Abstract
Objectives
ANCA-associated vasculitis (AAV) includes granulomatosis with polyangiitis (GPA) and microscopic polyangiitis (MPA). ANCA triggers neutrophil extracellular trap formation, which releases either mitochondrial (mt) DNA or nuclear DNA (n) DNA, contributing to inflammation. Our aim was to prospectively examine the extent and nature of circulating DNA in AAV and the clinical utility of DNA quantification.
Methods
DNA was isolated from platelet-free plasma of consecutive GPA and MPA patients and healthy controls (HCs). mtDNA and nDNA copy numbers were quantified by PCR. Clinical data, including the BVAS, were collected.
Results
Ninety-two HCs (median age 51 years, 58.7% female) and 101 AAV patients (80 GPA, 21 MPA, median age 64 years, 50.5% female, BVAS range: 0–30) were included. Median mtDNA copies were 13-fold higher in patients with AAV than in HCs; nDNA concentrations did not differ. Patients with active AAV (BVAS > 0) had 4-fold higher median mtDNA copies than patients in remission (P = 0.03). mtDNA, unlike nDNA, correlated with BVAS (r = 0.30, P = 0.002) and was associated with AAV activity at multivariable analysis. Receiver operating characteristic curve analysis indicated that mtDNA quantification differentiates patients with active AAV (BVAS > 0) from HCs with 96.1% sensitivity and 98.9% specificity (area under the curve 0.99). In 27 AAV patients with follow-up, mtDNA changes but not CRP or ANCA-titres correlated with BVAS changes (r = 0.56, P = 0.002).
Conclusions
mtDNA, unlike nDNA, is elevated in the plasma of AAV patients and may contribute to systemic inflammation. mtDNA could be superior to established biomarkers in the laboratory monitoring of AAV activity.
Keywords: ANCA-associated vasculitis, innate immunity, mitochondrial DNA, neutrophil extracellular traps, disease activity
Rheumatology key messages.
The quantification of cell-free mtDNA allows sensitive detection of patients with AAV, even those with quiescent disease.
Plasma mtDNA levels correlate cross-sectionally with AAV activity, and within individual AAV patients longitudinally.
mtDNA quantification could become clinically relevant in supporting AAV diagnosis and monitoring of disease activity.
Introduction
Granulomatosis with polyangiitis (GPA) and microscopic polyangiitis (MPA) are two closely related ANCA-associated vasculitides (AAVs) [1]. In GPA, ANCA are predominantly directed against the neutrophilic PR3 [2], whereas in MPA, ANCA are more frequently directed against MPO [3].
ANCA may have a direct role in the pathogenesis of GPA by activating primed neutrophils to release cell-free (cf) DNA in the form of neutrophil extracellular traps (NETs) [4, 5], a process in which DNA is externalized into the extracellular space [6]. It has been demonstrated recently that endothelial cell activation also promotes NET formation and that, conversely, NETs induce endothelial cell injury [5]. There is an increasing body of evidence supporting a fundamental role of NETosis and the consequent release of cfDNA in the pathogenesis of AAV [7–9]. Polymorphonuclear granulocytes may either release nuclear DNA (nDNA) or mitochondrial DNA (mtDNA) during NETosis. Still, it is so far unclear which of the two DNA species (nDNA or mtDNA) is more abundant in the systemic circulation and whether DNA quantification in plasma can be exploited clinically.
mtDNA is a dsDNA molecule phylogenetically derived from bacteria and rich in hypomethylated CpG sequences [10], and therefore is able to trigger a broad range of pro‐inflammatory signalling pathways, including the endosomal toll-like receptor (TLR)-9 and a series of cytosolic sensors [11–13]. mtDNA released by neutrophils in AAV could therefore be particularly well suited to promote disease flares and systemic immune-mediated organ injury [11, 13, 14]. The mechanisms governing the release of nDNA or mtDNA by NETosis are not well defined. AAV pathogenesis may therefore be driven by either circulating cf nDNA or mtDNA, or both DNA species in combination [15, 16].
To better understand the pathogenic and clinical role of cfDNA species in AAV, we aimed to quantify the abundance of mtDNA and nDNA in AAV patient plasma and, furthermore, to assess whether cell-free nucleic acid quantification may be helpful in the diagnosis and monitoring of AAV patients.
Methods
Study design and subjects
The study was approved by the ethics committee of Northwest and Central Switzerland (No. 2019–01693) and by the ethics committee of the University of Freiburg (No. 507/16). After ethics committee approval, adult patients classified as having AAV (GPA or MPA) [2, 3] and attending our tertiary care centres were consecutively recruited, provided that written informed consent was obtained. Active systemic infections, recent major trauma or surgery, malignancy, and concomitant secondary systemic autoimmune diseases were excluded as potential confounders of circulating DNA levels [17, 18]. AAV activity was scored by means of the BVAS (version 3) [19], and anti-PR3 and anti-MPO autoantibody levels were measured. We also prospectively collected patient plasma at follow-up visits scheduled independently from AAV activity. Adult volunteer blood donors, consecutively recruited from the Basel University Hospital blood bank and hospital staff deemed free of inflammatory disease, served as healthy controls (HCs). The study was approved by the regional ethical committees, and all procedures were under the Declaration of Helsinki [20]. Clinical information and reference standard results were available to the performers of the quantitative real-time (q) PCR testing. The Standards for Reporting of Diagnostic Accuracy (STARD) checklist for this study is provided in Supplementary Table S1, available at Rheumatology online [21].
