Summary
Studies in zebrafish can unravel the functions of cellular communication and thus identify novel bench-to-bedside drugs targeting cellular communication signaling molecules. Due to the incomplete annotation of zebrafish proteome, the knowledge of zebrafish receptors, ligands, and tools to explore their interactome is limited. To address this gap, we de novo predicted the cellular localization of zebrafish reference proteome using deep learning algorithm. We combined the predicted and existing annotations on cellular localization of zebrafish proteins and created repositories of zebrafish ligands, membrane receptome, and interactome as well as associated diseases and targeting drugs. Unlike other tools, our interactome atlas is based on both the physical interaction data of zebrafish proteome and existing human ligand-receptor pair databases. The resources are available as R and Python scripts. DanioTalk provides a novel resource for researchers interested in targeting cellular communication in zebrafish, as we demonstrate in applications studying synapse and axo-glial interactome. DanioTalk methodology can be applied to build and explore the ligand-receptor atlas of other non-mammalian model organisms.
Subject areas: Cell biology, Specialized functions of cells, Omics
Graphical abstract
Highlights
-
•
We de novo predicted the cellular localization of zebrafish reference proteome
-
•
We created zebrafish ligand-receptome interactome atlas & tools to explore them
-
•
Our tools also facilitate the identification of drugs against ligands & receptors
-
•
Our methodology can be applied to build and explore the interactome of other organisms
Cell biology; Specialized functions of cells; Omics.
Introduction
Ligand-receptor-mediated cellular communication is essential for diverse processes such as differentiation, homeostasis, tissue morphogenesis, animal development, and behavior.1,2,3,4 Dysregulated cellular communication can lead to a plethora of phenotypes ranging from uncontrolled cellular proliferation and developmental defects to disease states and drug resistance.5,6,7 Hence, receptors and ligands are among the leading drug targets for treating human disease conditions.8,9
With advances in cryoelectron microscopy and machine learning approaches that enable protein structure prediction, the number of drug targets is expected to increase.10,11,12 These developments can aid drug screenings directly using animal model organisms. Due to practical and ethical challenges in using mammalian models in drug screening and limitations of non-animal-based drug screens, larval zebrafish are being increasingly used in high-throughput drug screening.13,14,15,16,17 However, despite the importance of zebrafish in drug discovery, there are no actively maintained datasets of zebrafish ligands, receptors, and their interactions. Few existing predictions of secretome and membrane proteome are based on incomplete annotation data.18,19,20,21
With the extensive single-cell RNA sequencing (RNA-seq) datasets of zebrafish, cellular communication in zebrafish can be explored at a holistic level even for rare and transient cells.22,23 Several tools built on mammalian interactome records have been applied to map the zebrafish ligand-receptor interactions.24,25,26 However, tools that depend on ortholog records are likely to ignore zebrafish genes lacking mammalian orthologs or genes with incomplete ortholog records. Hence, it is necessary to build a zebrafish ligand and receptome database that also includes information on ortholog-less genes and genes with incomplete annotation data.
Here, we describe DanioTalk—a repository of zebrafish ligands, membrane receptome, and interactome and tools to explore them from zebrafish-based omics datasets. To address the limitations of existing tools, DanioTalk ligand and membrane receptome datasets are based on deep learning-based subcellular localization predictions allowing the incorporation of orphan zebrafish proteins. DanioTalk can identify ligand-receptor pairs based on physical interaction records of zebrafish proteome and orthologous interactions in human ligand-receptor databases. DanioTalk scripts are available on GitHub and allows exploration of ligand-receptor interactions in any zebrafish-based omics datasets. Finally, we present the application of DanioTalk on mapping the ligand-receptor interactions between synaptosome and postsynaptic density (PSD) and axo-glial interactions between oxytocin neurons and glial pituicytes from RNA-seq datasets. We identified novel ligand-receptor interaction pairs with potential functions in axo-glial interactions. DanioTalk provides a novel tool and opportunity for the zebrafish research community to explore ligand-receptor interactions.
Results
De novo prediction of zebrafish secretome and membrane proteome
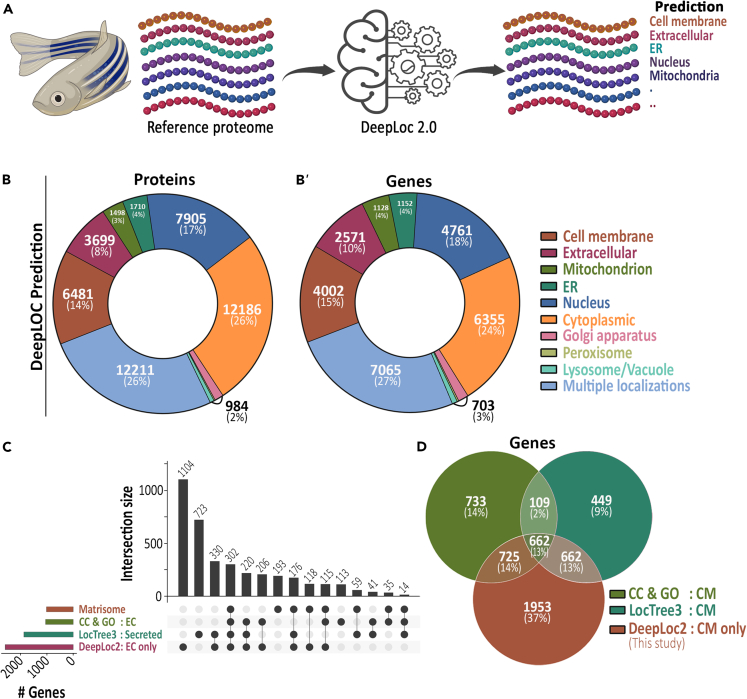
To build a zebrafish ligand and receptor database, we require either experimental or in silico knowledge on zebrafish protein localization. A bottleneck in using gene ontology (GO) terms or UniprotKB-based cellular localization records of the zebrafish proteome is the incompleteness of such annotations. We identified only 19,281 (41%) of zebrafish reference proteome records (UP000000437) in UniprotKB that contain annotations for protein localization (Figure S1A).27,28 To address this issue, we used DeepLoc 2.0, a sequence-based deep learning algorithm that can predict cellular localization of proteins with high accuracy29 (Figure 1A). We subjected the zebrafish reference proteome to DeepLoc 2.0-based prediction.30 The majority of proteins were predicted to be cytoplasmic (n = 12,186; 26%), nuclear (n = 7,905; 17%), and multilocalizing (n = 12,211; 26%) (Figures 1B, S1B, and Table S1). DeepLoc 2.0 predicted that 3,699 (8%) and 6,481 (14%) of proteins were exclusively extracellular- and cell membrane-localized, respectively (Figure 1B and Table S1). This corresponds to 2,571 (10%) and 4,002 (15%) genes that code extracellular- and membrane-localized proteins, respectively (Figure 1B’). We observe that out of the 2,571 genes whose proteins were predicted by DeepLoc 2.0 to be exclusively extracellular, 1,137 genes have been annotated or reported to be coding for extracellular proteins in UniprotKB and zebrafish matrisome database.21,28 Furthermore, DeepLoc 2.0 identified an additional 1,104 and 1,953 genes coding for extracellular and membranal proteins, respectively (Figures 1C and 1D).19,21,28
Figure 1.
Prediction of cellular localization of zebrafish reference proteome
(A) Graphic describing the DeepLoc 2.0-based prediction of cellular location of the reference zebrafish proteome.
(B) Zebrafish reference proteome cellular localization prediction. Donut chart showing the major DeepLoc 2.0 predictions of zebrafish proteome. The number of proteins (B) and protein-coding genes (B′) are shown.
(C) Upset plot comparing the number of genes with existing extracellular annotation records and DeepLoc 2.0-based 'Extracellular' predictions. CC & GO indicates cellular localization and GO term records, respectively, available in UniprotKB database.
(D) Venn diagram comparing genes with existing cell membrane localization annotation records and DeepLoc-based 'cell membrane' (CM) predictions. CC & GO indicates cellular localization and GO term records, respectively, available in UniprotKB database.
Next, we manually assessed how reliable are DeepLoc 2.0 predictions for a subset of proteins. DeepLoc 2.0 successfully predicted the most abundant secreted proteins comprising pancreatic enzymes (Prss, Cela, Cpa, Amy2a family) as extracellular proteins (coded by 24 genes)31 (Table S2). DeepLoc 2.0 prediction efficiency was also high for the Wnt family of secreted proteins (coded by 24 genes). Next, we assessed the DeepLoc 2.0 predictions for the Fgf protein family, which includes signal peptide-less secreted (Fgf9, Fgf16, Fgf20) and non-secreted Fgf11 subfamily.32 DeepLoc 2.0 successfully predicted 17 genes coding for canonical signal peptide-containing extracellular Fgf proteins (Fgf 3–8,10, 17–19, 21–23) and all 7 of the intracellular Fgf11 subfamily proteins as non-extracellular proteins. However, when classifying signal peptide-less extracellular Fgf’s, DeepLoc 2.0 only classified Fgf9 as extracellular and misclassified Fgf16, Fgf20a, and Fgf20b as cytoplasmic proteins (Table S2). Therefore, although DeepLoc 2.0 is highly reliable in predicting zebrafish secretome, manual curation is required to resolve the misclassified proteins.
