Summary
Background
For patients with early-stage breast cancers, neoadjuvant treatment is recommended for non-luminal tumors instead of luminal tumors. Preoperative distinguish between luminal and non-luminal cancers at early stages will facilitate treatment decisions making. However, the molecular immunohistochemical subtypes based on biopsy specimens are not always consistent with final results based on surgical specimens due to the high intra-tumoral heterogeneity. Given that, we aimed to develop and validate a deep learning radiopathomics (DLRP) model to preoperatively distinguish between luminal and non-luminal breast cancers at early stages based on preoperative ultrasound (US) images, and hematoxylin and eosin (H&E)-stained biopsy slides.
Methods
This multicentre study included three cohorts from a prospective study conducted by our team and registered on the Chinese Clinical Trial Registry (ChiCTR1900027497). Between January 2019 and August 2021, 1809 US images and 603 H&E-stained whole slide images (WSIs) from 603 patients with early-stage breast cancers were obtained. A Resnet18 model pre-trained on ImageNet and a multi-instance learning based attention model were used to extract the features of US and WSIs, respectively. An US-guided Co-Attention module (UCA) was designed for feature fusion of US and WSIs. The DLRP model was constructed based on these three feature sets including deep learning US feature, deep learning WSIs feature and UCA-fused feature from a training cohort (1467 US images and 489 WSIs from 489 patients). The DLRP model's diagnostic performance was validated in an internal validation cohort (342 US images and 114 WSIs from 114 patients) and an external test cohort (279 US images and 90 WSIs from 90 patients). We also compared diagnostic efficacy of the DLRP model with that of deep learning radiomics model and deep learning pathomics model in the external test cohort.
Findings
The DLRP yielded high performance with area under the curve (AUC) values of 0.929 (95% CI 0.865–0.968) in the internal validation cohort, and 0.900 (95% CI 0.816–0.953) in the external test cohort. The DLRP also outperformed deep learning radiomics model based on US images only (AUC 0.815 [0.719–0.889], p < 0.027) and deep learning pathomics model based on WSIs only (AUC 0.802 [0.704–0.878], p < 0.013) in the external test cohort.
Interpretation
The DLRP can effectively distinguish between luminal and non-luminal breast cancers at early stages before surgery based on pretherapeutic US images and biopsy H&E-stained WSIs, providing a tool to facilitate treatment decision making in early-stage breast cancers.
Funding
Natural Science Foundation of Guangdong Province (No. 2023A1515011564), and National Natural Science Foundation of China (No. 91959127; No. 81971631).
Keywords: Breast cancer, Ultrasound, Whole slide imaging, Deep learning
Research in context.
Evidence before this study
We searched PubMed for publications on artificial intelligence strategies to distinguish between luminal and non-luminal breast cancers using the search query (“deep learning” OR “machine learning” OR “artificial intelligence” OR “radiomics” OR “radiopathomics” OR “whole slide images” OR “pathological images”) AND (“molecular subtype” OR “luminal” OR “non-luminal” OR “hormonal”) AND “breast cancer”), without date or language restrictions. Nineteen original studies that either used radiomics- or pathomics-based algorithms to classify luminal and non-luminal breast cancers were found. Most of these studies targeted all stages of breast cancer rather than early-stage disease only. Among them, sixteen researches applied radiomics algorithms to distinguish between luminal and non-luminal cancers, but most of which were based on MRI with small sample size. Three researches applied pathomics based on deep learning algorithms and achieved good predictive performance in distinguishing between luminal and non-luminal breast cancers, with AUC of 0.90–0.92. However, none of these studies employed HE-stained slides of biopsied tissue samples to develop models for predicting final HR status based on IHC staining of surgical specimens, which may limit the clinical application of these model in aiding decision-making preoperatively.
Added value of this study
The proposed deep learning radiopathomics (DLRP) model had high performance in distinguishing between luminal and non-luminal breast cancers at early stages, with AUCs of 0.929 and 0.900 in internal validation corhort and external test corhort, respectively. The DLRP model outperformed single-modality models (deep learning radiomics model based on US images only and deep learning pathomics model based on WSIs only). This work could lead to a reduction in unnecessary neoadjuvant treatment of luminal breast cancer, thereby potentially enhancing personalized medical decision making and minimizing the burden of healthcare systems.
Implications of all the available evidence
Due to sampling errors in the context of intratumoral heterogeneity, inconsistent results may be found between immunohistochemical staining of biopsied samples and surgical resection specimens. By using deep learning strategies analysing preoperative gray-scale US images and H&E-stained tissue sections to distinguish between luminal and non-luminal breast cancers at early stages, a more accurate classification system can be established. In clinical practice, the DLRP has the potential to tailor neoadjuvant treatment for patients with early breast cancer. For patients predicted to have non-luminal breast cancer, neoadjuvant treatment should be considered according to the NCCN guideline. In contrast, for patients predicted to have luminal breast cancer, neoadjuvant treatment may not be necessary due to its typically good prognosis and relatively low rate of achieving a pathologic complete response after treatment.
Introduction
Breast cancer causes the leading cancer-related death in women worldwide.1 It is a biologically heterogeneous disease, with several recognized molecular subtypes corresponding to different responses to treatments and prognoses. Luminal subtypes are characterized by high expression of estrogen receptors (ER) and progesterone receptors (PR), which can be effectively treated with hormone therapy.2 Compared with luminal subtypes, non-luminal tumors including human epidermal growth factor receptor type 2 (HER2)-enriched and triple-negative subtypes, appear to be more aggressive with poorer prognoses.3,4 Fortunately, non-luminal tumors are more sensitive to neoadjuvant treatment, with a pathological complete response (pCR) rate of 20–40%5,6; patients who achieve pCR have a better prognosis.7 Thus, current guidelines of National Comprehensive Cancer Network for breast cancer recommend neoadjuvant therapy for clinical T1 and T2 non-luminal breast cancers to acquire the treatment response and pCR status.8 Therefore, preoperatively identifying luminal from non-luminal tumors for patients with early-stage breast cancer will facilitate treatment decision.
Currently, immunohistochemical (IHC) staining of breast cancer tissues acquired via preoperative core needle biopsy (CNB) is widely used for molecular subtyping of breast cancer. However, this process remains some limitations, including being time-consuming and expensive. Samples with poor quality may affect assessment of ER and PR levels.9,10 Besides, biopsy specimens fail to reflect the high intra-tumoral heterogeneity, which may result in inaccurate classification of IHC-based molecular subtypes preoperatively.11 Chen et al. reported moderate concordance rates for immunohistochemical results of ER (kappa values 0.52) and PR (kappa values 0.44) between the CNB and the surgical specimens.12 Thus, a more reliable approach is urgently required to identify molecular subtypes preoperatively.
