) |
| Methotrexate | 0.8 | 0.87 | 0.36 | 122.92 (−59.52, 1127.79) |
| Rituximab | −0.01 | 0.89 | 0.99 | −1.14 (−82.62, 462.23) |
| Belumimab | −2.17 | 1.23 | 0.08 | −88.59 (−98.97, 26.16) |
One unit change of log(IFNα2) is associated with a 71.53% (27.11%, 131.49%; P < 0.001) increase in the cSELENA-SLEDAI.
One unit change of log(anti-dsDNA) is associated with a 47.06% (18.77%, 82.08%; P < 0.001) increase in the cSELENA-SLEDAI.
Being from a white ethnic background is associated with a lower risk of having a high disease activity.
One unit change of log(disease duration) is associated with a 48.66% (62.32%, 30.04%; P < 0.001) decrease in the cSELENA-SLEDAI. cSELENA: clinical Safety of Estrogen in Lupus Erythematosus National Assessment.
Disease duration, ethnicity and medication use are associated with disease activity
Patient characteristics and treatment status can influence the association between disease activity and the assays that were included in our linear mixed effects models (Table 2, Supplementary Tables S2–S4, available at Rheumatology online). Disease duration, ethnicity and use of hydroxychloroquine were associated with disease activity. Patients of Caucasian descent had a lower risk of having high disease activity (Table 2). Moreover, a longer disease duration and use of hydroxychloroquine were both associated with lower disease activity in several models (Table 2, Supplementary Table S2–S4, available at Rheumatology online). These models thus highlight several factors that may impact the association between disease activity and the described assays.
Serum IFNα2 levels and IGS are equally able to characterize specific disease activity states
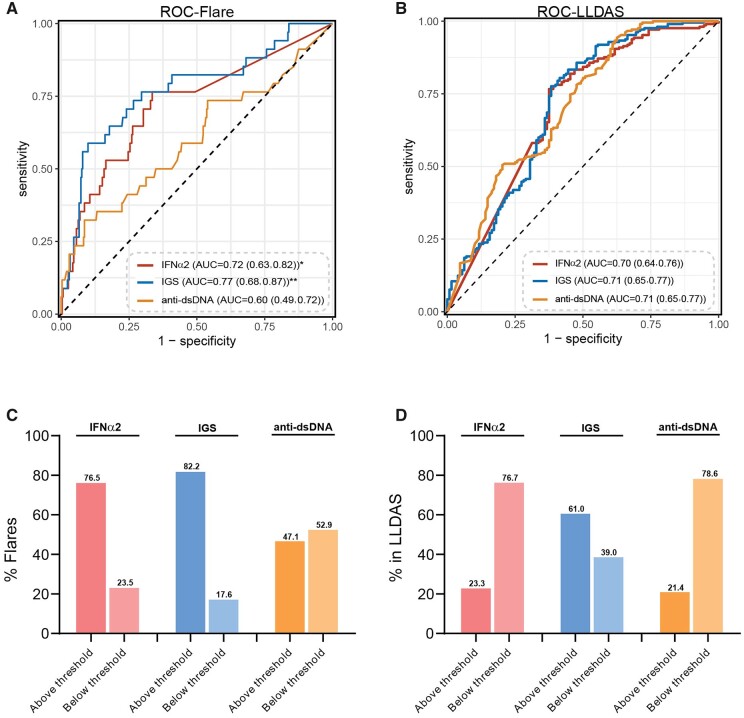
Serum anti-dsDNA level is currently the only serological marker used in the clinic to predict disease flares, despite conflicting results [22]. To assess and compare the accuracy by which the two IFN-I assays and anti-dsDNA levels were able to mark patients in LLDAS or having a disease flare, a ROC analysis was performed with data from all visits. First, when disease flares were studied, the AUC of the anti-dsDNA levels was significantly lower (AUC = 0.60) than the AUC of the serum IFNα2 levels (AUC = 0.72, P = 0.01) and IGS (AUC = 0.77, P = 0.005) (Fig. 2A). The maximum Youden’s J index was obtained at a threshold of 225.9 fg/ml, which gave the assay a sensitivity of 76.5%, specificity of 66.4%, NPV of 96.2% and PPV of 20.3% to identify patients with a disease flare. The IGS and anti-dsDNA had lower sensitivities (58.8% and 32.4%) but higher specificities (90.1% and 91.4%) than the serum IFNα2 to discriminate between patients with or without a flare (Supplementary Table S5, available at Rheumatology online).
