Summary
Background
The incidence of endometrial cancer is increasing worldwide. While delays in diagnosis reduce survival, case molecular misclassification might be associated with under- and over-treatment. The objective of this study was to evaluate genetic alterations to detect and molecularly classify cases of endometrial cancer using non-invasive samples.
Methods
Consecutive patients with incident endometrial cancer (N = 139) and controls (N = 107) from a recent Spanish case–control study were included in this analysis. Overall, 339 cervicovaginal samples (out of which 228 were clinician-collected and 111 were self-collected) were analysed using a test based on next-generation sequencing (NGS), which targets 47 genes. Immunohistochemical markers were evaluated in 133 tumour samples. A total of 159 samples were used to train the detection algorithm and 180 samples were used for validation.
Findings
Overall, 73% (N = 94 out of 129 clinician-collected samples, and N = 66 out of 90 self-collected samples) of endometrial cancer cases had detectable mutations in clinician-collected and self-collected samples, while the specificity was 80% (79/99) for clinician-collected samples and 90% (19/21) for self-collected samples. The molecular classifications obtained using tumour samples and non-invasive gynaecologic samples in our study showed moderate-to-good agreement. The molecular classification of cases of endometrial cancer into four groups using NGS of both clinician-collected and self-collected cervicovaginal samples yielded significant differences in disease-free survival. The cases with mutations in POLE had an excellent prognosis, whereas the cases with TP53 mutations had the poorest clinical outcome, which is consistent with the data on tumour samples.
Interpretation
This study classified endometrial cancer cases into four molecular groups based on the analysis of cervicovaginal samples that showed significant differences in disease-free survival. The molecular classification of endometrial cancer in non-invasive samples may improve patient care and survival by indicating the early need for aggressive surgery, as well as reducing referrals to highly specialized hospitals in cancers with good prognosis. Validation in independent sets will confirm the potential for molecular classification in non-invasive samples.
Funding
This study was funded by a competitive grant from Instituto de Salud Carlos III through the projects PI19/01835, PI23/00790, and FI20/00031, CIBERESP CB06/02/0073 and CIBERONC CB16/12/00231, CB16/12/00234 (Co-funded by European Regional Development Fund. ERDF: A way to build Europe). Samples and data were provided by Biobank HUB-ICO-IDIBELL, integrated into the Spanish Biobank Network, and funded by the Instituto de Salud Carlos III (PT20/00171) and by Xarxa de Bancs de Tumors de Catalunya (XBTC) sponsored by Pla Director d’Oncologia de Catalunya. This work was supported in part by the AECC, Grupos estables (GCTRA18014MATI). It also counts with the support of the Secretariat for Universities and Research of the Department of Business and Knowledge of the Generalitat de Catalunya, and grants to support the activities of research groups 2021SGR01354 and 2021SGR1112.
Keywords: Endometrial neoplasms, Early detection of cancer, Papanicolaou test, Mutation, Biomarkers
Research in context.
Evidence before this study
Endometrial cancer is the second most common gynaecological cancer, and is among the tumour types with the sharpest rise in incidence over the past 10 years. Current strategies for diagnosing this cancer involve pelvic ultrasonography and endometrial biopsy. Endometrial biopsies are invasive, uninformative, or impractical in a large proportion of women, and further investigations under general anaesthesia are required. This results in a significant financial burden on the healthcare system and increases diagnostic morbidities. Since the uterine cavity is connected to the cervix anatomically, routine Pap smears and vaginal sampling techniques can be used to extract biological material shed from the upper genital tract. This presents a distinctive opportunity for genomic analysis to identify endometrial cancer.
Added value of this study
In this study, we evaluated genetic alterations in cervicovaginal samples that can be obtained non-invasively and even self-collected. Furthermore, endometrial cancer can be molecularly classified into four molecular groups based on cervicovaginal samples, which is comparable to the classification obtained using tumour samples. The molecular classification of endometrial cancer (ProMisE) was incorporated into the guidelines to assist in clinical decision making. It more accurately reflects the diverse molecular characteristics of endometrial cancer and has a stronger prognostic value than the dualistic classification method. We classified cases into four molecular groups comparable to established tumour classifications using these non-invasive samples, which accordingly showed significant differences in progression-free survival. Consistent with the tumour samples, patients with mutations in POLE had an excellent prognosis, whereas patients with TP53 mutations had the poorest clinical outcome.
Implications of all the available evidence
The evaluation of somatic mutations using cervicovaginal samples offers a user-friendly tool for endometrial cancer detection and molecular classification. Self-collection of cervicovaginal samples can be especially beneficial in situations where access to specialists is limited. In addition, knowing the molecular group prior to hysterectomy or in the absence of endometrial tissue would offer valuable clinical insights. If confirmed in independent sets, the molecular classification of endometrial cancer in non-invasive samples could improve patient care and survival by indicating the early need for aggressive surgery, as well as reducing referrals to highly specialized hospitals in cancers with good prognosis.
Introduction
Endometrial cancer is the second most common gynaecologic cancer among women in very high human development index regions based on age-standardized incidence rates,1 and its incidence has sharply increased over the past 10 years.2,3 Endometrial cancer has been classically classified into two broad subtypes: types I and II, which are based on the histology of tumours. However, the lack of reproducibility of this classification yields heterogeneous molecular groups and impedes advances in precision medicine.3 Thus, the surrogates of The Cancer Genome Atlas (TCGA) consortium classification based on molecular features, which classified 373 endometrial cancer cases into four prognostically significant subgroups, are starting to be integrated into clinical practice.4, 5, 6, 7, 8 The molecular classification of endometrial cancer can potentially reduce over- and under-treatment and improve patient care. In particular, patients with mutations in POLE have an excellent prognosis; therefore, strategies are being explored to de-escalate adjuvant treatment of patients with POLE-mutated tumors.3,5 In contrast, patients with TP53 mutations have the poorest clinical outcome, independent of other prognostic variables, and adding adjuvant chemotherapy is particularly helpful for these patients.3,5
The standard strategy to diagnose endometrial cancer consists of pelvic ultrasonography and pipelle sampling in cases of increased endometrial thickness, while hysteroscopy is recommended when the diagnosis is uncertain.9 However, current diagnostic strategies suffer from several limitations, including a low specificity of ultrasonography, a considerable proportion of unsuccessful insertions of pipelle sampling or sample insufficiency, and a moderate agreement between pipelle and hysterectomy specimens on tumour histology and grade.10,11 Recently, we and other researchers have assessed molecular markers in cervicovaginal samples for endometrial cancer detection in order to overcome these limitations.12, 13, 14, 15 Routine clinician-collected cervical samples and self-collected vaginal specimens are used in cervical cancer screening programs in order to detect human papillomavirus (HPV).16 Although HPV has not been linked to the aetiology of endometrial cancer,17,18 there is potential for these clinician- and self-collected cervicovaginal specimens to detect markers of cancers other than cervical cancer. Molecular-based methods in these samples have shown great accuracy for detecting endometrial cancer, but most previous studies were affected by small sample sizes, lack of validation, and/or unmatched controls.13,15,19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 To date, none of these studies have demonstrated prognostic potential or the capacity to predict the molecular subtype of endometrial cancer. In this study, we evaluated a next-generation sequencing (NGS) approach in 339 cervicovaginal samples to detect endometrial cancer and developed a surrogate for the early molecular classification of this cancer.