Ultrasound (US) is widely used to characterize breast lesions preoperatively. Previous studies have demonstrated that certain ultrasonographic features on gray-scale images, such as margins and posterior acoustic features, were valuable for predicting molecular subtype of breast cancer.13,14 However, there is high inter- and intra-observer variability in radiologists' visual interpretation of US images for breast lesions.15 Deep learning method is an emerging technique for automatically extracting high-throughput phenotype (radiomics quantitative features) from medical images.16 Previous studies have shown that deep learning based on grey-scale US images can predict molecular subtyping of breast cancer preoperatively.17,18 But the area under the curve (AUC) for distinguishing between luminal and non-luminal tumors was only 0.83 and 0.8717, which is still insufficient in clinical application.
Morphological characteristics of breast cancer captured in the routine hematoxylin and eosin (H&E) staining may reflect the underlying molecular or genetic information.19,20 Digital pathology allows quantitative analysis of digitized whole slide imaging (WSI) of histological specimens using machine learning. Previous studies have shown that morphological features of H&E-stained WSI extracted by machine learning approaches can predict the expression of several single biomarkers, such as ER and HER2, in breast cancer.10,21 However, few studies have been conducted for molecular subtyping using biopsy WSI before surgery.
Emerging evidence suggested that integrating multimodal data could complement tumor heterogeneity at multiple scales and enhance the predictive power of existing models.22,23 A recent study showed that radiopathomics, combining the radiological information of the whole tumor at the macroscale and morphological features of local lesion at the microscale, could achieve better performance than that of the monomodal models based on radiological or pathological data in predicting the response of rectal cancer to neoadjuvant therapy.24 Thus, we hypothesized that combining breast US and biopsy WSI analysis might yield a promising efficiency in preoperatively distinguishing between luminal and non-luminal breast cancers at early stages. In this study, we aimed to develop and validate a deep learning radiopathomics model using pretreatment US images and H&E-stained WSI of biopsy specimens for distinguishing between luminal and non-luminal tumors in patients with early-stage breast cancer.
Methods
Ethics
This study population consisted of patients with early breast cancer from an ongoing, prospective study conducted by our research team and registered on the Chinese Clinical Trial Registry (ChiCTR1900027497). This study was approved by Sun Yat-sen University Cancer Center's Institutional Review Board (reference number: SL-G2022-193-02). Verbal informed consent was obtained from all patients and their families.
Study population
The same inclusion and exclusion criteria were used for all institutions. All eligible patients from the Sun Yat-Sen University Cancer Center (SYSUCC), between January 2019 and August 2021, were included as the training set and internal validation sets. From September 2020 to August 2021, the eligible patients from Peking University Shenzhen Hospital (PUSZH) and the First People's Hospital of Foshan (FPHFS), were included as the external test set.
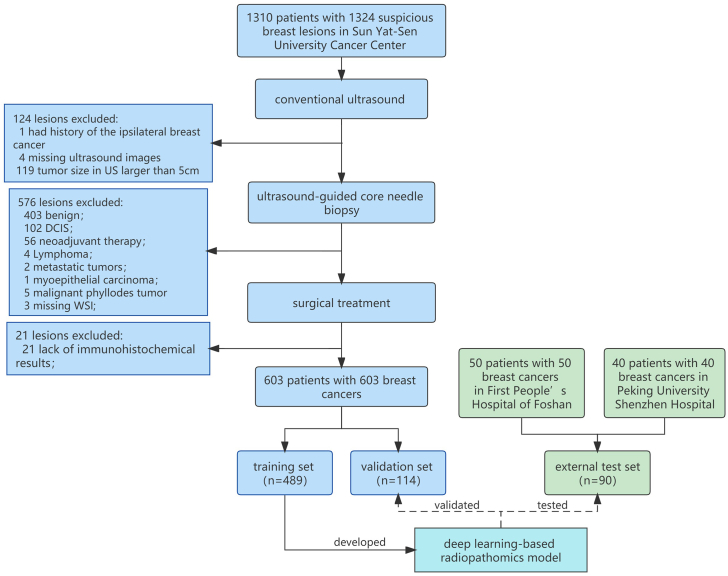
The inclusion criteria included: (a) Women who had suspicious breast cancer that can be detected by US; (b) Underwent US-guided CNB within one day after breast US examination; (c) Underwent breast surgery within 1–2 weeks after CNB. The exclusion criteria included: (a) Patients who had a history of the ipsilateral breast cancer, underwent preoperatively systemic therapy or radiotherapy; (b) The maximum tumor diameter exceeded the measurable range of US transducers (clinical T3-4 tumors); (c) Benign lesions or carcinoma in situ diagnosed by CNB or surgical pathology; (d) Lymphoma or metastatic tumor; (e) Lack of immunohistochemical results for the surgical specimen's ER/PR status; (f) Missing US images or WSI. The flowchart of patients' selection and process of dataset construction are shown in Fig. 1.
Fig. 1.
The flowchart of patients' selection. In total, 603 patients with 603 breast cancers were included as training set (489 lesions) and independent validation set (114 lesions), and 90 patients with 90 breast cancers were included as independent test set.
US examination
One of six experienced radiologists performed breast US examination following the standard practice protocol. Several ultrasound equipment from manufacturers including GE Healthcare, Mindray, Siemens, Philips, Samsung, Canon, HITACHI, and Supersonic Aixplorer, all equipped with a linear transducer (with a frequency range of 10–18 MHz), were used to generate breast US images. To ensure that the model was robust to subtle differences in the position among lesions, three grey-scale US images, including an unmarked image at the largest long–axis plane of the breast lesion and two unmarked parallel images with a distance of 2–10 mm from the largest long–axis plane, were obtained for each lesion by using one of the above US equipment. Tumor size was determined as the maximum diameter measured in the long axis of the lesion. All images were stored in DICOM format for subsequent analysis.
Biopsy and WSI acquisition
US-guided CNB was performed by using a 16-gauge core needle. Biopsy materials with two or three cores were obtained for the diagnosis of each lesion and immediately fixed in 10% neutral buffered formalin for 6–72 h. Samples were embedded in paraffin and then were sliced at 5 mm intervals. Each slide was stained with H&E. For each patient, all H&E-stained slides were evaluated by one senior breast pathologist and one slide with the maximum number of tumor-rich areas was chose for digital scanning using the Zeiss AxioScan Z1 (Carl Zeiss AG, Oberkochen, Germany) with a 20X or 40X objective to acquire WSI for analysis. A total of 304 WSIs were scanned by using 20X objectives, and the remainder were scanned by using 40X objectives.
