Abstract
Hepatitis C virus (HCV) initiates translation of its polyprotein under the control of an internal ribosome entry site (IRES) that comprises most of the 341-nucleotide (nt) 5′ nontranslated RNA (5′NTR). A comparative analysis of related flaviviral sequences suggested that an RNA segment for which secondary structure was previously ill defined (domain II, nt 44 to 118) forms a conserved stem-loop that is located at the 5′ border of the HCV IRES and thus may function in viral translation. This prediction was tested by a mutational analysis of putative helical structures that examined the impact of both covariant and noncovariant nucleotide substitutions on IRES activity in vivo and in vitro. Results of these experiments provide support for predicted base pair interactions between nt 44 to 52 and 111 to 118 and between nt 65 to 70 and 97 to 102 of the HCV 5′NTR. Substitutions at either nt 45 and 46 or nt 116 and 117 resulted in reciprocal changes in V1 nuclease cleavage patterns within the opposing strand of the putative helix, consistent with the predicted base pair interactions. IRES activity was highly dependent on maintenance of the stem-loop II structure but relatively tolerant of covariant nucleotide substitutions within predicted helical segments. Sequence alignments suggested that the deduced domain II structure is conserved within the IRESs of pestiviruses as well as the novel flavivirus GB virus B. Despite marked differences in primary nucleotide sequence within conserved helical segments, the sequences of the intervening single-stranded loop segments are highly conserved in these different viruses. This suggests that these segments of the viral RNA may interact with elements of the host translational machinery that are broadly conserved among different mammalian species.
Hepatitis C virus (HCV) is a positive-strand, enveloped RNA virus that is classified within the genus Hepacivirus of the family Flaviviridae (3). This virus establishes a persistent infection in most infected individuals, potentially leading to the development of chronic hepatitis, cirrhosis, or hepatocellular carcinoma (3, 12). It is thus a major cause of liver-specific morbidity and mortality in human populations. HCV isolates recovered from different patients demonstrate considerable genetic diversity (4, 21), and there is extensive quasispecies variation among HCV sequences recovered from individual infected patients (10, 31). However, the nucleotide sequence of the 5′ nontranslated RNA (5′NTR) is relatively well conserved among different genotypes of HCV. This conservation of primary structure likely reflects requirements for higher-ordered RNA structures that control translation and/or replication of the viral genome.
A number of previous studies have demonstrated the presence of an internal ribosome entry site (IRES) within the 5′NTR of HCV that directs the cap-independent initiation of virus translation (6, 11, 16, 17, 27, 30). Thus, the initiation of translation on HCV RNA occurs by a mechanism that is different from the cap-dependent translation initiation of yellow fever virus and other members of the genus Flavivirus (25). As an entity involved in highly specific macromolecular interactions (14), the IRES is a reasonable target for antiviral drug development. A detailed understanding of its structure is likely to contribute to such efforts.
Functional and structural studies of the HCV IRES have been carried out in a number of laboratories (1, 6, 9, 11, 14–19, 27–30). Most of these studies have drawn on a model of the secondary structure of the 5′NTR of HCV that was proposed by Brown et al. in 1992 (2). This model was modified by Wang et al. in 1995 (28) following the demonstration of a pseudoknot within the 5′NTR that is required for translation, and it was further refined by Smith et al. (24) in 1995 and Honda et al. (9) in 1996. To a considerable extent, the model is based on a comparative analysis of the sequences of multiple strains of HCV and members of the genus Pestivirus (bovine viral diarrhea virus [BVDV] and hog cholera virus [HoCV]) (2). Although the model has been validated by both physical probing of RNA structure and mutational analysis of IRES function, the assignment of structure has been problematic within the 5′ half of the 5′NTR (domain II). This is due the fact that there is considerable divergence of the nucleotide sequences of different genera of the family Flaviviridae in this region, despite strong conservation of this sequence among different HCV strains. This has made covariant sequence analysis difficult. Furthermore, there have been few attempts at mutational analysis of this part of the IRES structure. Thus, it is not surprising that quite different structures have been proposed in the past for these regions of the HCV and pestiviral 5′NTRs (2).
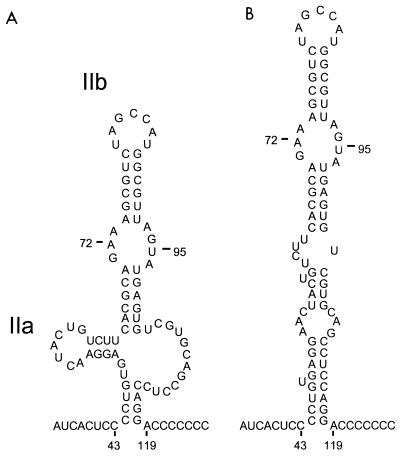
We recently presented a significantly different prediction of the secondary structure of this segment of the HCV 5′NTR based on a comparison of the HCV sequence with that of a newly discovered, hepatotropic mammalian virus, GB virus B (GBV-B) (9, 23). Unlike the more distantly related GB viruses A and C (22), GBV-B has a number of features in common with HCV, including many aspects of the secondary structure of its 5′NTR (9, 23). In the revised structural model, domain II was suggested to form a complex structure with two hairpins, extending from nucleotides (nt) 44 to 117 and containing a large, internal single-stranded bulge loop (nt 103 to 114) (Fig. 1A) (9). These latter structures were supported by previous nuclease mapping studies (2). However, computer-based, thermodynamic predictions of this domain, constrained with respect to predicted base pair interactions at its base, suggest that a more stable structure may be formed by alternative interactions between bases predicted previously to form hairpin IIa and those in the opposing bulge loop (Fig. 1B).
FIG. 1.
Alternative predictions of secondary RNA structure within domain II of the 5′NTR of HCV (N strain, genotype Ib). (A) Structure suggested by a recent comparison of the HCV and GBV-B sequences, comprising a complex stem-loop extending from nt 44 to 118, with two internal hairpin loops, IIa (nt 50 to 64) and IIb (nt 65 to 102), and a large, internal single-stranded bulge loop (nt 103 to 114) (9). Predicted ΔG = −8.6 kcal/mol. (B) Structure predicted by MFOLD when folding is constrained to include base pairing between nt 44 to 47 and nt 115 to 118, as suggested by the GBV-B sequence comparison. ΔG = −18.2 kcal/mol.
