Summary
Background
In Huntington's disease (HD), a CAG repeat expansion mutation in the Huntingtin (HTT) gene drives a gain-of-function toxicity that disrupts mRNA processing. Although dysregulation of gene splicing has been shown in human HD post-mortem brain tissue, post-mortem analyses are likely confounded by cell type composition changes in late-stage HD, limiting the ability to identify dysregulation related to early pathogenesis.
Methods
To investigate gene splicing changes in early HD, we performed alternative splicing analyses coupled with a proteogenomics approach to identify early CAG length-associated splicing changes in an established isogenic HD cell model.
Findings
We report widespread neuronal differentiation stage- and CAG length-dependent splicing changes, and find an enrichment of RNA processing, neuronal function, and epigenetic modification-related genes with mutant HTT-associated splicing. When integrated with a proteomics dataset, we identified several of these differential splicing events at the protein level. By comparing with human post-mortem and mouse model data, we identified common patterns of altered splicing from embryonic stem cells through to post-mortem striatal tissue.
Interpretation
We show that widespread splicing dysregulation in HD occurs in an early cell model of neuronal development. Importantly, we observe HD-associated splicing changes in our HD cell model that were also identified in human HD striatum and mouse model HD striatum, suggesting that splicing-associated pathogenesis possibly occurs early in neuronal development and persists to later stages of disease. Together, our results highlight splicing dysregulation in HD which may lead to disrupted neuronal function and neuropathology.
Funding
This research is supported by the Lee Kong Chian School of Medicine, Nanyang Technological University Singapore Nanyang Assistant Professorship Start-Up Grant, the Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 1 (RG23/22), the BC Children's Hospital Research Institute Investigator Grant Award (IGAP), and a Scholar Award from the Michael Smith Health Research BC.
Keywords: Huntington's disease, Alternative splicing, mRNA processing, Neurodegenerative disease, High-throughput RNA-sequencing, Proteomics
Research in context.
Evidence before this study
Widespread gene splicing dysregulation has been reported in brain tissue of human post-mortem HD patients. In addition, molecular biology studies demonstrated that mutant Huntingtin RNA and protein can sequester proteins involved in RNA processing, suggesting that splicing dysregulation partially contributes to HD pathology. However, post-mortem analyses are likely limited in terms of studying dysregulation in early pathogenesis, partly due to confounding factors related to cell type composition changes with extensive white matter loss in late stage HD.
Added value of this study
The findings in this study support the hypothesis that gene splicing dysregulation manifests early during neuronal development in HD and persists to later stages of the disease. Over-representation of the genes involved in RNA processing, neuronal function, and epigenetic modification was observed among HTT CAG length-dependent differential splicing events. This observation suggests that splicing dysregulation in early HD is associated with the disruption of healthy neuronal development. While previous studies have independently associated these processes to HD, here we propose that RNA processing dysregulation could be a critical factor underlying these HD pathological mechanisms.
Implications of all the available evidence
The results presented in this study suggest that the disruption of gene splicing by mutant HTT could play a significant role in the tissue-selective pathology of HD, particularly neuropathology in the striatum. Given that HD-associated splicing dysregulation occurs in embryonic stem cells, the physiological processes of cell differentiation and organogenesis may be adversely impacted in a tissue-specific manner, leading to pathology in selective tissue. Since the loss of specific neuronal subtypes in the striatum is a pathological presentation of HD neuropathology, our findings related to the dysregulation of splicing in neuronal development genes provide insight into early pathogenesis. Furthermore, these findings open up new avenues for potential therapeutic interventions.
Introduction
Huntington's disease (HD), a debilitating and fatal neurological disorder characterised by progressive motor function decline, cognitive impairment, and behavioural abnormalities, is caused by a hereditary CAG trinucleotide repeat expansion mutation in the Huntingtin (HTT) gene.1,2 The number of trinucleotide repeats in the HTT CAG tract is polymorphic and directly correlated with penetrance and onset of HD, where 6–35 CAG repeat genotypes represent healthy state, 36–39 repeat genotypes show incomplete penetrance, and 40 or more repeat genotypes are fully penetrant for HD.2,3 Furthermore, the length of HTT CAG repeat expansion is inversely correlated with age of onset, where 40–60 repeats is associated with adult onset HD and more than 60 repeats causes juvenile-onset HD. HD neuropathology is associated with prominent medium spiny neuron loss in the striatum and early changes in cortical thickness and cell loss.1,2,4, 5, 6
Transcriptomic profiling studies in post-mortem human tissue and mouse disease models have shown widespread transcriptional changes in the HD brain, particularly in genes associated with neurodevelopment, neuronal function, DNA damage repair, and mitochondrial activity.7, 8, 9, 10, 11 Despite the genetic cause being known and considerable progress made in uncovering the molecular mechanisms underlying HD pathogenesis, the direct causative pathways between the repeat mutation in HTT and HD neurological deficits are still poorly understood.1 Thus, studying how mutant HTT (mHTT) drives pathological alterations in molecular processes will help identify disease-causing molecular targets for the development of pharmacological treatment for HD.
In HD, the process of alternative splicing (AS) is also dysregulated in the brain, and gene mis-splicing likely contributes to HD neuropathology. AS is a ubiquitous RNA processing mechanism involving selective splicing of introns and exons in pre-mRNA transcripts to generate multiple isoforms from single genes.12, 13, 14 It is a crucial process in neurogenesis, brain development, and neuronal function, where neuron-specific splicing factors and RNA-binding proteins act together to regulate AS in genes associated with neuronal functions, such as differentiation, cell motility, and synaptogenesis.12, 13, 14 Widespread aberrant AS has been observed in post-mortem tissues from patients with neurodegenerative diseases like Alzheimer's disease,15, 16, 17 ALS,18 and HD.7,19,20 In addition, RNA-sequencing (RNA-seq) analysis in an isogenic human HD cell model (IsoHD) reported HTT CAG length-dependent transcriptional dysregulation of RNA binding-related genes in human embryonic stem cells (hESCs),21 suggesting that aberrant splicing might be involved in early HD pathogenesis. RNA-sequencing studies performed in post-mortem human brain cortex and striatum have demonstrated AS dysregulation in HD.7,19 However, it is challenging to determine if these observed mis-splicing signatures are associated with pathogenic processes. This difficulty arises largely due to the substantial change in cell composition as a result of considerable neuron loss and astrogliogenesis in late-stage HD.
To investigate early stage AS dysregulation in HD, we performed deep RNA-sequencing on the previously established IsoHD cell model, consisting of a panel of different CAG mutation lengths.21 We detected HTT CAG length-associated AS changes in genes related to neuronal function, RNA processing, and chromatin modification, many of which were also regulated during neuronal differentiation based on the differentiation stage comparison. By performing a proteogenomics analysis, we measured differential expression of protein isoforms of alternatively spliced genes and confirmed that several AS events observed at the gene transcript level translated to differential expression at the protein level. Finally, we compared our results with mouse models and post-mortem human HD brain datasets and confirmed mHTT-associated AS in several previously identified AS genes both in vitro and in vivo. In summary, we observed significant HD-associated AS changes in genes involved in neuronal function and epigenetic regulation processes, highlighting AS dysregulation as a major mechanism for post-transcriptional dysregulation in HD neuropathology. We have made available the AS analysis results in the form of a Shiny web app (https://vincenttano.shinyapps.io/2022_isohd_altsplicing).
Methods
Cell culture
The H9 female IsoHD hESC lines were previously generated, characterised and validated21 and are maintained by MA Pouladi. IsoHD hESC panel cells were cultured on matrigel-coated plates (BD Biosciences) in mTeSR 1 medium (STEMCELL Technologies) in a humidified environment with 5% CO2 at 37 °C. IsoHD hESCs were passaged by dissociating with dispase and seeding at a ratio of 1:6, as described.21
Neuron differentiation
hESCs were differentiated to neural progenitor cells (NPC) and mixed forebrain neurons as described.22 In short, for NPC differentiation, hESCs were cultured in N2B27 medium supplemented with 100 nM LDN193189 (Stemgent), 10 μM SB-431542 (Sigma Aldrich), 2 μM XAV929 (Stemgent) and 200 ng/mL SHH (R&D). For forebrain neuron differentiation, NPCs were cultured in N2B27 medium supplemented with 20 ng/mL BDNF and 20 ng/mL GDNF, 0.5 mM cAMP (Sigma Aldrich) and 0.2 M ascorbic acid (STEMCELL Technologies). Neurons were collected at 21 days in vitro.
Sample preparation for RNA-sequencing
RNA samples were prepared using a RNeasy mini kit (QIAGEN) according to the manufacturer's protocol. RNA quality was evaluated using Agilent 2100 Bioanalyser (Agilent) and all samples were confirmed to have RIN values of at least 7.4 (minimum of 7.4, maximum of 10, and median of 9.6) before library preparation. Sequencing libraries were prepared using TruSeq© Stranded mRNA sample preparation kit (Illumina) and paired end 150-bp sequencing using Illumina HiSeq 2000 was performed by Novogene. The sequencing experiment generated 2.9 billion pairs of 150 bp paired-end reads from 27 samples (three clones, three genotypes, three cell types), with 80–110 million read pairs per sample (GEO: GSE217469).
RNA-sequencing alignment
For quality control of the RNA-seq samples, FastQC (v0.11.9) (Babraham Institute) was used to assess the sequencing quality of each sample. Deep RNA-seq samples were confirmed to have at least ∼80 million reads per library. For gene expression analysis, read alignment and counting were performed using RSEM23 ‘rsem-calculate-expression’ with options for STAR (v2.7.0f)24 paired end alignment to the Ensembl hg38 reference genome assembly and gene annotation gtf (GRCh38 release 96 Homo sapiens).25 All libraries were confirmed to have >90% uniquely mapped alignment. The gene count matrix was then generated using RSEM ‘rsem-generate-data-matrix’ for gene-level counts. For alternative splicing (AS) and custom peptide proteomics analyses, reads were aligned using STAR (v2.7.0f) with options for the basic two-pass mode (‘--twopassMode Basic’) and filtering of non-canonical splice sites (‘--outFilterIntronMotifs RemoveNoncanonicalUnannotated’) to improve discovery of novel splice sites. Alignments were filtered to retain only uniquely mapped reads aligned to the chromosomal DNA using SAMBAMBA (v0.6.6).26 Alignment yielded 75 to 100 million uniquely mapped read pairs across all samples. Splice site read counting and annotation for each sample were then performed using RegTools.27
Differential gene expression analysis
To perform differential gene expression analysis of the IsoHD samples, the gene count matrix was imported into R and analysed using the DESeq2 R package (v1.30.0)28 (R v4.0.2). Briefly, gene expression analysis was performed using the ‘DESeq’ function with the option ‘test = ′Wald′' for testing differentiation stage (i.e., NPC vs hESC)-associated differences and ‘test = ′LRT′' for testing HTT CAG length-associated differences in each differentiation stage independently. The neurons (NEU) were generated in a separate differentiation and sequencing experiment (batch) and so were analysed independently from the hESCs and NPCs. Log fold changes (logFC) were corrected using the ‘lfcShrink’ function with the apeglm method. Batch effects were accounted for by including the sample replicate number in the design formula. Principal component analysis (PCA) was performed with the variance-stabilising transformed counts using prcomp in R. Finally, differentially expressed genes were filtered using the following cutoffs: mean counts ≥10, |logFC |≥1, and adj. P-value < 0.01.