Data analysis and annotation
Clinical data including patients’ age, tumor size, menopausal status and breast imaging reporting and data system (BI-RADS) category were recorded. Histopathologic data including histologic type and IHC results of ER status, PR status, HER2 status, and Ki-67 based on the whole tumor surgical specimen were also recorded. All breast tumors were classified into luminal and non-luminal subtypes according to the IHC results of hormonal receptor (HR) status based on the whole tumor surgical specimen. HR was deemed positive if ≥1% of tumor cells were stained for ER or PR. All tumors were identified as luminal subtype if HR was positive and as non-luminal subtype if HR was negative. All US images and WSI images of each tumor were assigned labels consistent with the tumor classification.
Data preprocessing
Before being input into the deep learning model, the paired US and WSI data of each case needs to be preprocessed. The region of interest (ROI) of US image is manually annotated by experienced radiologists, which is the minimum bounding rectangle covering the tumor area. The ROI was cropped from the original US image according to the annotation, and the minimum bounding rectangle expanded by 20% to include richer margins and posterior acoustic features. To avoid overfitting, online data augmentation pipeline was applied for each image during training, including resize, random crop, random horizontal flip, random brightness adjusting and normalize.
The preprocessing steps of WSI images include tissue segmentation, image patches extraction and feature extraction. Typical regions of invasive cancer on WSI were manually delineated by a trained doctor using the QuPath software (version 0.2.0) and rechecked by a senior breast pathologist. We performed automated segmentation of tissue using public CLAM repository for WSI analysis.25 As a comparison, an experienced pathologist simultaneously performed manual annotation to extract tissue area. Following segmentation, pyramidal multi-scale features were extracted by integrating WSI features from different magnifications. Specifically, a patch with a size of 256∗256 at 5X magnification contains 4 256∗256 sub-patches at 10X magnification and 16 256∗256 sub-patches at 20X magnification (Supplementary Fig. S1). Subsequently, following the idea of transfer learning, the pre-trained ResNet50 model was used to extract feature embedding of image patches, via spatial average pooling after 3rd residual block. Thus, for each 5X patch, we then concatenate the 5X feature vector (1024 dimensions) with each of the 10X features and 20X features and obtain 16 feature vectors with 3096 dimensions. After data preprocessing, each WSI image is converted into a bag of 3096-dimensional feature vectors. The dataset yields 0.78 million patches at 20X magnification with in average about 1124 patches per bag (Supplementary Methods 1 and Supplementary Table S1).
Model development
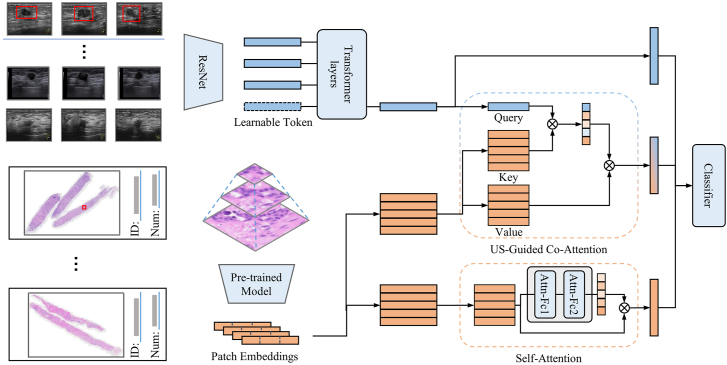
We randomly divided the development set into a training set and a validation set in the ratio of 8:2. All models were developed in the training set and were validated in an independent internal set and tested in an independent external set. The deep learning radiopathomics (DLRP) model that contains two pathways to process US images and pathology images in parallel was designed, and the features of both pathways were fused through a co-attention module. The overall architecture of the model is shown in Fig. 2. The inputs of the DLRP model are three view US images and WSI, and the output is the prediction of the luminal status. The information of the two modalities was separately encoded by their respective pathways to obtain feature vectors, while in the co-attention module, the two modal features interact with each other through attention to obtain the fused feature vector. Finally, the three features are input to the classifier by concatenating to obtain the final classification results. For comparison, a deep learning radiomics (DLR) model was generated with US features alone. A deep learning pathomics trained on automated segmentation of tissue (DLP) and a deep-learning based pathomics trained on manually annotated ROI (DLP-manual) were also generated with WSI features alone.
Fig. 2.
Workflow in developing DLRP model for distinguishing between luminal and non-luminal breast cancers at early stages. The DLRP model contains two pathways to process US images and pathology images in parallel, and the features of both pathways were fused through a co-attention module.
In the pathway of processing US images, we used the idea of transfer learning to encode US images using Resnet18 model pre-trained on ImageNet (Supplementary Methods 2 and Supplementary Table S2). Three US images from one patient were treated as a multi-view fusion problem and Resnet18 model with shared parameters was used as a feature extractor to extract features from each of them. The fully connected (FC) layer was discarded to adapt to our task. In particular, a multiresolution architecture was adopted to simulate the visual system of the human eye, which contained a context stream and a fovea stream that processed the raw US image and the center region of US image, respectively.
In the pathological pathway, a multi-instance learning based attention model was adopted for processing WSI images. Under the multi-instance learning framework, each WSI image was tiled in to a large number of patches, all of which as a whole had a slide-level label. The attention pool operation was used to aggregate the features of all patches into slide-level representation and perform supervised training with labels.26 This was a self-attention operation performed on the WSI image and the attention score was computed by:
where , and are the learnable weights of FC layers. Thus, the slide-level representation was calculated by weighted average of all image patch features by the attention score :
Due to the heterogeneity gap between the WSI representation and US features, it was limited to only incorporating late-fusion to fuse US and WSI features because it lacked the interaction between macroscopic US features and microscopic pathological features. We introduced an US-guided Co-Attention (UCA) module for feature fusion, which was analogous to the standard Transformer for cross-modal information interaction in VQA system.27 As shown in Fig. 2, the UCA uses US features to guide the aggregation of WSI features, where the US feature vectors are used as queries and the WSI features (, ) are used as key–value pairs. We keep each patch features as a Value, whose corresponding Key is computed by the patch features through a FC layer with trainable weights. The mapping function was defined by:
where , , are trainable weights of FC layer and is the co-attention vector for computing weighted average of WSI features.
Finally, we concatenate US features, self-attention pathological features and US-guided pathological features and input them into the FC layer for classification.