Figure 2.
Serum IFNα2 levels and IGS have the ability to mark specific disease activity states. (A) ROC curve of serum IFNα2, IGS and serum anti-dsDNA levels to discriminate between patients with or without a flare. (B) ROC curve of serum IFNα2, IGS and serum anti-dsDNA levels to discriminate between patients who were or were not in LLDAS. (C) Boxplots indicate the percentage of patients that had a disease flare with a value above/below threshold for the three assays (n = 34). (D) Boxplots indicate the percentage of patients that were in LLDAS with a value above/below threshold for the three assays (n = 210). DeLong’s test was used to test the correlation between two ROC curves. *P < 0.01, **P < 0.001. AUC: area under the curve; IGS: IFN type I gene signature; LLDAS: Low Lupus Disease Activity State; ROC: receiver operating characteristic
Next, when LLDAS was studied, the ROC analysis showed a comparable discriminative ability of serum IFNα2 levels (AUC = 0.70), IGS (AUC = 0.71) and anti-dsDNA levels (AUC = 0.71) to identify patients who were in LLDAS (Fig. 2B). The two IFN-I assays had a comparable sensitivity, specificity, PPV and NPV to discriminate between patients who were and were not in LLDAS. Serum anti-dsDNA levels had a higher sensitivity (94.3%) and lower specificity (37.5%) (Supplementary Table S5, available at Rheumatology online). Together, these data indicate that serum IFNα2 levels and IGS perform equally when used to characterize specific disease activity states.
Lastly, we used the previously calculated thresholds that discriminated between HC and cSLE patients to find out if these thresholds are helpful to identify patients in LLDAS and/or flare. For disease flares, 76.5% of the patients had a serum IFNα2 level above threshold (Fig. 2C). This was 82.2% for the IGS and only 47.1% for the serum anti-dsDNA levels (Fig. 2C). For LLDAS, comparable numbers of patients had a serum IFNα2 (76.7%) and anti-dsDNA (78.6%) level below threshold, while only 39% of the IGS were below the calculated threshold (Fig. 2D).
Discussion
Although there is significant evidence for the central role of IFN-I in the pathogenesis of SLE, the debate regarding its applicability as a biomarker for disease activity is complicated by conflicting results from primarily cross-sectional studies, and the scarce data on side-by-side comparison of multiple IFN-I assays in paired samples [10, 23–25]. To our knowledge this is the first longitudinal study comparing the performance of two different IFN-I assays simultaneously in paired samples in relation to disease activity in cSLE. We show that the ultrasensitive Simoa IFNα2 assay but not the IGS associates with disease activity while both can mark specific disease activity states.
Serum IFNα2 levels and IGS are both suitable assays to study IFN-I pathway activation. Although both assays measure different aspects of the pathway, previous work has shown a clear positive association between results obtained from both techniques [10, 24, 25]. We previously have shown that the commercially available Simoa kit that we have used to detect IFN-I proteins in this study primarily detects IFNα2 concentrations [24]. Since the genes with which the IGS is calculated are mainly driven by IFNα2, it is expected that there is a good overlap between the two assays [26]. A subset of patients, however, presents with low serum IFNα2 levels while having a high IGS (Supplementary Fig. S1D, available at Rheumatology online). This might be explained by the role of other IFN-I subtypes, but could also be a consequence of IFN-I independent signalling, which is able to enhance the expression of interferon-stimulated genes [27].
In our longitudinal data, we found that both IFN-I assays showed a decreasing trend over time, which might imply that these assays could reflect disease activity. However, these assays are only considered relevant if their value falls below a predefined threshold, since all values above are considered non-discriminative for clinical practice. In our study, only the serum IFNα2 levels were able to reach a mean value below the calculated threshold. Although the IGS showed a downward trend, its mean value remained above the calculated threshold, confirming previous results showing that the IGS is relatively stable over time and often remains positive even in quiescent disease [28–30]. These data illustrate the caveat of using cross-sectional studies to investigate relations between IFNα2/IGS levels and disease activity as in these studies both assays perform equally well [25]. Analysis of longitudinal data in a linear mixed model takes into account multiple confounding factors, like multiple sampling, the effect of time and medication use. Our linear mixed models showed a significant association between disease activity and serum IFNα2 concentrations but not with IGS. This finding is in line with several other studies that used a different analysis method [23, 28, 31].
