Abstract
The development and immune evasion of cancer stem cells (CSCs) limit the efficacy of currently available anticancer therapies. Recent studies have shown that epigenetic reprogramming regulates the expression of characteristic marker proteins and tumor plasticity associated with cancer cell survival and metastasis in CSCs. CSCs also possess unique mechanisms to evade external attacks by immune cells. Hence, the development of new strategies to restore dysregulated histone modifications to overcome cancer resistance to chemotherapy and immunotherapy has recently attracted attention. Restoring abnormal histone modifications can be an effective anticancer strategy to increase the therapeutic effect of conventional chemotherapeutic and immunotherapeutic drugs by weakening CSCs or by rendering them in a naïve state with increased sensitivity to immune responses. In this review, we summarize recent findings regarding the role of histone modifiers in the development of drug-resistant cancer cells from the perspectives of CSCs and immune evasion. In addition, we discuss attempts to combine currently available histone modification inhibitors with conventional chemotherapy or immunotherapy.
Subject terms: Cancer stem cells, Cancer microenvironment
Cancer drug resistance: Preventing modification of chromatin proteins
Understanding how modification of histones, structural proteins that support chromatin and affect gene expression, helps cancer stem cells interact with immune cells will help fight drug-resistant cancers. An important mechanism of cancer treatment resistance is histone modification. Ming Li Jin and Kwang Won Jeong at Gachon University in Incheon, South Korea, reviewed recent research into histone modifications in drug-resistant cancers. Much is known about how histone-modifying enzymes remodel the tumor micro-environment, allowing cancer stem cells to proliferate and block immune cell activity. Drugs that inhibit histone-modifying enzymes show great promise, but it is difficult to identify the precise mechanisms involved. Research should also address why inhibitors are less effective in some cancers, and how a patient’s unique tumor micro-environment could be analyzed to provide personalized precision medicine.
Introduction
Acquired resistance to anticancer drugs is commonly observed in most cancer types and is caused by both genetic and epigenetic intratumoral heterogeneity. Intratumoral heterogeneity refers to the formation of genetically diverse cell clusters, inducing the development of clones of drug-resistant cancer cells, including cancer stem cells (CSCs), after drug treatment. The development of anticancer drug resistance is also facilitated by heterogeneous complex and dynamic interactions in the tumor microenvironment (TME)1.
Previous studies have reported that epigenetic reprogramming regulates the expression of characteristic marker proteins and tumor plasticity associated with cancer cell survival and metastasis in CSCs2. Notably, CSCs have additional unique mechanisms to evade external immune cell attacks. The mechanisms used by the immune checkpoint (IC) and IC ligands to mediate immunosuppression and facilitate CSC stemness are well established. Moreover, an increased understanding of tumor-induced immune tolerance has led to innovative anticancer therapeutic strategies based on immune stimulation, such as cancer immunotherapy, including key drugs such as immune checkpoint inhibitors (ICIs) that positively regulate immune cell activation. ICIs have yielded promising results in melanoma treatment with subsequently positive effects in other cancer types3.
Self-renewal and tumor heterogeneity within subpopulations enables CSC survival against chemotherapy, leading to metastasis and tumor recurrence4. Immunotherapy has also yielded diverse results due to variable patient responses. Some patients treated with ICIs displayed a better prognosis due to tumor-specific immunological memory induction; however, a significant number of patients demonstrated ICI resistance, with recurrence even seen in patients with earlier responses5. Several theories have been postulated to explain the chemoresistance of CSCs and the unresponsiveness of ICIs. Epigenetic changes in tumor and immune cells are one of the key mechanisms proposed thus far6,7. DNA methylation and histone modifications can cause CSCs to increase stemness, reduce the expression of essential tumor-associated antigens on cancer cells recognized by the immune system, and modify IC expression8. Additionally, alterations in chemokine expression induced by histone-modifying enzymes lead to an immunosuppressive TME that contributes to the immune escape of CSCs9. Thus, targeting epigenetic regulators associated with the survival and immune evasion of CSCs is a novel therapeutic approach to effectively prevent cancer and has become a prominent field of interest, with a special focus on the regulation of histone modification, which could diversify therapeutic strategies for drug-resistant cancer cells. In this review, we summarize the latest findings on the roles of histone modifications in acquired drug resistance and immune evasion in CSCs and provide recent clinical treatment strategies for resistant cancer.
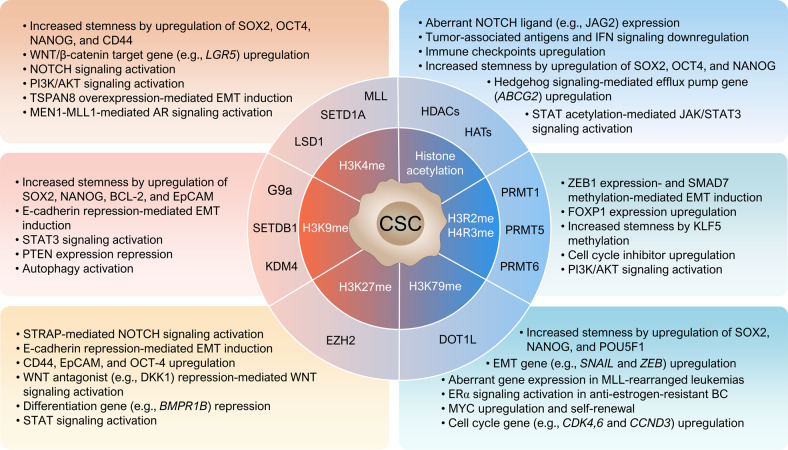
Aberrant signaling in CSCs regulated by histone modifications
Compared with progenitor cells, most CSCs exhibit increased drug resistance against chemotherapy and radiotherapy due to various factors, such as survival-promoting and anti-apoptotic signaling, overexpressed drug efflux pumps, intracellular drug-inactivating enzymes, enhanced DNA repair, and tumor niche2. Thus, a deeper understanding of CSCs leads to better implementation of existing anticancer therapies and the identification of new approaches to specifically target CSCs. In addition, CSCs modulate histone modifications to induce favorable conditions for cell survival and self-renewal through aberrant modification of various signaling pathways in normal cells, including the WNT, NOTCH, Janus kinase (JAK)/signal transducer and activator of transcription (STAT), hedgehog, and phosphatidylinositol-3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathways (Fig. 1).
