Abstract
In 1997, 18 confirmed cases of human influenza arising from multiple independent transmissions of H5N1 viruses from infected chickens were reported from Hong Kong. To identify possible phenotypic changes in the hemagglutinin (HA) and neuraminidase (NA) of the H5 viruses during interspecies transfer, we compared the receptor-binding properties and NA activities of the human and chicken H5N1 isolates from Hong Kong and of H5N3 and H5N1 viruses from wild aquatic birds. All H5N1 viruses, including the human isolate bound to Sia2-3Gal-containing receptors but not to Sia2-6Gal-containing receptors. This finding formally demonstrates for the first time that receptor specificity of avian influenza viruses may not restrict initial avian-to-human transmission. The H5N1 chicken viruses differed from H5 viruses of wild aquatic birds by a 19-amino-acid deletion in the stalk of the NA and the presence of a carbohydrate at the globular head of the HA. We found that a deletion in the NA decreased its ability to release the virus from cells, whereas carbohydrate at the HA head decreased the affinity of the virus for cell receptors. Comparison of amino acid sequences from GenBank of the HAs and NAs from different avian species revealed that additional glycosylation of the HA and a shortened NA stalk are characteristic features of the H5 and H7 chicken viruses. This finding indicates that changes in both HA and NA may be required for the adaptation of influenza viruses from wild aquatic birds to domestic chickens and raises the possibility that chickens may be a possible intermediate host in zoonotic transmission.
Global outbreaks of human influenza (pandemics) arise from influenza A viruses with novel neuraminidase (NA) and/or hemagglutinin (HA) molecules to which humans have no immunity. These pandemic strains derive from viruses of wild aquatic birds by interspecies transmission of the whole virus or by genetic reassortment between avian and human viruses (reviewed in reference 44).
Avian influenza viruses do not replicate efficiently in humans. For example, high doses of virus were found to be required for the replication of avian influenza virus strains in volunteers even at a limited level (3), and no cases of influenza virus infections were documented in workers exposed to highly pathogenic avian viruses during the 1985 outbreak in poultry in the United States (2). It is believed that a growth restriction of avian influenza viruses in humans limits the emergence of new pandemic strains. HA and NA are regarded as possible restrictive factors because of the differences between influenza virus receptors on the target cells of birds and humans. For example, the HAs of avian viruses bind to Sia2-3Gal-terminated sialylglycoconjugates and not to Sia2-6Gal-containing receptors, whereas those of human viruses display the opposite receptor-binding specificity (reviewed in reference 32; see also references 9, 18, and 27). The viral NA removes sialic acid from the HA and NA of progeny virus particles, intercellular glycoproteins, and host cell receptors, and thus facilitates virus release from infected cells and from intercellular inhibitors. The specificity of the NA has to match the specificity of the HA. Thus, the NA of the N2 avian virus cannot hydrolyze the Sia2-6Gal linkage, but it acquired this ability during evolution in humans (1). A poor match between the HA and NA can serve as a potential barrier to the reassortment of influenza viruses (35). Because of these host-range restrictions, it has not been clear whether an avian virus can be directly transmitted to humans and start a new pandemic. Scholtissek et al. (38) hypothesized that the pig may serve as a mixing vessel for the reassortment of avian and human viruses. Moreover, adaptation of the avian virus to pigs leads to changes in the specificity of receptor binding (22, 34), thus potentially facilitating transmission of the virus to humans.
In 1997, 18 Hong Kong residents were infected with H5N1 influenza viruses, which proved closely related to the H5N1 avian viruses that had caused influenza outbreaks in chickens in Hong Kong at the beginning of 1997 (7, 11, 41). Subsequent surveillance of live bird markets in Hong Kong indicated wide dissemination of H5N1 viruses, contributing to the decision to depopulate all birds in these markets. No additional human cases have been reported through July 1998. This incident demonstrated, for the first time, that avian viruses can cause severe disease in humans without reassortment with human viruses and without any apparent intermediate mammalian host.
The HA and NA of the human H5N1 isolates have been sequenced and compared with those of H5 chicken and duck viruses (7, 40). All avian and human isolates from live bird markets in Hong Kong 1997 contained multiple basic amino acids at the cleavage site of the HA, a feature known to be associated with high levels of virulence among avian influenza viruses. Two variants of the chicken and human viruses were identified that differed by the presence or absence of a glycosylation sequon at the top of the HA head (position 158 by H3 numbering), this sequon was shown to be glycosylated (40). The NA of chicken viruses and of HK/156/97 human virus contained a 19-amino-acid deletion in a stalk region (7).
In this study, we sought to identify and characterize possible phenotypic changes in the surface glycoproteins of H5N1 influenza viruses that accompanied their transmission from wild aquatic birds to chickens and from chickens to humans. In particular, we asked: Did H5N1 viruses from chickens and humans acquire the ability to recognize Sia2-6Gal determinants that are believed to be the major sialyloligosaccharide determinants on ciliated cells of human respiratory epithelia? What effect does the carbohydrate at position 158 have on the receptor-binding properties of the viruses? What is the effect of the deletion in the NA on the enzyme activity? Do additional glycosylation of the HA and deletion in the NA represent unique features of 1997 Hong Kong H5N1 virus lineage, or do they also occur in other influenza viruses? Thus, we compared the receptor-binding properties and NA activity of the first human virus isolate, A/Hong Kong/156/97, with those of chicken isolates identified in Hong Kong in 1997, and of the H5 isolates from aquatic birds. We also compared the HA and NA amino acid sequences of H5 and H7 chicken viruses with those of viruses from other hosts.
MATERIALS AND METHODS
Viruses.
The isolation and characterization of the 1997 H5N1 human and chicken influenza viruses (A/HK/156/97, ck/HK/258/97, ck/HK/220/97, ck/HK/728/97, ck/HK/786/97, and ck/HK/915/97) were described earlier (7, 40). Other influenza A virus strains were from the repository at St. Jude Children’s Research Hospital. All viruses were grown in 9- to 10-day-old chicken eggs. Because of their high pathogenicity, chicken and human H5N1 isolates were inactivated by treatment with β-propiolactone (BPL; 1/2,000) for 3 days at 4°C. To assess the effect of inactivation on the properties of the viruses, we prepared three nonpathogenic duck H5 viruses (dk/HK/205/77, dk/HK/698/79, and dk/Minnesota/1525/81) and divided them into two portions; one was treated with BPL, and the other was used as a control. None of other viruses used were inactivated. The allantoic fluids were clarified by low-speed centrifugation, and the viruses were pelleted by high-speed centrifugation, resuspended in 0.1 M Tris buffer (pH 7.2) containing 50% glycerol, and then stored at −20°C.