Heat map generation
US interpretability. Regions of US images were interpreted from spatial location that are relevant to molecular prediction. GradCAM algorithm was utilized to produce heat maps, which reflected the importance of different spatial locations on the US images for predicting results.28
WSI interpretability. The visual interpretation of the relative importance of different tissue regions for molecular prediction was performed by visualizing attention score. In the inference process, the model generated two attention scores for each 256∗256 patch during forward propagation, one was the pathological self-attention score, and the other is the US-guided co-attention score. For visualization, the attention scores were first normalized to 0 (low attention) to 1 (high attention), and spatially registered onto the corresponding patches. Then the normalized attention values were mapped to the RGB space using colormap and overlayed on the original WSIs with a transparency of 0.5.
Statistical analysis
The Kruskal-Wallis rank sum test and Chi-square test were used to compare the difference in non-normal continuous variables and categorical variables, respectively. Area under the receiver operating characteristic (ROC) curve (AUC) was used to estimate the probability of correctly distinguishing between luminal and non-luminal breast cancers at early stages. Difference between AUCs were compared using the Delong test. More comprehensive metrics including sensitivity, specificity, positive predictive values and negative predictive values were calculated. All statistics were two sided and p-values less than 0.05 indicate significant differences. SPSS software (V.21.0) and MedCalc software (V.11.4.2.0) were used for statistical calculations.
Role of the funding source
The funder of the study provided the financial support, but had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data and approved the final manuscript for submission.
Results
Baseline characters
A total of 1809 US images and 603 WSIs from 603 women (median age, 51 years; range, 27–83 years) in SYSUCC were finally obtained for model development. There were 1467 US images and 489 WSIs from 489 women (median age, 51; range, 27–83) in the training set and 342 US images and 114 WSIs from 114 women (median age, 52; range, 33–77) in the internal validation set. A total of 279 US images and 90 WSIs were obtained from 90 women (median age: 52 years; range: 26–73 years) in the PUSZH and FPHFS to form an independent test set. The median tumor size on US was 21 mm (range: 6–49 mm), 23 mm (range: 6–50 mm) and 20 mm (range: 7–48 mm) in training set, internal validation set, and independent test set, respectively.
All images have a mask that segments the tumor region. The baseline characteristics of all patients and breast cancers are provided in Table 1. No significant differences were found in the distribution of luminal and non-luminal breast cancers (p = 0.298) among the training set, internal validation set, and independent test set.
Table 1.
Baseline characteristics of patients and breast cancer.
| Characteristics | Training set | Validation set | External test set | P value |
|---|---|---|---|---|
| No. patients | 489 | 114 | 90 | |
| Age (y) | 51 (27–83) | 52 (33–77) | 53 (26–84) | 0.162 |
| Tumor size on US (mm) | 21 (6–49) | 23 (6–50) | 20 (7–48) | 0.003 |
| Clinical T stage | 0.144 | |||
| T1 (<2 cm) | 220 (45.0%) | 40 (35.1%) | 41 (45.6%) | |
| T2 (2–5 cm) | 269 (55.0%) | 74 (64.9%) | 49 (54.4%) | |
| Menopausal status | 0.804 | |||
| premenopausal | 235 (48.1%) | 53 (46.5%) | 40 (44.4%) | |
| postmenopausal | 254 (51.9%) | 61 (53.5%) | 50 (55.6%) | |
| BI-RADS | 0.260 | |||
| 4A | 6 (1.2%) | 1 (0.9%) | 1 (1.1%) | |
| 4B | 93 (19.0%) | 15 (13.2%) | 23 (25.6%) | |
| 4C | 226 (46.2%) | 55 (48.2%) | 23 (25.6%) | |
| 5 | 164 (33.5%) | 43 (37.7%) | 43 (47.8%) | |
| Histologic type | 0.206 | |||
| IDC | 428 (87.5%) | 98 (86.0%) | 76 (84.4%) | |
| ILC | 20 (4.1%) | 2 (1.8%) | 5 (5.6%) | |
| Mixed | 29 (5.9%) | 10 (8.8%) | 3 (3.3%) | |
| Other | 12 (2.5%) | 4 (3.5%) | 6 (6.7%) | |
| ER status | 0.814 | |||
| positive | 410 (83.8%) | 95 (83.3%) | 73 (81.1%) | |
| negative | 79 (16.2%) | 19 (16.7%) | 17 (18.9%) | |
| PR status | 0.130 | |||
| positive | 404 (82.6%) | 87 (76.3%) | 68 (75.6%) | |
| negative | 85 (17.4%) | 27 (23.7%) | 22 (24.4%) | |
| HER2 status | 0.343 | |||
| positive | 123 (25.2%) | 35 (30.7%) | 20 (22.2%) | |
| negative | 366 (74.8%) | 79 (69.3%) | 70 (77.8%) | |
| Ki-67 | 0.863 | |||
| ≥20% | 344 (70.3%) | 81 (71.1%) | 61 (67.8%) | |
| <20% | 145 (29.7%) | 33 (28.9%) | 29 (32.2%) | |
| Molecular subtypes | 0.298 | |||
| Luminal | 431 (88.1%) | 100 (87.7%) | 74 (82.2%) | |
| Non-luminal | 58 (11.9%) | 14 (12.3%) | 16 (17.8%) |
Note: Age and tumor size on US were presented as median (range). Other data were presented as number and percentages. The Kruskal–Wallis rank sum test was used to compare the difference in age, tumor size on US, clinical T stage and BI-RADS. Chi-square test was used to compare the difference in menopausal status, ER, PR, HER2, Ki-67 and molecular subtypes. US, ultrasound; BI-RADS, breast imaging reporting and data system; IDC, Invasive ductal cancer; ILC, Invasive lobular cancer; Mixed, more than one histologic type in one tumor; Other, just one histologic type other than the IDC or ILC; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Performance of DLRP and comparison with DLR and DLP
The DLRP model accurately distinguished between luminal and non-luminal breast cancers at early stages in both the internal validation set (AUC 0.929 [95% CI 0.865–0.968]; ACC 0.904) and independent external test set (AUC 0.900 [0.819–0.953]; ACC 0.878). The sensitivity of DLRP was extremely high in internal validation set (0.940 [0.874–0.978]) but dropped slightly to 0.892 [0.798–0.952] in independent external test set. The specificity of DLRP was relatively low (0.643 [0.351–0.872]) in internal validation set but could reach to 0.812 [0.544–0.960] in independent external test set. The PPV of DLRP was markedly high (0.949–0.957) in both sets, whereas the NPV was around 0.600.
