Abstract
Background
Antibiotic use is associated with collateral damage to the healthy microbiota. Afabicin is a first-in-class prodrug inhibitor of the FabI enzyme that, when converted to the pharmacologically active agent afabicin desphosphono, demonstrates a staphylococcal-specific spectrum of activity. An expected benefit of highly targeted antibiotics such as afabicin is microbiome preservation.
Objectives
To compare the effects of oral treatment with afabicin and standard-of-care antibiotics upon the murine gut microbiota, and to assess the effects of oral afabicin treatment on the human gut microbiota.
Methods
Gut microbiota effects of a 10 day oral course of afabicin treatment were monitored in mice and compared with clindamycin, linezolid and moxifloxacin at human-equivalent dose levels using 16S rDNA sequencing. Further, the gut microbiota of healthy volunteers was longitudinally assessed across 20 days of oral treatment with afabicin 240 mg twice daily.
Results
Afabicin treatment did not significantly alter gut microbiota diversity (Shannon H index) or richness (rarefied Chao1) in mice. Only limited changes to taxonomic abundances were observed in afabicin-treated animals. In contrast, clindamycin, linezolid and moxifloxacin each caused extensive dysbiosis in the murine model. In humans, afabicin treatment was not associated with alterations in Shannon H or rarefied Chao1 indices, nor relative taxonomic abundances, supporting the findings from the animal model.
Conclusions
Oral treatment with afabicin is associated with preservation of the gut microbiota in mice and healthy subjects.
Introduction
Widespread use of antibiotics has selected for bacterial strains that are antibiotic resistant and respond poorly to antibiotic chemotherapy. The problem is compounded by off-target effects, especially on the human gut microbiota. A healthy microbiota should maintain a complex, rich and balanced diversity, and this is thought to help maintain metabolic homeostasis and proper organ function, and to protect against infection and syndromes such as inflammatory bowel disease.1–3 Conversely, disruption of the microbiota, termed dysbiosis, can lead to colonization with antibiotic-resistant pathogens,4 antibiotic-associated infections [e.g. Clostridioides difficile infection (CDI)],5 and horizontal transfer of resistance genes across the microbiome.6,7 Thus, the development of new narrow-spectrum antimicrobials that preserve the microbiota, including those that target only a single pathogen, may provide broad, desirable impacts for human health.8–10
Effectively combatting antimicrobial resistance (AMR) likely requires a multimodal approach involving evidence-based prescription and use of currently approved antibiotics (i.e. antibiotic stewardship), and the development of new antibiotics with unique mechanisms of action that are effective against resistant bacterial strains and that, ideally, preserve the microbiome. De-escalation from broad- to narrow(er)-spectrum antibiotics is an essential approach for antibiotic stewardship.11–13 This is further reflected by recent guidance from regulatory bodies including the FDA, which have highlighted the potential utility of next-generation antibacterial drugs that target a limited number of species for patients with unmet medical needs.14
Staphylococcal infections are a significant global concern for multiple reasons, including the significant contribution of MRSA infections as the leading cause of AMR-related mortality,15 the increasing frequency of CoNS in infections associated with transiently or permanently inserted foreign bodies,16 and the lack of well-established anti-staphylococcal oral treatment options for infections that require long-term antibiotic therapy such as bone and joint infections or complicated bacteraemia.17,18
Afabicin (Debio 1450) is a first-in-class prodrug inhibitor of FabI, a key enzyme in bacterial fatty acid biosynthesis,19,20 which is being developed as a pathogen-specific anti-staphylococcal antibiotic available for oral and parenteral use.21 Whilst the prodrug, afabicin, has no antimicrobial activity, the active moiety afabicin desphosphono (Debio 1452, formerly AFN-1252) has potent activity against staphylococci including both coagulase-positive and coagulase-negative strains resistant to other antibiotic classes, but very limited activity against non-staphylococcal species.22,23 Afabicin meets each of the four WHO criteria for innovation (novel chemical class, novel target, no cross resistance, novel mechanism of action),24 and its staphylococcal-specific activity suggests it will produce limited deleterious off-target antimicrobial effects. In support of this, afabicin desphosphono was recently shown to induce only minor changes to the gut microbiota of mice.25 However, the effects of the newly developed oral formulation of afabicin, the prodrug, on the gut microbiota is yet to be determined.
The objectives of the present study were to compare the effect of a 10-day treatment with the oral formulation of afabicin to that of clindamycin, linezolid and moxifloxacin on mouse gut microbiota and to assess the effects of 20 day oral treatment with afabicin on the gut microbiota of healthy subjects.
Materials and methods
Bacterial strains and MIC testing
Bacterial strains used in the study are listed in Table 1. MIC values for afabicin desphosphono and the comparator antibiotic clindamycin were determined on Supplemented Brucella Agar (SBA) plates using the agar dilution method according to CLSI guidelines for anaerobic bacteria.26 Plates were incubated in a Bactron II anaerobic chamber for 48 h at 35°C–36°C prior to MIC determination.