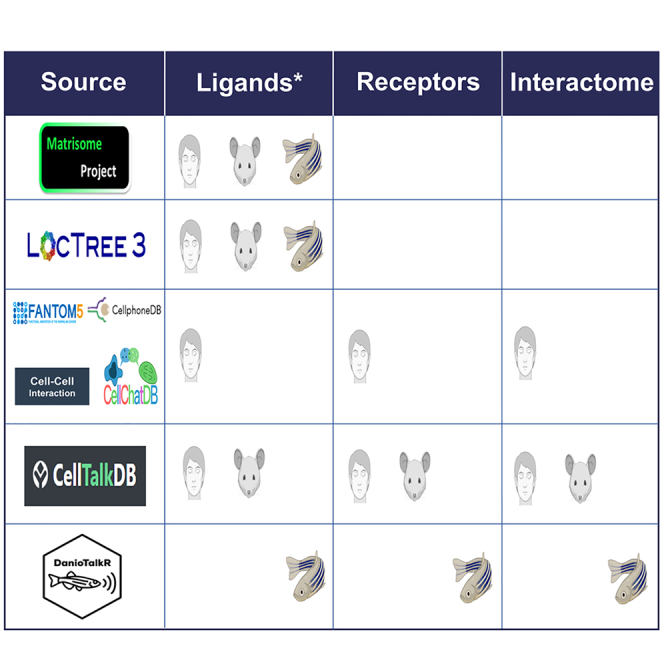
Curation of zebrafish ligands and membrane receptome
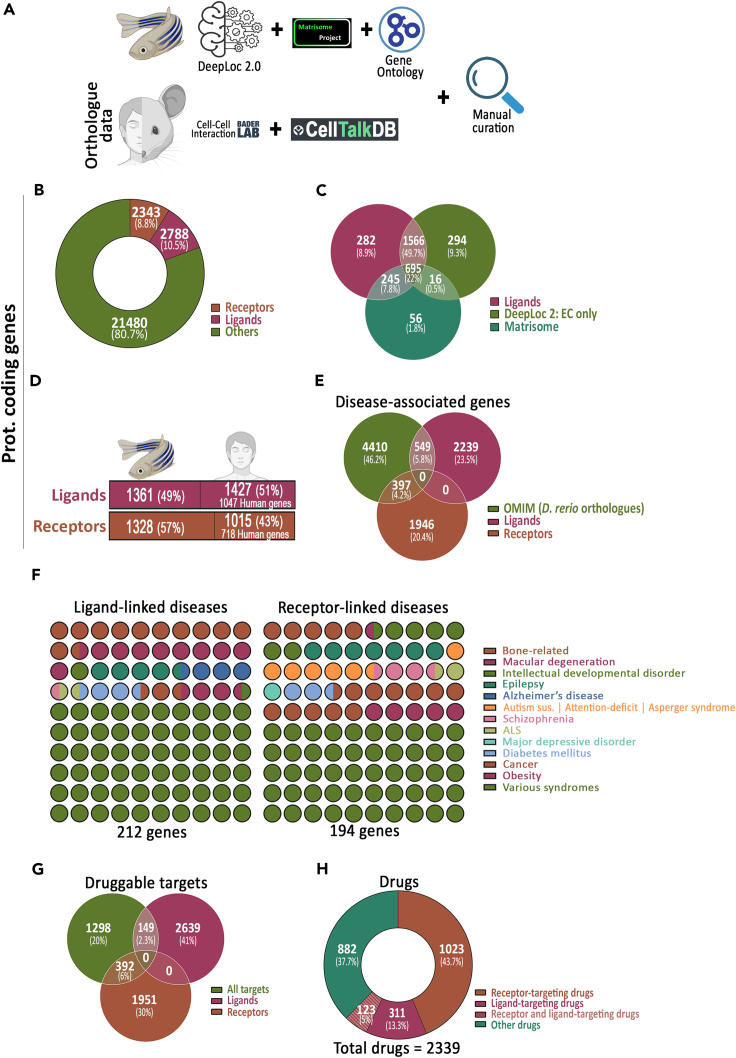
To create a list of zebrafish ligands and membrane receptome, we first combined our DeepLoc 2.0 predictions with annotations from other existing records (UniprotKB cellular localization, GO terms, and Matrisome).21,27,28 Furthermore, we added the zebrafish orthologs of curated mammalian ligand- and receptor-coding genes in Cell-Cell Interaction (baderlab.org/CellCellInteractions) and CellTalkDB databases (Figure 2A)33,34 and performed further manual curation (as detailed in STAR methods). Our curated ligands and membrane receptome database consist of 2,788 (10.5%) and 2,343 (8.8%) of protein-coding genes, respectively (Figure 2B and Table S3). In comparison to zebrafish matrisome dataset, our curated ligand lists contain 1,848 additional genes (1,566 DeepLoc 2 predictions and 282 manually curated extracellular-coding genes) (Figure 2C). In addition, we compared our curated ligand and receptor database with orthologs of ligand and receptor from human ligand-receptor databases (CellChatDB, CellPhoneDB, Ramilowski J.A. et al., CellTalkDB, and Cell-Cell Interactions database).33,35,36,37,38,39 Out of the 2,788 genes coding the curated ligands, 1,482, i.e., 53% of the genes are in common with human ligand databases (Figures S2A and S2C). Likewise, out of the 2,344 genes coding for receptor in our DanioTalk receptor database, 1,337, i.e., 57% are in common with human receptor databases (Figures S2B and S2D).
Figure 2.
Curated zebrafish ligands and membrane receptors
(A) Scheme describing the steps taken for the creation of curated zebrafish ligands and membrane receptors list.
(B) Donut chart showing the number and percentage of zebrafish genes coding for curated receptor- and ligand-coding genes.
(C) Venn diagram comparing curated ligands with DeepLoc 2.0 extracellular predicted proteins and previously reported Matrisome dataset.
(D) Conservation between zebrafish and human ligand- and membrane receptor-coding genes based on ZFIN records.
(E) Venn diagram comparing the number of curated ligand- and receptor-coding genes with zebrafish orthologs of human disease-linked genes in the OMIM database.
(F) Dot plot showing the diseases associated with human orthologs of some of the zebrafish ligands and membrane receptors.
(G) Venn diagram showing the number and percentage of zebrafish membrane receptors and ligands potentially targetable by drugs listed in the DrugCentralDB.
(H) Donut chart showing the number and percentage of drugs that can potentially target zebrafish membrane receptors and/or ligands.
Next, as ligands can be also membrane-bound, we analyzed the localization of the ligands in our curated database. Specifically, we compared our curated ligand database with 1) annotations for cell membrane localization in the Zebrafish UniprotKB database; 2) DeepLoc2 cell membrane predictions; and 3) zebrafish orthologs of CellphoneDB membrane-bound extracellular proteins.35 We identified that out of the 2,788 DanioTalk ligand-coding genes, 10.6%, i.e., 296 ligands are likely to be membrane-bound ligands (Figure S2E).
Disease-associated ligands and membrane receptome
Next, we explored the identity of human diseases in which zebrafish models can be used for understanding disease etiology and/or in drug screening. We cross-referenced our ligand and membrane receptor list with the human ortholog information available in ZFIN (Zebrafish Information Network) database.40 We identified 51% and 43% of the ligand- and cell membrane receptor-coding genes, respectively, having human ortholog gene records (Figure 2D and Table S3). Next, we cross-referenced our dataset with the zebrafish orthologs of human disease-associated genes in Online Mendelian Inheritance in Man (omim.org) database. We identified 549 ligand- and 397 receptor-coding genes containing records of human ortholog genes that are disease associated (Figure 2E and Table S4). Some of the major diseases associated with ligands and cell membrane receptors are related to bone diseases, macular degeneration, diabetes mellitus, and cancer (Figure 2F). Nevertheless, several of the ligands and receptors were also associated with numerous syndromes (e.g., Ehlers-Danlos syndrome, Loeys-Dietz syndrome, and Long QT syndrome) (Figure 2F).
