Abstract
Background
ESBL-producing Escherichia coli (ESBL-Ec) is considered a key indicator for antimicrobial resistance (AMR) epidemiological surveillance in animal, human and environment compartments. There is likelihood of ESBL-Ec animal–human transmission but proof of cross-compartment transmission is still unclear.
Objectives
To characterize ESBL-Ec genetic similarity in various compartments (humans, animals and environment) from a rural area of Madagascar.
Methods
We collected ESBL-Ec isolates prospectively from humans, animals and the environment (water) between April and October 2018. These isolates were subject to WGS and analysed with cutting-edge phylogenomic methods to characterize population genetic structure and infer putative transmission events among compartments.
Results
Of the 1454 samples collected, 512 tested positive for ESBL-Ec. We successfully sequenced 510 samples, and a phylogenomic tree based on 179 365 SNPs was produced. Phylogenetic distances between and amongst compartments were indistinguishable, and 104 clusters of recent transmission events between compartments were highlighted. Amongst a large diversity of ESBL-Ec genotypes, no lineage host specificity was observed, indicating the regular occurrence of ESBL-Ec transfer among compartments in rural Madagascar.
Conclusions
Our findings stress the importance of using a phylogenomic approach on ESBL-Ec samples in various putative compartments to obtain a clear baseline of AMR transmissions in rural settings, where one wants to identify risk factors associated with transmission or to measure the effect of ‘One Health’ interventions in low- and middle-income countries.
Introduction
Resistance to antimicrobial compounds (AMR) has evolved in bacterial species in response to the biosynthesis of these molecules by bacteria, fungi or plants present in their ecosystems.1 The genetic elements—antimicrobial-resistance genes (ARG)—conferring resistance have seen their selective advantage rise to an unprecedented level since humanity harnessed antibiotics in its pharmacopoeia: widespread antimicrobial use for treatment and prophylaxis of bacterial-related disease in clinical contexts, husbandry and agriculture has mechanistically increased the prevalence of antimicrobial resistant bacteria (ARB).2 Infections by ARB threaten to become one of the most critical public health issues in the near future.3 Although the extent of this threat is not accurately predictable,4 current figures on the prevalence and mortality related to AMR are already worthy of concern. Moreover, evidence of resistance transmission between different compartments of our ecosystems is accumulating.5 Circulation within and between human, animal and environment ecosystems can occur via transmission of ARB through direct contacts between organisms or via a vast array of dissemination pathways, including shared water sources, sewage, manure, soils, aerosols and pollution particles, and meat or plant consumption.6 Thus, many national and international stakeholders have recognized the urgency of addressing AMR in a concerted ‘One Health’ manner, involving the collaborative effort of multiple health science professions to attain optimal health for people, animals, plants and our environment.7
Escherichia coli is a leading cause of infections worldwide in hospitals and the community and is frequently found in asymptomatic carriers.8 The bacterium colonizes the gut of vertebrates and is ubiquitous in soil, plants and water.9 Cephalosporin resistance mediated by ESBLs has proliferated in E. coli since 2000 and globally has reached critical levels of prevalence.10 As such, ESBL-producing E. coli (ESBL-Ec) is considered a threat in healthcare11 and a key indicator for AMR trends using One-Health surveillance approaches.12,13 Epidemiology and transmission of ESBL-Ec have been intensively assessed and described within compartments such as hospitals,14,15 community,16,17 animals,18,19 food20,21 and the environment,22,23 although studies performed simultaneously amongst several such compartments remain scarce. Recent studies performed in Kenya,24 Réunion Island,25 the UK26 and the Netherlands27 have revealed distinct host-adapted ESBL-Ec lineages circulating with infrequent interspecies transmission. On the other hand, ESBL-Ec were hypothesized to disseminate from animals to the community in South-East Asia28,29 and India30 although the extent of such transmission remains highly uncertain. More recently, in a study examining the distribution of AMR enteric bacteria amongst people, animals and the environment in Tanzania, Subbiah et al.31 reported for the first time a lack of association between bacterial and host distributions. Such holistic approaches connecting the three compartments (humans, animals and the environment) in time and space are still scarce, particularly in low- and middle-income countries (LMIC), in which the threat is of particular concern.32
Herein, we aimed to characterize ESBL-Ec genomic diversity within and between human, animal and environmental compartments in a suburban rural area of Madagascar using WGS. We analysed bacterial population structure through phylogenetic reconstruction of 510 ESBL-Ec and assessed the diversity of their antibiotic resistance genes and accessory genomes in the three compartments. At such a scale, characterization of the genetic relatedness of ESBL-Ec allowed us to investigate different risk factors that might contribute to the transmission of ESBL-Ec in humans in Madagascar.
Methods
Study design, participants and survey
We implemented a cross-sectional population-based study from April to October 2018 (dry season), in Andoharanofotsy, Madagascar. Andoharanofotsy township is a rural area 12 km from Antananarivo, the capital of Madagascar. This township covers 7.4 km² and has around 60 000 inhabitants (census, 2020). Andoharanofotsy is composed of eight fokontanys (i.e. baseline administrative units), which are small districts of about 0.90 km² (Andoharanofotsy fokontany areas range from 0.4 km² to 1.5 km²).
We enrolled households owning at least three different animal species. A list of eligible households was compiled by the fokontany leaders and local healthcare workers before at least four households were randomly selected within each fokontany. Seventy households were visited at 7 AM before household members leave home and animals are released (e.g. pasture for cows). In each household, all consenting human individuals and all animals present (livestock and pets) were sampled using swabs. Children under 2 years old were excluded from the survey. When possible, 500 mL of humans’ and animals’ drinking water (if different) were also sampled resulting in 1368 samples (Figure 1) taken from 11 ‘hosts’ (human, horse, cat, cattle, chicken, dog, duck, goose, turkey, pig and water) within three ‘compartments’ (human, animal, environment). After sampling, all swabs and water samples were immediately maintained at 4°C and laboratory analyses done the same day.
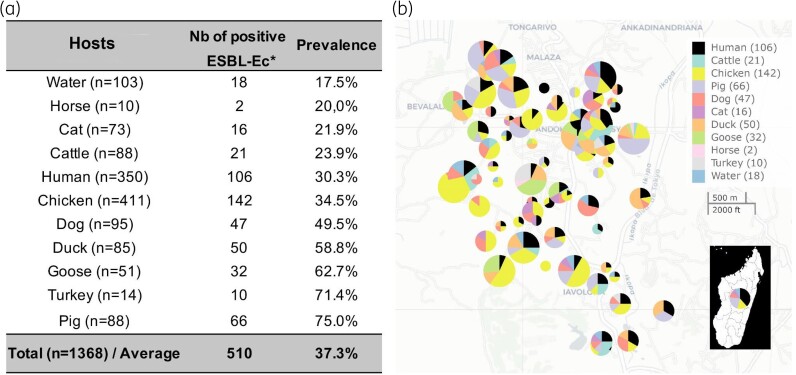
Figure 1.
