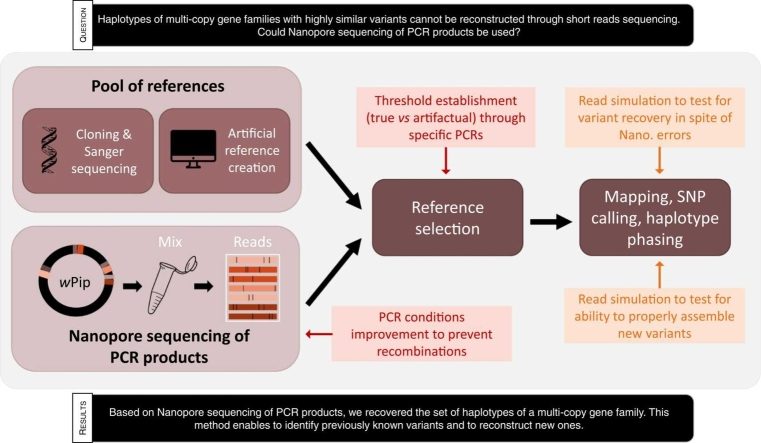
Abstract
The importance of gene amplifications in evolution is more and more recognized. Yet, tools to study multi-copy gene families are still scarce, and many such families are overlooked using common sequencing methods. Haplotype reconstruction is even harder for polymorphic multi-copy gene families. Here, we show that all variants (or haplotypes) of a multi-copy gene family present in a single genome, can be obtained using Oxford Nanopore Technologies sequencing of PCR products, followed by steps of mapping, SNP calling and haplotyping. As a proof of concept, we acquired the sequences of highly similar variants of the cidA and cidB genes present in the genome of the Wolbachia wPip, a bacterium infecting Culex pipiens mosquitoes. Our method relies on a wide database of cid genes, previously acquired by cloning and Sanger sequencing. We addressed problems commonly faced when using mapping approaches for multi-copy gene families with highly similar variants. In addition, we confirmed that PCR amplification causes frequent chimeras which have to be carefully considered when working on families of recombinant genes. We tested the robustness of the method using a combination of bioinformatics (read simulations) and molecular biology approaches (sequence acquisitions through cloning and Sanger sequencing, specific PCRs and digital droplet PCR). When different haplotypes present within a single genome cannot be reconstructed from short reads sequencing, this pipeline confers a high throughput acquisition, gives reliable results as well as insights of the relative copy numbers of the different variants.
Keywords: Multi-copy genes, Nanopore sequencing, PCR recombination, Wolbachia
Graphical Abstract
1. Introduction
Variation in DNA copy numbers have been described since the earliest days of molecular genetics (the first gene duplications and deletions being characterized as early as in the 1900s, e.g. [1], and polyploidy being described in natural populations, e.g. [2], [3]). It is more recently that the role of copy number variations (CNV) and multigenic families in rapid adaptation has been described, all over the tree of life, in eukaryotes (animals, fungi, plants or protozoans e.g. [4], [5], [6], [7], [8]) and bacteria (e.g. [9]). A role of gene amplification in rapid adaptation has also been demonstrated in monopartite viruses [10].
While the importance of gene amplification in adaptation is recognized, studying the structure of gene amplifications remains a challenge. Indeed, gene duplications result in the same piece of genetic material being present multiple times in a given genome: unlike nucleotide mutations (SNPs), no new sequence is created by such amplifications – with the exception of the insertion breakpoint. If this makes amplifications hard to find, they can be detected through coverage variations in next-generation sequencing (NGS, e.g. [11]) or through real-time quantitative PCR (qPCR) when targeting a specific locus. While multiple gene copies may be identical at first, sequence divergence can arise through time by mutations and/or recombinations, resulting in polymorphic but highly similar variants.
In that case, a further challenge is to identify all the different variants (or haplotypes) of a given gene. If variations are separated by a number of base pairs larger than the sequencing read length, haplotype reconstruction using short reads is complex, if not impossible. The recent expansion of long read sequencing is thus of key interest to sequence such gene families.
Yet, if long reads sequencing methods have been widely used for analyses of genomic structural variations, their high error rates (reported 6–8% for MinION sequencing in 2021 [12]) and/or financial cost (for Pacific Biosciences sequencing) have long been a major obstacle to their use for accurate variant identification. Lately, PacBio long reads have successfully enabled to identify new isoforms from RNAseq data, thanks to new tools [13]; improvement of the Nanopore basecallers with Nanopolish [14], Nanocaller [15] or the recent Bonito basecalling (https://github.com/nanoporetech/bonito) made possible the use of Nanopore sequencing for identification of inter-individual SNPs and variations in gene copy numbers [16]. Error correction enabled using Nanopore technology for reference-free transcriptome analysis [17]. Nanopore sequencing has also recently been used for multiplex amplicon sequencing, decreasing the financial cost by 200x as compared to Sanger sequencing [18].
A persistent problem for identification of genetic variants is the non-random distribution of long read sequencing errors, these errors being more frequent in homopolymer regions (representing approximately half of sequencing errors) and in GC-rich regions [12], making it hard to discriminate true mutations from sequencing errors even when increasing the depth of sequencing coverage. This can be mitigated by a previous knowledge of the within-gene polymorphism distribution, e.g. with a good pre-existing database. A further issue arises after having identified SNPs, when haplotype phasing is required to obtain the haplotypes. Indeed, common haplotype phasing tools (e.g. GATK, WhatsHap) require a previous knowledge on the expected number of different gene copies (copy number for multi-copy gene families, ploidy in most of the cases). This is an issue for multi-copy gene families, in which among individual CNVs are frequent.
Here, we studied the cidA and cidB genes, present in tandem in the genome of the endosymbiotic bacteria wPip infecting Culex pipiens mosquitoes. cid genes are respectively 1475 bp (cidA) and 3524 bp long (cidB). These genes are amplified and diversified in wPip with up to 6 different copies, named variants, described within a single Wolbachia genome [19]. The set of cidA/cidB gene variants present in an individual is called a repertoire [19], and cid genes copy numbers vary among strains of wPip.
These genes are of key interest, since they encode proteins involved in cytoplasmic incompatibility (CI) [20], [21], a well-studied reduction of hatching rates induced by Wolbachia in its arthropod hosts. This phenomenon, which can lead to crosses with null hatching rates, is currently implemented as a means of mosquito vectors and agricultural pests control [22], [23], [24], [25]. The identity of the cid variants present in a given genome correlates with incompatibility patterns [19], [26]. Being able to obtain individual’s cid repertoires is thus a prerequisite to understand and predict CI patterns and evolution in mosquito populations.