Here, we describe a mutational analysis of this segment of the 5′NTR of HCV. We present data supporting the existence of the structure shown in Fig. 1B and show that its integrity is essential to IRES-directed translation of the HCV open reading frame. An analysis of the nucleotide sequences of the pestiviruses and GBV-B indicates that this predicted structure is broadly conserved among those members of Flaviviridae that initiate translation of the polyprotein by internal ribosome entry.
MATERIALS AND METHODS
Plasmids.
Plasmid pCAT-N-CLuc contains a bicistronic, T7 transcriptional unit consisting of the chloramphenicol acetyltransferase (CAT) sequence fused at its 3′ end to the 5′NTR of the N strain of HCV (7), followed by 66 nt of the HCV open reading frame fused in frame with the luciferase sequence (8). pCAT-N-CΔE1 is a similar bicistronic plasmid in which the complete HCV capsid and 5′ E1 coding sequences represent the entire second cistron (8).
We constructed a series of pCAT-N-CLuc- and pCAT-N-CΔE1-related mutants (see Results) that contain nucleotide substitutions within segments of the domain II sequence that are predicted to participate in helix formation. To construct these plasmids, a shuttle vector, pSP73(1-279), was made by subcloning the SalI/StuI fragment (HCV nt 1 to 279) of pCAT-N-CLuc into the cloning vector pSP73 (Promega). Site-directed mutagenesis was carried out by a standard PCR-based strategy using selected oligonucleotide primers and Pfu (Pyrococcus furiosus) DNA polymerase. The mutated sequences were subsequently inserted into the pCAT-N-CLuc plasmid or used to replace homologous sequence in plasmid pCAT-N-CΔE1. Alternatively, some mutations were constructed directly in pCAT-N-CLuc by using a QuickChange site-directed mutagenesis kit (Stratagene). Plasmid DNAs were purified on Qiagen-tip 500 columns (Qiagen, Chatsworth, Calif.). The sequences of all PCR-manipulated regions and the presence of expected mutations were confirmed by DNA sequencing.
Cells.
Huh-T7 cells (20), derived from human hepatocellular carcinoma (Huh-7) cells, are stably transformed and constitutively express bacteriophage T7 RNA polymerase. BT7-H cells (32), derived from African green monkey kidney (BS-C-1) cells, similarly produce T7 RNA polymerase constitutively. Cell cultures were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and 400 μg (active compound) of geneticin (Gibco-BRL) per ml.
In vitro transcription and translation.
In vitro transcription-translation reactions were carried out with the Promega TnT coupled reticulocyte lysate system as specified by the manufacturer. Reaction mixtures (25 μl) contained 20 U of RNasin (Promega), 20 μCi of [35S]methionine (DuPont NEN), and 0.5 μg of plasmid template. After incubation at 30°C for 2 h, 3-μl aliquots of the translation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide–6 M urea gels, which were then subjected to autoradiography. The quantities of reporter proteins produced in these reactions were determined by enzymatic assay (see below) or by PhosphorImager analysis (Molecular Dynamics).
In vivo analysis of HCV translation in Huh-T7 cells.
Huh-T7 cells were seeded into six-well tissue culture plates 24 h before transfection. Since these cells have a relatively low level of endogenous T7 polymerase expression, they were infected when 90% confluent with a recombinant vaccinia virus, vTF7-3, that expresses the polymerase (5). The inoculum was in 500 μl of Opti-MEM (Gibco-BRL) at a multiplicity of infection of 5. After a 1-h incubation at 37°C, the inoculum was removed and replaced with a mixture containing 2 μg of plasmid DNA and 15 μl of Lipofectin (Gibco-BRL) in 200 μl of Opti-MEM at 37°C, followed 15 min later by an additional 800 μl of Opti-MEM (11). Cells were subsequently incubated at 37°C in a 5% CO2 atmosphere for 18 to 24 h. At harvest, 1 ml of 10 mM EDTA–phosphate-buffered saline was added to the cells, which were then incubated at 37°C for 10 min, collected, and divided into two equal aliquots. Following low-speed centrifugation, cells were resuspended in 500 μl of 1× lysis buffer (Promega) for luciferase assay or 500 μl of 0.25 M Tris-HCl for CAT assay. For the luciferase assay (Promega), 20-μl aliquots were assayed for light production. For the CAT assay, cells were lysed by freeze-thawing and lysates were further diluted to 1:500. CAT activity was determined by a phase extraction assay, which measures butyrylated [14C]chloramphenicol products by liquid scintillation counting, following xylene extraction (pCAT reporter gene system; Promega).
In vivo analysis of HCV translation in BT7-H cells.
BT7-H cells that constitutively express the T7 polymerase were seeded into six-well tissue culture plates and allowed to reach 90 to 95% confluency. Transfection was performed by the addition of a mixture containing 2 μg of plasmid DNA, 6 μl of FuGENE 6 transfection reagent (Boehringer Mannheim), and 92 μl of Opti-MEM (Gibco-BRL) and then added directly to cells in regular maintenance medium containing 10% fetal calf serum. Cells were incubated at 37°C in a 5% CO2 atmosphere for 24 h, after which cells were harvested and lysates were assayed for luciferase and CAT activity as described above.
Computer-based prediction of domain II RNA folding.
Thermodynamic predictions of RNA secondary structure at 37°C were developed by using the MFOLD 3.0 program with Turner energies and selected constraints on base pairing as indicated. The program was run on the server supporting the Zuker web site at the University of Washington (http://www.ibc.wustl.edu/∼zuker/mfold).
Nuclease mapping of RNA secondary structure.