Sampling metrics and prevalence of ESBL-Ec amongst hosts in a suburban rural zone of Antananarivo, Madagascar. (a) Total number of sampled individuals with observed prevalence of ESBL-Ec. (b) Geographical location of households (pies) with detected ESBL-Ec. Pie size is proportional to the number of samples collected. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
In addition, we performed a survey within each sampled household. We compiled a set of variables with the potential to be associated with selection and/or transmission of AMR bacteria in humans, animals and the environment (e.g. life traits of humans, recent hospitalization events and/or antimicrobial intake, caring of animals in households, husbandry practices, water and waste management; see File S2, available as Supplementary data at JAC Online).
This study was conducted in accordance with the Malagasy law and approved by the ethical committee on biomedical human research (Comité d´Ethique de la Recherche Biomédicale de Madagascar) under the reference N° 031-MSANP/CERBM.
Characterization of ESBL-Ec
Faecal swabs were suspended in LB broth (bioMérieux SA, Marcy l’Etoile, France) and incubated for 24 h at 35 ± 2°C with shaking. Then 100 µL of the enriched suspension was directly streaked onto selective chromogenic agar plates (CHROMagar ESBL; CHROMagar, Paris, France) and incubated overnight at 35 ± 2°C under aerobic conditions. Water samples were filtered onto a 0.45 µm membrane filter and directly cultured on selective chromogenic plates.
All presumptive ESBL-producer morphotypes were subcultured individually on LB agar plates and bacterial species identified using MALDI-TOF MS (Bruker Daltonics, Breme, Germany). Antimicrobial susceptibility testing was performed on one E. coli isolate according to the standard disc methods described in the 2015 ‘Comité de l'Antibiogramme de la Société Française de Microbiologie’ (CASFM)-EUCAST guidelines.33 Discs soaked with 20 µg amoxicillin, 75 µg ticarcillin, 75–10 µg ticarcillin/clavulanate, 20–10 µg amoxicillin/clavulanate, 10 µg ceftazidime, 30 µg cefoxitin, 30 µg cefalotin, 30 µg cefepime, 30 µg cefuroxime, 10 µg imipenem, 10 µg ertapenem, 5 µg ciprofloxacin, 10 µg gentamicin, 30 µg aztreonam, 5 µg cefotaxime and 30 µg nalidixic acid were tested. The presence of ESBL enzymes was confirmed by synergy of cefotaxime, ceftazidime and cefepime with amoxicillin/clavulanate or ticarcillin/clavulanic acid.
WGS
All ESBL-Ec were selected for WGS. DNA extraction was performed using the Cador Pathogen Extraction Kit (INDICAL Bioscience) on the Qiacube HT (QIAGEN, France) from 5 mL of liquid cultures grown overnight at 37°C in LB broth medium, following the manufacturer’s protocol for Gram-negative bacteria. DNA quantity and purity were assessed by using the Nanodrop 2000/200C (Thermo Fisher Scientific, Waltham, MA, USA). Library preparation was performed by the Mutualized Platform for Microbiology (Paris, France) using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and sequencing was performed on a NextSeq 500 platform (Illumina) using 2 × 150 bp runs. FqCleaner version 3.0 was used to eliminate adaptor sequences, reduce redundant or overrepresented reads, correct sequencing errors, merge overlapping paired reads and discard reads with Phred scores (measure of the quality of identification of nucleobases generated by automated DNA sequencing) <20.
Core genome analyses
Core genome analyses were performed by mapping the reads to the E. coli O157:H7 Sakai strain complete reference genome (NC_002695.1) using the ‘very-sensitive’ option of Bowtie2 aligner.34 PCR duplicates were removed using Picard tools (http://broadinstitute.github.io/picard/). SNPs were called with GATK UnifiedGenotyper35 and conserved only if: (i) the proportion of high-quality bases supporting the call was >90%; (ii) the coverage was >15; (iii) the mapping quality of reads was >20; and (iv) the distance from another SNP was >10 bp. Consensus bacterial genomes were then constructed by introducing high-confidence SNPs within the reference genome and replacing both filtered-out variants and uncovered sites (depth = 0) by ‘N’s. We used Gubbins36 to detect regions acquired via horizontal gene transfers and excluded them to generate a recombination-free SNP alignment from which a maximum likelihood phylogeny was constructed using RAxML 8.2.437 with a rapid bootstrap analysis, general time-reversible model of evolution with a four rate categories γ distribution (GTRGAMMA) and 1000 iterations. The tree was rooted using an Escherichia fergusonii strain isolated within the course of this study (but excluded from other analyses). Visualization of the phylogenetic tree, along with metadata was performed using custom scripts sourcing the ape38 and ggtree39 packages of the R software environment.40 STs and phylogroups were inferred for each strain using stringmlst41 and ClermonTyping42 software, respectively. Non-random distribution of STs and phylogroups amongst hosts was assessed using the chi-square test with simulated P values using the chisq.test R function. Pairwise genetic distances between each sequenced strain were computed using the ‘distTips’ function of the adephylo R package.43 Distributions of distances between strain pairs sampled ‘between’ and ‘within’ compartments, fokontanys or households were compared with Mann–Whitney U tests.
ESBL-Ec transmission clusters
Transmission clusters were inferred using a phylogenetic clustering tool designed to negate the need for arbitrarily defined cluster divergence thresholds.44 Requiring only the phylogenetic tree as input, Phydelity infers putative transmission clusters through the identification of groups of sequences that are more closely related than the ensemble distribution under a statistically principled framework. For each transmission cluster, we computed the number of SNPs between each pair of samples composing the cluster and the mean and maximum observed values. Because the total genetic diversity in our dataset was very high (mean number of SNPs between samples was 2914), Phydelity could group samples with a significant divergence in the same ‘transmission’ cluster. Thus, we filtered out transmission clusters harbouring a pair of samples with the number of SNPs >20 for analysis of their composition.45 Composition of transmission clusters including humans was further analysed statistically. We used multinomial tests to compare host composition of transmission clusters with random distributions based on (i) host frequency in the global dataset and (ii) host frequency in the household(s) where the cluster was observed.