Next, we explored the drug-targetable status of the zebrafish ligands and receptome. We cross-referenced our ligand and receptome list with DrugCentral database that contains an up-to-date list of drugs and their targets.41 We identified 149 ligands and 392 membrane receptors that can be potentially targeted by drugs (Figure 2G and Table S5). With respect to the choice of drugs, 311 (13%) and 1,023 (44%) of the drugs can potentially target zebrafish ligands and membrane receptors, respectively (Figure 2H). Moreover, zebrafish genes coding for the drug-targetable ligand or receptor are highly conserved in comparison to the orthologous human genes based on the orthology score in the Genome Alliance database (Figures S3A and S3B).42
DanioTalk—Putative ligand-membrane receptome interactome in zebrafish
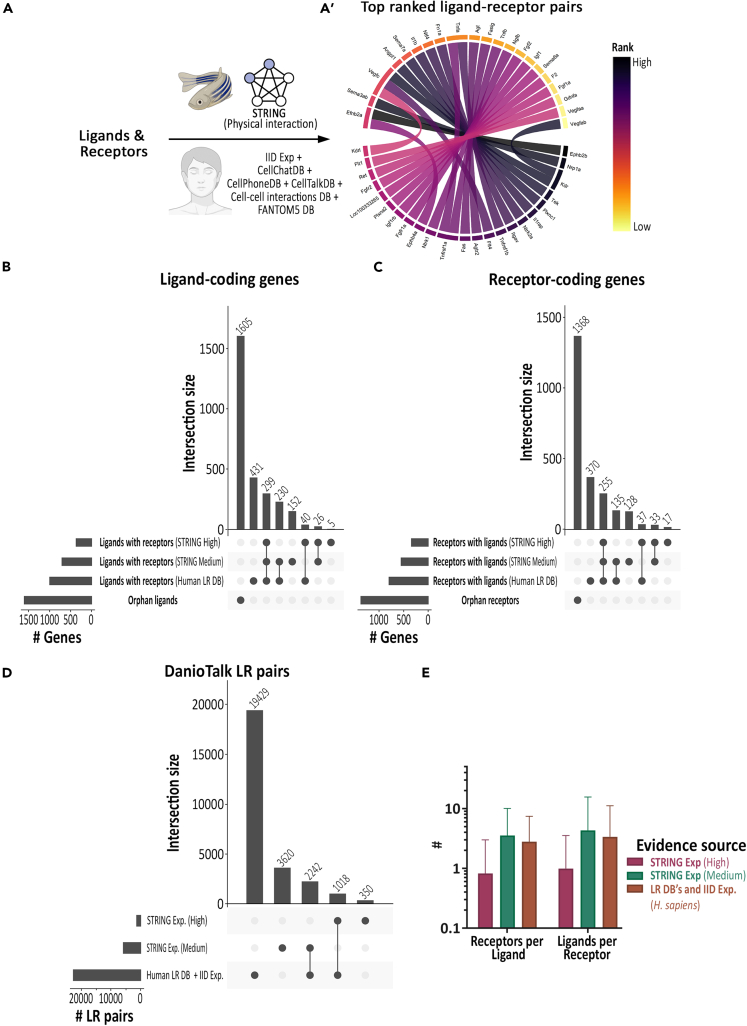
Next, to map the ligand-receptome interactions, we identified all pairwise interactions between our ligand and receptome proteins in the STRING (v11.5) physical protein-protein interaction (PPI) records of zebrafish (Figure 3A, Table S6, and Figure S4), a subset of which is shown in (Figure 3A’).43 The experimental score in STRING database predicts the confidence in the physical proximity of two proteins. Ligand-receptor pairs with high-confidence scores are very likely to physically interact and result in associated downstream signaling activities. We identified 70,476 ligand-receptor pairs out of which 1,368 pairs between 370 ligands and 342 receptors were above high confidence cutoff score (Figures 3B–3D).
Figure 3.
Zebrafish Ligand-Receptor Interactome
(A) Scheme describing steps undertaken to map the zebrafish ligands and membrane receptors interactome (A). Circle plot showing the top 25 ranked ligand-receptor pairs (A′).
(B) Orphan status of ligands. Upset plot of genes coding for ligands with or without potential receptors in the zebrafish STRING PPI database (High or Medium cutoff) or orthologous receptors in the human ligand-receptor database.
(C) Orphan status of receptors. Upset plot of genes coding for receptors with or without potential ligands in the zebrafish STRING PPI database (High or Medium cutoff) or orthologous ligands in the human ligand-receptor database.
(D) Upset plot of DanioTalk ligand-receptor pair database genes derived from zebrafish STRING PPI database (High or Medium cutoff) or orthologous ligand-receptor pairs in the human ligand-receptor databases.
(E) Zebrafish ligand-receptor interactome stats. Column chart comparing average number of ligand/receptor or receptor/ligand based on zebrafish STRING PPI database or orthologs in the human ligand-receptor database.
Next, to estimate the correlation between zebrafish and human ligand-receptor pairs, we compared zebrafish STRING physical interaction scores for the ligand-receptors pairs that have orthologous pairs in human ligand-receptor databases.33,34,35,36,37,38 We observed highly variable STRING physical interaction scores, and several hundred pairs were below the medium cutoff score (Figure S5A).
As setting a cutoff value below the medium confidence will result in increase in false-negative ligand-receptor pairs, we also incorporated orthologous human ligand-receptors pairs available in human PPI database IID (Integrated Interactions Database) and other human ligand-receptor databases (Figures S4 and S5B).33,34,35,36,37,38 This increased our database to 102,903 ligand-receptor pairs, out of which 22,689 pairs are present in human ligand-receptor databases, and IID experimentally validated PPI records (Figure 3D and Table S6). Even after the addition of orthologous ligand-receptor pairs, we observed 1,605 ligands and 1,368 receptors that lack interacting receptor or ligand, respectively, at medium confidence cutoff (Figures 3B and 3C). DanioTalk contains novel receptors for 183 ligands and novel ligands for 178 receptors (Figures 3B and 3C). DanioTalk contains 3,970 novel zebrafish-specific ligand-receptor pairs (350 high confidence + 3,620 medium confidence pairs) (Figure 3D), and we observe approximately 1–3 ligand per receptor and vice versa (Figure 3E).
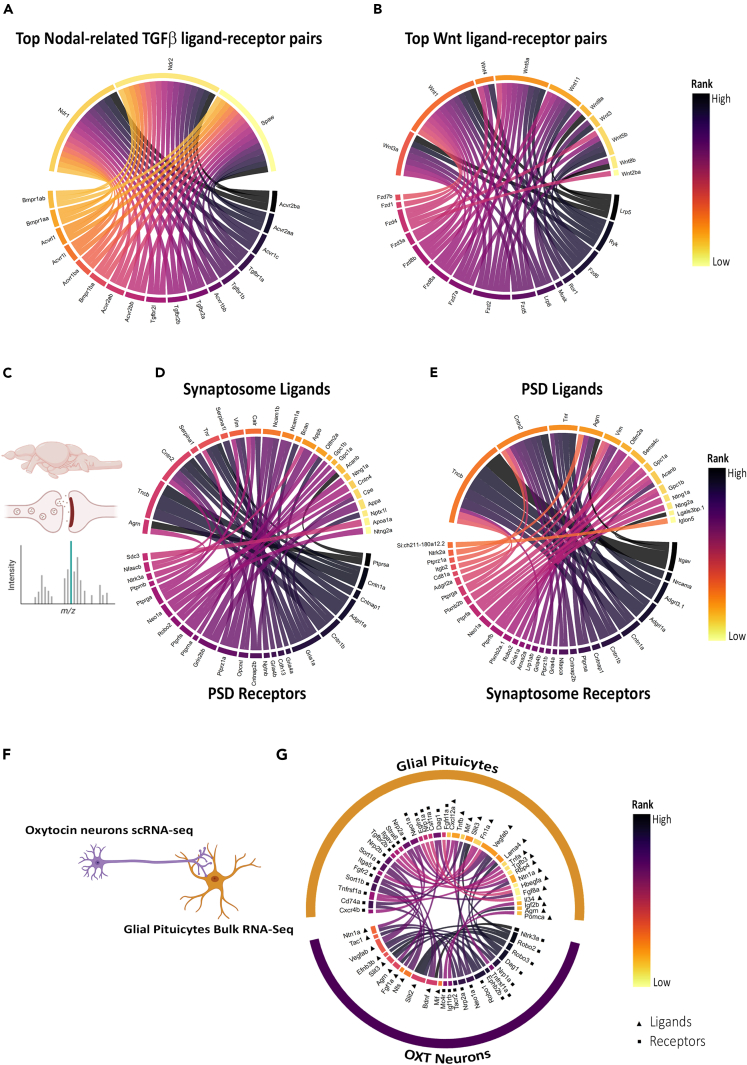
We next probed the accuracy of our ligand-receptor interactome database. We focused on the nodal-related transforming growth factor β (Tgfβ) ligands and Wnt ligands, key regulators of development.4,44,45 Our interaction database identified all the previously reported receptors for Ndr2/Cyclops, Ndr1/Squint, and Spaw/Southpaw ligands namely Acvr1b/Alk4, Acvr2a, and Acvr2b, respectively (Figure 4A and Table S7). We also noticed Acvr1c, Tgfbr1, and Tgfbr2 as top-ranked receptors. Next, we probed Wnt signaling ligand-receptor interactome. Our interaction database identified all the Wnt receptors (Lrp5, Ryk, Fzd, Ror1, and Lrp6) that can interact with the major Wnt ligands (Figure 4B and Table S7). Altogether, DanioTalk can accurately identify major ligand-receptor interactions for Notch and Wnt signaling pathways.
Figure 4.
Application of DanioTalk
(A) Receptors for Nodal-related Tgfβ ligands. Circle plot showing the top-ranked receptors for Ndr2, Ndr1, and Spaw ligands. The top 50 ranked interactions at medium confidence (>400) cutoff are plotted.
(B) Receptors for Wnt ligands. Circle plot showing the top-ranked receptors for Wnt ligands. The top 50 ranked interactions at high confidence (>700) cutoff are plotted.
(C) Scheme showing synaptosome-PSD from adult zebrafish brain. Mass spectrometry data were analyzed.46.
(D) Circle plot showing the top-ranked ligand-receptor interactions between the synaptosome ligands and PSD receptors. Interactions were ranked based on quasi-percentile of peptide expression. The top 50 ranked interactions at medium confidence (>400) cutoff are plotted. Ribbon width indicates interaction scores.
(E) Circle plot showing the top-ranked ligand-receptor interactions between the PSD ligands and synaptosome receptors. Interactions were ranked based on quasi-percentile of peptide expression. The top 50 ranked interactions at medium confidence (>400) cutoff are plotted. Ribbon width indicates interaction scores.
(F) Scheme showing axo-glial interaction between oxytocin neuron and glial pituicytes. Adult oxytocin neuron cluster pseudo-bulk data from scRNA-seq data and adult glial pituicyte bulk RNA-seq data were analyzed.47,48.
(G) Circle plot showing top-ranked ligand-receptor interactions between oxytocin neurons and glial pituicytes. Interactions were ranked based on quasi-percentile of gene expression. The top 50 ranked interactions at highest confidence cutoff (>900) are plotted.