Differential splicing analysis
Differentially splicing analysis was performed using the LeafCutter package.29 In brief, a unified splice site junction database was first generated by summarising sample junction read counts using bedtools.30 Splice junctions were then annotated using the Ensembl hg38 genome assembly and gtf as reference. Only splice junctions that have a non-repeat masked splice site base sequence and were identified in at least 3 samples were retained for downstream analysis. Splice junction clustering and read counting were performed using the ‘leafcutter_cluster_regtools.py’ script with the `-C′ option to include constitutively spliced junctions. Differential splice junction usage was then analysed using the ‘leafcutter_ds.R′ script with option `--min_coverage = 10′ and the sample replicate number included as a confounding factor to account for batch effects. For PCA and heatmap visualisation, junction usage ratios transformed using the logit function and analysed using prcomp in R. All pairs of biological groups-of-interest (differentiation stage: NPC vs hESC; CAG mutant hESC: 45Q vs control, 81Q vs control; CAG mutant NPC: 45Q vs control, 81Q vs control; and CAG mutant Neuron: 45Q vs control, 81Q vs control) were tested independently. The NPC vs hESC comparison was performed by grouping all CAG length genotypes in each differentiation stage. As above, the neurons were generated in a separate differentiation and sequencing experiment (batch) and were analysed independently from the hESCs and NPCs. Only highly used junction clusters, i.e., containing at least 1 junction with percent spliced in (PSI) ≥1% were retained for downstream analysis. The putative AS event type for each splicing cluster were annotated using in-house R scripts to match the intron-exon structure of each cluster to splicing event types (cassette exon (SE), alt. 5′ splice site (A5SS), alt. 3’ splice site (A3SS), mutually exclusive exons (MXE), and retained intron (RI)). Clusters containing multiple event types or no known types are classified as “Mixed” and “Unknown”, respectively. Mixed and Unknown event types are splice junction clusters that contain multiple canonical event types. The Mixed event type refers to clusters that are linear combinations of canonical events whereas the Unknown event type refers to clusters that cannot be resolved to separate complete canonical events. Differential usage in each splicing cluster were considered statistically significant using the following cutoffs: at least 1 junction with ΔPSI ≥1% (considered robust if ΔPSI ≥5%) and adj. P-value <0.05.
To facilitate downstream data integration and comparison, the differentially spliced junctions (DSJ) data table was further annotated using the VastDB AS atlas,31 a database of known AS events with event type annotation, functional association, and evolutionary conservation. Differentially spliced clusters putatively annotated as event types SE, A5SS, A3SS, MXE, or RI were matched to the VastDB EVENT INFORMATION table (Homo sapiens hg38) based on intron-exon structure coordinates, followed by the EVENT CONSERVATION table (Assembly: hg38) based on the VastDB hg38 event name. VastDB attributes including genomic coordinates, DNA sequence, and AS type, where available, were appended to the DSJ data table. Data integration between independent experiments were subsequently performed by matching the annotated VastDB event names.
Gene ontology (GO) functional enrichment analysis
The clusterProfiler R package (v3.18.0)32 was used to perform all GO term33 enrichment analysis with the ‘enricher ()` function. Ensembl gene IDs for the differential splicing and protein analyses were obtained using the ‘biomaRt’ R package (v2.46.0)34 “hsapiens_gene_ensembl” dataset. For GO term enrichment analysis, unless otherwise stated, the background gene list (“universe”) was set as the gene IDs of all features (junctions or peptides) detected in the corresponding experiment. Statistically significant enriched GO terms were filtered using BH-adjusted p-value <0.10.
Sample preparation for cDNA synthesis and real-time quantitative PCR
Approximately 1–2 million cells were lysed using FARB buffer with β-mercaptoethanol and RNA was purified using the FavorPrep Blood/Cultured Cell Total RNA Mini kit (Favorgen) according to the manufacturer's instructions. For all samples, cDNA was generated from 2 μg RNA in 20 μl reactions using High-Capacity Reverse Transcriptase kit (ABI, Thermo Fisher). Real-time quantitative PCR (RT-qPCR) reactions were performed in the Quant Studio 6 Flex Real Time PCR System (ABI, Thermo Fisher) using the SYBR Select PCR Master Mix (ABI, Thermo Fisher) with ten-fold dilution of cDNA and 200 nM of each primer pair as listed in Supplemental Table S1. Relative exon expression levels were calculated using the comparative ΔΔCT method and normalized against the control constitutive exon of the same gene35. Reactions were performed in technical triplicates and each CAG length of each differentiation stage were done in biological triplicates. Statistical significance was tested using Student's t-test with a P-value threshold of 0.05.
Immunoblot protein analysis
Protein lysates of NPC cell pellets (Control, 45Q, and 81Q) were prepared using RIPA lysis buffer (RIPA Buffer, 1 mM PMSF, 5 μm Z-VAD, 1 mM NaVan, and 1 × Complete Protease Inhibitor Mixture tablets). Protein lysates were sonicated for 2 cycles of 30 s on, 30 s off. 35 μg of protein lysates were separated on 10% TGX FastCast gel (Biorad) and transferred on nitrocellulose membranes (Biorad). The following primary antibodies were used: CUL4A (1:1000; Proteintech Cat# 14851-1-AP, RRID:AB_2261175), H3 (1:2500, Abcam Cat# ab1791, RRID:AB_302613), H3K27me3 (1:500, Millipore Cat# 07-449, RRID:AB_310624), H3K9me1 (1:500; Thermo Fisher Scientific Cat# MA5-33385, RRID:AB_2815523), H3K9me2 (1:500; Thermo Fisher Scientific Cat# 720092, RRID:AB_2532802), and β-actin (1:5000; Sigma-Aldrich Cat# A5441, RRID:AB_476744) (Supplemental Table S1). Antibody validation proved by venders. Membranes were incubated with primary antibodies at 4 °C overnight. The following secondary antibodies (1:5000) were used: IRDye® 680RD Goat anti-Rabbit IgG and IRDye® 800RD Goat anti-Mouse IgG (LI-COR). The membrane was imaged using the LI-COR Imaging System and Odyssey V3.0 software (LI-COR), followed by intensity analysis with GelAnalyzer. Statistical significance was tested using a two tailed t-test with a P-value threshold of 0.05.
Custom peptide database generation
To study alternative splicing-associated changes in protein expression and identify novel spliced proteins, a transcriptome-informed custom splice junction peptide database was generated using AS data measured by deep RNA-seq. This method was adapted from a proteogenomics workflow published by Sheynkman et al.36 In brief, splice junctions from the unified splice junction database that are highly used (PSI ≥0.5%), detected in at least 3 RNA-seq libraries, contain a non-repeat masked splice site base sequence, and have total coverage ≥10 were used to generate a transcript fragment (transfrag) database gtf using in-house scripts. Using the Ensembl hg38 gene annotation gtf as a reference, for known junctions, the 5′- and 3′-flanking exons and CDS, where available, were annotated as one transfrag. For novel junctions, the 51bp-flanking regions upstream and downstream of the splice site were annotated as one transfrag and the reference translation frame, where a CDS is available, of the 5′ splice site was included. Each transfrag corresponds to one unique splice junction which will be translated to unique junction peptides. To facilitate downstream filtering and subclass false discovery rate (FDR) analysis, the transfrag databases were split into four subclass databases: known junctions with CDS (knownWithCDS), known junctions with unknown frame (knownNoCDS), novel junctions with inferred frame (novelInferFrame), and novel junctions with unknown frame (novelNoFrame). GffRead37 and EMBOSS showorf38 were then used to generate the custom peptide databases. For junctions without a known frame, three-frame translation was performed. After translation, the N-terminal and C-terminal tails of the peptides were trimmed to the first tryptic site and any STOP codon, respectively, and only peptides longer than 7 amino acids were retained. Finally, BLASTP39 was used to remove identical or highly similar, with up to 2 mismatches at terminal ends, to prevent peptide-spectrum matching with highly similar sequences.
Proteomics
TMT10plex isobaric tagged MS/MS raw data for hESC and NPC IsoHD were downloaded from jPOSTrepo (JPST000243)21 and converted to the mzXML data format using Proteowizard msconvert40 for database search. Database search was performed using MSGF+41 with the following parameters: precursor mass tolerance 20 ppm, allow isotope peak errors −1 to 2, target-decoy strategy, tryptic cleavage, TMT protocol, minimum peptide length of 7, report 20 matches per spectrum, include additional features, maximum 2 missed cleavages, static modifications: Carbamidomethyl of C, TMT10plex; and variable modifications: oxidation of M, acetylation of N-terminal, deamidation of N, Q, and phosphorylation of S, T, and Y. To account for potential differences in subclass FDR, a sequential database search strategy was used. In short, MS/MS spectra were searched sequentially against each custom peptide database subclass in the following order: knownWithCDS, knownNoCDS, novelInferFrame, novelNoFrame. At each step, database search peptide-spectrum match results (q-value <0.01; FDR = 1%) were used to filter the input MS/MS spectra to obtain all “unmatched” spectra, which were then used as the input spectra for database search in the next step. Finally, database search results from all four steps were concatenated for combined post-processing peptide and protein confidence estimation using Percolator.42
Differential protein expression analysis
The Isobar R package (v1.36.0)43 was used to summarise spectrum isobaric reporter ion intensities to protein-level reporter intensities. Only proteins that are supported by spectrums measured in at least 3 samples were retained. Missing values were imputed using the impute R Bioconductor package (v1.64.0).44 Reporter intensities were scaled by total reporter abundance followed by batch correction using ComBat in the sva R package (v3.38).45 PCA was performed using prcomp in R. Differential protein expression to calculate differentiation stage- (NPC vs hESC) and HTT CAG length-associated differences was performed using limma R package (v3.44.1)45 with eBayes. The statistical significance threshold was set at BH-adjusted p-value <0.10.
Visualisation of protein domain
Ensembl gene and transcript annotation (EnsDb.Hsapiens.v99) was retrieved using the AnnotationHub R package (v2.22.1). The ensembldb R package (v2.14.1) was used to obtain transcript and Pfam protein domain tracks and Genome region tracks were plotted with the Gviz R package (v1.34.1).