Peroxidase-labeled glycoproteins.
Bovine fetuin and type III-O chicken egg white ovomucoid were purchased from Sigma. Pig α2-macroglobulin (PM) was isolated from total pig serum by gel chromatography on a Sephacryl S-300 column. To increase the binding affinity of ovomucoid to the virus, the glycoprotein was aggregated by heating of its 5% water solution at 95°C for 2 h. Conjugates with horseradish peroxidase (HRP) were prepared from PM, fetuin, and heat-aggregated ovomucoid by using the periodate method described by Boorsma and Streefkerk (5).
To selectively destroy minor amounts of Sia2-3Gal moieties present in PM-HRP conjugate, we treated it with the avian influenza virus N2 NA, which shows strict specificity for cleavage of this moiety and does not substantially affect the Sia2-6Gal sequence (1). Concentrated avian influenza virus A/mallard/NY/6750/78 (H2N2) was added to the PM-HRP conjugate in 0.1 M Tris buffer containing 10 mM CaCl2 to a final hemagglutination titer of 1:80. The mixture was incubated at 37°C for 4 h and centrifuged at 14,000 rpm for 10 min, and the precipitate was then discarded. Stocks of all conjugates were stored at −20°C in 50% glycerol–0.1 M Tris buffer (pH 7.3).
Binding of HRP-labeled sialylglycoproteins.
The binding of the HRP-labeled sialylglycoproteins to the viruses was evaluated in a solid-phase assay as described earlier (17). In brief, 96-well polyvinyl chloride microplates (Costar) were coated with fetuin (10 μg/ml in phosphate-buffered saline [PBS], 50 μl/well) at 4°C overnight, washed with water, and air dried. Purified viruses diluted with PBS to a hemagglutination titer of 1:20 were adsorbed to the wells of fetuin-coated plates at 4°C overnight (40 μl/well). After unbound virus was removed with washing buffer (WB, 0.01% Tween 80 in 0.2× PBS), twofold dilutions of the HRP-labeled sialylglycoproteins (30 μl/well) were added to the plate. The dilutions were prepared in reaction buffer (RB; PBS supplemented with 0.02% bovine serum albumin, 0.02% Tween 80, and 1 μM of the NA inhibitor zanamivir [2,3-didehydro-2,4-dideoxy-4-guanidino-N-acetyl-d-neuraminic acid; GG167], kindly provided by R. Bethell, Glaxo Wellcome). After incubation for 1 h at 4°C, the plates were washed with WB, and the amount of labeled sialylglycoprotein bound was quantified by evaluating the peroxidase activity present in the wells by using the standard o-phenylenediamine chromogenic substrate.
The binding data were converted to A490/C versus A490 Scatchard plots, where A490 represents the absorbancy of the colored product of peroxidase reaction and C represents the concentration of labeled sialylglycoprotein added to the corresponding wells. Because we were interested only in the relative binding affinity of different viruses for the same receptor analog, the concentration of the lowest dilution of each HRP-labeled sialylglycoprotein was arbitrarily taken to be 1 U, without any regard to the actual content of the protein, sialic acid, or peroxidase. The dissociation constants of the virus-sialylglycoprotein complexes were determined from the slopes of the Scatchard plots.
Binding of sialic acid and sialyloligosaccharides.
Free N-acetylneuraminic acid (Neu5Ac) and 3′-sialyllactose (3′-SL; Neu5Acα2-3Galβ1-4Glc) were purchased from Sigma. The virus-binding affinity for these receptor analogs was assessed with the solid-phase fetuin binding inhibition assay (17, 26). This assay is based on the competition for binding sites on the viral particle between nonlabeled sialic acid-containing compound and enzyme-labeled sialylglycoprotein fetuin. In brief, influenza viruses were adsorbed in the wells of fetuin-coated wells as described above. Twenty-five-microliter portions of solutions containing a fixed amount of fetuin-HRP conjugate and a variable amount of receptor analog in RB were added to the plate, which was then incubated for 1 h at 4°C. After this competitive binding step, the amount of fetuin-HRP bound was determined by using the o-phenylenediamine substrate.
The association constants of the virus-receptor analog complexes were calculated as described before (17) for each concentration of the compound used in the competitive reaction, and the results were averaged.
Virus elution from chicken erythrocytes.
The virus stocks were diluted with PBS to a hemagglutination titer of 1:32. Fifty-microliter portions of these solutions were mixed with equal volumes of 1% chicken erythrocytes (CRBCs) in U-bottomed microtiter plates. The plates were left on ice for 1 h and transferred to a water bath (37°C), and the precipitation of the agglutinated erythrocytes was monitored for 24 h to determine the time required for complete deagglutination (“elution”). In parallel, the NA activity and the agglutination of CRBC preparations treated with variable concentrations of Vibrio cholerae NA (see below) were determined for the same virus dilutions that were used in the elution experiment.
NA activity of the viruses.
Viral NA activity was determined with 4-methylumbelliferyl N-acetylneuraminic acid (4-MU-NANA; Sigma) used as a substrate (43). To 1 or 5 μl of virus dilutions in U-bottomed microtiter plates, we added 50 μl of a 40 μM solution of 4-MU-NANA in calcium-TBS buffer (6.8 mM CaCl2, 0.85% NaCl, 0.02 M Tris; pH 7.3). The mixtures were incubated for 10 to 30 min at 37°C in a water bath, and the reaction was stopped by the addition of 0.1 ml of 0.1 M glycine buffer (pH 10.7) containing 25% ethanol. The fluorescence of released 4-methylumbellyferone (4-MU) was determined with a Labsystems Fluoroskan II spectrophotometer (λexc = 355 nm, λem = 460 nm). The specific NA activity was expressed in moles of 4-MU liberated per hour per microliter of virus suspension.
Virus agglutination of CRBCs pretreated with NA.
