Abstract
The influenza A virus nucleoprotein (NP) is a multifunctional polypeptide which plays a pivotal role in virus replication. To get information on the domains and specific residues involved in the different NP activities, we describe here the preparation and characterization of 20 influenza A virus mutant NPs. The mutations, mostly single-amino-acid substitutions, were introduced in a cDNA copy of the A/Victoria/3/75 NP gene and, in most cases, affected residues located in regions that were highly conserved across the NPs of influenza A, B, and C viruses. The mutant NPs were characterized (i) in vivo (cell culture) by analyzing their intracellular localization and their functionality in replication, transcription, and expression of model RNA templates; and (ii) in vitro by analyzing their RNA-binding and sedimentation properties. The results obtained allowed us to identify both a mutant protein that accumulated in the cytoplasm and mutations that altered the functionality and/or the oligomerization state of the NP polypeptide. Among the mutations that reduced the NP capability to express chloramphenicol acetyltransferase protein from a model viral RNA (vRNA) template, some displayed a temperature-sensitive phenotype. Interestingly, four mutant NPs, which showed a reduced functionality in synthesizing cRNA molecules from a vRNA template, were fully competent to reconstitute complementary ribonucleoproteins (cRNPs) capable of synthesizing vRNAs, which in turn yielded mRNA molecules. Based on the phenotype of these mutants and on previously published observations, it is proposed that these mutant NPs have a reduced capability to interact with the polymerase complex and that this NP-polymerase interaction is responsible for making vRNPs switch from mRNA to cRNA synthesis.
Influenza A viruses contain a genome made up of eight negative-sense single-stranded RNA molecules. In the viral particle the genomic RNAs are found in the form of ribonucleoprotein (RNP) complexes which contain four virus-encoded polypeptides, the nucleoprotein (NP), which encapsidates the viral RNA (vRNA), and the three subunits (PB1, PB2, and PA) of the viral polymerase (18, 19).
The RNA segment 5 of influenza A virus codes for the NP, a basic protein that is 498 amino acids in length, which is phosphorylated in vivo (2, 15, 39). In vitro, the NP protein, purified from virions and devoid of RNA, assembles into polymeric forms ranging from trimers to large structures indistinguishable from authentic RNP complexes (42). In infected cells, the NP protein can form small oligomers (dimers and trimers) (40). These data, together with the observation that the RNP structure is maintained even when the vRNA is removed from viral nucleocapsids (14, 37), indicate that the protein contains an NP-NP binding domain (not yet identified) and that NP-NP interactions are critical for maintaining the structure of RNPs.
The NP protein displays RNA-binding activity, but no specificity for viral sequences has been demonstrated (1, 4, 13, 51). The N-terminal 180-amino-acid portion of the NP bears an RNA binding-domain, which can be subdivided into two smaller regions (residues 1 to 77 and 79 to 180) that also retained RNA-binding activity (1, 16).
At early times postinfection the newly synthesized NP is detected in the cell nucleus, and a sequence, located within amino acids 327 to 345 of the NP, was identified as required for nuclear accumulation of the protein in Xenopus oocytes (7). The significance of this sequence for nuclear accumulation of NP in mammalian cells has been questioned since NPs lacking this region accumulate efficiently in the cell nucleus (32, 50). Moreover, a nuclear localization signal has been identified within the 20 N-terminal residues of NP (32, 50).
The RNP complexes are the functional templates for replication and transcription of the viral genome (17, 19) and produce three different virus-specific RNA species: (i) mRNA molecules which are capped and polyadenylated, (ii) negative-sense vRNA molecules (found in the viral particle), and (iii) cRNA molecules which serve as templates for the synthesis of vRNA molecules. Although the three P proteins (PB1, PB2, and PA) constitute the RNA polymerase, biochemical and genetic evidences indicates that NP is involved in the RNA synthesis processes (3, 5, 11, 20, 25, 27, 45, 49). In fact, it has been shown that nucleocapsids in which most of the NP has been removed are unable to synthesize template-sized RNA transcripts (11) and that NP is required for the synthesis of vRNA and cRNA molecules (5, 45). In particular, experiments with mutants ts56, which contains an amino acid mutation at residue 314 of the NP (20), have been useful for demonstrating a role for NP in cRNA synthesis (45). In fact, it has been shown that nucleocapsids obtained from ts56-infected cells can synthesize mRNA but not cRNA templates at the nonpermissive temperature.
In a natural infection the incoming vRNPs give rise to mRNAs which are initiated by short capped RNA fragments derived from cellular heterogeneous nuclear RNAs. During elongation of mRNA molecules, the polymerase remains bound to the 5′ end of the vRNA and acts as an obstacle that prevents copying the end of the template (38, 48). As a consequence, the polymerase reiteratively copies a short oligo(U) stretch found 17 nucleotides from the 5′ end of the template, and the mRNA transcript becomes polyadenylated (21). Later in infection, the vRNPs switch from mRNA to cRNA synthesis. The cRNA molecules are also positive sense but they are initiated without a primer, and for its synthesis the polymerase passes through the oligo(U) stretch that serves as the polyadenylation signal. Although it has been demonstrated that free NP molecules are required for switching vRNPs from transcription to replication (5, 45) the precise mechanism by which NP carries this function is unknown.
NP is thus a multifunctional protein that plays a central role in influenza virus replication. To gain information on the protein regions (and specific residues) relevant for the various NP activities, we prepared 20 influenza virus mutant NPs by introducing specific mutations in a cDNA encoding the A/Victoria/3/75 NP protein. The recombinant proteins were expressed in mammalian cells and were characterized functionally and biochemically in a number of assays. The results obtained allowed the identification of specific residues that alter several of the activities associated to the NP protein and, in particular, mutants with altered transcriptional and replication capabilities.
MATERIALS AND METHODS
Biological materials and reagents.
COS-1 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal calf serum. The recombinant vaccinia virus vTF7-3 (9), which expresses T7 RNA polymerase, was kindly provided by B. Moss. Plasmids pGEM-PB1, pGEM-PB2, pGEM-PA, and pGEM-NP containing the influenza virus PB1, PB2, PA, and NP genes (of the A/Victoria/3/75 strain), respectively, cloned downstream from the T7 RNA polymerase promoter of plasmid pGEM-3 have been described (8, 29). Plasmid pIVACAT1/S (35) (a gift from P. Palese) was digested with HgaI and transcribed in vitro with T7 RNA polymerase to yield synthetic influenza virus-like chloramphenicol acetyltransferase (CAT) RNA molecules of negative polarity (hereafter designated NS-CAT RNA). Plasmids pNSZ and pcNSZ have been described (34). These plasmids, which derive from pIVACAT1/S and contain deletions in the CAT gene open reading frame, were used to generate model influenza virus RNA templates of negative (vNSZ RNA; 240 nucleotides [nt]) and positive polarity (cNSZ RNA; 240 nt). Cationic liposomes were prepared according to the procedure described by Rose et al. (41).
Antibodies and antisera.
Monoclonal antibody M/58/p44/E, which recognizes the A/Victoria/3/75 NP protein (23), and rabbit antisera raised against fusion proteins containing the N-terminal 77 amino acids or the C-terminal 120 amino acids of the NP have been described (2). A polyclonal antiserum against full-length NP was prepared by repeated immunization of rabbits with purified RNPs obtained from the A/Victoria/3/75 strain.