Mobilome and resistome analysis
De novo assembly of Illumina reads was performed using Unicycler46 with the option–normal for balance between completeness and reliability. Each contig was then analysed with the mob-typer component of the MOB suite software47 to retrieve putative loci of origin of replication, relaxase and incompatibility groups with a BLAST-based approach, followed by a clustering of plasmids based on the MOB database. The resistome of each sample was determined with Resfinder48 and its associated database using a mapping approach performed directly with raw Illumina reads. This direct mapping approach was chosen because de novo assembly in the absence of long reads technology is prone to misassembly even for small circular genomes.49 Statistical association between the distribution of ESBL resistance genes/alleles and the different hosts was tested using chi-square tests with simulated P values.
We also used a BLAST-like approach (Resfinder) on de novo assembled contigs annotated as a putative plasmid sequence (by Mob suite) to detect resistance genes carried by mobile elements. The contig-level association between hosts, STs, plasmid incompatibility groups and resistance gene counts was tested using chi-square tests with simulated P values and represented using a Sankey diagram. All association tests performed on the same variable were corrected for multiple tests with the false discovery rate method.
Results
Prevalence of ESBL-Ec
We identified 510 positive ESBL-Ec isolates amongst the 1368 collected samples (37.3%). ESBL-Ec prevalence was 17.5% in water, 30.3% in humans and 42.2% in the animal compartment (ranging from 20.0% in horses to 75.0% in pigs) (Figure 1). One isolate per sample was randomly selected for genetic analysis.
WGS
Sequencing generated 741.5 million paired-end reads with a vast majority (99.90%–99.96%) of bases scoring Q30 and above.
Core genome analyses
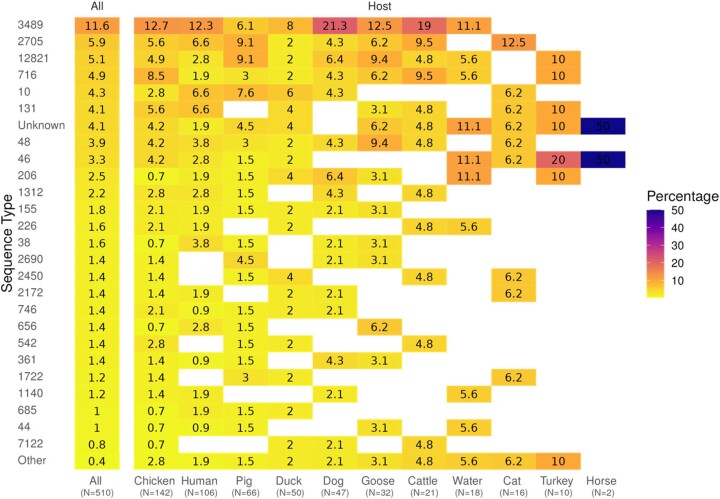
Sequencing all the 510 ESBL-Ec showed a prominent level of genetic diversity composed of seven phylogroups and 84 different known STs (Figure 2). Phylogroup A and ST3489 were the most represented (70.6% and 11.6%, respectively). Interestingly, 4.1% of strains were unknown from the searched ST database because they exhibited new allelic combinations unknown from the searched database (Table S1, available as Supplementary data at JAC Online). Those new STs accounted for most of the genetic diversity in the environment (water) and several animal hosts including pigs, ducks and geese. Phylogroups (chi-square test, P = 0.48) and STs (chi-square test, P = 0.17) were homogeneously distributed amongst hosts but marginally associated with households (chi-square test against phylogroups, P = 0.045; against ST, P = 0.02199) and significantly associated with fokotany (P = 0.0075 with phylogroups and P = 0.0005 with STs).
Figure 2.
Distribution of detected ESBL-Ec STs both in the total dataset and amongst hosts. Percentages are relative to the total number of samples in each host. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
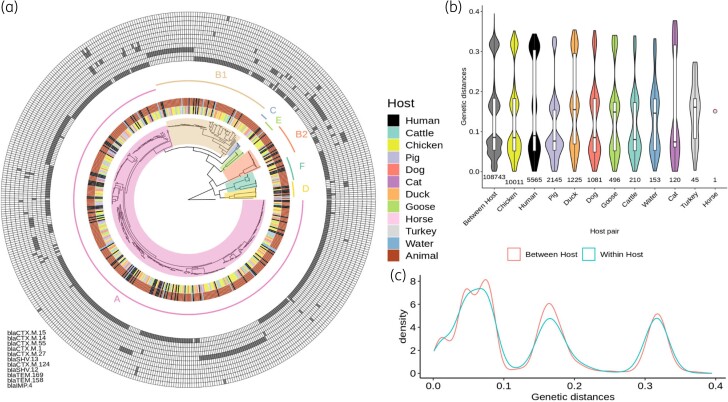
For the 4 563 001 sites in the O157:H7 Sakai strain reference genome, we obtained an average coverage of 75% (min = 71%, max = 88%) and an average depth of 45-fold (mean = 20×, max = 60×). A total of 205 139 sites (3.7%) were variable, among which 25 774 (12.5%) were found to fall within a recombining region, leaving 179 365 core and non-recombinant SNPs to build a robust maximum likelihood phylogenetic tree. The inferred phylogeny of the 510 ESBL-Ec represented a large genomic diversity with most strains distributed in many clades and subclades and some others being isolated on single branches (Figure 3a). Pairwise SNP number between strains (Figure S1) was 2914 on average and ranged from 0 (between 28 different strain pairs) to 8455. Human, animal and water isolates were intermixed amongst the whole phylogeny (Figure 3a). A similar pattern was observed for strains belonging to the same fokontany (administrative unit) and households (Figures S2 and S3), hence denoting the absence of compartment, host and spatial clear phylogenetic structure of the ESBL-Ec identified but evoking rather stochastic events. To illustrate further this pattern, we computed pairwise genetic distances between every single strain of the tree and compared within and between hosts distributions (Figure 3b and c). Interestingly, the distributions were not statistically different (Wilcoxon test, P > 0.05), which confirms the absence of genetic structuration by host. Comparison of the within and between household distributions showed a slight but significant reduction of the pairwise phylogenetic distances within households (Wilcoxon test, P < 0.001), suggesting a proximity of the ESBL-Ec strains sampled in some households (Figure S2) and thus confirming the existence of a moderate geographical structure.
Figure 3.