Comparison with human post-mortem and mouse model RNA-sequencing data
RNA-seq fastq data for post-mortem human HD BA4 motor cortex,19 post-mortem human HD striatum,7 mouse knock-in (KI) model allelic series striatum,9 mouse KI model allelic series cortex,9 and mouse R6/1 model striatum7 were downloaded from SRA and ENA (Supplemental Table S2). RNA-seq data from each experiment (human cortex HD, human striatum HD, mouse KI striatum, mouse KI cortex, and mouse R6/1 striatum) was processed independently. Sequencing quality of all samples was assessed using FastQC and processed for AS analysis as mentioned above. In short, RNA-seq reads were aligned to the Ensembl hg38 genome for human or the Ensembl mm10 genome (GRCm38 release 96 Mus musculus) for mouse using STAR basic two-pass mode filtering out non-canonical splice sites. Uniquely mapped chromosomal alignments were processed using RegTools to identify splice junctions and a unified splice site database was generated for each experiment. Junction clustering and differential usage analysis were then performed using LeafCutter for all splice junctions that contain a non-repeat masked splice site base sequence and detected in at least 3 libraries.
For the mouse KI model striatum and cortex data, differential junction usage was tested in pairwise comparisons between 80Q, 92Q, 111Q, 140Q, or 175Q vs 20Q in mice of 6 months age. For the mouse R6/1 model striatum data, differential junction usage was tested for R6/1 vs wild type mice at 3.5 months of age. Significant differential splicing in clusters were defined as: at least 1 junction with ΔPSI ≥1% and adj. P-value<0.05. For cross-species comparison, significant differential splicing in clusters were defined as: ΔPSI ≥1% and adj. P-value <0.05. Human and mouse gene homology information was downloaded from the Mouse Genome Database (MGD)46 (The Jackson Laboratory, Bar Harbor, Maine) (URL: http://www.informatics.jax.org/) [retrieved 13 Mar 2023].
For the human cortex HD data, the differential junction usage between HD grade 3–4 patient vs control samples were tested to ensure consistency with the striatum HD data which only included patients with grade 3–4 HD. For the human striatum HD data, differential junction usage was tested for HD vs control. AS event type and VastDB annotation were appended and used for comparison between data sets. For comparisons between human data, significant and robust differential splicing in clusters were defined as: at least 1 junction with ΔPSI ≥5% and adj. P-value<0.1.
Soft clustering analysis of junction inclusion levels was performed using the Mfuzz R package (v2.50.0)47 (R v4.0.2). In brief, logit-transformed PSI of junction usage levels were z-score transformed in each IsoHD cell differentiation stage independently (hESC, NPC, neuron) and the optimal number of 4 centers were determined using the ‘Dmin ()’ function elbow method with fuzzifier value determined by the ‘mestimate ()’ function. Soft clustering was then performed using the ‘mfuzz ()’ function and filtered for junctions with membership ≥0.6. Finally, junction usage levels in junctions of each IsoHD cluster in the cortex HD and striatum HD datasets were z-score transformed independently and compared.
Statistics
Box plots represent the median, upper and lower quartile range. Bar plots represent the mean ± standard error of the mean (SEM). Statistical testing methods in DESeq2 (Wald and likelihood ratio test), LeafCutter (likelihood ratio test) and LIMMA (moderated t-test) were used for high throughput RNA-seq and TMT10plex analyses, respectively. The Benjamini Hochberg procedure was used for multiple testing correction. For the RT-qPCR and immunoblot analyses, pairwise comparisons were assessed with Student's t-test. Differences were considered statistically significant when P-value <0.05 and adj. P-value <0.1 unless otherwise stated. The R software (v4.0.2) was used to analyse data for statistical significance.
Ethics
The animal and patient data were obtained from publicly available repositories; ethics approvals are not applicable for this study.
Role of funders
All funders have no direct involvement or impact on the study design, experimental procedures or data analyses.
Results
RNA-seq transcriptional profiling of hESC-derived isogenic HD neuronal cells
To study differentiation stage- and HTT CAG length-dependent AS changes, we performed deep RNA-sequencing (RNA-seq) at different differentiation stages, namely human embryonic stem cells (hESC), neural progenitor cells (NPC) and mature forebrain neurons (NEU), from the previously established IsoHD isogenic allelic panel.21 Deep RNA-seq (>80 million reads/sample in this study) allows us to perform differential splicing analysis with better accuracy and sensitivity, as the analysis utilises reads aligned to unique regions of alternatively spliced transcripts. In addition, deep sequencing improves the detection of unannotated splice junctions (referred to as novel events) that are lowly expressed. To match the published IsoHD isobaric tag proteomics data,21 we sequenced the IsoHD hESC and re-differentiated NPC cell lines containing HTT CAG repeat lengths representing the adult onset CAG length of 45 (45Q), the juvenile onset CAG length of 81 (81Q), and control CAG length of 27/30 (Control). For comparison, we re-sequenced samples from the IsoHD mixed forebrain neuron cells as previously described.22 As a preliminary analysis and validation of the deep RNA-seq transcriptome profiling, we first focused on gene-level differentiation stage- and HTT CAG length-dependent expression changes. A combined principal component analysis (PCA) of the current and original RNA-seq dataset21 confirmed high similarity of the deep sequencing gene expression profiles in all three differentiation stages (hESC, NPC, and NEU) with their corresponding cell types in the original study and show separation of libraries according to differentiation stages (Supplemental Fig. S1a). We further confirmed that both IsoHD NPCs and neurons showed decreased expression of stem cell marker gene POU5F1 (OCT3/4)48 and increased expression of neuronal marker genes MAP2,49 and POU3F2 (OCT7)50 (Supplemental Fig. S1b–d). NPCs also increased expression of the NPC marker PAX651 whereas neurons showed increased expression of neuronal FOXP252 and forebrain neuron marker genes BCL11B,53 CUX1,54 and PPP1R1B (DARPP32)55 (Supplemental Fig. S1b, d and e).
Differential gene expression analysis identified differentially expressed genes (DEGs) across CAG lengths (45Q/81Q vs control, n = 3 per group) in each differentiation stage independently (|logFC|≥1 and BH-adjusted P-value <0.01) (Supplemental Table S3). For CAG length-dependent differences, a total of 1,781, 1,390, and 39 DEGs were identified in hESCs, NPCs, and neurons, respectively. There were 349, 29, and 15 CAG length-dependent DEGs common between the current and original dataset in hESCs, NPCs, and neurons, respectively, all representing significant overlaps (P-value <1e-07, Fisher's exact test). We did not observe perfect concordance between the current and original dataset, due to differences in statistical cutoffs and experimental design, e.g., the original study was sequenced to a lower depth and included additional biological conditions.
Alternative splicing regulation in neuron differentiation and HTT CAG repeat expansion
To investigate AS regulation associated with mutant HTT, we performed differential splicing analysis using a custom pipeline to identify known and novel differential spliced events (Supplemental Fig. S2a). Splice junction usage, representing relative levels of intron inclusion in mRNA transcripts or Percent Spliced In (PSI) of introns, was first quantitated. PCA of junction usage levels (logit-transformed PSI) showed a clear separation of libraries by differentiation stages (Fig. 1a), similar to the gene-level expression analysis. Differential junction usage (i.e., differential splicing inclusion of introns) was next performed either across differentiation stages NPC vs hESC (all CAG length genotypes) or between CAG lengths (45Q/81Q vs control); referred to as CAG mutant) in hESC, NPC, and NEU independently (|ΔPSI|≥1% and BH-adjusted P-value <0.05). Since the NEU data set was independently generated as a separate experiment (differentiation and RNA-sequencing) from hESC and NPC, we did not directly test differences between NEU and either hESC or NPC due to confounding batch effects. For differentiation stage-dependent differential splicing, we identified 41,332 annotated and 13,140 novel differentially spliced junctions (DSJs) occurring in 10,714 genes in total (Supplemental Table S4; NPC vs hESC). A comparison between only hESC control and NPC control genotype cells yielded similar results (>80% overlap; Supplemental Table S4). For HTT CAG length-dependent differential splicing, a total of 31,904, 6,226, 16,281 annotated junctions (occurring in 8,244, 2,376, and 5,255 genes) were found to be differentially spliced in hESCs, NPCs, and NEU, respectively. We also identified an additional 11,504, 2,286, and 5,409 novel junctions which are differentially spliced in hESCs, NPCs, and NEU, respectively (Supplemental Table S4). We summarised and annotated the DSJs based on the canonical AS event types (Cassette/skipped exon, alternative 5′ splice site (A5SS), alternative 3’ splice site (A3SS), retained intron, and mutually exclusive exon (MXE)56) (Table 1; Fig. 1b). Since this analysis method is unable to determine independence between canonical AS events in either Mixed or Unknown types, only canonical AS event types are retained for further analysis.
Fig. 1.
Differential differentiation stage- and HTT CAG length-dependent splicing in the isogenic HD model. (a) Principal component analysis for Percent Spliced In (PSI) values of splice junctions in the isogenic HD model cell lines for hESC (red), NPC (blue), and neuron (magenta). (b) Alternative splicing (AS) event types of known and novel AS events showing differentiation stage- and HTT CAG length-associated differential splicing (LeafCutter likelihood ratio test: ΔPSI ≥1%, FDR<0.05, n = 3) (A5SS, Alternative 5′ splice site; A3SS, Alternative 3′ splice site; MXE, Mutually exclusive exons). (c) Scatter plots illustrating splicing changes in known AS event junctions showing robust changes in hESC, NPC, and NEU independently (ΔPSI ≥5%). Junctions that show significant splicing changes in 45Q only (red), in 81Q only (blue), or in both (purple) are indicated. (d) Top 25 significant functional enrichment Gene Ontology terms in differentiation stage-(NPC vs hESC) and CAG length-(summarised for hESC, NPC, and neuron independently) associated differentially spliced genes (FDR <0.1). (e) Overlap of CAG length-dependent known and canonical AS events in the isogenic HD lines (Cassette exon, A5SS, A3SS, Retained intron, MXE).
Table 1.