Construction of NP mutants.
Mutations were introduced in the NP gene of plasmid pGEM-NP by oligonucleotide-directed mutagenesis by using the Transformer site-directed mutagenesis kit (Clontech). In some cases the mutagenesis was carried out with mixtures of oligonucleotides that contained the desired nucleotide substitutions and that affected the same codon. All mutations introduced in the NP gene were confirmed by sequencing through the mutagenized region.
Expression of the NS-CAT RNA by recombinant influenza virus polymerase.
Experiments were basically carried out as previously described (29, 43). Briefly, COS-1 cells growing in 35-mm-diameter dishes were infected with vTF7-3 (multiplicity of infection [MOI] = 5) and transfected, by using cationic liposomes, with plasmids pGEM-PB1 (1 μg), pGEM-PB2 (1 μg), pGEM-PA (0.2 μg), and pGEM-NP (2.8 μg) (or the corresponding plasmid encoding a mutant NP). Five hours later, cells were transfected again with a mixture containing 0.5 μg of the synthetic NS-CAT RNA and 4.5 μg of yeast tRNA and then incubated for 18 h at 37°C. Cells were then scraped off the plates, separated into two identical aliquots, and pelleted by centrifugation. One of the aliquots was resuspended in sodium dodecyl sulfate (SDS) sample buffer and analyzed by Western blotting with anti-NP serum. The other aliquot was resuspended in 100 μl of 0.25 M Tris-HCl (pH 7.5), lysed by three cycles of freezing and thawing, and clarified by centrifugation for 5 min in a microcentrifuge. Aliquots of the clarified supernatant were used to determine the total protein content (with the Bio-Rad [Bradford] protein assay kit) and to determine CAT activity (with 0.1 μCi of [14C]chloramphenicol) and chromatography on thin-layer chromatography (TLC) plates. Routinely, different amounts (0.5 to 25 μl) of the cell extracts were incubated with radioactive chloramphenicol for 2 h to obtain CAT activity values in the linear range of the assay. Quantitation of the CAT activity was performed by phosphorimaging the acetylated spots detected on TLC plates (with a Fujix Bas 1000 equipment and the software PCBAS v2.09). The values obtained were corrected for the protein concentration of the cell extract and were expressed as a percentage of the CAT activity observed for cells expressing the wild-type NP. The detection limit was 1% of the value obtained for wild-type NP. To determine whether the NP mutants had a temperature-sensitive (Ts) phenotype, COS-1 cells were maintained at 33°C for 24 h. The cultures were then infected and transfected as indicated above, except that cells were always kept at 33°C and the cell extracts were prepared 30 h postinfection. Under these conditions, the levels of CAT expression yielded by wild-type NP were similar to those obtained in cell cultures transfected and maintained at 37°C for 18 h.
Accumulation of virus-specific RNA species in cells transfected with model RNA templates.
Assays were essentially done as described by Perales and Ortín (34). Briefly, COS-1 cells growing in 60-mm-diameter dishes were infected with vTF7-3 (MOI = 10) and transfected, by using cationic liposomes, with plasmids pGEM-PB1 (1.5 μg), pGEM-PB2 (1.5 μg), pGEM-PA (0.15 μg), and pGEM-NP (6 μg) (or plasmids encoding mutant NPs). After incubation for 5 h, cell cultures were transfected with a mixture containing a model RNA template (vNSZ, 300 ng; or cNSZ RNA, 100 ng) and 1 μg of carrier tRNA. Cells were harvested 16 h later, and total RNA was isolated by using the Ultraspec RNA isolation system (Biotecx). Total RNA was then fractionated into poly(A)+ and poly(A)− by oligo(dT)-cellulose chromatography. The poly(A)− fraction was self-annealed and treated with RNase A to select for vRNA-cRNA hybrids. Aliquots of the two samples [poly(A)+ and self-annealed poly(A)− fractions] were incubated with positive- or negative-polarity 32P-labeled RNA probes and analyzed by using the RNase protection assay. The protected fragments derived from the probes were visualized after electrophoresis in a 4% sequencing gel and autoradiography. The poly(A)+ fraction was hybridized to a negative-sense probe (vNSZ-L RNA), whereas the poly(A)− fraction was hybridized to a probe with the same polarity of the model RNA transfected into the cells. To synthesize the 32P-labeled probes, plasmid pNSZ or pcNSZ were digested with Asp718 or EcoRI, respectively, and transcribed in vitro in the presence of [α-32P]GTP to yield RNA probes vNSZ-L (264 nt; negative sense) and cNSZ-L RNA (269 nt; positive sense).
Sedimentation analysis in sucrose gradients.
COS-1 cells growing in 35-mm-diameter dishes were infected with vTF7-3 (MOI = 5) and transfected individually with the different pGEM-NP-derived plasmids (4 μg). After 5 h of incubation, the medium was replaced with 500 μl of a medium containing 30 μCi of Tran35S-label (ICN) and in 9:1 methionine-free DMEM–complete DMEM. All cell culture media contained cytosine β-d-arabinofuranosylcytosine (AraC) at 40 μg/ml. After incubation for 18 h, cells were scraped off the dish, collected by a brief spin in a microcentrifuge, washed twice with phosphate-buffered saline, and resuspended in 200 μl of buffer A (50 mM NaCl, 25 mM Tris-HCl [pH 7.5]). Cells were lysed by three consecutive freeze-thaw cycles, and the homogenates were clarified by centrifugation for 1 min at room temperature at ∼10,000 rpm in a microcentrifuge. The pellet of this centrifugation was resuspended in 200 μl of buffer A supplemented with 10 mM of MgCl2 and treated with DNase I. Aliquots (10 μl) of the pellet and of the supernatant fractions were analyzed by SDS polyacrylamide gel electrophoresis (PAGE) and autoradiography. The pooled supernatant (190 μl) was centrifuged through a linear 5 to 20% sucrose gradient (4.6 ml) at a value of w2t = 3 × 1011 s−1 (20,000 rpm for ∼19 h) in an SW50.1 rotor at 4°C. A total of 12 fractions (of 400 μl) were collected from the top and analyzed by SDS-PAGE and autoradiography.
RNA binding studies.