Global phylogeny of 510 ESBL-Ec and associated metrics. (a) Phylogenetic maximum likelihood tree of 510 ESBL-Ec genomes built from 179 365 core and non-recombinant SNPs. Hosts for each isolate and compartments (human, animal, water) are indicated in the two most central internal rings, respectively, whereas inferred phylogroups are designed both on the phylogenetic tree and on the next ring. A matrix of the presence of the most common resistance genes (prevalence >1%) is depicted in the outer rings, with the names of the genes listed from the inner to the outer lines in the matrix. (b) Comparison of pairwise phylogenetic distances computed between strains within each host. (c) Comparison of pairwise phylogenetic distances computed between versus within hosts. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
ESBL-Ec transmission clusters
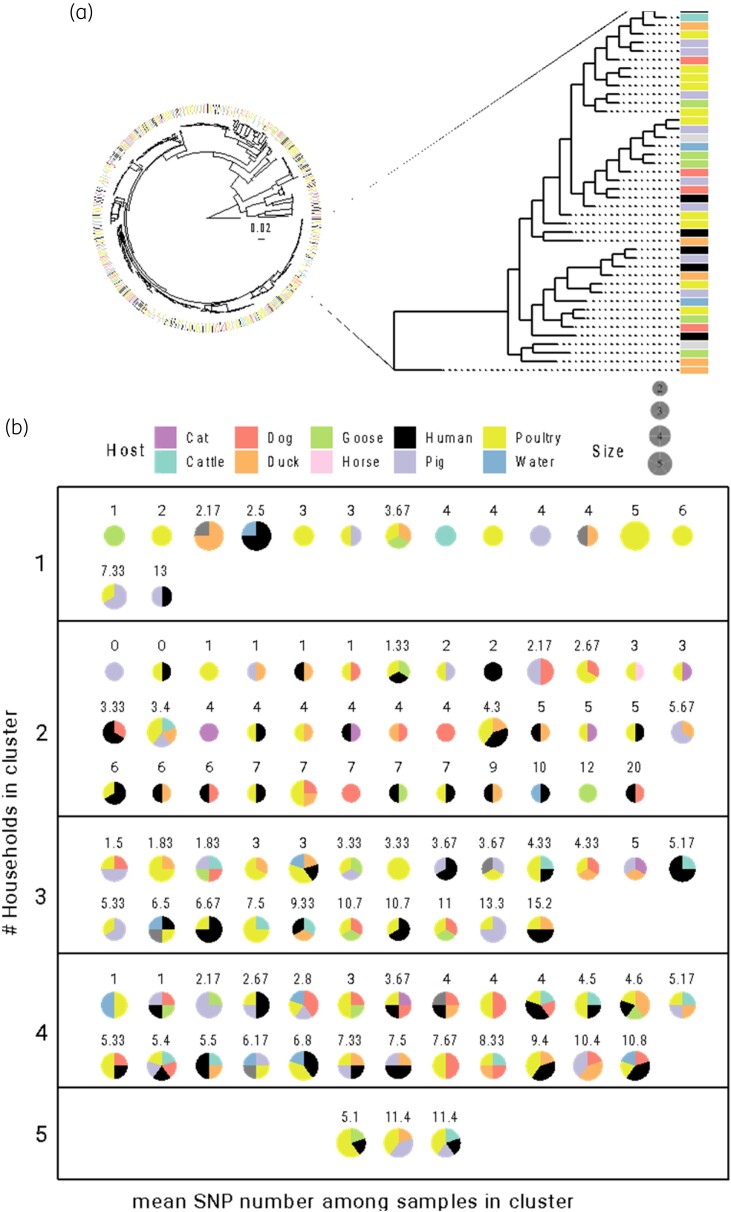
We inferred the presence of 104 clusters within the 510 ESBL-Ec phylogenetic tree with less than 20 SNPs between pairs of samples (Figure 4 and File S4). These clusters included two to five strains originating from one to five different hosts and households, including up to four different animal species. All transmission clusters were constituted of strains belonging to the same ST. Two clusters were composed of absolute clones with no SNPs between pairs (two pigs, one human and one chicken, both from two different households and belonging to an unknown ST and ST3489, respectively). In total, 49/104 clusters (47.1%) were composed of strains of both human and animal sources; 9/104 (8.7%) clusters were composed of water only; and 43/104 clusters (41.3%) were composed of at least two strains from the same household. Of those, 28/43 (65.1%) included pairs of strains both from animal sources, 7/43 (16.3%) from human/animal sources, 5/43 (11.6%) both from human sources, 2/43 (4.7%) from water/human sources and 1/43 (2.3%) from water/animal sources. Fifteen of 104 clusters (14.5%) were restricted to a single household, and 67/70 households (95.7%) hosted at least one sample involved in a cluster. Host distribution amongst clusters did not show statistical deviation from an expected random distribution calculated from the total dataset (multinomial test, P > 0.05). The composition of each cluster was independently compared with the global distribution and none showed significant departure from either global host or household composition (multinomial tests, all P > 0.05) (File S4).
Figure 4.
Clusters of transmission inferred from the global ESBL-Ec phylogeny. (a) Zoom on a random section of the global ESBL-Ec phylogeny for illustrative purposes. On the righthand phylogeny subset, highlighted clades represents different transmission clusters, most of them being composed of strains isolated from different host types. (b) Pie charts of the 104 identified clusters showing their composition in terms of host origins (colours) and number of different households included (boxes). Pie sizes are scaled to the number of strains within a cluster, and mean pairwise numbers of SNPs between strains of each cluster are reported above each chart. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Mobilome and resistome analysis
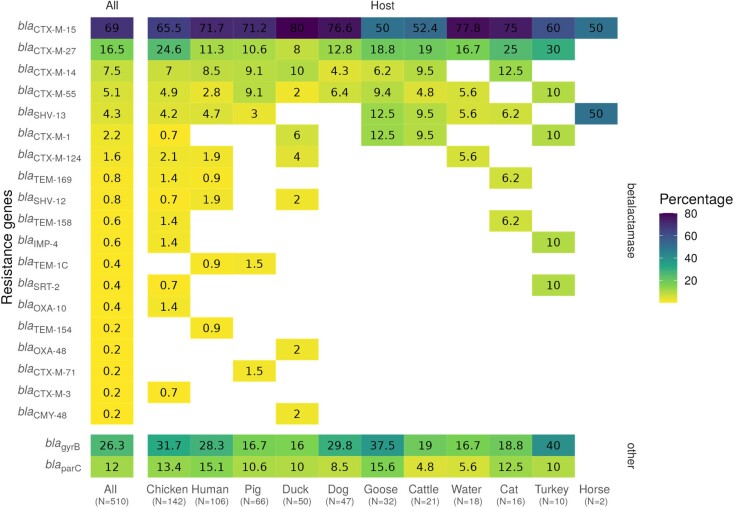
Among the samples 23 ESBL genes were identified (Figure 5). Among those, blaCTX-M-15 was the only one identified in all compartments (Figures 3a and 5) and represented 69.0% of all ESBL genes identified in the study. Multiple ESBL genes co-occurred on the same genomes, with up to four in a single sample (Files S1 and S3). We also screened for the presence of known antibiotic resistance-conferring mutations in gyrB and parC genes and found them in 26.3% and 12.0% of samples, respectively. Interestingly, we report the presence of SHV-13 in 20 ESBL-Ec isolated from the three compartments (File S1), an enzyme that has previously only been detected in clinical Klebsiella pneumoniae isolates. Globally we confirmed the absence of host-specific resistance genes structure in our dataset (chi-squared test with simulated P value = 0.55).
