Abstract
The multiple sclerosis-associated retrovirus (MSRV) isolated from plasma of MS patients was found to be phylogenetically and experimentally related to human endogenous retroviruses (HERVs). To characterize the MSRV-related HERV family and to test the hypothesis of a replication-competent HERV, we have investigated the expression of MSRV-related sequences in healthy tissues. The expression of MSRV-related transcripts restricted to the placenta led to the isolation of overlapping cDNA clones from a cDNA library. These cDNAs spanned a 7.6-kb region containing gag, pol, and env genes; RU5 and U3R flanking sequences; a polypurine tract; and a primer binding site (PBS). As this PBS showed similarity to avian retrovirus PBSs used by tRNATrp, this new HERV family was named HERV-W. Several genomic elements were identified, one of them containing a complete HERV-W unit, spanning all cDNA clones. Elements of this multicopy family were not replication competent, as gag and pol open reading frames (ORFs) were interrupted by frameshifts and stop codons. A complete ORF putatively coding for an envelope protein was found both on the HERV-W DNA prototype and within an RU5-env-U3R polyadenylated cDNA clone. Placental expression of 8-, 3.1-, and 1.3-kb transcripts was observed, and a putative splicing strategy was described. The apparently tissue-restricted HERV-W long terminal repeat expression is discussed with respect to physiological and pathological contexts.
A few years ago, the presence of extracellular particles associated with reverse transcriptase (RT) activity was observed in leptomeningeal cells (LM7) (42) and monocyte cultures (44) from patients with multiple sclerosis (MS). Viral RT-associated activity plus electron microscopy examination and the presence in MS patients of antibodies cross-reacting with human immunodeficiency virus type 1 and type 2 RT and human T-cell leukemia virus type 1 CA as well (41) led to the hypothesis of the retroviral nature of these viral particles. By mainly a PCR-based approach, a partial molecular identification of a novel retrovirus isolated from cerebrospinal fluid, plasma, or cell culture supernatant of MS patients was recently obtained (40). Retroviral RNAs with strong similarities to MS-associated retrovirus (MSRV) were also found in plasma of rheumatoid arthritis (RA) patients (17). Furthermore, expression of related mRNA sequences (8) was found in both MS and control brain tissues (29).
All together, these observations might result from three different situations. The extracellular particles may be produced by a replication-competent endogenous provirus. Alternatively, MSRV could represent a virion-producing exogenous member of an endogenous family, as described for the mouse mammary tumor virus and murine leukemia virus retroviral families of mice (23) and in the Jaagsiekte retroviral family of sheep (37). Lastly, a more complicated process would involve separated defective retroviral entities cooperating via trans-complementation.
MSRV was found to be related to the endogenous retroviral sequence ERV-9 (26), and Southern blot analysis with an MSRV pol probe showed hybridization with a multicopy endogenous family (40). Therefore, we have studied the MSRV-related human endogenous retroviruses (HERVs), which could represent either distant members of the ERV-9 family or an as yet undescribed HERV family. As (i) the HERV family was shown to be a multicopy family (40) and (ii) we focused on the hypothesis that MSRV could be a replication-competent HERV, we followed a strategy based on the characterization of mRNA isolated from a healthy human tissue, by using MSRV- and ERV-9-derived probes. We describe a new multicopy endogenous retroviral family tentatively named HERV-W. All members of the family are apparently not competent for replication. However, a functional U3 promoter and an open reading frame (ORF) putatively coding for an envelope protein were identified. HERV-W mRNA expression is restricted to placental tissue in Northern blot analysis. The strategy of transcription is addressed, and the promoter tissue specificity is discussed.
MATERIALS AND METHODS
Probe definition and labelling.
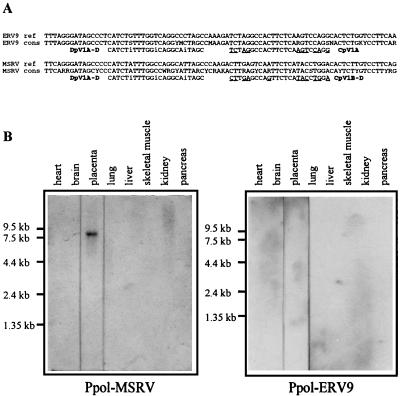
The 678-bp Ppol-MSRV probe was RT-PCR amplified from RNA extracted from particles collected from the supernatant of synoviocyte cultures of an RA patient. These particles were gradient concentrated as previously described (43), and RNA was extracted by the method of Chomczynski and Sacchi (10) and DNase treated as previously described (54). A first round of RT-PCR amplification was performed with primers AP2942 (5′-AGGAGTAAGGAAACCCAACGGAC-3′) (forward) and AP2152 (5′-TAAGAGTTGCACAAGTGCG-3′) (reverse). A nested PCR was then performed with primers AP2522 (5′-TCAGGGATAGCCCCCATCTAT-3′) (forward) and AP2510 (5′-AACCCTTTGCCACTACATCAATTTC-3′) (reverse). The 649-bp Ppol-ERV-9 probe was PCR amplified from a B-lymphocyte cDNA library of an MS patient with primers AP2522 and AP2510 mentioned above. Discrimination between the Ppol-MSRV- and Ppol-ERV-9-cloned PCR fragments was performed with an enzyme-linked oligosorbent assay (ELOSA) (35). Capture and detection oligonucleotide probes used in this nonisotopic sandwich technique are depicted in Fig. 1A.
FIG. 1.
(A) Capture and detection probes used in ELOSA to detect PCR-amplified ERV-9 and MSRV pol-related sequences. Discrimination between ERV-9 and MSRV cloned PCR fragments was performed at the capture step with CpV1A and CpV1B-D, respectively. Colorimetric detection was achieved with the DpV1A-D peroxidase-labelled probe located within the most conserved region of ERV-9 and MSRV pol genes. The selected PCR fragments were called Ppol-MSRV and Ppol-ERV-9 according to ELOSA characterization. The reference (ref) ERV-9 (gb X57147, gb M85205, and gb M37638) and MSRV (gb AF009668) sequences are indicated as well as the consensus (cons) sequences. These consensus sequences resulted from RT-PCR amplification of the conserved region of the pol gene present in all retroviruses (40, 50); the RNA material was obtained from MS patients. (B) Northern blot analysis of poly(A)+ cellular RNAs from eight human tissues with the Ppol-MSRV and Ppol-ERV-9 probes characterized by ELOSA.
The Pgag-LB19 (536 bp), Ppro-E (364 bp), and Penv-C15 (591 bp) probes corresponded to PCR fragments of the longest ORFs of LB19, E, and C15 clones, respectively. LB19 and E clones (37a) were obtained from RNA extracted from gradient-concentrated particles of B-lymphocyte cultures of an MS patient or from the plasma of an MS patient, respectively. The C15 clone (22a) was obtained from RNA extracted from the pellet of centrifuged supernatants of synoviocyte cultures of an RA patient.
Fifty nanograms of each probe was radioactively labelled by random priming with the Ready-to-Go DNA labelling kit from Pharmacia Biotech, Inc., according to the manufacturer’s protocol with [α-32P]dCTP (3,000 Ci/mmol). The specific activity of probes was >5.0 × 108 cpm/μg.
Placental cDNA library screening, 5′ rapid amplification of cDNA ends (RACE), and sequencing.
A placental cDNA library constructed in the bacteriophage lambda vector was used (human placenta 5′-Stretch Plus cDNA library from Clontech Laboratories, Inc., Palo Alto, Calif.). Screening of the library was performed according to the manufacturer’s instructions with the Ppol-MSRV probe for initial screening, and probes were derived from the resulting clones for additional screening. Briefly, for each screening the plating density was about 5 × 105 PFU for two 220- by 220-mm plates. After growth, plaques were transferred onto a nylon membrane (Hybond N+; Amersham) and hybridized with probes radioactively labelled as described above (106 cpm/ml of hybridization solution). Prehybridization was performed overnight at 42°C in a freshly prepared solution: 50% formamide, 5× SSPE (20× SSPE is 3 M NaCl, 0.2 M NaH2PO4, and 20 mM EDTA, pH 7.4), 5× Denhardt’s solution (100× Denhardt’s solution is 10 g of Ficoll 400, 10 g of polyvinylpyrrolidone, and 10 g of bovine serum albumin for 500 ml), and 0.1% sodium dodecyl sulfate (SDS) with 100 μg of heat-denatured herring sperm DNA per ml. Hybridization was performed overnight at the same temperature, in a renewed solution freshly prepared in the same way. After hybridization, filters were washed once for 20 min at room temperature with 2× SSC (20× SSC is 3 M NaCl and 0.3 M sodium citrate)–0.5% SDS followed by one wash for 25 min at 60°C with 1× SSC–0.1% SDS. Washed filters were exposed to X-ray films (Kodak) for 72 h at −80°C. Isolated positive plaques were picked and eluted in the appropriate buffer.