COS-1 cells growing in 35-mm-diameter dishes were infected with vTF7-3 and transfected with the different pGEM-NP-derived plasmids as described above. After incubation for 24 h, cells were scraped off the dishes, washed twice with phosphate-buffered saline, and harvested by low-speed centrifugation. Cells were resuspended in 60 μl of a buffer containing 1× TNE (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) and 1% Nonidet P-40. After incubation for 15 min on ice, with occasional vortexing, the cell lysates were supplemented with 60 μl of 1× TNE to bring the Nonidet P-40 concentration to 0.5%. This cell lysate was loaded onto a CsCl-glycerol step gradient (11, 28, 33). This gradient had four steps of 120 μl of: 3 M CsCl–43.5% glycerol, 2 M CsCl–34% glycerol, 1 M CsCl–26.1% glycerol, and 0.5 M CsCl–8.7% glycerol (all steps were buffered with 20 mM Tris-HCl (pH 7.5). The sample was centrifuged at 35,000 rpm in an SW50.1 rotor (in 0.8-ml tubes) for 24 h at 4°C. A total of 15 fractions of 50 μl were collected from the top, and the NP-containing fractions were identified by SDS-PAGE and Western blotting. Aliquots (5 μl) of the indicated fractions were incubated with 32P-labeled NS-CAT RNA (2 ng, corresponding to ∼105 cpm) for 30 min at 22°C in a buffer containing 10 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 100 mM NaCl. These mixtures were then irradiated for 15 min with UV light (250 nm) and treated with RNase A (1). The proteins were resolved by SDS-PAGE and electrotransfered onto Immobilon-P paper. This membrane was exposed to an X-ray film to detect the 32P-labeled RNA cross-linked to proteins, and then the same membrane was assayed by Western blotting with a rabbit antiserum against the NP C-terminal end.
Immunochemical techniques.
For Western blotting, cell extracts were resolved by SDS-PAGE, transferred to Immobilon-P paper, and developed with the appropriate antibody by using the ECL kit (Amersham) as previously described (2). For immunofluorescence, COS-1 cells on coverslips were infected with vTF7-3 (MOI = 1). These cultures were transfected with DNA mixtures that contained a total of 4 μg of plasmid DNA, including plasmid pGEM-4 and different amounts of the NP recombinant plasmids (60, 200, or 1,000 ng). Cells were maintained for 5 h with the DNA-liposome mixture, washed with DMEM, and incubated for another 18 h. At this time, cells were fixed in cold methanol for 20 min. To visualize the NP proteins, coverslips were incubated sequentially with the appropriate primary antibody (either monoclonal antibody M/58/p44/E or a polyclonal anti-RNP serum) and with a solution containing fluorescein-conjugated immunoglobulins and the nuclear Hoechst 33258 dye.
Nucleotide sequence accession number.
The nucleotide sequence of the NP gene of A/Victoria/3/75 strain cloned in pGEM-NP plasmid is available from GenBank under accession number AF072545.
RESULTS
Mutagenesis strategy.
The NP protein of influenza A virus is highly conserved (44, 46) and shares significant amino acid homology with the NPs of influenza B (38% homology) and C viruses (14% homology) (22, 31). Moreover, there are short regions (10 to 15 amino acids) remarkably conserved across virus types (with amino acid homologies greater than 50% (22, 31, and data not shown) that could constitute protein functional domains. To get information on the roles played by these conserved regions, it was decided to introduce mutations in the corresponding regions of a cDNA clone coding for the influenza A/Victoria/3/75 NP and then to characterize, both phenotypically and biochemically, the mutant proteins obtained.
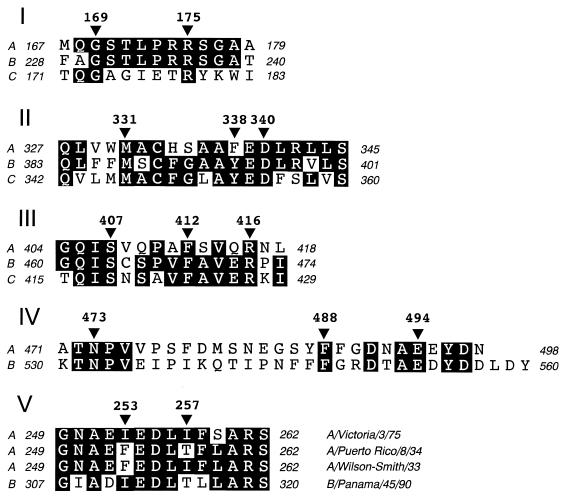
One region chosen for mutagenesis analysis extended from positions 169 to 178 of the influenza A virus NP (Fig. 1I), which corresponds the longest stretch of 100% amino acid identity between influenza A and B virus NPs. This region, which is not well conserved in influenza C virus, is located within the RNA-binding domain identified for influenza A virus NP (1, 16). In addition, we also decided to mutate residues located in the two regions that are most highly conserved across the NPs of the influenza A, B, and C viruses (more than 50% of residues conserved in the three NPs) (Fig. 1II and III). One of these regions, which extends from positions 331 to 340 of influenza A virus NP, partially overlaps with the region required for nuclear accumulation of NP in Xenopus oocytes (7), whereas the other region, which includes residues 405 to 416 of influenza A virus NP, had no assigned role. Mutations were also introduced at the C-terminal end of NP, a region which is peculiar in that the last 12 amino acids include four aromatic and four acidic residues (Fig. 1IV). A similar pattern is also observed for influenza B virus NP but not for the influenza C virus protein.
FIG. 1.
Conserved regions in the NPs of influenza virus and localization of mutagenized residues. The five panels (I to V) correspond to the different NP regions that were chosen for mutagenesis analysis (see the text for details). Sequences shown in panels I to IV correspond to the NP proteins of the influenza virus A/Victoria/3/75 (A) (GenBank accession number AF072545), B/Panama 45/90 (B) (12), and C/California/78 (C) (31). In panel V, the NP sequences correspond to the viral strains indicated on the right. The numbers at both sides of the amino acid sequences indicate the positions of the first and last residues in the corresponding NP protein. The positions that are totally conserved across the different NP sequences are highlighted. The residues, which were mutated in the A/Victoria/3/75 NP gene, are indicated by black triangles and a number above it that corresponds to the amino acid position.
Several single-amino-acid mutant proteins affecting conserved residues of the four above-mentioned regions were obtained (Fig. 1 and Table 1). Since NP is an RNA-binding protein and several of its activities imply interactions with other proteins, it was decided to replace preferentially aromatic and charged residues because these kinds of amino acids have been involved in protein-RNA and protein-protein interactions, respectively. Some of the mutations introduced were drastic substitutions (i.e., changing a basic for an acidic residue), whereas others were conservative changes (i.e., substituting Gly for Ala), on the expectation that these mutations would either abolish or reduce the normal role of that particular NP region, respectively. Two of the mutations were predicted to shorten the C-terminal end of the protein by 5 or 26 amino acids (proteins 473* and 494*), and one protein (494in), which was artifactually obtained during the preparation of plasmid pGNP-494*, was a frameshift mutant that contained 12 unrelated residues instead of the last five amino acids of the wild-type NP. In addition to all of the single mutants, protein DM containing two substitutions, at positions 253 and 257, was prepared (Fig. 1V). These two residues are included in an NP region that is totally conserved in all human strains except for the early isolates A/Puerto Rico/8/34 and A/Wilson-Smith/33 (44, 46).
TABLE 1.