Methods
Study population
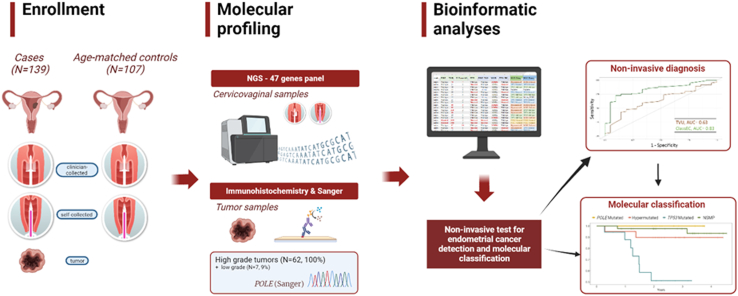
Participants were enrolled in a prospective case–control study (Screenwide, 2017–2021). Consecutive incident endometrial cancer cases and controls frequency-matched by age in ±5 years groups were also enrolled.15 Fig. 1 shows a graphical summary of the study. The exclusion criteria were pregnancy, puerperium, treatment with chemotherapy or radiotherapy during the last 6 months, and communication impediments that precluded the signing of informed consent. The inclusion criteria included having an intact uterus and, for the cases, having an incident diagnosis of endometrial cancer. Hospital controls were visited for benign gynaecologic conditions, including benign cysts, leiomyomas, polyps, and evaluation for abnormal bleeding. Clinical data, including endometrial thickness measured using transvaginal ultrasound (TVU), were extracted from electronic medical records using a predefined form. The participants donated blood samples, vaginal self-collected samples, clinician-collected cervical samples, endometrial aspirates, and, when available, tumour samples. The clinician collection of cervical samples was performed using a Cervex-Brush® (Rovers Medical Devices), and samples were suspended in 20 mL of ThinPrep liquid-based solution (Hologic). Cervical samples were processed using the Thinprep processor, assessment of cervical Pap tests slides was performed,30 as performed in the regular cervical cancer screening program in Catalonia. Once processed, samples were aliquoted and kept at room temperature, as indicated in the manufacturer's protocol. Vaginal self-samples were collected using an Evalyn® Brush device (Rovers Medical Devices), suspended in 5 mL of ThinPrep, and kept at room temperature. In total, 339 cervicovaginal samples from 139 patients with endometrial cancer and 107 controls were included in this study. Supplemental Figure S1 provides a STARD diagram. There were no significant differences between the included and non-included samples (data not shown). A 70% of the clinician-collected cervical samples (N = 159) were used to train the diagnostic algorithm. The remaining 30% of the clinician-collected samples, which were frequency matched by case–control status and histology, were used as validation samples (N = 69), and the complete set of vaginal self-samples (N = 111) also served to validate the diagnostic algorithm. Additionally, 133 endometrial tumour samples (65 of which were obtained from hysterectomy and 68 from Pipelle biopsy) out of 139 cases (95.6%) were used for molecular classification, as well as the full set of positive cervicovaginal samples (see “Molecular classification”). The reference standard for diagnosing cases consisted of histological data obtained following biopsy or hysterectomy. All cases and most controls had available biopsy or hysterectomy specimens and were histologically confirmed (Supplemental Figure S1).
Fig. 1.
Graphical summary of the study. Created with Biorender.com. NGS = Next generation sequencing, TVU = Transvaginal ultrasound, AUC = Area under the curve, NSMP = no specific molecular profile.
DNA mutation analyses
Panel
We used an NGS custom panel targeting exonic regions and intron-exon boundaries of 47 genes recurrently mutated in endometrial cancer, based on an analyses of The Cancer Genome Atlas (TCGA) dataset,31 to evaluate the variants in cervicovaginal samples and molecularly classify cancer cases (ClassEC test). The panel consisted of the following genes: POLE, TP53, PTEN, PIK3CA, PIK3R1, ARID1A, KRAS, CTNNB1, FBXW7, KMT2D, CHD4, CSMD3, KCNG4, NIPBL, PPP2R1A, RPL22, SETD1B, RNF43, JAK1, FGFR2, BCOR, DOCK3, SPOP, SOX17, ACVR2A, CTCF, SOS1, LZTR1, AP4E1, ARHGAP35, MYOM1, BAX, WWC3, SOX5, RACGAP1, TNRC6A, HOXD8, KRIT1, PAX2, AKT1, APC, BRAF, CDKN2A, EGFR, MAPK1, NRAS, and SPEN.
DNA isolation
DNA samples from various types of specimens were obtained using the following protocol. Approximately 5 mL of cervical clinician-collected and 3 mL of vaginal self-samples, were subjected to centrifugation at 11.000 rpm for 20 min. The resultant cell pellet was treated with proteinase K (Maxwell 16 Lev Blood kit, Promega Corporation) at 56 °C for 20 min before DNA extraction. The DNA from the cervical clinician-collected samples and vaginal self-sampling were extracted and finally eluted in 50 μl of nuclease-free water. The automated nucleic acid purification system, Maxwell® 16 Instrument (Promega Corporation), was used to isolate DNA from all samples, and the Qubit dsDNA Broad Range Assay Kit (Thermo Fisher Scientific) was utilized to determine DNA concentration. Only cervicovaginal samples with ≥250 ng DNA were included in the present analyses (Supplemental Figure S1).
Preparation of libraries, target enrichment and sequencing
Libraries and target enrichment were prepared in accordance with to the KAPA HyperCap Workflow v3.0 protocol (Roche Diagnostics) with certain variations to add duplex unique molecular identifiers (UMIs) (Integrated DNA Technologies) and blindly to case–control status. We created libraries utilizing UMIs intended to identify low-frequency allelic variants by adding a degenerate 3-nucleotide-long molecular barcode index. Further, the DNA fragments were captured with a custom panel to enrich the samples with the study target genes. This strategy was adopted due to the anticipated low quantity of tumour cells in the samples.
The preparation of libraries was performed with the KAPA HyperPlus Library Preparation kit (Roche Sequencing Solutions) and involved a range of input DNA concentrations from 250 ng to 500 ng. The input DNAs were enzymatically fragmented using the Frag Enzyme, and incubated at 37 °C to achieve 150–350 bp fragments. End-repair and A-tailing were then performed to DNA fragments before ligation of barcoded adaptors. The A-tailed fragments were ligated to the xGen CS Adapters (Integrated DNA Technologies), which contained an equimolar pool of 64 pairs of duplexed adapters, at an adapter:insert ratio of 20:1, for 20 min at 20 °C. Prior to amplification, libraries were subjected to clean-up and double-sided size selection using AMPure XP bead reagent (Beckman Coulter). Subsequently, libraries were PCR amplified using a final concentration of 4 μM of UDI Dup Seq amplification primers (Integrated DNA Technologies). After a clean-up step with AMPure XP bead reagent (Beckman Coulter), the eluted libraries were quantified using a Qubit fluorometer (Thermo Fisher Scientific), and the quality of DNA was assessed using the Agilent Bioanalyzer DNA 1000 assay (Agilent). Prior to capture, the libraries were multiplexed in groups of 10 samples, resulting in a final total DNA amount of 2 μg. The multiplexed libraries were blocked using COT Human DNA and xGen Universal Blockers TS-Mix (Integrated DNA Technologies). The hybridization of multiplexed libraries was performed with KAPA HyperChoice MAX 0.5 Mb T4 probes for 5 min at 95 °C and for 16–20 h at 55 °C. The captured DNA was washed and PCR amplified using xGen Library Amplification Primer Mix and purified with AMPure XP Bead reagent (Integrated DNA Technologies). The final captured pools were quantified using a Qubit fluorometer and their DNA quality was evaluated with the Agilent Bioanalyzer DNA 1000 assay (Agilent). High-depth deep sequencing was carried out on an Illumina NovaSeq 6000 (Illumina) using the SP flow cell and 150 bp paired-end sequencing protocol.
