Abstract
RNA interference mediated by small interfering RNAs (siRNAs) has been exploited for the development of therapeutics. siRNAs can be a powerful therapeutic tool because the working mechanisms of siRNAs are straightforward. siRNAs determine targets based on their sequence and specifically regulate the gene expression of the target gene. However, efficient delivery of siRNAs to the target organ has long been an issue that needs to be solved. Tremendous efforts regarding siRNA delivery have led to significant progress in siRNA drug development, and from 2018 to 2022, a total of five siRNA drugs were approved for the treatment of patients. Although all FDA-approved siRNA drugs target the hepatocytes of the liver, siRNA-based drugs targeting different organs are in clinical trials. In this review, we introduce siRNA drugs in the market and siRNA drug candidates in clinical trials that target cells in multiple organs. The liver, eye, and skin are the preferred organs targeted by siRNAs. Three or more siRNA drug candidates are in phase 2 or 3 clinical trials to suppress gene expression in these preferred organs. On the other hand, the lungs, kidneys, and brain are challenging organs with relatively few clinical trials. We discuss the characteristics of each organ related to the advantages and disadvantages of siRNA drug targeting and strategies to overcome the barriers in delivering siRNAs based on organ-specific siRNA drugs that have progressed to clinical trials.
Subject terms: RNAi therapy, RNAi
Drug development: Refining RNA-based therapeutics to target multiple organs
Refining the design and delivery of RNA-based drugs could improve the chances of targeting diseases in complex organs. Gene expression is regulated by small interfering RNAs (siRNAs), which bind to messenger RNA sequences, preventing subsequent gene expression. Drugs based on siRNA show promise as safe and specialised for multiple diseases. Jinju Han and co-workers at the Korea Advanced Institute for Science and Technology in Daejeon, South Korea, reviewed the current status of siRNA drugs. Five siRNAs approved by the US Food and Drug Administration target the liver, while drugs targeting eye conditions and wound healing in the skin are in clinical trials. Targeting complex organs like the brain and lungs remains challenging, because the size of siRNAs and their delivery mechanisms must be adjusted in order to pass safely into target regions.
Introduction
Antisense transcripts to perturb the expression of target mRNAs have been widely used for genetic analyses. DNA plasmids expressing antisense transcripts of target mRNAs have been introduced into mammalian cells1, and antisense transcripts synthesized in vitro have been injected into frog oocytes2,3. The strategy of suppressing gene expression using antisense transcripts has also worked well in C. elegans. However, it was revealed that double-stranded RNAs (dsRNAs) are more potent in suppressing target genes than antisense transcripts4. Exogenous long dsRNAs in worms are processed into short RNA duplexes of ~21-22 nt by RNase III and loaded onto ARGONAUTE proteins to suppress target gene expression. This biological process of RNA interference (RNAi) is well conserved in diverse organisms, including humans.
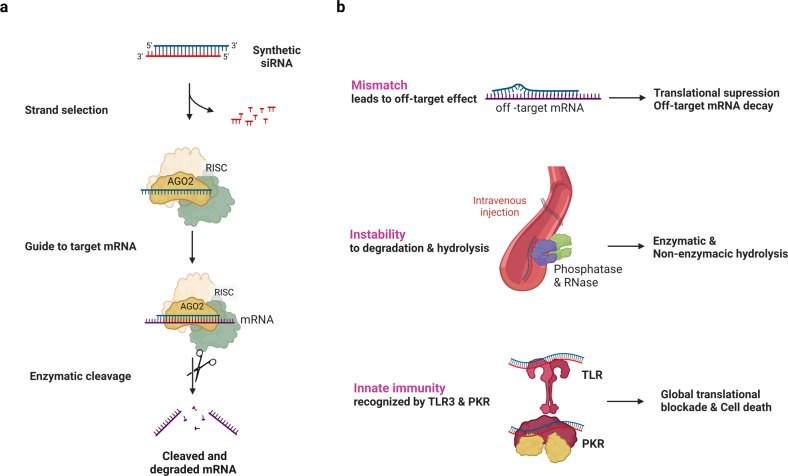
To apply RNAi to human cells, a short form of RNA duplexes should be delivered because long dsRNAs induce innate immune responses5. Exogenous ~21 nt RNA duplexes are incorporated into AGO proteins, and one of the strands is selected as a guide strand depending on the thermodynamic stability at the 5’ end of the RNA duplexes6. When the guide strand of siRNA forms a complex with AGO2 and binds to target RNAs with complementary sequences without mismatches, the ribonucleoprotein complex can cleave target RNAs (Fig. 1a). AGO2, among the four AGO proteins in humans, possesses endonuclease activity that cuts the phosphodiester bond of the target RNAs located between the 10th and 11th nucleotides from the 5’ end of the guide strand7.
Fig. 1. siRNA working mechanism and its features.
a General mechanism of action of synthetic small interfering RNA (siRNA). Exogenous RNA duplexes of ~21 nt are incorporated into an RNA-induced silencing complex (RISC), which includes ARGONAUTE2 (AGO2). AGO2 cleaves the passenger strand and liberates the guide strand of the siRNA. The guide strand then guides the RISC to the target mRNA and leads to the cleavage of mRNA. b Considerations that point to the development of siRNAs as drugs: off-target effects caused by binding of the guide strand to nontarget mRNA with mismatches, RNA stability decreased by degradation and hydrolysis, and immune responses induced by dsRNA recognition.
siRNAs have long been considered promising drug platforms because their working mechanisms have been well demonstrated, and siRNAs can be designed to target a specific RNA based on target sequences. Nevertheless, there are some factors to consider when developing siRNAs as drugs (Fig. 1b). siRNAs can bind to off-target RNAs with mismatches, which can result in translational suppression or decay of the off-target RNAs8. The off-targeting of siRNAs can be minimized by formulating siRNA sequences with computational algorithms9. The other issues that have arisen in using siRNAs as drugs are instability and potential immune reactions. RNAs are unstable and easily degraded by enzymatic and nonenzymatic hydrolysis. Unmodified naked siRNAs delivered into organisms through intravenous injection can be rapidly degraded because of vulnerability to RNases and phosphatases10. Moreover, a high concentration of unformulated and unmodified siRNAs that can be recognized by Toll-like receptor 3 (TLR3) or serine/threonine protein kinase (PKR)11–14 activate innate immune responses, leading to global translational blockade and cell death. To overcome these issues, the backbone and nucleotides of siRNAs are chemically modified15. Lastly, the delivery of negatively charged bulky siRNA to cells was improved by fusing molecules that can penetrate the lipid bilayers of cells or by encapsulating siRNAs into liposomes or lipid nanoparticles (LNPs). As a result of these tremendous efforts, the first siRNA-based drug, Patisiran, was approved by the United States Food and Drug Administration (US FDA) 20 years after the discovery of RNAi.
The approval of siRNA drugs has expanded the platforms used for oligonucleotide drug development. Before siRNA drugs were approved, only antisense oligonucleotide (ASO) drugs were used to control the expression of target genes that could not be targeted by traditional methods such as small molecules16. An ASO, a single-stranded oligonucleotide, binds to target RNAs and regulates gene expression in various ways. An ASO can perform RNase H1-mediated RNA cleavage, translational suppression, and splicing modulation17. The chemical modification of ASOs is indispensable for their stability and efficacy; however, the phosphorothioate or polyethylene glycol linkage backbone in ASOs can increase the binding affinity to unintended proteins, which is associated with toxicity18–20. siRNA drugs with less modified linkage backbones can be developed as alternatives to ASO drugs21,22.