Plasmids encoding mutant NPs of the influenza virus A/Victoria/3/75a
| Plasmid | Protein | Amino acid
|
Codon
|
|||
|---|---|---|---|---|---|---|
| Position | WT | Mut | WT | Mut | ||
| pGNP-169A | G169A | 169 | G | A | ggt | gct |
| pGNP-169D | G169D | 169 | G | D | ggt | gat |
| pGNP-175K | R175K | 175 | R | K | agg | aag |
| pGNP-175T | R175T | 175 | R | T | agg | acg |
| pGNP-331K | M331K | 331 | M | K | atg | aag |
| pGNP-331T | M331T | 331 | M | T | atg | acg |
| pGNP-338A | F338A | 338 | F | A | ttt | gca |
| pGNP-338E | F338E | 338 | F | E | ttt | gaa |
| pGNP-340H | D340H | 340 | D | H | gat | cat |
| pGNP-340R | D340R | 340 | D | R | gat | cgt |
| pGNP-407A | S407A | 407 | S | A | agt | gct |
| pGNP-412E | F412E | 412 | F | E | ttt | gaa |
| pGNP-416E | R416E | 416 | R | E | aga | gaa |
| pGNP-473R | N473R | 473 | N | R | aac | aga |
| pGNP-473* | 473* | 473 | N | STOP | aac | tga |
| pGNP-488G | F488G | 488 | F | G | ttc | gga |
| pGNP-494R | E494R | 494 | E | R | gag | aga |
| pGNP-494* | 494* | 494 | E | STOP | gag | tga |
| pGNP-494in | 494in | 494 | E | STTIKEKYPCFY | gag | ga− |
| pGNP-DM | DM | 253 | I | F | atc | ttc |
| 257 | I | T | ata | aca | ||
The name of the plasmid and of the mutant protein, as well as the codon and amino acid position in the wild-type (WT) and in the mutant (Mut) NP proteins, are indicated.
Activity of mutant NPs to express a synthetic CAT RNA.
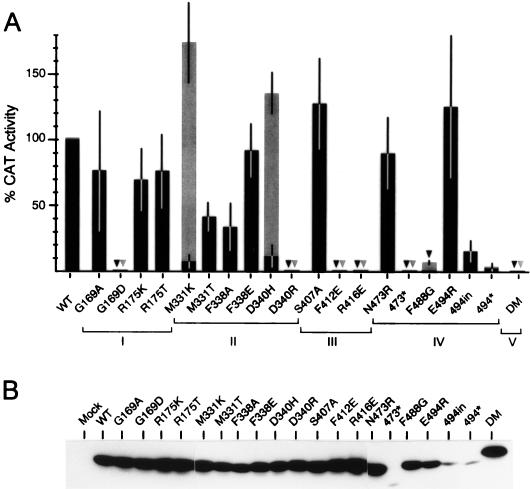
We have previously described a system in which expression in COS-1 cells of a synthetic influenza virus-like CAT RNA (negative sense) is driven by viral proteins (NP, PB1, PB2, and PA) expressed from recombinant plasmids (29). All mutant NPs were tested in this artificial system, and the results obtained in several independent experiments are summarized in Fig. 2A. Based on these results, the mutations could be classified into four different categories: (i) mutations that did not affect the NP function (CAT values between 50 and 100% of that obtained with the wild-type NP), (ii) mutations that moderately reduced CAT expression (between 20% and 50%), (iii) mutations that drastically reduced NP function (CAT values between 2 and 20%), and finally (iv) seven mutations that totally abolished NP function (less than 1% of CAT activity). Most of the mutant proteins classified within the last two categories were also tested for functionality in cultures that were maintained at 33°C instead of at 37°C. As can be seen in Fig. 2A and Table 2, most of these mutants did not change their phenotypes at this lower temperature. However, there were two proteins (M331K and D340H) which regained full functionality compared to wild-type NP and a third mutant protein (F488G) which had some activity when assayed at 33°C. The differences in CAT expression were not due to differences in the level of accumulation of the mutant proteins as determined by Western blotting (Fig. 2B). It should be noted, however, that proteins with mutations affecting the C-terminal end of the protein (473*, 494*, and 494in) were poorly recognized by a polyclonal serum that recognizes the last 121 amino acids of NP (Fig. 2B). However, these proteins accumulated to levels similar to that of the wild-type NP, as demonstrated by developing the Western blotting with a serum raised against the 77 N-terminal residues of NP (data not shown). As can be observed in Fig. 2B, the deletion mutants 473* and 494* migrated faster than NP in the acrylamide gel, a finding in good agreement with their predicted sizes. Strikingly, mutants N473R and DM showed an altered mobility in the gel despite having the length of the wild-type NP. The NP gene in mutant DM was fully sequenced to demonstrate that the altered mobility of this protein was exclusively due to the two substitutions indicated in Table 1.
FIG. 2.
Expression of a synthetic NS-CAT RNA by recombinant influenza virus polymerase. COS-1 cells were infected with vTF7-3 and transfected with the plasmids encoding the three P proteins and with either wild-type (WT) or mutant NP-encoding plasmids (as indicated). Cultures were then transfected with a synthetic CAT RNA, and cell extracts were prepared. Aliquots of these extracts were used to determine CAT activity (A) or to detect the recombinant NPs by immunoblotting (B) (with an antiserum raised against the C-terminal region of NP) as detailed in Materials and Methods. (A) The results depicted are the average values and the standard deviation calculated from two or three independent transfection experiments. In each experiment, the CAT expression level obtained with the wild-type NP was taken as 100%, and therefore the activity of the wild-type protein had no standard deviation. The symbols (bars and triangles) correspond to the CAT levels reached when the transfection experiments were carried out at 37°C (black symbols) or at 33°C (gray symbols). Triangles were used to indicate mutants that yielded less than 1% of the CAT activity compared with the wild-type NP.
TABLE 2.
Characteristics of the mutant NP proteinsa
| Region | Protein | CAT activityb at:
|
Synthesis of:
|
Solu-bilityc | Accumu-lationd | RNA binding | |||
|---|---|---|---|---|---|---|---|---|---|
| 37°C | 33°C | mRNA | cRNA | vRNA | |||||
| None | WT NP | +++ | +++ | + | + | + | Yes | Nuc | + |
| I | G169A | +++ | NA | + | + | + | Yes | Nuc | + |
| I | G169D | − | − | − | − | − | +/− | Nuc | NA |
| I | R175K | +++ | NA | NA | NA | NA | Yes | Nuc | + |
| I | R175T | +++ | NA | NA | NA | NA | Yes | Nuc | + |
| II | M331K | + | +++ | + | − | + | +/− | Nuc | NA |
| II | M331T | ++ | NA | NA | NA | NA | Yes | Nuc | NA |
| II | F338A | ++ | NA | NA | NA | NA | Yes | Nuc | NA |
| II | F338E | +++ | NA | NA | NA | NA | Yes | Nuc | NA |
| II | D340H | + | +++ | + | +/− | + | +/− | Nuc | NA |
| II | D340R | − | − | − | − | − | No | Nuc | NA |
| III | S407A | +++ | NA | NA | NA | NA | Yes | Nuc | NA |
| III | F412E | − | − | − | − | − | Yes | Nuc | + |
| III | R416E | − | − | − | − | − | No | Nuc | NA |
| IV | N473R | +++ | NA | NA | NA | NA | Yes | Nuc | NA |
| IV | 473* | − | − | NA | NA | NA | No | Nuc | NA |
| IV | F488G | − | + | + | − | + | Yes | Nuc | + |
| IV | E494R | +++ | NA | NA | NA | NA | Yes | Nuc | NA |
| IV | 494in | + | NA | NA | NA | NA | Yes | Nuc | NA |
| IV | 494* | + | + | + | +/− | + | Yes | Nuc | + |
| V | DM | − | − | − | NA | − | No | Cit | NA |
The symbols refer to the four categories discussed in the text.