Isolation of total DNA from plasma and quantification of circulating DNA copy numbers
In all subjects, 7 ml of peripheral venous blood was collected in an ethylenediaminetetraacetic acid (EDTA) tube and processed following previously established guidelines [22]. Briefly, platelet-poor plasma was obtained up to 4 h after blood collection using two centrifugation steps at room temperature (1200 g for 10 min followed by 16 000 g for another 10 min) [23]. The platelet-poor plasma was collected without disturbing or aspirating the buffy coat, and aliquoted and stored at –80°C until further processing.
Total DNA was extracted from 500 μl of platelet-poor plasma utilizing the QIAamp DNA Blood Mini kit (QIAgen, Hilden, Germany). The concentration and purity of the obtained total cell-free (cf) DNA were spectrophotometrically quantified with a NanoDrop ND‐1000 (Nano‐Drop Technologies, Wilmington, DE, USA). Eluates were stored at –20°C prior to analysis by qPCR.
mtDNA and nDNA copy numbers were quantified by qPCR as described [24]. Briefly, a PCR protocol utilizing SYBR Green DNA intercalating dye on an Applied Biosystems StepOne Plus Real-Time PCR system (Thermo Scientific, Wilmington, DE, USA) was applied in 96-well plates. The mtDNA ATP-6 gene was amplified between nucleotide positions 8981 and 9061. For the detection of nDNA, we selected exon number 8 of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene between nucleotide positions 4280 and 4342. The specificity of the PCR products was assessed with a melting curve. All measurements were performed in triplicates. A negative ‘no template’ control was included, and two standard curves were generated by amplifying mtDNA- and nDNA-containing vectors with known copy numbers. Absolute mtDNA and nDNA copy numbers in the PCR template were calculated from the standard curves. DNA plasma concentrations were calculated, accounting for the initial volume of plasma from which DNA was extracted, the elution volume, the DNA concentration in the eluate and the amount of DNA template used in each qPCR reaction. A DNA sample from the same specific HC donor was repeatedly included in each PCR plate to assess the inter-assay coefficient of variation (CV).
Statistical analysis
The required sample size was evaluated as previously described [25]. GraphPad Prism software for macOS, V.9.0 (GraphPad Software, Inc., La Jolla, CA, USA), was utilized for statistical analysis. Non-parametric values were expressed as the median and interquartile range (IQR). One-way analysis of variance combined with the Mann–Whitney U test or unpaired t test was used to evaluate statistically significant differences between groups, as appropriate. Spearman’s rank tests were applied to analyse correlations. Within groups, variables were assessed for their predictive power through linear regression analysis utilizing MATLAB R2018a (The Mathworks, Natick, MS, USA). Multiple variable analysis was performed by applying all variables with a P-value below 0.1 into a generalized linear regression model.
Results
Study subjects
One hundred and one patients with AAV were recruited. Eighty (79.2%) of these patients were classified as having GPA, and 21 as having MPA (20.8%) [2, 3]. The median age of the patients with AAV was 64 years (range 19–89 years), their median disease duration was 8 years (range 1–46 years), and 51 patients (50.5%) were female. The median BVAS at the time of blood collection was 0 (range: 0–30). Twenty-one patients presented with active AAV (BVAS > 0); in 8 patients (7.9%), the vasculitis was highly active at the time of blood sampling, as indicated by a BVAS of >10 [26]. Sixty-three patients (62.4%) were on prednisone therapy; these patients were receiving a median daily prednisone dose of 5.0 mg (range: 1–250 mg; IQR: 7.0 mg). Only 15 AAV patients had a daily prednisone dose of >10 mg. Moreover, a series of other immunosuppressive drugs was also administered (Table 1). Nineteen patients (18.7%) did not receive any additional immunosuppressive drug. At the time of plasma collection, 62 patients (61.4%) had anti-PR3 IgG antibodies above the upper limit of normal (ULN 3 IU/ml), and 15 patients (14.8%) had elevated anti-MPO IgG antibodies (ULN 5 IU/ml). Serum creatinine was above the ULN (107 μmol/l) in 21 patients (20.8%). Details about other study subjects’ demographics and disease characteristics are given in Table 1.
Table 1.
Demographics and disease characteristics of the study subjects
| Parameter (unit) | AAV (n = 101) | HC (n = 92) | |
|---|---|---|---|
| Age, years; median (IQR) | 64.0 (34.7) | 51.9 (26.0) | |
| Female, n (%) | 51 (50.5) | 36 (58.7) | |
| AAV duration, years; median (IQR) | 8 (11) | na | |
| BVAS, points; median (IQR) | 0 (1) | na | |
| Active organ manifestations at blood sampling | |||
| ENT involvement, n (%) | 16 (15.4) | na | |
| Lung involvement, n (%) | 12 (14.4) | na | |
| Kidney involvement, n (%) | 6 (10.6) | na | |
| Skin involvement, n (%) | 6 (6.7) | na | |
| Peripheral nervous system involvement, n (%) | 3 (3.8) | na | |
| Immunosuppression at blood sampling | |||
| Any prednisone, n (%) | 63 (62.4) | na | |
| Rituximab within the last 12 months, n (%) | 47 (46.5) | na | |
| CYC within the last 3 months, n (%) | 8 (7.9) | na | |
| MTX, AZA, LEF or MMF, n (%) | 27 (26.7) | na | |
| No immunosuppression other than prednisone | 19 (18.8) | na | |
| Laboratory parameters | RR | ||
| CRP, mg/l; median (IQR) | <10 | 3.1 (4.7) | nd |
| Serum creatinine, µmol/l; median (IQR) | 49–97 | 75 (87) | nd |
| ANCA positive, n (%) | na | 85 (86.1) | nd |
| anti-PR3 antibody titre, U/ml; median (IQR) | <3 | 10.0 (31.0) | nd |
| anti-MPO antibody titre, U/ml; median (IQR) | <5 | 2.5 (19.2) | nd |
AAV: ANCA associated vasculitis; HC: healthy controls; IQR: interquartile range; na: not applicable; nd: not done; RR: reference range.