Table 1.
Afabicin desphosphono MICs for representative bacteria from the human microbiota
| MIC (mg/L) | ||
|---|---|---|
| Bacterial isolate | Afabicin desphosphono | Clindamycin |
| Bifidobacterium bifidum 3965 (ATCC 15696) | >8 | ≤0.03 |
| Bifidobacterium breve 3967 (ATCC 15698) | >8 | ≤0.03 |
| Bifidobacterium infantis 3966 (ATCC 15702) | >8 | ≤0.03 |
| Bifidobacterium longum 3968 (ATCC 15707) | >8 | 0.06 |
| Clostridium perfringens 3414 | >8 | 2 |
| C. perfringens 3518 | >8 | >16 |
| C. difficile 3579 | >8 | 16 |
| C. difficile 3584 | >8 | 8 |
| Eggerthella lenta 1274 (ATCC 43055) | >8 | 0.12 |
| Lactobacillus acidophilus 0681 | >8 | 8 |
| Lactobacillus casei 1722 (ATCC 393) | >8 | 4 |
| Lactiplantibacillus plantarum 2791 (ATCC 39268) | >8 | 0.25 |
| Peptostreptococcus anaerobius 3526 | >8 | ≤0.03 |
| P. anaerobius 3531 | >8 | >16 |
| Peptostreptococcus micros 3432 | >8 | 0.25 |
| P. micros 3545 | >8 | 4 |
| Cutibacterium acnes 1713 | >8 | 0.12 |
| C. acnes 1267 | >8 | >16 |
| Streptococcus constellatus 1202 (ATCC 27823) | >8 | 0.25 |
| Streptococcus intermedius 1203 (ATCC 27335) | >8 | 0.25 |
| Bacteroides fragilis 3374 | >8 | 2 |
| B. fragilis 3479 | >8 | 4 |
| B. fragilis 123 (ATCC 25285) | >8 | 1 |
| Bacteroides ovatus 3503 | >8 | 2 |
| B. ovatus 3508 | >8 | ≤0.03 |
| Bacteroides thetaiotaomicron 3399 | >8 | 1 |
| B. thetaiotaomicron 3496 | >8 | >16 |
| Bacteroides vulgatus 3389 | >8 | 0.5 |
| B. vulgatus 3494 | >8 | 0.5 |
| Eikenella corrodens 1206 (ATCC 43278) | >8 | 0.5 |
| Fusobacterium necrophorum 3963 (ATCC 25286) | >8 | ≤0.03 |
| Fusobacterium nucleatum 3962 (ATCC 25586) | >8 | ≤0.03 |
| Porphyromonas asaccharolytica 3552 | >8 | 0.12 |
| P. asaccharolytica 3557 | >8 | 0.25 |
| Prevotella melaninogenica 3437 | >8 | >16 |
| P. melaninogenica 3443 | >8 | ≤0.03 |
| Prevotella spp. 3564 | >8 | 0.25 |
| Prevotella spp. 3568 | >8 | 4 |
| Veillonella parvula 1272 (ATCC 17745) | >8 | >16 |
Mouse model
All experiments were conducted in accordance with the current guidelines for animal welfare and were approved by the Institutional Animal Care and Use Committee at Michigan State University. To reduce the potential for stress-induced impacts on the microbiota, CD-1 female mice (Charles River Laboratories, Wilmington, MA, USA; 18–22 g; five mice per cage) were subject to a 5 day pre-treatment acclimatization period, where they were handled, received abdominal massage, and were daily oral mock-gavaged with a needle introduced into the oesophagus.
Five groups of five CD-1 mice were treated orally for 10 days, twice daily with either vehicle (0.5% methylcellulose), afabicin (65 mg/kg), clindamycin (100 mg/kg), linezolid (100 mg/kg) or once daily with moxifloxacin (65 mg/kg). Clindamycin, linezolid and moxifloxacin were chosen as comparators as they are each used as oral treatments for staphylococcal infections.27 Dose levels were selected based on the equivalent surface area dosage conversion factors in order to represent the human-equivalent dose levels (300 mg every 6 h for clindamycin; 600 mg twice daily for linezolid; 400 mg daily for moxifloxacin; 240 mg twice daily for afabicin, as used in prior human studies).21
An additional vehicle group was added to compensate for the death of two mice (unrelated to the study); the stool samples from the two vehicle groups were considered similar and were analysed as a unique vehicle group. For clindamycin and linezolid groups, samples from four mice were included in the analysis due to mouse death in each group (caused by the gavage procedure at baseline). In the afabicin group, one mouse died on Day 10 (gavage trauma); thus, the Day 8 sample was analysed in lieu of the Day 10 sample.
Faecal pellets were collected in an aseptic manner from each animal at four timepoints: Day −2 (baseline), Day 2 and Day 10 (during treatment), and Day 17 (7 days after the end of treatment) (Figure 1a). Abdominal massage was used when appropriate. The samples were placed on dry ice immediately after collection and stored at −80°C.