The clinical utility of IFN-I assays and anti-dsDNA levels can further be empowered by their ability to mark specific disease activity states. Anti-dsDNA levels have not been able to predict disease flare in serologically active clinically quiescent patients [32]. Serum IFNα2 levels have been shown to be lower in patients who are in disease remission and are considered as a marker to predict risk of relapse in SLE [12, 24]. Likewise, high baseline IGS has been associated to less time spent in LLDAS and a higher chance of disease flare [28, 33]. Our work supports these findings, yet also suggests caution when using the IGS for this matter. When looking at IGS data longitudinally, the IGS remains rather high and is often still elevated in patients in LLDAS (Figs 1C and 2D), which can cause false interpretation of associations. Hence, we recommend serum IFNα2 measurement as the most suitable assay to determine disease activity and disease activity states.
Auto-antibodies targeting IFNa2 can interfere with the Simoa IFNa2 measurement and theoretically lead to discordance between serum IFNa2 levels and IGS [34]. Anti-interferon autoantibodies have been reported in 5–27% of SLE patients [35]. As described, the IGS is driven by multiple IFN-I subtypes, indicating that antibodies that neutralize IFNα2 will not prevent the upregulation of the IGS caused by other IFN-I subtypes. Thus, the presence of auto-antibodies targeting IFNa2 might be an explanation for the discordance observed in Supplementary Fig. S1D, available at Rheumatology online.
This study has several strengths that support the robustness of our findings. These include the longitudinal design of this study, the detailed and prospectively collected disease characteristics including measuring both cSELENA-SLEDAI and BILAG-2004, and long-term follow-up of the patients. The relatively low number of patients is a potential caveat. In contrast to adult-onset SLE, cSLE is a rare disease with much lower prevalence [36]. However, statistical analysis revealed clear results underlining the robustness of the data. The Simoa is not approved for clinical practice yet and to date no clinical development is planned for this technology. More studies using the Simoa technique for measuring serum IFNa2 levels are needed first to confirm its value for clinical practice. Lastly, the patients in our cohort had relatively low disease activity (mean cSELENA-SLEDAI = 4), this highlights the need for a validation cohort with higher disease activity in the future.
In conclusion, serum IFNα2 is a good marker for monitoring of treatment responses and disease activity as it is susceptible to changes over time, associates with disease activity in a longitudinal cohort and marks disease activity states more accurately than the IGS and anti-dsDNA antibody levels. Hence, this technique deserves to be studied further and tested as a potential biomarker to study disease activity in clinical practice.
Supplementary Material
Contributor Information
M Javad Wahadat, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands; Department of Paediatric Rheumatology, Sophia Children’s hospital, Erasmus University Medical Center, Rotterdam, The Netherlands.
Hongchao Qi, Department of Biostatistics, Erasmus University Medical Center, Rotterdam, The Netherlands.
Cornelia G van Helden-Meeuwsen, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Erika Huijser, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Lotte van den Berg, Department of Paediatric Rheumatology, Sophia Children’s hospital, Erasmus University Medical Center, Rotterdam, The Netherlands.
Annette van Dijk-Hummelman, Department of Paediatric Rheumatology, Sophia Children’s hospital, Erasmus University Medical Center, Rotterdam, The Netherlands.
Jens C Göpfert, Department of Applied, Biomarkers and Immunoassays, NMI Natural and Medical Sciences Institute at the University of Tübingen, Reutlingen, Germany.
Anne Heine, Department of Applied, Biomarkers and Immunoassays, NMI Natural and Medical Sciences Institute at the University of Tübingen, Reutlingen, Germany.
Marleen Verkaaik, Department of Paediatric Rheumatology, Sophia Children’s hospital, Erasmus University Medical Center, Rotterdam, The Netherlands.