Circulating DNA concentrations
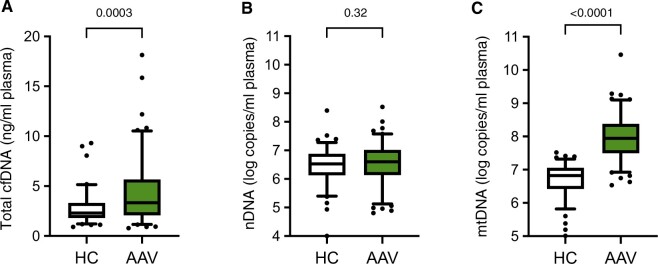
In HCs, the median total cfDNA plasma concentrations measured by spectrophotometry was 2.3 ng/ml (IQR: 1.5), ranging from 0.9 ng/ml to 13.4 ng/ml. In the AAV patients, the median cfDNA concentration was 3.3 ng/ml plasma (IQR: 3.7; range: 0.8–18.1 ng/ml), 43% higher than that in HCs, (P = 0.0003, Fig. 1A).
Figure 1.
AAV patients demonstrate high circulating mtDNA levels compared with healthy controls. Circulating nDNA copy numbers (B) are similar in patients diagnosed with AAV (n = 101) compared with healthy controls (HC, n = 92), whereas median total cfDNA levels (A) are elevated 3-fold, and mtDNA plasma concentrations (C) are elevated 13-fold. Boxes represent IQR, whiskers represent the 5th and 95th percentiles, and individual dots represent outliers. P values are indicated above each graph. AAV: ANCA-associated vasculitides; cfDNA: cell-free DNA; IQR: interquartile range; mtDNA: mitochondrial DNA; nDNA: nuclear DNA
To gain information about the biological nature of the cfDNA, we determined circulating nDNA and mtDNA copy numbers. The intra-assay CV for nDNA copy number measurements was 1.0%, while the inter-assay CV was 3.6%. Conversely, the intra-assay CV of mtDNA copy number quantification was 1.1%, and the inter-assay CV was 2.5%.
The median nDNA copy numbers in AAV plasma (4.1 × 106 copies/ml, IQR: 8.6 × 106) were not significantly different from those in HCs (3.3 × 106 copies/ml, IQR: 6.1 × 106, P = 0.32, Fig. 1B). mtDNA copy numbers in contrast were on average 13.0 times higher in AAV plasma (median 8.8 × 107 mtDNA copies/ml, IQR: 2.2 × 108) than in HC plasma (6.7 × 106 copies/ml, IQR: 8.8 × 106, P < 0.0001, Fig. 1C).
Diagnostic potential of quantifying plasma DNA concentrations
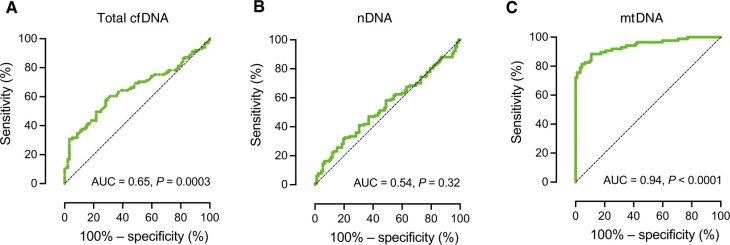
We next examined in our total study population of 193 subjects the utility of circulating plasma DNA quantification to discriminate between all AAV patients and HC individuals. For mtDNA quantification, the receiver operating characteristic (ROC) curve determined a sensitivity of 88.5% and a specificity of 89.1%, with an area under the curve (AUC) of 0.94 with an optimal cut-off value of 1.3 × 107 mtDNA copies/ml plasma (Fig. 2). Utilizing this cut-off, 90 out of 101 patients were correctly classified as having AAV, while 17 out of 92 HCs were falsely classified as having AAV (Supplementary Fig. S1, available at Rheumatology online). This performance (Supplementary Fig. S2, available at Rheumatology online) was notably superior to that of total plasma cfDNA concentrations (AUC 0.65; P = 0.0003) and nDNA copy numbers (AUC 0.54; P = 0.32, Fig. 2). Even when comparing HCs with AAV patients without any AAV activity (BVAS = 0), the ability of mtDNA quantification to identify patients with AAV remained high (sensitivity 85.3%, specificity 82.6%, AUC 0.92, P < 0.0001).