To estimate the ability of soluble exogenous NA to elute the virus from CRBCs and to compare the affinity of different viruses for the receptors on CRBCs, we assayed the effect of NA pretreatment on cell agglutination by the viruses (45). In brief, 10% suspensions of CRBCs in calcium-TBS buffer were incubated with twofold dilutions of V. cholerae NA (RDE; Center for Disease Control, Atlanta, Ga.) for 2 h at 37°C. The NA activity of the stock RDE preparation, determined as described above, was 3.2 nmol of 4-MU/h · μl; final dilutions of the stock during treatment ranged from 2 to 160. Treated erythrocytes were washed with PBS. To 50-μl virus suspensions in PBS with an HA titer of 1:32, 50 μl of 1% NA-treated CRBCs suspensions were added in U-bottomed microplate, and hemagglutination was scored after 1 h of incubation of mixtures on ice. The results were expressed as the lowest activity of RDE (nanomoles of 4-MU/μl) used for the treatment that completely prevented agglutination. Both assays that utilized CRBCs (adsorption/elution and agglutination of NA-treated cells) were done in duplicate and on the same day. As a rule, the HA titers of parallel probes were identical.
RNA extraction, PCR, and sequencing.
Viral RNA was extracted from allantoic fluid with the RNeasy Mini-Kit (Qiagen, Santa Clarita, Calif.). Amplification of the viral RNA was done by reverse transcription PCR (RT-PCR) as described previously (39). After being purified with the Quiquick PCR Purification Kit (Qiagen), the PCR products were subjected to sequencing. The sequencing reactions were performed by the Center for Biotechnology at St. Jude Children’s Research Hospital on template DNA with Prism BigDye terminator cycle sequencing ready reaction kits with Ampli-Taq DNA polymerase FS (Perkin-Elmer/Applied Biosystems, Inc. [PE/ABI], Foster City, Calif.) and synthetic oligonucleotides. Samples were electrophoresed, detected, and analyzed on PE/ABI model 373 and 377 DNA sequencers. The Wisconsin Sequence Analysis Package, version 9.0 (Genetic Computer Group, Inc., Madison, Wis.) was used for the analysis and translation of nucleotide sequence data. The HA1 sequences of H5 viruses determined in this study are available under GenBank accession numbers from AF082034 to AF082043.
Analysis of the HA and NA amino acid sequences.
The sequences of the influenza virus HAs and NAs were obtained from GenBank (release 105.0) and were studied with GeneDoc 2.3 software (28; K. B. Nicholas and H. B. Nicholas, Jr., 1997, GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the authors. http://www.cris.com/∼Ketchup/genedoc.shtml). The HA sequences were inspected for the presence of glycosylation sequons on the membrane distal end of the HA globular head (positions 90 to 260 of HA1). The H3 numbering system, in accord with the alignment of Nobusawa et al. (29), is used throughout this study. Two overlapping sequons (for example, NNSS) were counted as one; the sequences NPT and NPS were assumed to be nonglycosylated and were disregarded (14).
The NA sequences were analyzed for the presence of deletions of 10 or more amino acids in the stalk region of the enzyme, as, starting from residue 36 and ending with the conserved cysteine in position 92 (4). The N2 numbering system, in accord with the alignment of Colman et al. (8), is used here.
Phylogenetic relationships between the virus HAs and NAs were estimated with the PHYLIP 3.572 software package (15; J. Felsenstein, 1993, PHYLIP [Phylogeny Inference Package] version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle, Wash.; http://evolution.genetics.washington.edu/phylip.html). The trees shown on the figures were obtained for the nucleotide sequences of coding regions of the whole HA1 and NA genes, respectively, by using the neighbor-joining algorithm and the Jukes-Cantor distance. The TREEVIEW 1.5.2 program (31) was used to draw the trees.
RESULTS
H5 chicken and human viruses from Hong Kong display an avian-like receptor-binding phenotype and do not bind to Sia2-6Gal receptor determinants.
To evaluate possible changes in the receptor-binding specificity of chicken and human viruses from Hong Kong, we utilized two sialylglycoproteins, ovomucoid (Ovo) and PM, that contain Sia2-3Gal-determinants and Sia2-6Gal determinants, respectively (24, 36). Ovo and PM were labeled with peroxidase, and their binding to the viruses was determined in a solid-phase assay.
Table 1 shows the dissociation constants of complexes between different viruses and Ovo-HRP or PM-HRP conjugates. Neither virus isolate from chickens or virus isolate from wild aquatic birds bound PM-HRP (Kass <0.2), indicating the low affinity of these viruses for the terminal Sia2-6Gal moieties represented in PM. In contrast, two of the earliest human virus isolates from the pandemics of 1957 (RI/5+/57) and 1968 (Aichi/2/68) bound PM-HRP at least 20 times better than did avian viruses, in accord with the known preference of these human strains for Sia2-6Gal determinants (9). The H5N1 virus A/HK/156/97, isolated from a child in Hong Kong, did not bind PM-HRP, suggesting that this virus has not yet acquired the ability to bind Sia2-6Gal.
TABLE 1.
Binding of labeled Ovo (Ovo-HRP) and PM (PM-HRP) to influenza viruses
| Virus | Association constant (U−1)a
|
|
|---|---|---|
| Ovo-HRP (Sia2-3Gal) | PM-HRP (Sia2-6Gal) | |
| Chicken H5N1 | ||
| Ck/HK/220/97 (CHO158)bc | 0.4 | <0.2 |
| Ck/HK258/97 (CHO158)bc | 0.5 | <0.2 |
| Ck/HK/728/97c | 1 | <0.2 |
| Ck/HK/786/97 (CHO158)bc | 0.5 | <0.2 |
| Ck/HK/915/97c | 1 | <0.2 |
| Human H5N1 isolate HK/156/97c | 1.5 | <0.2 |
| Viruses from aquatic birds | ||
| Dk/HK/205/77 (H5N2)c | 2.6 | <0.2 |
| Dk/HK/308/78 (H5N2) | 3.1 | <0.2 |
| Dk/HK/698/79 (H5N2)c | 3.4 | <0.2 |
| Dk/MN/1525/81 (H5N1)c | 3.3 | <0.2 |
| Gull/PA/4175/83 (H5N1) | 2.2 | <0.2 |
| Dk/Bavaria/1/77 (H1N1) | 3.6 | <0.2 |
| Mallard/MT/61 (H2N2) | 2.3 | <0.2 |
| Dk/Hokkaido/7/82 (H3N8) | 7.6 | <0.2 |
| Human pandemic strains | ||
| Aichi/2/68 (H3N2) | <0.1 | 4 |
| RI/5+ (H2N2) | <0.1 | 60 |
The binding assay was performed, and the association constants of virus complexes with labeled sialylglycoproteins were calculated as described in Materials and Methods. Only the upper limit of Kass could be estimated when the association constants were below 0.1 to 0.2 U−1 because the absorbancy at most experimental points was close to the background level.
These viruses differ from the other H5N1 isolates from 1997 by the presence of a glycosylation sequon at positions 158 to 160 of HA1 (CHO158), which was shown to be glycosylated (40).