Illumina sequencing of these genes showed that they had a shared architecture, with two polymorphic regions separated by a monomorphic region of more than 500 bp [19], preventing haplotype reconstruction using short read sequencing. Thus, gene variants were previously identified using a 2-step process: (i) a portion of each gene of approximately 1.3 kb, containing the variable regions was amplified by PCR using generic primers amplifying all copies, and (ii) PCR products were cloned and Sanger sequenced, enabling to identify different gene variants present in the PCR product [19]. This method enabled to describe more than 30 variants of cidA and cidB in wPip infecting Culex pipiens mosquitoes from all around the world [19], [26], [27], [28]. Yet, this method is extremely time consuming to set up and thus inappropriate for large scale studies. In addition, it has limited descriptive power: due to time and money, a maximum of 48 clones were sequenced per individual PCR product, making it likely to miss a rare variant.
We sought to develop a faster and more efficient method, based on Nanopore sequencing of PCR products. We amplified the same 1.3 kb fragment encompassing the variable regions of each gene and sequenced the PCR products by Nanopore sequencing. Then, we set up a bioinformatics pipeline to identify all the different variants present in the Nanopore reads. This pipeline relies upon the 30 references previously identified through cloning and Sanger sequencing, and on previous knowledge of the genes mutation distribution. It assigns each read to its closest known reference through mapping, then uses a combination of SNP calling and haplotype phasing to identify new variants.
The pipeline was validated and fine-tuned using a combination of read simulations and molecular biology approaches. Read simulations were used to (i) confirm that the pipeline properly recovered variants in spite of Nanopore sequencing errors and (ii) ensure that it was possible to identify genetically distant new variants which were absent in the existing database. Furthermore, repertoires from the same strains were also obtained using the former cloning and Sanger sequencing method, giving concordant results and validating the pipeline. The rare discrepancies between Sanger and Nanopore sequencing were sorted out using PCRs targeting specific regions, showing that Nanopore sequencing results were correct. We highlighted that PCR and/or sequencing could induce frequent recombinations and implemented protocol changes to mitigate these recombinations. We established a coverage threshold, enabling to discriminate true variants from fake chimeric reads.
Overall, we developed an efficient and trustworthy method, enabling to easily sequence polymorphic multi-copy gene families at a wide scale. Moreover, using digital droplet PCR (ddPCR), we showed that such method could give access to the relative copy numbers of the different variants.
2. Material and methods
For all experiments, total DNA was extracted on adult mosquitoes following the acetyltrimethilammonium bromide (CTAB) protocol [29].
2.1. Mosquito lines used in this study
All the mosquito lines used were isofemale lines, i.e. lines obtained by rearing the progeny of a single female. cidA and cidB repertoires were acquired through Nanopore sequencing of PCR products for 13 isofemale lines reared at the laboratory (Table S1).
All isofemale lines were reared in 65 dm3 screened cages, in a single room maintained at 26 °C, under a 12 h light/ 12 h dark cycle. Larvae were fed with a mixture of shrimp powder and rabbit pellets, and adults were fed on honey solution. Females were fed with turkey blood, using a Hemotek membrane feeding system (Discovery Workshops, UK), to enable them to lay eggs.
2.2. Cloning and Sanger sequencing
cidA and cidB gene variants were obtained through cloning and Sanger sequencing of PCR products following the procedure from [19]. Polymorphism is located in 2 specific regions (named upstream and downstream regions) for both cidA and cidB. Generic primer pairs shown in Table S2 amplify all the variable regions of the cid genes [19]. The GoTaq polymerase (Promega) was used for all amplifications. The PCR products were then cloned using the TOPO TA cloning Kit pCR 2.1-TOPO Vector (Invitrogen), in order to separate the distinct variants present in the PCR product in distinct clones. Each clone was then Sanger sequenced.
2.3. Sequence acquisition through nanopore sequencing
cid genes were amplified using the same primer pairs as for Sanger sequencing (Table S2), on DNA extracted from a single adult mosquito. For each of the strains, repertoires were acquired for two distinct individuals. PCR products were purified in order to remove the PCR reagents using CleanPCR beads at 1.8X (CleanNA) and quantified using a Qubit fluorometer and Qubit DS DNA Broad Range kits (ThermoFisher). Purified PCR products of cidA and cidB genes were pooled in an equimolar mix. Preliminary tests of Nanopore sequencing (not shown here) were done using a sequencing protocol that involved an amplification step. Since we found that amplifications create artificial recombinations, the sequences used in this study were obtained using a PCR-free protocol, by the MGX platform (Montpellier GenomiX). The DNA amplicons were quantified using a Tecan infinite 500 Fluorometer (Tecan, Switzerland) with a dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific, Massachusetts, USA). The fragments size distribution was checked using a 5200 Fragment analyzer (Agilent, USA) system with a Sandard NGS kit (Agilent, USA). The amplicons libraries construction was done according to the Nanopore protocole NBA_9102_V109_revA_09Jul2020. Two hundred nanograms of DNA are end repaired and dA-tailed using NEBNext End repair/dA-tailing Module (E7546, New England Biolabs, Ipswich, Massachusetts, USA). The samples were then barcoded using EXP-NBD196 (barcodes 1–96) kits (Oxford Nanopore Technologies, Oxford, UK) and ligation Sequencing Kit 1D SQK-LSK109, (Oxford Nanopore Technologies, Oxford, UK). Up to 96 samples were pooled per run. Barcoded samples were pooled and purified using 0.4 vol of AMPure XP magnetic beads. The AMII sequencing adapter (Oxford Nanopore Technologies, Oxford, UK) was ligated to barcoded pools using Quick T4 DNA ligase (New England Biolabs, Ipswich, Massachusetts, USA) and the sequencing libraries were purified using 0.4 vol of AMPure XP magnetic beads. MinION sequencing was performed as per manufacturer’s guidelines using R9.4.1 flow cells FLO-MIN106, ONT and controlled using Oxford Nanopore Technologies MinKNOW software version v20.06.5. Flow cells were then transferred to Nanopore MinION Mk1b (Oxford Nanopore Technologies, UK) for Nanopore single molecular sequencing.
Base calling was performed after sequencing using the GPU-enabled guppy basecaller in high accuracy mode for 96 samples (version 6.5.7). Only reads with a PHRED quality above 9 were used for subsequent analyses.