Differential splicing events identified in the IsoHD cell model.
| Comparison | Cassette exon | Alt. 5′ splice site | Alt. 3′ splice site | Retained intron | Mixed | Unknown | Total |
|---|---|---|---|---|---|---|---|
| NPC vs hESC | 4053 (20.0%) | 1651 (8.2%) | 1818 (9.0%) | 1666 (8.2%) | 6948 (34.3%) | 4100 (20.3%) | 20236 |
| hESC 45Q vs control | 1703 (17.7%) | 710 (7.4%) | 965 (10.0%) | 1144 (11.9%) | 3382 (35.1%) | 1722 (17.9%) | 9626 |
| hESC 81Q vs control | 1966 (18.6%) | 852 (8.1%) | 955 (9.0%) | 1201 (11.4%) | 3588 (33.9%) | 2011 (19.0%) | 10573 |
| NPC 45Q vs control | 381 (16.6%) | 163 (7.1%) | 157 (6.9%) | 254 (11.0%) | 897 (39.2%) | 439 (19.2% | 2290 |
| NPC 81Q vs control | 108 (12.8%) | 55 (6.5%) | 63 (7.5%) | 89 (10.6%) | 365 (43.3%) | 162 (19.2%) | 842 |
| NEU 45Q vs control | 935 (16.7%) | 331 (5.9%) | 383 (6.8%) | 447 (8.0%) | 2406 (42.9%) | 1107 (19.7%) | 5609 |
| NEU 81Q vs control | 402 (15.1%) | 167 (6.3%) | 209 (7.8%) | 251 (9.4%) | 1117 (41.9%) | 522 (19.5%) | 2668 |
To further study HTT splicing regulation, we looked at HTT CAG length-dependent differential splicing across and in each differentiation stage. Focusing on known and canonical AS events (Cassette exon, A5SS, A3SS, retained intron, and MXE), we annotated splicing events using the VastDB AS event atlas31 and compared differential splicing events across the three different differentiation stages to evaluate if CAG length-associated AS regulation is differentiation stage-selective. We found 3,580, 567, and 1,243 CAG length-associated differential splicing events (corresponding to 2,532, 459, 999 genes) in hESC, NPC, and NEU, respectively. Among these, only 36 events (in 35 genes) were shared in all three differentiation stages (Fig. 1e), suggesting that mHTT-driven splicing regulation is largely modulated by differentiation stage specificity. Notably, neurodevelopmental- and neurodegeneration-related genes APBB2,57 NFASC,58 and VLDLR59 are differentially spliced in all three differentiation stages. A subset of CAG length-dependent DSJs further showed robust ΔPSI ≥5% in hESC (N = 1,908), NPC (N = 270), and NEU (N = 633) specifically (Fig. 1c). For these robust DSJs within each differentiation stage, scatter plots of the ΔPSI with respect to control show unique subsets of AS events that were significantly (FDR <0.05) associated with 45Q or 81Q repeat length (Fig. 1c, red and blue, respectively) and a subset that were found to be significantly (FDR<0.05) associated with both repeat lengths (Fig. 1c, purple). Not only do the hESCs have a larger number of splicing events (Table 1), but there is a marginally larger number of robust events identified in the 81Q (N = 1,368) as compared to the 45Q repeat length (N = 1,251) with an overlap of 730 that are significant in both repeat lengths. This is in contrast to the NPCs and NEUs that have over two-fold difference in the number of significant splicing events in the 45Q vs the 81Q repeat length. This non-monotonic relationship between splicing levels and CAG length is reflective of the previously observed gene-level transcriptional relationship with CAG length.9,21
Mutant HTT drives differentiation stage-specific aberrant splicing in neuronal development, mRNA splicing and epigenetic modifier genes
To investigate gene function of differentially spliced genes, we performed gene ontology (GO) term enrichment in the differentially spliced genes and found over-representation for genes related to neuronal development (e.g., “dendrite morphogenesis”), cell cycle (e.g., “cell cycle checkpoint”, “spindle organisation”), and mRNA splicing (e.g., “mRNA processing”) (Fig. 1d). KEGG pathway analysis also identified “Spliceosome” as the top enriched pathway (FDR<0.05) for CAG length-associated differentially spliced genes in hESC (Supplemental Fig. S3). In addition, we observed enrichment of the GO term “positive regulation of GTPase activity”, a biological process implicated in HD,60 being enriched in all three differentiation stages. Importantly, CAG length-associated functional terms were also found to be significantly enriched in differentiation stage-dependent differential splicing (i.e., NPC vs hESC), including “GTPase activity”, “neuron projection development”, and “mRNA processing” (Supplemental Table S5). This suggests that neuron differentiation-driven AS regulation was possibly disrupted by HTT CAG repeat expansion. Of the 4,082 differentially spliced events in NPC vs hESC, 1,424 (∼35%) showed CAG length-dependent differential splicing in at least one of the three differentiation stages, including genes previously identified as being mis-spliced in post-mortem HD tissue: SORBS1,7,19 PTPRD,7 PTPRF,7,19 PTBP2,7 MAP2,7,61 and TCERG1.7,62 GO terms related to epigenetic regulation, such as “covalent chromatin modification”, “histone lysine methylation”, and “regulation of chromatin organisation”, were also found to be enriched in both differentiation stage- and CAG length-dependent differentially spliced genes (Fig. 1d), suggesting that there is a relationship between HTT CAG length-driven mis-splicing and epigenetic dysregulation in HD. We assessed the total levels of three histone lysine methylation marks, H3K27me3, H3K9me1 and H3K9me2, in the NPCs and identified significant differences associated with CAG-length in H3K27me3 and H3K9me2, but no change in H3K9me1 (Supplemental Fig. S4).
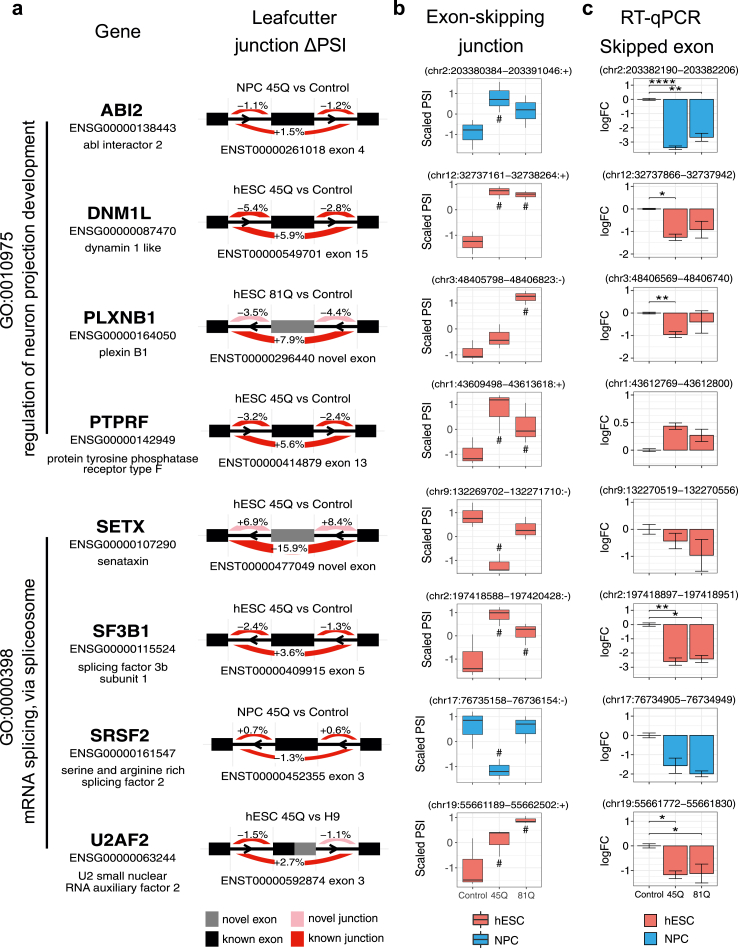
To validate the observed differentiation stage-selectivity of CAG length-driven differential splicing, we performed real-time qPCR exon expression analysis on CAG length-dependent differential splicing cassette exons selected based on the robustness of splicing changes. These include mRNA transcripts of GO term “regulation of neuron projection development”-related ABI2, DNM1L, PLXNB1, PTPRF, and GO term “mRNA splicing”-related SETX, SF3B1, SRSF2, and U2AF2 (Fig. 2a). We determined differential splicing regulation by ΔPSI of the exon-skipping junction from the deep RNA-seq data, representing relative levels of transcripts that do not include the corresponding cassette exon (exon skipped) (Fig. 2b), and tested the relative expression levels of the skipped exon (exon included), normalised to a neighbouring constitutively expressed exon to account for gene-level regulation35 (Fig. 2c). Using this approach, we expect to see an inverse correlation between the levels of exon-skipping junction (Fig. 2b) and the levels of skipped exon (Fig. 2c) for each transcript, e.g., increased level/higher PSI of an exon-skipping junction as measured by LeafCutter should be associated with a decreased level/negative logFC in the detection of the skipped exon as measured by RT-qPCR. For exons in ABI2, DNM1L, PLXNB1, and SRSF2, we observed differentiation stage-specific CAG length-dependent splicing changes (Fig. 2c). In PTPRF and SF3B1, there was a decrease in expression of cassette exons only in NPC and hESC, respectively, suggesting differentiation stage-specific mHTT-driven exon skipping. In addition, we tested RNA-binding proteins PUF60 and SNRNP70, epigenetic modifiers CARM1, transcriptional regulator FUBP1, and ubiquitin ligase MGRN1 (Supplemental Fig. S5). We were unable to validate HTT CAG length-dependent differential splicing in exons of CARM1, PTPRF, PUF60, and SETX, as detected by RNA-seq, possibly due to differences in sensitivity and specificity of the assays.
Fig. 2.
Differential splicing in mRNA splicing- and neurodevelopment-associated genes. (a) Select HTT CAG length-dependent alternatively spliced cassette exons in genes associated with enriched GO terms mRNA splicing and neuron development. (b) Scaled Percent spliced in (PSI) values of exon-skipping junction, i.e., cassette exon not included, showing CAG length-associated differential splicing in hESC (red) or NPC (blue). Data are represented as median and interquartile range. Significant splicing events relative to control in each differentiation stage (hESC and NPC tested independently) are indicated below (increase) or above (decrease) the box plots of each CAG length (LeafCutter likelihood ratio test, n = 3): #multiple testing-adjusted P-value <0.05. (c) Quantitative RT-PCR and fold change quantitation of select differential splicing cassette exons in IsoHD hESC (red) or NPC (blue). Data are represented as mean ± SEM. Significant splicing events relative to control in each differentiation stage (hESC and NPC tested independently) are indicated (Student's t-test; n = 3): ∗P-value <0.05; ∗∗P-value <0.01; ∗∗∗P-value <0.001; ∗∗∗∗P-value <0.0001.
Mutant HTT-driven aberrant expression in protein isoforms
Next, we investigated if the differentiation stage- and CAG length-dependent differential splicing in mRNA transcripts were associated with differential expression of their corresponding protein isoforms. To study differential protein expression associated with splice junction usage, we performed a proteogenomic analysis to measure relative protein expression of splice junction peptides in a published isobaric tag TMT-10 plex proteomics analysis of the IsoHD hESC and NPC lines each with control, 45Q, and 81Q HTT21 (Supplemental Fig. S2b). This proteogenomic approach allows the identification of junction peptides in tandem MS analysis corresponding to each splice junction detected in RNA-seq analysis by generating a custom junction peptide database for peptide-spectrum database search (Supplemental Fig. S6). The proteogenomic analysis of the IsoHD TMT-10plex data identified 17,514 quantifiable junction peptides (FDR <0.01), corresponding to 6,332 genes. Consistent with the original work, PCA showed clear separation between hESC and NPC along PC1 (Fig. 3a).
Fig. 3.