Synthetic HCV RNA was generated by in vitro transcription of AatII-linearized plasmid pCAT-N-CLuc with T7 RNA polymerase. Transcripts were heated to 65°C for 3 min and allowed to cool slowly to 4°C in the presence of 10 mM MgCl2–10 mM Tris (pH 7.6). Enzymatic modification of 1 μg of RNA was carried out with RNase V1 (Pharmacia) or RNase S1 (Pharmacia) at room temperature in the presence of 20 μg of carrier tRNA and 10 mM MgCl2–10 mM Tris (and 1 mM ZnSO4 for S1 nuclease) at pH 7.6 in a total volume of 40 μl for 10 min. Reactions were stopped by the addition of excess tRNA. The modified RNA was ethanol precipitated and analyzed by primer extension with a 32P-labeled, negative-strand HCV primer and 3 U of avian myeloblastosis virus reverse transcriptase (Life Sciences) at 42°C for 30 min. The primers used in these reactions were complementary to HCV sequences commencing at nt 110 and 190 of the viral genome. Reaction products were separated on a 6% polyacrylamide gel in parallel with dideoxynucleotide sequencing reactions of unmodified RNA.
RESULTS
Alternative predictions of the secondary structure of domain II of the HCV IRES.
The recent determination of the nucleotide sequence of GBV-B, a novel hepatotropic flavivirus (23), provided an additional HCV-related 5′NTR sequence which could be used in a search for covariant nucleotide changes indicative of conserved secondary RNA structure. This analysis led us to propose a significant revision of the structure of domain II of the 5′NTR of HCV, a segment of the 5′NTR which is known to contribute to IRES function (Fig. 1A) (2, 9). The most significant difference between this predicted structure (9) and the previous prediction (2) was the base pairing of nt 115 to 118 with nt 44 to 47, to form a putative helix located at the base of the structure. This potential base pairing is preserved in the sequence of GBV-B (with conservation of the primary sequence within the 3′ segment of the helix), unlike the previously predicted base pair arrangement in this region of the 5′NTR (9). We subsequently used the MFOLD program to predict an optimal structure for the intervening sequence, constraining the fold to include the nt 44–47/115–118 interaction. The result (Fig. 1B) was a structure in which the previously predicted hairpin IIa was no longer present and in which an extension of the basal helix included pairing between nt 49 to 52 and nt 111 to 114. This second structure is significantly more stable than that shown in Fig. 1A (ΔG = −16.8 versus −8.6 kcal/mol). However, since the involved sequences are highly conserved among HCV strains (24), comparative sequence analysis provides no clues as to which structure is correct. Furthermore, with few exceptions, both structures conform to previously identified sites of cleavage by single- and double-strand-specific nucleases (2).
Mutations which destabilize base pairing between nt 44 to 47 and nt 115 to 118 decrease IRES activity.
We carried out a mutational analysis to verify predicted helical segments that are common to both of the structures shown in Fig. 1, as well as to distinguish between the two predictions. We created mutations within the background of a dicistronic reporter construct, pCAT-N-CLuc, and assessed translational activities in a cell-free coupled transcription-translation system. Luciferase is translated from the second cistron of the transcripts derived from these plasmids, following internal entry of ribosomes on the 5′NTR sequence that is located within the intercistronic space. In contrast, CAT is translated by a ribosome scanning of the upstream cistron and serves as a control for transcription efficiency (see Materials and Methods).
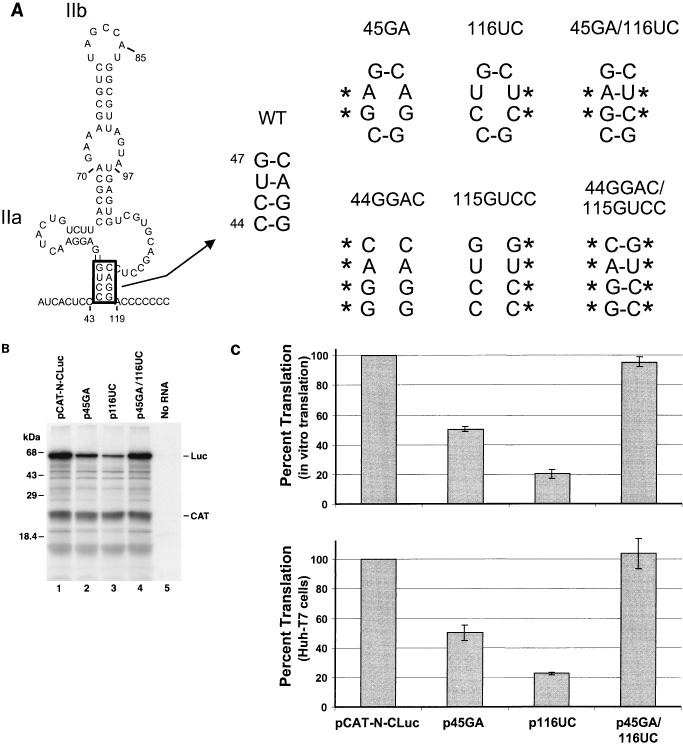
We first introduced a series of mutations into the putative helical segment (nt 44 to 47 and 115 to 118) that is located at the base of the domain II structure (Fig. 2A). This helical segment (which was predicted on the basis of a comparative analysis of the HCV and GBV-B sequences) (9) is common to the two predicted structures, as it was used to constrain the MFOLD prediction shown in Fig. 1B. Plasmid p45GA contained a CU-to-GA substitution involving nt 45 and 46 that should destabilize the helix, while p116UC contained a AG-to-UC substitution within the complementary strand of the helix, at nt 116 and 117. Plasmid p45GA/116UC contained both sets of mutations, potentially allowing reconstitution of the predicted secondary structure (Fig. 2A). Autoradiography of the products of translation demonstrated significant decreases in the quantities of luciferase expressed from p45GA and p116UC compared with pCAT-N-CLuc, despite relatively constant quantities of the CAT product (Fig. 2B; compare lanes 2 and 3 with lane 1). In contrast, there was little if any decrease in luciferase expressed from p45GA/116UC.
FIG. 2.