Figure 5.
Frequency of detected ESBL genes and mutations (gyrB and parC) both in the total dataset and amongst hosts. Percentages are relative to the total number of samples in each host. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
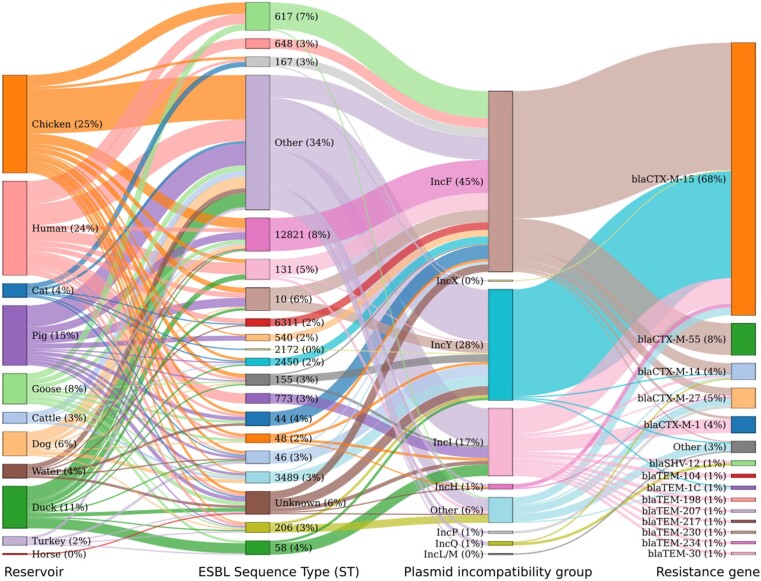
The most represented plasmid types, as identified from their incompatibility groups (Inc), were IncF (54.8%), IncY (26%) and IncI (8.9%). The most common associations between plasmid types and resistance genes were shown to be blaCTX-M-15/IncF (15.1%) and blaCTX-M-15/IncY (12.7%), as illustrated in Figure 6 and in File S3. Figure 6 illustrates the absence of association between host and bacterial STs, the ability of plasmids to be transferred among different bacterial genomic backgrounds (ST) and the plasticity of plasmid incompatibility groups to harbour a large variety of resistance genes.
Figure 6.
Sankey plot displaying association between hosts, STs, plasmids, incompatibility groups and resistance genes. Co-occurence of ESBL resistance gene, plasmid incompatibility locus and ST within each sampled host, highlighting the absence of host specificity in terms of genomic, plasmid and resistance features. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
Using a ‘One Health’ approach connecting humans, animals and the environment, we investigated the genetic relatedness and transmission of 510 ESBL-Ec isolated in a suburban rural area of Antananarivo, Madagascar. As previously suggested, performing such studies in LMICs is particularly relevant to address gaps in our understanding of AMR transmission drivers in humans.2,13,32,50–52 Among the ESBL-Ec isolated, genomic diversity was investigated without revealing any structure at the compartment level (water, animal, human). Our findings emphasize the multiple sources of ESBL-Ec acquisition.
Our results show a high level of ESBL-Ec carriage in human, animal and water compartments in the highlands of Madagascar. ESBL-Ec global prevalence is 37.3%, ranging from 17.5% in water, 30.3% in humans to 42.2% in animals (the highest prevalence was observed for pigs, with 75.0% of individual carriers). Previous findings focusing on human carriage in Madagascar reported similar prevalences (between 18.5% and 34% on average) although the data are not readily comparable because they were obtained from patients recruited in health centres or hospitals without any specific criterion about their contacts with animals.53–55 High levels of ESBL occurrence were previously reported in pigs, poultry and cattle farms in Madagascar.56 To our knowledge, ESBL-Ec presence in drinking water in Madagascar had not been tested before, but comparable values have been described in other LMIC countries, such as Bangladesh,57 the Democratic Republic of Congo58 and Tanzania.31
Sequencing the whole set of 510 detected ESBL-Ec allowed us to report a prominent level of genetic diversity for such a small spatial scale (7.4 km²). Most (70.6%) ESBL-Ec belonged to the A phylogroup, in accordance with a previous survey targeting humans in both community and hospital settings in Antananarivo, Madagascar.59 Our results also highlighted a large diversity of STs, with up to 84 different known STs and 4% of samples bearing new allelic combinations. The most prevalent, ST3489 (11.6%), has previously been observed in Madagascar as one of the most prevalent clones in human rectal carriage (N. Rabenandrasana, Pasteur Institute of Madagascar, personal communication). Few studies have characterized this clone60–62 but public data available on EnteroBase (https://enterobase.warwick.ac.uk/) describe its presence in several ecosystems (human, poultry, livestock, food, companion animal, environment, shellfish, wild animals) and several countries (Bangladesh, Cambodia, India, Japan, Vietnam, USA, the Netherlands, Denmark, Thailand, France and China). Our study confirms the broad host range of ST3489 and its ability to capture many resistance genes encoding ESBLs. Importantly, our reconstructed phylogeny showed a lack of association between genomic diversity and compartments, as illustrated by the comparison of pairwise phylogenetic distances between strains either within or between host groups. This finding is consistent with a recent study performed in Tanzania31 but contrasts strikingly with most studies conducted in high-income countries25–27,63,64 in which ESBL-Ec tended to be structured by compartment. Furthermore, our findings revealed frequent and multiple transmission events between all compartments in this rural area of Madagascar, with a total of 104 clusters of putative transmission events inferred. Our analyses did not show any statistical deviation from expected random distributions, suggesting effective transmission of ESBL-Ec between humans, animals and the environment without highlighting any specific behaviour or risk factor associated with this process. The observed cases of ESBL-Ec transmission across households could reflect the fact that keeping animals is not restricted to a single household and that animal/human movements are frequent at such a small spatiotemporal scale. It is also expected that alternative paths of dissemination are used by ARB to colonize their hosts. Notably, food routes of ESBL-Ec and ARB should be explored in LMIC to better enlighten patterns of transmission as hypothesized in Cambodia29 and in a systematic review.50
Screening the whole set of 510 detected ESBL-Ec revealed a high diversity of ESBL AMR genes in each compartment while highlighting the absence of any host-specific structure. Amongst the β-lactamase detected genes, blaCTX-M-15 identified in all compartments was the most frequent, in accordance with a previous report from Madagascar.59 As formerly described,65 we observed multiple ESBL genes co-occurring in the same genomes (File S3). Investigating our dataset at the plasmid level, we gathered information on putative associations between resistance genes, incompatibility loci, the bacterial ST and the EBSL-Ec carrier hosts. Interestingly, the most common association observed here, between the blaCTX-M-15 resistance gene and the IncF plasmid, has previously been reported in a study performed on ESBL-Ec strains isolated from healthy pregnant women in Madagascar.55 Importantly, no specific association was observed between hosts and bacterial and plasmidic features, highlighting the common occurrence of transfer between compartments, whatever the level of observation. A thorough analysis of the specific associations between ST, incompatibility loci and resistance genes would be interesting, but we chose not to delve into this subject for two reasons. First, it is beyond the scope of this article, which was to study transmission of ESBL-Ec between human, animal and environmental compartments. Second, caution should be exercised in this matter due to the complexity of de novo reconstruction of plasmid sequences using short reads.49 Although we can be confident in the overall observed content of resistance genes and incompatibility loci in each putative plasmid, we are far less confident in the reality of their associations when plasmid assemblies are fragmented. Nonetheless, we have provided extensive supplementary material for anyone willing to investigate these lower-level associations further.