Zebrafish brain synaptic ligand-membrane receptome interactome
Next, we tested the application of DanioTalk on existing datasets. We first focused on the zebrafish adult brain synaptosomal and PSD proteome dataset (Figure 4C).46 We identified protein products of 145 ligand- and 185 receptor-coding genes in the synaptosomal fraction. While in the PSD fraction, we identified protein products from 74 ligand- and 88 receptor-coding genes (Figures S6A, S6B and Table S8). The top-ranked ligand-receptor interacting pairs between synaptosome ligands and PSD receptors were Agrn-Ptprsa, Cntn2-Cntnap1/Cntn1a/Cntn1b, and Tncb/Adgrl1a (Figure 4D and Table S9). The top-ranked PSD ligand-synaptosome receptor pairs were Tncb-Itgav/Cntn1b and Cntn2-Nrcama/Cntnap1/Cntn1a (Figure 4E and Table S9).
Zebrafish oxytocin neuron-glial pituicytes axo-glial interactome
Axo-glial interactions contribute to synaptic morphogenesis and plasticity.49,50 However, the identity of all the axo-glial ligand-receptor interactions in neurohypophysis (NH), a major neuroendocrine interface has not been previously investigated.51,52 The NH-projecting axonal tracts, NH axo-glial cellular components, and their ultrastructure are conserved in zebrafish.47,53,54,55 We aimed to identify the axo-glial interactome of oxytocin neurons and glial pituicytes. Due to the lack of oxytocin neuronal axonal transcriptome, we used the oxytocin neuron scRNA-seq (single-cell RNA-sequencing) data and glial pituicyte bulk RNA-seq data (Figure 4F).47,48 Using the differentially expressed list of genes from these datasets, we identified only 3 ligand-receptor interactions between oxytocin neurons and glial pituicytes at high confidence (Nts-Sort1a, Bdnf-Ngrfb/Sort1a) (Figures S6C–S6E and Table S10). To incorporate all the genes expressed in oxytocin neurons, we used the pseudo-bulk expression data of oxytocin neuron-enriched cluster. This led to the identification of 282 ligands and 219 receptors in oxytocin neurons. Whereas in glial pituicytes transcriptome, we identified 363 ligands and 186 receptors with average read counts >50 (Figures S6F–S6G and Table S11). Analyzing the ligand-receptor pairs in this dataset led to identification of 38 oxytocin neuron-glial pituicyte ligand-receptor interactions. The top-ranked pituicyte ligand-oxytocin neuron receptor interactions were Slit3-Robo2/Robo1, Lama4-Dag1, and Vegfab-Nrp1a. While the top-ranked oxytocin neuron ligand-pituicyte receptors were Mif-Cd74a, Nts-Sort1a/Sort1b, and Bdnf-Sort1a/Sort1b (Figure 4G and Table S11). We also observed several autocrine signaling interactions in glial pituicytes (Cxcl12a-Cxcr4b, Tnfb-Tnfrsf1a, and Fgf8a/Fgf10a-Fgfr2) and in oxytocin neurons (Slit3/Slit2-Robo2/Robo1/Robo3, Bdnf-Ntrk3a/Sort1a, and Agrn-Dag1) (Figure 4G and Table S11).
Discussion
Zebrafish scRNA-seq datasets are also used to explore and study the ligand-receptor interactome using tools built for mammalian model organisms.24,25,26,56 However, a bottleneck in using such tools is in the dependence on incomplete orthologous mammalian ligand-receptor classifications and mammalian ligand-receptor interactome records.57,58 Thus, tools to study zebrafish ligands, receptome, and interactome independent of ortholog records are essential to help explore ligand-receptor interactions in zebrafish datasets.
In the lack of experimental records, recent studies reported the relatively high success of machine learning-based tools in predicting protein structures and cellular localizations.29,59,60 Here, we applied a deep learning algorithm to predict the cellular localization of the zebrafish proteome and created a curated repository of the zebrafish ligands and membrane receptome. This allows the incorporation of orphan zebrafish ligands or membranal receptor proteins that previously lacked cellular localization or ortholog records. DeepLoc 2.0 predictions of zebrafish proteome were largely accurate for predicting signal peptide-containing secreted, extracellular protein-based ligands; however, we noticed several instances of misclassified proteins based on literature knowledge on orthologous proteins. Hence, we had to manually curate the database. Our manual curation and inclusion of zebrafish matrisome and mammalian ligand databases resulted in resolving misclassified proteins and inclusion of membrane-bound ligands. Finally, our database of ligands and receptome-linked diseases will aid researchers interested in developing zebrafish models of human disease.
The DanioTalk algorithms are able to identify top interacting ligands and receptors in major pathways. This suggests that multiple published literature data has observed such PPIs leading to elevated physical interaction score in the zebrafish STRING database. Consequently, for these ligand-receptor pairs with high scores and high confidence, it is likely that the ligand-receptor pairs play a significant role in cellular signaling-mediated functions. High-confidence pairs that have not been previously reported in cellular signaling could be either novel ligand-receptor interactions or potential regulators of gradient formation. Currently, ligand-receptor pairs with low STRING PPI scores, mainly due to limited or less confident experimental evidence, will be assigned lower rankings by the DanioTalk algorithms. We anticipate that as the STRING PPI records are updated and PPI scores increase, the DanioTalk algorithms will be able to identify more high-confidence, zebrafish-specific ligand-receptor pairs.
Due to the inclusion of orthologous records of human ligand-receptor pairs in the DanioTalk ligand-receptor database, the majority of the putative ligand-receptor pairs are currently from human ligand-receptor databases. Hence, in addition to the zebrafish STRING PPI-based ligand-receptor pairs, DanioTalk output files also contain ligand-receptor pairs from multiple human ligand-receptor databases and human IID PPI database. The complementation of orthologous ligand-receptor pairs further strengthens the DanioTalk output files and provides users with the knowledge and database source of orthologous human ligand-receptor interactions. As some ligand-receptor signaling requires dimerization of receptors, to aid users, we also provide such information based on the orthologous records in human ligand-receptor pair databases.35,36,61 Finally, to provide users with multiple options to explore their datasets, DanioTalk output files contain multiple annotations including GO terms, type of ligand-receptor signaling (secreted vs. cell-contact), and membrane-bound status of the ligands.
One of the strengths of research on larval zebrafish is in the pharmacological perturbation experiments which we have also taken advantage of in the past.47 Some subscription-based tools built for mammalian model organisms can list drugs targeting mammalian proteins.62,63 Our DanioTalk repository and output files contain similar information on drugs that can likely target the zebrafish ligands and receptome. In addition, the output files also contain orthology scores and lists all the supporting algorithms. The availability of orthology score will give users the confidence on the drug choice but only at the level of gene conservation. For novel drugs that have not been tested in zebrafish, users will still have to manually determine the conservation of the drug target site at the protein sequence and/or structure level or assess the drug targeting efficiency by experimental analysis or by monitoring the associated phenotype and/or downstream signaling events.
The potential of DanioTalk was revealed in our analysis of the existing dataset on adult zebrafish brain synaptosome-PSD proteomes.46 Several top ligand-receptor pairs were previously studied for their synaptic role in other model organisms. For example, in mice CNTNAP1 (CASPR) is well known for its role in synaptic plasticity,64 and Contactin has been shown to promote synaptic CNTNAP1 surface localization via Cis-interaction.64 But the fact that zebrafish Cntn2 and Cntnap1 are enriched in synaptosome and PSD fractions suggests an experimental artifact or additional trans-synaptic roles for the Cntn2-Cntnap1 interaction.65,66 In addition, another top-ranking ligand-receptor pair was Tenascin (Tncb)-Adhesion G Protein-Coupled Receptor L1 (Adgrl1a). Both TNC and ADGRL1 regulate synaptic development and function, but the role of TNC-ADGRL1 interaction has not been addressed yet.67,68,69,70
The presence of several conserved ligand-receptor pairs in the zebrafish synapses further stresses the importance of zebrafish in understanding the role of ligand-receptor in synapse biology and in drugs targeting synapses.14,71,72 Although our synaptosome and PSD ligand-receptor pairs lack spatiotemporal accuracy, DanioTalk can be further combined with zebrafish single-cell brain transcriptome records to identify synaptosome-PSD ligand-receptor interactions in neuronal cell types of interest.57,73
In the absence of cell-specific proteome data, RNA-seq data in combination with protein interaction records are largely used to explore ligand-receptor-based cellular interactions.39,74 However, such data were not explored to analyze axo-glial ligand-receptor interactions in NH, a major neuroendocrine interface critical in maintaining water balance and reproductive functions.47,75,76 Except for few molecular players, the axo-glial interactions in NH were largely unknown.75,77,78,79
Our analysis identified the Slit3-Robo2 glial pituicyte-oxytocin neuron interaction, which has been reported to promote synaptic oxytocin levels in zebrafish.80 However, we also observed autocrine Slit-Robo interactions in oxytocin neurons. SLIT-ROBO autocrine signaling in mice motor neurons has been shown to promote axon fasciculation; however, such defects were not reported for NH-projecting oxytocin axons in zebrafish larvae.80,81 In vertebrates, central release of oxytocin and activity of oxytocin neurons play a key role in animal sociality and behavior.53,82 It is worth to mention that Robo-Slit expressions are conserved in mammalian oxytocin neurons and mutations in OXTR and ROBO2 are observed in children with social deficits and families with autism spectrum, respectively.76,83,84,85 And, in mice, although SLIT-ROBO signaling is essential for the differentiation and positioning of periventricular dopamine neurons, the role in oxytocin neuron development and oxytocin neuron-dependent animal behavior requires further studies.86
Neuro-glial signaling are bidirectional.49 We found oxytocin neuron-derived Neurotensin (Nts) can potentially signal to glial pituicytes via Sortilin (Sort1b) receptor. In rat and teleost brain, neurotensin has been shown to be localized in hypothalamic neurons and in pituitary.87,88,89 Neurotensin can regulate intracellular calcium levels in in vitro astrocyte cultures from rat ventral tegmental area.90 But the role of oxytocin neuron-derived Neurotensin in glial pituicytes function or intracellular calcium is unknown.91
We also observed potential Cxcl12a-Cxcr4b- and Fgf10a-Fgfr2-based autocrine interactions in glial pituicytes.92 CXCL12-CXCR4 functions as a chemokine and neurotransmitter during mice brain development and as mitogen for rat cortical astrocytes in vitro.93,94,95,96 Although CXCL12-CXCR4 interaction is known to modulate NH-projecting vasopressin neuron firing pattern in rats, the role of CXCL12-CXCR4 interaction in glial pituicytes has not been reported yet.97,98 We also observed Fgf10a-Fgfr2-based autocrine interactions in glial pituicytes, which suggest that Fgf10a could have additional autocrine functions in glial pituicytes such as reactivity and/or proliferation.99,100,101,102,103
Overall, DanioTalk offers a comprehensive repository of zebrafish ligands, membrane receptome, ligand-membrane receptome interaction atlas, and a novel data exploratory tool for zebrafish-based researchers. Our methodology can be also applied by other non-mammalian researchers to build ligand-receptor interactome atlas and tools for their model organism of interest.