Mutational analysis of the putative helical segment at the base of domain II that is common to the two structures shown in Fig. 1. (A) Mutant plasmid p45GA contains a CU- to GA-substitution at nt 45 and 46, while p116UC contains an AG-to-UC substitution within the predicted complementary strand at nt 116 and 117. Both mutations destabilize the helix at the base of domain II. Plasmid p45GA/116UC contains both of these mutations, potentially allowing restoration of the helix. Also shown are similar mutants containing 4-nt insertions (p44GGAC, etc.). WT, wild type. (B) SDS-PAGE of translation products from coupled transcription-translation reactions programmed with plasmids pCAT-N-CLuc (wild-type; lane 1), p45GA (lane 2), p116UC (lane 3), and p45GA/116UC (lane 4). No RNA, negative transcription control. Luc, core-luciferase fusion protein. (C) Mean relative IRES activities of mutants, measured as the ratio of luciferase to CAT activity produced in vitro in reticulocyte lysate (top) or in vivo in vTF7-3 infected Huh-T7 cells (bottom). The ratio of luciferase to CAT activity obtained with pCAT-N-CLuc was accorded a value of 100%. Error bars denote standard errors of values obtained in replicate experiments.
The amounts of luciferase and CAT expressed in coupled reactions programmed with these plasmids were determined quantitatively by enzymatic assay, and the ratio of luciferase to CAT activity was calculated for each construct as a measure of IRES strength (Fig. 2C, top). To facilitate comparisons, the luciferase/CAT ratio obtained with pCAT-N-CLuc in each experiment was set arbitrarily to a value of 1.0. Mean values obtained in separate experiments indicated that the strength of the IRES in p45GA was only 51% of that of the wild type, pCAT-N-CLuc, while IRES strength was 20% of the wild-type level in p116UC and 95% of that level in p45GA/116UC. These results provide strong support for the existence of the putative helical segment. They are indicative of impaired internal ribosome entry following destabilization of the helix, with nearly normal restoration of IRES function when the helical structure was reconstituted with complementary mutations in the opposing strands (Fig. 2A).
We constructed similar mutants (p45GA-CΔE1, p116UC-CΔE1, and p45GA/116UC-CΔE1) within the background of a dicistronic plasmid which contains as its second cistron the entire HCV capsid-coding sequence and part of the E1 sequence. The relative translational activities of these constructs were similar to those of pCAT-N-CLuc and its related mutant constructs in coupled transcription-translation reactions (data not shown). Thus, the nature of the second cistron did not influence the impairment of IRES activity imposed by these mutations.
To assess the translational activities of the mutated 5′NTR sequences in vivo, each of the plasmids depicted in Fig. 2A was transfected into Huh-T7 cells shortly after infection with the recombinant vaccinia virus vTF7-3 (17). CAT and luciferase activities were determined in lysates prepared 24 h following transfection. When relative luciferase/CAT ratios were calculated as a measure of IRES strength in these cells, the translational activities of the single mutants, p45GA and p116UC, were 50 and 23%, respectively, of that of the wild type, pCAT-N-CLuc (Fig. 2C, bottom). However, the IRES activity of the double mutant, p45GA/116UC, was equivalent to that of the wild type (104%). These in vivo results are remarkably similar to those obtained in the cell-free system in vitro.
These results confirm that the predicted helical segment involving nt 44 to 47 and 115 to 118 is required for optimal expression of IRES activity. However, they also show that substantial IRES activity is retained when the helix has been disrupted by the substitution of two nucleotides. To determine whether more extensive substitutions within this helix would completely extinguish IRES activity, we constructed plasmids in which four of the putative base pairs were altered (Fig. 2A). Thus, p44GGAC contained a CCUG-to-GGAC substitution at nt 44 to 47, while p115GUCC contained a GUCC substitution involving the complementary bases at nt 115 to 118. Each of these four-base mutants had approximately 50% of the IRES activity of the wild-type sequence, both in vivo and in vitro (data not shown). Again, the inclusion of both mutations in p44GGAC/115GUCC allowed restoration of the predicted helical segment, as it resulted in essentially normal translational activity (data not shown). These additional results thus confirm those obtained with the first set of mutants involving two base substitutions (Fig. 2). They indicate that the helical segment predicted at the base of the extended domain II stem-loop structure is required for optimal translational activity, but they confirm that disruption of this helix does not completely eliminate IRES activity. This could be due to local alternative base pair arrangements that are permissive for translation or to retention of essential IRES structure due to the presence of the four-base extension of this helical segment that is predicted within the structure shown in Fig. 1B.
Nuclease analysis of RNA secondary structure within domain II.
Previous work has shown that cobra venom nuclease V1 cleaves synthetic 5′NTR transcripts in the region from nt 116 to 118 (2). In addition, there are strong primer extension stops at nt 45 and at nt 118 and 119 in the absence of nuclease digestion (Fig. 3, lane 1). Since nuclease V1 preferentially cleaves double-stranded or helically stacked RNA substrates (13), these data are consistent with the base pairing predicted between nt 44 to 47 and nt 115 to 118. However, to confirm that these findings are due to these specific base pair interactions, RNA transcripts derived from pCAT-N-CLuc and its related mutants, p45GA, p116UC, and p45GA/116UC, were digested with RNases V1 and S1 and then used as templates in primer extension reactions.
FIG. 3.
Nuclease analysis of secondary RNA structure in the region of domain II of the HCV 5′NTR. Full-length 5′NTR RNAs transcribed from pCAT-N-CLuc, p45GA, and p116UC were digested with the double-strand-specific RNase V1 and used as the template in a primer extension reaction. Lanes 1 to 3 contain primer extension products derived from nondigested transcripts; lanes 4 to 6 contain primer extension products from V1-digested RNAs. Positions of specific nucleotides (marked on the right) were determined from a sequencing reaction run in parallel. Solid arrows indicate reciprocal changes in the V1 cleavage pattern caused by mutations at either nt 45 and 46 or nt 116 and 117, involving opposing strands within the putative helix located at the base of domain II; open arrows indicate V1 cleavage sites at nt 51 to 53 and 65 to 69 that are not affected by these mutations.