Figure 2.
mtDNA plasma quantification at enrolment exhibits exceptional diagnostic potential for AAV. Receiver operating characteristic (ROC) curve analyses for the ability of total cfDNA (A), nDNA (B) and mtDNA (C) plasma quantification to discriminate between HCs (n = 92) and patients with AAV (n = 101). AAV: ANCA-associated vasculitides; AUC: area under the curve; cfDNA: cell-free DNA; mtDNA: mitochondrial DNA; nDNA: nuclear DNA
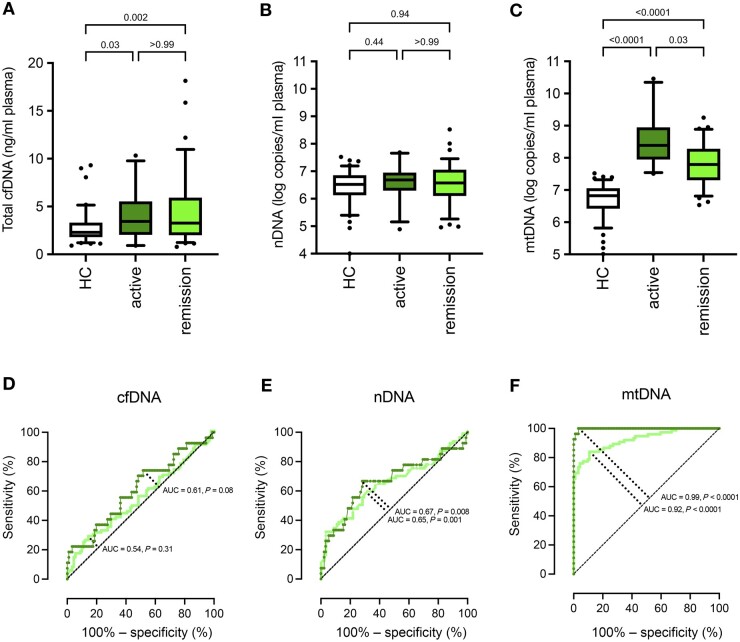
cfDNA and nDNA plasma levels did not differ between active and inactive AAV (Fig. 3A and B). nDNA copy numbers were 4.8 × 106/ml in active AAV and 3.7 × 106/ml in remission (P > 0.99). mtDNA plasma concentration in contrast was almost 4-fold elevated in patients with active AAV (BVAS ≥ 1, n = 24) compared with AAV patients in remission (BVAS = 0, n = 77) (Fig. 3C). Patients with active AAV had a median of 2.5 × 108 mtDNA copies/ml plasma, and AAV patients in remission had 6.2 × 107 copies/ml (P = 0.03). This significant difference between mtDNA levels in patients with active AAV and patients in remission was also reflected in the respective AUC calculated via ROC analyses for cfDNA, nDNA and mtDNA (Fig. 3D–F, respectively). mtDNA quantification differentiated between patients with active AAV and HCs with 96.3% sensitivity, 98.9% specificity and an AUC of 0.998 at a cut-off value of 2.9 × 107. According to this cut-off, 20 out of 21 patients were correctly classified as having active AAV. In contrast, 1 out of 92 HCs was falsely classified as such, resulting in a sensitivity of 95%, specificity of 99%, precision (positive predictive value) of 95%, negative predictive value of 99% and accuracy of 98%, in our study population (Supplementary Fig. S3, available at Rheumatology online).
Figure 3.
mtDNA plasma quantification at enrolment distinguishes patients with active AAV from patients with AAV in remission. Circulating mtDNA copy numbers (C), as well as total cfDNA levels (A), are elevated in patients diagnosed with active AAV (n = 24; dark green bar) compared with HCs (n = 92); nDNA copy numbers (B) are comparable between HCs and patients with AAV, regardless of disease activity. ROC curve analysis for the ability of total cfDNA (D), nDNA (E) and mtDNA (F) plasma quantification to discriminate between HCs and 24 AAV patients with active AAV (dark green), and between HCs and 77 AAV patients in remission (light green). Boxes represent IQRs, whiskers represent the 5th and 95th percentiles, and individual dots represent outliers. P values are indicated above each graph. AAV: ANCA-associated vasculitides; AUC: area under the curve; cfDNA: cell-free DNA; IQR: interquartile range; mtDNA: mitochondrial DNA; nDNA: nuclear DNA
mtDNA concentrations were associated with AAV activity
We subsequently analysed the clinical factors associated with the elevated DNA plasma concentrations. Within the AAV patients and the HC group, there were no differences in total plasma cfDNA concentrations, nDNA or mtDNA copy numbers between sexes, and no association with age (data not shown). Furthermore, the plasma mtDNA concentrations did not differ between patients treated with rituximab, patients receiving other immunosuppressive drugs, and patients receiving no DMARD (data not shown).
We next analysed factors associated with AAV activity. In Spearman rank correlations, plasma mtDNA levels correlated significantly with AAV activity in terms of BVAS (r = 0.30, P = 0.002). There were, however, no such correlations between BVAS and nDNA or cfDNA plasma concentrations. By univariable linear regression analysis (Table 2), mtDNA concentrations, unlike nDNA plasma levels, were associated with AAV activity, as were anti-PR3 antibody titres, CRP, neutrophil counts, and age. After multivariable linear regression analysis of all laboratory parameters with a P-value of below 0.1, the association of mtDNA copy numbers with BVAS persisted in addition to that of neutrophil counts and CRP serum concentration, with the highest statistical significance among variables for mtDNA plasma concentrations (P = 0.005, Table 2).
Table 2.