2.4. Pipeline details
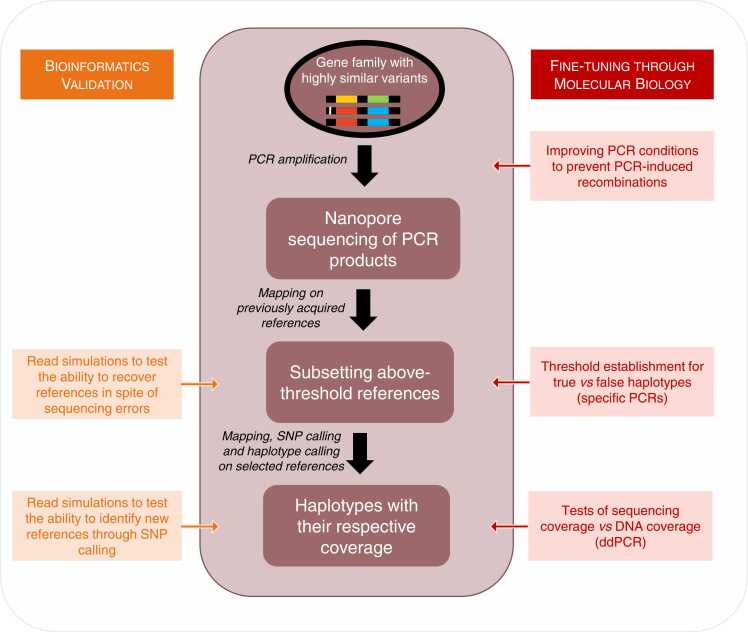
Throughout the whole pipeline, mapping was done using minimap2 [30], with parameters -ax map-ont, unless specified otherwise. We first separated cidA and cidB reads by mapping all reads on a short monomorphic sequence of each gene, corresponding to positions 228–327 and 1211–1309 for cidA and cidB respectively. After this step, there were around 15,000–20,000 reads per gene. The pipeline was then run separately for cidA and cidB reads, the following steps of the pipeline being identical for both genes.
For each gene, reads were mapped on the full reference database (the making of this database is described in Section 2.5) and secondary alignments were removed. Samtools coverage was then used (samtools 1.15, [31]) to obtain the coverage of each reference (Fig. 1). Using specific PCRs, we determined that selecting references which had a coverage above 10% of the total number of reads enabled to get all the true references, and excluded artifactual chimeric reads (detailed in Section 3.2). References above that threshold were extracted, along with reads mapping on these references. Since highly similar references artificially introduce INDELs, and decrease coverage in SNP calling when using bcftools mpileup, we looked for pairs of references differing by less than 3 SNPs and kept a single representative for each pair (Fig. 1). Exclusion was done by computing the raw distance among sequences using the dist.dna function in the R package ape [32].
Fig. 1.
Overview of the bioinformatics pipeline The different steps of the pipeline are shown, along with bioinformatics and molecular biology checks. cid gene variants are represented with their upstream and downstream variable regions shown by different colors. Some variants differ by few SNPs only, outside of colored variable regions. SNPs are represented by a white bar.
Reads which had mapped on at least a reference were then mapped on the reference subset, and SNPs were called using bcftools mpileup (with the ont config, a minimum mapping quality of 10, a disabled BAQ and a max depth of 75,000) and bcftools call (multiallelic caller, with an expected substitution rate of 0.5). If no SNPs were called, variants present and their respective coverage were extracted to assemble the repertoire. If SNPs were called, two alternative cases occurred: (i) cases where haplotypes could be directly deduced from the SNP calling or (ii) cases where haplotype calling was required to sort out haplotypes. In the first case, the consensus sequence was obtained using bcftools consensus (option --haplotype I), while in the second, WhatsHap phase [33] was used to phase the haplotypes. If the respective coverage of the references and alternative gene version can vary, we never found more than two distinct alleles at a given position, and thus used the default WhatsHap settings (in cases where more than 2 distinct variants can correspond to a given known reference, WhatsHap polyphase could be used).
2.5. Reference database: true and in silico references
The above-described pipeline is based on mapping on a cid (cidA or cidB) reference database. This database was made of two distinct sets of references: (i) references previously acquired through cloning and Nanopore sequencing of cid genes [19], following the protocol described in 2.2.; and (ii) references built in silico, using knowledge on the polymorphism profile in cid genes.
cid genes are composed of two variable regions separated by a monomorphic region, and recombination occurs between those two regions (Fig. 1, [19]). We thus completed the pool of references already sequenced by creating an in silico pool of references for each of the five wPip group [34] by combining all the previously sequenced upstream and downstream regions within each group.
2.6. Nanopore read simulation
Our read simulation script is based on the simulation of full-length transcripts used in [17] with modifications to fit our targeted sequencing scenario. We simulate reads as follows. The script takes as input a fasta file with a number of N starting reference sequences, an integer C (C> N) of targeted simulated references, a fraction S corresponding to the mutation rate of the references, and the mean error rate X of the reads. Furthermore, the script has two settings regarding mutation type. It can simulate substitutions only, or both SNPs and indels. The outputs of the script are a fasta file with the references (original and simulated) from which the reads were simulated, and a fastq file with the simulated reads. We now describe the workflow of the script.
First, C references are simulated from the N starting references as follows. The N original references are added to a pool of simulated references here denoted R. Then, a random reference r is sampled from R and mutated with the mean mutation rate S according to the mutation profile specified and placed back into the pool R which now contains N + 1 references. This procedure is repeated until the pool R contains C references. Note that a simulated reference can be selected from R and in turn be mutated into a new reference, creating a tree-like evolution structure.
Second, reads are sampled from the pool of C references in the same way as in [17] for full length transcripts. We briefly describe the procedure here, for details see Supplementary note 1 in [17]. To simulate a read we pick a reference in R at random. We simulate a quality value uniformly at random over each base pair in the sampled reference. The base is assigned the Phred score and we introduce an error at that position with a probability corresponding to the phred score. The error types are either deletion, substitution, or insertion with probabilities of 0.45, 0.35, and 0.2, respectively, which roughly mimics the error profile of ONT data although nanopore base calling algorithms changes rapidly. Our script is able to produce reads with a mean error of ∼3.9%, 7, and ∼11.4% error rate through different ratios of phred quality values.
2.7. Testing the coverage of specific variants using ddPCR
Prior to the experiment, cidA variants of the Lavar strains were cloned as described above and in [19]. Clones corresponding to each variant were used as controls for the PCR specificity, using pairs of specific primers that should amplify specifically a single clone. To that extent, we designed primer pairs (Table S2) and set up ddPCR protocols.