Figure 1.
Schematic representation of the experimental design. (a) Animals received oral afabicin (65 mg/kg) or clindamycin (100 mg/kg), linezolid (100 mg/kg), moxifloxacin (65 mg/kg) twice daily for 10 consecutive days (10d). (b) In the context of a DDI study, healthy volunteers received 20 days of oral treatment with afabicin 240 mg twice a day. For (a) and (b), faecal sampling was performed as indicated by triangles for 16S rDNA sequencing. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Human drug–drug interaction (DDI) study
Faecal sampling from healthy volunteers was performed to evaluate the effects of oral treatment with afabicin on the gut microbiota, as an exploratory endpoint, in the context of an open-label, fixed-sequence DDI study designed to assess the effect of afabicin on the pharmacokinetics of probe substrates of drug transporters, metformin, rosuvastatin and digoxin (Figure 1b; EudraCT: 2015-001525-17). Faecal sampling was performed prior to drug administration (baseline 1; BL1). Subjects received oral metformin (500 mg) followed by concomitant rosuvastatin (10 mg) and digoxin (0.5 mg) 48 h later. Faecal sampling was performed again following a ≥7 day washout period (baseline 2; BL2). Thereafter, subjects received oral afabicin 240 mg twice daily for 20 consecutive days,21 with an additional administration of DDI probes; single oral dose of metformin (500 mg) on treatment Day 14, and a concomitant oral dose of rosuvastatin (10 mg) and digoxin (0.5 mg) on treatment Day 16. Afabicin was administered in standardized fasting conditions. Faecal sampling was performed on treatment Day 7, Day 14 and Day 20, and again 7 to 14 days after the treatment period (end of study; EoS). Sixteen healthy subjects (mean age 48.7 years, 75% male) were included in the study with complete sample sets from 15 subjects analysed using 16S rDNA amplicon sequencing.
16S rDNA sequence analysis
DNA was extracted from 20 mg of mouse faecal pellets and 200 mg of human stool samples. Following bead beating, genomic DNA was isolated using phenol-chloroform methodology for mouse samples and using Maxwell 16 Tissue Purification kits (Promega Corporation, Madison, WI, USA) for human samples. PCR amplification was performed using 16S rDNA universal primers targeting the V3-V4 region of the bacterial 16S rRNA genes.28 All amplicons were purified with magnetic beads (Agencourt AMPure XP beads; Beckman Coulter, Brea, CA, USA) and the library was generated via addition of dual indices and Illumina sequencing adapters using Nextera XT Index kit (Illumina, San Diego, CA, USA). Each library was cleaned using magnetic beads and size was determined by capillary electrophoresis (2100 Bioanalyzer Instrument, Agilent Technologies). Libraries were quantified using the Qubit® 2.0 Fluorometer and the Qubit® dsDNA range assay (Thermo Fisher Scientific, Waltham, MA, USA), normalized to 4 nM, pooled and denatured before sequencing using the Illumina MiSeq platform with 2 × 250 paired-end MiSeq kit V2 (Illumina).
Sequences were analysed using an in-house bioinformatic pipeline adapted from mothur software.29 Sequences were trimmed and aligned to the V3-V4 region of the 16S rRNA gene of the Greengenes database formatted by mothur (gg_13_5_99 release). Chimera sequences were removed using the UCHIME algorithm.30 Reads were classified using naive Bayesian classifier31 against the Ribosomal Database Project 16S rRNA gene training set v9 formatted by mothur with a bootstrap cut-off of 60%. Sequences were then clustered into operational taxonomic units (OTUs) using furthest-neighbour clustering at a similarity threshold of 97%. For each sample, OTU-based microbial diversity and richness were estimated by calculating the Shannon and Chao1 indices, respectively, using the R package phyloseq.32,33 Chao1 index values were normalized by rarefaction to 10 000 reads. Shannon H indices were calculated as a measure of community diversity. Communities that are dominated by a small number of taxa exhibit low diversity and generate a low Shannon H Index. Conversely, communities where abundance is distributed across many taxa have high diversity and a high Shannon H index. Rarefied Chao indices were calculated as a measure of community richness. Communities consisting of many distinct taxa will produce high values, whereas those with few taxa will produce low values.
Statistical analysis
For each taxon, an analysis of variance for repeated measurements (repeated ANOVA) was conducted including treatment, days and treatment-by-day interaction as fixed factors in the statistical model. A rank transformation of data was applied in case of non-normal distribution. In this statistical model, post hoc tests were conducted (i) to perform pairwise between-group comparisons using a Tukey’s adjustment; and (ii) to conduct within-group analysis by comparing values before, during and after treatment in each animal group using a Dunnett’s adjustment.