Inactivated with BPL.
The binding affinity of the viruses for the Ovo-HRP conjugate varied. Isolates from aquatic birds bound Ovo-HRP with the highest affinity, while human H2N2 and H3N2 strains clearly displayed the lowest binding (if any), in accord with the known low affinity of human viruses for Sia2-3Gal-terminated sequences. Among the H5N1 viruses isolated in 1997, those with a carbohydrate at position 158 bound Ovo-HRP somewhat more weakly than did strains without this site (T160→A160 mutation), suggesting that a loss of the carbohydrate from the tip of the HA increased the binding affinity.
A known characteristic feature of the receptor-binding phenotype of avian viruses, a feature which clearly separates them from human virus strains, is the ability of the avian HA to bind to the galactose ring of the Sia2-3Gal moiety (27). To determine whether this feature is changed in H5 chicken viruses, we compared their binding to Neu5Ac and to 3′-SL (Neu5Ac2-3Gal1-4Glc) (Table 2). Like the duck viruses, two chicken strains and A/HK/156/97 bind 3′SL more than an order of magnitude more strongly than they bind free Neu5Ac, a finding indicative of favorable interactions in the receptor-binding site of these viruses with the penultimate galactose residue of 3′SL. This is in a marked contrast to the Aichi/2/68 human strain, which shows no binding to the 3-linked galactose (its affinity for 3′SL does not differ from its affinity for Neu5Ac). Thus, with respect to both their inability to bind Sia2-6Gal receptor determinants and their specific recognition of Sia2-3Gal determinants, H5N1 chicken viruses and the A/HK156/97 human isolate display a typical avian receptor-binding phenotype.
TABLE 2.
Binding of Neu5Ac and of 3′-SL (Neu5Ac2-3Gal-4Glc) to influenza viruses
| Virus | Association constant (mM−1)a
|
|
|---|---|---|
| Neu5Acb | 3′-SL | |
| Chicken H5N1 | ||
| Ck/HK/220/97 (CHO158) | 1.2 | 40 |
| Ck/HK/258/97 (CHO158) | 1.3 | 30 |
| Human H5N1 HK/156/97 | 0.5 | 30 |
| Duck viruses | ||
| Dk/HK/205/77 (H5N2) | 0.3 | 15 |
| Dk/HK/698/79 (H5N2) | 0.5 | 15 |
| Dk/Bavaria/1/77 (H1N1) | 0.5 | 10 |
| Mallard/MT/61 (H2N2) | 0.3 | 10 |
| Dk/Hokkaido/7/82 (H3N8) | 0.8 | 10 |
| Human pandemic strain Aichi/2/68 (H3N2) | 0.2 | 0.2 |
The association constants of virus complexes with Neu5Ac and 3′SL were determined in a competitive binding assay as described in Materials and Methods.
Free Neu5Ac in solution is a mixture of α- and β-anomers. The β-anomer is not present in the natural sialylglycoconjugates and does not bind to influenza virus (26). Represented here are the values for the α-anomer, which were calculated from the binding data (assuming a 5% content of α-anomer in the mixture).
Comparison of the binding affinity and NA activity of H5 viruses.
Partial sequencing of the NA gene of H5N1 chicken viruses from Hong Kong used in this study revealed that all of them contained the same 19-amino-acid deletion in the stalk that was previously identified in the NA of A/HK/156/97 human isolate (data not shown). To evaluate possible effects of this deletion on the NA activity, we prepared suspensions of different H5 viruses with the same hemagglutination titers and tested each suspension for (i) its ability to agglutinate CRBCs treated with V. cholerae NA, (ii) adsorption-elution from CRBCs, and (iii) NA activity against a low-molecular-weight substrate, 4-MU-NANA. The results of these experiments are summarized in Table 3.
TABLE 3.
Relative affinity, elution from CRBCs, and NA activity of H5 influenza viruses
| Virus | V. cholerae NA activity that abolishes hemagglutination (nmol of 4-MU/μl)a | Relative affinity for CRBCsb | Time to elute from CRBCs (h)c | Sp act of virus NA (nmol of 4-MU/μl · h)d |
|---|---|---|---|---|
| Chicken H5N1 | ||||
| Ck/HK/220/97 (CHO158)e | 0.16 | + | 12 | 0.66 |
| Ck/HK/258/97 (CHO158)e | 0.16 | + | 6 | 0.48 |
| Ck/HK/728/97e | 0.32 | ++ | >24 | 0.88 |
| Ck/HK/786/97 (CHO158)e | 0.16 | + | 3 | 0.8 |
| Ck/HK/915/97e | 0.64 | ++ | >24 | 1.6 |
| Human H5N1 HK/156/97e | 0.32 | ++ | >24 | 4 |
| Viruses from aquatic birds | ||||
| Dk/HK/205/77 (H5N2)e | 1.3 | +++ | 1.5 | 0.74 |
| Dk/MN/1525/81 (H5N1)e | 3.2 | +++ | 1.5 | 1.5 |
| Dk/MN/1525/81 (H5N1) | 3.2 | +++ | 1.5 | 1.8 |
| Gull/PA/4175/83 (H5N1) | 1.3 | +++ | 1.5 | 0.74 |
Minimal activity of V. cholerae NA used for the treatment of 10% CRBCs, which completely abrogated their agglutination by a particular virus preparation. The values were calculated from the activity of the stock NA solution assayed by using 4-MU-NANA as a substrate.
Relative affinity is based on the agglutination of V. cholerae-treated CRBCs (column 2), assuming that the higher the NA activity required to abolish hemagglutination, the higher the virus affinity for CRBCs (45). +, 0.16; ++, 0.32 to 0.64; +++, 1.3 to 3.2.
Elution was assayed as described in Materials and Methods by monitoring desaggregation at 37°C of agglutinated CRBCs.
Assayed as described in Materials and Methods for the same virus preparations that were used in the elution experiment.
Inactivated with BPL . The similarity of the data for the live and inactivated preparations of dk/MN/1525/81 virus indicates that inactivation had no significant effect on the HA and NA activities.
In the assay, in which CRBCs were treated with gradually increasing concentrations of bacterial NA, viruses from aquatic birds required the highest concentrations of NA to destroy receptors on the CRBCs, indicating that they possessed the highest affinity for the receptors (45). In the case of H5N1 viruses from Hong Kong, their affinity correlated with the presence of CHO158. Chicken virus strains that carried carbohydrate at this position had a lower affinity than did variants without the site. The relative affinity of H5 viruses for CRBC receptors correlated well with their affinity for a soluble labeled sialylglycoprotein, Ovo-HRP (Table 1).