Fig. 1. Aberrant histone modifications in cancer stem cells affecting drug resistance.
Cancer stem cells exhibit resistance to chemotherapy and radiation therapy through various intrinsic factors, including survival-promoting and anti-apoptotic signals and overexpression of drug efflux pumps. During resistance development, histone modification regulators create favorable conditions for CSC survival, self-renewal, and EMT regulation by modifying various signal transduction pathways, including the WNT, NOTCH, JAK/STAT, hedgehog, and PI3K/AKT/mTOR pathways in CSCs. Histone H3K4 methylation by SETD1A promotes the aberrant expression of stemness genes (e.g., SOX2 and OCT4). Epigenetic silencing of the CHD1 and PTEN genes by histone H3K9 and K27 methylation promotes EMT and induces the development of resistance against EGFR tyrosine kinase inhibitors. HDACs are involved in CSC stemness maintenance and the regulation of NOTCH, STAT3, and AKT signaling and are thus considered representative epigenetic target molecules for the treatment of cancers refractory to existing anticancer therapies.
The WNT signaling pathway is one of the most evolutionarily conserved signaling pathways, and its activation by WNT ligands plays critical roles in regulating stem cell differentiation and pluripotency10. Histone modification is a major mechanism underlying the dysregulated WNT/β-catenin pathway in cancer. For example, in glioblastoma, the decreased acetylation of histone H4K16 and increased trimethylation of histone H3K27 in the DKK1 gene promoter, a WNT antagonist, suppresses its expression, resulting in WNT signaling overactivation, which assists in CSC maintenance and tumorigenesis. Moreover, the recruitment of achaete-scute family bHLH transcription factor 1 (ASCL1) at H3K4me1 in the DKK1 gene enhancer region promotes repressive chromatin configuration in the form of a poised enhancer11,12. Thus, histone modifications alter the WNT signaling pathway.
Another mechanism of acquired chemoresistance in CSCs is via NOTCH signaling activation that induces self-renewal, epithelial-to-mesenchymal transition (EMT), and overexpression of efflux pumps such as ATP binding cassette subfamily C member 1 (ABCC1/MRP1)13,14. Jin et al. described the mechanism underlying the epigenetic activation of the NOTCH pathway by the serine-threonine kinase receptor-associated protein (STRAP) in CSCs to promote their proliferation and maintenance. STRAP reduces the histone methylation of H3K27 at the HES1 and HES5 promoter sites by inhibiting the association between the enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and SUZ12 subunits of the polycomb repressive complex (PRC)2, ultimately activating gene expression15. Moreover, abnormal overexpression of NOTCH ligands, including JAG2, due to histone acetylation at their promoter sites has been observed in multiple myeloma cells16.
The hedgehog (Hh) signaling pathway is activated by ligands including desert Hh, sonic Hh, and Indian Hh, and their association with Patched (PTCH) leads to the activation and nuclear translocation of downstream transcription factors, which induces the expression of genes involved in CSC survival, proliferation, and drug resistance17. The GLI1 and GLI2 proteins are regulatory transcription factors of Hh signaling and require deacetylation by HDAC1 to activate transcription18. Hh signaling directly regulates the efflux pump ABCG2 gene, where epigenetic histone modifications such as H3 acetylation, H3K4 trimethylation, and H3S10 phosphorylation upregulate its expression19. Additionally, in malignant rhabdoid tumors, the inactivation of SNF5, which directly interacts with GLI1 to suppress the expression of Hh target genes, including GLI1 and cyclin D, contributes to the activation of Hh signaling.
The JAK-STAT signaling pathway is the main axis of cytokine-mediated immune responses and is frequently dysregulated in CSCs, contributing to their stemness and self-renewal properties. Activation of the JAK/STAT3 signaling pathway also increases tumorigenesis and metastasis via enhanced EMT and CSC transition, leading to cancer chemoresistance20. The histone acetyltransferase p300 (KAT3B) plays a role in cytokine-induced STAT3 activation by acetylating it at Lys68521.
Aberrations in the PI3K/AKT/mTOR pathway have been shown to induce stemness, EMT, autophagy, and acquired resistance in CSCs22. Changes in the chromatin landscape caused by histone modifiers exert direct or indirect effects on CSC survival, and some of these modifiers are regulated by the PI3K/AKT signaling pathway. For example, AKT-mediated phosphorylation at Ser21 of EZH2 inhibits its activity. AKT also phosphorylates p300 and CREB-binding protein (CBP/KAT3A) to stimulate HAT activities, allowing them to participate in other signaling pathways in CSCs23.