Treatment of pCAT-N-CLuc transcripts with V1 nuclease generally confirmed earlier results obtained with wild-type HCV RNA, except that the V1 cleavage sites previously noted at nt 61 to 69 were confined to nt 65 to 69 (2) (Fig. 3; compare lanes 1 and 4). Interestingly, significantly different results were obtained with transcripts from p45GA and p116UC. Substitutions at nt 45 and 46 in p45GA not only altered V1 cleavage in this region of the RNA (loss of cleavage at nt 45) but also resulted in a change in the V1 cleavage pattern around nt 113 to 117 (reduced cleavage at nt 117 to 119, with new cleavage sites at nt 113 to 115) (Fig. 3; compare lanes 4 and 5). Similarly, the substitutions at nt 116 and 117 in p116UC altered both the nonspecific stop (Fig. 3; compare lanes 1 and 3) and the V1 cleavage pattern around nt 45 and 46 (compare lanes 4 and 6). As expected, the pattern obtained with the double mutant, p45GA/116UC, more closely resembled the wild-type cleavage pattern (data not shown). The reciprocal nature of the changes observed with these mutants strongly supports the proposed base pairing between nt 44 to 47 and nt 115 to 118 in the wild-type structure. However, the new V1 cleavage sites suggest the presence of continued interactions between these segments of the mutant RNAs that may preserve the general wild-type structure. Notably, the results of these experiments are most consistent with the structure shown in Fig. 1B. Thus, mutations at either nt 45 and 46 or nt 116 and 117 may allow the end of the helix at the base of domain II to “breath,” causing a shift in the V1 cleavage sites to adjacent base pairs further up the helix. This possibility is also consistent with the less than complete loss of IRES activity with these mutants.
In contrast to V1 cleavage sites within the helical segment, the p45GA and p116UC mutations did not affect V1 cleavage at nt 51 to 53 or nt 65 to 69, suggesting that these segments of the RNA preserve their wild-type structure in the mutant transcripts (Fig. 3). Results with RNase S1 generally confirmed previous findings (2) (data not shown).
Additional mutations which destabilize base pairing between nt 65 to 70 and nt 97 to 102 abolish IRES-directed translation.
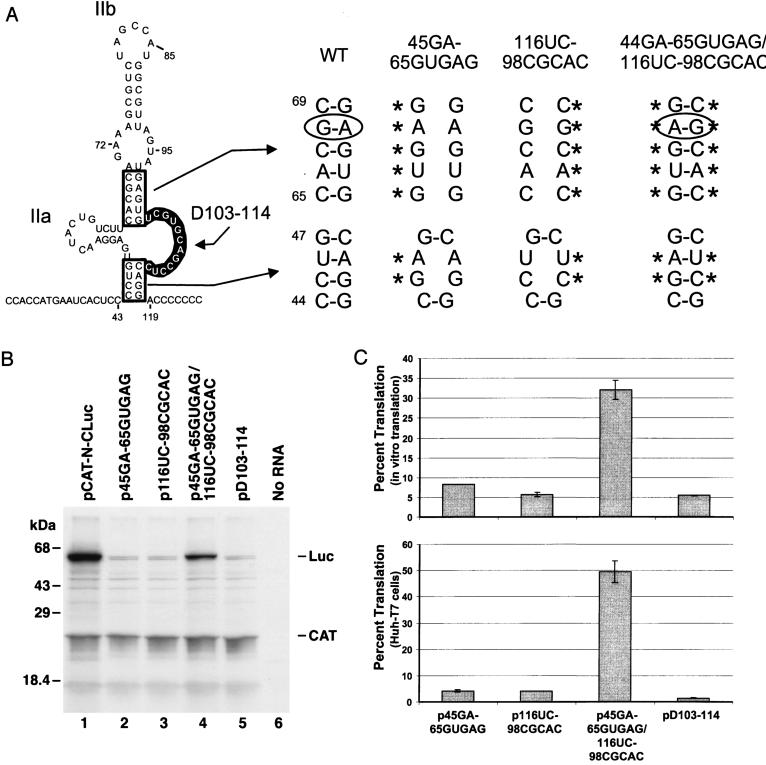
We next examined the predicted base pairing between nt 65 to 70 and nt 97 to 102, a helical segment that is also common to the two structures shown in Fig. 1. Mutations at nt 65 to 69 and/or nt 98 to 102 were created in the backgrounds of p45GA and p116UC, respectively (Fig. 4A). Thus, in addition to the CU-to-GA substitutions at nt 45 and 46, p45GA-65GUGAG contained a CACGC to GUGAG substitution at nt 65 to 69 that disrupts the second helical segment of the predicted structure. This mutated 5′NTR sequence demonstrated only 8.3% of the wild-type IRES activity in the cell-free translation system (Fig. 4B). Similarly, p116UC-98CGCAC, which had a GAGUG-to-CGCAC substitution at nt 98 to 102 as well as the AG-to-UC substitution at nt 116 and 117 (Fig. 4A), demonstrated only 5.7% of the wild-type IRES activity (Fig. 4B). The combination of these mutations in p45GA-65GUGAG/116UC-98CGCAC allowed reconstitution of the predicted secondary structure despite extensive primary sequence differences and restored the efficiency of IRES-directed translation in vitro to 32% of the wild-type level (Fig. 4A and B).
FIG. 4.
Further mutational analysis of domain II structure. (A) Nucleotide substitutions which destabilize the helix at the base of domain II as well as a putative internal helical segment of stem-loop IIb that is common to the two structures shown in Fig. 1. p45GA-65GUGAG contains a CACGC-to-GUGAG substitution at nt 65 to 69 and the CU-to-GA substitution at nt 45 and 46, while p116UC-98CGCAC contains a GAGUG-to-CGCAC substitution at nt 98 and 102 and the AG-to-UC substitution at nt 116 and 117. p45GA-65GUGAG/116UC-98CGCAC contains both of these complementary sets of mutations, potentially allowing restoration of the helical structures. Plasmid pD103-114 contains a 12-nt deletion within the large, internal bulge loop at the base of domain II in the structure shown in Fig. 1A. WT, wild type. (B) SDS-PAGE separation of the products of in vitro transcription-translation reactions programmed with plasmids pCAT-N-CLuc, p45GA-65GUGAG, p116UC-98CGCAC, p45GA-65GUGAG/116UC-98CGCAC, and pD103-114. No RNA, negative transcription control. (C) Mean IRES activities of mutants shown in Fig. 4A in vitro in reticulocyte lysates (top) or in vivo in Huh-T7 cells (bottom) relative to the activity of the wild-type IRES (pCAT-N-CLuc), taken as 100%. See the legend to Fig. 2C for further details.