A rapid method was used for subcloning of the cDNA clones. A PCR amplification using a primer generated according to the bacteriophage lambda vector sequence and a specific primer designated according to the probe sequence was performed in a 9600 Perkin-Elmer machine. PCR was carried out with initial denaturation at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 4 min; terminal extension was performed at 72°C for 7 min. After 1% agarose gel analysis, the longest fragments were subcloned into pCR 2.1 vector (TA cloning; Invitrogen, San Diego, Calif.) and sequenced.
A 5′ RACE technique was used to obtain the 5′ end of the retrovirus genome; the kit, used strictly according to the manufacturer’s recommendations, was provided by Life Technologies, Inc. (Bethesda, Md.). Reverse transcription, dC-tailing, and amplifications were performed on a commercially available placental mRNA [human placental poly(A)+ RNA; Clontech], and fragments were subcloned into pCR 2.1 vector (TA cloning).
Sequencing reactions were performed, in both directions for each clone, with the Prism Ready Reaction kit and Dye Deoxyterminator cycle sequencing kit (Applied Biosystems). Automatic sequence analysis was performed on an automatic sequencer (Applied Biosystems).
Hybridization of Northern blots and RNA dot blots.
Several Northern blots of different registration lot numbers were used (multiple-tissue Northern blot, catalog no. 7760-1 from Clontech Laboratories, Inc.). Prehybridization was performed overnight at 42°C in the following freshly prepared solution: 50% formamide, 5× SSPE, 10× Denhardt’s solution, 2% SDS, and heat-denatured herring sperm DNA (100 μg/ml). Hybridization was performed overnight at 42°C in a renewed solution with a probe labelled as described above (106 cpm/ml of hybridization solution). Filters were washed twice with 1× SSC–0.1% SDS at room temperature for 5 min and once with 0.1× SSC–0.1% SDS at 50°C for 10 min. Washed filters were exposed to X-ray films (Kodak) for 5 days at −80°C.
RNA dot blot (human RNA Master Blot from Clontech Laboratories, Inc.) was used according to the manufacturer’s instructions. Briefly, prehybridization was performed at 65°C in the ExpressHyb hybridization solution from Clontech for 30 min. Hybridization of a probe labelled as described above was performed overnight at 65°C in a renewed solution. The membrane was washed twice at 65°C with 2× SSC–1% SDS for 20 min and once at 55°C with 0.1× SSC–0.5% SDS and exposed to X-ray film (Kodak) for about 12 h.
Hybridization of Southern blots and DNA dot blots.
Human genomic DNA was digested with EcoRI, HindIII, and PstI. Ten micrograms of each reaction mixture was electrophoresed on a 0.8% agarose gel and transferred onto a charged nylon membrane (Hybond N+ [Amersham]). Prehybridization was performed overnight at 42°C in a freshly prepared solution: 50% formamide, 5× SSPE, 1% (wt/vol) nonfat dried milk, 1% SDS, and 50 μg of heat-denatured herring sperm DNA per ml. Hybridization was performed overnight at 42°C in a renewed solution, with a probe radioactively labelled as mentioned above (106 cpm/ml of hybridization solution). Filters were washed once with 1× SSC–0.1% SDS at room temperature for 5 min and once with 0.1× SSC–0.1% SDS at 50°C for 10 min. Washed filters were exposed to X-ray films (Kodak) for 5 days at −80°C.
For the DNA dot blot, successive dilutions of each probe (2.5, 5, 10, 25, 50, and 100 pg) and 0.5 μg of genomic DNA were blotted on a charged nylon membrane after denaturation. For each probe, the dot blot and the Southern blot were handled in the same vessel.
CAT assay.
The putative promoter region was cloned into pCAT3 Enhancer reporter vector purchased from Promega Biotec (Madison, Wis.). HeLa 60% confluent cells cultured in Dulbecco modified Eagle medium–10% fetal calf serum (Life Technologies) were transfected with the Superfect transfection kit (Qiagen GmbH) with 2 μg of purified recombinant plasmid. After a 48-h incubation, the cells were harvested in order to evaluate chloramphenicol acetyltransferase (CAT) activity by the use of the CAT enzyme assay system (Promega Biotec). For this purpose, the liquid scintillation counting protocol was followed as recommended by the manufacturer. Briefly, cell extracts were obtained by several freezes-thaws and a subsequent heating at 60°C for 10 min. Then, 10 μl of this extract was added to 0.25 M Tris-HCl, pH 8.0 (125 μl, final volume), containing 0.15 μCi of [14C]chloramphenicol (NEN, Boston, Mass.) and 5 μg of n-butyryl coenzyme A, and incubated for 4 h at 37°C. Five hundred microliters of a mix of xylene isomers was added, and the butyrylated chloramphenicol present in this organic phase was extracted twice with 100 μl of 0.25 M Tris-HCl. This 200 μl was mixed with 5 ml of scintillation fluid and counted in a liquid scintillation counter.
In vitro transcription-translation.
In vitro transcription-translation was performed with a kit from Promega Biotec (TNT coupled reticulocyte lysate systems). It was used strictly according to the manufacturer’s instructions. Briefly, DNA templates were obtained by PCR with a 5′ primer designed with a T7 promoter (5′-CATAATACGACTCACTATAGGGAGACCATGGCCCTCCCTTATCAT-3′). The radiolabelled amino acid was [35S]methionine, provided by Amersham. Glycosylation was studied by the translation in vitro in the presence of canine pancreatic microsomal membranes (also provided by Promega). Posttranslational analysis was performed by SDS-polyacrylamide gel electrophoresis, and the gel was exposed to X-ray films (Kodak), usually for 12 h.
Sequence analysis.
Individual cDNA clones were used for phylogenetic analysis. Sequences were filtered for low-complexity regions and repeat sequences (Alu-like and microsatellites) with XBLAST (12). A BLASTN (or BLASTX) (2) query for GenBank (release 101, June 1997) was performed, and sequence matches with scores greater than 200 were retained. Homologous fragments were extracted from GenBank and aligned with our cDNA clones. Alignments were performed semimanually, with the SEAVIEW multiple alignment editor (16) and the Clustal W (52), Dialign (36), or MABIOS (1) multiple alignment program. Regions corresponding to putative coding regions were determined with Blixem (51), a program allowing the visualization of protein similarities on a nucleic acid sequence from the alignment. We selected in the alignment three regions from long terminal repeat (LTR), Pol, and Env that are conserved in all members of the family of human endogenous viruses that we have identified. Phylogenetic trees were computed from these three regions with Phylo_win (16) with the neighbor-joining method (49). Five hundred bootstrap replicates were performed to evaluate the robustness of the trees.
Promoter prediction was performed with PROSCAN v1.7 (47) and SIGSCAN v4.05 (46). A searching for protein sequence similarities was performed with LFASTA (39). The prediction of leader peptide was made with GeneWorks 2.5.1 software. Prediction of the fusion domain and the transmembrane region was performed with the PHD package (48).
Nucleotide sequence accession numbers.
The sequences described in this paper have been submitted to GenBank under the following accession numbers. The probe numbers are as follows: Ppol-MSRV (AF072494), Ppol-ERV-9 (AF072495), Pgag-LB19 (AF072496), Ppro-E (AF072497), and Penv-C15 (AF072498). The placental cDNA clones numbers are as follows: cl.6A1 (AF072499), cl.6A2 (AF072500), cl.7A16 (AF072501), cl.Pi5T (AF072502), cl.Pi22 (AF072503), cl.44.4 (AF072504), cl24.4 (AF072505), cl.PH74 (AF072506), cl.PH7 (AF072507), and cl.C4C5 (AF072508).
RESULTS
Design of probes.
Pol probes were designed to evaluate the expression of MSRV-related sequences in healthy human tissues. The choice of Pol probes relied on the observations that (i) the previously described Pol region of MSRV was related to the ERV-9 HERV and (ii) ERV-9 sequences were occasionally detected with MSRV in plasma or cell culture supernatants of MS patients (40). Because (i) the 2,304-bp MSRV Pol region (40) was a consensus sequence resulting from overlapping clones obtained from different sources and (ii) the MSRV and ERV-9 codetected sequences were only 120 bp long, we chose to amplify two continuous 650-bp MSRV- and ERV-9-related fragments, defined by the same borders and including the 120-bp codetected region. MSRV- and ERV-9-related Pol regions were RT-PCR amplified as described in Materials and Methods, and mixed resulting fragments were cloned and checked (data not shown) with the ELOSA procedure (Fig. 1A), which was previously shown to discriminate between the MSRV and ERV-9 Pol 120-bp coamplified fragments (40). The ELOSA MSRV-related probe, which was called Ppol-MSRV, has 87 and 69% similarity with MSRV and ERV-9 reference sequences, respectively. The ELOSA ERV-9-related probe, which was called Ppol-ERV-9, has 70 and 87% similarity with MSRV and ERV-9, respectively.