Bioinformatics pipeline
We used a bioinformatics pipeline that combined Picard, fgbio, and BWA tools, as previously described.15 The variants were filtered based on quality and functional impact criteria, and all non-synonymous and consensus splicing variants were retained. Only variants with a population frequency of <0.1% were retained to exclude polymorphisms. The mean raw coverage was 21,224X for clinician-collected cervical samples and 21,969X for self-samples. The final coverage after deduplication and filtering was 3384X for clinician-collected samples and 2515X for self-samples. We estimated that a minimum coverage of 1000X was required to detect variants at a frequency of 0.5% or more. All samples had coverage >1000X, except for seven clinician-collected samples from cases and ten from controls (range 219X-995X), and one self-sample from a case (978X). Each gene was classified as a tumour suppressor, oncogene, ambiguous, or non-driver based on the classification from IntOgen32 and the literature review. Mutations were considered hotspots if they were found in the TCGA dataset in five endometrial cancer patients or more. If all mutations of a gene in the TCGA dataset were found in less than five patients, we considered the mutations that were observed in at least three patients, and a literature review was carried out to confirm whether these mutations were hotspots. Additionally, all mutations found in the same codon as the hotspot were retained, and all consensus splice sites as well as nonsense and frameshift variants were selected as tumour suppressor genes. Finally, all the variants with variant allele frequency (VAF) ranging from 15% to 40% were selected independent of the type of mutation or gene role, given that the clonal variants in this range of VAFs were only observed in endometrial cancer cases, while the variants with higher VAFs (>40%) could correspond to germline variants. In summary, we included variants at VAF <15% according to their functional impact, and variants at VAF 15%–40%, regardless of their functional impact. Samples with at least one selected variant were classified as positive and those without variants were classified as negative. The list of variants can be found in Supplemental Material.
Molecular classification
Tumours
Immunohistochemical markers (mismatch repair-MMR- and p53) were available for 133 (95.6%) tumour samples. All high-grade 3 tumours (N = 62) and 7 low-grade tumours had POLE Sanger sequencing information and were classified according to the ProMisE (Proactive Molecular Risk Classifier for Endometrial Cancer) algorithm.7 Endometrial cancer cases were classified into four molecular groups based on POLE sequencing and immunohistochemical markers using tumour samples obtained from hysterectomy pieces (N = 30) or endometrial aspirate samples (N = 32). Samples harbouring pathogenic mutations in POLE exonuclease domain as in León-Castillo et al.33 were classified as POLE-mutated. For grade 1–2 tumour samples (N = 77), only immunohistochemical markers, but not POLE sequencing, were available.34 The complete loss of expression of one or more MMR proteins is used to diagnose MMR-deficient (MMRd) endometrial cancer cases. Then, p53 immunostaining is used to classify endometrial cancer as p53-abn, excluding POLE-mutated and MMRd cases.
Cervicovaginal samples
An adaptation of the ProMisE algorithm was used to classify the cases using unique NGS data from cervicovaginal samples. All positive samples from cases were used to train the molecular classification algorithm. Samples harbouring pathogenic mutations in POLE exonuclease domain as in León-Castillo et al.33 were classified as POLE-mutated, in accordance with tumour samples. MMRd tumours are characterized by a high number of mutations and enrichment of frameshift mutations in microsatellites.5,35 Therefore, the total number of variants and number of frameshift mutations per sample was used as a surrogate for MMRd. The cut-off values were established based on receiver-operating characteristic (ROC) curve analyses for each type of sample to discriminate samples based on tumour immunohistochemical results (Supplemental Figure S2). The selected cut-offs for the number of variants per Mb for clinician-collected and self-collected samples were 65 and 85, respectively, whereas the cut-off values for the number of frameshift variants for clinician-collected and self-collected samples were 4 and 8, respectively. The samples were classified as hypermutated (as a surrogate for the MMRd ProMiSe group) if any of these cut-off values were exceeded. Among those not classified as POLE-mutated or hypermutated, mutations identified in TP53 were used to define the TP53-mutated and the no specific molecular profile (NSMP; TP53 wild-type) groups.
Statistical analyses
Descriptive analyses were performed using medians and interquartile range (IQR) per participant for continuous data and counts and percentages for categorical data. Chi-square tests were used for categorical variables, as long as the main assumptions underlying this test were met, and Fisher exact tests otherwise. The test performance was evaluated using sensitivity, specificity, negative and positive predictive values (NPV and PPV, respectively), and the area under the ROC curve. ROC curves were estimated using age-adjusted (in 5-years groups) logistic regression models. Residual age, which represents the distance between each participant's age and the centre of their age-matching category in years, was also included in the logistic models.36 The age-adjusted (5-years groups and residual age) area under the ROC curve values were computed using the plotROC R package (version 2.3.0).37 The PPV and NPV adjusted for disease prevalence (9% based on the pooled risk estimate in a meta-analysis of prevalence in women with postmenopausal bleeding38), as well as their 95% confidence intervals, were calculated using the bdpv R package (version 1.3).39 Kappa statistics to estimate concordance between tumour and cervicovaginal molecular classifications were calculated with the irr R package (version 0.84.1). Survival analyses were performed using the survival R package (version 3.4.0). In particular, we analysed survival using Kaplan–Meier curves with log-rank tests and Cox proportional hazards models adjusted for age (in tertiles). Proportional-hazards assumption was confirmed by using cox.zph function in survival R package. Adjustments were chosen according to directed acyclic graphs (Supplemental Figure S3). The start time was defined as the time at which each individual was enrolled and had their samples collected. For overall survival, time to death was computed as time from sample collection until death of any cause. Patients who are still alive prior to the censoring date are censored at the time of last follow up. For disease-free survival, time to event was computed from sample collection until there was evidence of recurrent or progressive disease or if they died of the disease. Patients who are alive and disease-free prior to the censoring date or if they died of an unrelated cause, are censored at the time of last follow-up. Censoring included 118 (91.5%), 122 (94.6%) of women with clinician-collected samples, and 83 (92.2%) and 85 (94.4%) of women with self-samples, for disease-free survival and overall survival, respectively. A Firth bias reducing correction was applied to obtain estimates using coxphf R package.
We determined that a sample size of 136 cases and 96 controls was necessary to achieve estimates of 85% sensitivity and 90% specificity, with a 95% confidence level and 0.20 precision,40 taking into account a 9% disease prevalence among symptomatic women.38 For survival analyses, we estimated a sample size of 65 cases, assuming a hazard ratio of 9.14 (p53abnormal) and a percentage of events of 11.8%,7 and 15% of losses (false negatives) at a 95% confidence level.
Ethics statement
This study was approved by the Ethics Committee for Clinical Research of Bellvitge University Hospital (reference: PR128/16). All eligible participants signed an informed consent form after receiving information about the study before participating in any study-related activities. The study followed the national and international directives on ethics and data protection (Declaration of Helsinki and subsequent amendments; EU Reglament 2016/679) and Spanish laws on data protection (Organic Law 3/2018; Law 14/2007 biomedical research). This study was registered in the National Register of Biobanks/Collections (C.0004389).
Role of funders
The funders did not have any role in the study design, data collection, data analyses, interpretation, or writing of this article.
Results
The age, stage, and other epidemiological and clinical information collected from 139 patients with endometrial cancer and 107 controls included in this study are provided in Table 1.
Table 1.