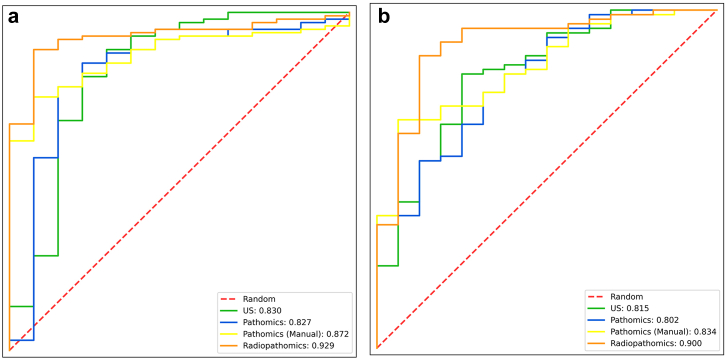
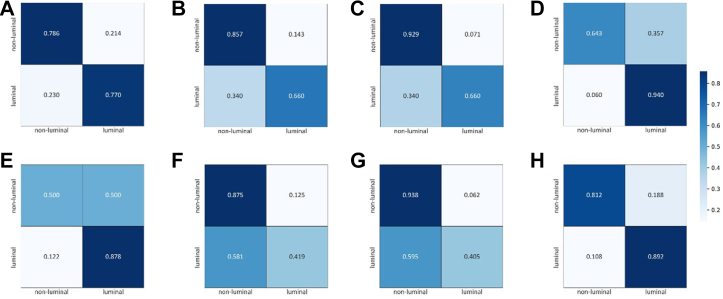
The DLRP model has superior performance in distinguishing between luminal and non-luminal breast cancers at early stages compared with all single-modality models (Table 2). The AUC (0.90 [0.819–0.953]) of the DLRP model was significantly higher than DLR (AUC 0.815 [0.719–0.889], p < 0.027), DLP (AUC 0.802 [0.704–0.878], p < 0.013) and DLP-manual (AUC 0.834 [0.740–0.904], p < 0.023) models in independent external test set. The ROCs showed that DLRP significantly improved AUC for distinguishing between luminal and non-luminal breast cancers at early stages compared with the three single-modality models (Fig. 3). The normalized confusion matrix of all models was showed in Fig. 4. The sensitivity of DLRP model (0.892 [0.798–0.952]) was similar to that of DLR model (0.878 [0.782–0.943], p = 1.00) and significantly higher than that of DLP model (0.419 [0.305–0.539], p < 0.0001) and DLP-manual model (0.405 [0.293–0.526], p < 0.0001) in independent external test set. Although the specificity of DLRP model (0.812 [0.544–0.960]) was lower than that of DLP-manual model (0.937 [0.698–0.998]) in independent external test set, but no significant difference was found between these two groups (p = 0.50). The DLRP, DLP, and DLP-manual models displayed an excellent PPV exceeded 0.9. The NPV of DLRP model was much higher than that of all single-modality models.
Table 2.
Performance and Comparison of DLR, DLP, DLP-manual and DLRP for distinguish between luminal and non-luminal breast cancers at early stages.
| models | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| DLR model | |||||
| Training set (n = 489) | 0.909 (0.879, 0.933) | 0.895 (0.862, 0.923) | 0.763 (0.634, 0.864) | 0.965 (0.942, 0.981) | 0.500 (0.392, 0.608) |
| Validation set (n = 114) | 0.830 (0.748, 0.894) | 0.770 (0.675, 0.848) | 0.786 (0.492, 0.953) | 0.962 (0.894, 0.992) | 0.324 (0.174, 0.505) |
| Test set (n = 90) | 0.815a (0.719, 0.889) | 0.878 (0.782, 0.943) | 0.500 (0.247, 0.753) | 0.890 (0.795, 0.951) | 0.471 (0.230, 0.722) |
| DLP model | |||||
| Training set (n = 489) | 0.882 (0.850, 0.909) | 0.730 (0.686, 0.772) | 0.915 (0.813, 0.972) | 0.984 (0.964, 0.995) | 0.318 (0.248, 0.394) |
| Validation set (n = 114) | 0.827 (0.745, 0.892) | 0.660 (0.558, 0.752) | 0.857 (0.572, 0.982) | 0.971 (0.898, 0.996) | 0.261 (0.143, 0.411) |
| Test set (n = 90) | 0.802b,d (0.704, 0.878) | 0.419 (0.305, 0.539) | 0.875 (0.617, 0.984) | 0.939 (0.798, 0.993) | 0.246 (0.141, 0.378) |
| DLP-manual model | |||||
| Training set (n = 489) | 0.889 (0.857, 0.915) | 0.754 (0.710, 0.794) | 0.881 (0.771, 0.951) | 0.979 (0.957, 0.991) | 0.329 (0.257, 0.408) |
| Validation set (n = 114) | 0.872 (0.797, 0.927) | 0.660 (0.558, 0.752) | 0.929 (0.661, 0.998) | 0.985 (0.920, 1.000) | 0.277 (0.156, 0.426) |
| Test set (n = 90) | 0.834c (0.740, 0.904) | 0.405 (0.293, 0.526) | 0.937 (0.698, 0.998) | 0.968 (0.833, 0.999) | 0.254 (0.150, 0.384) |
| DLRP model | |||||
| Training set (n = 489) | 0.975 (0.956, 0.987) | 0.907 (0.875, 0.933) | 1.000 (0.939, 1.000) | 1.000 (0.991, 1.000) | 0.596 (0.493, 0.693) |
| Validation set (n = 114) | 0.929 (0.865, 0.968) | 0.940 (0.874, 0.978) | 0.643 (0.351, 0.872) | 0.949 (0.886, 0.984) | 0.600 (0.323, 0.837) |
| Test set (n = 90) | 0.900 (0.819, 0.953) | 0.892 (0.798, 0.952) | 0.812 (0.544, 0.960) | 0.957 (0.878, 0.991) | 0.619 (0.384, 0.819) |
Note: data in parentheses are 95% confidence intervals. DLR, deep learning radiomics; DLP, deep learning pathomics; DLP-manual, deep learning pathomics trained on manually annotated WSI; DLRP, deep learning radiopathomics; AUC, area under the receiver operating characteristic curve; PPV, positive predictive value; NPV, Negative predictive value.
Indicates P = 0.027, Delong et al. in comparison with DLRP model in independent test set.
Indicates P = 0.013, Delong et al. in comparison with DLRP model in independent test set.
Indicates P = 0.023, Delong et al. in comparison with DLRP model in independent test set.
Indicates P = 0.352, Delong et al. in comparison with DLP-manual model in independent test set.