E-cadherin, encoded by the CDH1 gene, regulates the TME as an adherent junction protein that maintains cells in an epithelial state, and its downregulation induces EMT24. E-cadherin expression is regulated by SNAIL, TWIST, and zinc finger E-box binding homeobox 1 (ZEB1/ZEB2) transcription factors25. The EZH2 and PRC2 complex binds to SNAI1 for recruitment to the CDH1 promoter, and methylation of H3K27 suppresses E-cadherin expression26. KDM7B (PHF8) induces EMT by enhancing SNAIL1 and ZEB1 expression by removing the repressive histone marks H3K9me1/2, H3K27me2, and H4K20me1927. Another mechanism of EMT regulation via histone modification is by LSD1 (KDM1A), which is overexpressed in various cancer types. By removing the mono- and dimethyl groups from histones K4 and K9, LSD1 regulates chromatin configuration and gene expression28. LSD1-mediated AKT activation promotes EMT in a CRC cell line with a PIK3CA mutation29, and another study using a different CRC cell line showed that LSD1 reduced the level of H3K9me2 in the promoter region of the EMT gene TSPAN8, leading to its overexpression30.
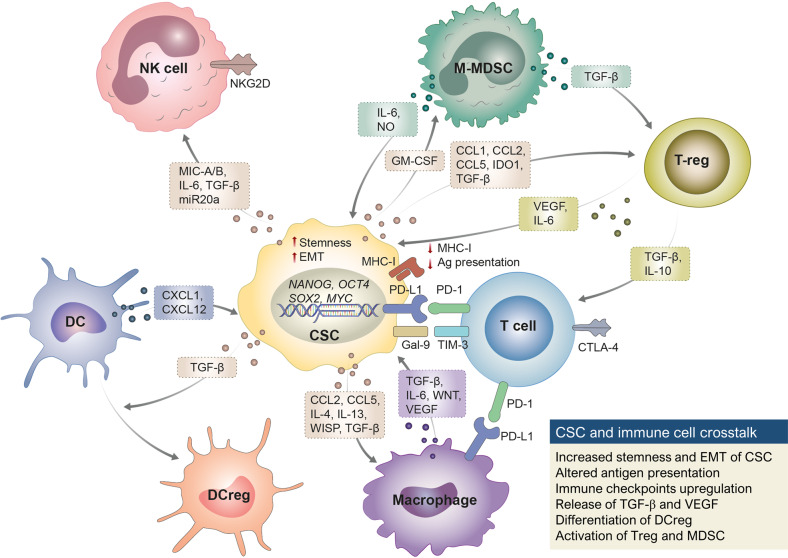
CSC-immune cell crosstalk in immune evasion
CSCs evade T-cell-mediated antitumor immune surveillance through various mechanisms, including suppression of T-cell activation, altered expression of major histocompatibility complex (MHC)-I at the transcriptional and posttranscriptional levels, downregulation of tumor-associated antigens (TAAs), and overexpression of ICs such as programmed cell death 1 ligand 1 (PD-L1) and Galectin-9 (Gal-9)8. MHC-I downregulation affects CD8+ T-cell activation that targets TAAs expressed by CSCs. WNT/β-catenin signaling acts on STT3, which is responsible for the glycosylation and stabilization of PD-L1, resulting in increased PD-L1 levels in CSCs, thereby contributing to CSC evasion of T-cell immune surveillance31. In hypoxic TME conditions, the expression of PD-L1 and vascular endothelial growth factor (VEGF) in CSCs is upregulated, and the expression of the T-cell inhibitory receptor TIM-3 is promoted via VEGF32. The overexpression of immune checkpoints such as PD-L1 and Gal-9 results in their association with corresponding receptors (e.g., PD-1 and TIM-3) in T cells to suppress T-cell proliferation and cytokine production and induce T-cell exhaustion, leading to CSC evasion of cytotoxic T cells8.
Mature dendritic cells (DCs) present TAAs and express costimulatory molecules that activate T-cell-mediated immune responses33. CSCs inhibit DC antitumor responses through various mechanisms, including transforming growth factor (TGF)-β release. Specifically, CSCs prevent the recruitment of CD103+ DCs to tumors34 and inhibit their maturation while promoting differentiation into immunosuppressive regulatory DCs (DCreg)35 and initiating the development of PD-1+ DCs that inactivate CD8+ T cells36. As an upregulated IC in chronic lymphocytic leukemia, breast cancer (BC), non-small-cell lung cancer (NSCLC), and CRC, CD200 induces immunological tolerance by negatively regulating DCs, macrophages, and T cells that express its receptors37.
The crosstalk between tumor-associated macrophages (TAMs) and CSCs supports the survival of CSCs and the formation of an immunosuppressive TME38. The niche of CSCs involves a unique TME with various cells, including fibroblasts and endothelial and immune cells, and is enriched in C-C motif chemokine 2/5 (CCL2/5), interleukins (ILs), periostin (osteoblast-specific factor, OSF-2), TGF-β, and colony-stimulating factor 1 (CSF1)39, which induce the conversion of protumorigenic macrophages into the immunosuppressive M2 or TAM phenotype40. TAM-released factors, including WNT, TGF-β, and VEGF, induce cancer stemness, immunosuppressive TME, EMT, and cancer metastasis41. TAMs also stimulate the expression of PD-L1 in CSCs and PD-1 in T cells to reduce T-cell-mediated cytotoxicity42.
Myeloid-derived suppressor cells (MDSCs) secrete cytokines and chemokines in the TME to assist in the formation of an immunosuppressive niche and reduce the efficacy of immunotherapy43. The mTOR signaling pathway is used by CSCs to promote the release of granulocyte-macrophage colony-stimulating factors (GM-CSFs) to induce tumor infiltration of MDSCs44. MDSCs in the TME release IL-6 and nitric oxide to induce the expression of CSC markers such as NANOG, OCT4, and SOX2 through epigenetic regulation45 while promoting the activation of regulatory T cells (Tregs) through the release of TGF-β46.