Descriptive characteristics of participants.
| Cases (N = 139) |
Controls (N = 107) |
P-value | |||
|---|---|---|---|---|---|
| N | (%) | N | (%) | ||
| Age (years) | |||||
| <60 | 38 | (27.3) | 23 | (21.5) | 0.335a |
| 60–69.9 | 50 | (35.9) | 48 | (44.9) | |
| 70+ | 51 | (36.7) | 36 | (33.6) | |
| BMI (kg/m2) | |||||
| <25 | 22 | (15.8) | 31 | (28.9) | <0.001a |
| 25–29.9 | 40 | (28.8) | 38 | (35.5) | |
| 30+ | 72 | (51.8) | 29 | (27.1) | |
| Menopausal status | |||||
| Premenopausal | 6 | (4.3) | 9 | (8.4) | 0.092b |
| Perimenopausal | 7 | (5.0) | 1 | (0.9) | |
| Postmenopausal | 126 | (90.7) | 97 | (90.7) | |
| Abnormal bleeding | |||||
| Yes | 124 | (89.2) | 43 | (40.2) | <0.001a |
| No | 15 | (10.8) | 47 | (43.9) | |
| MMR status | |||||
| Proficient | 110 | (79.1) | NA | NA | |
| Deficient | 24 | (17.3) | NA | ||
| TVU | |||||
| Normal | 13 | (9.4) | 41 | (38.3) | <0.001a |
| Abnormal | 119 | (85.6) | 47 | (43.9) | |
| Stage | |||||
| I-II | 111 | (79.9) | NA | NA | |
| III-IV | 27 | (19.4) | NA | ||
| Grade | |||||
| 1 | 61 | (43.9) | NA | NA | |
| 2 | 16 | (11.5) | NA | ||
| 3 | 62 | (44.6) | NA | ||
| Histology | |||||
| Endometrioid | 98 | (70.5) | NA | NA | |
| Non-endometrioid | 41 | (29.5) | NA | ||
| Collection | |||||
| Clinician-collected | 129 | (92.8) | 99 | (92.5) | <0.001a |
| Self-collected | 90 | (64.7) | 21 | (19.6) | |
NA = Not Applicable; BMI = Body mass index; MMR = Mismatch repair; TVU = Transvaginal ultrasound.
Numbers are not up to the total due to missing values.
Chi-squared, comparing cases and controls.
Fisher exact test.
Variant detection in clinician-collected cervical samples
In the validation set of clinician-collected cervical samples, 69% (27/39) of the patients had detectable variants and 80% (24/30) of the controls did not (Table 2). Overall, the sensitivity of the ClassEC test for diagnosing endometrial cancer was 73% (94/129), and the specificity was 80% (79/99). Assuming a prevalence of 9% in symptomatic women, PPV and NPV were 26% and 97%, respectively (Table 2, Supplemental Figure S4). The results were similar for postmenopausal women (88/117, 75% sensitivity). Variants were observed in 77% (71/92) of patients with endometrioid cancers and 62% (23/37) of patients with non-endometrioid cancers. Moreover, the sensitivity was 69% (72/105) in patients with early disease (i.e., stages I and II) and 91% (21/23) in patients with advanced disease (i.e., stages III and IV; Supplemental Figure S5a). MMR proficient (MMRp) and MMRd tumours (defined using immunohistochemistry of tumour samples) yielded sensitivities of 73% (77/105) and 65% (13/20), respectively. Among women with abnormal bleeding symptoms, sensitivity and specificity were 77% (89/115) and 82% (32/39), respectively. Contrarily, only 36% (5/14) of women without bleeding symptoms had mutations (Supplemental Figure S5a), while specificity was similar among women without bleeding symptoms (81%, 35/43).
Table 2.
Diagnostic performance of the ClassEC test.
| Clinician-collected samples |
Self-collected samples |
|||||||
|---|---|---|---|---|---|---|---|---|
| Training |
Validation |
Overall |
Validation |
|||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Test positive | 67 | 14 | 27 | 6 | 94 | 20 | 66 | 2 |
| Test negative | 23 | 55 | 12 | 24 | 35 | 79 | 24 | 19 |
| Sensitivity, % (95% CI) | 74 (64, 83) | 69 (52, 83) | 73 (64, 80) | 73 (63, 82) | ||||
| Specificity, % (95% CI) | 79 (68, 88) | 80 (61, 92) | 80 (71, 87) | 90 (70, 99) | ||||
| PPV, % (95% CI)a | 27 (18, 37) | 25 (14, 42) | 26 (20, 35) | 43 (17, 74) | ||||
| NPV, % (95% CI)a | 97 (96, 98) | 96 (94, 97) | 97 (96, 98) | 97 (96, 98) | ||||
CI = Confidence interval; PPV = Positive predictive value; NPV = Negative predictive value.
Assumed population prevalence = 9%.38
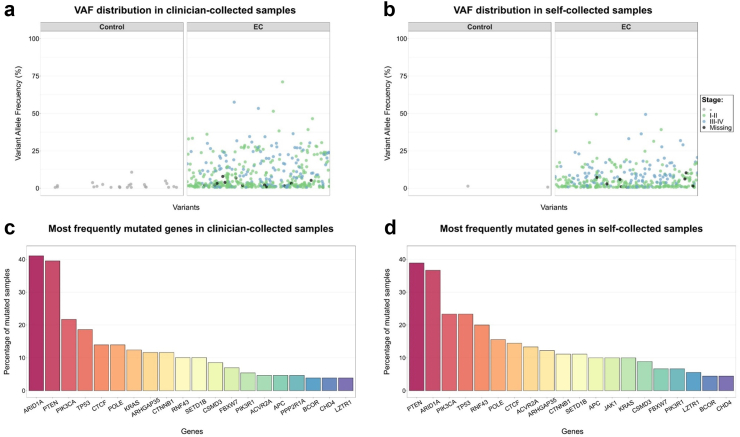
In a clinical setting, negative samples with low coverage (<1000X) would be considered uninformative, and repetition of the test would be advised. Excluding these uninformative samples yielded a sensitivity of 74% and specificity of 78%. The most commonly mutated genes in clinician-collected samples from cases were ARID1A (41%), PTEN (40%), PIK3CA (22%), TP53 (19%), CTCF (14%), POLE (14%), and KRAS (12%; Fig. 2c; Supplemental Figure S6a), which was in accordance with the previous data using tumour specimens.5 The median VAF in clinician-collected samples from cases was 3.8% (Fig. 2a). Sensitivity increased and specificity decreased with increasing number of evaluated genes, and a plateau was reached at approximately 20 genes (Supplemental Figure S7). The ClassEC test on clinician-collected samples showed a higher area under the curve (AUC) than transvaginal ultrasound (TVU; 0.81 vs 0.75), although this difference was not statistically significant (P = 0.20, DeLong). The ClassEC test outperformed cervical cytology significantly (0.81 vs 0.67, P < 0.001, DeLong) (Supplemental Figure S8).
Fig. 2.
Variant allele frequency (VAF) and frequently mutated genes in clinician- and self-collected samples. (a) VAF distribution in clinician-collected samples. (b) VAF distribution in self-collected samples. (c) Most frequently mutated genes in clinician-collected samples. (d) Most frequently mutated genes in self-collected samples.
Variant detection in self-collected vaginal samples
The test was further validated using 111 vaginal self-samples, which yielded 73% sensitivity and 90% specificity, although we counted with a low sample size of controls (21; Table 2). Similar patterns to those of cervical samples were observed in stratified analyses by stage, histology, and MMR status (Supplemental Figure S5b). Sensitivity and specificity were 77% (60/78) and 90% (9/10) in women with bleeding symptoms, respectively. Analyses restricted to women without bleeding symptoms showed a sensitivity of 50% (6/12; Supplemental Figure S5b) and specificity of 91% (10/11).
The most commonly mutated genes detected in the self-collected samples from cases were PTEN (39%), ARID1A (37%), PIK3CA (23%), TP53 (23%), RNF43 (20%), POLE (16%), and CTCF (14%; Fig. 2d; Supplemental Figure S6b). The median VAF in self-samples from these cases was 3.6% (Fig. 2b). Paired clinician-collected and self-collected samples were available for 80 cases, and 52 of them had common variants in both samples, totalling 234 variants in common (Supplemental Figure S9). Thirty-five women were assisted by health care professionals with collection using the self-sampling Evalyn-brush device. Excluding those assisted by clinicians yielded similar results (Supplemental Table S1). The AUC was 0.83 for ClassEC and 0.63 for TVU (P < 0.01; Supplemental Figure S8).