Similar results were obtained with these mutants in vivo after their transfection into vTF7-3-infected Huh-T7 cells (Fig. 4C). Either of the single sets of mutations in p45GA-65GUGAG or p116UC-98CGCAC resulted in a 25-fold reduction of IRES activity. However, the combination of these mutations in p45GA-65GUGAG/116UC-98CGCAC restored translation to nearly half of that of the wild type. These data indicate that the predicted internal helical segment is essential for IRES activity. It is important to note that this helical segment contains a noncanonical A-G base pair involving nt 68 and 99 (Fig. 4A). This is preserved in the double mutant, p45GA-65GUGAG/116UC-98CGCAC, but in the opposite orientation, a feature that may account for the less than complete restoration of IRES activity.
Secondary structure involving nt 50 to 64 and nt 103 to 114.
The preceding results support the presence of helical segments that are predicted to be present in both of the structures shown in Fig. 1, but they do not distinguish between these two alternative foldings of the RNA. In an initial step to address this issue, we determined that the sequence comprising the large bulge loop located near the base of domain II in the structure shown in Fig. 1A is required for IRES activity. RNA transcripts with a deletion spanning nt 103 to 114 (pD103-114 [Fig. 4A]) demonstrated negligible IRES activity (Fig. 4B), indicating that retention of this sequence is essential for the translation.
Additional mutations were then constructed to evaluate potential base pairing in this region (Fig. 5A). In both of the predicted structures shown in Fig. 1, nt 50 to 52 are involved in base pair interactions leading to short helical segments. However, in the model shown in Fig. 1A, nt 50 to 53 pair with nt 61 to 64 to form the stem of hairpin IIa, while in the alternate model (Fig. 1B), nt 49 to 53 pair with nt 111 to 114 to form an extended helix joining the ends of domain II. Consistent with either prediction, a AGG-to-UUC substitution at nt 50 to 52 reduced translation to 32% of the wild-type level in the cell-free translation system and to 7.5% of the wild-type level in transfected BT7-H cells (Fig. 5B and C).
FIG. 5.
Mutational analysis aimed at discriminating between the two alternative structures shown in Fig. 1. (A) Mutant plasmid p50UUC contains an AGG-to-UUC mutation involving nt 50 to 52, p62GGA contains a CUU-to-GGA mutation involving nt 62 to 64, and p111GGA contains a CCU-to-GGA mutation involving nt 111 to 113. Alternative base pairing arrangements of these RNA segments in the two predicted structures are as shown. (B) SDS-PAGE of translation products from coupled transcription-translation reactions programmed with plasmids pCAT-N-CLuc (wild-type; lane 1), p50UUC (lane 2), p62GGA (lane 3), p111GGA (lane 4), p50UUC/62GGA (lane 5), and p50UUC/111GGA (lane 6). Luc, core-luciferase fusion protein. (C) IRES activities of mutants shown in Fig. 5A in vitro in a coupled transcription-translation reaction (top) or in vivo in BT7-H cells (bottom) relative to the activity of the wild-type IRES (pCAT-N-CLuc), taken as 100%. See the legend to Fig. 2 for further details.
We next examined the effects of additional substitutions within those segments of the RNA that are predicted to base pair with nt 50 to 52 in the two alternative models (Fig. 5A). By itself, a CUU-to-GGA substitution at nt 62 to 64 had little effect on translation in vitro (88% of the wild-type level), although it reduced translation to 25% of the wild-type level in BT7-H cells (Fig. 5C). The addition of the 62GAA substitution to p50UUC did not significantly rescue the defect in translation due to the 50UUC substitution: 34% translation efficiency in vitro, versus 20% in vivo (Fig. 5C). Thus, these data do not provide support for the predicted IIa stem structure shown for this region of the RNA in Fig. 1A. In contrast, a CCU-to-GGA substitution at nt 111 to 113 was capable of rescuing the defect in translation in p50UUC. By itself, the 111GGA substitution resulted in only 14% of wild-type translation efficiency both in vitro and in vivo (Fig. 5C). Combined with the 50UUC mutation, 111GA restored translation to approximately 50% of the wild-type level both in the cell-free system and in transfected BT7-H cells, a level of translation that is significantly greater than that observed with either mutation alone. These results thus strongly support the existence of the extended stem-loop structure shown for domain II in Fig. 1B. The requirement for this structure also explains the abrogation of translational activity observed with mutant pD103-114 (Fig. 4).
DISCUSSION
A considerable body of evidence supports the existence of the secondary and tertiary structures that have been proposed previously for the 3′ half (domains III and IV) of the HCV 5′NTR. These data include the results of extensive comparative sequence analyses, the probing of secondary structure by nucleases or chemicals, and mutational analyses of IRES function (2, 9, 24, 27–29). Of these various approaches to determination of RNA structure, the most powerful has proven to be phylogenetic searches for covariant nucleotide substitutions that are indicative of conserved helical elements (2, 27). The results of such analyses allow critical constraints to be entered into computer-based RNA folding programs and permit rationally designed mutational studies. However, until the sequencing of GBV-B, it was difficult to apply this strategy to domain II of the HCV 5′NTR because of the absence of related sequences with an appropriate degree of nucleotide sequence divergence.
The recent discovery of GBV-B (23) provided such a sequence and has allowed a more accurate prediction of the domain II structure of HCV. This segment of the 5′NTR was predicted to form a complex structure spanning nt 44 to 117 and containing two hairpin loops (stem-loops IIa and IIb), a single large, internal bulge loop comprising nt 103 to 114, and several smaller internal bulge loops (Fig. 1A) (9). The existence of stem-loop IIa was supported by previous nuclease probing studies (2, 9). However, subsequent thermodynamic analyses suggested that a more stable structure could be formed by the collapse of the nucleotides involved in the putative IIa hairpin into the main stem structure (Fig. 1B). The mutational analysis described in this communication supports the existence of this latter structure and demonstrates that it is essential for efficient functioning of the HCV IRES.