As multiple hypotheses were tested simultaneously, a false discovery rate (FDR) adjustment was used to correct the statistical significance (P values) of the ANOVA model. To investigate the compositional distribution patterns based on relative abundance, principal coordinate analysis (PCoA) using Bray–Curtis dissimilarities was generated using R software.32 For all statistical tests (two-sided), P ≤ 0.05 was considered statistically significant. Outliers were defined as <25th percentile minus 1.5 times the IQR or as >75th percentile plus 1.5 times IQR.
Ethics
Animal studies were approved by the ethics committee of Michigan State University in vivo facility. The human study was conducted in accordance with the Declaration of Helsinki and national and international standards.
Results
In vitro activity of afabicin desphosphono against a panel of bacteria from the human microbiota
The active moiety of afabicin, afabicin desphosphono, has potent antimicrobial activity against staphylococcal isolates in vitro and has shown limited activity against various non-staphylococcal species.22,23 In the current report, we extended this analysis to include a panel of bacteria that are common representatives of the human microbiota (n = 39 isolates). Afabicin desphosphono was not active against any of the microbiota representatives tested (MIC > 8 mg/L for each; Table 1). In contrast, the control antibiotic clindamycin showed antimicrobial activity against most isolates in vitro; the concentration of clindamycin that inhibited 50% of the isolates was 0.5 mg/L and only six isolates (∼15%) had an MIC of ≥16 mg/L (Table 1).
Analysis of the effect of treatment with afabicin upon the faecal microbiota of mice compared with clindamycin, linezolid and moxifloxacin
Baseline comparisons
PCoA of the gut microbiota composition showed that all baseline samples (Day −2) clustered together (Figure 2). No differences in the Shannon H diversity (Figure 3a) and rarefied Chao1 richness indices (Figure 3b) were determined between the five groups at baseline. No statistically significant differences in relative abundance at the phylum and family level were observed across the five groups following randomization and prior to treatment, except for a lower TM7 phylum relative abundance in the linezolid treated group compared with the afabicin-treated group (P < 0.05; Table S1, available as Supplementary data at JAC Online). Together, these data indicate there were negligible intergroup differences prior to antibiotic treatment.
Figure 2.
PCoA of 16S rDNA sequence data from antibiotic-treated mice. The compositional distribution pattern (Bray–Curtis distance) was calculated at baseline (Day −2, squares, each group presented in grey) and after 10 days of treatment (triangles) for each treatment group (represented by different colours).
Figure 3.
Longitudinal effects of antibiotic treatment upon the gut microbiota of mice. (a) Shannon H diversity was determined for each sample as a measure of diversity for mice treated with afabicin or comparators (clindamycin, linezolid or moxifloxacin). (b) Rarefied Chao1 was calculated as a measure of sample richness for mice treated with afabicin, or comparators. Note, rarefied Chao1 was only determined for samples with library size >10 000 reads. (c) Relative abundance percentages for major bacterial families from the murine gut microbiota. For (a) and (b), statistical significance was determined using a mixed-effects model with Dunnett’s multiple comparisons test relative to Day −2 (baseline), * P < 0.05, ** P < 0.01.
Vehicle control
Longitudinal vehicle control samples clustered together on PCoA (Figure 2) and no differences in Shannon H diversity (Figure 3a) or rarefied Chao1 richness indices (Figure 3b) were identified. No statistically significant differences in relative abundance at the phylum and family level were observed compared with baseline at any timepoint except for a decrease in Porphyromonadaceae at Day 17 (29.28% to 18.31%). Together, these data indicate that the murine microbiota remained stable in the vehicle controls throughout the experiment.
Afabicin treatment
Afabicin-treated samples clustered together with the baseline samples on PCoA (Figure 2). Supporting the findings of the PCoA, no significant differences in Shannon H (Figure 3a) or rarefied Chao1 (Figure 3b) indices were observed between afabicin-treated mice during or after treatment (compared with baseline), indicating a limited impact on overall gut microbial composition.
Considering within-group differences compared with baseline, during treatment, there was a decrease in Erysipelotrichaceae (baseline 2.2%, Day 2 0.75%, Day 10 0.85%) and increase in Peptococcacae_1 in the afabicin group on Day 10 (baseline 0.57%, Day 10 1.09%), each of which had returned to baseline levels by Day 17 (Table S1; Figure 3c). The only significant differences between the baseline and Day 17 samples for the afabicin-treated group was an increase in the Firmicutes phyla (from 44.3% to 61.9%), more specifically the Lachnospiraceae family for the latter (from 10.5% to 32.1%). Importantly, however, these relative abundances were not statistically different from the vehicle control group at Day 17 (Firmicutes phyla vehicle 61.23%, afabicin-treated 61.93%; Lachnospiraceae family vehicle 29.55%, afabicin-treated 32.06%; Table S1).
Clindamycin, linezolid and moxifloxacin treatment
In contrast to the afabicin-treated group, clindamycin, linezolid and moxifloxacin each had substantial effects on the murine gut microbiota. Samples from animals treated with each comparator did not cluster with respective baseline samples or vehicle control samples on PCoA (Figure 2). Clindamycin and linezolid samples clustered together, and each were separate from moxifloxacin. Clindamycin and linezolid treatment were each associated with statistically significant reductions in Shannon H diversity during and after treatment (Figure 3a) and a statistical reduction in rarefied Chao1 for at least one timepoint compared with baseline (Figure 3b).