The solubility of the proteins after centrifugation in a microcentrifuge are indicated as follows: Yes, most of the protein was soluble; No, most of the protein was found in the pellet; or +/−, proteins with an intermediate solubility phenotype.
Nuc and Cit indicate that the protein accumulated in the nucleus or cytoplasm, respectively.
Accumulation of virus-specific RNAs in cells expressing recombinant influenza virus core proteins.
The CAT expression system allowed us to identify mutations that alter the NP function with regard to the expression of a synthetic CAT RNA. However, this system did not provide information on the RNA synthesis step(s) (synthesis of mRNA, vRNA, or cRNA) affected by the mutations. To get information on this issue, the experimental approach developed by Perales and Ortín (34) was used. This approach is identical to the CAT expression system except that cells were transfected with short (240-nt) model RNA templates (of positive or negative polarity) instead of the NS-CAT RNA. Total RNA isolated from transfected cells was fractionated into poly(A)+ and poly(A)−, and these fractions were analyzed for the presence of mRNA and replicative products (cRNA or vRNA) by the RNase protection assay. The wild-type NP, a mutant protein (G169A) functioning as the wild-type NP in the CAT expression system, and most mutant NPs (nine proteins) that strongly compromised NP function in the same assay were chosen for these analyses.
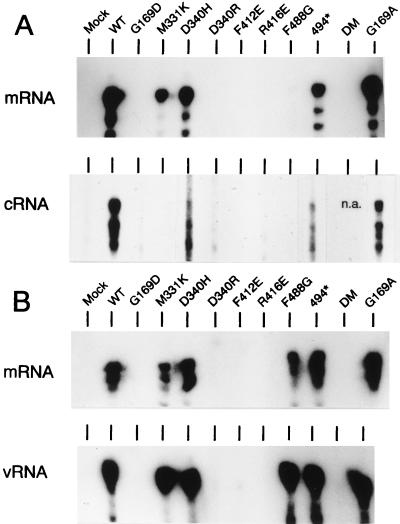
When a model vRNA was transfected and as expected from the results obtained in the CAT expression system in which a negative-sense RNA template was used, only those proteins which yielded levels of CAT expression higher than 1% produced clear mRNA bands (Fig. 3A, mRNA). cRNA molecules were readily detected in cells expressing wild-type and G169A proteins and clear, lower-intensity signals were observed in cultures expressing proteins D340H and 494*. No signal above background was observed in the samples from the other mutants analyzed (Fig. 3A, cRNA).
FIG. 3.
Accumulation of virus-specific RNA products in cells expressing recombinant core proteins. COS-1 cells expressing the P proteins and the NP polypeptides indicated at the top were transfected with synthetic model RNAs of either negative (vNSZ RNA) (A) or positive (cNSZ RNA) (B) polarity. Total RNA was collected at 24 h postinfection with vTF7-3 and fractionated into poly(A)+ and poly(A)− samples. The poly(A)+ samples were then analyzed for the presence of mRNA derived from the transfected RNA by the RNase protection assay by using a negative-sense labeled probe (panels labeled mRNA). The poly(A)− samples were analyzed for the presence of cRNA (panel A, cRNA) or vRNA (panel B, vRNA) by the same procedure with 32P-labeled probes of predetermined polarity. In the mock sample, plasmid pGEM-NP was omitted. The protected labeled fragments were resolved in a sequencing gel and visualized by autoradiography. In all panels the mobility of the slowest-migrating band had the expected mobility as determined by comparison with DNA makers included in the gel (not shown).
In a different experiment, a model cRNA was transfected, and the cultures were examined for the presence of mRNA and vRNA (Fig. 3B). Mutants that yielded less than 1% CAT activity (except for protein F488G) were negative for synthesis of vRNA and mRNA. All other mutants were positive in the assays and yielded similar high-intensity vRNA and mRNA signals. Strikingly, mutants M331K, D340H, F488G, and 494*, which showed a reduced functionality in cRNA synthesis from a vRNA template (Fig. 3A), appeared to function as efficiently as the wild-type NP in vRNA synthesis from a model cRNA (Fig. 3B). Dramatic examples are mutants M331K and F488G, which yielded undetectable levels of cRNA but were practically as competent as the wild-type NP in the synthesis of vRNA from cRNA.
Mutant F488G, which allowed detection of mRNA synthesis when cRNA was used as a template (Fig. 3B), produced only a minor mRNA signal, which is barely detectable in Fig. 3A, when the cells were transfected with a vRNA template. To explain this apparently contradictory result, it should be mentioned that the mRNA signal obtained in the absence of vRNA synthesis is below the level of detection of the assay (34). Therefore, since protein F488G was defective in cRNA synthesis, the transfected vRNA would not be amplified and thus no mRNA signal was detected in the assay (Fig. 3A). However, when transfecting cRNA templates, the F488G protein allowed efficient synthesis of vRNA molecules (Fig. 3B). Thus, in this latter case the intracellular concentration of the vRNA molecules would be higher than when transfecting a vRNA template, and therefore mRNA synthesis was observed (Fig. 3B).
Sedimentation analysis of mutant NPs.
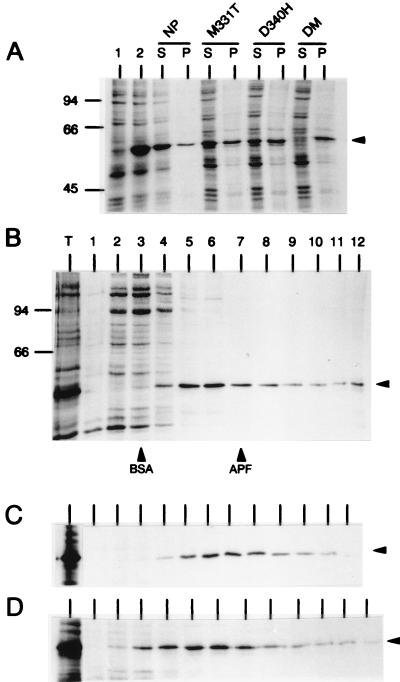
To determine whether the wild-type NP expressed from a cDNA was monomeric or it formed multimeric aggregates, COS-1 cells transfected with plasmid pGEM-NP were labeled with [35S]methionine-[35S]cysteine. Cell lysates were then separated into pellet and supernatant fractions by a brief centrifugation (1 min in a microcentrifuge). The supernatant fraction, which contained virtually all labeled proteins as well as most of the NP (Fig. 4A), was centrifuged through a linear 5-to-20% sucrose gradient. In this gradient, NP was found through fractions 4 to 12, with a peak in fractions 5 and 6, which represented ∼30% of the NP detected by autoradiography (Fig. 4B). By considering the mobility of the cellular proteins (in the same gradient), as well as that of bovine serum albumin (BSA) (66 kDa) and apoferritin (443 kDa) (loaded in independent sucrose gradients) (Fig. 4B), it was clear that the NP was not predominantly found as an unassembled monomeric protein; rather, NP appears to form (i) very high molecular weight aggregates, which were sedimented by a low-speed centrifugation in a microcentrifuge, and (ii) large heterogeneous complexes, which could be resolved in the sucrose gradient. No attempt was made to determine whether the complexes detected contained RNA and/or other cellular proteins. However, it was considered, based on the fact that NP, free of RNA, assembles into multimeric structures (42) and by analogy to the situation found with the N protein of other negative-strand RNA viruses (reference 30 and references therein), that the complexes detected in the sucrose gradient correspond to NP multimers.