As of December 2022, five siRNA drugs have been approved by the US FDA: Patisiran (Onpattro)23, Givosiran (Givlaari)24, Lumasiran (Oxlumo)25, Inclisiran (Leqvio)26, and Vutrisiran (Amvutta)27 (Table 1). All five approved siRNA drugs target mRNAs expressed in the liver. This is not very surprising because siRNAs delivered into an animal rapidly accumulate in the liver, a major organ for detoxifying exogenous materials. However, siRNA drugs targeting mRNAs expressed in other organs are also under development. In this review, we discuss the features of organs from the perspective of siRNA targeting. We focus on a few organs that are targeted by currently available siRNA drugs and siRNAs in phase 2 and 3 clinical trials.
Table 1.
siRNA drugs approved by the FDA as of 2022.
| Drug/Trade name | Date of Approval | siRNA Carrier | Routes of administration | Indication and usage | Target organ | Target gene | Reference |
|---|---|---|---|---|---|---|---|
| Patisiran/Onpattro | August 10, 2018 | Lipid nanoparticles | intravenous | Adult patients with hereditary transthyretin mediated (hATTR) amyloidosis | Liver | transthyretin (TTR) | 23 |
| Givosiran/Givlaari | November 20, 2019 | GalNAc-conjugation | subcutaneous | Adult patients with acute hepatic porphyria (AHP) | Liver | aminolevulinate synthase 1 (ALAS1) | 24 |
| Lumasiran/Oxlumo | November 23, 2020 | GalNAc-conjugation | subcutaneous | Adult and pediatric patients with primary hyperoxaluria type 1 (PH1) | Liver | hydroxy acid oxidase 1 (HAO1) | 25 |
| Inclisiran/Leqvio | December 21, 2021 | GalNAc-conjugation | subcutaneous | Adult patients with heterozygous familial hypercholesterolemia or clinical atherosclerotic cardiovascular disease. | Liver | proprotein convertase subtilisin/kexin type 9 (PCSK9) | 26 |
| Vutrisiran/amvuttra | June 13, 2022 | GalNAc-conjugation | subcutaneous | Adult patients with hereditary transthyretin mediated (hATTR) amyloidosis | Liver | transthyretin (TTR) | 27 |
Preferred organs targeted by siRNAs: Liver, eye and skin
Different strategies of siRNA delivery are being developed to target various organs precisely. Among the many organs, we categorized the liver, eye, and skin as preferred organs targeted by siRNAs. The first siRNA drug to enter clinical trials was AGN211745 (siRNA-027)13, targeting the eye. All siRNA drugs approved by the FDA target the liver23–27. Moreover, additional siRNA candidates targeting the liver, eye, and skin have progressed to phase 2 or 3 clinical trials. This section will discuss organs that are the preferred targets of siRNAs.
Liver
The liver is an essential organ responsible for numerous functions, including protein synthesis, detoxification, and the production of necessary biochemicals for sustaining life. Most drugs are metabolized in the liver. In this process, enzymes located in the endoplasmic reticulum of liver cells convert lipid-soluble metabolites into water-soluble metabolites to excrete the metabolites originating from drugs through the kidneys28.
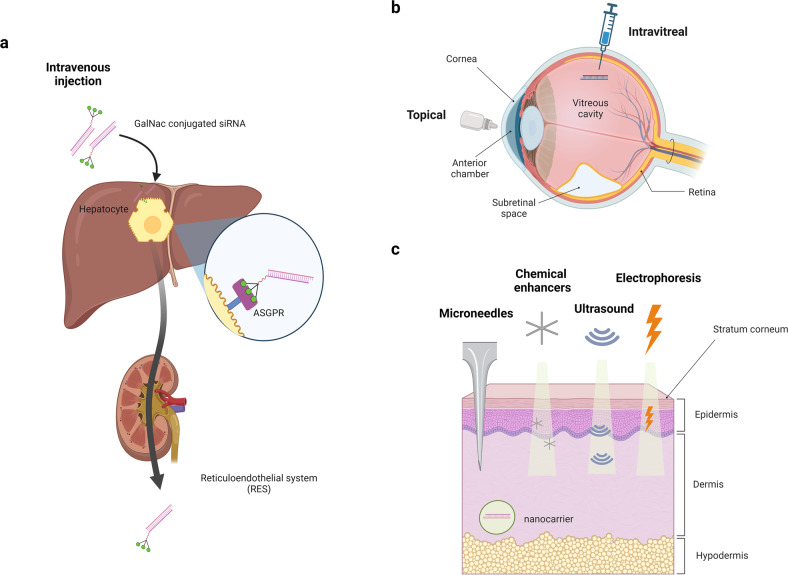
Both passive and targeted siRNA delivery can be used for the liver (Fig. 2a). Passive delivery is determined by the intrinsic properties and anatomy of a specific tissue or cell type29. Recognition moieties or drug carriers are not necessary for passive delivery (also known as physiology-based targeting). The reticuloendothelial system (RES), a part of the immune system, preferentially captures vesicles and removes foreign bodies found in the blood circulation to protect the body from harmful effects. Therefore, siRNAs encapsulated by liposomes or LNPs tend to accumulate through passive delivery in the liver, spleen, lymph nodes, and kidneys, which are filtering organs belonging to the RES30,31. For targeted siRNA delivery, the asialoglycoprotein receptor (ASGPR) is utilized. The expression of ASGPR is negligible in other tissues but very high in parenchymal hepatocytes, which comprise 70~85% of the liver volume. siRNAs conjugated with N-acetylgalactosamine (GalNAc), a carbohydrate moiety, specifically bind to ASGPR with a high affinity that results in hepatocyte-specific uptake of the conjugates32. Four siRNA drugs among the five FDA-approved drugs, except for patisiran, are conjugated with GalNAc. GalNAc conjugation also provides additional stability to siRNAs (Table 1).
Fig. 2. Preferred organs targeted by siRNAs: liver, eye and skin.
The characteristics of the liver, eye, and skin as target organs of siRNA drugs are described. a siRNAs injected intravenously are accumulated in the liver through the reticuloendothelial system (RES). siRNAs conjugated with N-acetylgalactosamine (GalNAc) bind to hepatocyte-specific asialoglycoprotein receptor (ASGPR) with high affinity. b The eye is a clinically accessible organ with immune-privileged regions: the vitreous cavity, anterior chamber, and subretinal space. siRNA drugs can be administered topically or by intravitreal injections into the eye. c The skin is known to be the largest organ in the human body. Microneedles, chemical enhancers, ultrasound, electrophoresis, and nanocarrier delivery systems can be used to deliver siRNAs to the skin.
Patisiran, the first FDA-approved siRNA drug, is delivered to the liver by LNPs to cure transthyretin-mediated amyloidosis (ATTR). ATTR is a fatal disease caused by the accumulation of misfolded TTR as amyloid fibrils in various tissues, including the heart, nerves, and gastrointestinal tract. While TTR proteins are expressed in all tissues of the human body, TTR mRNAs are primarily expressed in the liver. Vutrisiran, approved in 2022, also targets TTR mRNAs. Vutrisiran has advantages in the dosing method and interval. While patisiran is delivered by intravenous injection once every three weeks, vutrisiran is delivered subcutaneously only once every 3 months27. Givosiran was developed to treat acute hepatic porphyria (AHP), which is caused by the accumulation of neurotoxic intermediates called aminolevulinic acid (ALA) and porphobilinogen (PBG). ALA and PBG primarily accumulate in the liver and circulate through the body, resulting in neurological damage. Givosiran suppresses the expression of 5’-aminolevulinate synthase 1 (ALAS1), an enzyme required for ALA production, and reduces ALA and PBG levels. Lumasiran is used to treat primary hyperoxaluria type 1 (PH1). PH1 is caused by a deficiency in liver alanine glyoxylate-aminotransferase (AGT), which is responsible for the detoxification of glyoxylate. In the absence of AGT activity, glyoxylate is converted to oxalate, which forms insoluble calcium oxalate crystals in the kidney and other organs. Lumasiran cleaves hydroxy acid oxidase 1 (HAO1), an enzyme involved in oxalate synthesis25. Inclisiran was developed to treat heterozygous familial hypercholesterolemia and clinical atherosclerotic cardiovascular disease (ASCVD). Extremely high levels of low-density lipoprotein (LDL) and cholesterol (LDL-C) in plasma lead to premature ASCVD. Inclisiran delivered to the liver represses the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9), a protein necessary for LDL-C metabolism26.