Considering within-group differences compared with baseline, the comparator antibiotics produced statistically significant changes in the relative abundance of at least five of the six major phyla assessed (Table S1; differences at the family taxonomic level are represented graphically in Figure 3c), indicating widespread dysbiosis. As expected based on the PCoA, similar changes in relative taxa abundances were observed for clindamycin- and linezolid-treated animals, and these profiles were distinct from animals treated with moxifloxacin. The most noteworthy change for clindamycin- and linezolid-treated animals was the increased relative abundance of Enterobacteriaceae (from 0.23% and 0.08% at baseline, respectively, to up to 84.94% for clindamycin at Day 2 and up to 97.56% for linezolid at Day 10).
Evaluation of the treatment effects of afabicin on the gut microbiota of healthy subjects
In the context of an open-label, Phase I DDI study, healthy volunteers received 20 days of oral treatment with afabicin 240 mg twice daily. Unrelated to the current report, subjects also received a single dose of DDI probes (metformin, rosuvastatin, digoxin) in two treatment periods; first, prior to receiving afabicin, and second, during afabicin treatment (Figure 1b). Two baseline faecal samples were collected; the first, prior to DDI probe administration, and the second, after the probe washout period and prior to afabicin treatment. No statistical differences were determined between baseline samples, indicating negligible impact of these probes on human microbiota composition (Figure 4).
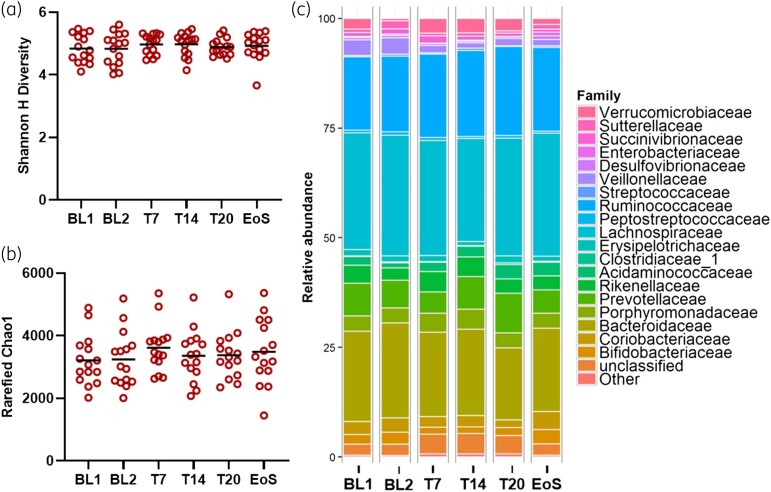
Figure 4.
Longitudinal effects on the gut microbiota of healthy human subjects treated with afabicin. (a) Shannon H diversity was determined for each sample as a measure of diversity for healthy subjects receiving afabicin. (b) Rarefied Chao1 was determined for each sample as a measure of richness for healthy subjects receiving afabicin. (c) Relative abundance percentages for major bacterial families from the murine gut microbiota. For (a) and (b), statistical significance was assessed using a one-way ANOVA with Dunnett’s multiple comparisons test. No statistical differences were observed. T7, treatment day 7; T14, treatment day 14; T20, treatment day 20.
Mirroring the findings from the murine model (Figure 3), afabicin treatment did not alter Shannon H diversity (Figure 4a) or rarefied Chao1 indices (Figure 4b) compared with baseline controls. Further, afabicin was not associated with any statistically significant changes in mean relative microbial abundance at the phylum, family (Figure 4c) or genus level at any of the timepoints assessed. Relative abundances at the phylum, family and genus level for each longitudinal sample from each individual subject are presented graphically in Figure S1, Figure S2 and Figure S3, respectively.