Univariable and multivariable predictors of BVAS in patients with active AAV (n = 24)
| Univariable analysis | ||||||
|---|---|---|---|---|---|---|
| Parameter (unit) | BVAS |
|||||
| β (beta) | 95% CI | P | R 2 | |||
| Age (years) | –0.008 | –0.013, –0.002 | 0.008 | 0.07 | ||
| Female sex | 0.08 | –0.08, –0.25 | 0.32 | 0.01 | ||
| Laboratory parameters | ||||||
| Neutrophil count (109 cells/ml) | 0.050 | 0.025, 0.075 | 0.0001 | 0.14 | ||
| CRP (mg/l) | 0.0075 | 0.0022, 0.0127 | 0.005 | 0.07 | ||
| Serum creatinine (µmol/l) | 0.0005 | –0.00037, –0.00138 | 0.25 | 0.01 | ||
| ANCA titre | 0.002 | 0.0004, –0.003 | 0.01 | 0.06 | ||
| anti-PR3 Ab titre (IU/ml) | 0.0015 | 9.32 × 10–6, 0.0031 | 0.04 | 0.04 | ||
| anti-MPO Ab titre (IU/ml) | 0.002 | –0.0008, 0.0054 | 0.15 | 0.02 | ||
| Total cfDNA (ng/μl) | –0.008 | 0.034, 0.017 | 0.51 | 0.004 | ||
| mtDNA (copies/ml plasma) | 3.11 × 10–11 | 2.19 × 10–10, 6.01 × 10–10 | 0.03 | 0.04 | ||
| nDNA (copies/ml plasma) | –3.32 × 10–10 | –2.77 × 10–5, 2.10 × 10–8 | 0.78 | 0.0007 | ||
| Multivariable analysis | ||||||
| (Intercept) | 0.26 | –0.15, 0.68 | 0.21 | |||
| Age (years) | –0.005 | –0.011, –0.0001 | 0.06 | |||
| CRP (mg/l) | 0.005 | 0.0008, 0.01 | 0.02 | |||
| ANCA titre | 0.0009 | –0.0006, 0.002 | 0.23 | |||
| Neutrophil count (109 cells/ml) | 0.035 | 0.009, 0.06 | 0.009 | |||
| mtDNA (copies/ml plasma) | 3.68 × 10–11 | 0, 0 | 0.005 | |||
| Adjusted R²: 0.27, P-value: <0.0001 | ||||||
The linear regression coefficient β represents the additive increase or decrease in the outcome variable per unit increase of BVAS. Significant parameters are highlighted in bold. AAV: ANCA-associated vasculitides; cfDNA: cell-free DNA; mtDNA: mitochondrial DNA; nDNA: nuclear DNA.
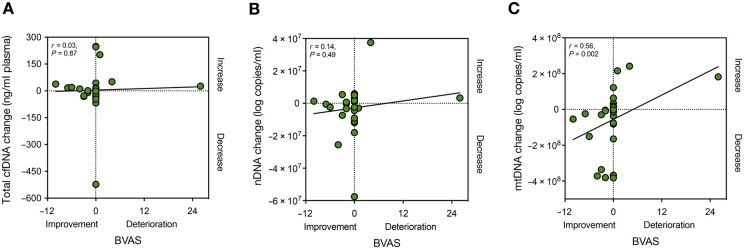
Follow-up visits were organized for 27 AAV patients (median follow-up time: 6.5 months; IQR: 6.3 months). At the time of follow-up, 5 out of 24 patients with GPA and 2 out of 3 patients with MPA had a BVAS > 0. The immunosuppressive drugs administered to the patients during follow-up in addition to glucocorticosteroids were rituximab (11/27), CYC (1/27), MTX (6/27), AZA (3/27) and LEF (1/27), while 5 out of 27 patients (18.8%) did not receive any additional treatment. In this context, we examined whether or not longitudinal changes of mtDNA levels in AAV patients’ plasma correlated with the evolution of the BVAS at follow-up. Indeed, a strong correlation was present between the shift in BVAS and the change in mtDNA (r = 0.56, P = 0.002, Fig. 4), while cfDNA and nDNA changes did not correlate with the shift in BVAS over time (Fig. 4).
Figure 4.
mtDNA plasma levels correlate to AAV disease activity at follow-up. Changes (delta) in total cfDNA (A) or nDNA (B) levels in AAV patients’ plasma (n = 27) do not correlate with the evolution of AAV activity at follow-up (delta BVAS). In contrast, delta-mtDNA levels (C) correlate significantly with delta BVAS. AAV: ANCA-associated vasculitides; AUC: area under the curve; cfDNA: cell-free DNA; mtDNA: mitochondrial DNA; nDNA: nuclear DNA; r: Spearman’s rank correlation coefficient
Interestingly, the changes in CRP serum concentrations and ANCA titres did not connect with the evolution of BVAS at follow-up (P = 0.24 and P = 0.23, respectively), indicating that mtDNA measurement could offer benefits as a biomarker to monitor AAV activity.
Finally, we assessed whether additional clinically helpful information could be obtained by calculating the ratio of mtDNA and nDNA copy numbers in each plasma sample. The median mtDNA/nDNA copy ratio was 2.0 (IQR: 4.9) in HC individuals and 22.6 (IQR: 41.3) in AAV patients (P < 0.0001). The mtDNA/nDNA ratio distinguished AAV patients from HCs with a sensitivity of 81.5% and a specificity of 83.7% (AUC: 0.89; cut-off: 6.4), but there was no correlation between the ratio and the AAV activity. These results indicate that the determination of the mtDNA/nDNA copy ratio provides no diagnostic advantage over measuring mtDNA copy numbers alone.