Molecular classification
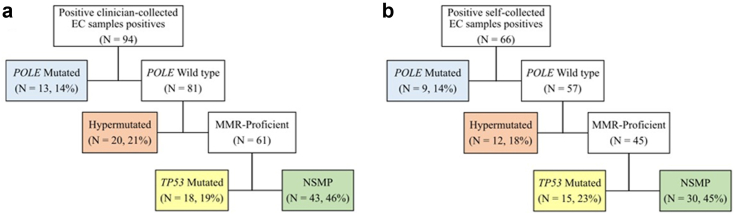
Of the 94 cases with variants in clinician-collected samples, 13 (13.8%) were classified as POLE-mutated, 20 (21.3%) as hypermutated, 43 (45.7%) as NSMP, and 18 (19.1%) as TP53-mutated (Fig. 3a). The proportions of the molecular groups were similar in the self-collected samples (Fig. 3b). The concordance between the molecular classification obtained from cervicovaginal samples and the classification obtained through immunohistochemistry and Sanger sequencing for POLE in paired tumour samples was moderate (kappa = 0.70 for clinician-collected samples, and kappa = 0.74 for self-samples, Table 3). Two POLE-mutated (V411L and P286R) cases identified with Sanger sequencing in tumour samples were not observed using cervicovaginal samples (one was classified as NSMP using a clinician-collected sample, and another as TP53 mutated using a self-sample), while one POLE-mutated (V411L) case was identified in a cervical sample but not in the tumour sample. Similarly, the concordance between the molecular classification using only immunohistochemistry data of tumour samples among the full set of cancers (without POLE sequencing data) and NGS data from cervicovaginal samples was also moderate (kappa = 0.59 for clinician-collected samples; kappa = 0.63 for self-samples). Excluding POLE-mutated samples yielded similar results (kappa = 0.64 for clinician-collected samples; kappa = 0.71 for self-samples).
Fig. 3.
Molecular classification using clinician- and self-collected samples. a) Clinician-collected samples, b) Self-collected samples. MMR = mismatch repair; NSMP = No specific molecular profile.
Table 3.
Concordance in molecular classification between cervicovaginal and tumor samples.
| Tumour samples | Clinician-collected samples |
Self-collected samples |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POLE mutated | Hyper-mutated | TP53 mutated | NSMP | Total | % Correct | Kappa |
POLE mutated | Hyper-mutated | TP53 mutated | NSMP | Total | % Correct | Kappa |
|
| (95% CI) | (95% CI) | |||||||||||||
| POLE Sanger availablea | POLE Sanger availableb | |||||||||||||
| POLE mutated | 5 | 0 | 0 | 1 | 6 | 83% | 0.703 | 4 | 0 | 1 | 0 | 5 | 80% | 0.739 |
| MMR-deficient | 0 | 10 | 0 | 0 | 10 | 100% | (0.543–0.863) | 0 | 9 | 0 | 0 | 9 | 100% | (0.552–0.927) |
| p53 abnormal | 0 | 1 | 16 | 5 | 22 | 73% | 0 | 0 | 13 | 4 | 17 | 76% | ||
| NSMP | 1 | 2 | 0 | 6 | 9 | 67% | 0 | 0 | 1 | 2 | 3 | 67% | ||
| Total | 6 | 13 | 16 | 12 | 47 | 79% | 4 | 9 | 15 | 6 | 34 | 82% | ||
| All | All | |||||||||||||
| MMR-deficient | NA | 11 | 1 | 1 | 13 | 85% | 0.585 | NA | 10 | 1 | 2 | 13 | 77% | 0.625 |
| p53 abnormal | NA | 1 | 17 | 6 | 24 | 71% | (0.436–0.734) | NA | 0 | 14 | 5 | 19 | 74% | (0.456–0.794) |
| NSMP | NA | 8 | 5 | 39 | 52 | 75% | NA | 2 | 5 | 26 | 33 | 79% | ||
| Total | NA | 20 | 23 | 46 | 89 | 75% | NA | 12 | 20 | 33 | 65 | 77% | ||
| All, excluding POLEmut casesc | All, excluding POLEmut casesd | |||||||||||||
| MMR-deficient | NA | 11 | 0 | 1 | 12 | 92% | 0.636 | NA | 10 | 0 | 2 | 12 | 83% | 0.714 |
| p53 abnormal | NA | 1 | 16 | 6 | 23 | 70% | (0.484–0.787) | NA | 0 | 13 | 5 | 18 | 72% | (0.553–0.876) |
| NSMP | NA | 8 | 1 | 32 | 41 | 78% | NA | 2 | 1 | 23 | 26 | 88% | ||
| Total | NA | 20 | 17 | 39 | 76 | 78% | NA | 12 | 14 | 30 | 56 | 82% | ||
NA = Not available; CI = Confidence interval; NSMP = No specific molecular profile.
Bold indicates the number of concordant classifications.
40 High-grade cancers and 7 low-grade cancers.
31 High-grade cancers and 3 low-grade cancers.
Excluding 5 POLE mutated cases jointly identified in tumor and cervicovaginal samples, 1 POLE mutated identified in tumor samples, and 7 POLE mutated cases identified in cervicovaginal samples.
Excluding 4 POLE mutated cases jointly identified in tumor and cervicovaginal samples, 1 POLE mutated identified in tumor samples, and 4 POLE mutated cases identified in cervicovaginal samples.
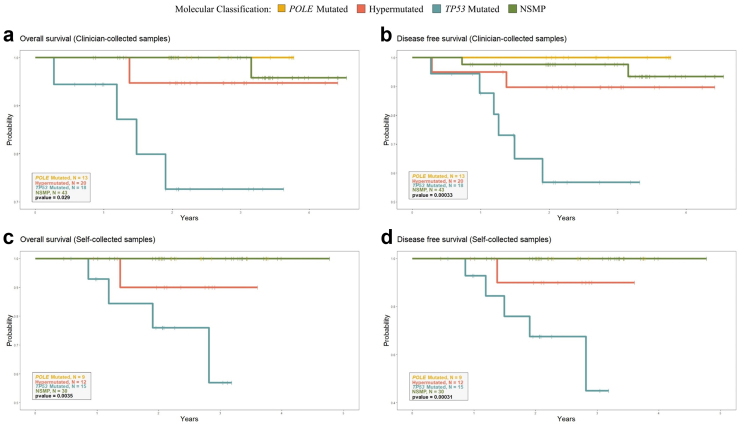
The median follow-up duration was 33.8 months (IQR = 24.6–40.6) among the cases. Importantly, molecular classification using non-invasive samples was predictive of both overall (P = 0.029 using clinician-collected samples and P = 0.004 using self-samples, log-rank test) and disease-free survival (P < 0.001 using clinician-collected samples and P < 0.001 using self-samples, log-rank test, Fig. 4). In Cox models adjusted for age, P-values for overall survival were 0.005 and 0.003 for clinician-collected and self-collected samples, respectively, and <0.001 and 0.001 for disease-free survival (Anova). Survival was high in POLE-mutated cases (0% recurrences in all instances) and poor in TP53-mutated cases (39% recurrences in clinician-collected samples and 33% in self-collected specimens).
Fig. 4.
Overall and disease-free survival according to molecular groups defined using clinician- and self-collected samples. (a) Overall survival (clinician-collected samples). (b) Disease free survival (clinician-collected samples). (c) Overall survival (self-collected samples). (d) Disease free survival (self-collected samples). NSMP = No specific molecular profile.