In order to improve the likelihood of finding sequences containing ORFs, consistent with the hypothesis that MSRV could be a replication-competent HERV, three other probes were derived from putatively packaged extracellular mRNAs detected in pathological contexts. These mRNAs were RT-PCR amplified with primers designed according to the obtained placental cDNA clones (see below) and/or to the conserved region of retroviruses and/or to 5′ and 3′ RACE protocols. Although no amplified fragments supported coding capacities compatible with a replication-competent retrovirus, Pgag-LB19, Ppro-E, and Penv-C15 probes were designed, corresponding to the longest ORFs found in PCR products derived from gag, pro, and env, respectively.
Expression of a novel HERV in placenta.
Ppol-MSRV, Ppol-ERV-9, Pgag-LB19, and Penv-C15 probes were then used for hybridization of a multiple-tissue Northern blot under stringent conditions. The Ppol-MSRV probe revealed an 8-kb transcript that was expressed in placenta but not in heart, brain, lung, liver, skeletal muscle, kidney, or pancreas (Fig. 1B). No signal was detected with the Ppol-ERV-9 probe (Fig. 1B). The placenta-restricted expression was confirmed for Pgag-LB19 and Penv-C15 probes (data not shown). Furthermore, 48 tissue samples including cerebral, muscular, endocrine, exocrine, lymphoid, visceral, and fetal tissues were not revealed by Pgag-LB19 and Penv-C15 probes by RNA dot blot, but both placental and kidney mRNAs were found to be positive (data not shown). As the kidney mRNA expression was not revealed by Northern blotting, with four different commercial lots, we interpreted the kidney mRNA positive signal as an artifact resulting from the method per se, including tissue-specific mRNA preparation or DNA contamination.
A BLASTN query on the EST (expressed sequence tag) database, with placental cDNA clones (see below) derived from the above probes, showed hundreds of related transcripts in the human tissues, most in the opposite direction. With a search criterion of 90% identity over 100 nucleotides (nt) or more, these sequences were found predominantly expressed in the placenta (53%), but also in fetal liver-spleen (28%). The relative abundance of HERV-W sequences among placenta ESTs was found to be three times higher than that among fetal liver-spleen ESTs.
Isolation and sequencing of overlapping cDNA fragments.
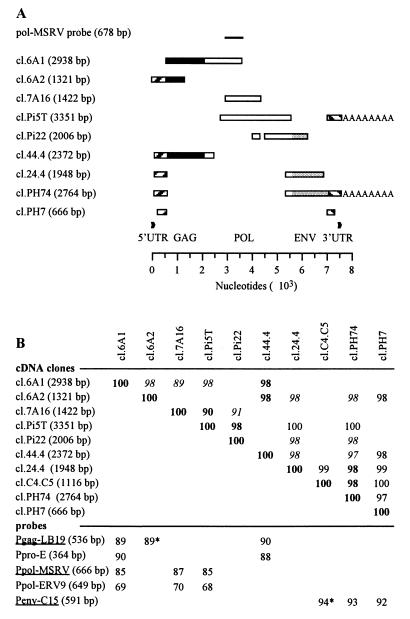
A placental cDNA library was screened with Ppol-MSRV, Pgag-LB19, and Penv-C15 probes. Probes derived from the resulting clones were then used for additional screening. The clones obtained with all three screenings exhibited the same characteristics. All together, the overlapping cDNA clones covered 7.6 kb. Nine overlapping clones obtained with the Ppol-MSRV screening are shown in Fig. 2A. The percentages of similarity among the overlapping parts of these Ppol-MSRV-derived cDNA clones, a polyadenylated clone (cl.C4C5) probed with Penv-C15, and the probes used for the screening as well are presented in Fig. 2B. The similarity between cDNA clones ranged from 97 to 100% with the exception of the cl.7A16 clone, which was about 90%. The similarity between the cDNA clones and probes derived from MSRV-related extracellular RNA sequences ranged from 85 to 94%. Similarly, this percentage was near 90% with the MSRV Pol region previously described (40) but was lower than 70% with the ERV-9 Pol probe.
FIG. 2.
(A) Clones isolated from a placental cDNA library first with the Ppol-MSRV probe and probes derived from the resulting clones for additional screening. The length of the clone is indicated in parentheses. Black, white, and grey boxes represent regions showing extensive homology with gag, pol, and env genes, respectively; AAAAAAAA indicates poly(A) tail; flanking striped boxes represent UTRs; and repeats are indicated by black arrowheads. (B) Percentages of similarity between the overlapping regions of placental cDNA clones and between the cDNA clones and the probes used in this study. For the cDNA clone analysis, percentages are indicated in boldface and italics for overlapping fragments larger than 1 kb and 490 bp, respectively. The smallest overlapping fragment is 205 bp long. For the comparison between cDNA clones and probes, percentages labelled with asterisks indicate a partial overlap. Probes used for the placental cDNA library screening are underlined.
With BLASTX, sequences of the different fragments showed extensive homology with Gag, Pol, and Env retroviral proteins. However gag and pol genes did not support ORFs corresponding to functional proteins. An ORF putatively coding for a retroviral envelope protein was observed on the 3′ cDNA clones (cl.24.4, cl.C4C5, and cl.PH74). This Env ORF was totally contained in the cl.PH74 cDNA clone. Flanking untranslated regions (UTRs) were observed upstream and downstream from gag and env genes, respectively, containing repeated sequences (R) as described for LTRs. The length of R was estimated as about 120 bp by comparison of the 5′ end of the 5′ most cDNA clone (cl.6A2) and the 3′ end of a polyadenylated clone (cl.PH74), both clones showing 98% similarity in their 5′ UTR overlap. Three clones contained 3′ UTRs followed by a poly(A) tail (cl.Pi5T, cl.PH74, and cl.C4C5).
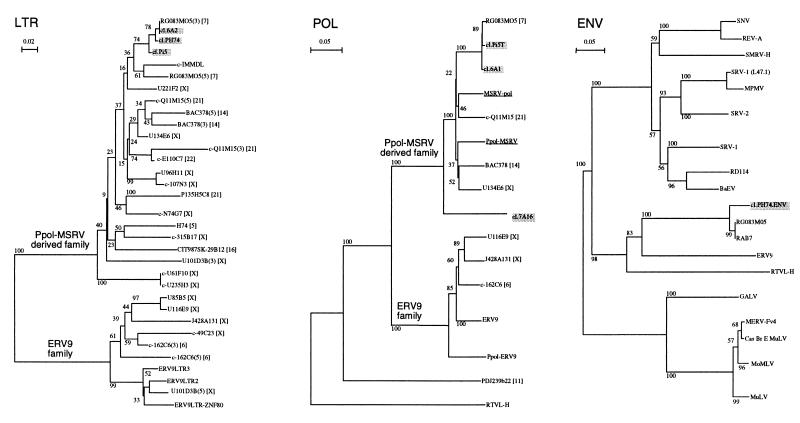
Phylogenetic analysis.
A phylogenetic analysis was performed at the nucleic acid level on 11 different subregions of the region spanned by the cDNAs and at the protein level on two different Env subregions, as described in Materials and Methods. All the trees had the same topology regardless of the region addressed. Notably, the human BAC clone RG083M05 was represented in all trees. These results were illustrated at the nucleic acid level within the most conserved regions of the LTR (R-U5) and the pol gene (RT catalytic domain) between the obtained sequences and ERV-9 and RTVL-H (Fig. 3). The trees clearly showed that those sequences described a new family related to, but distinct from, ERV-9 and even more distant from RTVL-H as highlighted by the bootstrap analysis. Those sequences were found on several chromosomes, including 5, 7, 14, 16, 21, 22, and X, with an apparent high concentration of LTR in chromosome X.
FIG. 3.