To compare the abilities of the NA of different H5 viruses to induce virus release from CRBCs, we adsorbed the viruses to erythrocytes at 4°C and monitored their elution at 37°C. The results indicated three distinct groups. H5 viruses from aquatic birds eluted most readily, with elution being complete after 1.5 h at 37°C. All Hong Kong 1997 viruses eluted at a slower rate. For these viruses, the elution rate correlated with the presence of a glycosylation site at position 158 of the HA. Isolates with this site were able to elute during 3 to 12 h of observation, whereas strains without this site failed to elute even after 24 h of incubation. Thus, the rate of elution from CRBCs of chicken and human H5N1 viruses corresponds to their affinity for CRBC receptors: the lower the affinity the faster the elution. However, there is an apparent discrepancy between the highest binding affinity of duck and gull viruses and their highest elution rate. To explain this finding, we attributed the slower elution rate of chicken viruses to the deletion in the NA stalk, which appears to have decreased the ability of the enzyme to destroy receptors on the erythrocytes.
To test this hypothesis, we determined the NA activity against a low-molecular-weight substrate, 4-MU-NANA, in the same virus preparations that were used in the elution experiments (Table 3). The NA activity of chicken and human virus preparations against this substrate was not lower than that of viruses from aquatic birds, indicating that a lower activity of the chicken virus NA against virus receptors on CRBCs most likely results from the steric hindrance of the enzyme active site. This effect is consistent with the presence of the deletion in the NA stalk of chicken viruses.
Chicken influenza viruses of the H5 and H7 subtypes often carry additional carbohydrates at the head of their HA and contain a deletion in their NA.
To evaluate whether the carbohydrate at position 158 of the HA and the deletion in the NA stalk were unique to the H5N1 influenza viruses from Hong Kong, we analyzed the HA and NA amino acid sequences of different avian viruses available from GenBank.
Table 4 summarizes data on the glycosylation of the HA globular head of H1, H2, H3, and H4 viruses from wild aquatic birds. Generally, only one glycosylation sequon is present on the upper part of the HA1 of these viruses.
TABLE 4.
Glycosylation sequons on the HA1 globular head (positions 90 to 260) of avian influenza virusesa
| HA subtype | Total no. of sequences analyzed | Positions of glycosylation sequonsb |
|---|---|---|
| H1 | 11 (1 turkey) | 94* |
| H2 | 12 (1 guinea fowl) | 94 [1], 169/170* |
| H3 | 15 | 165* |
| H4 | 9 (1 seal, 1 turkey, 1 chicken) | 165*, 216 [1] |
The analysis was performed by using the sequences of avian viruses from GenBank. Most viruses were from the wild aquatic birds (waterfowl, gulls, shorebirds, and shearwaters); the number and host of virus isolates from other species are indicated in parentheses.
The number of sequences with a sequon at a particular position is shown in brackets. An asterisk indicates that all of the sequences analyzed bear a glycosylation sequon at that position.
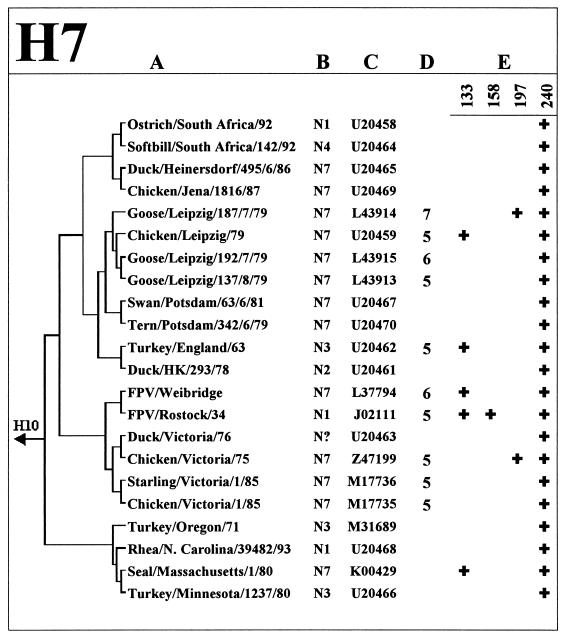
Figure 1 shows the HA glycosylation patterns of the H5 and H7 subtype viruses from different avian species, including chickens. Some viruses of these two subtypes are known to be highly pathogenic in poultry, a property that has been attributed to the presence of multiple basic amino acids at the HA cleavage site (reviewed in reference 23). None of the 11 nonpathogenic avian H7 viruses (three or fewer basic amino acids at the HA cleavage site) isolated mainly from aquatic birds contained more than one glycosylation sequon. Remarkably, among the seven other strains that carry one or two additional glycosylation sequons, six were highly pathogenic, four were isolated directly from chickens, and one belongs to the avian-like virus lineage that caused disease in salts (19). Phylogenetic analysis of the HA sequences (Fig. 1, H7) shows that these H7 viruses with additional glycosylation of the HA do not belogn to the same phylogenetic lineage but emrged through independent evolution from nonpathogenic precursors.
FIG. 1.
Analysis of glycosylation sites on the HA1 globular head of H5 and H7 influenza viruses. Columns: A, Phylogenetic relationships based on the HA1 nucleotide sequences of the virus strains; B, NA subtype; C, GenBank accession number; D, number of basic amino acids at the HA cleavage site, if there were more than three such amino acids at this site; E, positions of glycosylation sequons by H3 numbering (see Fig. 2 for the location of sequons on the three-dimensional model of the HA). The asterisks next to the GenBank accession number mark the H5 HA sequences determined in this study.
Similarly, none of nonpathogenic H5 isolates from aquatic birds has more than one glycosylation sequon on the HA head, and the presence of additional sequons clearly correlates with isolation of such H5 viruses from chickens. As with H7 strains, H5 strains with additional glycosylation sites belong to different lineages that emerged from the viruses of aquatic birds. However, unlike the H7 strains analyzed, not all H5 viruses with additional glycosylation of the HA are highly pathogenic. For example, all H5N2 Mexican viruses isolated in 1994 and 1995 contain an additional site at position 240, but only some late isolates from 1995 have insertions of basic amino acids at the cleavage site. This feature may indicate that additional glycosylation of the HA precedes the development of pathogenicity of the influenza viruses during their evolution in chickens.