The digital PCR assays were then set up performed using the Naica digital PCR system (Stilla Technologies). The dPCR reaction mixture (25 μL) contained 5 μL of Quantabio PerfeCTa Multiplex qPCR ToughMix 5x (Quantabio), 1 μL of Dextran Alexa Fluor 647 10,000 MW (Thermofisher), 1.9 μL of Evagreen 20x (Biotium), 3.125 μL of the target primer set (final concentration of 125 nM) and nuclease-free water up to 25 μL. DNA was added in a quantity sufficient to get enough positive droplets (for full mosquitoes, DNA amount could not be quantified as infection levels of the endosymbiotic Wolbachia vary and extracted DNA is always mixed with host DNA). The reaction mixtures were loaded into wells of Sapphire chip and were subsequently emulsified (20,000–30,000 droplets/sample) and amplified in a geode thermocycler (Stilla technologies). The ddPCR conditions used were 10 min initial denaturation at 95 °C, followed by 45 cycles of 95 °C for 30 s, 58 °C for 15 s and 72 °C for 30 s. After template amplification, the chips were transferred to the reader. Extracted fluorescence values for each droplet were analyzed using the Crystal Miner software (Stilla Technologies).
2.8. Code availability
All scripts used for simulating datasets, to run the pipeline and its evaluation are found at https://github.com/alnam3/nano_seq. Specifically, the code to simulate Nanopore reads under https://github.com/alnam3/nano_seq/Nano-read-simulator, and the suggested awk script to test for missed SNPs under https://github.com/alnam3/nano_seq/Missed_SNPs.
3. Results
3.1. Pipeline overview
Our goal was to create a bioinformatics pipeline to reconstruct multi-copy gene families with variable copy numbers through Nanopore sequencing of PCR products. To this aim, we worked on the cidA and cidB genes of wPip Wolbachia infecting various Culex pipiens isofemales lines.
We successfully established a pipeline which is made of the following steps: (i) amplification of the target gene(s) through a generic PCR, amplifying all gene copies present in the genome, regardless of the variant; (ii) mapping of the reads on a reference base; (iii) selection of the references covered above a pre-determined threshold (detailed below) and of the reads which mapped on them (called cleaned reads); (iv) filtering the references to keep only references differing by at least three SNPs (final references); (v) mapping and SNP calling of useful reads final references; (vi) haplotype reconstruction through haplotype calling (Fig. 1).
The filtering of references before the step of mapping and SNP calling is required, as references which are highly similar are an issue for downstream SNP calling, causing spurious indels. Note that different PCR products can be pooled for sequencing and separated for downstream analyses (here, we pooled cidA and cidB PCR products). Furthermore, while the pipeline requires a pre-existing database, new variants can be recovered through SNP calling.
3.2. Definition of the threshold for true variants by specific PCRs
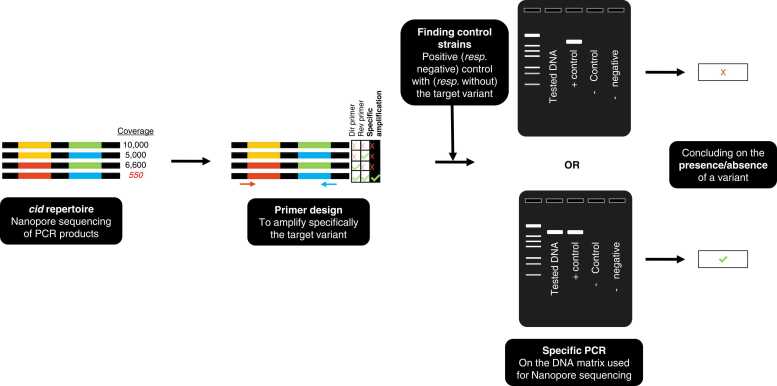
For all the wPip strains analyzed, the coverage distribution across all references showed that most references had no coverage, few had a high (more than 10% of reads) coverage, and some had a low-to-intermediate coverage (exemplified in Fig. S1). One of the key steps was to determine if all the covered variants were truly present in the genome of the studied wPip strain or if some were artifactual. In order to determine whether those “low-to-intermediate coverage” variants were truly present in the sample, we designed multiple specific primers to test the presence of these variants by PCR, in the exact same DNA matrix used for Nanopore sequencing.
To do so, we chose cases for which specific primers of the questioned variant could be designed (i.e. primers amplifying solely the target variant and no other variants present in the repertoire, Fig. 2). We restricted tests to cases for which both a negative control (i.e. an infected Culex mosquito for which the primers should not amplify any variant), and a positive control (i.e. an infected mosquito for which the primers should amplify a variant, Fig. 2) could be used. We considered that a variant was truly present in a strain, and that the strain could thus be used as a control, when the variant was highly covered in Nanopore sequencing and previously found by Sanger sequencing [19], [26], [27], [28]. This design enabled to eliminate the potential non-specificity of primers, a problem that can be frequent on cid variants due to the recombinant nature of the genes.
Fig. 2.
Method developed to test for the presence/absence of a specific variant using specific PCRs Primers amplifying this specific variant and not others have to be found, then true positive and negative controls are used (other strains in which this specific variant is specifically present or absent).
Overall, we successfully tested the presence/absence of 8 “low to intermediate-coverage” variants, in 8 different wPip strains repertoires and found that none of them could be amplified by PCR (shown in Table S3, either in dark blue or in orange, depending on whether the variant had been found in Sanger or not). Since the number of reads differed among wells, we established a relative coverage threshold, expressed as a percentage of the total number of reads. Using specific PCRs, we found that truly present references were those covered by at least 10% of the total number of reads, or a depth above 1500 reads.
3.3. Subsetting the putative references to prevent SNP calling issues
We used samtools 1.15 [31] for SNP calling, and found that keeping references which are really close (differing by a few SNPs) caused SNP calling problems, by artificially introducing INDELs and diminishing the coverage. The conclusion that SNP calling issues are involved, rather than mapping issues was reached because using samtools depth and samtools mpileup (also giving the depth at each position for each reference, along with calling SNPs) on the same.bam file and with the same options (setting the minimum quality at 13 for both, and increasing the maximum depth value to 70,000 for mpileup) gave drastically different results: the coverage obtained with samtools mpileup can be below 100 reads, when samtools coverage outputted a coverage of several hundred reads). This issue was solved by keeping a single representative of each pair of highly similar references. An important note is that here, since we are using targeted long reads, the reference and the reads have the same length. We thus expect the average coverage to reflect the depth at each position.