As an immunosuppressive T-cell-derived subpopulation, Tregs promote CSC immune evasion by interacting with them in the immunosuppressive TME to inhibit the effects of cancer immunotherapy. TGF-β produced by CSCs mediates tumor infiltration by Tregs, which is associated with a poor survival rate47. In addition, CSCs increase CCL1 expression to recruit Tregs to the TME via epigenetic mechanisms that reduce the level of H3K27me3 at the CCL1 promoter site48. CSCs also evade T-cell-induced apoptosis by differentiating uncommitted CD4+ T cells into Tregs49. In particular, Tregs in the hypoxic TME release VEGF to induce CSC stemness and angiogenesis, ultimately promoting EMT50,51.
Natural killer (NK) cells express killer cell lectin-like receptor K1 (NKG2D), Fas ligand (FASL), and TNF-related apoptosis-inducing ligand (TRAIL), which are activated by their respective binding factors to selectively kill MHC-I-negative CSCs52. NK cell-mediated cell lysis has been reported in MHC-I-negative colon CSCs (or cancer-initiating cells) that express NKG2DL53 and in MHC-1-negative ovarian CSCs that express ligands for the activating receptors NKp30 and NKp4454. However, CSCs of some ovarian and renal carcinoma patients display upregulated MHC-I molecules, making them less susceptible to NK cell-mediated cell lysis55,56. Interestingly, latent competent cancer cells expressing SOX2/SOX9 increase dormant CSCs (or latency-competent cancer cells) that downregulate NKG2DL through a unique mechanism that produces the WNT inhibitor DKK1, thus evading NK cell-mediated immunity57.
Interferon (IFN) is a potent antitumor cytokine that prevents CSC expansion and suppresses tumor-initiating properties. In addition, IFN-stimulated genes suppress the chemoresistance of CSCs58. However, CSCs develop mechanisms to resist the antitumor effect of IFN and induce the expression of CSC markers and immune evasion using IFN signaling59,60. This dual role of IFNs may depend in part on the duration and concentration of IFN exposure61,62. Suboptimal type-I IFN signaling induced by immunogenic cell death (ICD) did not lead to therapeutic anticancer immunity but rather promoted tumor progression through the expansion of the CSC population with characteristic immune evasion63. Similarly, while a high concentration of IFN-γ induced apoptosis in NSCLC through the JAK1/STAT1/caspase pathway, a low concentration of IFN-γ enhanced the properties of CSCs through the ICAM1/PI3K/AKT/NOTCH1 pathway64. As described above, the crosstalk between various immune cells and CSCs contributes to the formation of an immunosuppressive TME and immune evasion of CSCs, ultimately leading to adaptive resistance against cancer immunotherapy (Fig. 2).
Fig. 2. Immune evasion by crosstalk between immune cells and CSCs in the tumor microenvironment.
CSCs inhibit T-cell immune-mediated cytotoxicity by downregulating MHC-I and TAA expression and overexpressing immune checkpoints, such as PD-L1. The overexpression of immune checkpoints results in their association with corresponding receptors (e.g., PD-1 and TIM-3) in T cells to suppress T-cell proliferation and cytokine production and induce T-cell exhaustion, leading to CSC evasion of cytotoxic T cells. CSC-derived TGF-β/CCL interferes with the recruitment of DCs to the tumor site, promotes differentiation into DCregs, and induces the differentiation of macrophages from M1 to M2. VEGF, WNT, and TGF-β secreted from tumor-associated macrophages promote the stemness and EMT of CSCs. MDSCs secrete IL-6 and NO to induce the expression of CSC markers (e.g., NANOG, OCT4, and SOX2) and TGF-β to promote the activation of regulatory T cells (Tregs). Treg-derived VEGF induces the survival and EMT of CSCs. CSC-derived IL-6 and TGF-β downregulate NK-activating receptors, and CSCs inhibit NK cell activation through the release of MIC A/B and CD155 inhibitory activating receptors.
Histone modifications in CSC drug resistance
Histone lysine methylation
The functions of SET domain family proteins in drug-resistant or refractory cancers are well documented65. The mixed-lineage leukemia gene (MLL1/KMT2A) fused by chromosomal translocation is a prominent cause of aggressive leukemia in both children and adults66. Mechanistically, the MLL1 fusion protein in acute myeloid leukemia (AML) causes abnormal recruitment of histone modification enzymes, including DOT1L (KMT4) and protein arginine methyltransferase (PRMT) 1, at target genes for epigenetic reprogramming, conferring stem cell-like properties67,68. Recently, developed MLL inhibitors prevent the interaction between MEN1 and MLL1 to downregulate the expression of their target genes, thereby exerting a therapeutic effect on leukemia69. The MEN1-MLL1 association also regulates androgen receptor signaling in castration-resistant prostate cancer (CRPC). MI-136, a selective MEN1-MLL1 inhibitor, blocks this signaling to suppress tumor growth70. In addition, MLL1 regulates the expression of LGR5, a target gene of WNT/β-catenin and a stem cell gene in intestinal stem cells. LGR5+ intestinal stem cells and human colon carcinoma with elevated MLL1 and β-catenin expression levels are associated with poor survival71. In contrast, MLL3 (KMT2C), which is frequently lost in myelodysplastic syndrome (MDS), AML, aggressive nasopharyngeal carcinoma, and CRC, acts as a myeloma inhibitor that suppresses the self-renewal of hematopoietic stem cells and engages in differentiation via IL-1 stimulation72. MLL4 (KMT2D) is the gene that most frequently exhibits mutations in oral squamous cell carcinoma, and MLL4 knockdown decreases the expression of CD133 and β-catenin, colony formation, metastasis, and invasion to delay tumor growth73. In hematopoietic tissues, MLL5 (KMT2E) is involved in terminal myeloid differentiation and hematopoietic stem cell self-renewal74.