Differential spliced junction-associated peptide expression in the isogenic HD model. (a) Principal component analysis for junction peptide TMT10plex intensities in the isogenic HD model cell lines for hESC (red) and NPC (blue) from the jPOST repository (ID:JPST000243). (b) Heatmap and hierarchical clustering of junction peptide expression associated with differentiation stage-dependent differential splicing (LIMMA moderated t-test: FDR<0.1, n = 12). (c and d) Heatmap of junction peptide expression associated with HTT CAG length-dependent differential splicing in hESC (c) and NPC (d) showing significant differential expression at the protein level (FDR<0.1, n = 4). (e) Top 10 significant functional enrichment Gene Ontology terms in differentiation stage-(NPC vs hESC) associated differentially spliced junction peptides. (f) Immunoblot analysis of CUL4A levels in NPCs expressing control, 45Q, and 81Q CAG length HTT with representative blot. Protein expression values are normalized to control and represented as median and interquartile range. Statistically significant changes are indicated (two-tailed t-test; n = 6) ∗P-value <0.05.
We identified 2,022 junction peptides differentially expressed in NPC vs hESC, as well as 43 and 447 junction peptides differentially expressed in hESC and NPC, respectively, when comparing expanded HTT CAG length (45Q, and 81Q) to control (BH-adjusted P-value <0.1; Supplemental Table S6; Supplemental Fig. S7). We next looked at differentially expressed junction peptides (DEPs) that were also differentially spliced at the transcript level by comparing the differential peptide expression results with the differential junction usage. Out of a total of 92,255 DSJs (|ΔPSI|≥0.01 and BH-adjusted P-value <0.01) across all comparisons (differentiation stage- and CAG length-dependent splicing), 4,615 (∼5%) junctions were quantified at the protein level. Out of these quantified junctions, we identified DEPs that displayed differential splicing at the transcript level in NPC vs hESC (N = 445, Fig. 3b), hESC CAG length-associated (N = 11, Fig. 3c), and NPC CAG length-associated (N = 97, Fig. 3d). Among differentiation stage-associated DEPs, peptides corresponding to HD-associated genes7 APP, MAP2, NCOR1, and SORBS1 were found to be differentially expressed in NPC vs hESC (Supplemental Table S6). For CAG length-dependent DEPs, we found only four peptides commonly regulated in hESC and NPC, namely C1orf53, CUL4A, COG8, and TSEN54. Differentiation stage-associated DEPs were enriched in GO terms that include “cell–cell junction” and “neuronal development” (Fig. 3e). We assessed the total levels of CUL4A in the NPCs and identified a small but significant difference between control and 81Q (Fig. 3f).
Impact of splicing changes on functional protein domains
To predict functional outcomes of HTT CAG length-associated differential splicing in HD neuropathology, we evaluated the putative impact of alternative splicing events on protein sequence and domains in select genes. For this analysis, we focused on HTT CAG length-dependent differential splicing events in neuronal function-associated genes, namely AKT2, DNM1L, MACF1, PTBP2, and PTPRD. For AKT2, DNM1L, and PTBP2, differential splicing events are associated with changes in transcripts annotated as nonsense-mediated decay (NMD) on the Ensembl genome database, suggesting that these splicing events drive post-transcriptional regulation through the NMD pathway and do not directly impact protein structure and function.
For MACF1 and PTPRD, differential splicing cassette exon events were related to protein coding alternative splice forms showing differences in CDS encoding functional protein domains in the MACF1 and PTPRD proteins (Supplemental Fig. S8). For MACF1, we observed CAG length-dependent increased skipping of cassette exons associated with an increase in expression of the transcript encoding the non-canonical MACF1 isoform 2 protein (ENST00000361689; Q9UPN3-2), and a corresponding decrease in the transcript encoding the canonical MACF1 isoform 1 (ENST00000564288; Q9UPN3-1). MACF1 isoforms 1 and 2 have different protein sequences in the Plectin repeats, Spectrin repeats, and Growth-Arrest-Specific Protein 2 (GAR) domain regions (Supplemental Fig. S8a). In the PTPRD gene, CAG length-dependent splicing changes were associated with an increase in transcript encoding the non-canonical PTPRD isoform 4 (ENST00000397606; P23468-4). Compared to the canonical PTPRD isoform 1 (ENST00000381196; P23468-1), PTPRD isoform 4 contain missing sequences in the Fibronectin type III (FN III) domain and Immunoglobulin (Ig) domain regions (Supplemental Fig. S8b). Notably, the expression of MACF1 and PTPRD isoforms are tissue-selective, particularly in the brain63,64 and we detected differential junction peptide expression for MACF1 (Supplemental Table S6). Taken together, these results suggest that cell type-selective disruption of the alternative splicing process in early HD driven by HTT CAG expansion mutation may cause functional outcomes in protein isoform expression changes leading to neurological pathogenesis.
HTT CAG length-dependent splicing dysregulation in HD mouse models and human HD post-mortem tissue
Knock-in (KI) and transgenic mouse models of HD serve as valuable tools to study early HD pathogenesis,65 and transcriptomic profiling of brain tissue in pre-symptomatic mice which have not yet exhibited neuronal loss allows for the study of early gene expression changes in the brain. To determine if the IsoHD CAG length-dependent splicing changes can also be observed in the early symptomatic HD mouse model, we compared our results to three published RNA-seq studies: a CAG allelic series KI mouse model cortex tissue dataset, a CAG allelic series KI mouse model striatum tissue dataset, and a transgenic R6/1 zQ175 mouse model striatum tissue dataset. We analysed the mouse RNA-seq data sets independently using the pipeline described above to identify significant AS events (|ΔPSI|≥1% and BH-adjusted P-value <0.05). For the CAG allelic series cortex and striatum data sets, we identified CAG length-dependent differential splicing events between HD (80Q, 92Q, 111Q, 140Q, or 175Q) and control (20Q) in pairwise comparisons (n = 8). For the transgenic R6/1 striatum data set, we identified HD-associated differential splicing between R6/1 and wild-type mice (n = 3). In total, we identified 12, 107, and 627 differential splicing events (in 12, 100, and 542 mouse genes) in allelic cortex, allelic striatum, and R6/1 striatum, respectively (Fig. 4a; Supplemental Table S7). We compared differentially spliced gene human orthologs to the IsoHD CAG length-dependent differentially spliced genes and found an overlap of 5, 37, and 228 genes in allelic cortex, allelic striatum, and R6/1 striatum, respectively (Supplemental Fig. S9). We found a larger number of differentially spliced genes in the mouse HD striatum than in the mouse HD cortex (Fig. 4a, Supplemental Table S7) and this is particularly pronounced in the longer lengths of 111Q, 140Q, and 175Q KI models of the mouse allelic series,9,66, 67, 68 where 36 mis-spliced genes with human orthologs were detected in the striatum vs 2 in the cortex (Supplemental Table S7). The difference in the magnitude of splicing alterations between the two mouse models may be driven by both the sequencing depth between dataset as well as by the mouse models themselves, given that the R6/1 is a faster progression model.65 Among the overlapping mouse HD striatum and IsoHD differentially spliced genes, we identified an enrichment of GO terms “Cell Morphogenesis Involved In Differentiation” (P-value = 0.0004), including genes DNM1L and MACF1, and “RNA Splicing” (P-value = 0.002), including genes CELF2, SRSF2 and TCERG1.
Fig. 4.
Splice event intersection between isogenic HD model, mouse HD models, and human post-mortem HD brain tissue. (a) Number of differential splicing events in mouse transgenic R6/1 striatum (left, n = 3), mouse KI allelic series striatum and cortex (center, n = 8), and human HD post-mortem cortex (n = 5) and striatum (n = 3) (right) (LeafCutter likelihood ratio test: FDR<0.1). (b) Overlap of CAG length- and HD-associated differential splicing events in IsoHD, post-mortem HD cortex, and post-mortem HD striatum. (c) Soft clustering of PSI values of intersect splice junctions in common AS events between IsoHD model and either HD cortex or HD striatum datasets. The three columns of line plots represent differentially spliced junctions in hESC (left), NPC (center), and neuron (right) cells. Only junctions with clustering membership ≥0.6 were retained. Samples are ordered by CAG length (Control, 45Q, 81Q) from left to right. The black lines represent the mean PSI value across samples for all differentially spliced junctions in each cluster. (d) Box plot of PSI values of soft clustering intersect splice junctions in post-mortem HD cortex (left) and striatum (right) tissue. Samples are ordered by HD grade (Control, HD grades 2-4) from left to right. Data are represented as mean ± SEM.
As our analysis of HTT CAG length-associated differential splicing uncovered a number of genes previously reported to be mis-spliced in post-mortem HD, we compared our IsoHD cell data to published human post-mortem brain HD transcriptome profiling datasets to evaluate if these differential splicing events occur in human HD in vivo. Two published deep RNA-seq datasets, one in post-mortem human grade 3–4 HD motor cortex (BA4)19 and one in post-mortem human grade 3–4 HD striatum,7 described widespread transcriptome-wide alternative splicing dysregulation in HD in the brain. To ensure consistent integration across the three datasets, we analysed the deep RNA-seq data sets independently using the pipeline described above and annotated differential splicing events using VastDB AS event atlas31 for comparison. Additionally, we compared AS events with |ΔPSI|≥5% and BH-adjusted P-value <0.1 which was equivalent to thresholds initially used in both post-mortem studies.
The IsoHD differential splicing events represent HTT CAG expansion-associated (either 45Q or 81Q vs control) splicing changes, and HD cortex and striatum events represent HD patient vs healthy control splicing changes. We identified 1,827, 104, and 1,684 robust AS events in IsoHD, HD cortex (n = 5) and HD striatum (n = 3), respectively (Fig. 4a; Supplemental Table S8). Eight genes (from eight AS events) were commonly mis-spliced in all three datasets (Fig. 4b), including mRNA processing factors CPSF7 and RPRD2; developmental genes CAMK2G and PTPRM; transcriptional regulator BCOR; as well as EHBP1, KCNMA1, and TBC1D5. In addition, six and 272 IsoHD AS events, in six and 261 genes, were found to be commonly differentially spliced only in HD cortex or HD striatum, respectively (Fig. 4b). Overall, we detected a higher number of differential splicing events in HD striatum as compared to HD cortex. Genes that were differentially spliced both in vitro and in vivo were also enriched in GO terms “covalent chromatin modification” and “regulation of neuron projection development” (Supplemental Fig. S10), including neuronal function gene DNM1L and mRNA processing genes PTBP2 and TCERG1.