Above-threshold references were thus subsetted to keep a single representative of each pair of references differing by less than three SNPs. We found that a 3-SNP threshold was sufficient to get rid of SNP calling issues.
3.4. Simulations confirm pipeline’s ability to recover variants
Although Nanopore sequencing quality has drastically improved in the last years, the rate of sequencing errors was still estimated to be around 7% in 2021 [12]. We tested, using simulated reads, the ability of the pipeline to recover all variants present in spite of Nanopore errors. To our knowledge, Nanopore read simulators such as NanoSim [35], DeepSimulator [36], SimLord [37], and SNaReSim [38] are designed for genomic data and do not mimic targeted sequencing where the majority of reads covers the whole amplicon. We therefore wrote our own read simulator for targeted data based on the full-length transcriptome read simulation pipeline in [17] (isONcorrect) (details of the simulation script in the method section Nanopore read simulation). Using our simulation script, we simulated 20,000 Nanopore reads from existing references. We tested the influence of (i) the read error rate, (ii) the number of distinct variants and (iii) the repertoire’s complexity on the pipeline’s ability to recover the references. To do so, we simulated reads from variants in eight different settings. The eight settings were all combinations of varying error rates (4 or 12%), gene copy numbers (2 or 6 distinct copies) and repertoire complexity (starting from existing variants which are either highly similar or strongly different). The pipeline was run on reads simulated from all these cases, and the right repertoire was recovered each time. This simulation confirmed that our pipeline was robust at error rates higher than the typical Nanopore error rate of about 7%.
3.5. SNP calling and haplotype calling enable to properly recover new variants
We also tested the ability of the pipeline to recover true new variants (i.e. not yet present in the complete database). To that extent, we used our Nanopore read simulator, which enables to create simulated mutants with a user-set mutation rate. We created new variants by introducing a fixed number of SNPs in the existing ones. Nanopore reads were then simulated from (i) simulated variants only and (ii) a mixture of simulated and existing variants. For this experiment, reads were simulated with a 7% error rate, as we showed above that the pipeline correctly recovered the references with error rates up to 12%, and because 7% is close to recent estimates of the realistic Nanopore sequencing error rates [12]. Simulated SNPs were introduced in the existing variants in such a way that the new simulated variants differed from the existing ones by a distance equal to the median pairwise raw (Hamming) distance among true references (the database being already wide, finding a new variant differing from all others by more than this distance was highly unlikely). No INDELs were introduced since previous work showed they were unlikely in cid genes [19], [26], [28], even if the read simulation script used enables to introduce INDELs. Simulating sequences enabled us to know exactly where the SNPs were located and thus to check our pipeline’s performance in a controlled scenario.
Using 100 different simulations, we computed the true number of SNPs (ranging from 41 to 71), along with the number of SNPs recovered by SNP calling. We found that on average, 3.4 ± 3.1 SNPs were missed (mean ± standard deviation, ranging from 0 to 18). There were two distinct cases for missed SNPs: (i) SNPs located in repeated regions (e.g. AATTAATA -> AATTTAAT, likely considered by the caller as a spurious SNP resulting from a sequencing error); (ii) SNPs not called in spite of a high coverage of the alternative base (e.g. at some positions, 6500 reads supporting the alternative base, yet the SNP was not called). This last issue was not solved by increasing the prior on the substitution rate. We thus wrote an awk script to detect such issues of missed SNPs (in spite of a high number of supported reads). Using this script on our true sequencing data, we found no occurrence of such issue, suggesting it might only occur when SNPs are numerous.
3.6. Comparison with Sanger sequencing results
Repertoires obtained through Nanopore sequencing were compared with repertoires previously obtained by Sanger sequencing [19], [26], [27], [28]. We found an overall agreement between Nanopore-obtained repertoires and Sanger-obtained repertoires (Table S3). A total of 54 variants were detected for cidA (excluding variants which were only found in low coverage in Nanopore sequencing, highlighted in dark blue in Table S3). Among those, only 4 differed between Sanger and Nanopore sequencing (excluding variants shown in dark blue, i.e. counting pink, light blue and orange cells). For cidB, 41 variants were detected in total, and 8 disagreements among sequencing methods were found. Out of the 12 total disagreements, 4 variants were only found in Nanopore sequencing (highlighted in pink), and 8 were only found in Sanger sequencing (or found with a low coverage in Nanopore sequencing, which we determined to be spurious, highlighted in light blue or orange). While it was expected to find some additional variants in Nanopore sequencing, since this method gives a coverage of up to 40,000 reads per gene, as compared to the analysis of 24–48 clones used in Sanger sequencing, the opposite case was not expected.
In order to solve these discrepancies, we used the same approach as for threshold establishment: we identified a target region which could be diagnostic of the presence/absence of a given cidA variant, then designed specific PCR primers and checked for its presence/absence (Fig. 2). This was done on a total of 9 variants whose presence/absence differed between Sanger and Nanopore sequencing. All variants which were found only in Nanopore with a high coverage were detected through specific PCR. Conversely, none of the variants only found by Sanger sequencing were found (Table S3).
3.7. Artifactual variants are due to PCR-induced recombinations
Close examination of artifactual variants’ sequences found both by Sanger and Nanopore sequencings, revealed that they were all putative recombinant variants that could result from recombination of variants present in the repertoire of the same wPip strain. It seemed that more artifactual variants were observed when the wPip strain had a high number of cid variants (for example Slab, [27]). Sanger and Nanopore sequencings were both run on PCR products, and PCR amplification has previously been described as a potential cause of artificial recombinations, especially when using a high number of PCR cycles [39], [40], [41]. To test the putative link between artifactual variants and number of cycles used for PCR, we chose the Wolbachia from the Slab isofemale line because it shows many cid variants [27]. We amplified the cidA genes in 4 individuals using either 31 or 35 PCR cycles (on the same DNA for both PCRs), then sequenced the amplicons through the same Nanopore sequencing protocol. Slab had also been previously Sanger sequenced using 35 PCR cycles. We found more artifactual, chimeric reads when 35 cycles were used compared to 31, coverage of the corresponding references being up to almost 10% at 35 cycles. The cidA repertoire has 4 different variants, and two additional artificially created variants were repeatedly sequenced, by both Sanger and Nanopore sequencing (namely cidA‐III‐gamma(3)‐25 and cidA‐III‐beta(2)‐12, which can come from recombinations between existing variants). Similarly, two additional artifactual cidB variants had been found in Sanger sequencing in Slab (using more PCR cycles), suggesting this strain may be prone to recombinations possibly due to its high number of cid variants.