Our study has several limitations. First, for each pair of samples, we measured the genetic distance between two randomly drawn ESBL-Ec among the gut microbiota, potentially missing any direct link between samples harbouring recently transmitted bacteria. This could lead to a systematic underestimation of the transmission events between and among compartments. Although the effect of this bias on the observed absence of genetic structure in our dataset is not straightforward to assess, we hypothesize that our observations are an underestimation of the bacterial network’s connectedness. Exhaustive sampling of ESBL-Ec in each sample would alleviate this bias and could be interesting to conduct in the future, albeit representing a tremendous amount of sequencing to fund and process. Second, we specifically chose a study design susceptible to favouring the observation of transmission among compartments, with a household inclusion criterion of at least three species (human and animals), in a limited geographical setting where direct contacts of humans with animals are frequent. Replication of our protocol in other areas of Madagascar as well as in other LMIC would provide a broader view of the phenomenon. Third, we designed this study at the household scale, gathering epidemiological data with questionnaires in order to discriminate between households where transmission occurred versus the others. Unexpectedly, we observed a large amount of potential transmission between individuals sampled in different households, partially impairing our ability to analyse risk factors associated with transmission at the household level as hypothesized at first. Although it would be interesting to analyse the potential determinants of transmission between households, we lack information on potential contacts of individuals and animals through food markets, commercial exchange of animals, and animal and human movements in the area. Overall, we recommend using contrasting settings (e.g. rural versus periurban or even urban) in order to gather sufficient samples that could be analysed using case-control and longitudinal designs to infer risk factors, while acknowledging that the variety of possible transmission paths for resistant bacteria among compartments could render such a design difficult to draw (e.g. sanitation,66 soil,67 wildlife13). Finally, we acknowledge that defining transmission solely through genetic proximity of ESBL-Ec samples using a transversal survey, albeit possible using the cutting-edge clustering tool chosen in this study, is unorthodox. Epidemiological studies based on phylodynamic approaches need a longitudinal design in order to infer rates of transmission in a canonical sense50 and document the direction of AMR transmission.13 Thus, we advocate that future studies construct sampling schemes allowing the deployment of phylodynamic methodologies in order to alleviate any doubt on our ability to discriminate between direct, recent transmission and older events. Future work aiming to further characterize the typology of main transmission routes between the three compartments and investigating patterns at a larger spatial scale in Madagascar and LMIC will be essential. If antibiotic stewardship intervention in LMIC is highly recommended, these interventions targeting antibiotic use might be insufficient to curtail AMR, as observed in England.68 Actions to control the main AMR transmission routes between humans, animals and the environment should be implemented. This survey is a first step toward those actions in LMIC.
Supplementary Material
Acknowledgements
We thank Dr Laurence Baril, head of the Epidemiological Unit of Institut Pasteur de Madagascar, for ethics committee support, and the Andoharanofotsy community (mayors, heads of fokontanys) and Dr Lys Hélène Rahantanirina for help with household participant inclusion. Computational work was performed on the CIRAD HPC data centre of the SouthGreen bioinformatics platform (http://www.southgreen.fr/).
Contributor Information
Noellie Gay, Epidemiology Unit, Institut Pasteur de Nouvelle-Calédonie, Nouméa, Nouvelle-Calédonie; UMR ASTRE, French Agricultural Research Centre for International Development, Montpellier, France.
Mamitina Alain Noah Rabenandrasana, Experimental Bacteriology Unit, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Harielle Prisca Panandiniaina, Experimental Bacteriology Unit, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Marie Florence Rakotoninidrina, Epidemiology Unit, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Ilo Tsimok’Haja Ramahatafandry, Veterinary Direction, Ministry of Agriculture, Antananarivo, Madagascar.
Vincent Enouf, Mutualized Platform of Microbiology, Pasteur International Bioresources Network, Institut Pasteur, Paris, France.
François Roger, UMR ASTRE, French Agricultural Research Centre for International Development, Montpellier, France.
Jean-Marc Collard, Experimental Bacteriology Unit, Institut Pasteur de Madagascar, Antananarivo, Madagascar.
Eric Cardinale, UMR ASTRE, French Agricultural Research Centre for International Development, Montpellier, France.
Adrien Rieux, UMR PVBMT, French Agricultural Research Centre for International Development, Réunion Island.
Etienne Loire, UMR ASTRE, French Agricultural Research Centre for International Development, Montpellier, France.
Data availability
Raw genetic data are available from NCBI under the accession number PRJNA787774. Scripts and bioinformatic pipelines used to produce results are available at https://github.com/loire/AMR_mada2020.
Funding
This project was funded by the Indian Ocean Health Agency (N.G., PhD fellowship), the INTERREG FEDER TROI 2018–2020 (E.C.) and the grant L’Oréal-UNESCO For Women in Science 2019 (N.G.). A.R. was financially supported by l’Agence Nationale pour la Recherche (JCJC MUSEOBACT contrat ANR-17-CE35-0009-01), the European Regional Development Fund (ERDF contract GURDT I2016-1731-0006632) and Région Réunion.
Transparency declarations
None to declare.