Moreover, fifteen siRNA therapeutics have progressed to phase 2 and 3 clinical trials (Table 2). The potential siRNA drugs in phase 3 include nedosiran (DCR-PHXC), ARO-APOC3, fitusiran (ALN-AT3SC), and revusiran (ALN-TTRSC), which target LDH, APOC3, SERPINC1, and TTR, respectively. Eleven siRNAs, cemdisiran, zilebesiran (ALN-AGT01), olpasiran (AMG 890), ARO-ANG 3, ARO-HBV (JNJ-3989), AB-729, SLN360, ARO-AAT, ARC-520 and ND-L02-s0201, are in phase 2. ARO-HBV, AB-729, and ARC-520 are for infectious diseases; fitusiran is for hematology diseases, and revusiran and ARO-AAT are for hereditary diseases. ND-L02-s0201 treats fibrotic diseases by inhibiting the expression of heat shock protein 47 (HSP47). All the other drugs are for metabolic diseases. Only ARC-520 and ND-L02-s0201 use a nanoparticle system to deliver siRNAs, while the rest of the drugs use the GalNAc conjugate delivery system.
Table 2.
List of siRNA drug candidates in phase 2 and 3 clinical trials targeting the preferred organs: liver, eye, and skin.
| Target organ | Drug name | Delivery system | Target gene | Disease | status |
|---|---|---|---|---|---|
| Liver | Nedosiran (DCR-PHXC) | GalNAc conjugate | LDH | Primary hyperoxaluria type 1,2 | Phase 3, enrolling by invitation, NCT04042402 |
| Fitusiran (ALN-AT3SC) | GalNAc conjugate | SERPINC1 | Hemophilia A and B | Phase 3, completed, NCT03549871 | |
| ARO-APOC3 | GalNAc conjugate | APOC3 | Familial chylomicronemia syndrome | Phase 3, recruiting, NCT05089084 | |
| Cemdisiran | GalNAc conjugate | C5 | Paroxysmal nocturnal hemoglobinuria | Phase 2, active, not recruiting, NCT03841448 | |
| Zilebesiran(ALN-AGT01) | GalNAc conjugate | AGT | Mild-to-moderate hypertension | Phase 2 (KARDIA-1), recruiting, NCT04936035 | |
| Olpasiran(AMG 890) | GalNAc conjugate | LPA | Cardiovascular disease, patients with elevated serum lipoprotein A | Phase 2, active, not recruiting, NCT04270760 | |
| ARO-HBV (JNJ-3989) | GalNAc conjugate | HBV RNAs | Hepatitis B | Phase 2, completed, NCT03365947 | |
| AB-729 | GalNAc conjugate | Viral antigens | Hepatitis B,D | Phase 2, active, not recruiting, NCT04820686 | |
| ARO-ANG3 | GalNAc conjugate | ANGPTL3 | Mixed dyslipidemia | Phase 2, recruiting, NCT04832971 | |
| Revusiran (ALN-TTRSC) | GalNAc conjugate | TTR | Transthyretin (TTR)-mediated amyloidosis | Phase 3, completed, NCT02319005 | |
| SLN360 | GalNAc conjugate | LPA | Cardiovascular diseases, atherosclerosis | Phase 2, Not yet recruiting, NCT05537571 | |
| ALN-PCSSC | GalNAc conjugate | PCSK9 | Homozygous familial hypercholesterolemia | Phase 2, completed, NCT02963311 | |
| ARO-AAT | GalNAc conjugate | AAT | Alpha-1 antitrypsin deficiency | Phase 2, Active, not recruiting, NCT03946449 | |
| ARC-520 | Nanoparticle | HBV RNAs | Hepatitis B | Phase 2, terminated NCT02738008 | |
| ND-L02-s0201 | Nanoparticle | HSP47 | Fibrosis | Phase 2, completed, NCT03538301 | |
| Eye | AGN211745 (sirna-027) | naked siRNA | VEGFR1 (FLT1) | Neovascular AMD | Phase 2, terminated, NCT00395057 |
| Bevasiranib | naked siRNA | VEGF | Neovascular AMD | Phase 3, Terminated, NCT00499590 | |
| Tivanisiran (SYL1001) | naked siRNA | TRPV1 | Dry eye disease with Sjogren syndrome | Phase 3, completed, NCT03108664 | |
| Bamosiran(SYL040012) | naked siRNA | ADRB2 | Elevated intraocular pressure | Phase 2, completed, NCT02250612 | |
| Codosiran(QPI-1007) | naked siRNA | CASP2 | Acute primary angle closure glaucoma | Phase 2a, completed, NCT01965106 | |
| PF-0423655 | naked siRNA | RTP801 (DDIT4) | Diabetic macular edema, choroidal neovascularization, diabetic | Phase 2, completed, NCT01445899 | |
| RXI-109 | naked siRNA | CTGF (CCN2) | Wet AMD | Phase 2, Unknown NCT02599064 | |
| SYL1801 | naked siRNA | NRARP | Wet AMD | Phase 2, recruiting, NCT05637255 | |
| Skin | Cotsiranib (STP705) | Nanoparticle | TGFB1 and COX-2 (PTGS2) | Hypertrophic scarring | Phase 2, recruiting, NCT04669808 |
| BMT101(cp-asiRNA) | naked siRNA | CTGF (CCN2) | Prevention of hypertrophic scarring | Phase 2a, recruiting, NCT04012099 | |
| OLX10010 | naked siRNA | CTGF (CCN2) | Reducing recurrence of hypertrophic scarring | Phase 2a, recruiting, NCT04877756 | |
| RXI-109 | naked siRNA | CTGF (CCN2) | Hypertrophic scarring | Phase 2, Completed NCT02030275 |
Resource: http://clinicaltrials.gov.
Eye
The ocular system is located outside the cranium; thus, it is clinically accessible, unlike other central nervous system tissues. In particular, local delivery of siRNA to the ocular tissue is less complicated than delivery to other tissues. Local and near-direct delivery to the eye avoids the difficulties of systemic administration and minimizes systemic toxic effects. The additional advantage of delivering siRNA drugs to the eye is in the immune characteristics of the eye. The eye is an immune-privileged organ with limited local immune and inflammatory responses to maintain vision. This eye immune privilege is achieved through anatomical and biochemical mechanisms and is maintained in the vitreous cavity, anterior chamber, and subretinal space33. These features make the eye an excellent candidate for siRNA therapy. Indeed, the first siRNA drug that entered clinical trials was AGN211745 (sirna-027), targeting the eye.