3.8. Nanopore sequencing coverage predicts relative variant copy numbers
Nanopore sequencing of different wPip strains gave consistent differences in coverage between the variants (some variants being more covered than others). To test whether these differences in coverage may reflect variations in copy number among the different variants, we analyzed the three cidA variants (cidA-II-alpha-15, cidA-II-alpha-7, and cidA-II-beta-15) of the Wolbachia from the Lavar isofemale line, which were identified both by Sanger and Nanopore sequencings. Since specific amplification of the different variants requires a sequence of approximately 600 bp, which is too long for classic quantitative PCR (qPCR), we used digital droplet (ddPCR) to amplify specifically the variants. Since different PCR primers were used for each variant, we first investigated whether these PCRs had similar efficiencies, to ensure that differences in variant quantifications resulted from true differences in copy number rather than distinct PCR efficiencies. We first cloned each variant then designed specific primer pairs for each of the three variants, and validated their specificity (a given primer pair must amplify the corresponding clone only to be validated). To ensure that differences in PCR efficiency did not affect differences in nanopore coverages, we ran the three specific PCRs on the same amount of DNA from each clone. We obtained similar concentrations, showing differences in Nanopore coverage indeed reflected concentration variations (Table S4A).
Since quantifying the different copies present in wPip-Lavar genome by ddPCRs required the use of total DNA containing a mixture of all cidA present, we had to ensure that we were able to quantify all cidA copies. To do so, we tested three generalist cidA primer pairs located in a monomorphic region of the cidA gene that should amplify all gene variants (Table S2). We tested each generalist pair of primers using an equimolar mixture of the isolated clones. Surprisingly, and although ddPCR is supposed to be devoid of efficiency effects associated with amplified fragment size, we found that concentration (number of copies) obtained were widely different when the fragment size was 150, 580 or 1300 basepairs (bp) (715.8, 535.4 and 307.7 copies/μL, respectively), showing that the fragment size strongly influences the obtained concentration (Table S4B). For further experiments, we chose the 580 bp generalist pair of primers as it was the pair giving (i) amplified fragments with a size close to the specific fragments (∼600 bp), and (ii) a concentration close to the sum of individual copy numbers (i.e. 586 copies/μL) in the equimolar mix of the three specific clones.
We then quantified the different variants in the DNA obtained from a single Lavar mosquito. Comparing the relative quantifications of the different variants using ddPCR with relative coverage obtained in Nanopore sequencing, we found an overall agreement with fewer copies of the cidA-II-gamma-15 variant compared with the two others, suggesting that Nanopore sequencing coverage could give proper insights in relative copy numbers (Table S5). Combined with a qPCR giving the total number of cidA copies, such relative coverage could be used to obtain variants’ copy numbers.
4. Discussion
Within a genome, there can be multiple highly similar copies of a multi-copy gene family. For cid genes, our focal example here, we called these different copies ‘variants’, and the full set of copies per genome is called a ‘repertoire’. Sequencing and assembling such families, with highly similar copies and copy number variations represents an issue. Indeed, long reads are highly error-prone, making it difficult to differentiate true polymorphism from sequencing errors, and short reads fail to reconstruct haplotypes. Here, we developed a bioinformatics pipeline enabling to reconstruct the full repertoire of a gene family within a single genome through long read Nanopore sequencing of PCR products, based on an existing reference database of Sanger sequences. We validated the pipeline using a combination of read simulation and molecular biology approaches. These approaches were combined in order to ensure that (i) all variants present in a given host were recovered and (ii) putative artifactual variants were identified as such. We confirmed that all variants were recovered by comparing repertoires obtained with the current method (Nanopore sequencing of PCR products) and with the previous method (Sanger sequencing of PCR products). Furthermore, we could compare repertoires obtained with our method with those from 2 full genome long reads, previously acquired [19]. We also performed several specific PCRs, targeting random variants, showing that these variants truly existed in the individuals. We found that some chimeric variants were present, likely resulting of the PCR step. To sort these variants from “true” ones, we set up a coverage threshold through numerous specific PCRs. Additionally, repertoires were acquired for two distinct individuals for most of the strains.
While it was possible to discriminate true from artifactual variants, chimeric variants were regularly detected, both in Sanger and in Nanopore sequencing, likely resulting from PCR-induced recombinations. Our results confirm previous results showing that the probability of chimera formation increases with the number of cycles ([39], here with 31 vs 35 cycles), and when similar template sequences are amplified in the same PCR reaction [39], [42], [43]. Recently, along with the increased sequencing of PCR products with the rise of metabarcoding, and of other methods such as MPRAs (massively parallel reporter assays), some studies examined the impact of such chimeras on multiplexing results, showing that up to 28% of the sequences correspond to mistags, resulting from tag-switching events [44]. Finding such high occurrences of PCR-linked errors made previous studies seek for the optimal PCR conditions to reduce chimera formation, examining for instance the consequences of the type of polymerase used [45] or various factors such as the number of cycles used or the amount of DNA template [46], [47]). Here, we further stress the urge to consider such chimeras, especially when working on recombinant genes, since the genetic architecture of those genes makes harder to discriminate true variants and chimeras.
PCR-induced recombinations being an issue regardless of the sequencing method, Nanopore sequencing of multigene families has massive advantages compared to cloning and Sanger sequencing of PCR products: it enables to sequence longer fragments (up to 3 kb here) which could hardly be inserted into plasmids, and it is much more time effective as multiple genes can be sequenced in the same sequencing well. While we sequenced just two genes at the same time here, simultaneous acquisition of multiple PCR products through Nanopore sequencing has already been validated, increasing the cost-effectiveness of the method.
Since we found that Nanopore sequencing coverage strongly differed among variants, we sought to see if these coverage differences could be reproduced using other methods, or if they likely resulted from Nanopore sequencing biases – those biases being numerous, see [12] for a review. We confirmed that coverage variations in Nanopore sequencing could give hints of copy number variations using digital droplet PCR (ddPCR). To go from coverage variation to variant copy number, the total number of cid copies is required. This can be easily obtained with a generic quantitative PCR normalized on a single-copy gene. While ddPCR mirrors Nanopore sequencing coverage, it has to be kept in mind that both methods involve a PCR, and that PCRs have been shown to skew template-to-product ratios, with a bias towards some gene versions [48]. Some gene variants may be more easily amplifiable, resulting in higher coverage. In making controls to ensure that all gene copies were amplified in ddPCR, we found that fragment size, and by extent reaction efficacy, unexpectedly influenced the outcomes of ddPCR, which could result in some biases if one was to compare concentrations of different genes using amplicons differing in size. This bias can be counterbalanced by ensuring that all amplified fragments have approximately the same size, but has to be carefully taken into account when using ddPCR.