Limitations of the study
One limitation of our study is that for the axo-glial ligand-receptor pairs, we used datasets derived from works that used different fish, tissue dissociation protocols, and technologies for generating transcriptome data.47,48 This could have led to some artifactual ligand-receptor pairs, at least in the case of paracrine ligand-receptor pairs. Another limitation is the dependence of DanioTalk on the zebrafish STRING physical PPI pair database for generating zebrafish-specific ligand-receptor pairs. For zebrafish, in comparison to Pathway Commons, IntAct, and BioGrid, STRING database is the only database with extensive zebrafish physical PPI records.43,104,105,106 Another limitation is due to the manual curation of ligands and membrane receptors. Currently, DanioTalk ligand-receptor pair-finder scripts are dependent on the manually curated list of ligands and membrane receptors. However, as we provide the original scripts, it is possible for users to edit the manual list of ligands and membrane receptors and rebuild the interaction database required for executing DanioTalk LR finder scripts. Finally, DanioTalk scripts will ignore receptors localized in other organelles and receptors that only require non-protein- or non-peptide-based ligands for signaling. This will be a disadvantage for researchers interested in such signaling.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| DanioTalk | This manuscript | https://github.com/DanioTalk and https://doi.org/10.5281/zenodo.7966578 |
| R | R Core Team | Team et al.107 |
| DeepLoc 2.0 | Thumuluri et al.29 | Thumuluri et al.29 |
| Circlize R package | Gu et al.108 | Gu et al.108 |
| Other | ||
| Zebrafish reference proteome (UP000000437) | UniprotKB | UniProt Consortium28 |
| Zebrafish gene symbols, gene descriptions, orthologue records, ZFIN IDs, OMIM disease information | Zebrafish Information Network | Bradford et al.40 |
| Zebrafish Matrisome | Nauroy et al.21 | Nauroy et al.21 and https://github.com/Matrisome/MatrisomeAnalyzeR/raw/main/data/matrisome.list.rda |
| Zebrafish protein protein physical interaction data | STRING (v11.5) | Szklarczyk et al.43 |
| Zebrafish synaptosome and postsynaptic density proteome | Bayés et al.46 | Bayés et al.46 |
| Zebrafish oxytocin neuron transcriptome | Shafer et al.48 | Shafer et al.48 |
| Zebrafish glial pituicyte transcriptome | Anbalagan et al.47 | Anbalagan et al.47 |
| Zebrafish GO terms | Zebrafish Information Network | Gene Ontology Consortium, Bradford et al.27,40 and http://current.geneontology.org/annotations/zfin.gaf.gz |
| Zebrafish protein localization predictions | LocTree3 | Goldberg et al.19 |
| Human Matrisome | Shao et al.109 | Shao et al.109 and https://github.com/Izzilab/MatrisomeAnalyzeR/blob/main/data/matrisome.list.rda |
| Human protein protein physical interaction data | IID database (v2021-05) | Kotlyar et al.37 |
| Human LR pair database and annotations | CellPhoneDB_v4.1.0 | Garcia-Alonso et al.35 and https://github.com/ventolab/cellphonedb-data/archive/refs/tags/v4.1.0.tar.gz |
| Human LR pair database and annotations | CellChat_1.5.0 | Jin et al.36 and https://github.com/sqjin/CellChat/archive/refs/tags/v1.5.0.tar.gz |
| Human LR pair database | Ramilowski J.A. et al.,38 FANTOM5 | Ramilowski et al.38 |
| Human LR and LR pair database | Cell-Cell Interactions | Ximerakis et al.34 and https://zenodo.org/record/7589953/files/receptor_ligand_interactions_mitab_v1.0_April2017.txt.gz |
| Human LR and LR pair database | CellTalkDB | Shao et al.33 and https://github.com/ZJUFanLab/CellTalkDB/releases/download/v1.0/scsrctdb-1.0.tar.gz |
| Human GO terms | Gene Ontology | Gene Ontology Consortium27 and http://geneontology.org/gene-associations/goa_human.gaf.gz |
| List of drugs | DrugCentralDB | Avram et al.41 and https://unmtid-shinyapps.net/download/DrugCentral/2021_09_01/drug.target.interaction.tsv.gz |
Resource availability
Lead contact
Further information and requests for scripts should be directed to and will be fulfilled by the lead contact, Dr. Savani Anbalagan (savanb@amu.edu.pl).
Materials availability
Software code generated in this study have been deposited to Github: https://github.com/DanioTalk and Zenodo https://doi.org/10.5281/zenodo.7966578.
Method details
Zebrafish, mouse and human datasets
A complete list of datasets used are listed in the key resources table. Zebrafish reference proteome (UP000000437) sequences along with GO-term and cellular localization annotations were downloaded from UniprotKB.28 Gene symbols, ortholog gene symbols, aliases/synonyms, gene descriptions, and ZFIN IDs were downloaded from the ZFIN database.40 The extracellular protein-coding gene was identified based on the annotation terms 'extracellular' or 'secreted' or 'ligands'. Cell membrane protein-coding genes were identified based on the terms “plasma membrane”. Human ligand-receptor databases were downloaded from Cell-cell InteractionDB, CellChatDB, CellPhoneDB, CellTalkDB and Ramilowski et al.33,34,35,36,38 Human PPI database was downloaded from IID.37
Subcellular localization prediction
For machine learning-based cellular localization data, zebrafish reference proteome was subjected to DeepLoc 2.0 algorithm with default parameters to obtain protein localization predictions and associated scores.29
Curated zebrafish ligands and membrane receptome database
For an initial list of zebrafish ligands, the extracellular protein-coding genes were identified based on DeepLoc 2.0-based extracellular protein coding gene predictions. The list was supplemented with genes with GO terms or subcellular location annotation matching 'extracellular' or 'secreted' or 'ligands'. These lists were further expanded with secreted and extracellular matrix-associated genes from zebrafish Matrisome database, extracellular proteins from LocTree3 database, zebrafish orthologs from human matrisome database, CellTalkDB ligands, curated Cell-Cell Interaction database ligands.19,21,33,34,109
For an initial list of zebrafish membrane receptors, membrane protein-coding genes were identified on DeepLoc 2.0-based membranal protein-coding gene predictions. The list was supplemented with genes with GO terms or subcellular location annotation matching “plasma membrane”. These lists were further expanded with cell membrane proteins from LocTree3 database, zebrafish orthologs from human CellTalkDB receptors and in curated Cell-Cell Interaction database receptors.19,33,34
Both the initial ligand and membrane receptor lists were manually curated based on gene descriptions and localization records of human orthologs available in the Compartments database and the Human Protein Atlas.31,110
To attest the credibility of ZFIN listed human orthologs of zebrafish genes, we also incorporated Genome Alliance database provided human orthology score and identity of algorithms that support the orthology.42
Ligand-receptor interactome database
The ligand-receptor (LR) interaction database was generated based on zebrafish protein-protein interaction in STRING physical interaction database (v11.5). To supplement human LR pairs, we also added orthologous pairs from multiple human LR databases namely CellChatDB, CellPhoneDB, CellTalkDB, Cell-Cell Interactions DB, Ramilowski_FANTOM5 database and also orthologous human PPI data available in the IID database (version 2021-05).33,34,35,36,37,38 Human LR pairs annotated to be interacting in complexes in CellPhoneDB and CellChatDB were split into individual interactions.