Recently, we reported the critical role of SETD1A (KMT2F) in CSC stemness in tamoxifen-resistant BC and CRPC. In tamoxifen-resistant BC, SETD1A expression is upregulated, which directly regulates the expression of SOX2, which plays a crucial role in the acquisition of stemness and resistance to tamoxifen75. Similarly, SETD1A expression is higher in metastatic CRPC (mCRPC) cells than in primary prostate cancer cells, whereas the expression of the stem cell factor OCT4 and transcription factor FOXM1 is upregulated76. Additionally, SETD1A allows NSCLC cells to maintain the characteristics of CSCs through WNT/β-catenin signaling and to develop resistance against anticancer drugs such as cisplatin77.
SETDB1 (KMT1E) is overexpressed in melanoma, BC, and lung cancer, is involved in stem cell maintenance and is considered an effective target for cancer therapy. Zhang et al. showed that SETDB1 is closely associated with EMT in BC stem cells, as it directly regulates STAT3 signaling78. In addition, the blockade of SETDB1 significantly increased the sensitivity of KRAS-mutant CRC to cetuximab treatment, demonstrating that it could be a potential anticancer target79. However, the lack of selective inhibitors remains an additional obstacle for studies on SETDB1.
G9a (EHMT2/KMT1C), which is responsible for mono- and dimethylation of H3K9, is a representative histone methylation enzyme that induces the epigenetic silencing of tumor suppressor genes, including TP53, CDH1, DUSP5, and PTEN, contributing to cancer metastasis and the maintenance of the malignant phenotype80. G9a interacts with SNAI1 at the CDH1 promoter and promotes EMT and lymph node metastasis by suppressing the expression of E-cadherin via H3K9 methylation81. In addition, G9a in lung cancer increases H3K9me2 levels at the PTEN promoter to suppress its transcription, thereby activating the AKT signaling pathway and contributing to the development of resistance against EGFR tyrosine kinase inhibitors (EGFR-TKIs). These results confirmed that the G9a inhibitor UNC0638 suppressed the growth of drug-resistant cancer cells82. In glioma stem-like cells, which are generally resistant to conventional therapy, autophagy plays a critical role in stemness, and G9a acts on the promoters of genes associated with autophagy (MAP1LC3B and WIPI1) and differentiation (GFAP and TUBB3). The G9a-specific inhibitor BIX-01294 upregulated the genes associated with autophagy and differentiation in glioblastoma CSCs to induce autophagy-dependent differentiation of glioma stem-like cells83.
Tazemetostat, which was recently approved as an inhibitor of EZH2, a histone H3K27 selective methyltransferase, has shown positive effects not only in epithelioid sarcoma but also in a variety of drug-resistant cancers. In SMARCB1 (a.k.a. SNF5)-deleted malignant rhabdoid tumors, tazemetostat induces the expression of genes related to neuronal differentiation and cell cycle inhibition while suppressing the Hh pathway genes. Notably, malignant rhabdoid tumors in animal models displayed dose-dependent degeneration after treatment with tazemetostat, and tumor regrowth was prevented even after the discontinuation of administration84. In papillary thyroid cancer, abnormal H3K27 trimethylation plays an important role in BRAFV600E-MAPK-induced differentiation and drug resistance. Combination treatment with tazemetostat and a MAPK inhibitor resulted in a reduction in global H3K27 trimethylation, leading to the differentiation of papillary thyroid cancer with BRAFV600E 85. Resistance to receptor tyrosine kinase inhibitors (RTKis) is a major obstacle in the treatment of clear cell renal cell carcinoma. Overexpression of EZH2 promotes global kinase phosphorylation, which induces acquired and intrinsic resistance. Treatment with the EZH2 inhibitor EPZ011989 reduced kinase phosphorylation and activated tumor suppressors to reverse resistance to sunitinib86.
DOT1L, a histone H3K79 methyltransferase, plays an important role in the proliferation, self-renewal, and metastasis of BC cells resistant to antihormone therapy87. In anti-estrogen-resistant BC, blocking DOT1L affects ERα-dependent transcription, including silencing of ESR1 and FOXA1 genes, with a consequent inhibitory effect on tumor growth both in vitro and in vivo88. A recent study demonstrated that DOT1L is the main regulator of CSCs, with roles in the expression of MYC in ALDH1+ triple-negative BC (TNBC) and the maintenance of self-renewal. The DOT1L inhibitor EPZ-5676 reduced tumorsphere formation and ALDH1+ cells in vitro and inhibited tumor-initiating stem cells and metastasis of ALDH1+-derived tumor xenografts in vivo89.
Histone arginine methylation
PRMT1, a type I protein arginine methyltransferase, catalyzes the production of asymmetric dimethylarginine and plays a role in various cellular processes, including the generation of hematopoietic and tumor cells90. PRMT1 promotes EMT and CSC properties by activating ZEB1 gene transcription via the asymmetric dimethylation of H4 (H4R3me2as) at the ZEB1 promoter in BC cells91. Additionally, PRMT1 acts as a crucial mediator of TGF-β signaling and promotes TGF-β-induced EMT and the generation of epithelial stem cells through SMAD7 methylation92.
PRMT5 catalyzes the production of symmetric dimethyl arginine and is involved in various cellular processes, including pluripotency and tumorigenesis. In patient-derived BC stem cells, PRMT5 generates H3R2me2s at the FOXP1 promoter, which is recognized by WDR5, a component of the SET1 complex that consequently promotes gene expression93. Treatment of liver CSCs with a PRMT5 inhibitor (DW14800) results in the inhibition of HNF4A gene expression by reducing H4R3me2, resulting in the reconstruction of hepatocyte-specific properties and antitumor effects94. In addition, PRMT5 in basal-like BC cells promotes the maintenance and proliferation of BC stem cells through arginine methylation of KLF595. Recently, PRMT5 has emerged as a promising target for glioblastoma treatment. The critical influence of PRMT5 on CSC growth in BC and glioblastoma has been verified using a PRMT5 inhibitor (e.g., GSK3203591 or LLY-283)93,96.