Phylogenetic analyses were performed at the nucleic acid level for LTR and pol regions and at the protein level for Env. The included sequences were selected as described in Materials and Methods. Experimental clones are indicated in shaded boxes, and MSRV sequences are underlined. LTR, POL, and ENV neighbor-joining trees based upon 123-nt LTR (nt 6 to 143 of clone cl.6A2), 575-nt pol (nt 2309 to 2933 of clone cl.6A1), and 113-aa Env (translation of nt 1852 to 2211 of clone cl.PH74) fragments are presented from left to right. Pairwise gap removal was used for the LTR tree, and all-gap removal was used for POL and ENV trees. Bootstrap values for 500 replicated trees are indicated. “c-” means that the sequence is presented in reverse orientation. Chromosomal assignments are indicated in brackets. Upstream (5) and downstream (3) LTRs are indicated when required. Sequences used in the LTR tree are as follows: clone RG083M05 (AC000064); IMMDL, DNA sequence of the human immunoglobulin D segment locus (X97051 S64822); cosmid U221F2 (Z75893); cosmid cU96H11 (Z70042); PAC 107N3 (Z75741); cosmid E110C7 (Z68223); P135H5C8, H. sapiens DNA sequence on chromosome 21 (L35660); cosmid cN74G7 (Z69715); cosmid U101D3 (Z85998); BAC clone CIT987SK-29B12 (U95738); PAC clone H74 (AC000352); cosmid 315B17 (Z73967); cosmid U61F10 (Z75895); cosmid U85B5 (Z69724); cosmid U116E9 (Z95333); human DNA sequence from clone J428A131 (Z82209); PAC 49C23 (Z93019); PAC 162C6 containing endogenous retrovirus pHE.1 (ERV-9) (Z84475); ERV-9LTR3 (M92648); ERV-9LTR2 (M92647); and ERV-9LTR-ZNF80, H. sapiens DNA for ZNF80-linked ERV-9 LTR (X83497). Additional sequences included in the POL tree are as follows: MSRV-pol reconstructed consensus (AF009668); Ppol-MSRV probe; ERV-9, HERV pHE.1 mRNA sequence (X57147, M85205, and M37638); Ppol-ERV-9 probe; PAC pDJ239b22 (AC003969 and U90583); and RTVL-H, HERV type H (D11078). Sequences appearing in the ENV tree are as follows: SNV, spleen necrosis virus (EMBL M87666); REV-A, reticuloendotheliosis virus strain A (X01455 K02537); SMRV-H, simian sarcoma virus (M23385); SRV-1 (L47.1), simian SRV-1 type D retrovirus (M11841); MPMV, simian Mason-Pfizer type D retrovirus (M12349); SRV-2, simian SRV-2 type D retrovirus (M16605); SRV-1, simian type D virus 1 (U85505); RD114, cat endogenous retrovirus RD114 env gene (X87829); BaEV, baboon endogenous virus virion (M16550); GALV, gibbon ape leukemia virus (M26927); MERV-Fv4, mouse endogenous retrovirus in Fv4 locus (M33884); Cas Br E MuLV, murine leukemia virus (Cas-Br-E MuLV) (M14702); MoMLV, Moloney murine leukemia virus (J02255, J02256, J02257, and M76668); MuLV, murine leukemia virus (M93052); and human sequences RTVL-H (NBRF B44282), ERV-9 (M85205 and M37638), and RAB7 (H. sapiens mRNA) (X93499).
Comparison at the protein level between the most conserved regions of Env retroviral proteins (immunosuppressive to transmembrane [TM] domain) resulted in trees showing a sequence distribution similar to the one observed at the nucleic acid level (Fig. 3). The two proximal coding sequences corresponded to the translated env ORF of RG083M05 and a truncated ORF located downstream from the GTP-binding protein RAB7 on the same mRNA. This group of sequences was more closely related to simian type D and reticuloendotheliosis avian retroviruses than to type C mammalian retroviruses.
Genomic complexity of the family.
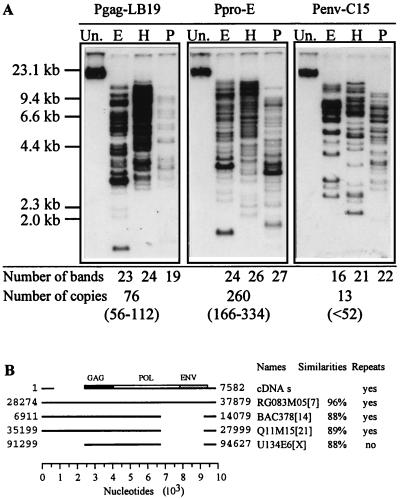
In order to characterize the complexity of this new endogenous retroviral family, we used several probes spanning the entire genome. Southern blot analysis of human DNA digested with three different restriction enzymes showed complex patterns with an apparent increase of complexity from env to gag and protease regions (Fig. 4A). A more precise determination of the copy number of each region was done by a dot blot analysis of serial dilutions of the plasmids that contained the corresponding probes (data not shown). The calculated number of copies per haploid genome (Fig. 4A) emphasized the heterogeneous distribution of the retroviral subregions. A 6- and 20-fold increase of Gag and protease signals, respectively, versus the Env signal confirmed the gradation suspected in Southern blot analysis.
FIG. 4.
(A) Southern blot analysis of human genomic DNA digested separately with EcoRI (E), HindIII (H), and PstI (P) and probed with Pgag-LB19, Ppro-E, and Penv-C15. The number of bands detected on the blot is indicated at the bottom for each restriction enzyme. The absolute copy numbers deduced from dot blot analysis, as well as confidence intervals, are also indicated at the bottom. Un., undigested. (B) Alignment of the placental cDNA clones (cDNAs; schematic representation of the 7.6-kb region defined with the overlapping cDNA clones) with the four sequences showing the higher scores with BLASTN query. Shown are human BAC clone RG083M05 (gb AC000064), human BAC378 (gb U85196 and gb AE000660), H. sapiens cosmid Q11M15 (gb AF045450), and human DNA sequence from cosmid U134E6 (EMBL Z83850). Locations of the aligned region of each clone are indicated. Chromosomal assignments are indicated in brackets. The percentages of similarity (without gaps) between the four sequences and the consensus sequence (cDNAs) are given (values for individual clones are indicated in the text). The presence of repeats at both ends of the genome is stated.
To characterize some genomic sequences at the molecular level, a BLASTN query on several databases was performed, with the placental cDNA clones. The four most significant sequence hits are depicted in Fig. 4B. They consisted of the human BAC clone RG083M05 from 7q21-7q22, the human BAC378 corresponding to Homo sapiens T-cell receptor alpha delta locus whose chromosomal location is 14q11-12, the H. sapiens chromosome 21q22.3 cosmid Q11M15, and the human DNA sequence from cosmid U134E6 on chromosome Xq22 containing NIK-like and thyroxin-binding globulin precursor. Repeated sequences were found located at both ends of three of these clones. All the cDNA sequences fell entirely within the clone RG083M05 (10.2 kb) with a similarity of about 98 to 99%, except the clone cl.7A16 (90%). However, in addition to gag, pol, and env genes, RG083M05 exhibited a 2-kb insert located just downstream from the 5′ UTR, this insert being found also in clones BAC378 and Q11M15. These two clones and a third one (U134E6) presented a strictly conserved 2.3-kb deletion just upstream from the 3′ UTR. No clone contained all three gag, pol, and env ORFs. The clone RG083M05 exhibited a 538-amino-acid ORF corresponding to a full-length envelope. The cosmid Q11M15 contained two large contiguous ORFs of 413 (frame 0) and 305 (frame +1) aa corresponding to a truncated Pol polyprotein. All together, these data suggested a quite complex family from which the RG083M05 clone could represent a genomic prototype.
RNA elements: LTR, polypurine tract, and primer binding site (PBS) characterization.
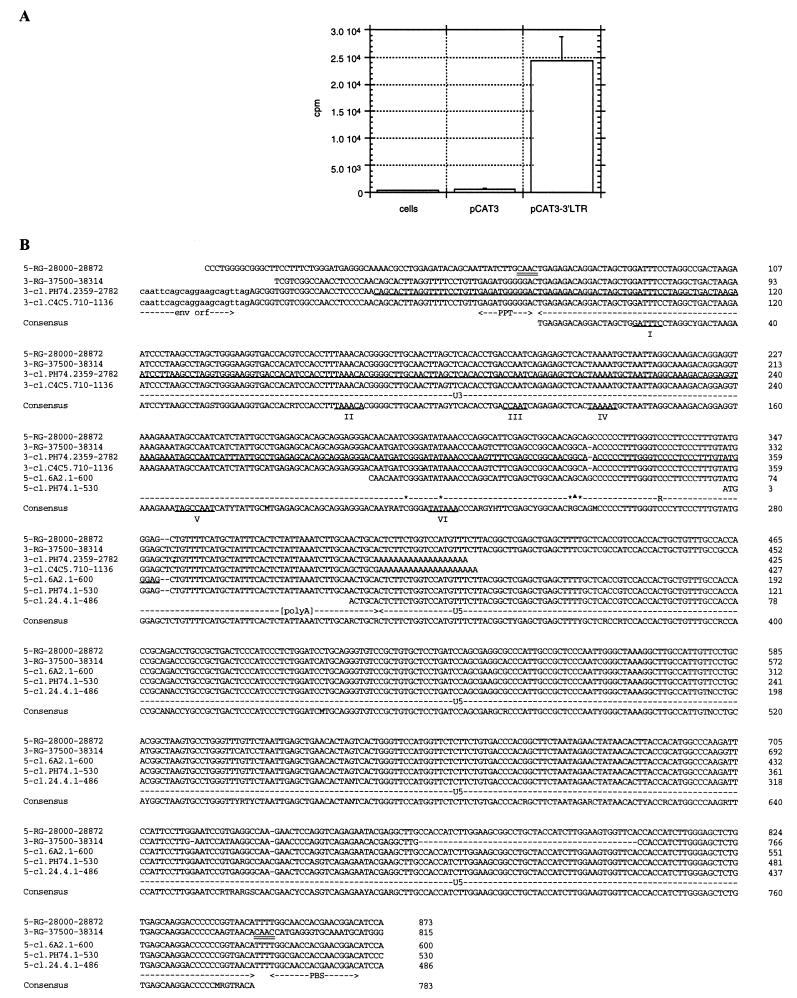
In order to identify a potential promoter downstream from the cl.PH74 putative env ORF, a predictive analysis was performed. Proscan 1.7 did not predict any obvious promoter, but Signalscan 4.05 indicated the presence of putative transcription factor binding sites, including a CCAAT box and a TATA box. The 2,364- to 2,720-nt corresponding region of the cl.PH74 clone (2,764 bp) was PCR amplified and subcloned into pCAT3 reporter vector. The CAT assay showed a significant expression level (Fig. 5A). A promoter activity was also found in BeWo human choriocarcinoma cells and Jurkat human T cells (data not shown).