Annotations of LR interactome database
Ligand and receptor type (Secreted or ECM or membrane-bound and etc.) were annotated based on the annotations available in matrisome database or GO terms or zebrafish orthologs of human genes in CellChat database.21,27,28,36,109 LR signaling that work via dimers or complexes were annotated based on zebrafish orthologs of human genes in CellChat and CellPhoneDB database. In addition, the type of LR interactions (Secreted signaling vs. Cell-Cell contact) was also annotated based on zebrafish orthologs of human genes in CellChat database.36
Scoring of LR pairs in datasets
LR pairs were scored either according to the STRING physical interaction score (Highest ≥900, High ≥700, Medium ≥400) or evidence of orthologous human LR interaction in any of the human databases listed in the key resources table. For scoring purposes, LR pairs reported in human LR pair databases (CellTalkDB, CellPhoneDB, CellChatDB, Ramilowski et al.) were given a score of 500. Pairs from human IID PPI with “exp” (individual or in combination with “ortho” or “pred”) were given a score of 400. Pairs from Cell-Cell interactions or IID PPI without “exp” were given a score of 250. These scores were chosen to prioritize the zebrafish high ranking STRING PPI-based LR pairs (>700).
Application of DanioTalk on zebrafish datasets
For identifying Nodal-related TGFβ and Wnt receptors, DanioTalk LR finder script were run for selected ligands and across all curated receptors (using pseudo-cell type label, fold change and p-adj values in the input files).
Zebrafish synapse and PSD proteome were obtained from quantitative mass spectrometry data previously reported by the Seth Grant group (University of Edinburgh, Scotland).46 For peptide expression, the average peptide count for synaptosome and post synaptic density was utilized (count >0.05).
Zebrafish oxytocin neuron marker genes and pseudo-bulk expression were downloaded from Supplemental_data/3-marker_gene_lists/Drerio_zebrafish.markers.sub.csv (Neuronal_04_2 cluster, avg. Log2FC > 0.25, p-adj. < 0.05) and Supplemental_data/4-pseudobulk_expression/Subclusters-Danio_rerio.csv (Neuronal_04_2 cluster, expression >0.05) respectively.48 Glial pituicytes genes were downloaded from the Table S1 (For differentially expressed genes: Log2FC > 1, p-adj. < 0.05, average AMCA+ read count ≥50 and for all expressed genes average: AMCA+ read count ≥50).47 Gene symbols were updated based on ZFIN alias database. The ligand and receptors were compared and ranked based on DanioTalk interaction database and plotted using Circlize R package (Data S1–S6).108,107
Acknowledgments
We thank Emilia Wysocka, Iwona Kanonik-Jędrzejak, and Arleta Kucz for technical and administrative support. We thank Damian Szklarczyk (Swiss Institute of Bioinformatics, Zurich) for discussion and advice. We thank the BioRender.com team. S.A. is supported by National Science Centre grants (SONATA-BIS 2020/38/E/NZ3/00090 and SONATA 2021/43/D/NZ3/01798). The funding agency was not involved in the design of the study.
Author contributions
S.A. conceived and designed the study. A.Z. contributed to study design and performed DeepLoc 2.0 predictions. M.C. wrote all the scripts. M.C. and S.A. performed the analysis. S.A. performed manual curation of ligands and receptors, wrote the manuscript, and prepared figures. All authors read and approved of the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location.
Published: July 12, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107309.
Supplemental information
Data and code availability
-
•
The DanioTalk scripts are open-source and publicly available from Zenodo https://doi.org/10.5281/zenodo.7966578 Or Github: https://github.com/DanioTalk
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Codes use to analyze the data are available in this paper’s supplemental information.
References
- 1.Blockus H., Chédotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 2.Dufour S., Quérat B., Tostivint H., Pasqualini C., Vaudry H., Rousseau K. Origin and Evolution of the Neuroendocrine Control of Reproduction in Vertebrates, With Special Focus on Genome and Gene Duplications. Physiol. Rev. 2020;100:869–943. doi: 10.1152/physrev.00009.2019. [DOI] [PubMed] [Google Scholar]
- 3.Pires-daSilva A., Sommer R.J. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 4.Steinhart Z., Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;145:dev146589. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczyk W., Romanelli L., Atkins M., Hillen H., Bravo González-Blas C., Jacobs J., Xie J., Soheily S., Verboven E., Moya I.M., et al. Hippo signaling instructs ectopic but not normal organ growth. Science. 2022;378 doi: 10.1126/science.abg3679. [DOI] [PubMed] [Google Scholar]
- 6.Nisar S., Bhat A.A., Masoodi T., Hashem S., Akhtar S., Ali T.A., Amjad S., Chawla S., Bagga P., Frenneaux M.P., et al. Genetics of glutamate and its receptors in autism spectrum disorder. Mol. Psychiatry. 2022;27:2380–2392. doi: 10.1038/s41380-022-01506-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z.R., Du J., Davis A., Mongare M.M., Gould J., Frederick D.T., et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attwood M.M., Jonsson J., Rask-Andersen M., Schiöth H.B. Soluble ligands as drug targets. Nat. Rev. Drug Discov. 2020;19:695–710. doi: 10.1038/s41573-020-0078-4. [DOI] [PubMed] [Google Scholar]
- 9.Hauser A.S., Attwood M.M., Rask-Andersen M., Schiöth H.B., Gloriam D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Congreve M., de Graaf C., Swain N.A., Tate C.G. Impact of GPCR Structures on Drug Discovery. Cell. 2020;181:81–91. doi: 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Lees J.A., Dias J.M., Han S. Applications of Cryo-EM in small molecule and biologics drug design. Biochem. Soc. Trans. 2021;49:2627–2638. doi: 10.1042/BST20210444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cagan R.L., Zon L.I., White R.M. Modeling Cancer with Flies and Fish. Dev. Cell. 2019;49:317–324. doi: 10.1016/j.devcel.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman E.J., Turner K.J., Fernandez J.M., Cifuentes D., Ghosh M., Ijaz S., Jain R.A., Kubo F., Bill B.R., Baier H., et al. Estrogens Suppress a Behavioral Phenotype in Zebrafish Mutants of the Autism Risk Gene, CNTNAP2. Neuron. 2016;89:725–733. doi: 10.1016/j.neuron.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patton E.E., Zon L.I., Langenau D.M. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021;20:611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ségalat L. Invertebrate animal models of diseases as screening tools in drug discovery. ACS Chem. Biol. 2007;2:231–236. doi: 10.1021/cb700009m. [DOI] [PubMed] [Google Scholar]
- 17.Anbalagan S. ‘Blind men and an elephant’, the need for animals in research and drug safety studies in developing countries. OSF Preprints. 2023 doi: 10.31219/osf.io/dwsvt. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Csályi K., Fazekas D., Kadlecsik T., Türei D., Gul L., Horváth B., Módos D., Demeter A., Pápai N., Lenti K., et al. SignaFish: A Zebrafish-Specific Signaling Pathway Resource. Zebrafish. 2016;13:541–544. doi: 10.1089/zeb.2016.1277. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg T., Hecht M., Hamp T., Karl T., Yachdav G., Ahmed N., Altermann U., Angerer P., Ansorge S., Balasz K., et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014;42:W350–W355. doi: 10.1093/nar/gku396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klee E.W. The zebrafish secretome. Zebrafish. 2008;5:131–138. doi: 10.1089/zeb.2008.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nauroy P., Hughes S., Naba A., Ruggiero F. The in-silico zebrafish matrisome: A new tool to study extracellular matrix gene and protein functions. Matrix Biol. 2018;65:5–13. doi: 10.1016/j.matbio.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov D., Türei D., Garrido-Rodriguez M., Burmedi P.L., Nagai J.S., Boys C., Ramirez Flores R.O., Kim H., Szalai B., Costa I.G., et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 2022;13:3224. doi: 10.1038/s41467-022-30755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schier A.F. Single-cell biology: beyond the sum of its parts. Nat. Methods. 2020;17:17–20. doi: 10.1038/s41592-019-0693-3. [DOI] [PubMed] [Google Scholar]
- 24.Campbell N.R., Rao A., Hunter M.V., Sznurkowska M.K., Briker L., Zhang M., Baron M., Heilmann S., Deforet M., Kenny C., et al. Cooperation between melanoma cell states promotes metastasis through heterotypic cluster formation. Dev. Cell. 2021;56:2808–2825.e10. doi: 10.1016/j.devcel.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holler K., Neuschulz A., Drewe-Boß P., Mintcheva J., Spanjaard B., Arsiè R., Ohler U., Landthaler M., Junker J.P. Spatio-temporal mRNA tracking in the early zebrafish embryo. Nat. Commun. 2021;12:3358. doi: 10.1038/s41467-021-23834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu B., Lelek S., Spanjaard B., El-Sammak H., Simões M.G., Mintcheva J., Aliee H., Schäfer R., Meyer A.M., Theis F., et al. Origin and function of activated fibroblast states during zebrafish heart regeneration. Nat. Genet. 2022;54:1227–1237. doi: 10.1038/s41588-022-01129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thumuluri V., Almagro Armenteros J.J., Johansen A.R., Nielsen H., Winther O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022;50:W228–W234. doi: 10.1093/nar/gkac278. gkac278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevers Y., Jones T.E.M., Jyothi D., Yates B., Ferret M., Portell-Silva L., Codo L., Cosentino S., Marcet-Houben M., Vlasova A., et al. The Quest for Orthologs orthology benchmark service in 2022. Nucleic Acids Res. 2022;50:W623–W632. doi: 10.1093/nar/gkac330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 32.Ornitz D.M., Itoh N. The Fibroblast Growth Factor Signaling Pathway. Wiley Interdiscip Rev. Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao X., Liao J., Li C., Lu X., Cheng J., Fan X. CellTalkDB: a manually curated database of ligand-receptor interactions in humans and mice. Brief. Bioinform. 2021;22:bbaa269. doi: 10.1093/bib/bbaa269. [DOI] [PubMed] [Google Scholar]