Authors’ contributions
N.G., J.-M.C., F.R. and E.C. designed the study. N.G., M.A.N.R., H.P.P., M.F.R. and I.T.R. performed sampling and fieldwork. N.G., M.A.N.R., H.P.P. and M.F.R. performed lab work and V.E. assisted in managing the sequencing work. M.A.N.R., A.R. and E.L. performed genomic analyses. N.G., A.R. and E.L. prepared the first draft of the manuscript. All authors commented on the data and their interpretation, revised the content critically, and approved the final version.
Supplementary data
Figures S1 to S4 and Table S1 are available as Supplementary data at JAC Online.
References
- 1. D’Costa VM, McGrann KM, Hughes DW et al. Sampling the antibiotic resistome. Science 2006; 311: 374–7. 10.1126/science.1120800 [DOI] [PubMed] [Google Scholar]
- 2. Holmes AH, Moore LSP, Sundsfjord A et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016; 387: 176–87. 10.1016/S0140-6736(15)00473-0 [DOI] [PubMed] [Google Scholar]
- 3. O’Neill J. Tackling drug-resistant infections globally: Final report and recommandations. Review on Antimicrobial Resistance, 2016. [Google Scholar]
- 4. Brogan DM, Mossialos E. A critical analysis of the review on antimicrobial resistance report and the infectious disease financing facility. Glob Health 2016; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manaia CM. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol 2017; 25: 173–81. 10.1016/j.tim.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 6. Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. MBio 2016; 7: e02227-15. 10.1128/mBio.02227-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McEwen SA, Collignon PJ. Antimicrobial resistance: a one health perspective. Microbiol Spectr 2018; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vihta KD, Stoesser N, Llewelyn MJ et al. Trends over time in Escherichia coli bloodstream infections, urinary tract infections, and antibiotic susceptibilities in Oxfordshire, UK, 1998–2016: a study of electronic health records. Lancet Infect Dis 2018; 18: 1138–49. 10.1016/S1473-3099(18)30353-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van den Bogaard A, Stobberingh EE. Epidemiology of resistance to antibiotics: links between animals and humans. Int J Antimicrob Agents 2000; 14: 327–35. 10.1016/S0924-8579(00)00145-X [DOI] [PubMed] [Google Scholar]
- 10. Rawat D, Nair D. Extended-spectrum ß-lactamases in gram negative bacteria. J Glob Infect Dis 2010; 2: 263–74. 10.4103/0974-777X.68531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cassini A, Högberg LD, Plachouras D et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis 2019; 19: 56–66. 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matheu J, Aidara-Kane A, Andremont A. The ESBL tricycle AMR surveillance project: a simple, one health approach to global surveillance. AMR Control 2017. [Google Scholar]
- 13. Hassell JM, Ward MJ, Muloi D et al. Clinically relevant antimicrobial resistance at the wildlife–livestock–human interface in Nairobi: an epidemiological study. Lancet Planet Health 2019; 3: e259–69. 10.1016/S2542-5196(19)30083-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stoesser N, Sheppard AE, Pankhurst L et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 2016; 7: e02162. 10.1128/mBio.02162-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagel S, Makarewicz O, Hartung A et al. ESBL colonization and acquisition in a hospital population: the molecular epidemiology and transmission of resistance genes. PLoS One 2019; 14: e0208505. 10.1371/journal.pone.0208505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoshide RR, Chung H, Tokeshi J. Emergence of community-acquired extended-spectrum beta-lactamase Escherichia coli (ESBLEC) in Honolulu: a case series of three individuals with community-acquired ESBLEC bacteriuria. Hawaii Med J 2011; 70: 193–5. [PMC free article] [PubMed] [Google Scholar]
- 17. Doi Y, Park YS, Rivera JI et al. Community-associated extended-spectrum -lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis 2013; 56: 641–8. 10.1093/cid/cis942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palmeira J D, Ferreira HMN. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—a threat around the world. Heliyon 2020; 6: e03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carattoli A. Animal reservoirs for extended spectrum β-lactamase producers. Clin Microbiol Infect 2008; 14: 117–23. 10.1111/j.1469-0691.2007.01851.x [DOI] [PubMed] [Google Scholar]
- 20. Ye Q, Wu Q, Zhang S et al. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front Microbiol 2018; 9: 1709. 10.3389/fmicb.2018.01709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Börjesson S, Ny S, Egervärn M et al. Limited dissemination of extended-spectrum β-lactamase- and plasmid-encoded AmpC-producing Escherichia coli from food and farm animals, Sweden. Emerg Infect Dis 2016; 22: 634–40. 10.3201/eid2204.151142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu H, Zhou H, Li Q et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli isolated from the rivers and lakes in Northwest China. BMC Microbiol 2018; 18: 125. 10.1186/s12866-018-1270-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fagerström A, Mölling P, Khan FA et al. Comparative distribution of extended-spectrum beta-lactamase–producing Escherichia coli from urine infections and environmental waters. PLoS One 2019; 14: e0224861. 10.1371/journal.pone.0224861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muloi DM, Wee BA, McClean DMH et al. Population genomics of Escherichia coli in livestock-keeping households across a rapidly developing urban landscape. Nat Microbiol 2022; 7: 581–9. 10.1038/s41564-022-01079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miltgen G, Martak D, Valot B et al. One health compartmental analysis of ESBL-producing Escherichia coli on Réunion Island reveals partitioning between humans and livestock. J Antimicrob Chemother 2022; 77: 1254–62. 10.1093/jac/dkac054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ludden C, Raven KE, Jamrozy D et al. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. MBio 2019; 10: e02693-18. 10.1128/mBio.02693-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dorado-García A, Smid JH, Van Pelt W et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother 2018; 73: 339–47. 10.1093/jac/dkx397 [DOI] [PubMed] [Google Scholar]
- 28. Trung NV, Jamrozy D, Matamoros S et al. Limited contribution of non-intensive chicken farming to ESBL-producing Escherichia coli colonization in humans in Vietnam: an epidemiological and genomic analysis. J Antimicrob Chemother 2019; 74: 561–70. 10.1093/jac/dky506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nadimpalli M, Vuthy Y, de Lauzanne A et al. Meat and fish as sources of extended-spectrum β-lactamase–producing Escherichia coli, Cambodia. Emerg Infect Dis 2019; 25: 126–31. 10.3201/eid2501.180534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hussain A, Shaik S, Ranjan A et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front Microbiol 2017; 8: 1–13. 10.3389/fmicb.2017.02120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subbiah M, Caudell MA, Mair C et al. Antimicrobial resistant enteric bacteria are widely distributed amongst people, animals and the environment in Tanzania. Nat Commun 2020; 11: 228. 10.1038/s41467-019-13995-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rousham EK, Unicomb L, Islam MA. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and one health approaches. Proc R Soc B Biol Sci 2018; 285: 20180332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jehl F. Comité de l’antibiogramme de la Société Française de Microbiologie. Société Française de Microbiologie, 2020.