Fig. 3.
Comparison of receiver operating characteristic (ROC) curves between different models for distinguishing between luminal and non-luminal breast cancers at early stages in (a) the internal validation set and (b) the external test set. Data in parentheses are areas under the receiver operating characteristic curves (AUCs).
Fig. 4.
Normalized confusion matrix of models with the validation set (A–D) and test set (E–H). (A, E) DLR model. (B, F) DLP model. (C, G) DLP-manual model. (D, H) DLRP model. True and predicted subtype classifications are shown on the y- and x-axes, respectively, such that the correct predictions are shown on the diagonal from the top left to the bottom right of each matrix. The blue gradient color represents the model accuracy for detecting each subtype. The darker the blue color, the better the model performance.
Ablation analysis for DLRP
The feature pyramid and co-attention module in the DLRP model were investigated to illustrate their effects. The results of the ablation study are summarized in Table 3. Without the pyramidal feature and co-attention module, the baseline1 model achieved an AUC of 0.849 [0.769–0.909] in predicting luminal and non-luminal breast cancers, which was significantly lower than the DLRP model (AUC 0.929 [0.865–0.968], p = 0.016). The baseline2 model (AUC 0.863 [0.786–0.920], p = 0.356) has a slight performance improvement compared to the baseline1 model after using the pyramidal feature. The baseline3 model (AUC 0.899 [0.828–0.947], p = 0.041) uses the co-attention module on the basis of the baseline1 model, and the performance has been greatly improved compared with the baseline1 model.
Table 3.
The performance of modules utilized in the ablation study on the internal validation set.
| Pyramid feature | Co-attention | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|
| Baseline 1 | 0.849 (0.769, 0.909) | 0.930 (0.861, 0.971) | 0.571 (0.289, 0.823) | 0.940 (0.874, 0.978) | 0.571 (0.289, 0.823) | ||
| Baseline 2 | √ | 0.863 (0.786, 0.920) | 0.920 (0.848, 0.965) | 0.643 (0.351, 0.872) | 0.941 (0.875, 0.978) | 0.583 (0.277, 0.848) | |
| Baseline 3 | √ | 0.899 (0.828, 0.947) | 0.930 (0.861, 0.971) | 0.643 (0.351, 0.872) | 0.922 (0.852, 0.966) | 0.615 (0.316, 0.861) | |
| DLRP | √ | √ | 0.929 (0.865,0.968) | 0.940 (0.874, 0.978) | 0.643 (0.351, 0.872) | 0.949 (0.886, 0.984) | 0.600 (0.323, 0.837) |
Visual interpretation of the DLRP model
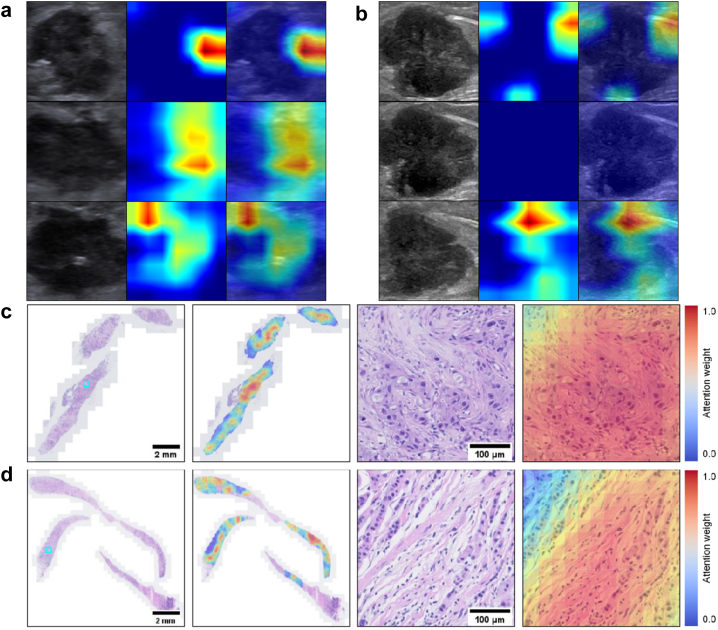
The corresponding heat maps of multiple greyscale US images and WSI visualized the most predictive areas and image features that the DLRP model focus on by displaying different color distributions, as shown in Fig. 5. The most valuable locations on US images for distinguishing between luminal and non-luminal breast cancers at early stages were the boundary of the tumor and low echo area inside the tumor. The US images from different views have complementary contributions to the differentiate results for each lesion. The areas on WSI that were most valuable in distinguishing between luminal and non-luminal tumors were those that had both apparently invasive carcinoma and abundant collagenous stroma.
Fig. 5.
Visualization of two patient examples. The corresponding heat maps of three greyscale US (a) and biopsy WSIs (c) in a non-luminal tumor from a 48-year-old woman with invasive ductal carcinoma. And the corresponding heat maps of three greyscale US (b) and biopsy WSIs (d) in a luminal tumor from a 55-year-old woman with invasive ductal carcinoma mixed with 15% micropapillary carcinoma. The most valuable locations on US images were the boundary of the tumor and low echo area inside the tumor. The US images from different views provided complementary information which contributed to differentiate results for each lesion. In terms of WSIs, the areas that were deemed most valuable were the ones containing both apparently invasive carcinoma and abundant collagenous stroma.
Discussion
In this study, we established a DLRP model to preoperatively distinguish between luminal and non-luminal breast cancers at early stages by incorporating pretherapeutic quantitative radiomics and pathomics features. The DLRP model accurately distinguished between luminal and non-luminal breast cancers at early stages with favorable AUC, high sensitivity, and high PPV in the independent internal and external test sets. The DLRP model proposed in this study also had superior performance to the radiomics model based on US features only and the pathomics model based on histomorphological features only. The superior performance of our DLRP model highlighted the potential for application in distinguishing between luminal and non-luminal cancers preoperatively. Since patients with the non-luminal lesions were recommended to undergo neoadjuvant treatment, our model may be capable of tailoring neoadjuvant treatment for patients with early breast cancer.