- 34.Ximerakis M., Lipnick S.L., Innes B.T., Simmons S.K., Adiconis X., Dionne D., Mayweather B.A., Nguyen L., Niziolek Z., Ozek C., et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019;22:1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Alonso L., Lorenzi V., Mazzeo C.I., Alves-Lopes J.P., Roberts K., Sancho-Serra C., Engelbert J., Marečková M., Gruhn W.H., Botting R.A., et al. Single-cell roadmap of human gonadal development. Nature. 2022;607:540–547. doi: 10.1038/s41586-022-04918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotlyar M., Pastrello C., Ahmed Z., Chee J., Varyova Z., Jurisica I. IID 2021: towards context-specific protein interaction analyses by increased coverage, enhanced annotation and enrichment analysis. Nucleic Acids Res. 2022;50:D640–D647. doi: 10.1093/nar/gkab1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., Forrest A.R.R. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 40.Bradford Y.M., Van Slyke C.E., Ruzicka L., Singer A., Eagle A., Fashena D., Howe D.G., Frazer K., Martin R., Paddock H., et al. Zebrafish information network, the knowledgebase for Danio rerio research. Genetics. 2022;220 doi: 10.1093/genetics/iyac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avram S., Bologa C.G., Holmes J., Bocci G., Wilson T.B., Nguyen D.-T., Curpan R., Halip L., Bora A., Yang J.J., et al. DrugCentral 2021 supports drug discovery and repositioning. Nucleic Acids Res. 2021;49:D1160–D1169. doi: 10.1093/nar/gkaa997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alliance of Genome Resources Consortium Harmonizing Model Organism Data in the Alliance of Genome Resources. Genetics. 2022;220:iyac022. doi: 10.1093/genetics/iyac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schier A.F., Shen M.M. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 45.Shen M.M. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 46.Bayés À., Collins M.O., Reig-Viader R., Gou G., Goulding D., Izquierdo A., Choudhary J.S., Emes R.D., Grant S.G.N. Evolution of complexity in the zebrafish synapse proteome. Nat. Commun. 2017;8 doi: 10.1038/ncomms14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anbalagan S., Gordon L., Blechman J., Matsuoka R.L., Rajamannar P., Wircer E., Biran J., Reuveny A., Leshkowitz D., Stainier D.Y.R., Levkowitz G. Pituicyte Cues Regulate the Development of Permeable Neuro-Vascular Interfaces. Dev. Cell. 2018;47:711–726.e5. doi: 10.1016/j.devcel.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 48.Shafer M.E.R., Sawh A.N., Schier A.F. Gene family evolution underlies cell-type diversification in the hypothalamus of teleosts. Nat. Ecol. Evol. 2022;6:63–76. doi: 10.1038/s41559-021-01580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen N.J., Eroglu C. Cell Biology of Astrocyte-Synapse Interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen N.J., Lyons D.A. Glia as architects of central nervous system formation and function. Science. 2018;362:181–185. doi: 10.1126/science.aat0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson C.A., Placzek M. Development of the medial hypothalamus: forming a functional hypothalamic-neurohypophyseal interface. Curr. Top. Dev. Biol. 2013;106:49–88. doi: 10.1016/B978-0-12-416021-7.00002-X. [DOI] [PubMed] [Google Scholar]
- 52.Wittkowski W. Tanycytes and pituicytes: morphological and functional aspects of neuroglial interaction. Microsc. Res. Tech. 1998;41:29–42. doi: 10.1002/(SICI)1097-0029(19980401)41:1<29::AID-JEMT4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 53.Grinevich V., Neumann I.D. Brain oxytocin: how puzzle stones from animal studies translate into psychiatry. Mol. Psychiatry. 2021;26:265–279. doi: 10.1038/s41380-020-0802-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutnick A., Blechman J., Kaslin J., Herwig L., Belting H.-G., Affolter M., Bonkowsky J.L., Levkowitz G. The hypothalamic neuropeptide oxytocin is required for formation of the neurovascular interface of the pituitary. Dev. Cell. 2011;21:642–654. doi: 10.1016/j.devcel.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herget U., Gutierrez-Triana J.A., Salazar Thula O., Knerr B., Ryu S. Single-Cell Reconstruction of Oxytocinergic Neurons Reveals Separate Hypophysiotropic and Encephalotropic Subtypes in Larval Zebrafish. eNeuro. 2017;4 doi: 10.1523/ENEURO.0278-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herpelinck T., Ory L., Nasello G., Barzegari M., Bolander J., Luyten F.P., Tylzanowski P., Geris L. 2022. An Integrated Single-Cell Atlas of the Skeleton from Development through Adulthood. 03.14.484345. [DOI] [Google Scholar]
- 57.Farnsworth D.R., Saunders L.M., Miller A.C. A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 2020;459:100–108. doi: 10.1016/j.ydbio.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrell J.A., Wang Y., Riesenfeld S.J., Shekhar K., Regev A., Schier A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360 doi: 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Z., Akin H., Rao R., Hie B., Zhu Z., Lu W., Smetanin N., Verkuil R., Kabeli O., Shmueli Y., et al. 2022. Evolutionary-scale Prediction of Atomic Level Protein Structure with a Language Model. 07.20.500902. [DOI] [PubMed] [Google Scholar]
- 61.Olayioye M.A., Neve R.M., Lane H.A., Hynes N.E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krämer A., Green J., Pollard J., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinformatics. 2016;54:1. doi: 10.1002/cpbi.5. 30.1-1.30.33. [DOI] [PubMed] [Google Scholar]
- 64.Murai K.K., Misner D., Ranscht B. Contactin Supports Synaptic Plasticity Associated with Hippocampal Long-Term Depression but Not Potentiation. Curr. Biol. 2002;12:181–190. doi: 10.1016/S0960-9822(02)00680-2. [DOI] [PubMed] [Google Scholar]
- 65.Dubessy A.L., Mazuir E., Rappeneau Q., Ou S., Abi Ghanem C., Piquand K., Aigrot M.S., Thétiot M., Desmazières A., Chan E., et al. Role of a Contactin multi-molecular complex secreted by oligodendrocytes in nodal protein clustering in the CNS. Glia. 2019;67:2248–2263. doi: 10.1002/glia.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruegg M.A., Stoeckli E.T., Kuhn T.B., Heller M., Zuellig R., Sonderegger P. Purification of axonin-1, a protein that is secreted from axons during neurogenesis. EMBO J. 1989;8:55–63. doi: 10.1002/j.1460-2075.1989.tb03348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurevicius K., Kuang F., Stoenica L., Irintchev A., Gureviciene I., Dityatev A., Schachner M., Tanila H. Genetic ablation of tenascin-C expression leads to abnormal hippocampal CA1 structure and electrical activity in vivo. Hippocampus. 2009;19:1232–1246. doi: 10.1002/hipo.20585. [DOI] [PubMed] [Google Scholar]
- 68.Vitobello A., Mazel B., Lelianova V.G., Zangrandi A., Petitto E., Suckling J., Salpietro V., Meyer R., Elbracht M., Kurth I., et al. ADGRL1 haploinsufficiency causes a variable spectrum of neurodevelopmental disorders in humans and alters synaptic activity and behavior in a mouse model. Am. J. Hum. Genet. 2022;109:1436–1457. doi: 10.1016/j.ajhg.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J., Shalev-Benami M., Sando R., Jiang X., Kibrom A., Wang J., Leon K., Katanski C., Nazarko O., Lu Y.C., et al. Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse. Cell. 2018;173:735–748.e15. doi: 10.1016/j.cell.2018.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gottschling C., Wegrzyn D., Denecke B., Faissner A. Elimination of the four extracellular matrix molecules tenascin-C, tenascin-R, brevican and neurocan alters the ratio of excitatory and inhibitory synapses. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hutson L.D., Chien C.-B. Wiring the zebrafish: axon guidance and synaptogenesis. Curr. Opin. Neurobiol. 2002;12:87–92. doi: 10.1016/s0959-4388(02)00294-5. [DOI] [PubMed] [Google Scholar]
- 72.Oprişoreanu A.M., Smith H.L., Krix S., Chaytow H., Carragher N.O., Gillingwater T.H., Becker C.G., Becker T. Automated in vivo drug screen in zebrafish identifies synapse-stabilising drugs with relevance to spinal muscular atrophy. Dis. Model. Mech. 2021;14 doi: 10.1242/dmm.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raj B., Farrell J.A., Liu J., El Kholtei J., Carte A.N., Navajas Acedo J., Du L.Y., McKenna A., Relić Đ., Leslie J.M., Schier A.F. Emergence of Neuronal Diversity during Vertebrate Brain Development. Neuron. 2020;108:1058–1074.e6. doi: 10.1016/j.neuron.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Türei D., Valdeolivas A., Gul L., Palacio-Escat N., Klein M., Ivanova O., Ölbei M., Gábor A., Theis F., Módos D., et al. Integrated intra- and intercellular signaling knowledge for multicellular omics analysis. Mol. Syst. Biol. 2021;17 doi: 10.15252/msb.20209923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown C.H. Magnocellular Neurons and Posterior Pituitary Function. Compr. Physiol. 2016;6:1701–1741. doi: 10.1002/cphy.c150053. [DOI] [PubMed] [Google Scholar]