Inhibition of PRMT6 in cancer cells upregulates tumor suppressor genes. The depletion of PRMT6 results in increased p21, p27, and CD44 and downregulated MMP-9 expression and PI3K/AKT/mTOR signaling in CRPC, thus inducing sensitivity to chemotherapy, and the methylation of p21 by PRMT6 (R156) reduces chemosensitivity in CRC cells97,98. Therefore, this strategy can be used to improve the efficacy of conventional therapies for drug-resistant cancers. In addition, PRMT6 assists in the maintenance of CSC characteristics in glioblastoma through methylation of the regulator of chromosome condensation 1 (RCC1) protein. Hence, EPZ020411, a PRMT6-specific inhibitor, inhibits the methylation of the arginine residue of RCC1 to maximize the positive effects of radiotherapy in a glioblastoma xenograft model99.
Histone demethylation
LSD1 interacts with OCT4 in CSCs to turn the enhancer site of the pluripotency gene (PpG) into a ‘primed’ state with sensitivity to reactivation, and abnormally increased PpG expression leads to the increased tumorigenicity of CSCs100. Liu et al.101 showed that the increased LSD1 expression in CSCs of hepatocellular carcinoma is critical for maintaining self-renewal and tumorigenicity through activated NOTCH signaling and that LSD1 overexpression in non-CSCs is sufficient to induce self-renewal. In contrast, inactivation of LSD1 leads to the downregulation of SOX2 and OCT4 and the induction of differentiation genes such as BMP2102. In BC cells, LSD1 regulates BC stem cells by modulating self-renewal, EMT, and anticancer drug resistance103. Hence, combined treatment with an LSD1 inhibitor and chemotherapy in an in vivo BC model abolished the mesenchymal signature and promoted an innate M1 macrophage-like immune response104. PKC-θ is a determinant of the regulation of epigenetic mesenchymal gene expression in LSD1. Phosphorylation of the serine-111 residue of LSD1 (LSD1-s111p) by protein kinase C-theta (PKC-θ) is critical for LSD1 demethylase activity, and phosphorylated LSD1 is elevated in chemoresistant cancer111. The enhancer region of SOX2, which is overexpressed in luminal-B BC cells, is a target site for LSD1. Iadademstat, a known LSD1 inhibitor, suppresses SOX2 expression and mammosphere formation in patient-derived CSCs of multidrug-resistant luminal-B BC105. Interestingly, LSD1 in gastric cancer patients can migrate to other gastric cancer cells via small extracellular vesicles106. In acceptor cells, the delivered LSD1 not only promoted cancer stemness by inducing the expression of NANOG, OCT4, SOX2, and CD44 but also produced resistance to oxaliplatin. Ongoing research is addressing the prevalence of sEV-derived cancer resistance in other carcinomas, which would be a new mechanism of acquired drug resistance by cancer cells.
In ovarian cancer, KDM3A induces the expression of SOX2, NANOG, and the anti-apoptotic protein Bcl-2 and suppresses the expression of p21WAF1/CIP1, a cyclin-dependent kinase inhibitor, thereby regulating stemness and cisplatin resistance107. Recently, a KDM4 inhibitor, TACH101, displayed promising results in various tumor models. For instance, the administration of TACH101 reduced the rate of CRC growth and the ratio of cells exhibiting CSC characteristics (CD44high EpCAM+), and in different xenograft models of lymphoma, esophageal, gastric, and breast cancers, TACH101 inhibited up to 100% of tumor growth (2022 ASCO Annual Meeting Abstract).
Histone acetylation
The expanded knowledge of the functions of other HAT family proteins in CSCs has prompted the development of inhibitors for the treatment of drug-resistant cancers. For instance, WM-3835 can chemically inhibit the acetyl-CoA binding site of HBO1 (KAT7 or MYST2), ultimately inhibiting leukemia stem cell growth108. CPTH6, an inhibitor of PCAF and GCN5, can reduce CSC markers such as CD133 and ALDH and engage in the autophagy pathway to inhibit the growth of lung cancer stem-like cells109. WM-8014 and WM-1119 inhibit KAT6A, an acetyltransferase subunit in the MOZ complex, to suppress proliferation and induce senescence in lymphoma via the p16INK4a and p19ARF pathways110. In TNBC, these inhibitors block SMAD3 acetylation, suppress cytokine expression, inhibit stemness, and prevent the recruitment of MDSCs to substantially increase sensitivity to anti-PD-L1 therapy111. In addition, the inhibition of KAT6A in ovarian cancer could increase sensitivity to cisplatin, which implies its potential use in combination therapy for the treatment of drug-resistant cancers112.
Histone deacetylation
HDAC1 induces the deacetylation and ubiquitination of SMAD7 required for the maintenance of the epithelial phenotype of CSCs in ovarian cancer, thereby reducing SMAD7 stability113. In CSCs in head and neck cancer, HDAC1 controls the acetylated state of GRP78 to increase the CD44high/CD24low phenotype and assist in CSC maintenance114. Additionally, HDAC1 and HDAC7 increase CSC phenotypes, such as miR-34a, CD44, and CD166, assisting with chemotherapy drug resistance, metastasis, and recurrence in BC and ovarian cancer115,116. Indeed, high levels of HDAC1 and HDAC7 have been observed in residual tumor cells with low sensitivity to chemotherapy117.