Drugs are usually delivered to the eye directly by topical application (eye drops) or injection34 (Fig. 2b). For siRNA therapeutics, topical and intravitreal injection methods have likewise been used35. Topical administration is a noninvasive method and can be self-applied. However, topical administration is limited to diseases related to the anterior segment because there are multiple barriers to the back of the eye. Only approximately 10% of the applied dose is absorbed due to the physical barrier consisting of the corneal and conjunctival epithelium, nasolacrimal duct, and tears. Cell-penetrating peptide and silicon-based delivery approaches have been developed as siRNA delivery systems to increase the corneal permeability of drugs36. Various siRNAs using topical routes are under development37,38. The success of intravitreal injections of the anti-vascular endothelial growth factor (VEGF) “bevasiranib”12 has made the injection method common in drug delivery to the eye. Although piercing the retina can cause a physical break in blood tissue barriers and increase the risk of a systemic immune response39,40, injection of drugs into the vitreous humor is the most efficient method to deliver drugs to the posterior segments of the eye because it can bypass the natural barriers of the eyes. By using this advantage, siRNA drugs based on intravitreal injection are under development to treat diseases occurring in the posterior segment, such as age-related macular degeneration (AMD) and diabetic retinopathy (DR)13,41.
Currently, among siRNAs targeting the eye, eight drugs have progressed to phase 2 and 3 clinical stages: AGN211745 (sirna-027), bevasiranib, tivanisiran (SYL1001), bamosiran (SYL040012), codosiran (QPI-1007), PF-0423655, RXI-109, and SYL1801 (Table 2). Both AGN211745 (sirna-027) and bevasiranib have been developed to inhibit the VEGF signaling pathway for treating neovascular AMD. AGN211745 (sirna-027) targets VEGF receptor 1 (VEGFR1/FLT1), while bevasiranib targets VEGF itself. Tivanisiran (SYL1001) was developed to reduce ocular pain/discomfort in patients with dry eye disease by targeting Transient Receptor Potential Vanilloid 1. (TRPV1), a cation channel that acts as a nociceptive transducer. Bamosiran (SYL040012) targets the β2-Adrenergic Receptor (ADRB2) to reduce aqueous humor production and lower elevated intraocular pressure. Codosiran (QPI-1007) inhibits the loss of retinal ganglion cells (RGCs) and prevents optic neuropathy by targeting Caspase-2 (CASP2), which is highly expressed in RGCs during optical injury. PF-0423655 was developed for AMD patients by targeting RTP801 (DDIT4), a hypoxia-inducible gene overexpressed in choroidal neovascularization and diabetic retinopathy. SYL1801 targets NOTCH Regulated Ankyrin Repeat Protein (NRARP) and reduces the effects of the VEGF signaling pathway. Tivanisiran, bamosiran, and SYL1801 are administered topically, whereas AGN211745, bevasiranib, codosiran (QPI-1007), and PF-0423655 are administered by intravitreal injection.
Skin
Human skin consists of three main layers, the epidermis, dermis, and hypodermis42, and it prevents excessive transepidermal water loss and protects the human body from the external environment, such as ultraviolet rays (UV), and from the entry of xenobiotics and microbes. The skin is the largest and most accessible organ in our body. In addition, it is relatively easy to apply treatment to local areas, monitor modified areas, perform tissue biopsies, and remove abnormal areas surgically for the skin. Thus, the skin is an attractive organ for the development of therapeutics.
Topical administration is a noninvasive drug delivery method commonly used for skin, but it is difficult to bypass a barrier called the stratum corneum (SC), which is the outer layer of the epidermis43,44. For transdermal drug delivery, physical methods including microneedles, chemical enhancers, ultrasound, electroporation, and iontophoresis have been developed43 (Fig. 2c). In addition to these general drug administration routes, delivering siRNA drugs by another approach using nanocarriers is being explored because nanocarriers have the advantages of biocompatibility, biodegradability, and versatility45,46.
Currently, among the siRNA studies that target the skin, four drugs, STP705 (cotsiranib), BMT101 (cp-asiRNA), OLX10010, and RXI-109, have progressed to clinical trials. The goal of these siRNA drugs is the same, treating hypertrophic scars caused by the excessive production of collagen from myofibroblasts during wound healing. All these siRNAs are delivered to the skin by intradermal injection. BMT101 (cp-asiRNA), OLX10010, and RXI-109 target connective tissue growth factor (CTGF), which is involved in the formation of hypertrophic scars and keloids. STP705 targets TGFβ1 and Cyclooxygenase-2 (COX-2/PTGS2), which modulate signaling pathways related to hypertrophic scars47.
Challenging organs to deliver siRNAs: the lungs, kidneys and brain
siRNA targeting the lung, kidney, and brain has rarely progressed to the clinical stage because there are many barriers to targeting the lung, kidney, and brain. This section will discuss organs that are challenging to target with siRNAs.
Lungs
The main function of the lungs is the process of gas exchange called respiration. The lung is divided into a conducting region responsible for air conductance and a respiratory region where gaseous exchange takes place. The conducting region contains the nasal cavity, pharynx, trachea, bronchi, and bronchioles, while the respiratory region contains the respiratory bronchioles and alveoli. To deliver drugs more efficiently, intratracheal, intranasal, and inhalational drug delivery methods can be applied rather than systemic drug delivery methods. Drug loss is low by avoiding first-pass metabolism, and siRNA stability is better maintained because the airway contains fewer nucleases than the serum.
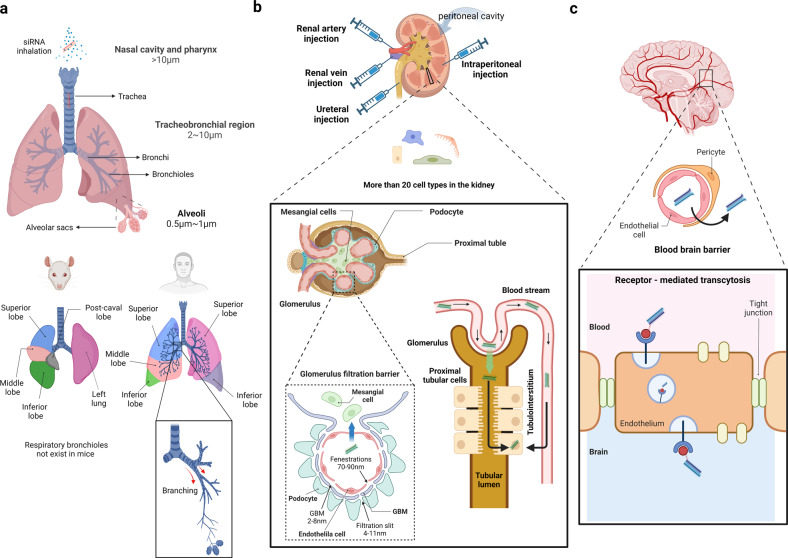
The primary barrier in the lungs that siRNAs have to pass is the extracellular barrier48 formed by anatomical, physiological, and metabolic features of the lungs. Extracellular barriers include the reticulate pulmonary architecture from the trachea to the alveoli (Fig. 3a). The active clearance processes in this area, such as mucociliary clearance, cough clearance, and effective immune responses, inhibit the invasion of foreign material into the lungs. In addition, the presence of respiratory mucus in the upper airways and the airway surface liquid (surfactant) in the lower airways act as major physical and chemical barriers, reducing the drug penetration and diffusion rate48. To avoid the extracellular barrier, the proper size and density of injected particles are essential. As a drug delivery strategy, aerosolized particles are usually delivered to the lungs by inhalation. In this case, the distance at which the drug is deposited depends on the size and density of the particles. When the aerodynamic diameter is >10 µm, the drug will deposit in the nasal cavity and pharynx; particles between 2 and 10 µm will deposit in the tracheobronchial region; and finally, particles between 0.5 to 1 μm will deposit on lower bronchioles and alveoli49. Therefore, it is necessary to use the optimal scale for the pulmonary route. Because it is known that viral vectors increase cell uptake and siRNA efficacy, attempts have been made to overcome barriers using them. However, applying viral vectors to human therapeutics presents problems in terms of uncontrolled viral replication, immunogenicity, tumorigenicity, and toxicity50,51. For these reasons, the size aspect is met by utilizing naked siRNA aerosol delivery or nanoparticles52,53, which are nonviral vectors. Naked siRNA generally fails due to the lack of a significant gene silencing effect, but surprisingly, local delivery in the lung has shown success54–56.