Discussion
Principal findings
We designed an NGS-based test (ClassEC) for the detection and molecular classification of endometrial cancer. We applied this tool to 339 cervicovaginal samples, which were obtained in a minimally invasive manner or through convenient self-collection. Overall, the ClassEC test identified 73% of endometrial cancer cases using either clinician-collected or self-collected samples. Specificity was 80% for clinician-collected samples and 90% for self-collected samples. Importantly, the ClassEC test classified endometrial cancer cases into four molecular groups that demonstrated significant differences in disease-free survival using both clinician-collected and self-collected samples. Consistent with tumour samples in previous studies,4, 5, 6, 7, 8 endometrial cancer patients with mutations in POLE had an excellent prognosis, whereas patients with TP53 mutations had the poorest clinical outcome.
Previous results
The sensitivity of the ClassEC test was comparable to that of previous studies that used a NGS panel in cervicovaginal samples and achieved sensitivities ranging from 67% to 81%.13,41, 42, 43 However, we obtained relatively low sensitivity compared to epigenomic markers in cervicovaginal samples,12,20,21 and it was lower for non-endometrioid histologies. Studies evaluating proteins in cervicovaginal samples are also promising, although based on low sample sizes (22 and 9 cases),44,45 low performance (<60% specificity),23 or did not provide diagnostic accuracy estimates.28 Other promising pilot studies (≤30 cases) using other molecular methods on cervicovaginal samples22,24, 25, 26, 27,46 have been summarized elsewhere.14 The specificities of studies using a NGS panel were unclear because the controls of the largest study were considerably younger than endometrial cancer cases (mean age 34 vs. 62 years, respectively)41 and aging is strongly associated with the accumulation of somatic mutations.47 The remainder of the NGS studies were based on a small sample size of controls (i.e., 11 and 31) without validation13,42 or no controls at all.43 We used matched controls and observed 80% specificity using clinician-collected samples. Notably, none of the studies that evaluated genetic markers in cervicovaginal samples13,41,43 or other molecular alterations to detect endometrial cancer12,15,19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 provided survival or prognostic data. Interestingly, Kim et al. applied a surrogate of molecular classification in 26 endometrial cancer cases using cervical swab-based genomic DNA from a 50-genes NGS panel; however, they did not evaluate the survival of these patients to assess the validity of this classification.42
The molecular classification better reflects the heterogeneous molecular features of endometrial cancer and has a better prognostic value than dualistic classification.5,48 Several publications have demonstrated that surrogate markers can be used for a molecular classification similar to TCGA in routine surgical pathology, based on immunohistochemical markers and targeted sequencing of tumour samples.4,6, 7, 8 A previous study evaluated the concordance between the immunohistochemical-based classification (ProMisE) and a classification using a NGS panel of 36 genes in tumour samples, observing good concordance.49 Our study molecularly classified endometrial cancer by using non-invasive samples and evaluate survival. We found moderate concordance between the ProMisE classification of tumour samples and NGS classification of non-invasive gynaecologic samples. Importantly, this non-invasive classification yielded significant differences in disease-free survival using both clinician-collected and self-collected cervicovaginal samples. We used 159 samples to train the detection algorithm and 180 cervicovaginal samples for the validation. However, all positive samples from cases were used to develop a molecular classification algorithm. Therefore, validation in independent sets is needed to confirm the potential of molecular classification in non-invasive samples.
Clinical implications
TVU shows high sensitivity for detecting endometrial cancer but very poor specificity (≈50%), which implies that a considerable proportion of women require additional invasive tests to rule out endometrial cancer. In contrast, the ClassEC test demonstrated better specificity than the TVU test did. Endometrial sampling is associated with painful procedures and unsuccessful sampling in a considerable proportion of women due to cervical stenosis or inadequate samples. Moreover, the failure rate of endometrial sampling was 11% (ranging from 1 to 53%), whereas insufficient samples were found in 31% of women (ranging from 7 to 76%).50 In our study, insufficient DNA yields were observed in 13.7% (36/263) and 21.8% (31/142) of the clinician-collected and self-collected samples, respectively. Low yield is a frequent problem with non-invasive samples, which can be solved with repeated sampling, as they are painless and acceptable to women, and optimized extraction protocols.
The ClassEC test is based on non-invasive samples and has the potential for self-collection. This could be particularly beneficial in situations where access to specialists is limited, including pandemics and long referral times. Moreover, cervicovaginal samples can be easily collected before surgery; thus, the molecular classification obtained from these minimally invasive samples could help guide aggressive treatment in TP53-mutated cases. Additionally, this non-invasive classification can help reduce the need for referral to highly specialized hospitals for cancers with good prognosis, such as POLE-mutated cases. The assessment of somatic mutations in consecutive plasma samples has exhibited potential for monitoring the response to treatment.51 Further research is required to determine whether repeated cervicovaginal sampling could be used for detecting relapses and monitoring treatment response among cases.
The results hold potential, especially for postmenopausal women who experience bleeding symptoms. Early detection strategies that target women with postmenopausal bleeding could potentially identify up to 90% of endometrial cancers.38 Molecular methods can aid in better discriminating symptomatic women, as only 9% of postmenopausal women with bleeding are diagnosed with endometrial cancer.38 Furthermore, a recent cost-effectiveness study evaluated the introduction of molecular testing in cervicovaginal samples to detect endometrial cancer in this population compared with current strategies. The authors found that, assuming a molecular test cost of 310€, the molecular strategy was more effective and less expensive than the current strategy.52 The molecular detection of endometrial cancer could also hold promise in screening scenarios among asymptomatic women. However, our data showed that the test had a low diagnostic performance among asymptomatic women. Also, there are still several gaps in knowledge among the general population, regarding the natural history of the disease and the cost-effectiveness of testing in this population,14 which need to be understood prior any implementation of novel technologies in a routine screening setting.
Strengths and limitations
We used well-defined reference tests for endometrial cancer and evaluated all consecutive cases within a defined period range, minimizing the potential for selection bias. Our sample size was large, and we included both clinician-collected and self-collected samples to increase generalizability and to evaluate the potential of self-sampling. The disadvantages of the ClassEC test are related mainly to the unavailability of NGS facilities in some pathology departments and the cost of sequencing at high depth. The cost of the ClassEC test is comparable to that of colonoscopy or computed tomography, but it is more expensive than mammography or a routine Pap test. A panel spanning a smaller genomic region would be cheaper without losing its performance in detecting endometrial cancer, as shown in Supplemental Figure S7. However, the molecular classification would be poorer, as MMRd cases would be less likely to be detected using a smaller genomic panel. Adding microsatellite instability (MSI) loci to the gene panel can help to better define MMRd cases, as shown in recent studies on uterine aspirates.53,54 p53 status in tumour samples was assessed using immunohistochemistry, which is commonly used in pathology laboratories as an acceptable surrogate marker for TP53 mutation status. Around 8% of cases were misclassified comparing p53 immunohistochemistry and TP53 mutations in a study evaluating 168 cases from five centres after consensus review.55 Misclassification of p53 status in tumour samples using immunohistochemistry may have contributed to the lower concordance observed in this study with cervicovaginal samples, which were evaluated using NGS. We adjusted our analyses for age, as this is a variable associated with the appearance of somatic mutations, and with endometrial cancer risk and survival. While we considered other potential variables, we did not include them in the analysis as they were not clearly linked to somatic mutations or endometrial cancer, or were considered mediator variables. Nevertheless, we cannot rule out the possibility of residual confounding from unknown variables that may affect the estimates. The calculation of PPVs and NPVs were derived from the assumed prevalence observed in a systematic review.38 Therefore, changes in the assumed prevalence could alter the estimated predictive values. We found that while changes in the estimated prevalence had an impact on the PPV, the NPV remained relatively consistent, as shown in Supplemental Figure S4. We do not expect verification bias to overestimate our detection rates given that we evaluated consecutive incident endometrial cases.