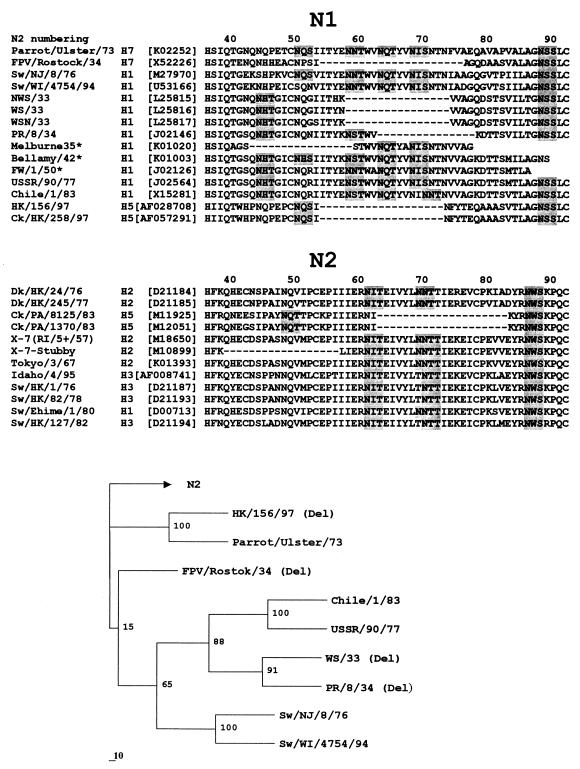
Table 5 presents the results of the analysis of the influenza virus NA sequences for the presence of deletions in the stalk region that would be comparable in size to the 19-amino-acid deletion in the NA of H5N1 strains from Hong Kong. We noticed that the length of the stalk does not vary significantly between NA subtypes: the N9 NA has the longest stalk, the stalks of most other type A and type B virus NAs were only 1 to 3 amino acids shorter (not shown). Nine of more than 100 NA sequences from GenBank were found to contain deletions in the stalk that ranged from 15 to 22 amino acids in length (Fig. 3). One of them (X-7-Stubby) was a laboratory mutant (13). Five NAs with shortened stalks belong to H1N1 human virus isolates from 1933 to 1935. Remarkably, three of the five available NA sequences of the chicken viruses (FPV/Rostock/34, ck/PA/8125/83, and ck/PA/1370/83) contain deletions in the NA stalk that are similar in size and position to that of H5N1 viruses from Hong Kong. Thus, although only a small number of NA sequences of chicken viruses were analyzed in this study, we think the above finding is striking. Moreover, like some of the Hong Kong H5N1 viruses, all three chicken H5N2 and H7N1 strains with deletions in their NAs have additional glycosylation sites on the top of their HAs (Fig. 1), suggesting an interrelationship between these features.
TABLE 5.
Occurrence of the deletion in the NA stalk of influenza virusesa
| NA subtype or type | No. of NA sequences analyzed (no. of those with deletion found)
|
||||
|---|---|---|---|---|---|
| Avian | Equine | Swine | Human | Chicken | |
| N1b | 1 | 2 | 17 (5) | 1 (1) | |
| N2 | 2 | 12 | 36 (1) | 2 (2) | |
| N3 | 1 | 1 | |||
| N4 | |||||
| N5 | 1 | ||||
| N6 | 1 | ||||
| N7 | 1 | 2 | 1 | 2 | |
| N8 | 15 | 7 | |||
| N9 | 3 | ||||
| NB | 13 | ||||
| Total | 25 | 10 | 15 | 66 (6) | 5 (3) |
The NA amino acid sequences from the GenBank database were inspected for the presence of deletions in the stalk region of 10 or more amino acids.
FIG. 3.
Amino acid sequences of the stalk region of N1 and N2 influenza virus NAs (top) and a phylogenetic tree for some of the N1 NAs (bottom). The positions of deletions are shown by dashes; potential glycosylation sites are shaded. Only a partial sequence was available for the NA stalk region of the three human H1N1 strains marked with an asterisk (∗). Numbers at the tree nodes show bootstrap values (100 replications); the horizontal lengths of the branches are proportional to these values.
In the case of the N1 NA, deletions were observed in three distinct groups of viruses (FPV/Rostock.34 [H7N1]; H5N1 viruses from Hong Kong, 1997; and the earliest human H1N1 strains). Phylogenetic analysis (Fig. 3, tree) suggested that these groups belong to separate evolutionary lineages. The region of the NA stalk that is deleted in these viruses is conserved among all N1 NAs that lack deletions (Fig. 3, N1). This feature indicates that deletions in the stalk of the of ancestral N1 NA (rather than stalk insertions) occurred independently in each of the three lineages after their diversion.
DISCUSSION
Poor binding of avian viruses to Sia2-6Gal-terminated sialyloligosaccharide receptor determinants on human respiratory cells is thought to be one of the factors that limit the replication of avian influenza viruses in humans. We found in this study that the H5N1 virus isolate from a human A/HK/156/97 displays the receptor-binding properties that are typical of avian but not of human viruses. This finding indicates that an avian virus can replicate and cause a fatal disease in humans without a significant change in its binding specificity for sialyloligosaccharide receptor determinants. In contrast, the earliest available H2 and H3 virus strains from the previous human pandemics of 1957 and 1968 clearly differ from avian viruses in their ability to bind Sia2-6Gal-specific receptors (9; the present study). The difference between these pandemic viruses and the H5N1 viruses isolated in Hong Kong in 1997 is that the former were isolated when the pandemic was already under way, that is, when the viruses had been present in humans for some time and were effectively being transmitted from human to human. This scenario contrasts with the situation in Hong Kong, where cases of H5N1 influenza in humans resulted from independent and direct introductions of the virus from chickens into humans without evidence of human-to-human transmission (7, 41). Two conclusions can be drawn. First, the receptor-binding specificity of the avian HA does not change immediately after the HA is introduced into humans but rather evolves only after a certain number of replications in this host. Second, alterations of receptor-binding specificity appear necessary for effective virus transmission among humans. Therefore, changes of the receptor-binding phenotype of the avian HA in humans could serve as a marker of ongoing enhancement of the epidemiologic potential of the virus.
Wild aquatic birds are regarded as the source of all human pandemic viruses (reviewed in reference 44). The H5N1 human cases in Hong Kong demonstrate that chickens can serve as an intermediate host for the introduction of these viruses into humans. Indeed, our study indicates that influenza viruses from aquatic birds undergo significant selective pressure in chickens, leading to definite changes in both the HA and the NA during the adaptation process. Thus, although the number of isolates available for the study of HA and NA sequences of chicken influenza viruses was limited, our analysis indicates a marked correlation between the isolation of the virus from chickens and (i) the presence of a deletion in the stalk of the NA and (ii) increased glycosylation of the HA globular head. These features of the HA and NA clearly separate chicken viruses from the viruses of wild aquatic birds.