Fig. 3. Challenging organs to deliver siRNAs: lungs, kidneys, and brain.
Characteristics of the pulmonary system, kidneys, and brain as targeting organs of siRNA drugs are described. The lungs, kidneys, and brain have complex structures that limit a drug’s size. a siRNA is usually delivered to the lungs through inhalation, and the location of the drug distribution differs depending on the size of the drug. Mouse and human respiratory systems are very different in terms of anatomy. The mouse lung consists of four right lobes and one left lobe, whereas human lungs consist of three right and two left lobes. Respiratory bronchioles do not exist in mice, whereas humans have many bronchioles branched from bronchi. b The glomerular filtration barrier exists in the glomerulus, one of the components of the nephron. siRNAs can be delivered to proximal tubular cells from the apical or basolateral side. siRNAs can be directly delivered to the kidney through the renal artery, renal vein, ureteral, or intraperitoneal injection. c The brain has the most exceptional barrier, the blood‒brain barrier (BBB). Most siRNA nanoparticles cross the BBB using receptor-mediated transcytosis (RMT) strategies.
Regarding strategies for delivering siRNAs to the lungs, it is difficult to apply animal studies directly to human studies57 because of the anatomical differences in the lungs between animals and humans58. The numbers of lobes on each side of the lungs and patterns of airway branching differ between humans and rodents. In addition, human lungs contain small intrasegmental bronchi and respiratory bronchioles that are absent or rare in rodents. Thus, the administration routes used in animal studies are unsuitable for humans, and it is difficult to measure the efficiency before starting clinical research. Even if an animal study is conducted, human lung delivery must be considered.
Among many siRNA studies to target the pulmonary system, there are two candidates that have reached the clinical stage: ALN-RSV01 and MIR 19 (siR-7-EM/KK-46) (Table 3). ALN-RSV01, developed to be administered by nasal spray as an antiviral drug, silences the nucleocapsid protein transcripts of respiratory syncytial virus (RSV). It was confirmed that RSV infection was reduced in a phase 2 clinical trial. However, there was no further progress in phase 2a and 2b trials, and the clinical trial could not proceed to the end. MIR 19 was developed with inhalation administration as a COVID-19 treatment. MIR 19 inhibits viral replication by targeting SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). It has progressed to the completion of a phase 2 clinical trial.
Table 3.
List of siRNAs in phase 2 and 3 clinical trials targeting the challenging organs: lungs, kidneys, and brain.
| Target organ | Drug name | Delivery system | Target gene | Disease | Status |
|---|---|---|---|---|---|
| Lung | ALN-RSV01 | naked siRNA | RSV nucleocapsid messenger RNA | RSV01 | Phase 2, completed, NCT00658086 discontinued |
| MIR 19(siR-7-EM/KK-46) | Nanoparticle | RdRp | COVID-19 | Phase 2, completed, NCT05184127 | |
| Kidney | Teprasiran (I5NP, QPI-1002) | naked siRNA | P53 | AKI | Phase 3, completed NCT02610296 |
Resource: http://clinicaltrials.gov.
Kidneys
The kidneys have an important role in filtering blood and eliminating wastes generated in the body. They also play a homeostatic role by regulating electrolytes and water to maintain the acid-base balance and blood pressure59. The diverse cell types60 and structural complexity61 are the major barriers for siRNA drugs to target the kidneys. While the liver and ocular tissue consist of 4 and 5 cell types, respectively, the kidneys consist of at least 26 cell types60. These diverse cell types of the kidneys make it difficult to optimize and deliver drugs to specific cell types.
The glomerulus, one of the major constituents of nephrons, has a glomerular filtration barrier that acts as a barrier to the delivery of siRNA drugs (Fig. 3b). The glomerular filtration barrier comprises the endothelial fenestration, glomerular basement membrane, and podocyte extension filtration slits that are 70–90, 2–8, and 4–11 nm in diameter, respectively. Therefore, the size of the drug is critical for crossing this glomerular filtration barrier. Only small molecules with diameters less than 6 nm can pass through the glomerular filtration barrier62. Glomerular mesangial cells are the major targets of siRNA delivery in the glomerulus63–65. For targeting mesangial cells, siRNA drugs should be larger than 6 nm to prevent filtering by the urinary tract but smaller than 70–90 nm to be captured in the glomerulus and pass through the endothelial fenestration66. Polyethylene glycol (PEG)-poly L-lysine (PLL) nanocarriers65 and LNPs64 were used as vehicles for siRNA delivery. siRNAs in PEG-PLL carriers and LNPs have been delivered to the glomerulus via intraperitoneal65 and intravenous injections64. Naked siRNAs through the renal artery have also been tried to target mesangial cells63.
The tubular system, the other part of the nephrons, reabsorbs endogenous compounds. Most RNA-based studies have attempted to treat kidney disease by targeting proximal tubular cells67–69. Proximal tubular cells can be targeted from either the apical side (facing the tubular lumen) or the basolateral side (facing the interstitium). Only naked siRNA, which is 3–6 nm in size, can be applied from the apical side because siRNA drugs need to pass through the glomerular filtration barrier (6–7 nm) and be reabsorbed into the proximal tubular cells. Particles of siRNA carriers with larger sizes can access tubular cells from the basolateral side. They enter the tubulointerstitium by capillary pressure and are absorbed into proximal epithelial cells70. Many different strategies for delivering siRNAs to tubular cells have been investigated71–73. If oligonucleotides are accommodated in nanoparticles, they can pass through the glomerular filtration barrier only in the condition of glomerular injury. This has led to extensive studies on developing various strategies for delivering siRNAs to tubular cells71–74. However, direct injection into the kidneys, such as intraparenchymal injection65, retrograde ureter injection67, renal vein injection71, and renal artery injection73, has been mostly used in many preclinical studies because it can avoid the size restriction caused by the glomerular filtration barrier. The direct injection method also has the advantage of being able to target the kidney locally, avoiding accumulation in the liver. However, it has not been well applied to humans because the method is too invasive and difficult to administer75.
Among many studies on siRNA targeting the kidneys, one candidate has reached the clinical stage (Table 3). Teprasiran (I5NP, QPI-1002), a naked p53 siRNA, was investigated to treat acute kidney injury delayed graft function (DGF) after transplantation and cardiac surgery. Teprasiran reduces the expression of the proapoptotic protein P53 to protect the kidneys from cell death resulting from acute ischemia‒reperfusion injury and to maintain tissue and organ integrity76. Additionally, it was designated an orphan drug because of its efficacy in a phase 3 pivotal trial for DGF.
Brain
The brain is an exceptional and extremely friable organ in the human body. Because the CNS is an essential system that monitors and coordinates the functions of internal organs and responds to changes in the environment, it must be protected from both endogenous and exogenous threats.