- 34. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McKenna A, Hanna M, Banks E et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Croucher NJ, Page AJ, Connor TR et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–13. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paradis E, Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019; 35: 526–8. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 39. Yu G, Smith DK, Zhu H, et al. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 2017; 8: 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- 40. R Core Development Team . R: a language and environment for statistical computing, 3.2.1. 2020. http://www.r-project.org
- 41. Gupta A, Jordan IK, Rishishwar L. stringMLST: a fast k-mer based tool for multilocus sequence typing. Bioinformatics 2017; 33: 119–21. 10.1093/bioinformatics/btw586 [DOI] [PubMed] [Google Scholar]
- 42. Beghain J, Bridier-Nahmias A, Le NH et al. Clermontyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genomics 2018; 4: 1–8. 10.1099/mgen.0.000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jombart T, Dray S. Adephylo: exploratory analyses for the phylogenetic comparative method. Bioinformatics 2010; 26: 1907–9. [DOI] [PubMed] [Google Scholar]
- 44. Han AX, Parker E, Maurer-Stroh S et al. Inferring putative transmission clusters with Phydelity. Virus Evol 2019; 5: vez039. 10.1093/ve/vez039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ludden C, Coll F, Gouliouris T et al. Defining nosocomial transmission of Escherichia coli and antimicrobial resistance genes: a genomic surveillance study. Lancet Microbe 2021; 2: e472–80. 10.1016/S2666-5247(21)00117-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wick RR, Judd LM, Gorrie CL et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genomics 2018; 4: e000206. 10.1099/mgen.0.000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zankari E, Hasman H, Cosentino S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arredondo-Alonso S, Willems RJ, van Schaik W et al. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genomics 2017; 3: e000128. 10.1099/mgen.0.000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chatterjee A, Modarai M, Naylor NR et al. Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect Dis 2018; 18: e368–78. 10.1016/S1473-3099(18)30296-2 [DOI] [PubMed] [Google Scholar]
- 51. Mendelson M, Brink A, Gouws J et al. The one health stewardship of colistin as an antibiotic of last resort for human health in South Africa. Lancet Infect Dis 2018; 18: 288–94. 10.1016/S1473-3099(18)30119-1 [DOI] [PubMed] [Google Scholar]
- 52. Robinson TP, Bu DP, Carrique-Mas J et al. Antibiotic resistance is the quintessential one health issue. Trans R Soc Trop Med Hyg 2016; 110: 377–80. 10.1093/trstmh/trw048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chereau F, Herindrainy P, Garin B et al. Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother 2015; 59: 3652–5. 10.1128/AAC.00029-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Herindrainy P, Randrianirina F, Ratovoson R et al. Rectal carriage of extended-spectrum beta-lactamase-producing gram-negative bacilli in community settings in Madagascar. PLoS One 2011; 6: e22738. 10.1371/journal.pone.0022738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Milenkov M, Rasoanandrasana S, Rahajamanana LV et al. Prevalence, risk factors, and genetic characterization of extended-spectrum beta-lactamase Escherichia coli isolated from healthy pregnant women in Madagascar. Front Microbiol 2021; 12: 786146. 10.3389/fmicb.2021.786146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gay N, Leclaire A, Laval M et al. Risk factors of extended-spectrum β-lactamase producing Enterobacteriaceae occurrence in farms in Réunion, Madagascar and Mayotte islands, 2016–2017. Vet Sci 2018; 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mahmud ZH, Kabir MH, Ali S et al. Extended-spectrum beta-lactamase-producing Escherichia coli in drinking water samples from a forcibly displaced, densely populated community setting in Bangladesh. Front Public Health 2020; 8: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de Boeck H, Miwanda B, Lunguya-Metila O et al. ESBL-positive enterobacteria isolates in drinking water. Emerg Infect Dis 2012; 18: 1019–20. 10.3201/eid1806.111214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rakotonirina HC, Garin B, Randrianirina F et al. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol 2013; 13: 85. 10.1186/1471-2180-13-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hasan B, Islam K, Ahsan M et al. Fecal carriage of multi-drug resistant and extended spectrum β-lactamases producing E. coli in household pigeons, Bangladesh. Vet Microbiol 2014; 168: 221–4. 10.1016/j.vetmic.2013.09.033 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Q, Lv L, Huang X et al. Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob Agents Chemother 2019; 63: 573–9. 10.1128/AAC.00573-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wei X, Wang W, Lu N et al. Prevalence of multidrug-resistant CTX-M extended spectrum beta-lactamase-producing Escherichia coli from different bovine faeces in China. Front Vet Sci 2022; 9: 738–44. 10.3389/fvets.2022.738904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Day MJ, Hopkins KL, Wareham DW et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 2019; 19: 1325–35. 10.1016/S1473-3099(19)30273-7 [DOI] [PubMed] [Google Scholar]
- 64. Ludden C, Moradigaravand D, Jamrozy D et al. A one health study of the genetic relatedness of Klebsiella pneumoniae and their mobile elements in the east of England. Clin Infect Dis 2020; 70: 219–26. 10.1093/cid/ciz174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Silago V, Kovacs D, Samson H et al. Existence of multiple ESBL genes among phenotypically confirmed ESBL producing Klebsiella pneumoniae and Escherichia coli concurrently isolated from clinical, colonization and contamination samples from neonatal units at Bugando Medical Center, Mwanza, Tanzania. Antibiotics (Basel) 2021; 10: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Berendes DM, de Mondesert L, Kirby AE et al. Variation in E. coli concentrations in open drains across neighborhoods in Accra, Ghana: the influence of onsite sanitation coverage and interconnectedness of urban environments. Int J Hyg Environ Health 2020; 224: 113433. 10.1016/j.ijheh.2019.113433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Montealegre MC, Roy S, Böni F et al. Risk factors for detection, survival, and growth of antibiotic-resistant and pathogenic Escherichia coli in household soils in rural Bangladesh. Appl Environ Microbiol 2018; 84: e01978-18. 10.1128/AEM.01978-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aliabadi S, Anyanwu P, Beech E et al. Effect of antibiotic stewardship interventions in primary care on antimicrobial resistance of Escherichia coli bacteraemia in England (2013–18): a quasi-experimental, ecological, data linkage study. Lancet Infect Dis 2021; 21: 1689–700. 10.1016/S1473-3099(21)00069-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw genetic data are available from NCBI under the accession number PRJNA787774. Scripts and bioinformatic pipelines used to produce results are available at https://github.com/loire/AMR_mada2020.