Many efforts have been put into predicting molecular subtypes of breast cancer using pretherapeutic radiographic images because the immunohistochemical results from the preoperative biopsy are not sufficiently accurate.29,30 Ma et al. constructed a radiomics model based on conventional machine learning to distinguish between luminal and non-luminal cancers using mammography images but only achieved a moderate AUC of 0.752.31 Radiomics model based on the convolutional neural network with transfer learning to extract high-dimensional features related to molecular classification using MRI images improved the accuracy with an AUC of 0.836 in distinguishing between luminal and non-luminal breast cancers.32 Nevertheless, this study was limited by its relatively small sample size and lack of external validation, thus leading to potential over-fitting and uncertain generalizability of these findings. Besides, breast MRI is expensive and time-consuming. More importantly, it is not commonly used for preoperative evaluation of early breast cancer. In this study, we developed a DLR model to distinguish between luminal and non-luminal breast cancers at early stages. The AUC for validation and independent external test sets were 0.830 and 0.815 respectively, which was similar to a previous study.17 The US images in this study were acquired in multiple institutions using various devices, establishing the breadth and generality of these results. However, the performance of DLR model is insufficient (AUC <0.90), suggesting that combining radiomic-derived features with additional features is necessary for achieving a higher predictive value.
Unlike radiographic images that can provide the spatial macrostructure information of the tumor and its adjacent tissue, histopathological images capture morphology information at the microscale. The morphological-based features of H&E-stained images extracted using deep learning algorithms have been shown to be valuable for assessing ER status, achieving the accuracy of 0.80–0.92.10,19,33 These results were noninferiority to that of traditional IHC for ER status (0.82–0.88).9,34 In this study, we used a multiple-instance learning approach to explore morphological features on H&E-stained WSI of biopsy that were valuable for predicting the HR expression. Although the AUC reached 0.834, the performance of DLP-manual model developed in this study is still inferior to that of a previous model (AUC, 0.92).10 The underlying reasons might be that Naik Nikhil et al.10 developed their model using H&E images and IHC-based labels from the same tumor tissue blocks, while we used H&E-stained WSI of biopsied tissue to predict HR status determined by IHC based on surgical specimen. Although the core biopsy may not be morphologically representative of the whole resected tumor in some cases, the use of CNB images that are routinely available before treatment enables the clinical application of our model in aiding decision-making for neoadjuvant chemotherapy. The study of Greer et al. showed that concordance rates of IHC results for CNB and surgical specimens increased with increasing numbers of core biopsies.35 Shamai Gil et al. also reported that using multiple H&E-stained images yielded better results than using a single image for one patient in the training model to assess ER status.19 In the future, we may be able to further improve the prediction performance by using multiple WSIs to train the model.
The biopsy can provide insights into specific subregions or areas with unique characteristics of the tumor tissue such as cellular composition and the morphological features under a microscope. By fusing biopsy data with imaging, complementary information from both the microscopic and macroscopic levels representative of the tumor's heterogeneity can be provided. The superior performance of our DLRP model is likely due to the integration of heterogeneous radiomics and pathomics features, suggesting that the comprehensive capture of microstructural and macrostructural features can effectively predict the HR expression of breast cancer.
The DLRP model can assist clinicians to classify luminal and non-luminal lesions rapidly. Input the routinely available images of breast US and biopsies to the DLRP model would obtain a prediction result (luminal vs. non-luminal). Current NCCN guidelines recommend preoperative systemic treatment for patients with non-luminal T1 and T2 cancers.11 The addition of neoadjuvant therapy to surgery is associated with benefit for patients with non-luminal tumors; however, little benefit is evident for patients with luminal tumors. We do note that the established DLRP model yielded extremely high PPV, meaning that luminal cases were filtered out to avoid ineffective (or at least redundant) treatments when applying the model. In this study, about 89.2% (66/74) early-stage luminal breast cancers can be identified by DLRP in test cohort, potentially avoiding the need for neoadjuvant therapy, while about 81.2% (13/16) non-luminal tumors were identified by DLRP in test cohort and would be benefited from neoadjuvant chemotherapy. Therefore, decisions regarding whether neoadjuvant therapy should be given can be at the discretion of the model.
Our study still has some limitations. Firstly, the training and testing data are both highly imbalanced. The reason for this may be that non-luminal subtypes (HER2-enriched and TNBC) take up only a small proportion of breast cancer, and only clinical T1 and T2 tumors were included in this study. Given that, we handled class imbalance by oversampling non-luminal cases or by using focal loss to train the models in this study. Secondly, all the slides were scanned by using a digital scanner. The influence of different scanners with different parameters on model building should be investigated in future studies. Thirdly, deep learning models still exhibited some overfitting. For this phenomenon, several measures have been taken in the various links to reduce the risk of overfitting. Although the model performance in external test cohort was reduced compared to the training set, it was still in a satisfactory range overall.
In conclusion, a DLRP model was developed and validated to distinguish between luminal and non-luminal breast cancers at early stages by integrating pretherapeutic grey-scale US images and biopsy whole slide images. The superior performance of the DLRP model showed the potential for application in tailoring neoadjuvant treatment for patients with early breast cancer.
Contributors
Jianhua Zhou and Yini Huang designed the study; Yini Huang, Rushuang Mao, Weijun Huang, Zhengming Hu, Yixin Hu and Yun Wang collected the data; Jianhua Zhou, Rongzhen Luo, Yini Huang, Zhao Yao, Ruohan Guo, Xiaofeng Tang and Liang Yang analyzed the data; Zhao Yao, Yuanyuan Wang and Jinhua Yu proposed the model; Yini Huang wrote the original draft; Yini Huang, Zhao Yao, Lingling Li, Jianhua Zhou and Jinhua Yu revised the manuscript. Yini Huang, Zhao Yao, Lingling Li and Rushuang Mao contributed equally to this work. Yini Huang, Zhao Yao, Jianhua Zhou and Jinhua Yu directly accessed and verified the underlying data reported in the manuscript.
Data sharing statement
Due to the privacy of patients, the data related to patients cannot be available for public access but can be obtained from the corresponding author (zhoujh@sysucc.org.cn) on reasonable request approved by the institutional review board of Sun Yat-sen University Cancer Center.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
This work was funded by the Natural Science Foundation of Guangdong Province (No. 2023A1515011564), National Natural Science Foundation of China (No. 91959127; No. 81971631).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104706.
Contributor Information
Rongzhen Luo, Email: luorzh@sysucc.org.cn.
Jinhua Yu, Email: jhyu@fudan.edu.cn.