The biggest obstacle in targeting the brain with siRNA is the blood‒brain barrier (BBB) (Fig. 3c). The BBB separates the cerebrospinal fluid and blood, protecting the brain from pathogens such as viruses and various harmful substances, and selectively controls the movement of ions and molecules to regulate brain homeostasis. Brain capillary endothelial cells (BCECs), known as the thin layer of the BBB, have tight junctions (TJs) for molecules that strongly inhibit the passage of hydrophilic substances over 300 Da between cells, which is called a “physical barrier function.” Therefore, almost 98% of molecules cannot pass through the BBB, with the sole exception of lipid-soluble small molecules with a molecular weight <400 Da77. Because siRNAs are hydrophilic and highly negatively charged molecules with a molecular weight of ~14 kDa, it is challenging for them to pass through the BBB78,79.
Even if siRNAs pass through the BBB and reach the brain, the endocytosis efficiency of highly negatively charged siRNAs is very low80. In addition, the drug cannot be controlled to target specific areas of the brain or specific cell types in the brain. The durability of siRNAs in the brain is another issue. When siRNAs were injected into the brain parenchyma, the silencing effect was observed only in cells close to the injection site for a short time81 Increasing the siRNA dose may provide sufficient effects of target gene suppression in the brain. However, a high dose of siRNAs can cause strong side effects in nontargeted cells in the brain82. Recent studies have reported that long-lasting siRNAs are expressed in a broad area of the brain when delivered into the brain through the cerebrospinal fluid (CSF)83,84. A divalent siRNA (di-siRNA), in which two siRNAs are chemically conjugated, contains enough phosphorothioates in its backbone to help cellular uptake and promote a broad distribution83. siRNA conjugated with 2′-O-hexadecyl (C16) is broadly distributed and efficiently suppresses a target gene for a long time84.
Although injection methods have been utilized in preclinical research to deliver siRNAs to the brain, nanoparticles (NPs) have been used in clinical trials to deliver siRNAs to the brain. Receptor-mediated transcytosis (RMT), cell-mediated transport, carrier-mediated transport, adsorptive-mediated transcytosis, and a method for breaking the integrity of tight junctions are being used as strategies to allow nanoparticles to pass through the BBB85,86 (Fig. 3c). Most of the siRNA nanoparticles shown to cross the BBB use the RMT strategy, which is known to transport a wide range of proteins using the vesicular trafficking machinery in brain endothelial cells. Among them, transferrin (Tf) and rabies viral glycoprotein (RVG) tags are the most widely used87. Tf and RVG bind to the transferrin receptor (TfR) and nicotinic acetylcholine receptor (nAchR), respectively, both of which are located in the endothelial cells of the brain88,89. TfR is widely expressed throughout the human body, including the brain, but because nAchR is expressed only in the brain, brain-specific targeting is possible. This RVG strategy has been applied to suppress HMGB1, mHTT, and BACE1, which are genes related to ischemic stroke, Huntington’s disease, and Alzheimer’s disease, respectively90–92. In addition to RVG and Tf, apolipoprotein E3-reconstituted high-density lipoprotein (ApoE-rHDL)93, angiopep-294, leptin95, and T7 peptides96 are also promising candidates for RMT.
Delivery systems for siRNA targeting the brain have been continually developed. One drug, NU-0129, has progressed to a clinical trial. This drug has not yet entered phase 2, but it is the only brain-targeting siRNA that has progressed to a clinical trial. NU-0129 is a spherical nucleic acid (SNA) siRNA that targets the glioblastoma oncogene BCL2L12 and crosses the BBB using the RMT strategy. SNA consists of nanoparticles in which the siRNA duplex is densely bound to the spherical gold surface. BCL2L12 is overexpressed in glioblastoma, inhibiting apoptosis and P53, thereby promoting cancer growth. An early phase 1 study was conducted in 2020, and its safety profile was confirmed97.
Conclusion
RNA therapy is a promising treatment for a wide range of diseases, including cancer, cardiovascular diseases, neurodegenerative diseases, inflammatory conditions, viral infections, and eye diseases. siRNAs have the advantages of higher specificity than chemical drugs and a high degree of safety. siRNAs are also good in terms of efficiency, and candidate groups of siRNA drugs can be developed quickly and easily. All siRNA drugs approved as of 2022 target the liver, but understanding and researching siRNAs and their delivery to diverse organs besides the liver continue to be a goal. The liver has the hepatocyte-specific receptor ASGPR to which GalNAc-conjugated siRNAs can bind with high affinity. In addition, the eye and skin have a structure with good accessibility for drugs. The lungs, kidneys, and brain have complex structures that limit drug size, and the brain, in particular, has the most exceptional barrier, the BBB. Nevertheless, to overcome these barriers, siRNA drugs using various RNA modifications, conjugation systems, and delivery systems are being tested in the preclinical and clinical stages. Through understanding and researching each organ in terms of siRNA delivery, siRNA drugs targeting other organs beyond the liver are expected to emerge.
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program (NRF-2022M3E5F1016556), Basic Science Research Program (NRF-2019R1C1C1010482), and Basic Research Laboratory Program (NRF-2021R1A4A3032789) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT. I. Ahn was supported by the KAIST Short-Term Innovative Research for Graduate students. C. Kang was supported by the KAIST long-term Undergraduate Research Participation program. The figures were created with the BioRender online platform.
COMPETING INTERESTS
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Izant JG, Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984;36:1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- 2.Harland R, Weintraub H. Translation of mRNA injected into Xenopus oocytes is specifically inhibited by antisense RNA. J. Cell Biol. 1985;101:1094–1099. doi: 10.1083/jcb.101.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melton DA. Injected anti-sense RNAs specifically block messenger RNA translation in vivo. Proc. Natl Acad. Sci. USA. 1985;82:144–148. doi: 10.1073/pnas.82.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19:639–648. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 9.Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J. Am. Chem. Soc. 2008;130:10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol. Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Garba AO, Mousa SA. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol. Eye Dis. 2010;2:75–83. doi: 10.4137/OED.S4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser PK, et al. RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027. Am. J. Ophthalmol. 2010;150:33–39.e32. doi: 10.1016/j.ajo.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Puthenveetil S, et al. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broering R, et al. Chemical modifications on siRNAs avoid Toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro. Int. Immunol. 2014;26:35–4. doi: 10.1093/intimm/dxt040. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, Zhu L, Wang X, Jin H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 2022;13:644. doi: 10.1038/s41419-022-05075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhuri K, et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. 2020;9:2004. doi: 10.3390/jcm9062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W, et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019;37:640–650. doi: 10.1038/s41587-019-0106-2. [DOI] [PubMed] [Google Scholar]