Discussion
The present study documents a marked increase in circulating cfDNA in the plasma of patients with AAV compared with HCs, mainly due to a substantial increase in mtDNA, while nDNA copy numbers did not differ significantly. Our data are well matched with the observation that neutrophils release mtDNA as a structural component of NETs [27, 28], which were also reported in kidney biopsies and thrombotic vessels of patients with AAV [8, 29, 30]. The lack of a difference in nDNA plasma levels between AAV patients and HCs argues against the possibility that patients with AAV patients have impaired DNA scavenging mechanisms similar to those described in SLE [31] and reasonably supports an enhanced mtDNA release as the pathobiological mechanism in AAV.
Although mtDNA plasma concentration measurements were able to differentiate patients with active AAV from HCs with extraordinary sensitivity, elevated mtDNA plasma levels are not specific to AAV. Recent studies have also observed high mtDNA amounts in the blood circulation of patients with SLE, trauma, malignancy, preeclampsia, and infections, the extent of which was mostly moderate [24, 32, 33]. Significantly, however, some of these reports are confounded by the use of serum instead of plasma, with serum being an inadequate analyte due to the in vitro release of mtDNA from platelets during coagulation [23]. As a technical annotation, the platelet-poor plasma isolation procedure used in our study removes platelets, mitochondria and microaggregates; it, however, does not remove microparticles and exosomes. Therefore, it is unclear whether some mtDNA may also originate from these extracellular vesicles.
It was previously shown that in vitro stimulation of primary human leukocytes with anti-PR3 and anti-MPO antibodies boosts TLR9 expression and promotes the release of cytokines upon stimulation with microbial components [34]. The direct pathogenic role of ANCA is also supported by animal models [35] and in vitro studies demonstrating that PR3- and MPO-ANCA can activate neutrophils to produce reactive oxygen species (ROS) and proteolytic enzymes [36]. Anti-PR3 and anti-MPO-ANCA are found in the majority of patients with active GPA or MPA [37], rendering them essential as diagnostic tools. ANCA levels, however, do not conclusively predict AAV relapse [38], and there is an unmet need for biomarkers for this purpose.
Because anti-PR3 and anti-MPO antibodies prime neutrophils for NETosis in vitro, we were somewhat surprised that we failed to detect a strong relationship between mtDNA and ANCA titres in our study. The lack of such correlation could be merely attributed to low patient numbers, suggest additional regulators of mtDNA release, or also support non-neutrophil sources of mtDNA liberation [39]. Platelets, for example, contain mtDNA, are also activated in AAV [40] and represent another possible source of mtDNA in AAV [41].
The involvement of mitochondria in the pathobiology of autoimmunity is lately gaining attention. These organelles resemble bacteria by carrying a circular genome with hypomethylated CpG islands, formylated proteins, and cardiolipin-containing membranes [42]. Under certain cellular stress, mitochondria and their inner components are released into the extracellular space, potentially eliciting proinflammatory autoimmune responses [42, 43]. Moreover, it was recently demonstrated that during ANCA-induced NET formation, the regulated opening of mitochondrial pores might release mitochondrial ROS and induce cellular and nuclear membrane vulnerability [5, 44]. In this context, mtDNA could be differentially released, acting as a proinflammatory damage-associated molecular pattern (DAMP) [10].
While only a relatively minor proportion of patients in our cohort had high AAV activity, mtDNA levels strongly and independently of other markers corresponded to disease activity at inclusion and follow-up. These observations also support the proinflammatory role of circulating mtDNA in the pathogenesis of AAV. mtDNA is a ligand of both TLR9 and cyclic GMP-AMP synthase (cGAS), which appear to trigger the type I IFN signature and autoantibody formation in autoimmunity [10, 15, 45, 46]. Neutrophils can be activated per se by circulating mtDNA via TLR9 and the cGAS-STING-pathway, closing a vicious circle that perpetuates the delivery of cfDNA [46, 47] and elicits systemic inflammation and organ injury [10, 48].
Lastly, the observed correlation of the longitudinal changes of mtDNA levels with the changes in BVAS also suggests that circulating mtDNA levels may become a useful adjunct in the long-term monitoring of AAV activity. More extensive studies must, however, replicate the utility of clinical mtDNA testing and also evaluate whether mtDNA measurements could support the prediction of AAV flares.
In summary, we demonstrate that circulating mtDNA copy numbers are vastly elevated in AAV patient plasma. Even more importantly, plasma mtDNA copy numbers are associated with vasculitis activity. In contrast, other indicators of disease activity, such as CRP and PR3-ANCA levels, either have a weaker or no correlation with BVAS [49, 50]. Despite the limitation of our study due to the small sample size, circulating mtDNA concentrations distinguish AAV patients from non-inflammatory controls with high sensitivity and represent a novel independent biomarker of AAV activity. Future work is needed to determine and evaluate the origin of mtDNA in AAV plasma.
Supplementary Material
Acknowledgements
We would like to thank all enrolled patients, the medical teams and supporting personnel of the Rheumatological Polyclinic of the University of Basel and the Immunology-Rheumatology Biobank (IR-B) of the Department of Rheumatology and Clinical Immunology of the University Medical Center Freiburg, and the Basel Blood Donation Center for sample provision, collection, and storage.
Contributor Information
Stavros Giaglis, Laboratory for Experimental Rheumatology, Department of Biomedicine, University of Basel, Basel, Switzerland; Department of Rheumatology, University Hospital Basel, Basel, Switzerland.