To determine if IsoHD HTT CAG length-dependent differential splicing and HD brain mis-splicing signatures are correlated, we performed soft clustering analysis on IsoHD DSJs that were also found to be differentially spliced in either HD cortex or HD striatum. In total, 1,012 IsoHD DSJs overlapped with either cortex or striatum HD-associated DSJs. We performed Fuzzy clustering with four centers on the IsoHD logit-transformed PSI values of the HD-associated mis-spliced junctions which assigned 107, 129, 129, and 146 junctions (membership ≥0.6) to clusters 1, 2, 3, and 4, respectively. Based on the expression patterns of the cluster centers, we infer that cluster 1 represents junctions with HTT CAG length-associated up-regulation in junction PSI in both hESC and NPC, cluster 2 represents up-regulation in junction PSI in hESC but not NPC, cluster 3 represents down-regulation in junction PSI in both hESC and NPC, whereas cluster 4 represents down-regulation in junction PSI in hESC but not NPC (Fig. 4c). Interestingly, differential splicing in NPC showed a largely non-monotonic relationship with HTT CAG length, where CAG length-dependent differential splicing occurs mostly in 45Q but not in 81Q. We did not observe any clear CAG length-associated expression patterns in NEU. When comparing IsoHD cluster expression patterns to the post-mortem HD cortex and HD striatum DSJs, we observed a correlated up- and down-regulation of junction PSI in the HD striatum for clusters 1 and 3, respectively, and to a lesser extent clusters 2 and 4 (Fig. 4d). This correlation in junction PSI pattern, representing relative levels of intron inclusion, between IsoHD CAG length-associated differential splicing and human HD striatum suggests that HTT CAG repeat expansion-associated splicing regulation patterns we observed in the IsoHD model corresponds to in vivo HD-associated mis-splicing in the human striatum. However, this correlation was not observed in HD cortex, possibly due to the smaller number of HD-associated mis-splicing events identified in the cortex indicating neuronal subtype-specific HTT CAG length-dependent effects. Overall, we found that differential splicing events in the IsoHD hESC, and to a lesser extent NPC, that overlapped with events in the post-mortem HD dataset showed a direct correlation in terms of intron inclusion changes in the HD striatum, but not in the HD motor cortex.
Discussion
In this study, we identified mutant HTT-associated AS changes using the IsoHD panel in differentiated neuronal cells.21,22 We performed deep transcriptome profiling in hESC, NPC, and NEU carrying different CAG lengths corresponding to control, adult onset (45Q), and juvenile onset (81Q), and reported disease-associated AS events. Using a proteogenomics approach, we identified altered protein isoforms arising from AS events in the hESCs and NPCs. In comparison with two mouse models of HD, we showed that splicing is altered in a disease dependent manner in the prodromal stages of the disease. We elucidated patterns of AS which are present in both embryonic stem cells and in human HD post-mortem tissue, with a specificity shown in human striatal tissue. Throughout these analyses, we identified several molecular processes that are enriched in AS events and may represent feedback loops of RNA processing dysregulation. Although these molecular processes have been previously observed to be disrupted in Huntington's disease, this study highlights mHTT-driven splicing dysregulation as a potential major contributing factor to multiple HD pathological mechanisms. This disruption may instigate malfunctional RNA processing, neurogenesis, neuron maintenance, and epigenetic regulation in early pathogenesis and progression of HD neuropathology.
Here, we identified HD-associated altered splicing in mRNA processing genes, where RNA binding protein (RBP) and splicing factor genes were found to be differentially spliced in the disease state. This is consistent with our previous work with the IsoHD panel where we identified CAG length-associated transcriptional changes in RNA processing genes,21 suggesting both transcriptional and post-transcriptional regulation of RNA processing. Since AS is extremely sensitive to changes in the spliceosome complex due to the low specificity and transient nature of RNA-RBP interactions,69 dysregulated splicing of these genes likely further disrupts global AS in HD.20 Within the IsoHD panel, we identified several AS events in RNA processing genes, including SRSF2, PTBP2, TSEN54, and TCERG1. SRSF2 and PTBP2 are both RBPs involved in splicing machinery assembly and have been previously identified as upstream regulators of HD-associated splicing dysregulation in post-mortem HD tissue.7,19 As such, the dysregulated splicing of these RBPs likely further exacerbate changes in AS of their downstream targets. While the dysregulation of splicing of RBPs and splicing factors has previously been shown in post-mortem HD tissue,7,19 our findings here indicate that these AS events occur as early as the embryonic stem cell stage and may persist through the late stages of the disease. We reported splicing changes and differential junction peptide expression in TSEN54, which encodes for a subunit of the tRNA splicing endonuclease complex involved in both tRNA and mRNA processing,70 and identified differential expression of protein isoforms from the splicing events in both the hESCs and NPCs. Missense mutations in TSEN54 result in the rare neurodegenerative disorder Pontocerebellar hypoplasia type 2, which is characterized by severe cognitive and motor deficits in infancy and early childhood.71 We also detected dysregulated splicing in the TCERG1 gene, a regulator of transcriptional elongation and pre-mRNA splicing, in IsoHD, HD mouse striatum, and HD human striatum. TCERG1 has been identified as a genetic modifier of age of onset in HD, where the length of a quasi-tandem repeat (QTR) hexamer in exon 4 of TCERG1 is inversely correlated with age at the onset of symptoms.62,72 In our results, we found that the decreased inclusion of an adjacent retained intron and exon 6 in the TCERG1 transcripts is associated with CAG length, although not monotonically. While the mechanism of TCERG1 QTR hexamer's modulation of HD onset is still unknown, we postulate that mHTT-induced differential splicing of TCERG1 might be associated with the loss of TCERG1-mediated neuroprotection in HD. This study, along with the previously observed transcriptional dysregulation, global splicing alterations and the enrichment of AS events in key splicing factors, highlights the complex relationship between the RNA processing and genetic disease modifiers.
AS is a key regulatory mechanism for many functional processes and dysregulation of AS in HD may have widespread downstream effects. Splicing is an important regulatory process in neurogenesis and neuronal maintenance, with AS of splicing factors themselves being tightly regulated.13 Since AS regulation is a crucial process in brain development and maintenance,12,14 HD neuropathology could be caused in part by cell type- and CAG-specific disruption of AS regulation during brain development. We observe dysregulated splicing of genes relating to the “regulation of neuron projection” and “neuron development”, but also in “cell cycle regulation” and “DNA replication”, which are important factors for neurogenesis and cell fate decisions.12 In genes with CAG length-associated AS events, we saw a significant enrichment in the functional terms of “cell cycle”, “regulation of G1/S transition”, and “replication”. This aligns with previous work in IsoHD-derived cortical organoids, where Zhang et al. showed premature neuronal differentiation in 81Q-mHTT organoids as a result of reduced symmetrical cell division and extended cell cycle arrest (preprint73). Our in-depth analysis of splicing changes in neuronal development genes MACF1 and PTPRD also reveal putative functional outcomes in neurogenesis. MACF1, a regulator of actin and microtubule cytoskeleton function, plays a critical role in neuronal cell migration and neurite extension63,74 while PTPRD, a phosphatase involved in cell adhesion and cell–cell interaction, is important in promoting neurite outgrowth and cortical interconnectivity with other regions in the brain.64,75 Although these two genes play multiple roles in the development of various organs, they are both essential for brain development.75, 76, 77 In particular, mutations in the MACF1 gene GAR domain has been linked to impairment of neuronal migration leading to malformation of cerebral convolutions.78 For PTPRD, Pulido et al. proposed that alternatively splicing of exons encoding peptides of the FN III and Ig domains regulates protein–protein interaction of PTPRD which impacts axonal growth in neurons during brain development.79,80
In addition to neurogenesis, CAG length-dependent AS events observed in the IsoHD panel further suggest splicing dysregulation may drive neuronal pathology. Through functional enrichment analysis of AS events, we identified enriched terms related to GTPase activity, shown previously to be associated with HD neuropathology.60,81,82 For example, the dysregulation of GTPase-regulated mitochondrial fission can directly cause neuronal cell death.82,83 Mitochondrial abnormalities are a prominent pathological feature in HD83,84 and also confirmed in the IsoHD panel.21 Here, we observed CAG length-dependent splicing dysregulation in several GTPase-regulated genes, including the DNM1L gene that encodes the DRP1 protein involved in mitochondrial fission. Importantly, DRP1 directly interacts with mHTT protein which can disrupt its physiological function, and subsequent restoration of DRP1 regulatory activity can partially rescue the diseased phenotype.82 Our results indicate that mHTT could additionally affect DNM1L gene function through the alternative splicing of its mRNA transcript, and could thus contribute to HD neuropathology. However, additional molecular and cellular investigation is needed to further determine whether these changes in splicing might be neuroprotective or neurotoxic.
Our IsoHD AS analysis also identified the CAG length-dependent splicing changes in epigenetic modifier genes. Epigenetic modification, a process where epigenetic modifiers chemically alter chromatin to change its structure thereby regulating gene expression, is critical in the maintenance of neuronal function85 and is also known to be altered in HD.86, 87, 88 Extensive changes in DNA methylation and histone modifications, including H3K4 trimethylation and H3K27 acetylation, were found in cell and animal models as well as in HD patient post-mortem brains.86, 87, 88 In particular, genes down-regulated in HD were associated with progressive decrease in euchromatic histone acetylation in a brain region-specific manner.88 In our results, we observed an enrichment of epigenetic modifier genes that display mHTT-associated splicing dysregulation, such as histone deacetylase HDAC7 and methyltransferases CARM1, EHMT1, and EHMT2, in both the IsoHD panel and post-mortem HD striatum, suggesting that mHTT-associated dysregulated splicing may influence epigenetic changes in HD. However, given the extensive epigenetic changes that occur during differentiation,89,90 we cannot exclude the possibility that the enrichment of epigenetic modifiers may be a downstream effect of CAG length-associated disruption to the differentiation process.
Finally, given that AS is a major mechanism in neuronal cell fate decisions,12 HD neuronal toxicity,20 and epigenetic regulation of neuronal function,85 cell type-specific AS regulation may be a fundamental contributor to the regionality of brain tissue vulnerability in the HD disease progression. We observed a considerable level of differentiation stage selectivity in CAG length-associated splicing dysregulation, where only 35 genes (∼1% of the total differentially spliced genes) were differentially spliced in all three IsoHD differentiation stages. We also identified a higher number of AS events in the post-mortem striatum as compared to the post-mortem cortex and observed correlated CAG length-associated splicing events between both IsoHD hESC and NPC and the striatum. The minimal correlation between the IsoHD NEU and striatum may be due to the neuronal differentiation used here, which generates a mixed forebrain culture instead of a medium spiny neuron-specific culture. This region specificity of AS dysregulation was consistent when comparing HD mouse model data sets at a disease stage where the mice have started displaying behavioural symptoms but no detectable brain cell loss.9 Out of 2,006 post-mortem HD striatum AS genes, 269 overlapped with genes containing the same AS event in at least one of the IsoHD differentiation stages, representing ∼13.4% of all striatum AS genes. Combined, these results indicate that dysregulation of splicing occurs at early stages, well before the onset of symptoms, and that there may be continued alterations to a subset of genes through to the end stages of the disease. Our analysis highlights a striking observation that the shared pattern of splicing between the IsoHD lines and post-mortem brain tissue datasets are more evident in the striatum, but not in the cortex. However, neuronal cell fate determination and brain region specification during brain development is a very dynamic process with rapid changes in both transcriptional and post-transcriptional gene regulation. As such, it is challenging to deconvolute development- and disease-associated splicing changes in HD during neurogenesis, particularly with a cell-based experimental model. Nevertheless, the differences in the splicing patterns and overall overlap of IsoHD AS genes with post-mortem cortex HD and striatum HD datasets points to brain region-, and neuronal subtype-selective dysregulation in mHTT-driven mis-splicing.