Jianhua Zhou, Email: zhoujh@sysucc.org.cn.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ignatiadis M., Sotiriou C. Luminal breast cancer: from biology to treatment. Nat Rev Clin Oncol. 2013;10(9):494–506. doi: 10.1038/nrclinonc.2013.124. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Yang J., Peng L., et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161(2):279–287. doi: 10.1007/s10549-016-4059-6. [DOI] [PubMed] [Google Scholar]
- 4.Rubin I., Yarden Y. The basic biology of HER2. Ann Oncol. 2001;12(Suppl 1):S3–S8. doi: 10.1093/annonc/12.suppl_1.s3. [DOI] [PubMed] [Google Scholar]
- 5.Gianni L., Pienkowski T., Im Y.H., et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 6.Barton M.K. Bevacizumab in neoadjuvant chemotherapy increases the pathological complete response rate in patients with triple-negative breast cancer. CA Cancer J Clin. 2014;64(3):155–156. doi: 10.3322/caac.21223. [DOI] [PubMed] [Google Scholar]
- 7.Rastogi P., Anderson S.J., Bear H.D., et al. Preoperative chemotherapy: updates of national surgical adjuvant breast and bowel project protocols B-18 and B-27. J Clin Oncol. 2008;26(5):778–785. doi: 10.1200/JCO.2007.15.0235. [DOI] [PubMed] [Google Scholar]
- 8.NCCN clinical practice guidelines in oncology. Breast cancer. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf Version 4.2022.
- 9.Hammond M.E., Hayes D.F., Dowsett M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch pathol Lab Med. 2010;134(7):e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 10.Naik N., Madani A., Esteva A., et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun. 2020;11(1):5727. doi: 10.1038/s41467-020-19334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pölcher M., Braun M., Tischitz M., et al. Concordance of the molecular subtype classification between core needle biopsy and surgical specimen in primary breast cancer. Arch Gynecol Obstet. 2021;304(3):783–790. doi: 10.1007/s00404-021-05996-x. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Wang Z., Lv Q., et al. Comparison of core needle biopsy and excision specimens for the accurate evaluation of breast cancer molecular markers: a report of 1003 cases. Pathol Oncol Res. 2017;23(4):769–775. doi: 10.1007/s12253-017-0187-5. [DOI] [PubMed] [Google Scholar]
- 13.Irshad A., Leddy R., Pisano E., et al. Assessing the role of ultrasound in predicting the biological behavior of breast cancer. AJR Am J Roentgenol. 2013;200(2):284–290. doi: 10.2214/AJR.12.8781. [DOI] [PubMed] [Google Scholar]
- 14.Rashmi S., Kamala S., Murthy S.S., Kotha S., Rao Y.S., Chaudhary K.V. Predicting the molecular subtype of breast cancer based on mammography and ultrasound findings. Indian J Radiol Imaging. 2018;28(3):354–361. doi: 10.4103/ijri.IJRI_78_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sultan L.R., Bouzghar G., Levenback B.J., et al. Observer variability in BI-RADS ultrasound features and its influence on computer-aided diagnosis of breast masses. Adv Breast Cancer Res. 2015;4(1):1–8. doi: 10.4236/abcr.2015.41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Lu X., Zhou H., et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68(4):729–741. doi: 10.1136/gutjnl-2018-316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang M., Zhang D., Tang S.C., et al. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: a multicenter retrospective study. Eur Radiol. 2021;31(6):3673–3682. doi: 10.1007/s00330-020-07544-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhou B.Y., Wang L.F., Yin H.H., et al. Decoding the molecular subtypes of breast cancer seen on multimodal ultrasound images using an assembled convolutional neural network model: a prospective and multicentre study. eBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shamai G., Binenbaum Y., Slossberg R., Duek I., Gil Z., Kimmel R. Artificial intelligence algorithms to assess hormonal status from tissue microarrays in patients with breast cancer. JAMA Netw Open. 2019;2(7) doi: 10.1001/jamanetworkopen.2019.7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Xu W.D., Shang Z.H., et al. Breast cancer molecular subtype prediction on pathological images with discriminative patch selection and multi-instance learning. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.858453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bychkov D., Linder N., Tiulpin A., et al. Deep learning identifies morphological features in breast cancer predictive of cancer ERBB2 status and trastuzumab treatment efficacy. Sci Rep. 2021;11(1):4037. doi: 10.1038/s41598-021-83102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehm K.M., Khosravi P., Vanguri R., Gao J., Shah S.P. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22(2):114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao L., Liu Z., Feng L., et al. Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: a multicenter radiopathomic study. Ann Surg Oncol. 2020;27(11):4296–4306. doi: 10.1245/s10434-020-08659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L., Liu Z., Li C., et al. Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health. 2022;4(1):e8–e17. doi: 10.1016/S2589-7500(21)00215-6. [DOI] [PubMed] [Google Scholar]
- 25.Lu M.Y., Williamson D.F.K., Chen T.Y., Chen R.J., Barbieri M., Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5(6):555–570. doi: 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilse M., Tomczak J.M., Welling M. Attention-based deep multiple instance learning. Pr Mach Learn Res. 2018;80:2127–2136. [Google Scholar]
- 27.Vaswani A., Shazeer N., Parmar N., et al. Advances in neural information processing systems. Vol. 30. 2017. Attention is all you need. [Google Scholar]
- 28.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Ieee I Conf Comp Vis. 2017:618–626. [Google Scholar]
- 29.Davey M.G., Davey M.S., Boland M.R., Ryan É.J., Lowery A.J., Kerin M.J. Radiomic differentiation of breast cancer molecular subtypes using pre-operative breast imaging – a systematic review and meta-analysis. Eur J Radiol. 2021;144 doi: 10.1016/j.ejrad.2021.109996. [DOI] [PubMed] [Google Scholar]
- 30.Li C., Huang H., Chen Y., et al. Preoperative non-invasive prediction of breast cancer molecular subtypes with a deep convolutional neural network on ultrasound images. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma W., Zhao Y., Ji Y., et al. Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol. 2019;26(2):196–201. doi: 10.1016/j.acra.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun R., Hou X., Li X., Xie Y., Nie S. Transfer learning strategy based on unsupervised learning and ensemble learning for breast cancer molecular subtype prediction using dynamic contrast-enhanced MRI. J Magn Reson imaging. 2022;55(5):1518–1534. doi: 10.1002/jmri.27955. [DOI] [PubMed] [Google Scholar]
- 33.Couture H.D., Williams L.A., Geradts J., et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer. 2018;4:30. doi: 10.1038/s41523-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regan M.M., Viale G., Mastropasqua M.G., et al. Re-evaluating adjuvant breast cancer trials: assessing hormone receptor status by immunohistochemical versus extraction assays. J Natl Cancer Inst. 2006;98(21):1571–1581. doi: 10.1093/jnci/djj415. [DOI] [PubMed] [Google Scholar]
- 35.Greer L.T., Rosman M., Mylander W.C., et al. Does breast tumor heterogeneity necessitate further immunohistochemical staining on surgical specimens? J Am Coll Surg. 2013;216(2):239–251. doi: 10.1016/j.jamcollsurg.2012.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.