FIG. 5.
Definition of a putative LTR. (A) The 2,364- to 2,720-nt region of the cl.PH74 clone, identified as a putative promoter region with Signalscan 4.05, was PCR amplified and subcloned into pCAT3 reporter vector. The resulting pCAT3-3′LTR plasmid and pCAT3 control plasmid were used to transfect HeLa cells prior to CAT activity determination. Cells, HeLa cell extract; pCAT3 and pCAT3-3′LTR, extract of HeLa cells transfected with control plasmid and promoter plasmid, respectively. (B) Alignment of 5′ and 3′ UTRs of placental experimental clones with the 5′ (5-RG-28000-28872) and 3′ (3-RG-37500-38314) repeated sequences of the 28,000- to 38,314-nt fragment of human DNA sequence RG083M05. The end of the 3′-most ORF is indicated (env orf). The tandemly repeated CAAC flanking sequences are doubly underlined on DNA sequences. The 783-bp LTR consensus sequence is positioned at the bottom. The polypurine tract (PPT) upstream from the 5′ end of the LTR and the tRNATrp PBS downstream from the 3′ end of LTR are indicated. U3, R, and U5 subparts are indicated, except for the U3-R junction. Transcription factor sites determined with Signalscan 4.05 are underlined and labelled from I to VI. The sites V and VI may correspond to CCAAT box and TATA box, respectively. The 3′ region of clone cl.PH74 used in the CAT assay is underlined on the cl.PH74 sequence. ∗ and ▴, potential cap sites obtained in 5′ RACE experiments with human placental poly(A)+ RNA and total RNA extracted from pCAT-3′LTR-transfected HeLa cells, respectively. [polyA], polyadenylation signal.
In order to characterize the retroviral LTR, the 5′ and 3′ UTRs of cDNA experimental clones were aligned with the 5′ and 3′ repeated sequences of the most proximal human DNA sequence, RG083M05. Nonretroviral flanking cellular sequences allowed us to delineate a 783-bp putative LTR (Fig. 5B). A duplicated 4-bp sequence (CAAC) was found flanking 5′ and 3′ LTRs. The characteristic TG-CA base pairs were found juxtaposed with nonretroviral sequences at each end of the integrated provirus. The 5′ end of U3 was confirmed by the presence, on the cl.PH74 and cl.C4C5 clones, of a polypurine tract located immediately upstream from U3 and 46 bp downstream from the env stop codon. Determination of the U3-R junction was quite complex. Several possible cap sites were found by 5′ RACE experiments on placental mRNA (data not shown), but surprisingly, all these sites were found included within the cl.6A2 clone (Fig. 5B). More surprisingly, the predicted TATA box fell within the R sequence defined according to the cl.6A2 clone. To simplify the analysis, a similar procedure was applied to a single U3-R region, with total RNA extracted from pCAT3-3′LTR-transfected HeLa cells (data not shown). It indicated a potential cap site downstream from the TATA box (Fig. 5B). The 3′ end of R was precisely defined according to the presence of a poly(A) tail on cl.PH74 and cl.C4C5 clones. The polyadenylation signal was found within the R region, 13 bp upstream of U5. The U5 LTR remaining sequence was found to be 455 bp long. The 3′ end of U5 was confirmed by the presence, on the cl.6A2 (gag), cl.PH74, and cl.24.4 (env) clones, of a putative PBS located 4 bp downstream. This PBS showed extensive homology with the avian retrovirus PBS (53) used by tRNATrp for minus-strand DNA synthesis (34). As the tRNATrp was not described for humans and this PBS sequence did not correspond to any other tRNA, we tentatively named this new family HERV-W.
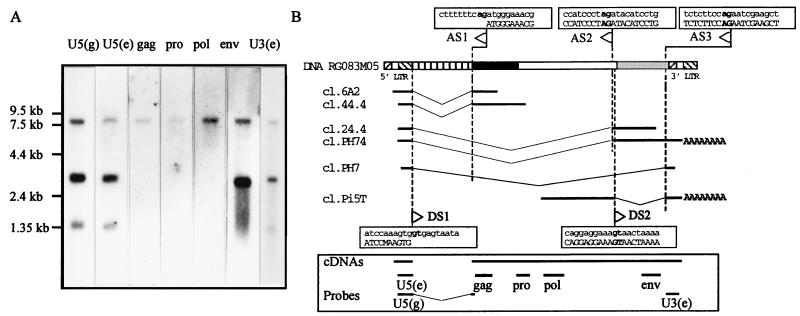
Transcription and putative splicing strategy.
The expression of the HERV-W family in placenta was analyzed by Northern blotting (Fig. 6A) with a set of probes spread over the 7.6-kb region containing the cDNA clones (Fig. 6B) and consisting of two U5 probes, U5(g) and U5(e), derived from an RU5-gag cDNA clone (cl.6A2) and a U5-env cDNA clone (cl.24.4), respectively, as well as gag, pro, pol, and env probes as described in Materials and Methods, and a U3-R probe, U3(e), derived from an env–U3-R–poly(A)+ clone (cl.C4C5). A simple pattern consisting of three bands at 8, 3.1, and 1.3 kb was observed. Probes from the gag, pro, and pol genes hybridized only to the 8-kb transcript which was also revealed by env, U5, and U3 probes and thus may correspond to a putative full-length transcript. Probe from the env gene detected both the full-length transcript and an abundant 3.1-kb transcript which also hybridized to the U5 and U3 probes. Thus, this 3.1-kb transcript exhibited the characteristic features of a singly spliced subgenomic retroviral env mRNA. The U5 and U3-R probes revealed an additional transcript of 1.3 kb, lacking detectable gag, pro, pol, and env sequences, which may result from alternative splicing or transcription from defective HERV-W provirus.
FIG. 6.
(A) Northern blot analysis of poly(A)+ RNAs from placental tissues with the U5(g) (nt 115 to 717 of cl.6A2) and U5(e) (nt 1 to 491 of cl.24.4) probe, gag (Pgag-LB19), pro (Ppro-E), pol (Ppol-MSRV), and env (Penv-C15) probes as described in Materials and Methods and the U3(e) (nt 732 to 1116 of cl.C4C5) probe. (B) Hypothetical splicing strategy. The RG083M05 prototype DNA clone as well as six experimental clones (cl.6A2 and cl.44.4, cl.24.4 and cl.PH74, cl.PH7, and cl.Pi5T) isolated from the cDNA library is depicted. The nucleotide sequences of regions overlapping putative splice sites are boxed. Nucleic acid sequences of RG083M05 and all placental cDNA clones are indicated with lowercase and capital letters, respectively. AG and GT bordering splice sites are in boldface. Splice donor (DS) and acceptor (AS) sites are indicated by rightward and leftward arrows, respectively. U3 (hatched from lower left to upper right), R (light grey), U5 (hatched from upper left to lower right), 2-kb insert (vertically striped), Gag (black), Pol (white), and Env (dark grey) are shown as boxes. The positioning of the seven probes used in the Northern blot analysis and the placental region spanned by the cDNAs (cDNAs) are illustrated at the bottom of the figure.