- 76.Romanov R.A., Zeisel A., Bakker J., Girach F., Hellysaz A., Tomer R., Alpár A., Mulder J., Clotman F., Keimpema E., et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 2017;20:176–188. doi: 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leng G., Pineda R., Sabatier N., Ludwig M. 60 YEARS OF NEUROENDOCRINOLOGY: The posterior pituitary, from Geoffrey Harris to our present understanding. J. Endocrinol. 2015;226:T173–T185. doi: 10.1530/JOE-15-0087. [DOI] [PubMed] [Google Scholar]
- 78.Miyata S. Advances in Understanding of Structural Reorganization in the Hypothalamic Neurosecretory System. Front. Endocrinol. 2017;8:275. doi: 10.3389/fendo.2017.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosso L., Mienville J.-M. Pituicyte modulation of neurohormone output. Glia. 2009;57:235–243. doi: 10.1002/glia.20760. [DOI] [PubMed] [Google Scholar]
- 80.Anbalagan S., Blechman J., Gliksberg M., Gordon L., Rotkopf R., Dadosh T., Shimoni E., Levkowitz G. Robo2 regulates synaptic oxytocin content by affecting actin dynamics. Elife. 2019;8 doi: 10.7554/eLife.45650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaworski A., Tessier-Lavigne M. Autocrine/juxtaparacrine regulation of axon fasciculation by Slit-Robo signaling. Nat. Neurosci. 2012;15:367–369. doi: 10.1038/nn.3037. [DOI] [PubMed] [Google Scholar]
- 82.Grinevich V., Ludwig M. The multiple faces of the oxytocin and vasopressin systems in the brain. J. Neuroendocrinol. 2021;33 doi: 10.1111/jne.13004. [DOI] [PubMed] [Google Scholar]
- 83.Leng G., Leng R.I., Ludwig M. Oxytocin-a social peptide? Deconstructing the evidence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022;377 doi: 10.1098/rstb.2021.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anitha A., Nakamura K., Yamada K., Suda S., Thanseem I., Tsujii M., Iwayama Y., Hattori E., Toyota T., Miyachi T., et al. Genetic analyses of roundabout (ROBO) axon guidance receptors in autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1019–1027. doi: 10.1002/ajmg.b.30697. [DOI] [PubMed] [Google Scholar]
- 85.Xu C., Fan C.-M. Expression of Robo/Slit and Semaphorin/Plexin/Neuropilin family members in the developing hypothalamic paraventricular and supraoptic nuclei. Gene Expr. Patterns. 2008;8:502–507. doi: 10.1016/j.gep.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romanov R.A., Tretiakov E.O., Kastriti M.E., Zupancic M., Häring M., Korchynska S., Popadin K., Benevento M., Rebernik P., Lallemend F., et al. Molecular design of hypothalamus development. Nature. 2020;582:246–252. doi: 10.1038/s41586-020-2266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goedert M., Lightman S.L., Mantyh P.W., Hunt S.P., Emson P.C. Neurotensin-like immunoreactivity and neurotensin receptors in the rat hypothalamus and in the neurointermediate lobe of the pituitary gland. Brain Res. 1985;358:59–69. doi: 10.1016/0006-8993(85)90948-5. [DOI] [PubMed] [Google Scholar]
- 88.Batten T.F., Marivoet S., Vandesande F. Neurotensin-like immunoreactivity in the pituitary and hypothalamus of bony fishes. Peptides. 1987;8:135–143. doi: 10.1016/0196-9781(87)90177-x. [DOI] [PubMed] [Google Scholar]
- 89.Muraki K., Okahata H., Nishi Y., Usui T., Yamada H., Fujita S., Miyachi Y., Yanaihara N., Yajima H. Distribution of neurotensin-like immunoreactivity in the hypothalamus, pituitary gland, and gastro-intestinal tract of rats. Acta Endocrinol. 1985;110:1–5. doi: 10.1530/acta.0.1100001. [DOI] [PubMed] [Google Scholar]
- 90.Trudeau L.E. Neurotensin regulates intracellular calcium in ventral tegmental area astrocytes: evidence for the involvement of multiple receptors. Neuroscience. 2000;97:293–302. doi: 10.1016/s0306-4522(99)00597-7. [DOI] [PubMed] [Google Scholar]
- 91.Hatton G.I., Bicknell R.J., Hoyland J., Bunting R., Mason W.T. Arginine vasopressin mobilises intracellular calcium via V1-receptor activation in astrocytes (pituicytes) cultured from adult rat neural lobes. Brain Res. 1992;588:75–83. doi: 10.1016/0006-8993(92)91346-g. [DOI] [PubMed] [Google Scholar]
- 92.Hickey K.N., Grassi S.M., Caplan M.R., Stabenfeldt S.E. Stromal Cell-Derived Factor-1a Autocrine/Paracrine Signaling Contributes to Spatiotemporal Gradients in the Brain. Cell. Mol. Bioeng. 2021;14:75–87. doi: 10.1007/s12195-020-00643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barbero S., Bajetto A., Bonavia R., Porcile C., Piccioli P., Pirani P., Ravetti J.L., Zona G., Spaziante R., Florio T., Schettini G. Expression of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1 in human brain tumors and their involvement in glial proliferation in vitro. Ann. N. Y. Acad. Sci. 2002;973:60–69. doi: 10.1111/j.1749-6632.2002.tb04607.x. [DOI] [PubMed] [Google Scholar]
- 94.Lazarini F., Tham T.N., Casanova P., Arenzana-Seisdedos F., Dubois-Dalcq M. Role of the alpha-chemokine stromal cell-derived factor (SDF-1) in the developing and mature central nervous system. Glia. 2003;42:139–148. doi: 10.1002/glia.10139. [DOI] [PubMed] [Google Scholar]
- 95.Bhattacharyya B.J., Banisadr G., Jung H., Ren D., Cronshaw D.G., Zou Y., Miller R.J. The chemokine stromal cell-derived factor-1 regulates GABAergic inputs to neural progenitors in the postnatal dentate gyrus. J. Neurosci. 2008;28:6720–6730. doi: 10.1523/JNEUROSCI.1677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miyasaka N., Knaut H., Yoshihara Y. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134:2459–2468. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- 97.Callewaere C., Banisadr G., Desarménien M.G., Mechighel P., Kitabgi P., Rostène W.H., Mélik Parsadaniantz S. The chemokine SDF-1/CXCL12 modulates the firing pattern of vasopressin neurons and counteracts induced vasopressin release through CXCR4. Proc. Natl. Acad. Sci. USA. 2006;103:8221–8226. doi: 10.1073/pnas.0602620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Callewaere C., Fernette B., Raison D., Mechighel P., Burlet A., Calas A., Kitabgi P., Parsadaniantz S.M., Rostène W. Cellular and subcellular evidence for neuronal interaction between the chemokine stromal cell-derived factor-1/CXCL 12 and vasopressin: regulation in the hypothalamo-neurohypophysial system of the Brattleboro rats. Endocrinology. 2008;149:310–319. doi: 10.1210/en.2007-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gómez-Pinilla F., Vu L., Cotman C.W. Regulation of astrocyte proliferation by FGF-2 and heparan sulfate in vivo. J. Neurosci. 1995;15:2021–2029. doi: 10.1523/JNEUROSCI.15-03-02021.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goodman T., Nayar S.G., Clare S., Mikolajczak M., Rice R., Mansour S., Bellusci S., Hajihosseini M.K. Fibroblast growth factor 10 is a negative regulator of postnatal neurogenesis in the mouse hypothalamus. Development. 2020;147 doi: 10.1242/dev.180950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haan N., Goodman T., Najdi-Samiei A., Stratford C.M., Rice R., El Agha E., Bellusci S., Hajihosseini M.K. Fgf10-expressing tanycytes add new neurons to the appetite/energy-balance regulating centers of the postnatal and adult hypothalamus. J. Neurosci. 2013;33:6170–6180. doi: 10.1523/JNEUROSCI.2437-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu F., Pogoda H.-M., Pearson C.A., Ohyama K., Löhr H., Hammerschmidt M., Placzek M. Direct and indirect roles of Fgf3 and Fgf10 in innervation and vascularisation of the vertebrate hypothalamic neurohypophysis. Development. 2013;140:1111–1122. doi: 10.1242/dev.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor Specificity of the Fibroblast Growth Factor Family: THE COMPLETE MAMMALIAN FGF FAMILY. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodchenkov I., Babur O., Luna A., Aksoy B.A., Wong J.V., Fong D., Franz M., Siper M.C., Cheung M., Wrana M., et al. Pathway Commons 2019 Update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020;48:D489–D497. doi: 10.1093/nar/gkz946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Del Toro N., Shrivastava A., Ragueneau E., Meldal B., Combe C., Barrera E., Perfetto L., How K., Ratan P., Shirodkar G., et al. The IntAct database: efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022;50:D648–D653. doi: 10.1093/nar/gkab1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oughtred R., Rust J., Chang C., Breitkreutz B.-J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F., et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Team R.C. R Foundation for Statistical Computing; 2013. R: A Language and Environment for Statistical Computing.http://www.R-project.org/ [Google Scholar]
- 108.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 109.Shao X., Taha I.N., Clauser K.R., Gao Y.T., Naba A., Naba A. MatrisomeDB: the ECM-protein knowledge database. Nucleic Acids Res. 2020;48:D1136–D1144. doi: 10.1093/nar/gkz849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O’Donoghue S.I., Schneider R., Jensen L.J. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database. 2014;2014:bau012. doi: 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The DanioTalk scripts are open-source and publicly available from Zenodo https://doi.org/10.5281/zenodo.7966578 Or Github: https://github.com/DanioTalk
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Codes use to analyze the data are available in this paper’s supplemental information.