Discussion
The active moiety of afabicin, afabicin desphosphono, displays potent activity against diverse isolates of staphylococci, and only limited activity against other bacteria.23 The drug targets an enoyl-acyl carrier protein reductase FabI, which in staphylococci plays an essential role in fatty acid synthesis.34 FabI, however, is not essential in all bacterial species, and can be replaced by isoforms such as FabK, FabV or FabL, with some species producing multiple functionally redundant isoforms (e.g. FabI along with FabL in Bacillus subtilis),35,36 which largely defines the spectrum of activity of FabI inhibitors. Further, bacteria that can efficiently scavenge exogenous fatty acids, including those present in serum, can overcome FabI inhibition.34,37 Lastly, afabicin desphosphono is generally inactive against Gram-negative bacteria.38 Together, these mechanisms explain the limited impact that afabicin, which is rapidly converted to afabicin desphosphono when administered systemically,20 had on the gut microbiota of mice and healthy subjects. Accordingly, findings from the current study support those of Yao et al.,25 which revealed no significant changes in murine gut diversity measures attributable to afabicin desphosphono treatment. The minor differences in taxonomic abundance changes observed between this study and that of Yao et al. can be explained by the distinct baseline microbiota of the two different animal species used in each.39
The minimal effects of afabicin treatment on the gut microbiota contrasted with that of clindamycin, linezolid and moxifloxacin, each of which caused extensive dysbiosis in the murine model, supporting findings from previous animal studies.25,40,41 Further, the global microbiota response to clindamycin and linezolid treatment, each protein synthesis inhibitors, was similar, and distinct from that of moxifloxacin (a fluoroquinolone targeting DNA gyrase), which is in accordance with previous findings.25 The most striking change for clindamycin- and linezolid-treated animals was the increase in Enterobacteriaceae; this family contains important opportunistic pathogens known to cause antibiotic-resistant infections including Escherichia coli and Klebsiella pneumoniae.42 Of note, clindamycin and linezolid treatment have each been shown to select for Enterobacteriaceae in human subjects.43,44
It is important to consider the translatability of murine microbiota studies for human disease. Whilst the gastrointestinal tract of mice and men share some similarities, the actual composition of the microbiota in terms of relative taxa abundance is quite distinct.39 The main objective of the current study was to assess global microbiota dysbiosis due to antibiotic therapy, which has a now well-recognized link to metabolic and inflammatory diseases, as well as secondary bacterial infections.45 Collectively, our findings, and the findings of others, suggest that the relative sensitivity of the murine gut microbiota to antibiotic-induced dysbiosis is similar to that of humans. Ten-day courses of oral clindamycin (either 150 mg four times per day or 500 mg twice daily) significantly altered human gut microbiota diversity (Shannon H) and richness measures, with changes persisting for up to two months,46,47 mirroring findings from the current murine study. In addition, a 5 day course of oral moxifloxacin (400 mg once daily) was associated with a median maximal Shannon H loss of 27.5% in healthy subjects,48 which was recapitulated here using the murine model (26.0% median loss at Day 10). Importantly, minimal changes to the murine microbiota due to afabicin treatment were also observed in healthy subjects in the current study. These findings have clinical relevance, as antibiotic-induced dysbiosis in mice has been shown to underpin important diseases including C. difficile colitis,40,49 which is well described in clinical studies,50 and VRE colonization, which was shown to precede bloodstream infection in patients undergoing allogeneic HSCT.51
The current study has notable limitations. Firstly, four to five animals per group is adequate to identify large statistical shifts in relative taxa abundance, as shown for clindamycin, linezolid and moxifloxacin; however, it may not adequately detect subtle changes that may have been caused by afabicin treatment. Secondly, OTU based analysis of 16S rDNA sequencing was performed in the current study, which may not have had the appropriate sensitivity to detect small differences in relative taxa abundance. Future studies should consider using amplicon sequence variant (ASV) analyses as this approach is less prone to bias introduced by aligning to sequence databases,52 as well as the use of shallow shotgun sequencing, which may provide more accurate and higher-resolution taxonomic profiling.53 Finally, it would have been useful to measure the concentration of afabicin/afabicin desphosphono and comparator antibiotics in the longitudinal faecal samples, and to relate these data to effects upon the microbiota. Taken together, it is reasonable to conclude that afabicin produced significantly less microbiota disruption compared with clindamycin, linezolid and moxifloxacin; however, subtle signs of dysbiosis may not have been detected due to the limitations described above.
To summarize, afabicin is a member of a short list of microbiota-sparing antibiotics currently in clinical development.10 Further efforts are warranted to expand this list through reorientation of research towards highly targeted pathogen-specific antibiotics with very narrow spectrum of activity and low ecological impact;3,9,54–56 clinical implementation of highly targeted antimicrobials should limit dysbiosis, thereby helping to control AMR.
Supplementary Material
Acknowledgements
We acknowledge the efforts of M. Barbier, K. Stanczewska, A. Parks and P. Vaissié. The MIC study was performed at Micromyx (Kalamazoo, MI, USA).
Contributor Information
J Nowakowska, Translational Medicine Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
D R Cameron, Translational Medicine Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
A De Martino, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
J Kühn, Translational Medicine Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
S Le Fresne-Languille, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
S Leuillet, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
Y Amouzou, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
F Wittke, Clinical Development Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
T Carton, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
F Le Vacon, Research and Development Department, Biofortis SAS, 3 route de la Chatterie, 44800 Saint-Herblain, France.
R L Chaves, Clinical Development Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
V Nicolas-Metral, Translational Medicine Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
G Vuagniaux, Translational Medicine Department, Debiopharm International SA, Chemin Messidor 5-7, 1006 Lausanne, Switzerland.
Funding
Findings from this study were financed by Debiopharm International SA.