- 19.Crooke ST, Vickers TA, Liang X-H. Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 2020;48:5235–5253. doi: 10.1093/nar/gkaa299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X-h, et al. Solid-phase separation of toxic phosphorothioate antisense oligonucleotide-protein nucleolar aggregates is cytoprotective. Nucleic Acid Ther. 2021;31:126–144. doi: 10.1089/nat.2020.0923. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, et al. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich M, Aigner A. Therapeutic siRNA: state-of-the-art and future perspectives. BioDrugs. 2022;36:549–571. doi: 10.1007/s40259-022-00549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristen AV, et al. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 2019;9:5–23. doi: 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- 24.Scott LJ. Givosiran: first approval. Drugs. 2020;80:335–339. doi: 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- 25.Scott LJ, Keam SJ. Lumasiran: first approval. Drugs. 2021;81:277–282. doi: 10.1007/s40265-020-01463-0. [DOI] [PubMed] [Google Scholar]
- 26.Ray KK, et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 2020;382:1507–1519. doi: 10.1056/NEJMoa1912387. [DOI] [PubMed] [Google Scholar]
- 27.Mullard A. FDA approves fifth RNAi drug - Alnylam’s next-gen hATTR treatment. Nat. Rev. Drug Discov. 2022;21:548–549. doi: 10.1038/d41573-022-00118-x. [DOI] [PubMed] [Google Scholar]
- 28.Almazroo OA, Miah MK, Venkataramanan R. Drug metabolism in the liver. Clin. Liver Dis. 2017;21:1–20. doi: 10.1016/j.cld.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Holm A, Lovendorf MB, Kauppinen S. Development of siRNA therapeutics for the treatment of liver diseases. Methods Mol. Biol. 2021;2282:57–75. doi: 10.1007/978-1-0716-1298-9_5. [DOI] [PubMed] [Google Scholar]
- 30.Hirsjarvi S, Passirani C, Benoit JP. Passive and active tumour targeting with nanocarriers. Curr. Drug Discov. Technol. 2011;8:188–196. doi: 10.2174/157016311796798991. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y, et al. Overcoming the reticuloendothelial system barrier to drug delivery with a “Don’t-Eat-Us” strategy. ACS Nano. 2019;13:13015–13026. doi: 10.1021/acsnano.9b05679. [DOI] [PubMed] [Google Scholar]
- 32.Debacker AJ, Voutila J, Catley M, Blakey D, Habib N. Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol. Ther. 2020;28:1759–1771. doi: 10.1016/j.ymthe.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Streilein JW. Ocular immune privilege: therapeutic opportunities from an experiment of nature. Nat. Rev. Immunol. 2003;3:879–889. doi: 10.1038/nri1224. [DOI] [PubMed] [Google Scholar]
- 34.Novack GD. Ophthalmic drug delivery: development and regulatory considerations. Clin. Pharmacol. Ther. 2009;85:539–543. doi: 10.1038/clpt.2008.297. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Zhang X, Tang Y, Li S, Chen J. Progress on ocular siRNA gene-silencing therapy and drug delivery systems. Fundam. Clin. Pharmacol. 2021;35:4–24. doi: 10.1111/fcp.12561. [DOI] [PubMed] [Google Scholar]
- 36.Bachu RD, Chowdhury P, Al-Saedi ZHF, Karla PK, Boddu SHS. Ocular drug delivery barriers-role of nanocarriers in the treatment of anterior segment ocular diseases. Pharmaceutics. 2018;10:28. doi: 10.3390/pharmaceutics10010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benitez-Del-Castillo JM, et al. Safety and efficacy clinical trials for SYL1001, a novel short interfering RNA for the treatment of dry eye disease. Invest. Ophthalmol. Vis. Sci. 2016;57:6447–6454. doi: 10.1167/iovs.16-20303. [DOI] [PubMed] [Google Scholar]
- 38.Zahir-Jouzdani F, et al. Corneal chemical burn treatment through a delivery system consisting of TGF-beta(1) siRNA: in vitro and in vivo. Drug Deliv. Transl. Res. 2018;8:1127–1138. doi: 10.1007/s13346-018-0546-0. [DOI] [PubMed] [Google Scholar]
- 39.Chong DY, Anand R, Williams PD, Qureshi JA, Callanan DG. Characterization of sterile intraocular inflammatory responses after intravitreal bevacizumab injection. Retina. 2010;30:1432–1440. doi: 10.1097/IAE.0b013e3181dc04da. [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Durairaj C, Lin T, Liu Y, Burke J. Ocular pharmacokinetics of intravitreally administered brimonidine and dexamethasone in animal models with and without blood-retinal barrier breakdown. Invest. Ophthalmol. Vis. Sci. 2014;55:1056–1066. doi: 10.1167/iovs.13-13650. [DOI] [PubMed] [Google Scholar]
- 41.Jiang S, Chen X. HMGB1 siRNA can reduce damage to retinal cells induced by high glucose in vitro and in vivo. Drug Des. Devel. Ther. 2017;11:783–795. doi: 10.2147/DDDT.S129913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilaberte, Y., Prieto-Torres, L., Pastushenko, I. & Juarranz, Á. Anatomy and Function of the Skin. Nanoscience in Dermatology 1–14 (2016).
- 43.Prausnitz MR, Mitragotri S, Langer R. Current status and future potential of transdermal drug delivery. Nat. Rev. Drug Discov. 2004;3:115–124. doi: 10.1038/nrd1304. [DOI] [PubMed] [Google Scholar]
- 44.Benson HAE, Grice JE, Mohammed Y, Namjoshi S, Roberts MS. Topical and transdermal drug delivery: from simple potions to smart technologies. Curr. Drug Deliv. 2019;16:444–460. doi: 10.2174/1567201816666190201143457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geusens B, et al. Flexible nanosomes (SECosomes) enable efficient siRNA delivery in cultured primary skin cells and in the viable epidermis of ex vivo human skin. Adv. Funct. Mater. 2010;20:4077–4090. doi: 10.1002/adfm.201000484. [DOI] [Google Scholar]
- 46.Bracke S, et al. Targeted silencing of DEFB4 in a bioengineered skin-humanized mouse model for psoriasis: development of siRNA SECosome-based novel therapies. Exp. Dermatol. 2014;23:199–201. doi: 10.1111/exd.12321. [DOI] [PubMed] [Google Scholar]
- 47.Colwell AS, Phan TT, Kong W, Longaker MT, Lorenz PH. Hypertrophic scar fibroblasts have increased connective tissue growth factor expression after transforming growth factor-beta stimulation. Plast. Reconstr. Surg. 2005;116:1387–1390. doi: 10.1097/01.prs.0000182343.99694.28. [DOI] [PubMed] [Google Scholar]
- 48.Sanders N, Rudolph C, Braeckmans K, De Smedt SC, Demeester J. Extracellular barriers in respiratory gene therapy. Adv. Drug Deliv. Rev. 2009;61:115–127. doi: 10.1016/j.addr.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thakur, A. K., Kaundle, B. & Singh, I. in Targeting Chronic Inflammatory Lung Diseases Using Advanced Drug Delivery Systems 475–491 (2020).
- 50.Raper SE, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to Gene Ther.apy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11:S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 52.Bai X, et al. Inhaled siRNA nanoparticles targeting IL11 inhibit lung fibrosis and improve pulmonary function post-bleomycin challenge. Sci. Adv. 2022;8:eabn7162. doi: 10.1126/sciadv.abn7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keil TWM, Baldassi D, Merkel OM. T-cell targeted pulmonary siRNA delivery for the treatment of asthma. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1634. doi: 10.1002/wnan.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulton A, et al. Effective treatment of respiratory alphaherpesvirus infection using RNA interference. PLoS ONE. 2009;4:e4118. doi: 10.1371/journal.pone.0004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li BJ, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 57.Hofmann W, Koblinger L, Martonen TB. Structural differences between human and rat lungs: implications for Monte Carlo modeling of aerosol deposition. Health Phys. 1989;57:41–46. doi: 10.1097/00004032-198907001-00005. [DOI] [PubMed] [Google Scholar]
- 58.Meyerholz, D. K., Suarez, C. J., Dintzis, S. M. & Frevert, C. W. in Compar. Anatom. Histol. 147–162 (2018).