5. Conclusion
While the method presented here requires a good pre-existing reference database, the recent development of methods to work on multigene families based on less error-prone PacBio sequencing data [13], along with the improvement of both Nanopore basecalling [15], [49] and error-correction [17] suggest that de novo assembly of multigene families will be possible with Nanopore sequencing in the coming years. Overall, in spite of some limitations due to the PCR step itself, this method enables a quick acquisition of numerous variants, which could be further increased by acquiring more than two genes at the same time. This paves the way to wide scale multigene family acquisitions and, with the present study, to a better understanding of links between cid genes and cytoplasmic incompatibility phenotypes.
Authors’ contributions
AN, MS and MW conceived the experiments. KS, KB and IB participated in the pipeline development and/or bioinformatics validation. Molecular assays were conceived by AN, PM and MW, and performed by AN and PM. MW acquired funding. AN, MS and MW wrote the paper. AN, KS, MS, KB and MW revised the manuscript. All authors read and approved the final manuscript.
CRediT authorship contribution statement
Alice Namias: Methodology, Validation, Investigation, Writing, Reviewing and editing. Kristoffer Sahlin: Methodology, Validation, Editing. Patrick Makoundou: Methodology, Investigation. Iago Bonnici: Methodology. Mathieu Sicard: Conceptualization, Writing, Reviewing and editing. Khalid Belkhir: Methodology, Editing. Mylène Weill: Conceptualization, Funding acquisition, Supervision, Writing, Reviewing and editing.
Declaration of Competing Interest
The authors have declared no competing interests.
Acknowledgements
We thank the MBB platform for their help on code-related issues. Digital PCRs were run on the ISEM qPCR platform. Qubit were run using GenSeq platform facilities. We thank the MGX platform for Nanopore sequencing, and especially Dany Severac and Anaïs Louis. This project was funded by the French MUSE project with the reference ANR-16-IDEX-0006. Kristoffer Sahlin was supported by the Swedish Research Council (SRC, Vetenskapsrådet) under Grant No. 2021–04000.
We thank Nicole Pasteur and three anonymous reviewers for thorough reading and helpful comments on previous versions of this manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.012.
Contributor Information
Alice Namias, Email: alice.e.namias@gmail.com.
Mylène Weill, Email: mylene.weill@umontpellier.fr.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
References
- 1.Sturtevant A.H. The effects of unequal crossing over at the bar locus in Drosophila. Genetics. 1925;10:117–147. doi: 10.1093/genetics/10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avise J.C., Kitto G.B. Phosphoglucose isomerase gene duplication in the bony fishes: an evolutionary history. Biochem Genet. 1973;8:113–132. doi: 10.1007/BF00485540. [DOI] [PubMed] [Google Scholar]
- 3.Patrick Gage L. Polyploidization of the silk gland of Bombyx mori. J Mol Biol. 1974;86:97–108. doi: 10.1016/S0022-2836(74)80010-0. [DOI] [PubMed] [Google Scholar]
- 4.Iskow R.C., Gokcumen O., Lee C. Exploring the role of copy number variants in human adaptation. Trends Genet. 2012;28:245. doi: 10.1016/J.TIG.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardigan M.A., Crisovan E., Hamilton J.P., Kim J., Laimbeer P., Leisner C.P., et al. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell. 2016;28:388–405. doi: 10.1105/TPC.15.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steenwyk J.L., Rokas A. Copy number variation in fungi and its implications for wine yeast genetic diversity and adaptation. Front Microbiol. 2018;9:288. doi: 10.3389/FMICB.2018.00288/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauer S., Gresham D. An evolving view of copy number variants. Curr Genet. 2019;65:1287–1295. doi: 10.1007/s00294-019-00980-0. [DOI] [PubMed] [Google Scholar]
- 8.Nair S., Miller B., Barends M., Jaidee A., Patel J., Mayxay M., et al. Adaptive copy number evolution in malaria parasites. PLOS Genet. 2008;4 doi: 10.1371/JOURNAL.PGEN.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schirrmeister B.E., Dalquen D.A., Anisimova M., Bagheri H.C. Gene copy number variation and its significance in cyanobacterial phylogeny. BMC Microbiol. 2012;12:1–15. doi: 10.1186/1471-2180-12-177/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer A., Brennan G., Geballe A.P. Adaptation by copy number variation in monopartite viruses. Curr Opin Virol. 2018;33:7–12. doi: 10.1016/J.COVIRO.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assogba B.S., Milesi P., Djogbénou L.S., Berthomieu A., Makoundou P., Baba-Moussa L.S., et al. The ace-1 Locus is amplified in all resistant Anopheles gambiae mosquitoes: fitness consequences of homogeneous and heterogeneous duplications. PLoS Biol. 2016;14:1–26. doi: 10.1371/journal.pbio.2000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delahaye C., Nicolas J. Sequencing DNA with nanopores: troubles and biases. PLoS One. 2021:16. doi: 10.1371/journal.pone.0257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahlin K., Tomaszkiewicz M., Makova K.D., Medvedev P. Deciphering highly similar multigene family transcripts from Iso-Seq data with IsoCon. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-06910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;128:733–735. doi: 10.1038/nmeth.3444. 2015;12. [DOI] [PubMed] [Google Scholar]
- 15.Ahsan M.U., Liu Q., Fang L., Wang K. NanoCaller for accurate detection of SNPs and indels in difficult-to-map regions from long-read sequencing by haplotype-aware deep neural networks. Genome Biol. 2021;22:1–33. doi: 10.1186/s13059-021-02472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak A., Murik O., Mann T., Zeevi D.A., Altarescu G. Detection of single nucleotide and copy number variants in the Fabry disease-associated GLA gene using nanopore sequencing. Sci Rep. 2021;11:1–7. doi: 10.1038/s41598-021-01749-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahlin K., Medvedev P. Error correction enables use of Oxford Nanopore technology for reference-free transcriptome analysis. Nat Commun. 2021;12:1–13. doi: 10.1038/s41467-020-20340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitford W., Hawkins V., Moodley K.S., Grant M.J., Lehnert K., Snell R.G., et al. Proof of concept for multiplex amplicon sequencing for mutation identification using the MinION nanopore sequencer. Sci Rep. 2022;12:1–9. doi: 10.1038/s41598-022-12613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonneau M., Atyame C.M., Beji M., Justy F., Cohen-Gonsaud M., Sicard M., et al. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun. 2018;9:319. doi: 10.1038/s41467-017-02749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckmann J.F., Ronau J.A., Hochstrasser M.A. Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2017:2. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LePage D.P., Metcalf J.A., Bordenstein S.R., On J., Perlmutter J.I., Shropshire J.D., et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zabalou S., Riegler M., Theodorakopoulou M., Stauffer C., Savakis C., Bourtzis K. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atyame C.M., Pasteur N., Dumas E., Tortosa P., Tantely M.L., Pocquet N., et al. Cytoplasmic incompatibility as a means of controlling Culex pipiens quinquefasciatus mosquito in the islands of the South-Western Indian Ocean. PLoS Negl Trop Dis. 2011;5:20–22. doi: 10.1371/journal.pntd.0001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolouli K., Colinet H., Renault D., Enriquez T., Mouton L., Gibert P., et al. Sterile insect technique and Wolbachia symbiosis as potential tools for the control of the invasive species Drosophila suzukii. J Pest Sci. 2004;91:489–503. doi: 10.1007/s10340-017-0944-y. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utarini A., Indriani C., Ahmad R.A., Tantowijoyo W., Arguni E., Ansari M.R., et al. Efficacy of Wolbachia-infected mosquito deployments for the control of dengue. N Engl J Med. 2021;384:2177–2186. doi: 10.1056/nejmoa2030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonneau M., Caputo B., Ligier A., Caparros R., Unal S., Perriat-Sanguinet M., et al. Variation in Wolbachia cidB gene, but not cidA, is associated with cytoplasmic incompatibility mod phenotype diversity in Culex pipiens. Mol Ecol. 2019;28:4725–4736. doi: 10.1111/mec.15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonneau M., Landmann F., Labbé P., Justy F., Weill M., Sicard M. The cellular phenotype of cytoplasmic incompatibility in Culex pipiens in the light of cidB diversity. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sicard M., Namias A., Perriat-Sanguinet M., Carron E., Unal S., Altinli M., et al. Cytoplasmic incompatibility variations in relation with Wolbachia cid genes divergence in Culex pipiens. MBio. 2021;12:e02797–20. doi: 10.1128/mBio.02797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers S.O., Bendich A.J. Plant Mol. Biol. Man. Springer,; Netherlands: 1994. Extraction of total cellular DNA from plants, algae and fungi; pp. 183–190. [DOI] [Google Scholar]
- 30.Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:1–4. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradis E., Blomberg S., Bolker B., Brown J., Claramunt S., Claude J., et al. Package “ape”. R Top Doc 2022.
- 33.Martin M., Patterson M., Garg S., Fischer S.O., Pisanti N., Gunnar W., et al. WhatsHap: fast and accurate read-based phasing 2016:1–18.
- 34.Atyame C.M., Delsuc F., Pasteur N., Weill M., Duron O. Diversification of Wolbachia endosymbiont in the Culex pipiens mosquito. Mol Biol Evol. 2011;28:2761–2772. doi: 10.1093/molbev/msr083. [DOI] [PubMed] [Google Scholar]
- 35.Yang C., Chu J., Warren R.L., Birol I. NanoSim: Nanopore sequence read simulator based on statistical characterization. Gigascience. 2017;6:1–6. doi: 10.1093/gigascience/gix010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Han R., Bi C., Li M., Wang S., Gao X. DeepSimulator: a deep simulator for Nanopore sequencing. Bioinformatics. 2018;34:2899–2908. doi: 10.1093/bioinformatics/bty223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stöcker B.K., Köster J., Rahmann S. SimLoRD: Simulation of Long Read Data. Bioinformatics. 2016;32:2704–2706. doi: 10.1093/bioinformatics/btw286. [DOI] [PubMed] [Google Scholar]
- 38.Faucon P.C., Balachandran P., Crook S. SNaReSim: Synthetic Nanopore Read Simulator. Proc - 2017 IEEE Int Conf Healthc Informatics, ICHI 2017 2017:338–44. 10.1109/ICHI.2017.98. [DOI]
- 39.Smyth R.P., Schlub T.E., Grimm A., Venturi V., Chopra A., Mallal S., et al. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene. 2010;469:45–51. doi: 10.1016/j.gene.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Di Giallonardo F., Zagordi O., Duport Y., Leemann C., Joos B., Künzli-Gontarczyk M., et al. Next-Generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. PLoS One. 2013;8 doi: 10.1371/JOURNAL.PONE.0074249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Song H., Liu D., Zuo T., Lu F., Zhuang H., et al. Extensive recombination due to heteroduplexes generates large amounts of artificial gene fragments during PCR. PLoS One. 2014;9 doi: 10.1371/JOURNAL.PONE.0106658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Judo M.S.B., Wedel A.B., Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819. doi: 10.1093/NAR/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fonseca V.G., Nichols B., Lallias D., Quince C., Carvalho G.R., Power D.M., et al. Sample richness and genetic diversity as drivers of chimera formation in nSSU metagenetic analyses. Nucleic Acids Res. 2012;40 doi: 10.1093/NAR/GKS002. e66–e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esling P., Lejzerowicz F., Pawlowski J. Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res. 2015;43:2513–2524. doi: 10.1093/nar/gkv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagai S., Sildever S., Nishi N., Tazawa S., Basti L., Kobayashi T., et al. Comparing PCR-generated artifacts of different polymerases for improved accuracy of DNA metabarcoding. Metabarcoding Metagenomics. 2022;6 doi: 10.3897/MBMG.6.77704. 6:e77704- [DOI] [Google Scholar]
- 46.Potapov V., Ong J.L. Examining sources of error in PCR by single-molecule sequencing. PLoS One. 2017;12:1–19. doi: 10.1371/journal.pone.0169774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omelina E.S., Ivankin A.V., Letiagina A.E., Pindyurin A.V. Optimized PCR conditions minimizing the formation of chimeric DNA molecules from MPRA plasmid libraries. BMC Genom. 2019;20:1–10. doi: 10.1186/S12864-019-5847-2/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polz M.F., Cavanaugh C.M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724. doi: 10.1128/AEM.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rang F.J., Kloosterman W.P., de Ridder J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:1–11. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.