Although we identified differentiation stage-specific AS changes, a caveat of our results presented here is that these AS changes may be confounded by some level of cellular heterogeneity. Our analyses were performed on bulk cellular populations and non-homogenous tissue, and not purified cell populations. It is of great interest to extend this work to the HD vulnerable medium-sized spiny projection neurons, but it is challenging to optimise current MSN differentiation protocols for the efficiency and cell heterogeneity needed in such RNA-seq analyses.91 As such, we did not investigate the selective vulnerability of neuronal subtypes to cell death in HD, particularly striatal MSNs.1,2,4,10 In addition, even though we have presented evidence that splicing dysregulation in early HD is associated with post-transcriptional gene regulation in biological processes related to HD neuropathology, the extent in which splicing dysregulation contributes directly to pathogenesis remains to be evaluated. Further mechanistic studies using alternative splicing modulators92,93 could be used to explore the mechanisms of action of how splicing alteration directly impacts brain development and function. However, given the widespread alterations, coupled with the differences we observe between cell stages and brain regions, it will remain a challenge to translate these mechanistic insights into clinical applications. For example, a clinical trial for the application of a splicing modulator to HD patients has recently been paused due to potential detrimental off-target effects (ClinicalTrials.gov Identifier: NCT05111249).
Our results show a high level of differential splicing at the adult onset-associated CAG length of 45Q as compared to the juvenile onset length of 81Q, especially in the NPCs and neurons. This non-monotonic relationship has been observed at the gene expression level,9,21 despite the correlation between the CAG length and the age of onset and disease progression.94 Previous studies have shown that the age of onset and rate of disease progression can be modified by different rates of somatic expansion in the HTT CAG repeats in HD.94, 95, 96 This has led to the hypothesis that HD is a two-stage process, with distinct disease mechanisms operating at the shorter CAG lengths as compared with the longer, somatically expanded, CAG lengths.97,98 In this two-stage hypothesis, disease-causing CAG lengths undergo somatic CAG tract expansion until a threshold where CAG expanded mutant HTT triggers distinct pathogenic pathways in susceptible cells.97 While not explicitly tested in this study, the disparity in the magnitude of altered splicing, and the overall transcriptional patterns, between the CAG lengths may be reflective of this two-stage mechanistic model.
Lastly, even though we were able to annotate splicing events based on the five defined canonical event types (Cassette exon, A5SS, A3SS, MXE, or RI56), a large number of differential splicing events were annotated as “Mixed” or “Unknown” events as they cannot be resolved into any single specific canonical event type. This is due to a technical limitation in the AS analysis approach,29 where the independence of overlapping events cannot be determined. Importantly, individual AS event types are associated with distinct splicing regulation mechanisms involving unique RNA processing regulatory factors that possibly relate to specific biological processes.56,99 In the context of HD, the selective usage of splice sites by specific RNA-binding proteins and splice factors has been hypothesised to be a mediator of splicing dysregulation in HD pathogenesis.7 As such, in-depth investigations into these non-canonical events could further elucidate other molecular mechanisms contributing to HD neuropathology.
In summary, we have identified mHTT-associated widespread AS dysregulation of genes that are integral to RNA processing, neuronal differentiation and function, and epigenetic state. Our results highlight aberrant splicing as a possible major mechanism underlying early HD neuropathology and suggest a potential mHTT-driven regulatory feedback loop that results in transcriptional and post-transcriptional dysregulation in HD pathogenesis. Furthermore, our proteogenomics analysis also reveals that mHTT-induced splicing alterations can yield protein isoforms, with potential, yet unappreciated, downstream pathogenic effects. By disrupting biological processes crucial to early development of striatal neurons and other implicated cell types, mHTT could impair cell survival and function eventually leading to selective vulnerability and cell death. We have made our results available at https://vincenttano.shinyapps.io/2022_isohd_altsplicing to encourage and facilitate future efforts focusing on the role of these key mis-spliced genes in HD pathogenesis, with the prospects of leading to potential targets for therapeutic intervention to mitigate HD.
Contributors
S.R.L. conceived and provided overall supervision in this project; V.T. and S.R.L. designed the experiments; V.T. conducted the bioinformatics analyses of the data; V.T. and S.R.L. verified the underlying data for the sequencing, qPCR, and proteomics results presented in this study. S.R.L. and M.A.P. verified the underlying data for the immunoblot analysis. M.A.P. contributed ideas to the experimental design. K.H.U. and N.A.B.M.Y. conducted the cell culture and RNA extraction for RNA-sequencing; K.H.U. and J.B. conducted the molecular biology experiments; W.W.L.T. assisted in the RNA-seq gene expression analysis. V.T. and S.R.L. were involved in the interpretation of the results and wrote the manuscript with feedback from the other authors. All authors have read and approved the final manuscript.
Data sharing statement
The RNA-seq data presented in this paper is deposited in the GEO under the accession number GSE217469 and will be made available upon publication. The AS analysis results is available in the form of a Shiny web app (https://vincenttano.shinyapps.io/2022_isohd_altsplicing). The TMT10plex proteomics data used for the proteogenomics analysis is available in jPOST (accession number JPST000243), the post-mortem human and mouse model RNA-seq are available in SRA (accession numbers PRJNA274985,9 PRJNA274989,9 and PRJNA31662519) and ENA (accession number PRJEB441407). The full IsoHD alternative splicing result has been made available at Mendeley Data (https://doi.org/10.17632/njfhnpf7dh.1) and the R analysis scripts are available through GitHub (https://github.com/langleylab/Tano_et_al_IsoHD_alt_splicing). Additional data and analysis scripts can be made available upon request.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
The authors thank Nevin Tham (Lee Kong Chian School of Medicine), Nathan Harmston (Yale-NUS) and Toh Hean Ch'ng (Lee Kong Chian School of Medicine) for assisting in proof-reading and editing of the manuscript, and Glen Sequiera (University of British Columbia) for technical assistance. We would like to acknowledge the members of the Integrative Biology of Disease group at the Lee Kong Chian School of Medicine for their helpful comments and suggestions on this work. The computational work for this article was partially performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg). M.A.P. is the recipient of a BC Children’s Hospital Research Institute Investigator Grant Award (IGAP), and a Scholar Award from the Michael Smith Health Research BC. S.R.L. is supported by the Lee Kong Chian School of Medicine, Nanyang Technological University Singapore Nanyang Assistant Professorship Start-Up Grant and by the Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 1 (RG23/22).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104720.
Appendix A. Supplementary data
References
- 1.Bates G.P., Dorsey R., Gusella J.F., et al. Huntington disease. Nat Rev Dis Primers. 2015;1 doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.Ross C.A., Aylward E.H., Wild E.J., et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10(4):204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 3.Langbehn D.R., Hayden M.R., Paulsen J.S., and the PREDICT-HD Investigators of the Huntington Study Group CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):397–408. doi: 10.1002/ajmg.b.30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergonzoni G., Döring J., Biagioli M. D1R- and D2R-medium-sized spiny neurons diversity: insights into striatal vulnerability to Huntington's disease mutation. Front Cell Neurosci. 2021;15 doi: 10.3389/fncel.2021.628010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosas H.D., Salat D.H., Lee S.Y., et al. Cerebral cortex and the clinical expression of Huntington's disease: complexity and heterogeneity. Brain. 2008;131(Pt 4):1057–1068. doi: 10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thu D.C.V., Oorschot D.E., Tippett L.J., et al. Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington's disease. Brain. 2010;133(Pt 4):1094–1110. doi: 10.1093/brain/awq047. [DOI] [PubMed] [Google Scholar]
- 7.Elorza A., Márquez Y., Cabrera J.R., et al. Huntington's disease-specific mis-splicing unveils key effector genes and altered splicing factors. Brain. 2021;144(7):2009–2023. doi: 10.1093/brain/awab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges A., Strand A.D., Aragaki A.K., et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 9.Langfelder P., Cantle J.P., Chatzopoulou D., et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19(4):623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malla B., Guo X., Senger G., Chasapopoulou Z., Yildirim F. A systematic review of transcriptional dysregulation in Huntington's disease studied by RNA sequencing. Front Genet. 2021;12 doi: 10.3389/fgene.2021.751033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yildirim F., Ng C.W., Kappes V., et al. Early epigenomic and transcriptional changes reveal Elk-1 transcription factor as a therapeutic target in Huntington's disease. Proc Natl Acad Sci U S A. 2019;116(49):24840–24851. doi: 10.1073/pnas.1908113116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furlanis E., Scheiffele P. Regulation of neuronal differentiation, function, and plasticity by alternative splicing. Annu Rev Cell Dev Biol. 2018;34:451–469. doi: 10.1146/annurev-cellbio-100617-062826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazin P., Xiong J., Liu X., et al. Widespread splicing changes in human brain development and aging. Mol Syst Biol. 2013;9:633. doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su C.H., D D., Tarn W.Y. Alternative splicing in neurogenesis and brain development. Front Mol Biosci. 2018;5:12. doi: 10.3389/fmolb.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinrich A.J., Jodelka F.M., Chang J.L., et al. Therapeutic correction of ApoER2 splicing in Alzheimer's disease mice using antisense oligonucleotides. EMBO Mol Med. 2016;8(4):328–345. doi: 10.15252/emmm.201505846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh Y.C., Guo C., Yalamanchili H.K., et al. Tau-mediated disruption of the spliceosome triggers cryptic RNA splicing and neurodegeneration in Alzheimer's disease. Cell Rep. 2019;29(2):301–316.e10. doi: 10.1016/j.celrep.2019.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj T., Li Y.I., Wong G., et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer's disease susceptibility. Nat Genet. 2018;50(11):1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold E.S., Ling S.C., Huelga S.C., et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013;110(8):E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin L., Park J.W., Ramachandran S., et al. Transcriptome sequencing reveals aberrant alternative splicing in Huntington's disease. Hum Mol Genet. 2016;25(16):3454–3466. doi: 10.1093/hmg/ddw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schilling J., Broemer M., Atanassov I., et al. Deregulated splicing is a major mechanism of RNA-induced toxicity in Huntington's disease. J Mol Biol. 2019;431(9):1869–1877. doi: 10.1016/j.jmb.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Ooi J., Langley S.R., Xu X., et al. Unbiased profiling of isogenic Huntington disease hPSC-derived CNS and peripheral cells reveals strong cell-type specificity of CAG length effects. Cell Rep. 2019;26(9):2494–2508.e7. doi: 10.1016/j.celrep.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Xu X., Tay Y., Sim B., et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-Mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Rep. 2017;8(3):619–633. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A., Gingeras T.R. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;51 doi: 10.1002/0471250953.bi1114s51. 11.14.1-11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates A.D., Achuthan P., Akanni W., et al. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotto K.C., Feng Y.Y., Ramu A., et al. Integrated analysis of genomic and transcriptomic data for the discovery of splice-associated variants in cancer. Nat Commun. 2023;14(1):1589. doi: 10.1038/s41467-023-37266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y.I., Knowles D.A., Humphrey J., et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50(1):151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tapial J., Ha K.C.H., Sterne-Weiler T., et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27(10):1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smedley D., Haider S., Ballester B., et al. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey S.E., Cheng C. Methods for characterization of alternative RNA splicing. Methods Mol Biol. 2016;1402:229–241. doi: 10.1007/978-1-4939-3378-5_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheynkman G.M., Shortreed M.R., Frey B.L., Smith L.M. Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol Cell Proteomics. 2013;12(8):2341–2353. doi: 10.1074/mcp.O113.028142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea G., Pertea M. GFF utilities: GffRead and GffCompare. F1000Res. 2020;9 doi: 10.12688/f1000research.23297.2. ISCB Comm J-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Chambers M.C., Maclean B., Burke R., et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30(10):918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 43.Breitwieser F.P., Müller A., Dayon L., et al. General statistical modeling of data from protein relative expression isobaric tags. J Proteome Res. 2011;10(6):2758–2766. doi: 10.1021/pr1012784. [DOI] [PubMed] [Google Scholar]