- 59.Ruggiero A, et al. Paradoxical glomerular filtration of carbon nanotubes. Proc. Natl Acad. Sci. USA. 2010;107:12369–12374. doi: 10.1073/pnas.0913667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schumacher A, et al. Defining the variety of cell types in developing and adult human kidneys by single-cell RNA sequencing. NPJ Regen. Med. 2021;6:45. doi: 10.1038/s41536-021-00156-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jourde-Chiche N, et al. Endothelium structure and function in kidney health and disease. Nat. Rev. Nephrol. 2019;15:87–108. doi: 10.1038/s41581-018-0098-z. [DOI] [PubMed] [Google Scholar]
- 62.Huang J, Gretz N. Light-emitting agents for noninvasive assessment of kidney function. ChemistryOpen. 2017;6:456–471. doi: 10.1002/open.201700065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takabatake Y, Isaka Y, Imai E. In vivo transfer of small interfering RNA or small hairpin RNA targeting glomeruli. Methods Mol. Biol. 2009;466:251–263. doi: 10.1007/978-1-59745-352-3_18. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, et al. Co-delivery of p38alpha MAPK and p65 siRNA by novel liposomal glomerulus-targeting nano carriers for effective immunoglobulin a nephropathy treatment. J. Control. Release. 2020;320:457–468. doi: 10.1016/j.jconrel.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 65.Shimizu H, et al. siRNA-based therapy ameliorates glomerulonephritis. J. Am. Soc. Nephrol. 2010;21:622–633. doi: 10.1681/ASN.2009030295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Masehi-Lano JJ, Chung EJ. Peptide and antibody ligands for renal targeting: nanomedicine strategies for kidney disease. Biomater. Sci. 2017;5:1450–1459. doi: 10.1039/C7BM00271H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia Z, et al. Suppression of renal tubulointerstitial fibrosis by small interfering RNA targeting heat shock protein 47. Am. J. Nephrol. 2008;28:34–46. doi: 10.1159/000108759. [DOI] [PubMed] [Google Scholar]
- 68.Alidori S, et al. Targeted fibrillar nanocarbon RNAi treatment of acute kidney injury. Sci. Transl. Med. 2016;8:331ra339. doi: 10.1126/scitranslmed.aac9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morishita Y, et al. siRNAs targeted to Smad4 prevent renal fibrosis in vivo. Sci. Rep. 2014;4:6424. doi: 10.1038/srep06424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dolman ME, Harmsen S, Storm G, Hennink WE, Kok RJ. Drug targeting to the kidney: advances in the active targeting of therapeutics to proximal tubular cells. Adv. Drug Deliv. Rev. 2010;62:1344–1357. doi: 10.1016/j.addr.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 71.Hamar P, et al. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc. Natl Acad. Sci. USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng X, et al. Attenuating ischemia-reperfusion injury in kidney transplantation by perfusing donor organs with siRNA cocktail solution. Transplantation. 2016;100:743–752. doi: 10.1097/TP.0000000000000960. [DOI] [PubMed] [Google Scholar]
- 73.Yang B, Hosgood SA, Nicholson ML. Naked small interfering RNA of caspase-3 in preservation solution and autologous blood perfusate protects isolated ischemic porcine kidneys. Transplantation. 2011;91:501–507. doi: 10.1097/TP.0b013e318207949f. [DOI] [PubMed] [Google Scholar]
- 74.Liu L, et al. Small interfering RNA targeting Toll-like receptor 9 protects mice against polymicrobial septic acute kidney injury. Nephron Exp. Nephrol. 2012;122:51–61. doi: 10.1159/000346953. [DOI] [PubMed] [Google Scholar]
- 75.Bondue T, van den Heuvel L, Levtchenko E, Brock R. The potential of RNA-based therapy for kidney diseases. Pediatr. Nephrol. 2022;38:327–344. doi: 10.1007/s00467-021-05352-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson JD, et al. Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 2012;22:255–264. doi: 10.1089/nat.2012.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pardridge WM. A historical review of brain drug delivery. Pharmaceutics. 2022;14:1283. doi: 10.3390/pharmaceutics14061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 79.Khvorova A, Osborn MF, Hassler MR. Taking charge of siRNA delivery. Nat. Biotechnol. 2014;32:1197–1198. doi: 10.1038/nbt.3091. [DOI] [PubMed] [Google Scholar]
- 80.Zhang W, Mehta A, Tong Z, Esser L, Voelcker NH. Development of polymeric nanoparticles for blood-brain barrier transfer-strategies and challenges. Adv. Sci. (Weinh) 2021;8:2003937. doi: 10.1002/advs.202003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomes MJ, Martins S, Sarmento B. siRNA as a tool to improve the treatment of brain diseases: mechanism, targets and delivery. Ageing Res. Rev. 2015;21:43–54. doi: 10.1016/j.arr.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 82.Murthy SK. Nanoparticles in modern medicine: state of the art and future challenges. Int. J. Nanomed. 2007;2:129–141. [PMC free article] [PubMed] [Google Scholar]
- 83.Alterman JF, et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 2019;37:884–894. doi: 10.1038/s41587-019-0205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown KM, et al. Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nat. Biotechnol. 2022;40:1500–1508. doi: 10.1038/s41587-022-01334-x. [DOI] [PubMed] [Google Scholar]
- 85.Chen Y, Liu L. Modern methods for delivery of drugs across the blood-brain barrier. Adv. Drug Deliv. Rev. 2012;64:640–665. doi: 10.1016/j.addr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 86.Saraiva C, et al. Nanoparticle-mediated brain drug delivery: overcoming blood-brain barrier to treat neurodegenerative diseases. J. Control. Release. 2016;235:34–47. doi: 10.1016/j.jconrel.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 87.Lajoie JM, Shusta EV. Targeting receptor-mediated transport for delivery of biologics across the blood-brain barrier. Annu. Rev. Pharmacol. Toxicol. 2015;55:613–631. doi: 10.1146/annurev-pharmtox-010814-124852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clark AJ, Davis ME. Increased brain uptake of targeted nanoparticles by adding an acid-cleavable linkage between transferrin and the nanoparticle core. Proc. Natl Acad. Sci. USA. 2015;112:12486–12491. doi: 10.1073/pnas.1517048112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 90.Zhang L, et al. Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs. Brain. 2021;144:3421–3435. doi: 10.1093/brain/awab354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim M, Kim G, Hwang DW, Lee M. Delivery of high mobility group box-1 siRNA using brain-targeting exosomes for ischemic stroke therapy. J. Biomed. Nanotechnol. 2019;15:2401–2412. doi: 10.1166/jbn.2019.2866. [DOI] [PubMed] [Google Scholar]
- 92.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 93.Huang JL, et al. Lipoprotein-biomimetic nanostructure enables efficient targeting delivery of siRNA to Ras-activated glioblastoma cells via macropinocytosis. Nat. Commun. 2017;8:15144. doi: 10.1038/ncomms15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao S, et al. A non-viral suicide gene delivery system traversing the blood brain barrier for non-invasive glioma targeting treatment. J. Control. Release. 2016;243:357–369. doi: 10.1016/j.jconrel.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 95.Liu Y, et al. A leptin derived 30-amino-acid peptide modified pegylated poly-L-lysine dendrigraft for brain targeted gene delivery. Biomaterials. 2010;31:5246–5257. doi: 10.1016/j.biomaterials.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 96.Wei L, et al. Brain tumor-targeted therapy by systemic delivery of siRNA with Transferrin receptor-mediated core-shell nanoparticles. Int. J. Pharm. 2016;510:394–405. doi: 10.1016/j.ijpharm.2016.06.127. [DOI] [PubMed] [Google Scholar]
- 97.Kumthekar P, et al. A first-in-human phase 0 clinical study of RNA interference-based spherical nucleic acids in patients with recurrent glioblastoma. Sci. Transl. Med. 2021;13:eabb3945. doi: 10.1126/scitranslmed.abb3945. [DOI] [PMC free article] [PubMed] [Google Scholar]