Transparency declarations
D.R.C., F.W., J.K., J.N., R.L.C., V.N.M. and G.V. are or were employees of Debiopharm International SA at the time of the study conduct. S.L.F.L., S.L., Y.A., A.D.M., T.C. and F.L.V. are or were employees of Biofortis SAS.
Supplementary data
Figures S1 to S3 and Table S1 are available as Supplementary data at JAC Online.
References
- 1. Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016; 535: 85–93. 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016; 22: 458–78. 10.1016/j.molmed.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhalodi AA, van Engelen TSR, Virk HS et al. Impact of antimicrobial therapy on the gut microbiome. J Antimicrob Chemother 2019; 74: i6–i15. 10.1093/jac/dky530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anthony WE, Burnham CD, Dantas G et al. The gut microbiome as a reservoir for antimicrobial resistance. J Infect Dis 2021; 223: S209–S13. 10.1093/infdis/jiaa497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Worley J, Delaney ML, Cummins CK et al. Genomic determination of relative risks for Clostridioides difficile infection from asymptomatic carriage in intensive care unit patients. Clin Inf Dis 2021; 73: e1727–e36. 10.1093/cid/ciaa894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Modi SR, Lee HH, Spina CS et al. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013; 499: 219–22. 10.1038/nature12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamberte LE, van Schaik W. Antibiotic resistance in the commensal human gut microbiota. Curr Opin Microbiol 2022; 68: 102150. 10.1016/j.mib.2022.102150 [DOI] [PubMed] [Google Scholar]
- 8. Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science 2016; 352: 544–5. 10.1126/science.aad9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diamantis S, Retur N, Bertrand B et al. The production of antibiotics must be reoriented: repositioning old narrow-spectrum antibiotics. developing new microbiome-sparing antibiotics. Antibiotics 2022; 11: 924. 10.3390/antibiotics11070924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Avis T, Wilson FX, Khan N et al. Targeted microbiome-sparing antibiotics. Drug Discov Today 2021; 26: 2198–203. 10.1016/j.drudis.2021.07.016 [DOI] [PubMed] [Google Scholar]
- 11. Barlam TF, Cosgrove SE, Abbo LM et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62: e51–77. 10.1093/cid/ciw118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Waele JJ, Schouten J, Beovic B et al. Antimicrobial de-escalation as part of antimicrobial stewardship in intensive care: no simple answers to simple questions—a viewpoint of experts. Intensive Care Med 2020; 46: 236–44. 10.1007/s00134-019-05871-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garnacho-Montero J, Gutiérrez-Pizarraya A, Escoresca-Ortega A et al. De-escalation of empirical therapy is associated with lower mortality in patients with severe sepsis and septic shock. Intensive Care Med 2014; 40: 32–40. 10.1007/s00134-013-3077-7 [DOI] [PubMed] [Google Scholar]
- 14. FDA . Antibacterial Therapies for Patients with an Unmet Medical Need for the Treatment of Serious Bacterial Diseases - Questions and Answers (Revision 1). 2022. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/antibacterial-therapies-patients-unmet-medical-need-treatment-serious-bacterial-diseases-questions.
- 15. Jernigan JA, Hatfield KM, Wolford H et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med 2020; 382: 1309–19. 10.1056/NEJMoa1914433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Widerström M. Significance of Staphylococcus epidermidis in health care-associated infections, from contaminant to clinically relevant pathogen: this is a wake-up call! J Clin Micro 2016; 54: 1679–81. 10.1128/JCM.00743-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li HK, Rombach I, Zambellas R et al. Oral versus intravenous antibiotics for bone and joint infection. N Engl J Med 2019; 380: 425–36. 10.1056/NEJMoa1710926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouijzer IJE, van Leerdam EJ, Gompelman M et al. Intravenous to oral switch in complicated Staphylococcus aureus bacteremia without endovascular infection: a retrospective single-center cohort study. Clin Inf Dis 2021; 73: 895–8. 10.1093/cid/ciab156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao J, Rock CO. Bacterial fatty acid metabolism in modern antibiotic discovery. Biochim Biophys Acta Mol Cell Biol Lipids 2017; 1862: 1300–9. 10.1016/j.bbalip.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hafkin B, Berg JK, Kaplan N et al. Single-dose escalation study to evaluate the safety, tolerability, and pharmacokinetics of a FabI inhibitor, the prodrug Debio 1450 and its active moiety Debio 1452, administered intravenously in healthy subjects. Open Forum Infect Dis 2015; 2: 797. 10.1093/ofid/ofv133.514 [DOI] [Google Scholar]
- 21. Wittke F, Vincent C, Chen J et al. Afabicin, a first-in-class antistaphylococcal antibiotic, in the treatment of acute bacterial skin and skin structure infections: clinical noninferiority to vancomycin/linezolid. Antimicrob Agents Chemother 2020; 64: e00250-20. 10.1128/AAC.00250-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaplan N, Albert M, Awrey D et al. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrob Agents Chemother 2012; 56: 5865–74. 10.1128/AAC.01411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karlowsky JA, Kaplan N, Hafkin B et al. AFN-1252, a FabI inhibitor, demonstrates a Staphylococcus-specific spectrum of activity. Antimicrob Agents Chemother 2009; 53: 3544–8. 10.1128/AAC.00400-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. WHO . 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. 2022. https://www.who.int/publications/i/item/9789240047655.