The stalk region of the influenza virus NA is often regarded as being rather variable, both with respect to its amino acid sequence and with respect to its length. However, as revealed by our analysis, natural variation in the stalk length is limited to a few distinct virus lineages that have large deletions in the stalk compared to the stalks of most other NAs. Three of these lineages belong to chicken viruses with N1 and N2 NAs (FPV/Rostock/34 [H7N1]; H5N1 isolates from Hong Kong; and chicken H5N2 isolates from Pennsylvania, 1983). The fourth known lineage includes several human H1N1 viruses isolated from 1933 to 1935 (4). Partial sequencing of the NA of the A/North Carolina/1/18 strain, which has been implicated in the “Spanish flu” pandemic, indicates that the NA of this strain belongs to the same human virus lineage (42). Given that a deletion in the NA stalk is a characteristic feature of chicken influenza viruses, one can speculate that this human virus lineage originated from a chicken virus precursor.
The mechanisms for the selection in chickens with a shortened NA stalk is unclear. One possibility could be that there is a defect in a formation of enzymatically active NA in chicken cells, which is somehow corrected by the deletion. For example, a deletion in the N1 and N2 NAs of different viruses leads to a loss of several glycosylation sites in the same position of the stalk (Fig. 3), and it can be speculated that this feature may influence the efficiency of co- and posttranslational protein folding (20). Alternatively, the variant with the deletion could be selected because the NA enzymatic activity of the progenitor virus from aquatic birds is too high for its effective multiplication and/or transmission in chickens. Previous studies showed that deletions in the stalk impaired the ability of the enzyme to release influenza virus from erythrocytes (6, 13, 25). We found that the same is true for a deletion in the NA of H5N1 Hong Kong viruses. This means that the chicken virus NA destroys sialylglycoconjugates on the surface of cells less effectively than does the NA of the virus from wild aquatic birds. The selective advantage that could be conferred by this property to the chicken virus remains unresolved.
Inkster et al. (21) were the first to notice that avian H1 influenza viruses differ from human strains by virtue of reduced glycosylation of the HA. Our analysis confirms the notion about low glycosylation of the HA for the other influenza virus subtypes from wild aquatic birds. In contrast, H5 and H7 chicken strains had additional carbohydrates on the top of the HA globular head in close proximity to the receptor-binding site (Fig. 2). Numerous studies indicated that glycans attached at the tip of the HA decrease the binding of the virus to solid-phase exposed receptors, erythrocytes, and cells (10, 12, 16, 27, 30), presumably because of steric hindrance of the receptor-binding site. In accord with these observations, we found that the carbohydrate attached at position 158 of the H5N1 virus HA decreased the virus affinity for soluble sialylglycoproteins and for CRBCs. It can be suggestd, therefore, that additional glycosylation of the HA of chicken viruses modifies the receptor-binding properties of the progenitor aquatic bird virus in chickens. This adjustment could be connected to the deficiency in the NA activity of the chicken virus. Thus, in the case of H5N1 strains, we found that carbohydrate at position 158 compensates for the decreased enzymatic activity of the viral NA and significantly improves elution of the virus from erythrocytes. Alternatively, it cannot be excluded that a deletion in the NA does not occur as a result of an adjustment of the NA activity to match a decreased affinity of additionally glycosylated HA.
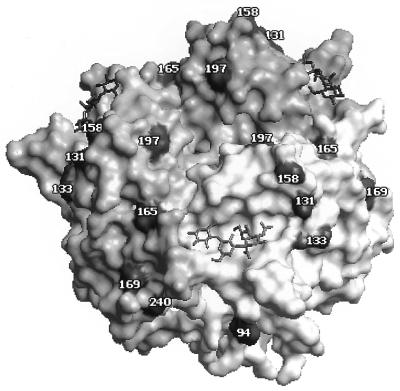
FIG. 2.
Positions of glycosylation sites on the upper part of the HA1 subunit (amino acid residues 90 to 260) of avian influenza viruses shown on a model of the complex between X31 virus HA and 3′-SL (1HGG structure; Brookhaven Protein Databank [37]). The HA monomers are depicted in shades of gray. Positions corresponding to the asparagine residues in the N-linked glycosylation triplets are shown in black and numbered. The sialyllactose molecules are shown as stick models. The figure was generated with WebLab ViewerPro 3.1 (Molecular Simulations, Inc., San Diego, Calif.).
Our observation that the HAs of highly pathogenic H5 and H7 viruses (in poultry) often carry additional glycosylation sites agrees with the finding of Perdue et al. (33), who showed that a single substitution in the HA resulting in the addition of a new glycosylation site at the tip of the HA in position 197 (see Fig. 2) renders the virus highly pathogenic in chickens. It seems reasonable to suggest that this mutation increases the release of virus from cells, facilitating its spread and replication in different tissues of chickens.
Our data introduce the notiion that chickens represent a separate natural reservoir of influenza viruses, with the functional properties of their HAs and NAs being substantially different from those of influenza viruses from wild aquatic birds. Further study is required to understand the relation of these distinctions to the biologic characteristics of chicken viruses, incuding the potential for their transmission to humans.
ACKNOWLEDGMENTS
We thank Richard Bethell of Glaxo Wellcome Research & Development for providing the NA inhibitor zanamivir, Scott Krauss and Melissa Norwood for the technical assistance, and John Gilbert for editing the manuscript.
This work was supported by Public Health Service research grants AI08831 and AI29680 from the National Institute of Allergy and Infectious Diseases, Cancer Center Support (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC). M. N. Matrosovich was supported by a Karnofsky fellowship from St. Jude Children’s Research Hospital.