FIG. 4.
Sedimentation analysis of mutant NPs. COS-1 cells were infected with vTF7-3 and transfected individually with the different pGEM-NP-derived plasmids. Cultures were metabolically labeled with [35S]methionine-[35S]cysteine, and cell lysates were prepared by freezing and thawing. (A) The lysates were then fractionated into supernatant (S) and pellet (P) fractions by centrifugation for 1 min in a microcentrifuge, and the proteins in these fractions were resolved by SDS-PAGE and analyzed by autoradiography. The proteins present in these two fractions in cells expressing the wild-type NP protein and three mutant NPs are shown. Total cell extracts (not fractionated by centrifugation) from mock-transfected cells or from cells expressing wild-type NP are shown in lanes 1 and 2, respectively. (B) An aliquot of the supernatant fraction from a culture expressing the wild-type NP was centrifuged through a 5-to-20% sucrose gradient, and the fractions harvested from the gradient (from top to bottom, lanes 1 to 12) were analyzed by SDS-PAGE and fluorography. Lane T corresponds to an aliquot of the sample loaded in the gradient. Fractions containing the peak of BSA and apoferritin loaded in parallel sucrose gradients are indicated. Panels C and D correspond to the same analysis presented in panel B, except that the cell extracts were obtained from cultures expressing proteins R175K and F412E, respectively. Only the relevant part of the gels are shown. Position of NP is indicated by an arrow at the right of each gel. In parts A and B, the molecular weights of protein standards are indicated on the left in thousands.
To test the effect of the different NP mutations on the oligomerization state of NP, all mutant proteins were expressed and analyzed as described above. There were four proteins (D340R, R416E, 473*, and DM) that could not be analyzed by sucrose gradient centrifugation since virtually all recombinant protein was found in the pellet of the microcentrifuge centrifugation step (as shown for the DM mutant, as a representative example, in Fig. 4A). Three other mutations (G169D, M331K, and D340H) also altered the ratio of soluble to highly aggregated protein, since the corresponding recombinant protein was equally distributed in both the pellet and the supernatant fractions (see protein D340H in Fig. 4A). For the proteins G169D, M331K, and D340H, there was, however, enough labeled soluble protein to be analyzed in the sucrose gradients. The distribution of these three proteins, as well as that of the rest of the mutant proteins (see Table 2), in the sucrose gradient was indistinguishable from that observed for the wild-type NP. Representative examples of these analyses are shown in Fig. 4C and D. From these studies, it was concluded that none of the mutant proteins had alterations that resulted in the accumulation of a monomeric protein in mammalian cells, but that seven of the mutations diminished the solubility of the NP polypeptide.
Intracellular localization of mutant NPs.
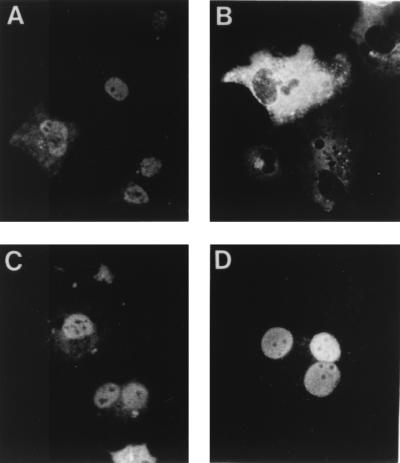
To determine whether the different mutations affected the intracellular localization of the NP protein, COS-1 cells were infected with vTF7-3, transfected with the different NP-derived plasmids, and analyzed by indirect immunofluorescence. To minimize the effect of NP concentration on the intracellular localization of the protein (32), independent cultures were transfected with three different doses of each mutant plasmid (see Materials and Methods for details). It was considered that a mutant protein accumulated in the cell nucleus if, at the lowest dose of transfected plasmid, which yielded 10 to 20% of transfected cells, there were more than 90% of the cells showing exclusively nuclear staining. All mutant proteins except protein DM, which was found in the cytosol of transfected cells at all doses of plasmids tested, fulfilled this criterion (representative results are shown in Fig. 5). It was thus concluded that the mutations in protein DM preclude its nuclear accumulation, although it cannot be determined whether the mutations in the protein actually prevented nuclear entry or promoted nuclear export.
FIG. 5.
Cellular localization of mutant NPs. COS-1 cells were infected with vTF7-3 and transfected with 60 ng of the plasmids expressing the wild-type NP (A) or proteins DM (B), D340R (C), or E494R (D). Cells were fixed with methanol, and the NP was visualized by indirect immunofluorescence.
RNA-binding studies.
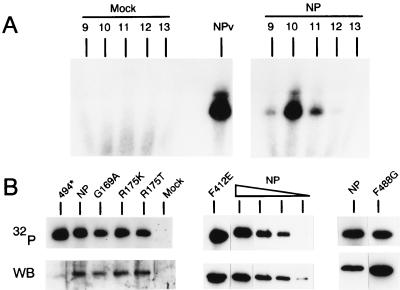
It has been shown that centrifugation in a CsCl-glycerol step gradient of either RNPs (11, 28, 33) or extracts from cells that express a recombinant NP (data not shown) yields fractions (found at the middle of the gradient) highly enriched in NP. These NP-containing fractions are practically depleted of other proteins, which are found in the top half of the gradient, and RNAs, which sediment in the bottom fractions. To test whether the recombinant wild-type NP isolated from this gradient was suitable for RNA-binding studies, aliquots of the gradient fractions were analyzed by their capability to cross-link a 32P-labeled RNA probe after UV light irradiation. As observed in Fig. 6A, the wild-type NP protein present in fractions 9, 10, and 11 of the gradient was efficiently cross-linked to RNA, whereas no labeled protein was present in the corresponding fractions prepared from a mock-transfected culture. Proteins showing solubility properties similar to those of the wild-type NP and that contained either mutations in region I (which is included within the NP RNA-binding domain) or mutations that reduced by more than 80% the functionality of NP in the CAT expression system (F412E, F488G, and 494*) were chosen for this analysis. For each of the selected mutants, fractions 9 to 11 of the CsCl-glycerol gradients were pooled, and this sample was analyzed by UV cross-linking to a 32P-labeled RNA as described above (Fig. 6B). All proteins tested behaved as wild-type NP in that similar amounts of recombinant protein and cross-linked RNA were observed in the pooled fraction. It was thus concluded that these mutations did not alter the RNA-binding capacity of NP.
FIG. 6.