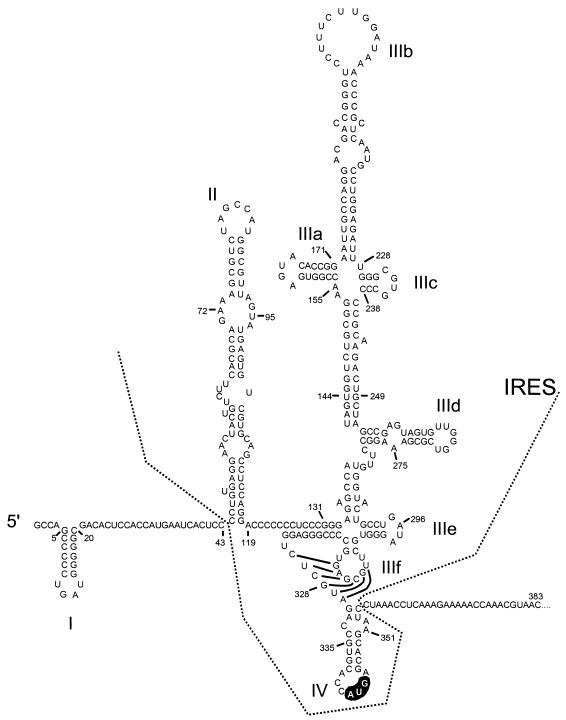
These data are important, since an appreciation of the higher-ordered structures that contribute to the IRES is essential to understanding the mechanisms by which this RNA element controls cap-independent viral translation. Moreover, such data will be helpful in designing small-molecule inhibitors of IRES function that could become useful antiviral therapeutics. The analysis of the domain II structure reported here allows a more complete picture of the secondary structure of the entire HCV IRES (Fig. 6).
FIG. 6.
Secondary and tertiary RNA structure within the complete 5′NTR of HCV and immediately downstream open reading frame. The region comprising the IRES extends from domain II to domain IV. The initiator AUG codon is highlighted. The structure is modified from that shown in reference 9 by inclusion of the revised domain II structure.
The data summarized in Fig. 2, 3, and 5 confirm the existence of two of the predicted helical segments that form the core of this structure. Mutations in plasmids p45GA and p116UC destabilized the helix at the base of the domain and resulted in a decrease in IRES-directed translation to about 50 and 20% of the wild-type (pCAT-N-CLuc) level. In contrast, p45GA/116UC, which contains both mutations, demonstrated nearly normal IRES activity. This reflects the restoration of the helical segment through base pairing of the mutated residues (Fig. 2A) and indicates that the helical structure is much more important for IRES activity than the primary nucleotide sequence within these segments of the RNA. Similar results were obtained in vitro and in vivo, with larger, 4-nt substitutions within this helical segment (data not shown) and with substitutions involving nt 50 to 52 and 111 to 113 (Fig. 5). Additional support for the basal helix of domain II comes from the analysis of V1 nuclease cleavage sites near the substitutions in these mutants. Remarkably, nucleotide substitutions within one strand of the helix affected V1 cleavage activity on the opposing strand as well (Fig. 3).
The results obtained in these experiments and the model shown in Fig. 1B are in general agreement with previous nuclease mapping of the secondary structure of domain II, as are the results of V1 nuclease mapping shown in Fig. 3. The major exception to this statement is the cleavage between nt 57 and 60 that we observed previously using nucleases with preferences for single-stranded substrates (2). This finding suggests that the short helical segment that is present between the two lower internal bulge loops in the model shown in Fig. 1B either may not form or may be subject to breathing. It is worth noting that this helical segment contributes little to the folded energy of the proposed domain II structure. The calculated free energies of the structure shown in Fig. 1B are −16.8 kcal/mol with and −15.5 kcal/mol without these base pair interactions. Two prominent primer extension stops that occurred in the absence of V1 nuclease cleavage (at nt 75 to 78 and 88 to 90 [Fig. 3]) are located within the apical helical segment of the proposed structure. These stops most likely result from the proposed secondary structure.
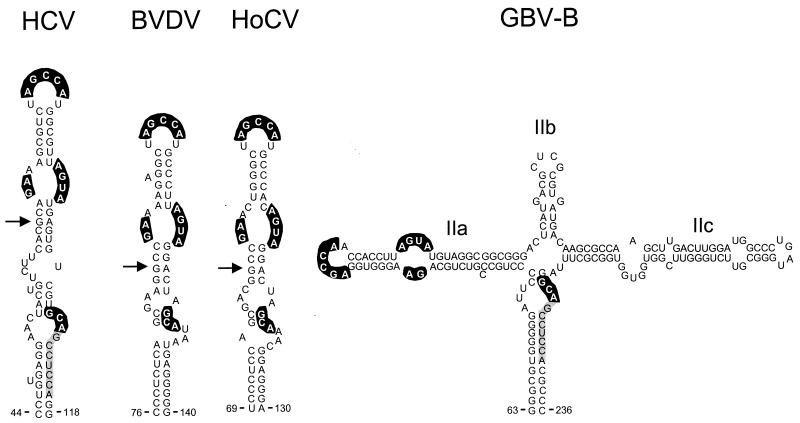
The 5′NTRs of the pestiviruses BVDV and HoCV, as well as the novel flavivirus GBV-B, also contain IRES elements. These IRESs contain structures that are remarkably similar to the domain III structure of HCV, including the centrally located pseudoknot that is present in HCV (18, 28). Following identification of the elements of the domain II structure that are conserved in HCV and GBV-B (9), we examined the pestivirus sequences for the possible presence of similar conserved structures within analogous regions of the 5′NTR. This search led to identification of putative structures within the BVDV and HoCV RNAs which are somewhat simpler than that present in HCV or GBV-B but which nonetheless share a number of characteristics with the HCV structure (Fig. 7). The most notable feature of these proposed domain II structures is the conservation of primary nucleotide sequences within the terminal loop and internal bulge loops (highlighted in Fig. 7), despite extensive variation within sequences that contribute to helical segments of the structure. The highly conserved nature of these loop sequences among flaviviruses that infect very different host species and diverse target tissues is remarkable. It suggests the possibility that these sequences interact with highly conserved elements of the host translational apparatus.
FIG. 7.