REFERENCES
- 1.Baum L G, Paulson J C. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology. 1991;180:10–15. doi: 10.1016/0042-6822(91)90003-t. [DOI] [PubMed] [Google Scholar]
- 2.Bean W J, Kawaoka Y, Wood J M, Pearson J E, Webster R G. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J Virol. 1985;54:151–160. doi: 10.1128/jvi.54.1.151-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 4.Blok J, Air G M. Sequence variation at the 3′ end of the neuraminidase gene from 39 influenza type A viruses. Virology. 1982;121:211–229. doi: 10.1016/0042-6822(82)90162-3. [DOI] [PubMed] [Google Scholar]
- 5.Boorsma D M, Streefkerk J G. Periodate or glutaraldehyde for preparing peroxidase conjugates? J Immunol Methods. 1979;30:245–255. doi: 10.1016/0022-1759(79)90098-x. [DOI] [PubMed] [Google Scholar]
- 6.Castrucci M R, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claas E C J, Osterhaus A D M E, Vanbeek R, de Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N2 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 8.Colman P M, Hoyne P A, Lawrence M C. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol. 1993;67:2972–2980. doi: 10.1128/jvi.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 10.Crecelius D M, Deom C M, Schulze I T. Biological properties of a hemagglutinin mutant of influenza virus selected by host cells. Virology. 1984;139:164–177. doi: 10.1016/0042-6822(84)90337-4. [DOI] [PubMed] [Google Scholar]
- 11.De Jong J C, Claas E C J, Osterhaus A D M E, Webster R G, Lim W L. A pandemic warning. Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deom C M, Caton A J, Schulze I T. Host cell-mediated selection of a mutant influenza A virus that has lost a complex oligosaccharide from the tip of the hemagglutinin. Proc Natl Acad Sci USA. 1986;83:3771–3775. doi: 10.1073/pnas.83.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Els M C, Air G M, Murti K G, Webster R G, Laver W G. An 18-amino acid deletion in an influenza neuraminidase. Virology. 1985;142:241–247. doi: 10.1016/0042-6822(85)90332-0. [DOI] [PubMed] [Google Scholar]
- 14.Feldmann H, Kretzschmar E, Klingeborn B, Rott R, Klenk H D, Garten W. The structure of serotype H10 hemagluttinin of influenza A virus: comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology. 1988;165:428–437. doi: 10.1016/0042-6822(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—phylogeny inference package (verison 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Gambaryan A S, Marinina V P, Tuzikov A B, Bovin N V, Rudneva L A, Sinitsyn B V, Shilov A A, Matrosovich M N. Effects of host-dependent glycosylation of hemagglutinin on receptor-binding properties of H1N1 human influenza A virus grown in MDCK cells and in embryonated eggs. Virology. 1998;247:170–177. doi: 10.1006/viro.1998.9224. [DOI] [PubMed] [Google Scholar]
- 17.Gambaryan A S, Matrosovich M N. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J Virol Methods. 1992;39:111–123. doi: 10.1016/0166-0934(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 18.Gambaryan A S, Tuzikov A B, Piskaev V E, Yamnikova S S, Lvov D K, Robertson J S, Bovin N V, Matrosovich M N. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine) Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 19.Geraci J R, St. Aubin D J, Barker I K, Webster R G, Hinshaw V S, Bean W J, Ruhnke H L, Prescott J H, Early G, Baker A S, Madoff S, Schooley R T. Mass mortality of harbor seals: pneumonia associated with influenza A virus. Science. 1982;215:1129–1131. doi: 10.1126/science.7063847. [DOI] [PubMed] [Google Scholar]
- 20.Hebert D N, Zhang J X, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inkster M D, Hinshaw V S, Schulze I T. The hemagglutinins of duck and human H1 influenza viruses differ in sequence conservation and in glycosylation. J Virol. 1993;67:7436–7443. doi: 10.1128/jvi.67.12.7436-7443.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T, Couceiro J N S S, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7443. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Kawaoka Y. Avian influenza. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. London, England: Blackwell Science; 1998. pp. 126–136. [Google Scholar]
- 24.Kato A, Hirata S, Kobayashi K. Nature of the carbohydrate side chains and their linkage to the protein in chicken egg white ovomucin. Part III. Structure of the sulfated oligosaccharide chain of ovomucin. Agric Biol Chem. 1978;42:1025–1029. [Google Scholar]
- 25.Luo G, Chung J, Palese P. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res. 1993;29:141–153. doi: 10.1016/0168-1702(93)90055-r. [DOI] [PubMed] [Google Scholar]
- 26.Matrosovich M N, Gambaryan A S, Tuzikov A B, Byramova N E, Mochalova L V, Golbraikh A A, Shenderovich M D, Finne J, Bovin N V. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology. 1993;196:111–121. doi: 10.1006/viro.1993.1459. [DOI] [PubMed] [Google Scholar]
- 27.Matrosovich M N, Gambaryan A S, Teneberg S, Piskarev VE, Yamnikova S S, Lvov D K, Robertson J S, Karlsson K A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas K B, Nicholas H B, Jr, Deerfield D W. GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14–14. [Google Scholar]
- 29.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 30.Ohuchi M, Ohuchi R, Feldmann A, Klenk H D. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 1997;71:8377–8384. doi: 10.1128/jvi.71.11.8377-8384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 32.Paulson J C. Interactions of animal viruses with cell surface receptors. In: Conn M, editor. The receptors. Vol. 2. Orlando, Fla: Academic Press; 1985. pp. 131–219. [Google Scholar]
- 33.Perdue M L, Latimer J W, Crawford J M. A novel carbohydrate addition site on the hemagglutinin protein of a highly pathogenic H7 subtype avian influenza virus. Virology. 1995;213:276–281. doi: 10.1006/viro.1995.1571. [DOI] [PubMed] [Google Scholar]
- 34.Rogers G N, D’Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 35.Rudneva I A, Sklyanskaya E I, Barulina O S, Yamnikova S S, Kovaleva V P, Tsvetkova I V, Kaverin N V. Phenotypic expression of HA-NA combinations in human-avian influenza A virus reassortants. Arch Virol. 1996;141:1091–1099. doi: 10.1007/BF01718612. [DOI] [PubMed] [Google Scholar]
- 36.Ryan-Poirier K A, Kawaoka Y. Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology. 1993;193:974–976. doi: 10.1006/viro.1993.1208. [DOI] [PubMed] [Google Scholar]
- 37.Sauter N K, Hanson J E, Glick G D, Brown J H, Crowther R L, Park S J, Skehel J J, Wiley D C. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 38.Scholtissek C, Burger H, Kistner O, Shortridge K F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 39.Shu L L, Bean W J, Webster R G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparison of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X Y, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 42.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 43.Warner T G, O’Brien J S. Synthesis of 2′-(4-methylumbelliferyl)-alpha-d-N-acetylneuraminic acid and detection of skin fibroblast neuraminidase in normal humans and in sialidosis. BIochemistry. 1979;18:2783–2787. doi: 10.1021/bi00580a014. [DOI] [PubMed] [Google Scholar]
- 44.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yewdell J A, Caton A J, Gerhard W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J Virol. 1986;57:623–628. doi: 10.1128/jvi.57.2.623-628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]