Conclusions
In conclusion, the ClassEC could represent a non-invasive test to detect endometrial cancer and its prognosis with potential for self-collection. We classified cases of endometrial cancer into four molecular groups based on the analysis of cervicovaginal samples that showed significant differences in overall and disease-free survival. If confirmed in independent sets, the molecular classification of endometrial cancer in non-invasive samples could improve patient care and survival by indicating the early need for aggressive surgery, as well as reducing referrals to highly specialized hospitals in cancers with good prognosis.
Contributors
L.C. interpreted the data and wrote the manuscript. L.C., X.M.G., J.P., J.B., S.S., and F.X.B., conceptualized the study, and obtained funding for the study. S.P., A.C., and E.D. performed experiments. B.P. and F.M. performed the bioinformatics analyses. B.P. and L.C. performed the statistical analysis. Y.B. supervised the statistical analysis. B.P., F.M., and L.C. verified the underlying data. M.L.Q., J.F.G., J.M.M, P.P.T, M.B., S.F.G, J.d.F, V.C, A.V., A.Z., L.P. recruited the study participants and collected relevant samples and clinical information. All authors provided critical comments and read and agreed to the final version of the manuscript.
Data sharing statement
Data are available upon reasonable request from the corresponding author at lcostas@iconcologia.net.
Declaration of interests
L.C. received supplies from Integrated DNA Technologies (IDT) and Roche Diagnostics at a 50% discount for the pilot study of this project and received a speaker's honoraria from Roche. Idibell and Roche signed a contract to collaborate in the development of the bioinformatics pipeline. L.C. has received Colli-Pee® devices (Novosanis) for a research project free of charge. X.B. and L.A. received funds from Merck & Co, Elsevier, Hologic, GlaxoSmithKline (GSK) and SP received funds to attend a meeting from Seegene. P.P.T received funds from Merck & Co and Werfen. JHcispoly and IDIBELL signed a service agreement for another project on endometrial cancer.
Acknowledgements
We thank all the participants of this study. We thank Sara Tous, Alexandra Montoliu, Yolanda Florencia, Vanesa Camón, and Nati Patón for their valuable contributions. We thank the CERCA Programme/Generalitat de Catalunya for institutional support. This study was funded by a competitive grant from Instituto de Salud Carlos III through the projects PI19/01835, PI23/00790, and FI20/00031, CIBERESP CB06/02/0073 and CIBERONC CB16/12/00231, CB16/12/00234 (Co-funded by European Regional Development Fund. ERDF: A way to build Europe). Samples and data were provided by Biobank HUB-ICO-IDIBELL, integrated into the Spanish Biobank Network, and funded by the Instituto de Salud Carlos III (PT20/00171) and by Xarxa de Bancs de Tumors de Catalunya (XBTC) sponsored by Pla Director d’Oncologia de Catalunya. This work was supported in part by the AECC, Grupos estables (GCTRA18014MATI), and the Spanish Ministry of Science and Innovation (PID2019-111254RB-I00). It also counts with the support of the Secretariat for Universities and Research of the Department of Business and Knowledge of the Generalitat de Catalunya, and grants to support the activities of research groups 2021SGR01354 and 2021SGR1112.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104716.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Lortet-Tieulent J., Ferlay J., Bray F., Jemal A. International patterns and trends in endometrial cancer incidence, 1978-2013. J Natl Cancer Inst. 2018;110(4):354–361. doi: 10.1093/jnci/djx214. [DOI] [PubMed] [Google Scholar]
- 3.Oaknin A., Bosse T.J., Creutzberg C.L., et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2022;33(9):860–877. doi: 10.1016/j.annonc.2022.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Talhouk A., McConechy M.K., Leung S., et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer. 2015;113(2):299–310. doi: 10.1038/bjc.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Kandoth C., Schultz N., et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelloo E., Nout R.A., Osse E.M., et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin Cancer Res. 2016;22(16):4215–4224. doi: 10.1158/1078-0432.CCR-15-2878. [DOI] [PubMed] [Google Scholar]
- 7.Kommoss S., McConechy M.K., Kommoss F., et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann Oncol. 2018;29(5):1180–1188. doi: 10.1093/annonc/mdy058. [DOI] [PubMed] [Google Scholar]
- 8.León-Castillo A., de Boer S.M., Powell M.E., et al. Molecular classification of the PORTEC-3 trial for high-risk endometrial cancer: impact on prognosis and benefit from adjuvant therapy. J Clin Oncol. 2020;38(29):3388–3397. doi: 10.1200/JCO.20.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosbie E.J., Kitson S.J., McAlpine J.N., Mukhopadhyay A., Powell M.E., Singh N. Endometrial cancer. Lancet. 2022;399(10333):1412–1428. doi: 10.1016/S0140-6736(22)00323-3. [DOI] [PubMed] [Google Scholar]
- 10.Visser N.C.M., Reijnen C., Massuger L.F.A.G., Nagtegaal I.D., Bulten J., Pijnenborg J.M.A. Accuracy of endometrial sampling in endometrial carcinoma: a systematic review and meta-analysis. Obstet Gynecol. 2017;130(4):803–813. doi: 10.1097/AOG.0000000000002261. [DOI] [PubMed] [Google Scholar]
- 11.Reijnen C., Visser N.C.M., Bulten J., Massuger L.F.A.G., van der Putten L.J.M., Pijnenborg J.M.A. Diagnostic accuracy of endometrial biopsy in relation to the amount of tissue. J Clin Pathol. 2017;70(11):941–946. doi: 10.1136/jclinpath-2017-204338. [DOI] [PubMed] [Google Scholar]
- 12.Herzog C., Marín F., Jones A., et al. A simple cervicovaginal epigenetic test for screening and rapid triage of women with suspected endometrial cancer: validation in several cohort and case/control sets. J Clin Oncol. 2022;40(33):3828–3838. doi: 10.1200/JCO.22.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reijnen C., Putten L.J.M., Bulten J., et al. Mutational analysis of cervical cytology improves diagnosis of endometrial cancer: a prospective multicentre cohort study. Int J Cancer. 2020;146(9):2628–2635. doi: 10.1002/ijc.32686. [DOI] [PubMed] [Google Scholar]
- 14.Costas L., Frias-Gomez J., Guardiola M., et al. New perspectives on screening and early detection of endometrial cancer. Int J Cancer. 2019;145(12):3194–3206. doi: 10.1002/ijc.32514. [DOI] [PubMed] [Google Scholar]
- 15.Peremiquel-Trillas P., Paytubi S., Pelegrina B., et al. An integrated approach for the early detection of endometrial and ovarian cancers (screenwide study): rationale, study design and pilot study. J Pers Med. 2022;12(7):1074. doi: 10.3390/jpm12071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano B., Ibáñez R., Robles C., Peremiquel-Trillas P., de Sanjosé S., Bruni L. Worldwide use of HPV self-sampling for cervical cancer screening. Prev Med. 2022;154 doi: 10.1016/j.ypmed.2021.106900. [DOI] [PubMed] [Google Scholar]
- 17.Olesen T.B., Svahn M.F., Faber M.T., et al. Prevalence of Human Papillomavirus in endometrial cancer: a systematic review and meta-analysis. Gynecol Oncol. 2014;134(1):206–215. doi: 10.1016/j.ygyno.2014.02.040. [DOI] [PubMed] [Google Scholar]
- 18.Castle P.E., Locke A., Tergas A.I., et al. The relationship of human papillomavirus and cytology co-testing results with endometrial and ovarian cancer diagnoses. Gynecol Oncol. 2021;161(1):297–303. doi: 10.1016/j.ygyno.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakkum-Gamez J.N., Wentzensen N., Maurer M.J., et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol Oncol. 2015;137(1):14–22. doi: 10.1016/j.ygyno.2015.01.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.C., Wang H.C., Liao Y.P., et al. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J Gynecol Oncol. 2018;29(1):e17. doi: 10.3802/jgo.2018.29.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R.L., Su P.H., Liao Y.P., et al. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017;23(1):263–272. doi: 10.1158/1078-0432.CCR-16-0863. [DOI] [PubMed] [Google Scholar]