Conserved domain II structural motifs in four genetically diverse flaviviruses: HCV, the pestiviruses BVDV (ΔG = −19.1 kcal/mol) and HoCV (ΔG = −22.2 kcal/mol), and the novel flavivirus GBV-B (ΔG = −79.2 kcal/mol). Bases shown with heavy highlighting are absolutely conserved among these viruses; lightly highlighted bases indicate an extended region of primary sequence identity shared by HCV and GBV-B only. The structures shown represent output of the MFOLD program, with folding constrained as follows: forced base pair interactions between nt 88 and 89 and nt 123 and 124 of BVDV (GenBank accession no. M31182), nt 84 and 85 and nt 112 and 113 of HoCV (accession no. D49532), and nt 63 to 67 and 232 to 237 and nt 80 to 84 and 128 to 132 of the GBV-B genome (accession no. U22304); nt 116 to 119 of GBV-B were forced to remain unpaired.
The extensive nucleotide substitutions in plasmids p45GA-65GUGAG and p116UC-98CGCAC almost completely abrogated translation directed by the HCV IRES, in vitro and in vivo (Fig. 4). On the other hand, p45GA-65GUGAG/116UC-98CGCAC, which contains a combination of these mutations, demonstrated 32 to 50% of the translational activity of the wild-type construct (Fig. 4B). This impressive restoration of IRES activity in the face of extensive changes in the primary nucleotide sequence of domain II reinforces the importance of the domain II structure to translation. However, the failure of p45GA-65GUGAG/116UC-98CGCAC to demonstrate translational activity that is equivalent to that of the wild-type IRES also indicates that the primary sequence within the internal helix is important for translation. It is tempting to relate this to the noncanonical A-G base pair within the internal helix of stem-loop IIb. An identical A-G base pair is located within the analogous helical segments of BVDV and HoCV (Fig. 7). Indeed, the sequences of this base-paired helical segment are remarkably conserved among these viruses. The conservation of this noncanonical base pair in these diverse viruses suggests that it may have functional importance. The double mutant p45GA-65GUGAG/116UC-98CGCAC contains a G-A base pair within the reconstituted helix, but in orientation opposite that of the A-G pair in the wild-type sequence. A-G/G-A base pairs may form through four different types of hydrogen bond interactions, each with a unique structure (26). Thus, the inversion of the A-G base pair may well have altered the conformation of the helix, creating either a change in the orientation of the apical segment of the stem-loop or a bubble within the stem itself. This may account for the less than complete restoration of translational activity observed with the double mutant.
Data from other laboratories support the importance of the terminal helical segment of stem-loop II. Rijnbrand et al. (19) and Reynolds et al. (15) investigated the role of the AUG triplet that is located within this stem-loop at nt 85 to 87. Nucleotide substitutions within this triplet led to loss of IRES activity. However, substitutions of the G at nt 87 were tolerated, provided that a compensatory substitution preserving the terminal helical segment was made at nt 79 (15).
Pestova et al. (14) recently demonstrated that the HCV and pestiviral IRESs are able to form binary complexes with purified 40S ribosome subunits in the absence of any canonical or noncanonical translation factors. Eucaryotic initiation factor 3 (eIF3) contributes to this process by binding both domain III of the IRES and the 40S subunit. This distinguishes these RNA elements from the picornaviral IRESs that form 48S complexes only in the presence of eIF2, eIF3, and eIF4. A reasonable hypothesis is that domain II of the HCV and pestiviral IRESs contributes to the process of ribosome subunit recognition through specific interactions of its conserved single-stranded loop segments with ribosome proteins or unpaired segments of the 18S RNA.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institute of Allergy and Immunology (U19-AI40035 and RO1-AI32599) and the Advanced Technology Program of the Texas Higher Education Coordinating Board.
We are grateful to R. Rijnbrand for comments and helpful criticism and to T. Zuker for making the MFOLD program available.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A, Seed C R, Krusius T, Lin C, Medgyesi G A, Kiyokawa H, Olim G, Duraisamy G, Cuypers T, Saeed A A, Teo D, Conradie J, Kew M C, Lin M, Nuchaprayoon C, Ndimbie O K, Yap P L. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 5.Fuerst T, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi N, Higashi H, Kaminaka K, Sugimoto H, Esumi M, Komatsu K, Hayashi K, Sugitani M, Suzuki K, Tadao O, Nozaki C, Mizuno K, Shikata T. Molecular cloning and heterogeneity of the human hepatitis C virus (HCV) genome. J Hepatol. 1993;17:S94–S107. doi: 10.1016/s0168-8278(05)80432-5. [DOI] [PubMed] [Google Scholar]
- 8.Honda, M., G. Abell, D. Kim, and S. M. Lemon. Natural variation in translational activities of the 5′ nontranslated RNAs of genotypes 1a and 1b hepatitis C virus: evidence for a long range RNA-RNA interaction outside of the internal ribosomal entry site. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 9.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 10.Honda M, Kaneko S, Sakai A, Unoura M, Murakami S, Kobayashi K. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology. 1994;20:1144–1151. [PubMed] [Google Scholar]
- 11.Honda M, Ping L-H, Rijnbrand R, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 12.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell R H, Alter H J. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 13.Lowman H B, Draper D E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- 14.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds J E, Kaminski A, Carroll A R, Clarke B E, Rowlands D J, Jackson R J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds J E, Kaminski A, Kettinen H, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6016–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 18.Rijnbrand R, van der Stratten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rijnbrand R C, Abbink T E, Haasnoot P C, Spaan W J, Bredenbeek P J. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 20.Schultz D E, Honda M, Whetter L E, McKnight K L, Lemon S M. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 22.Simons J N, Desai S M, Schultz D E, Lemon S M, Mushahwar I K. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications on genome organization. J Virol. 1996;70:6126–6135. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, Van Sant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P-L, Simmonds P International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 25.Strauss J H, Strauss E G, Hahn C S, Rice C M. The genomes of alphaviruses and flaviviruses: organization and translation. In: Rowlands D J, Mayo M A, Mahy B W J, editors. The molecular biology of the positive strand RNA viruses. London, England: Academic Press; 1987. pp. 75–104. [Google Scholar]
- 26.Tinoco I. Structure of base pairs involving at least two hydrogen bonds. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1993. pp. 603–7. [Google Scholar]
- 27.Tsukiyama K K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Le S Y, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, et al. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whetter L E, Day S P, Elroy-Stein O, Brown E A, Lemon S M. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol. 1994;68:5253–5263. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]