- 25. Yao J, Carter RA, Vuagniaux G et al. A pathogen-selective antibiotic minimizes disturbance to the microbiome. Antimicrob Agents Chemother 2016; 60: 4264–73. 10.1128/AAC.00535-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CLSI . Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria—Ninth Edition: M11. 2018.
- 27. Kim B-N, Kim ES, Oh M-D. Oral antibiotic treatment of staphylococcal bone and joint infections in adults. J Antimicrob Chemother 2014; 69: 309–22. 10.1093/jac/dkt374 [DOI] [PubMed] [Google Scholar]
- 28. Klindworth A, Pruesse E, Schweer T et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013; 41: e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schloss PD, Westcott SL, Ryabin T et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75: 7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edgar RC, Haas BJ, Clemente JC et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27: 2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q, Garrity GM, Tiedje JM et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73: 5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. R Core Team . R: A Language and Environment for Statistical Computing. 2015. https://www.r-project.org/.
- 33. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8: e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balemans W, Lounis N, Gilissen R et al. Essentiality of FASII pathway for Staphylococcus aureus. Nature 2010; 463: E3, discussion E4. 10.1038/nature08667 [DOI] [PubMed] [Google Scholar]
- 35. Heath RJ, Su N, Murphy CK et al. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J Biol Chem 2000; 275: 40128–33. 10.1074/jbc.M005611200 [DOI] [PubMed] [Google Scholar]
- 36. Heath RJ, Rock CO. A triclosan-resistant bacterial enzyme. Nature 2000; 406: 145–6. 10.1038/35018162 [DOI] [PubMed] [Google Scholar]
- 37. Brinster S, Lamberet G, Staels B et al. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 2009; 458: 83–6. 10.1038/nature07772 [DOI] [PubMed] [Google Scholar]
- 38. Parker EN, Drown BS, Geddes EJ et al. Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat Microbiol 2020; 5: 67–75. 10.1038/s41564-019-0604-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 2018; 75: 149–60. 10.1007/s00018-017-2693-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buffie CG, Jarchum I, Equinda M et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 2012; 80: 62–73. 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hertz FB, Budding AE, van der Lugt-Degen M et al. Effects of antibiotics on the intestinal microbiota of mice. Antibiotics 2020; 9: 191. 10.3390/antibiotics9040191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antibiotic Resistance Collaborators . Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nyberg SD, Osterblad M, Hakanen AJ et al. Long-term antimicrobial resistance in Escherichia coli from human intestinal microbiota after administration of clindamycin. Scand J Infect Dis 2007; 39: 514–20. 10.1080/00365540701199790 [DOI] [PubMed] [Google Scholar]
- 44. Lode H, Von der Höh N, Ziege S et al. Ecological effects of linezolid versus amoxicillin/clavulanic acid on the normal intestinal microflora. Scand J Infect Dis 2001; 33: 899–903. 10.1080/00365540110076714 [DOI] [PubMed] [Google Scholar]
- 45. Carding S, Verbeke K, Vipond DT et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015; 26: 26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rashid MU, Zaura E, Buijs MJ et al. Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin Inf Dis 2015; 60 Suppl 2: S77–84. 10.1093/cid/civ137 [DOI] [PubMed] [Google Scholar]
- 47. Zaura E, Brandt BW, Teixeira de Mattos MJ et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio 2015; 6: e01693-15. 10.1128/mBio.01693-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burdet C, Nguyen TT, Duval X et al. Impact of antibiotic gut exposure on the temporal changes in microbiome diversity. Antimicrob Agents Chemother 2019; 63: e00820-19. 10.1128/AAC.00820-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bassis CM, Theriot CM, Young VB. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother 2014; 58: 2767–74. 10.1128/AAC.02262-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vardakas KZ, Trigkidis KK, Boukouvala E et al. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: a systematic review and meta-analysis. Int J Antimicrob Agents 2016; 48: 1–10. 10.1016/j.ijantimicag.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 51. Ubeda C, Taur Y, Jenq RR et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120: 4332–41. 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 2017; 11: 2639–43. 10.1038/ismej.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hillmann B, Al-Ghalith GA, Shields-Cutler RR et al. Evaluating the information content of shallow shotgun metagenomics. mSystems 2018; 3: e00069-18. 10.1128/mSystems.00069-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest 2014; 124: 4212–8. 10.1172/JCI72333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paharik AE, Schreiber HLT, Spaulding CN et al. Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med 2017; 9: 110. 10.1186/s13073-017-0504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Melander RJ, Zurawski DV, Melander C. Narrow-spectrum antibacterial agents. MedChemComm 2018; 9: 12–21. 10.1039/C7MD00528H [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.