- 22.Doufekas K., Zheng S.C., Ghazali S., et al. DNA methylation signatures in vaginal fluid samples for detection of cervical and endometrial cancer. Int J Gynecol Cancer. 2016 doi: 10.1097/IGC.0000000000000739. [DOI] [PubMed] [Google Scholar]
- 23.Calis P., Yuce K., Basaran D., Salman C. Assessment of cervicovaginal cancer antigen 125 levels: a preliminary study for endometrial cancer screening. Gynecol Obstet Invest. 2016;81(6):518–522. doi: 10.1159/000444321. [DOI] [PubMed] [Google Scholar]
- 24.De Strooper L.M.A., van Zummeren M., Steenbergen R.D.M., et al. CADM1, MAL and miR 124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;67(12):1067–1071. doi: 10.1136/jclinpath-2014-202616. [DOI] [PubMed] [Google Scholar]
- 25.Kinde I., Bettegowda C., Wang Y., et al. Evaluation of DNA from the papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5(167):167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones A., Teschendorff A.E., Li Q., et al. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 2013;10(11) doi: 10.1371/journal.pmed.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G.E., Kweon S.S., Lee J.S., Lee J.H., Nam J.H., Choi C. Quantitative assessment of DNA methylation for the detection of cervical and endometrial adenocarcinomas in liquid-based cytology specimens. Anal Quant Cytopathol Histpathol. 2012;34(4):195–203. [PubMed] [Google Scholar]
- 28.He S.M., Xing F., Sui H., et al. Determination of CA-125 levels in the serum, cervical and vaginal secretions, and endometrium in Chinese women with precancerous disease or endometrial cancer. Med Sci Monit. 2011;17(11):618–625. doi: 10.12659/MSM.882046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiegl H., Gattringer C., Widschwendter A., et al. Methylated DNA collected by tampons--a new tool to detect endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(5):882–888. [PubMed] [Google Scholar]
- 30.Frias-Gomez J., Tovar E., Vidal A., et al. Sensitivity of cervical cytology in endometrial cancer detection in a tertiary hospital in Spain. Cancer Med. 2021;10(19):6762–6766. doi: 10.1002/cam4.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costas L., Palomero L., Benavente Y., et al. Defining a mutational signature for endometrial cancer screening and early detection. Cancer Epidemiol. 2019;61:129–132. doi: 10.1016/j.canep.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Jiménez F., Muiños F., Sentís I., et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 33.León-Castillo A., Britton H., McConechy M.K., et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol. 2020;250(3):323–335. doi: 10.1002/path.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrede S.W., Kasius J., Bulten J., et al. Relevance of molecular profiling in patients with low-grade endometrial cancer. JAMA Netw Open. 2022;5(12) doi: 10.1001/jamanetworkopen.2022.47372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortes-Ciriano I., Lee S., Park W.Y., Kim T.M., Park P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8 doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansournia M.A., Jewell N.P., Greenland S. Case-control matching: effects, misconceptions, and recommendations. Eur J Epidemiol. 2018;33(1):5–14. doi: 10.1007/s10654-017-0325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs M.C. plotROC: a tool for plotting ROC curves. J Stat Softw. 2017;79 doi: 10.18637/jss.v079.c02. Code Snippet 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke M.A., Long B.J., Del Mar Morillo A., Arbyn M., Bakkum-Gamez J.N., Wentzensen N. Association of endometrial cancer risk with postmenopausal bleeding in women: a systematic review and meta-analysis. JAMA Intern Med. 2018;178(9):1210–1222. doi: 10.1001/jamainternmed.2018.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercaldo N.D., Lau K.F., Zhou X.H. Confidence intervals for predictive values with an emphasis to case-control studies. Stat Med. 2007;26(10):2170–2183. doi: 10.1002/sim.2677. [DOI] [PubMed] [Google Scholar]
- 40.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 2014;48:193–204. doi: 10.1016/j.jbi.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Li L., Douville C., et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018;10(433) doi: 10.1126/scitranslmed.aap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim N., Kim Y.N., Lee K., et al. Feasibility and clinical applicability of genomic profiling based on cervical smear samples in patients with endometrial cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.942735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimura R., Kondo E., Yoshida K., et al. Cancer-associated gene analysis of cervical cytology samples and liquid-based cytology significantly improve endometrial cancer diagnosis sensitivity. Oncol Lett. 2022;24(4):376. doi: 10.3892/ol.2022.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Garcia E., Coll-de la Rubia E., Lesur A., et al. Cervical fluids are a source of protein biomarkers for early, non-invasive endometrial cancer diagnosis. Cancers. 2023;15(3):911. doi: 10.3390/cancers15030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Njoku K., Chiasserini D., Geary B., et al. Comprehensive library generation for identification and quantification of endometrial cancer protein biomarkers in cervico-vaginal fluid. Cancers. 2021;13(15):3804. doi: 10.3390/cancers13153804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heng S., Stephens A.N., Jobling T.W., Nie G. Measuring PC activity in endocervical swab may provide a simple and non-invasive method to detect endometrial cancer in post-menopausal women. Oncotarget. 2016;7(29):46573–46578. doi: 10.18632/oncotarget.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milholland B., Auton A., Suh Y., Vijg J. Age-related somatic mutations in the cancer genome. Oncotarget. 2015;6(28):24627–24635. doi: 10.18632/oncotarget.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bokhman J. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15(1):10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Feng J., Zhao C., et al. A new strategy in molecular typing: the accuracy of an NGS panel for the molecular classification of endometrial cancers. Ann Transl Med. 2022;10(16):870. doi: 10.21037/atm-22-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Hanegem N., Prins M.M.C., Bongers M.Y., et al. The accuracy of endometrial sampling in women with postmenopausal bleeding: a systematic review and meta-analysis. Eur J Obstet Gynecol Reprod Biol. 2016;197:147–155. doi: 10.1016/j.ejogrb.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Ashley C.W., Selenica P., Patel J., et al. High-sensitivity mutation analysis of cell-free DNA for disease monitoring in endometrial cancer. Clin Cancer Res. 2023;29(2):410–421. doi: 10.1158/1078-0432.CCR-22-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peremiquel-Trillas P., Gómez D., Martínez J.M., et al. Cost-effectiveness analysis of molecular testing in minimally invasive samples to detect endometrial cancer in women with postmenopausal bleeding. Br J Cancer. 2023;129:325. doi: 10.1038/s41416-023-02291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casas-Arozamena C., Moiola C.P., Vilar A., et al. Non-invasive detection of microsatellite instability in patients with endometrial cancer. Int J Cancer. 2023;152(10):2206–2217. doi: 10.1002/ijc.34435. [DOI] [PubMed] [Google Scholar]
- 54.Canet-Hermida J., Marín F., Dorca E., et al. Highly sensitive microsatellite instability and immunohistochemistry assessment in endometrial aspirates as a tool for cancer risk individualization in lynch syndrome. Mod Pathol. 2023;36(7) doi: 10.1016/j.modpat.2023.100158. [DOI] [PubMed] [Google Scholar]
- 55.Singh N., Piskorz A.M., Bosse T., et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol. 2020;250(3):336–345. doi: 10.1002/path.5375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.