No trivial correspondence between the observed transcripts and the isolated cDNA clones could be found, as the 5′ end of R was not unambiguously defined. However, the cl.PH74 clone containing an env ORF and repeats at both ends was a good candidate for the 3.1-kb subgenomic mRNA. Likewise, the cl.PH7 clone which contained U5 and U3 sequences at the 5′ and 3′ ends, respectively, but no structural gene, might represent a large spliced subgenomic mRNA. In addition, if we assumed (i) that the cap site location was as defined by the 3′ LTR transfection experiment and (ii) the region spanned by the cDNAs reflects a genomic RNA organization as suggested by the Northern blot analysis, the lengths of genomic, subgenomic, and small mRNAs deduced from the cDNA clones would be 7,541, 2,812, and 1,148 nt, consistent with the 8-, 3.1-, and 1.3-kb observed transcripts, respectively.
Although no band larger than 8 kb was observed in the Northern blot, the alignment of the experimental clones with the 10.2-kb DNA sequence of the RG083M05 clone was performed in order to understand whether these RNA transcripts result from splicing events or indicate expression of defective HERV-W proviruses. This strategy permitted us to identify several apparently well-conserved splice donor and acceptor sites (Fig. 6B). The comparison of the 5′ cDNA clones cl.44.4 and cl.6A2 with the RG083M05 sequence identified a splice donor (DS1) and a splice acceptor (AS1) site, strictly flanking the 2,075-bp insertion previously observed (Fig. 4). They were found on RG083M05, just 564 and 2,640 nt, respectively, downstream from R, as defined above. The absence of a major splice donor site in the 5′ cDNA clones cl.44.4 and cl.6A2 suggested that the 8-kb apparent genomic RNA could result from a splicing strategy. The comparison of cl.PH74 and cl.PH7 clones with the RG083M05 sequence identified two putative splice acceptor sites, AS2 and AS3, located 7,338 and 9,001 bp downstream from R, respectively (Fig. 6B). AS3 was also found in the cl.Pi5T clone, which contained, in addition, a second putative donor site (DS2), 7,545 bp downstream from R and 207 bp downstream from the env acceptor site but upstream from the env ATG codon. The occurrence of DS1/AS1, DS1/AS2, and DS1/AS3 splicing events on a putative RG083M05 precursor RNA would lead to 7,491-, 2,793-, and 1,130-nt transcripts whose sizes are remarkably close to those of genomic, subgenomic, and small mRNAs deduced from the cDNA clones.
Coding capacities.
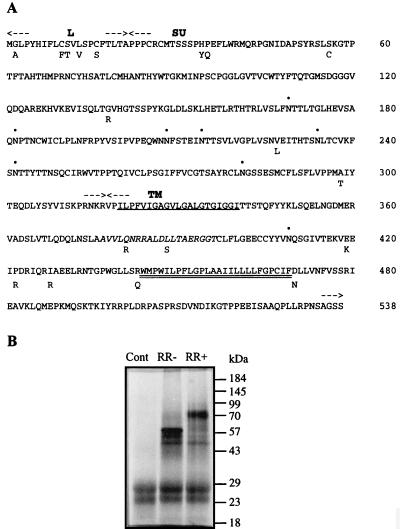
On the cl.PH74 clone, the initiation codon of the long ORF putatively encoding the envelope protein was found 227 bp downstream from the putative DS1/AS2 splice junction. It was the first ATG downstream from this junction, although not the first from the 5′ end of the subgenomic RNA as five ATGs preceding five small ORFs (less than 27 aa) were situated ahead. Nevertheless, this ATG was in a relatively favorable context (CCCATGG) although slightly different from the known (A/G)CCATGG favorable context for translation (24, 25). Figure 7A presents the amino acid sequence of a putatively functional envelope protein. This sequence was derived from three clones: cl.PH74 presented a 5′ LTR, a splice junction, a coding sequence of 538 aa, and a 3′ LTR poly(A)+; cl.24.4 presented a 5′ LTR, a splice junction, and a coding sequence of 410 aa; and cl.C4C5 presented a coding sequence of 242 aa, a 3′ LTR, and poly(A)+. This ORF exhibited the characteristic features of the precursor polypeptide of retroviral envelope proteins. It presented a leader peptide at the amino terminus and a carboxy-terminal hydrophobic segment which could anchor the protein in the membrane. A furin cleavage site (RNKR) separated the two characteristic subdomains consisting of the surface protein (SU) and the transmembrane protein (TM). The TM contained, in addition to the membrane-spanning segment, a hydrophobic fusion domain and a putative immunosuppressive region homologous to the immunosuppressive p15E retroviral peptide conserved among murine, feline, and human retroviruses (11). The SU and TM regions contained seven and one potential glycosylation site, respectively. Furthermore, these potential glycolysation sites are conserved among the three considered clones and in the RG083M05 clone, which might code for the same protein too. An in vitro transcription-translation assay has been performed on this env gene. The results are shown in Fig. 7B. The measured molecular mass of the precursor is 60 kDa, in agreement with the calculated molecular mass of 59,565 Da of the Env ORF based on its amino acid composition. Glycosylation of the Env precursor was observed, consistent with the prediction of carbohydrate addition sites. The measured molecular mass of the glycosylated protein is 80 kDa.
FIG. 7.
(A) Sequence of the putative HERV-W envelope polypeptide derived from three different placental cDNA clones. The leader peptide (L), the surface protein (SU), and the transmembrane protein (TM) are indicated between arrows. The hydrophobic fusion peptide and the carboxy-transmembrane region are singly and doubly underlined, respectively. The immunosuppressive region is indicated by italics. Potential glycosylation sites are indicated by dots. Divergent amino acids are indicated on the lower line. (B) In vitro transcription-translation assay for HERV-W envelope gene product, without (RR−) or with (RR+) canine microsomes, and control (Cont) without template.
Two initiation codons potentially directing the synthesis of 52-aa (ORF1) and 48-aa (ORF2) peptides were found 22 and 95 bp downstream from the putative AS3 splice acceptor site, respectively. Both ATGs were not in a favorable context. ORF1 consisted of the carboxy-terminal part of Env, and ORF2 was translated in a different but overlapping frame. No obvious homology was found by using BLAST query. However, with an LFASTA query on the restricted Retroviridae subdatabase, ORF1 and ORF2 showed about 35% identity with Rex from primate and human T-lymphotropic virus and Tat from simian immunodeficiency virus, respectively (data not shown). Although unusual, the presence of such regulatory proteins in HERVs has been described elsewhere for HERV-K10 (33).
DISCUSSION
Here we report the molecular characterization of HERV-W, a new multicopy family of HERVs whose expression in healthy tissues seemed restricted to the placenta. The phylogenetic trees within the Pol region showed that the HERV-W family is related to ERV-9 and RTVL-H families and thus belongs to the class I endogenous retroviruses (5). Phylogenetic analysis of the env ORF showed that it was closer to simian type D and avian reticuloendotheliosis retroviruses than to murine type C retroviruses. The homologies within the pol and env genes with the murine type C and simian type D retroviruses, respectively, suggest a chimeric genome structure as described for baboon endogenous virus (22). Based on the size criteria, such a chimerism seemed to exist within the LTR: the 247-nt U3 and the 79- to 81-nt R elements were comparable to avian or type D retrovirus U3 and mammalian type C R elements, respectively, although the 410- to 455-nt U5 element remained unclassified as unusually long (57). The bush-like topology that is observed in the phylogenetic trees for the HERV-W and ERV-9 sequences suggests that in both families most elements were fixed in the germ line during a relatively short period of evolution and probably derive from one or a few active elements. The phylogenetic tree, supported by high bootstrap values, shows that ERV-9 and HERV-W families derive from two independent bursts of insertions. Thus, the active element(s) at the origin of the HERV-W family is distinct from the one(s) from which the ERV-9 family is derived. Moreover, HERV-W PBS is predicted to use tRNATrp, whereas ERV-9 probably uses tRNAArg. Finally, members of the HERV-W family are expressed in the placenta, whereas we have not detected ERV-9 RNAs in this tissue. However, by RNase protection assay, the detection of protected fragments smaller than the expected one suggested that ERV-9-related sequences could be expressed at a low level in placenta (26).
The persistence of flanking duplicated sequences generated during the integration process suggests either that a selection process persists within the LTR or that the integration occurred recently. The 5′ and 3′ LTRs of RG083M05 HERV-W DNA prototype showed 4% divergence and 6% gaps. This is comparable to the 0.2 to 4.3% divergence described for HERV-K, but smaller than the 5 to 12% divergence described for a number of cloned proviruses (5). Given an evolution rate of 3.5 × 10−9 per site per year (30), the 4% divergence corresponds to a relatively recent integration event 6 million years ago. The HERV-W distribution in different primates will be studied to determine the first introduction into the germ line.