STAT3 signaling mediates the maintenance of CSCs by IL-6 in various cancer types118, and deacetylation of the STAT3 Lys685 residue by HDAC3 facilitates the phosphorylation of the Tyr705 residue to activate STAT3119. HDAC11, which is upregulated in the stem-like population of NSCLC, interacts with the transcription factor GLI1 to activate the expression of SOX2. Moreover, selective HDAC11 inhibitors (FT234 and FT895) efficiently inhibit the growth of drug-resistant lung adenocarcinoma stem cells120.
SIRT1, another HDAC, is overexpressed in stem-like cells in CRC to increase the CD133+ cell population, expression of stemness-related genes including TDGF1, NANOG, OCT4, TERT, and LIN28, CSC sphere formation, and tumorigenesis121. Inhibition of p53 is involved in this process because SIRT1 can deacetylate the C-terminal Lys120, Lys164, and Lys382 residues of p53 to inhibit its activity122. Inhibition of SIRT1 activity has been highlighted as an effective alternative for the removal of imatinib-resistant quiescent leukemia stem cells (BCR-ABL tyrosine kinase inhibitor). Indeed, a SIRT1 inhibitor (tenovin-6) increased the expression of the p53 target gene in chronic myeloid leukemia CD34+ cells by activating p53 transcriptional activity123.
The HDACs described above are representative epigenetic target molecules for the treatment of cancers refractory to existing anticancer therapies. Accordingly, extensive trials of HDAC inhibitor monotherapy or combination therapy with other drugs for various resistant carcinomas have been conducted, and the role of HDAC inhibitors in reducing CSC aggressiveness has been promising. Early HDAC inhibitors, such as vorinostat and panobinostat, exhibited higher effects than anticipated in a single or combined administration in preclinical models of various drug-resistant cancers, thereby providing a sufficient rationale to initiate clinical trials. Vorinostat reverses cisplatin resistance and reduces the self-renewal ability of CSCs by suppressing NOTCH signaling via miR-34a re-expression in head and neck and pancreatic cancer cells124,125. For a more effective application of HDAC inhibitors, a detailed in-depth understanding of their differential mechanisms of action in different cancers refractory to HDAC inhibitors is needed. However, new mechanisms for overcoming resistant cancers have been revealed through various combination therapy attempts (Table 1). Thus, the vast accumulated epigenetic information will provide optimal conditions and combination therapies for the treatment of resistant cancer in the near future, along with the development of immuno-oncology, which will be described later.
Table 1.
Mechanisms of action of histone modification inhibitors used in single or combination therapy for overcoming drug resistance in cancer.
| Histone modifier | Inhibitor | Resistant cancer cell type | Combination therapy | Proposed mechanism of overcoming cancer resistance | Refs. |
|---|---|---|---|---|---|
| PRMT5 | GSK3203591 | Breast cancer stem cell | NA | Downregulation of FOXP1 expression by inhibiting H3R2 methylation at the FOXP1 promoter | 93 |
| DW14800 | Liver cancer stem cell | NA | Upregulation of HNF4α expression by reducing H4R3me2s levels | 94 | |
| LLY-283 | Glioblastoma stem cell | NA | Deregulation of alternative splicing, affecting regulators of cell cycle | 96 | |
| PRMT6 | EPZ020411 | Glioblastoma stem cell | Radiation therapy | Inhibition of arginine methylation of RCC1 | 99 |
| G9a | UNC0638 | Neuroblastoma (MYCN-amplified) | EPZ011989 | Upregulation of IFN-γ-induced expression of CXCL9 and CXCL10 | 131 |
| UNC0638 | NSCLC (EGFR-TKI resistance) | EGFR-TKI (Erlotinib) | Upregulation of PTEN expression and inhibition of the AKT signaling pathway | 82 | |
| BIX-01294 | Glioma stem-like cell | NA | Upregulation of autophagy and differentiation genes | 83 | |
| EZH2 | Tazemetostat | Papillary thyroid cancer (radioiodine-refractory) | Dabrafenib, Selumetinib | Upregulation of iodine-metabolizing gene expression and promotion of radioiodine uptake | 85 |
| Malignant rhabdoid tumor (SMARCB1-deleted) | NA | Reductions in global H3K27me3 levels, upregulation of genes related to neuronal differentiation and cell cycle inhibition, and silencing of hedgehog pathway genes | 84 | ||
| GSK503 | Melanoma | IL-2cx/anti-CTLA-4 Ab | Downregulation of PD-L1 expression and accumulation of IFN-γ-producing CD8+ T cells | 135 | |
| CPI-1205 | Bladder cancer, Melanoma | Ipilimumab | Inhibition of EZH2 expression induced by anti–CTLA-4 | 140 | |
| EPZ011989 | Clear cell renal cell carcinoma (sunitinib-resistant) | Sunitinib | Re-expression of tumor suppressors (e.g., DAB2IP and PTPN3) and attenuation of the global kinome reprogramming | 86 | |
| DOT1L | EPZ004777 | Breast cancer (anti-estrogen resistant) | NA | Silencing of ERα and FOXA1 and inhibition of ER-dependent signaling | 88 |
| EPZ-5676 | Triple-negative breast cancer | NA | Downregulation of MYC expression | 89 | |
| LSD1 | GSK-LDS1 | Melanoma (ICI-refractory) | Anti-PD-(L)1 Ab | Increase in endogenous retroviral elements (ERVs) and activation of the dsRNA-IFN pathway | 144 |