- 44.Huber W., Carey V.J., Gentleman R., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blake J.A., Baldarelli R., Kadin J.A., et al. Mouse genome database (MGD): knowledgebase for mouse-human comparative biology. Nucleic Acids Res. 2021;49(D1):D981–D987. doi: 10.1093/nar/gkaa1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar L., E Futschik M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda J., Seino S., Bell G.I. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res. 1992;20(17):4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goedert M., Crowther R.A., Garner C.C. Molecular characterization of microtubule-associated proteins tau and MAP2. Trends Neurosci. 1991;14(5):193–199. doi: 10.1016/0166-2236(91)90105-4. [DOI] [PubMed] [Google Scholar]
- 50.He X., Treacy M.N., Simmons D.M., Ingraham H.A., Swanson L.W., Rosenfeld M.G. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340(6228):35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 51.Larsen K.B., Lutterodt M., Rath M.F., Møller M. Expression of the homeobox genes PAX6, OTX2, and OTX1 in the early human fetal retina. Int J Dev Neurosci. 2009;27(5):485–492. doi: 10.1016/j.ijdevneu.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Konopka G., Bomar J.M., Winden K., et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462(7270):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desplats P.A., Lambert J.R., Thomas E.A. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington's disease. Neurobiol Dis. 2008;31(3):298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li N., Zhao C.T., Wang Y., Yuan X.B. The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brené S., Lindefors N., Ehrlich M., et al. Expression of mRNAs encoding ARPP-16/19, ARPP-21, and DARPP-32 in human brain tissue. J Neurosci. 1994;14(3 Pt 1):985–998. doi: 10.1523/JNEUROSCI.14-03-00985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126(1):37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 57.Golanska E., Sieruta M., Gresner S.M., et al. Analysis of APBB2 gene polymorphisms in sporadic Alzheimer's disease. Neurosci Lett. 2008;447(2–3):164–166. doi: 10.1016/j.neulet.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Monfrini E., Straniero L., Bonato S., et al. Neurofascin (NFASC) gene mutation causes autosomal recessive ataxia with demyelinating neuropathy. Parkinsonism Relat Disord. 2019;63:66–72. doi: 10.1016/j.parkreldis.2019.02.045. [DOI] [PubMed] [Google Scholar]
- 59.Boycott K.M., Flavelle S., Bureau A., et al. Homozygous deletion of the very low density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am J Hum Genet. 2005;77(3):477–483. doi: 10.1086/444400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tourette C., Li B., Bell R., et al. A large scale Huntingtin protein interaction network implicates Rho GTPase signaling pathways in Huntington disease. J Biol Chem. 2014;289(10):6709–6726. doi: 10.1074/jbc.M113.523696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabrera J.R., Lucas J.J. MAP2 splicing is altered in Huntington's disease. Brain Pathol. 2017;27(2):181–189. doi: 10.1111/bpa.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arango M., Holbert S., Zala D., et al. CA150 expression delays striatal cell death in overexpression and knock-in conditions for mutant huntingtin neurotoxicity. J Neurosci. 2006;26(17):4649–4659. doi: 10.1523/JNEUROSCI.5409-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu L., Su P., Li R., et al. Isoforms, structures, and functions of versatile spectraplakin MACF1. BMB Reports. 2016;49(1):37–44. doi: 10.5483/BMBRep.2016.49.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhl G.R., Martinez M.J. PTPRD: neurobiology, genetics, and initial pharmacology of a pleiotropic contributor to brain phenotypes. Ann N Y Acad Sci. 2019;1451(1):112–129. doi: 10.1111/nyas.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pouladi M.A., Morton A.J., Hayden M.R. Choosing an animal model for the study of Huntington's disease. Nat Rev Neurosci. 2013;14(10):708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 66.Jacobsen J.C., Gregory G.C., Woda J.M., et al. HD CAG-correlated gene expression changes support a simple dominant gain of function. Hum Mol Genet. 2011;20(14):2846–2860. doi: 10.1093/hmg/ddr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menalled L.B., Kudwa A.E., Miller S., et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington's disease: zQ175. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 69.Fredericks A.M., Cygan K.J., Brown B.A., Fairbrother W.G. RNA-binding proteins: splicing factors and disease. Biomolecules. 2015;5(2):893–909. doi: 10.3390/biom5020893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paushkin S.V., Patel M., Furia B.S., Peltz S.W., Trotta C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell. 2004;117(3):311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 71.Budde B.S., Namavar Y., Barth P.G., et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet. 2008;40(9):1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 72.Lobanov S.V., McAllister B., McDade-Kumar M., et al. Huntington's disease age at motor onset is modified by the tandem hexamer repeat in TCERG1. NPJ Genom Med. 2022;7(1):53. doi: 10.1038/s41525-022-00317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J., Ooi J., Utami K.H., et al. Developmental Biology; 2019. Expanded huntingtin CAG repeats disrupt the balance between neural progenitor expansion and differentiation in human cerebral organoids.http://biorxiv.org/lookup/doi/10.1101/850586 [cited 2022 Aug 30]. Available from: [Google Scholar]
- 74.Moffat J.J., Ka M., Jung E.M., Smith A.L., Kim W.Y. The role of MACF1 in nervous system development and maintenance. Semin Cell Dev Biol. 2017;69:9–17. doi: 10.1016/j.semcdb.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uetani N., Chagnon M.J., Kennedy T.E., Iwakura Y., Tremblay M.L. Mammalian motoneuron axon targeting requires receptor protein tyrosine phosphatases σ and δ. J Neurosci. 2006;26(22):5872–5880. doi: 10.1523/JNEUROSCI.0386-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ka M., Kim W.Y. Microtubule-actin crosslinking factor 1 is required for dendritic arborization and axon outgrowth in the developing brain. Mol Neurobiol. 2016;53(9):6018–6032. doi: 10.1007/s12035-015-9508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goryunov D., He C.Z., Lin C.S., Leung C.L., Liem R.K.H. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol Cell Neurosci. 2010;44(1):1–14. doi: 10.1016/j.mcn.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobyns W.B., Aldinger K.A., Ishak G.E., et al. MACF1 mutations encoding highly conserved zinc-binding residues of the GAR domain cause defects in neuronal migration and axon guidance. Am J Hum Genet. 2018;103(6):1009–1021. doi: 10.1016/j.ajhg.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pulido R., Krueger N.X., Serra-Pagès C., Saito H., Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem. 1995;270(12):6722–6728. doi: 10.1074/jbc.270.12.6722. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez-Brito M.R., Bixby J.L. Differential activities in adhesion and neurite growth of fibronectin type III repeats in the PTP-delta extracellular domain. Int J Dev Neurosci. 2006;24(7):425–429. doi: 10.1016/j.ijdevneu.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casella C., Lipp I., Rosser A., Jones D.K., Metzler-Baddeley C. A critical review of white matter changes in Huntington's disease. Mov Disord. 2020;35(8):1302–1311. doi: 10.1002/mds.28109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song W., Chen J., Petrilli A., et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17(3):377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J., Moody J.P., Edgerly C.K., et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum Mol Genet. 2010;19(20):3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu X., Ng B., Sim B., et al. pS421 huntingtin modulates mitochondrial phenotypes and confers neuroprotection in an HD hiPSC model. Cell Death Dis. 2020;11(9):809. doi: 10.1038/s41419-020-02983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feng J., Fouse S., Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61(5 Pt 2):58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- 86.Hervás-Corpión I., Guiretti D., Alcaraz-Iborra M., et al. Early alteration of epigenetic-related transcription in Huntington's disease mouse models. Sci Rep. 2018;8(1):9925. doi: 10.1038/s41598-018-28185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ng C.W., Yildirim F., Yap Y.S., et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110(6):2354–2359. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sadri-Vakili G., Bouzou B., Benn C.L., et al. Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum Mol Genet. 2007;16(11):1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 89.Srinageshwar B., Maiti P., Dunbar G.L., Rossignol J. Role of epigenetics in stem cell proliferation and differentiation: implications for treating neurodegenerative diseases. Int J Mol Sci. 2016;17(2):199. doi: 10.3390/ijms17020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Atlasi Y., Stunnenberg H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet. 2017;18(11):643–658. doi: 10.1038/nrg.2017.57. [DOI] [PubMed] [Google Scholar]
- 91.Le Cann K., Foerster A., Rösseler C., et al. The difficulty to model Huntington's disease in vitro using striatal medium spiny neurons differentiated from human induced pluripotent stem cells. Sci Rep. 2021;11(1):6934. doi: 10.1038/s41598-021-85656-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keller C.G., Shin Y., Monteys A.M., et al. An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion. Nat Commun. 2022;13(1):1150. doi: 10.1038/s41467-022-28653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Relizani K., Goyenvalle A. The use of antisense oligonucleotides for the treatment of duchenne muscular dystrophy. Methods Mol Biol. 2018;1687:171–183. doi: 10.1007/978-1-4939-7374-3_12. [DOI] [PubMed] [Google Scholar]
- 94.Swami M., Hendricks A.E., Gillis T., et al. Somatic expansion of the Huntington's disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet. 2009;18(16):3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciosi M., Maxwell A., Cumming S.A., et al. A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. eBioMedicine. 2019;48:568–580. doi: 10.1016/j.ebiom.2019.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kacher R., Lejeune F.X., Noël S., et al. Propensity for somatic expansion increases over the course of life in Huntington disease. Elife. 2021;10 doi: 10.7554/eLife.64674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Donaldson J., Powell S., Rickards N., Holmans P., Jones L. What is the pathogenic CAG expansion length in Huntington's disease? J Huntingtons Dis. 2021;10(1):175–202. doi: 10.3233/JHD-200445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaplan S., Itzkovitz S., Shapiro E. A universal mechanism ties genotype to phenotype in trinucleotide diseases. PLoS Comput Biol. 2007;3(11):e235. doi: 10.1371/journal.pcbi.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zavolan M., Kondo S., Schonbach C., et al. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13(6B):1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.