Like other HERVs, most (if not all) current members of the HERV-W family are defective for replication, due to mutations that disrupt one or more of the gag, pol, and env ORFs (38, 58). The observation of more gag-pro than env sequences in the human genome may reflect the loss of the env gene. Several genomic clones seemed to reflect such a loss, which appears to be a common feature among endogenous elements (5). However, a complete env ORF was observed on the cl.PH74 placental cDNA clone and the genomic RG083M05 clone which was nevertheless defective for replication. These results are not compatible with the hypothesis that particles in cell culture supernatants of MS patients (42, 44) or the presence of specifically packaged mRNA in plasma of MS and RA patients (17, 40) may result from a replication-competent HERV. However, although no replication-competent genome has been observed in the databases, it cannot be excluded that a nonidentified one exists. Furthermore, no HERV single genetic unit has yet been shown experimentally to be a source of retroviral particles (58), even in the HERV-K family, which corresponds to the HTDV particles (6, 32). On the other hand, a trans-complementation process between distinct but individually defective loci cannot be excluded. Indeed, queries on nucleic acid databases showed that some large regions were conserved and confirmed a widespread distribution on numerous chromosomes. To address these two hypotheses, a new approach will be developed to detect potential coding sequences, irrespective of the existence of a putative single genetic element. A procedure based on probe hybridization coupled with in vitro transcription-translation will be applied to isolated human chromosomes in heterokaryon somatic hybrid cells.
Although HERV-W is a quite complex family at the genomic level, a relatively simple pattern of mRNA expression was observed by Northern blot analysis, consisting of three major bands of 8, 3.1, and 1.3 kb. This pattern resembles the one of lentiviruses or oncoviruses such as mouse mammary tumor virus and human T-cell leukemia virus (45). It is also similar to the one described for ERV-9 in undifferentiated embryonal carcinoma cells (28) and the HTDV/HERV-K family in teratocarcinoma cell lines (32), in which three to four transcripts were derived from a single locus by alternative usage of splicing signals. Whether the observed transcripts resulted from a splicing strategy of one or several genetic entities or originated from different defective entities remains unresolved. Due to (i) the overall behavior of U5, gag, pro, pol, env, and U3 probes in Northern blot analysis and (ii) the identification of conserved splice sites on the RG083M05 DNA prototype and the cDNA clones, it is probable that the three observed bands represent genomic, subgenomic, and large spliced mRNAs. The situation resulting from the proposed splicing strategy was quite unusual as the genomic mRNA would result from a splicing event. Furthermore, no 9.6-kb precursor mRNA was detectable by Northern blotting, which may reflect a very short half-life of such a transcript. A query of the EST database with the 2-kb insert of the RG083M05 clone showed only smaller internal fragments, suggesting either a complex splicing strategy or the presence of shorter unidentified genomes. The existence of an alternative splicing strategy was confirmed by the isolation of clone cl.Pi5T, for which no trivial corresponding transcript was revealed on the Northern blot; thus, this clone may represent a poorly expressed mRNA. Nevertheless, we cannot exclude that some if not most of the placental cDNA clones, although highly homologous, were obtained from different genetic entities. The most divergent clone, Pol cl.7A16, may reflect such a situation. The contradiction between the 5′ end of the cl.6A2 clone and the positioning of the predicted TATA box may also result from such a context. This clone may be derived from transcription driven by a nonretroviral promoter upstream from a complete or altered LTR and an extension through the R polyadenylation site as described for HERV-R (ERV-3) (20). Taken as a whole, this suggests that the complex HERV-W family observed at the DNA level could include well-conserved genomic elements (subset) coexpressed in the placenta.
According to the results of the Northern blot and the RNA dot blot analyses, a significant LTR functionality among healthy tissues seemed restricted to the placenta. Thus, the expression of HERV-W could be regulated in a hormone-dependent manner as suggested for ERV-9 with the ZNF80 zinc finger gene (15) or for HERV-R with the H-plk gene (21). However, by more sensitive techniques, it may be possible to find much lower expression of HERV-W in nonplacental tissue. Such related mRNA sequences were found in brains of healthy and MS individuals (8, 29), by an RT-PCR-based method, and a complex mRNA expression was observed in various tissues by Northern blotting with the amplified product as a probe (29). This discrepancy with our experimental data, obtained with six probes spanning all the regions of a retroviral genome, may result from different experimental conditions, including the length of the probe and the hybridization stringency. Furthermore, the EST query showed a significant concentration of HERV-W expression in fetal liver-spleen, in addition to placenta. As the RNA dot blots were negative for both fetal liver and fetal spleen, one may suspect the influence of the differentiation stage or a low level of expression. Partial LTR tags were found in other fetal, adult, and tumoral tissues, most in the opposite direction. This may reflect expression driven by solitary LTRs or nonretroviral promoters. Interestingly, the 85 to 94% similarities observed between the placental cDNA clones and the probes derived from extracellular RNA isolated in a pathological context compared with the 90 to 100% similarities obtained between overlapping placental cDNAs indicate that at least some of the probes were derived from transcriptional units distinct from those expressed in the placenta. This interpretation was supported by the reproducible isolation of env interrupted ORFs from extracellular mRNA in MS and RA patients (22b) versus an env-containing ORF from the placental cDNA library screening. This suggests a placenta-restricted expression of an HERV-W subset versus a different or probably broader HERV-W expression in a pathological context (or in other tissues). In agreement with this, the expression in different tissues at different times of different subsets of one given family of elements was described, e.g., the expression of HTDV/HERV-K (27), ERV-9 (31), and HERV-R (13, 19) mRNAs differs quantitatively and qualitatively with respect to the tissue considered. In pathological situations, silent LTRs could be reactivated by several factors such as virus, e.g., members of the herpesvirus family (3, 9, 18), or by local immune activation.
The finding in placental cDNA clones of a large ORF coding for a putatively functional envelope protein suggests some physiological functions related to pregnancy, such as a role in the creation of the syncytiotrophoblast layer of the placenta (7, 55) and a role in suppressing the maternal immune response against the fetal allograft (7, 55, 56) as suggested for the envelope protein of HERV-R. In pathological situations, an HERV Env might protect against exogenous retroviral infections by a receptor interference mechanism (4) or alter the local cellular immunity via, for example, a superantigen-encoded region, as was recently proposed for type I diabetes (insulin-dependent diabetes mellitus) (14). Conversely, HERV-W expression may represent only a consequence of physiological or pathological events. Whatever the situation is, the finding of HERV-W sequences both in a physiological tolerogenic immune context such as pregnancy and in autoimmune diseases such as MS and RA deserves further investigations.
ACKNOWLEDGMENTS
We thank F. L. Cosset (UCB-Lyon I, Lyon, France), C. Guillon (Erasmus University, Rotterdam, The Netherlands), and Doran Spencer for critical reading of the manuscript. We also thank C. Gautier and all the staff of the Biometry Laboratory (UCB-Lyon I, Lyon, France) for helpful discussions and assistance with nucleic acid analysis and F. Penin and C. Gourgeon (IBCP, Lyon, France) and F. Horn (EMBL, Heidelberg, Germany) for helpful assistance in protein analysis. Most of the sequence analyses were performed in the PBIL (Pôle Bio-Informatique Lyonnais) facilities (http://pbil.univ-lyon1.fr). We are grateful to Florence Komurian-Pradel and Glaucia Parhanos-Baccala for providing C15 and LB19 and E clones, respectively.
J.-L.B. was supported by a doctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche and bioMérieux.