RNA-binding activities of mutant NPs. (A) COS-1 cells were infected with vTF7-3 and transfected with either plasmid pGEM-NP (NP) or mock-transfected (mock). Cell extracts were prepared and resolved by centrifugation in a CsCl-glycerol gradient, and fractions were harvested from the top (sample 1) to the bottom (sample 15). Aliquots of fractions 9 to 13 (as indicated in the figure) were incubated with a 32P-labeled RNA, irradiated with UV light, treated with RNase A, resolved by SDS-PAGE, and the proteins containing residual cross-linked radioactive nucleotides were visualized by autoradiography. Lane NPv corresponds to a sample containing NP purified from virions which was also included in the cross-linking analysis. (B) The NPs indicated in each panel were expressed and resolved by CsCl-glycerol centrifugation as described in panel A. Fractions 9 to 11 of each gradient were pooled and cross-linked to a labeled RNA. The mixtures were then resolved by SDS-PAGE and electroblotted onto Immobilon-P paper. In each panel, the autoradiography of the membrane (32P), as well as the result of developing the same membrane by using the ECL kit with a rabbit serum, which recognizes the C-terminal region of NP (WB), are shown. Lane NP corresponds to the wild-type NP protein. In the central panel, in addition to the standard wild-type NP sample, three serial twofold dilutions prepared from this sample were also included in the gel.
DISCUSSION
We have described here the preparation and characterization of 20 mutant influenza A virus NPs. The properties of these proteins are summarized in Table 2. Mutations that drastically compromised NP function were found in all five regions chosen for mutagenesis, a result which suggests that these regions may be included within NP functional domains. There were seven mutations (G169A, R175K, R175T, F338E, S407A, N473R, and E494R) that did not significantly alter NP function in expressing a synthetic CAT RNA and that therefore may be affecting NP activities different from those involved in RNA synthesis.
Four mutant NPs (M331K, D340H, F488G, and 494*) showed a reduced functionality in reconstituting vRNPs competent for cRNA synthesis. However, these same proteins were fully competent to reconstitute cRNPs capable of synthesizing vRNPs, which in turn yielded mRNA molecules. It has been demonstrated that the synthesis of full-length virus-specific transcripts requires fully encapsidated templates (10, 11). Since proteins M331K, D340H, F488G, and 494* allowed synthesis of full-length mRNAs and vRNAs, it is concluded that they are functional in encapsidating vRNA and cRNA templates, respectively. Thus, it is suggested that the defect of these mutants in cRNA synthesis is due to a reduced capability of the NP proteins in interacting with a factor(s) required for cRNA synthesis but not for mRNA nor for vRNA synthesis. We propose that such an interaction involves a contact between free NP molecules with one or several of the P proteins associated with a vRNP complex, and we suggest that this interaction is responsible for making vRNAs switch from mRNA to cRNA synthesis. Two sets of data support the proposed model: (i) it has been shown that NP is required for the switching the vRNPs from mRNA to cRNA synthesis (5, 45) (see Introduction), (ii) and there is evidence suggesting that there are specific interactions between NP and P proteins (3, 6, 12, 25, 27, 45, 47). Indirect evidence of such an interaction has also been provided (i) by analysis of a virus with a Ts defect in the NP gene which can be extragenically suppressed by a defect in the PB2 gene (27), (ii) by studies showing that anti-NP monoclonal antibodies interfere with the initiation step of mRNA synthesis (3), and (iii) by experiments showing that influenza A and B virus NPs cannot substitute for each other to reconstitute functional RNPs (12, 47). While this study was in preparation, Biswas et al. (6) reported direct evidence showing that NP can interact with the PB2 and PB1 proteins but not with the PA subunit. We have also experimental data that support the same interactions (unpublished observations), although under our experimental conditions, and unlike the data reported by Biswas et al. (6), a fraction of the NP-PB1 and NP-PB2 complexes can be dissociated after RNase treatment.
It is conceivable that free NP also interacts with the P proteins associated with cRNPs to allow vRNA synthesis. However, to accommodate the results obtained here with the mutants, such an interaction should be different from the regulatory interaction (which allows switching the vRNPs from transcription to replication) of NP with vRNPs. In this regard it should be mentioned that cRNPs serve as a template for only one virus-specific RNA species (vRNA) and therefore no NP regulatory binding would be needed to alter the specificity of the cRNP-associated polymerase complex. It is possible that the binding of the P proteins to the cRNA or vRNA promoter determines different configurations of the polymerase complex so that the regulatory binding of NP only takes place when the complex is bound to a vRNA promoter.
To understand the molecular basis of the phenotype of the 11 mutant NPs that showed a reduced function in CAT expression, these NPs were analyzed for a series of activities. Seven of these NPs contained substitutions located in different NP conserved regions which increased the aggregation state of NP (Fig. 4 and Table 2), and four of these proteins (D340R, R416E, 473*, and DM) were exclusively found in high-molecular-weight aggregates that were sedimented by low-speed centrifugation. Further experiments are required to show whether these latter complexes represent structures like those detected in low amounts in cells expressing the wild-type NP or aberrant complexes. Most likely, the proteins with altered solubility properties contain drastic alterations in their structure. This would explain why protein DM, which contains mutations outside of the karyophilic sequences identified in NP (7, 32, 50), did not accumulate in the cell nucleus. It should be mentioned that the mutations in the DM protein are found in the human isolate A/Puerto Rico/8/34 (Fig. 1). The NP of this strain contains 32 substitutions compared to the A/Victoria/3/75 NP, and therefore in the A/Puerto Rico/8/34 NP some of these mutations should compensate for the deleterious effect of the substitutions present in the DM protein. There were several mutant proteins (G169D, M331K, F338E, D340H, R416E, and 473*) that were found in large aggregates but accumulated in the cell nucleus. These results indicate that the mutation introduced in the protein did not prevent the exposure of an NP nuclear import signal. However, the fact that these proteins were not soluble suggests that the mutation altered the conformation of the NP so as to promote abnormal self-association and/or interactions of NP with other proteins or nucleic acids present in the cell nucleus.
Although residues 169 and 175 (region I) are included within the NP-RNA binding domain, we could not find conclusive evidence on the importance of these residues for RNA binding. None of the mutations introduced in region II prevented the nuclear localization of NP, a result which is in agreement with recent reports (32, 50) that question the importance of this region for nuclear accumulation of NP in mammalian cells. No conclusive evidence on the specific roles played by regions II, III, and IV was obtained. However, as mentioned above, there were mutations in regions II and IV that affected virus-specific RNA synthesis, and we identified three mutations (F412E, F488G, and 494*) in regions III and IV that strongly compromised NP function in the CAT system but that did not alter the behavior of the protein in the biochemical assays described here. It may be suggested that these mutations identified regions that affect the association of NP with cellular or viral proteins required for virus-specific RNA synthesis. In this context, it is worth mentioning that it has recently been shown that the C-terminal region of NP appears to regulate the stability of the NP-PB2 interaction (6).
In summary, we have identified mutations that alter the functionality of NP in RNA replication and mutations that diminish NP function or confer a Ts phenotype to the protein. It would be interesting to rescue, by reverse genetics technology (24), these mutations together with others identified previously (6, 20, 25, 26, 27, 36) in an infectious virus, since such a virus may display characteristics desirable for an attenuated vaccine.
ACKNOWLEDGMENTS
I. Mena and E. Jambrina contributed equally to the experiments described in this study.
This work was supported by the Fondo de Investigaciones Sanitarias (grant 98/0315). I. Mena and E. Jambrina were supported by fellowships from Comunidad Autónoma de Madrid.