Douglas Daoudlarian, Laboratory for Experimental Rheumatology, Department of Biomedicine, University of Basel, Basel, Switzerland; Department of Rheumatology, University Hospital Basel, Basel, Switzerland.
Jens Thiel, Department of Rheumatology and Clinical Immunology, University Medical Center Freiburg, Freiburg, Germany.
Marta Rizzi, Department of Rheumatology and Clinical Immunology, University Medical Center Freiburg, Freiburg, Germany.
Diego Kyburz, Laboratory for Experimental Rheumatology, Department of Biomedicine, University of Basel, Basel, Switzerland; Department of Rheumatology, University Hospital Basel, Basel, Switzerland.
Nils Venhoff, Department of Rheumatology and Clinical Immunology, University Medical Center Freiburg, Freiburg, Germany.
Ulrich A Walker, Laboratory for Experimental Rheumatology, Department of Biomedicine, University of Basel, Basel, Switzerland; Department of Rheumatology, University Hospital Basel, Basel, Switzerland.
Supplementary material
Supplementary material is available at Rheumatology online.
Data availability
The data generated and analysed in the present study can be shared upon reasonable request to the corresponding author.
Contribution statement
S.G. performed experiments and the data analysis required for this study and read and approved the final manuscript. D.D. performed statistical analyses and also read and approved the final manuscript. D.K., U.A.W., J.T. and M.R. contributed samples and read and approved the final manuscript. U.A.W. and N.V. conceived the study, contributed samples, helped design the experiments, interpreted clinical data, and read and approved the manuscript. In addition, S.G. and U.A.W. wrote the manuscript.
Funding
This work is supported by the Swiss National Science Foundation (Project No. 185300).
Disclosure statement: Freiburg University holds a patent for mtDNA quantification in the diagnosis and follow-up of autoimmune diseases, coinvented by U.A.W. and N.V. Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research. All other authors have declared no conflicts of interest.
References
- 1. Kitching AR, Anders HJ, Basu N et al. ANCA-associated vasculitis. Nat Rev Dis Primers 2020;6:71. [DOI] [PubMed] [Google Scholar]
- 2. Robson JC, Grayson PC, Ponte C et al. ; DCVAS Study Group. 2022 American College of Rheumatology/European Alliance of Associations for Rheumatology classification criteria for granulomatosis with polyangiitis. Arthritis Rheumatol 2022;74:393–9. [DOI] [PubMed] [Google Scholar]
- 3. Suppiah R, Robson JC, Grayson PC et al. ; DCVAS INVESTIGATORS. 2022 American College of Rheumatology/European Alliance of Associations for Rheumatology classification criteria for microscopic polyangiitis. Ann Rheum Dis 2022;81:321–6. [DOI] [PubMed] [Google Scholar]
- 4. Schreiber A, Rousselle A, Becker JU et al. Necroptosis controls NET generation and mediates complement activation, endothelial damage, and autoimmune vasculitis. Proc Natl Acad Sci USA 2017;114:E9618–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kudo T, Nakazawa D, Watanabe-Kusunoki K et al. Regulation of NETosis by cyclophilin D in myeloperoxidase-positive antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol 2023;75:71–83. [DOI] [PubMed] [Google Scholar]
- 6. West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 2017;17:363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giaglis S, Hahn S, Hasler P. “The NET Outcome”: are Neutrophil Extracellular Traps of any relevance to the pathophysiology of autoimmune disorders in childhood? Front Pediatr 2016;4:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kessenbrock K, Krumbholz M, Schonermarck U et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med 2009;15:623–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kessler N, Viehmann SF, Krollmann C et al. Monocyte-derived macrophages aggravate pulmonary vasculitis via cGAS/STING/IFN-mediated nucleic acid sensing. J Exp Med 2022;219:e20220759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q, Raoof M, Chen Y et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010;464:104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lood C, Blanco LP, Purmalek MM et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 2016;22:146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hajizadeh S, DeGroot J, TeKoppele JM, Tarkowski A, Collins LV. Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res Ther 2003;5:R234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caielli S, Athale S, Domic B et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med 2016;213:697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Romo GS, Caielli S, Vega B et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 2011;3:73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lande R, Ganguly D, Facchinetti V et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med 2011;3:73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol 2011;7:691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Margraf S, Logters T, Reipen J et al. Neutrophil-derived circulating free DNA (cf-DNA/NETs): a potential prognostic marker for posttraumatic development of inflammatory second hit and sepsis. Shock 2008;30:352–8. [DOI] [PubMed] [Google Scholar]
- 18. Logters T, Paunel-Gorgulu A, Zilkens C et al. Diagnostic accuracy of neutrophil-derived circulating free DNA (cf-DNA/NETs) for septic arthritis. J Orthop Res 2009;27:1401–7. [DOI] [PubMed] [Google Scholar]
- 19. Mukhtyar C, Lee R, Brown D et al. Modification and validation of the Birmingham Vasculitis Activity Score (version 3). Ann Rheum Dis 2009;68:1827–32. [DOI] [PubMed] [Google Scholar]
- 20. [The Helsinki Declaration of the World Medical Association (WMA). Ethical principles of medical research involving human subjects.]. Pol Merkur Lekarski 2014;36:298–301. https://apps.who.int/iris/handle/10665/268312. [PubMed] [Google Scholar]