Marco W J Schreurs, Laboratory Medical Immunology, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Willem A Dik, Laboratory Medical Immunology, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Sylvia Kamphuis, Department of Paediatric Rheumatology, Sophia Children’s hospital, Erasmus University Medical Center, Rotterdam, The Netherlands.
Marjan A Versnel, Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Supplementary material
Supplementary material is available at Rheumatology online.
Data availability
All data that support the findings of this study, including R-scripts used, de-identified patient-level data, results from assays used etc., are available on request from the corresponding author.
Contribution statement
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published.
Funding
This work was made possible by the support of the Sophia Children’s Hospital Fund [B18-04], NVLE (Dutch patient organization for Lupus, APS, Scleroderma and MCTD) [BP12-1-261] and Dutch Arthritis Society [CO-19-001]. The research was performed within the framework of the Erasmus Postgraduate School Molecular Medicine. N.M.I. received financial support from the State Ministry of Baden-Wuerttemberg for Economic Affairs, Labour and Tourism.
Disclosure statement: The authors have declared no conflicts of interest.
Study approval: Written informed consent was obtained from all participants in compliance with the Declaration of Helsinki. The study was approved by the medical ethics review committee of the Erasmus University Medical Center, Rotterdam, the Netherlands (MEC-2019-0412).
References
- 1. Bengtsson AA, Ronnblom L. Role of interferons in SLE. Best Pract Res Clin Rheumatol 2017;31:415–28. [DOI] [PubMed] [Google Scholar]
- 2. Banchereau R, Hong S, Cantarel B et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 2016;165:551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morand EF, Furie R, Tanaka Y et al. ; TULIP-2 Trial Investigators. Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med 2020;382:211–21. [DOI] [PubMed] [Google Scholar]
- 4. Vital EM, Merrill JT, Morand EF et al. Anifrolumab efficacy and safety by type I interferon gene signature and clinical subgroups in patients with SLE: post hoc analysis of pooled data from two phase III trials. Ann Rheum Dis 2022;81:951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munroe ME, Lu R, Zhao YD et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann Rheum Dis 2016;75:2014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hasni SA, Gupta S, Davis M et al. Phase 1 double-blind randomized safety trial of the Janus kinase inhibitor tofacitinib in systemic lupus erythematosus. Nat Commun 2021;12:3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgs BW, Liu Z, White B et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis 2011;70:2029–36. [DOI] [PubMed] [Google Scholar]
- 8. Bave U, Nordmark G, Lovgren T et al. Activation of the type I interferon system in primary Sjögren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum 2005;52:1185–95. [DOI] [PubMed] [Google Scholar]
- 9. Rissin DM, Kan CW, Campbell TG et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 2010;28:595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodero MP, Decalf J, Bondet V et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med 2017;214:1547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathian A, Mouries-Martin S, Dorgham K et al. Monitoring disease activity in systemic lupus erythematosus with single-molecule array digital enzyme-linked immunosorbent assay quantification of serum interferon-α. Arthritis Rheumatol 2019;71:756–65. [DOI] [PubMed] [Google Scholar]
- 12. Mathian A, Mouries-Martin S, Dorgham K et al. Ultrasensitive serum interferon-α quantification during SLE remission identifies patients at risk for relapse. Ann Rheum Dis 2019;78:1669–76. [DOI] [PubMed] [Google Scholar]
- 13. Kirou KA, Lee C, George S et al. Activation of the interferon-α pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum 2005;52:1491–503. [DOI] [PubMed] [Google Scholar]
- 14. Petri M, Orbai AM, Alarcon GS et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petri M, Kim MY, Kalunian KC et al. ; OC-SELENA Trial. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med 2005;353:2550–8. [DOI] [PubMed] [Google Scholar]
- 16. Isenberg DA, Rahman A, Allen E et al. ; BILAG 2004. Development and initial validation of an updated version of the British Isles Lupus Assessment Group's disease activity index for patients with systemic lupus erythematosus. Rheumatology (Oxford) 2005;44:902–6. [DOI] [PubMed] [Google Scholar]
