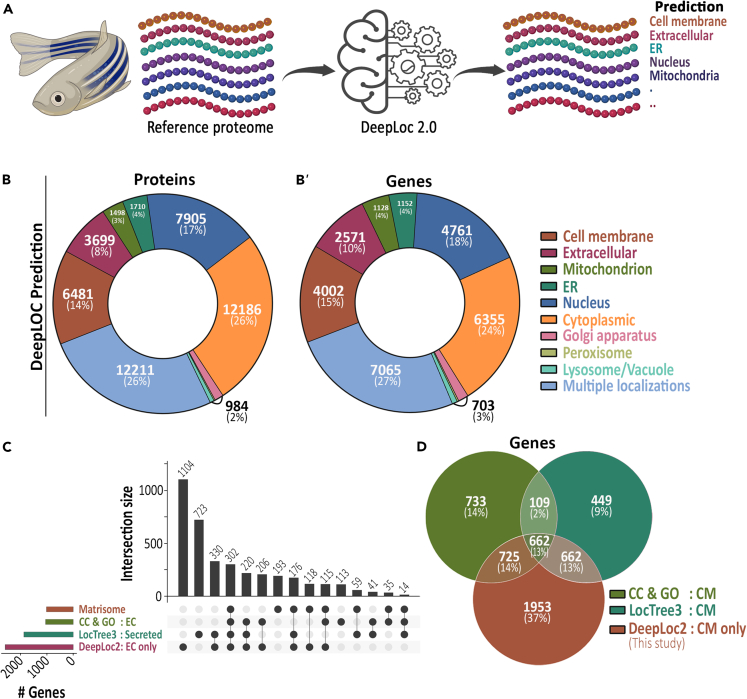
Figure 1.
Prediction of cellular localization of zebrafish reference proteome
(A) Graphic describing the DeepLoc 2.0-based prediction of cellular location of the reference zebrafish proteome.
(B) Zebrafish reference proteome cellular localization prediction. Donut chart showing the major DeepLoc 2.0 predictions of zebrafish proteome. The number of proteins (B) and protein-coding genes (B′) are shown.
(C) Upset plot comparing the number of genes with existing extracellular annotation records and DeepLoc 2.0-based 'Extracellular' predictions. CC & GO indicates cellular localization and GO term records, respectively, available in UniprotKB database.
(D) Venn diagram comparing genes with existing cell membrane localization annotation records and DeepLoc-based 'cell membrane' (CM) predictions. CC & GO indicates cellular localization and GO term records, respectively, available in UniprotKB database.