- 21. Bossuyt PM, Reitsma JB, Bruns DE et al. ; STARD Group. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem 2015;61:1446–52. [DOI] [PubMed] [Google Scholar]
- 22. Kerachian MA, Azghandi M, Mozaffari-Jovin S, Thierry AR. Guidelines for pre-analytical conditions for assessing the methylation of circulating cell-free DNA. Clin Epigenetics 2021;13:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meddeb R, Dache ZAA, Thezenas S et al. Quantifying circulating cell-free DNA in humans. Sci Rep 2019;9:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giaglis S, Daoudlarian D, Voll RE et al. Circulating mitochondrial DNA copy numbers represent a sensitive marker for diagnosis and monitoring of disease activity in systemic lupus erythematosus. RMD Open 2021;7:e002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones SR, Carley S, Harrison M. An introduction to power and sample size estimation. Emerg Med J 2003;20:453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biscetti F, Carbonella A, Parisi F et al. The prognostic significance of the Birmingham Vasculitis Activity Score (BVAS) with systemic vasculitis patients transferred to the intensive care unit (ICU). Medicine (Baltimore) 2016;95:e5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yipp BG, Kubes P. NETosis: how vital is it? Blood 2013;122:2784–94. [DOI] [PubMed] [Google Scholar]
- 28. Yousefi S, Simon D, Stojkov D et al. In vivo evidence for extracellular DNA trap formation. Cell Death Dis 2020;11:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang S, Zhang Y, Yin SW et al. Neutrophil extracellular trap formation is associated with autophagy-related signalling in ANCA-associated vasculitis. Clin Exp Immunol 2015;180:408–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakazawa D, Tomaru U, Yamamoto C, Jodo S, Ishizu A. Abundant neutrophil extracellular traps in thrombus of patient with microscopic polyangiitis. Front Immunol 2012;3:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hakkim A, Furnrohr BG, Amann K et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci USA 2010;107:9813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reznik E, Miller ML, Şenbabaoğlu Y et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016;5:e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun J, Longchamps RJ, Piggott DA et al. Association between HIV infection and mitochondrial DNA copy number in peripheral blood: a population-based, prospective cohort study. J Infect Dis 2019;219:1285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uehara A, Sato T, Iwashiro A, Yokota S. PR3-ANCA in Wegener’s granulomatosis prime human mononuclear cells for enhanced activation via TLRs and NOD1/2. Diagn Pathol 2009;4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Little MA, Al-Ani B, Ren S et al. Anti-proteinase 3 anti-neutrophil cytoplasm autoantibodies recapitulate systemic vasculitis in mice with a humanized immune system. PLoS One 2012;7:e28626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Falk RJ, Terrell RS, Charles LA, Jennette JC. Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci USA 1990;87:4115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kallenberg CG. Pathogenesis and treatment of ANCA-associated vasculitides. Clin Exp Rheumatol 2015;33:S11–4. [PubMed] [Google Scholar]
- 38. Kemna MJ, Damoiseaux J, Austen J et al. ANCA as a predictor of relapse: useful in patients with renal involvement but not in patients with nonrenal disease. J Am Soc Nephrol 2015;26:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yousefi S, Gold JA, Andina N et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med 2008;14:949–53. [DOI] [PubMed] [Google Scholar]
- 40. Tomasson G, Lavalley M, Tanriverdi K et al. ; Wegener’s Granulomatosis Etanercept Trial (WGET) Research Group. Relationship between markers of platelet activation and inflammation with disease activity in Wegener’s granulomatosis. J Rheumatol 2011;38:1048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melki I, Allaeys I, Tessandier N et al. Platelets release mitochondrial antigens in systemic lupus erythematosus. Sci Transl Med 2021;13:eaav5928. [DOI] [PubMed] [Google Scholar]
- 42. Becker Y, Marcoux G, Allaeys I et al. Autoantibodies in systemic lupus erythematosus target mitochondrial RNA. Front Immunol 2019;10:1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Becker YLC, Gagne JP, Julien AS et al. Identification of Mitofusin 1 and Complement Component 1q Subcomponent Binding Protein as Mitochondrial Targets in Systemic Lupus Erythematosus. Arthritis Rheumatol 2022;74:1193–203. [DOI] [PubMed] [Google Scholar]
- 44. Kim J, Gupta R, Blanco LP et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 2019;366:1531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 2006;25:383–92. [DOI] [PubMed] [Google Scholar]
- 46. West AP, Khoury-Hanold W, Staron M et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015;520:553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dorner T, Furie R. Novel paradigms in systemic lupus erythematosus. Lancet 2019;393:2344–58. [DOI] [PubMed] [Google Scholar]
- 48. McIlroy DJ, Jarnicki AG, Au GG et al. Mitochondrial DNA neutrophil extracellular traps are formed after trauma and subsequent surgery. J Crit Care 2014;29:1133.e1–5. [DOI] [PubMed] [Google Scholar]
- 49. Suppiah R, Mukhtyar C, Flossmann O et al. A cross-sectional study of the Birmingham Vasculitis Activity Score version 3 in systemic vasculitis. Rheumatology (Oxford) 2011;50:899–905. [DOI] [PubMed] [Google Scholar]
- 50. Tervaert JW, van der Woude FJ, Fauci AS et al. Association between active Wegener’s granulomatosis and anticytoplasmic antibodies. Arch Intern Med 1989;149:2461–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analysed in the present study can be shared upon reasonable request to the corresponding author.