- 17. Cresswell L, Yee CS, Farewell V et al. Numerical scoring for the Classic BILAG index. Rheumatology (Oxford) 2009;48:1548–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franklyn K, Lau CS, Navarra SV et al. ; Asia-Pacific Lupus Collaboration. Definition and initial validation of a Lupus Low Disease Activity State (LLDAS). Ann Rheum Dis 2016;75:1615–21. [DOI] [PubMed] [Google Scholar]
- 19. Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32–5. [DOI] [PubMed] [Google Scholar]
- 20. Robin X, Turck N, Hainard A et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Hoyos M, Cabeza R, Martinez-Taboada VM et al. Clinical disease activity and titers of anti-dsDNA antibodies measured by an automated immunofluorescence assay in patients with systemic lupus erythematosus. Lupus 2005;14:505–9. [DOI] [PubMed] [Google Scholar]
- 22. Gensous N, Marti A, Barnetche T et al. ; FHU ACRONIM. Predictive biological markers of systemic lupus erythematosus flares: a systematic literature review. Arthritis Res Ther 2017;19:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landolt-Marticorena C, Bonventi G, Lubovich A et al. Lack of association between the interferon-α signature and longitudinal changes in disease activity in systemic lupus erythematosus. Ann Rheum Dis 2009;68:1440–6. [DOI] [PubMed] [Google Scholar]
- 24. Huijser E, Gopfert J, Brkic Z et al. Serum interferon-α2 measured by single-molecule array associates with systemic disease manifestations in Sjögren's syndrome. Rheumatology (Oxford) 2022;61:2156–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chasset F, Mathian A, Dorgham K et al. Serum interferon-α levels and IFN type I-stimulated genes score perform equally to assess systemic lupus erythematosus disease activity. Ann Rheum Dis 2022;81:901–3. [DOI] [PubMed] [Google Scholar]
- 26. Moll HP, Maier T, Zommer A, Lavoie T, Brostjan C. The differential activity of interferon-α subtypes is consistent among distinct target genes and cell types. Cytokine 2011;53:52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Xu L, Brandsma JH et al. Convergent transcription of interferon-stimulated genes by TNF-α and IFN-α augments antiviral activity against HCV and HEV. Sci Rep 2016;6:25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Northcott M, Gearing LJ, Nim HT et al. Glucocorticoid gene signatures in systemic lupus erythematosus and the effects of type I interferon: a cross-sectional and in-vitro study. Lancet Rheumatol 2021;3:e357–70. [DOI] [PubMed] [Google Scholar]
- 29. Petri M, Fu W, Ranger A et al. Association between changes in gene signatures expression and disease activity among patients with systemic lupus erythematosus. BMC Med Genomics 2019;12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tesser A, de Carvalho LM, Sandrin-Garcia P et al. Higher interferon score and normal complement levels may identify a distinct clinical subset in children with systemic lupus erythematosus. Arthritis Res Ther 2020;22:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petri M, Singh S, Tesfasyone H et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus 2009;18:980–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steiman AJ, Urowitz MB, Ibanez D et al. Anti-dsDNA and antichromatin antibody isotypes in serologically active clinically quiescent systemic lupus erythematosus. J Rheumatol 2015;42:810–6. [DOI] [PubMed] [Google Scholar]
- 33. Mai L, Asaduzzaman A, Noamani B et al. The baseline interferon signature predicts disease severity over the subsequent 5 years in systemic lupus erythematosus. Arthritis Res Ther 2021;23:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slavikova M, Schmeisser H, Kontsekova E et al. Incidence of autoantibodies against type I and type II interferons in a cohort of systemic lupus erythematosus patients in Slovakia. J Interferon Cytokine Res 2003;23:143–7. [DOI] [PubMed] [Google Scholar]
- 35. Mathian A, Breillat P, Dorgham K et al. Lower disease activity but higher risk of severe COVID-19 and herpes zoster in patients with systemic lupus erythematosus with pre-existing autoantibodies neutralising IFN-α. Ann Rheum Dis 2022;81:1695–1703. [DOI] [PubMed] [Google Scholar]
- 36. Levy DM, Kamphuis S. Systemic lupus erythematosus in children and adolescents. Pediatr Clin North Am 2012;59:345–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study, including R-scripts used, de-identified patient-level data, results from assays used etc., are available on request from the corresponding author.