REFERENCES
- 1.Abdeddaim S. Proceedings of the IEEE International Joint Symposia on Intelligence and Systems, 4 to 5 November 1996. N.Mex: Los Alamos; 1996. Fast and sound two-step algorithms for multiple alignment of nucleic sequences; pp. 4–11. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bergström T, Andersen O, Vahlne A. Isolation of herpes simplex virus type 1 during first attack of multiple sclerosis. Ann Neurol. 1989;26:283–285. doi: 10.1002/ana.410260218. [DOI] [PubMed] [Google Scholar]
- 4.Best S, Le Tissier P R, Stoye J P. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 1997;5:313–318. doi: 10.1016/S0966-842X(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 6.Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 7.Boyd M T, Bax C M R, Bax B E, Bloxam D L, Weiss R A. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology. 1993;196:905–909. doi: 10.1006/viro.1993.1556. [DOI] [PubMed] [Google Scholar]
- 8.Brahic M, Bureau J F. Multiple sclerosis and retroviruses. Ann Neurol. 1997;42:984–985. doi: 10.1002/ana.410420625. [DOI] [PubMed] [Google Scholar]
- 9.Challoner P B, Smith K T, Parker J D, MacLeod D L, Coulter S N, Rose T M, Schultz E R, Bennett J L, Garber R L, Chang M, Schad P A, Stewart P M, Nowinski R C, Brown J P, Burmer J P. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci USA. 1995;92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Cianciolo G J, Copeland T D, Oroszlan S, Snyderman R. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope protein. Science. 1985;230:453–455. doi: 10.1126/science.2996136. [DOI] [PubMed] [Google Scholar]
- 12.Claverie J M, States D J. Information enhancement methods for large scale sequence analysis. Comput Chem. 1993;17:191–201. [Google Scholar]
- 13.Cohen M, Kato N, Larsson E. ERV3 human endogenous provirus mRNAs are expressed in normal and malignant tissues and cells, but not in choriocarcinoma tumor cells. J Cell Biochem. 1988;36:121–128. doi: 10.1002/jcb.240360203. [DOI] [PubMed] [Google Scholar]
- 14.Conrad B, Weissmahr R N, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 15.Di Cristofano A, Strazzullo M, Longo L, La Mantia G. Characterization and genomic mapping of the ZNF80 locus: expression of this zinc-finger gene is driven by a solitary LTR of ERV9 endogenous retroviral family. Nucleic Acids Res. 1995;23:2823–2830. doi: 10.1093/nar/23.15.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galtier N, Gouy M, Gautier C. SEA VIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 17.Gaudin P, Perron H, Favre G, Mandrand B, Juvin R, Marcel F, Beseme F, Bedin F, Mallet F, Mougin B, Fauconnier J, Seigneurin J M, Phelip X. Detection of retrovirus RNA in plasma from rheumatoid arthritis. Arthritis Rheum. 1997;40:S245. [Google Scholar]
- 18.Haahr S, Sommerlund M, Christensen T, Jensen A W, Hansen H J, Moller-Larsen A. A putative new retrovirus associated with multiple sclerosis and the possible involvement of Epstein-Barr virus in this disease. Ann N Y Acad Sci. 1994;724:148–156. doi: 10.1111/j.1749-6632.1994.tb38903.x. [DOI] [PubMed] [Google Scholar]
- 19.Kato N, Larsson E, Cohen M. Absence of expression of a human endogenous retrovirus is correlated with choriocarcinoma. Int J Cancer. 1988;41:380–385. doi: 10.1002/ijc.2910410310. [DOI] [PubMed] [Google Scholar]
- 20.Kato N, Pfeifer-Ohlsson S, Kato M, Larsson E, Rydnert J, Ohlsson R, Cohen M. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J Virol. 1987;61:2182–2191. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato N, Shimotohno K, VanLeeuwen D, Cohen M. Human proviral mRNAs down regulated in choriocarcinoma encode a zinc finger protein related to Krüppel. Mol Cell Biol. 1990;10:4401–4405. doi: 10.1128/mcb.10.8.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato S, Matsuo K, Nishimura N, Takahashi N, Takano T. The entire nucleotide sequence of baboon endogenous virus DNA: a chimeric genome structure of murine type C and simian type D retroviruses. J Genet. 1987;62:127–137. [Google Scholar]
- 22a.Komurian-Pradel, F. Unpublished data.
- 22b.Komurian-Pradel, F. Personal communication.
- 23.Kozak C A, Ruscetti S. Retroviruses in rodents. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 405–481. [Google Scholar]
- 24.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. A consideration of alternative models for the initiation of translation in eukaryotes. Crit Rev Biochem Mol Biol. 1992;27:385–402. doi: 10.3109/10409239209082567. [DOI] [PubMed] [Google Scholar]
- 26.La Mantia G, Maglione D, Pengue G, Di Cristofano A, Simeone A, Lanfrancone L, Lania L. Identification and characterization of novel human endogenous retroviral sequences preferentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 1991;19:1513–1520. doi: 10.1093/nar/19.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Mantia G, Pengue G, Maglione D, Pannuti A, Pascucci A, Lania L. Identification of new human repetitive sequences: characterization of the corresponding cDNAs and their expression in embryonal carcinoma cells. Nucleic Acids Res. 1989;17:5913–5922. doi: 10.1093/nar/17.15.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lania L, Di Cristofano A, Strazzullo M, Pengue G, Majello B, La Mantia G. Structural and functional organization of the human endogenous retroviral ERV9 sequences. Virology. 1992;191:464–468. doi: 10.1016/0042-6822(92)90211-7. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre S, Hubert B, Tekaia F, Brahic M, Bureau J F. Isolation from human brain of six previously unreported cDNAs related to the reverse transcriptase of human endogenous retroviruses. AIDS Res Hum Retroviruses. 1995;11:231–237. doi: 10.1089/aid.1995.11.231. [DOI] [PubMed] [Google Scholar]
- 30.Li W H, Graur D. Fundamentals of molecular evolution. Sunderland, Mass: Sinauer Associates, Inc.; 1991. pp. 67–73. [Google Scholar]
- 31.Lindeskog M, Medstrand P, Blomberg J. Sequence variation of human endogenous retrovirus ERV9-related elements in an env region corresponding to an immunosuppressive peptide: transcription in normal and neoplastic cells. J Virol. 1993;67:1122–1126. doi: 10.1128/jvi.67.2.1122-1126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Mueller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retrovirus HTDV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallet F, Cros P, Mandrand B. Enzyme-linked oligosorbent assay for detection of PCR-amplified HIV-1. In: Becker Y, Darai G, editors. PCR: protocols for diagnosis of human and animal virus diseases. Berlin, Germany: Springer-Verlag; 1995. pp. 19–28. [Google Scholar]
- 36.Morgenstern B, Dress A, Werner T. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc Natl Acad Sci USA. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmarini M, Cousens C, Dalziel R G, Bai J, Stedman K, DeMartini J C, Sharp J M. The exogenous form of Jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Parhanos-Baccala, G. Unpublished data.
- 38.Patience C, Wilkinson D A, Weiss R A. Our retroviral heritage. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 39.Pearson W R. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 40.Perron H, Garson J A, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke P W, Voisset C, Blond J L, Lalande B, Seigneurin J M, Mandrand B The Collaborative Research Group on Multiple Sclerosis. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc Natl Acad Sci USA. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perron H, Geny C, Genoulaz O, Pellat J, Perret J, Seigneurin J M. Antibody to reverse transcriptase of human retroviruses in multiple sclerosis. Acta Neurol Scand. 1991;84:507–513. doi: 10.1111/j.1600-0404.1991.tb05004.x. [DOI] [PubMed] [Google Scholar]
- 42.Perron H, Geny C, Laurent A, Mouriquand C, Pellat J, Perret J, Seigneurin J M. Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particules. Res Virol. 1989;140:551–561. doi: 10.1016/s0923-2516(89)80141-4. [DOI] [PubMed] [Google Scholar]
- 43.Perron H, Gratacap B, Lalande B, Genoulaz O, Laurent A, Geny C, Mallaret M, Innocenti P, Schuller E, Stoebner P, Seigneurin J M. In vitro transmission and antigenicity of a retrovirus isolated from a multiple sclerosis patient. Res Virol. 1992;143:337–350. doi: 10.1016/s0923-2516(06)80122-6. [DOI] [PubMed] [Google Scholar]
- 44.Perron H, Lalande B, Gratacap B, Laurent A, Genoulaz O, Geny C, Mallaret M, Schuller E, Stoebner P, Seigneurin J M. Isolation of a retrovirus from patients with multiple sclerosis. Lancet. 1991;337:862–863. doi: 10.1016/0140-6736(91)92579-q. [DOI] [PubMed] [Google Scholar]
- 45.Petropoulos C. Retroviral taxonomy, protein structures, sequences, and genetic maps. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 757–805. [Google Scholar]
- 46.Prestridge D S. SIGNALSCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- 47.Prestridge D S. Predicting polII promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- 48.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Shih A, Misra R, Rush M G. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J Virol. 1989;63:64–75. doi: 10.1128/jvi.63.1.64-75.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonnhammer E L L, Durbin R. A workbench for large-scale sequence homology analysis. CABIOS. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- 52.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoraval P, Savatier P, Xiao J H, Mallet F, Samarut J, Verdier G, Nigon V. Partial nucleotide sequence of the avian erythroblastosis virus (AEV ES4) Nucleic Acids Res. 1987;15:9612. doi: 10.1093/nar/15.22.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuke P W, Perron H, Bedin F, Beseme F, Garson J A. Development of a pan-retrovirus detection system for multiple sclerosis studies. Acta Neurol Scand. 1997;95:16–21. doi: 10.1111/j.1600-0404.1997.tb08145.x. [DOI] [PubMed] [Google Scholar]
- 55.Venables P J W, Brookes S M, Griffiths D, Weiss R A, Boyd M T. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology. 1995;211:589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 56.Villareal L P. On viruses, sex, and motherhood. J Virol. 1997;71:859–865. doi: 10.1128/jvi.71.2.859-865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 58.Wilkinson D A, Mager D L, Leong J A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]