We thank J. A. Melero for critically reading the manuscript and A. del Pozo for the artwork.
REFERENCES
- 1.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrese M, Portela A. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J Virol. 1996;70:3385–3391. doi: 10.1128/jvi.70.6.3385-3391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bárcena J, Ochoa M, de la Luna S, Melero J A, Nieto A, Ortín J, Portela A. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin F, Bach C, Cusak S, Ruigrok R W H. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaton A R, Krug R M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas S K, Boutz P L, Nayak D P. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey J, Dimmock N J, Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985;40:667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- 8.de la Luna S, Martínez C, Ortín J. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 1989;13:143–155. doi: 10.1016/0168-1702(89)90012-9. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen M, Chung T D Y, Butcher A, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda A, Uéda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 12.Jambrina E, Bárcena J, Uez O, Portela A. The three subunits of the polymerase and the nucleoprotein of influenza B virus are the minimum set of viral proteins required for expression of a model RNA template. Virology. 1997;235:209–217. doi: 10.1006/viro.1997.8682. [DOI] [PubMed] [Google Scholar]
- 13.Kingsbury D W, Jones I M, Murti K G. Assembly of influenza ribonucleoprotein in vitro using recombinant nucleoprotein. Virology. 1987;156:396–403. doi: 10.1016/0042-6822(87)90419-3. [DOI] [PubMed] [Google Scholar]
- 14.Kingsbury D W, Webster R G. Some properties of influenza virus nucleocapsids. J Virol. 1969;4:219–225. doi: 10.1128/jvi.4.3.219-225.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kistner O, Müller H, Becht H, Scholtissek C. Phosphopeptide fingerprints of nucleoproteins of various influenza A virus strains grown in different host cells. J Gen Virol. 1985;66:465–472. doi: 10.1099/0022-1317-66-3-465. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi M, Toyoda T, Adyshev D M, Azuma Y, Ishihama A. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol. 1994;68:8433–8436. doi: 10.1128/jvi.68.12.8433-8436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krug R M, Alonso-Caplen F V, Julkunem I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- 18.Lamb R A. Genes and proteins of influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–88. [Google Scholar]
- 19.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1353–1395. [Google Scholar]
- 20.Li R, Palese P, Krystal M. Complementation and analysis of an NP mutant of influenza virus. Virus Res. 1989;12:97–112. doi: 10.1016/0168-1702(89)90057-9. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Palese P. Characterization of the polyadenylation signal of influenza virus RNA. J Virol. 1994;68:1245–1249. doi: 10.1128/jvi.68.2.1245-1249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londo D R, Davis A R, Nayak D P. Complete nucleotide sequence of the nucleoprotein gene of influenza B virus. J Virol. 1983;47:642–648. doi: 10.1128/jvi.47.3.642-648.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López J A, Guillen M, Sánchez-Fauquier A, Melero J A. An antigen-binding assay to determine the specificity of monoclonal antibodies against influenza virus and mapping of epitopes. J Virol Methods. 1986;13:255–264. doi: 10.1016/0166-0934(86)90019-4. [DOI] [PubMed] [Google Scholar]
- 24.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Amplification, expression, and packaging of a foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 25.Mahy B W J. Mutants of influenza virus. In: Palese P, Kingsbury D W, editors. Genetics of influenza viruses. Vienna, Austria: Springer; 1983. pp. 192–242. [Google Scholar]
- 26.Mandler J, Scholtissek C. Localisation of the temperature-sensitive defect in the nucleoprotein of an influenza A/FPV/Rostock/34 virus. Virus Res. 1989;12:113–121. doi: 10.1016/0168-1702(89)90058-0. [DOI] [PubMed] [Google Scholar]
- 27.Mandler J, Müller K, Scholtissek C. Mutants and revertants of an avian influenza A virus with temperature-sensitive defects in the nucleoprotein and PB2. Virology. 1991;181:512–519. doi: 10.1016/0042-6822(91)90883-d. [DOI] [PubMed] [Google Scholar]
- 28.Martín J, Albo C, Ortín J, Melero J A, Portela A. In vitro reconstitution of active influenza virus ribonucleoprotein complexes using viral proteins purified from infected cells. J Gen Virol. 1992;73:1855–1859. doi: 10.1099/0022-1317-73-7-1855. [DOI] [PubMed] [Google Scholar]
- 29.Mena I, de la Luna S, Albo C, Martín J, Nieto A, Ortín J, Portela A. Synthesis of biologically active influenza virus core proteins using a vaccinia virus-T7 RNA polymerase expression system. J Gen Virol. 1994;75:2109–2114. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 30.Myers T M, Pieters A, Moyer S A. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology. 1997;229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 31.Nakada S, Creager R S, Krystal M, Palese P. Complete nucleotide sequence of the influenza C/California/78 virus nucleoprotein gene. Virus Res. 1984;1:433–441. doi: 10.1016/0168-1702(84)90001-7. [DOI] [PubMed] [Google Scholar]
- 32.Neumann G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perales B, Ortín J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piccone M E, Fernánzdez-Sesma A, Palese P. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 1993;28:99–112. doi: 10.1016/0168-1702(93)90129-b. [DOI] [PubMed] [Google Scholar]
- 36.Pleschka S, Jaskunas S R, Engelhardt O G, Zürcher T, Palese P, García-Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pons M W, Schulze I T, Hirst G K, Hauser R. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969;39:250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- 38.Pritlove D C, Poon L L M, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Privalsky M L, Penhoet E E. Influenza virus proteins: identity, synthesis, and modification analyzed by two-dimensional gel electrophoresis. Proc Natl Acad Sci USA. 1978;75:3625–3629. doi: 10.1073/pnas.75.8.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prokudina-Kantorovich E N, Semenova N P. Intracellular oligomerization of influenza virus nucleoprotein. Virology. 1996;223:51–56. doi: 10.1006/viro.1996.0454. [DOI] [PubMed] [Google Scholar]
- 41.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 42.Ruigrok R W H, Baudin F. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J Gen Virol. 1995;76:1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
- 43.Sanz-Ezquerro J J, de la Luna S, Ortín J, Nieto A. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol. 1995;69:2420–2426. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholtissek C, Ludwig S, Fitch W M. Analysis of influenza A virus nucleoproteins for the assessment of molecular genetic mechanisms leading to new phylogenetic virus lineages. Arch Virol. 1993;131:237–250. doi: 10.1007/BF01378629. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro G I, Krug R M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu L L, Bean W J, Webster R G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens M P, Barclay W S. The N-terminal extension of the influenza B virus nucleoprotein is not required for nuclear accumulation or the expression and replication of a model RNA. J Virol. 1998;72:5307–5312. doi: 10.1128/jvi.72.6.5307-5312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Wyke K L, Bean W J, Jr, Webster R G. Monoclonal antibodies to the influenza A virus nucleoprotein affecting RNA transcription. J Virol. 1981;39:313–317. doi: 10.1128/jvi.39.1.313-317.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P, Palese P, O’Neill R E. The NPI-1/NPI-3 (Karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]