| HCI-2509 | Triple-negative breast cancer | Anti-PD-1 Ab | Induction of the expression of chemokines (CCL5, CXCL9,10) and PD-L1 | 145 | |
| Iadademstat | Luminal-B breast cancer (multidrug-resistant) | Downregulation of SOX2 expression | 105 | ||
| GSK2879552 | HCC stem-like cells (sorafenib-resistant) | Sorafenib | Derepression of WNT antagonist expression | 146 | |
| KDM4D | TACH101 | Colorectal cancer stem cell | NA | Decrease in CSC signature (CD44High EpCAM+) | Meeting Abstract |
| KAT6A | WM-1119 | Triple-negative breast cancer | Anti-PD-L1 Ab | Inhibition of SMAD3 acetylation, reduction of cytokine expression, inhibition of stemness, inhibition of myeloid-derived suppressor cell (MDSC) recruitment | 111 |
| KAT7 | WM-3835 | Leukemia stem cell | NA | Removal of acetyl-CoA binding site of HBO1 (KAT7) and repression of HOXA9,10 expression | 108 |
| HDACs | Vorinostat | Pancreatic cancer | NA | Inhibition of Notch signaling by upregulating miR-34a expression | 125 |
| Relapsed lymphoma (rituximab–chemotherapy-resistant) | NA | Increase in p21 and acetylation of histone H3 leading to G1 cell cycle arrest | 169 | ||
| Small-cell lung cancer (navitoclax-resistant) | Navitoclax | Upregulation of BH3-only pro-apoptotic BCL-2 family protein (e.g., NOXA) | 170 | ||
| Pancreatic cancer | Sorafenib, anti-PD-1 Ab | Reduction of HDAC protein levels, promotion of autophagy, reduction of PD-L1 and IDO-1 levels | 171 | ||
| Squamous cancer cell | 5-FU/Cisplatin | Reverted 5FU/cisplatin-induced EGFR nuclear translocation, and induction of cisplatin uptake | 172 | ||
| Lung cancer (EGFR-C797S-mutated) | Brigatniib | Reduction of total EGFR-3M (L858R/T790M/C797S) proteins through STUB1-mediated ubiquitination and degradation | 173 | ||
| Castration resistant prostate cancer | Docetaxel | Suppression of the expression and nuclear translocation of AR-FL and AR-Vs | 174 | ||
| Triple-negative breast cancer | Anti-PD-L1 Ab | Increase in sensitivity to anti-PD-L1 therapy by increasing PD-L1 expression and downregulation of CD4+ FOXP3+ Treg | 156 | ||
| Belinostat | NSCLS (cisplatin-resistant) | Cisplatin | Increase in the association of a transcriptional repressor to the ABCC2 gene | 175 | |
| Advanced HCC | Anti-CTLA4 ab | Enhanced IFN-γ production by antitumor T cells and a decrease in regulatory T cells | 176 | ||
| Acute promyelocytic leukemia | All-trans-retinoic acid | Downregulation of EZH2, SUZ12, HDAC-1, HDAC-2, and PCAF expression | 177 | ||
| Panobinostat | DLBCL | Ibrutinib | Decreased phosphorylated STAT3 binding to the MyD88 promotor, downregulation of MyD88 expression, and inhibition of NF-κB activity | 178 | |
| Acute myeloid leukemia (adriamycin-resistant) | Bortezomib | Induction of apoptosis by inhibiting the PI3K/AKT and NF-κB pathway | 179 | ||
| Triple-negative breast cancer | NA | Upregulation of CDH1 protein expression and reversal of the mesenchymal phenotype | 180 | ||
| Neuroblastoma (MYCN-amplified) | JQ1 | Downregulation of LIN28B gene and N-MYC protein expression | 181 | ||
| Breast cancer (trastuzumab-refractory) | Trastuzumab | Abrogation of AKT signaling and induction of CXCR3-reactive ligand expression | 182 | ||
| NSCLC (KRAS-mutated) | Gefitinib | Downregulation of TAZ downstream targets, including EGFR and EGFR ligand | 183 | ||
| Castration resistant prostate cancer | Bicalutamide | Reduction of AR-mediated resistance to bicalutamide | 184 | ||
| EGFR-Mutant NSCLC (osimertinib-resistant) | Osimertinib | Elevation of BIM in osimertinib-resistant cells | 185 | ||
| Melanoma | Anti-PD-(L)1 Ab | Increase in sensitivity to anti-PD-L1 therapy by induction of PD-L1 and PD-L2 expression | 154 | ||
| Class I HDACs | Entinostat | Bladder, colorectal, and breast cancers | Anti-PD-1 Ab | Promotion of immune editing of tumor neoantigens, effectively remodeling the tumor immune microenvironment | 159,161 |
| Romidepsin | Peripheral T-cell lymphoma | Tamoxifen | Induction of apoptosis by activating FOXO1 expression | 186 | |
| Lung cancer | Anti-PD-1 Ab | Upregulation of chemokine (CXCL9, CXCL10), PD-1L, and STAT1 expression. | 152 | ||
| Triple-negative breast cancer Clear cell renal cell carcinoma | Decitabine | Re-expression of the tumor suppressor gene (sFRP1) | 187 | ||
| Class I/II HDACs | Givinostat (ITF-2358) | NSCLC | Aazacitidine (DNMTi) | Depletion of MYC in NSCLC and enhancement of NSCLC sensitivity to HDACi, Upregulation of T-cell chemoattractant (CCL5) expression | 158 |
| HDAC6 | Nexturastat A | Melanoma | Anti-PD-1 Ab | Complete neutralization of the upregulation of PD-L1 and other immunosuppressive pathways induced by the treatment with anti-PD-1 blockade | 162 |
| Citarinostat | Multiple myeloma | NA | Upregulation of B7 (CD80, CD86) and MHC-I,II expression in tumor and dendritic cells | 163 | |
| HDAC11 | FT234 | NSCLC (erlotinib-resistant) | NA | Suppression of GLI1-mediated SOX2 expression | 120 |
| SIRT1 | Tenovin-6 | Chronic myeloid leukemia | Imatinib | Promotion of p53 acetylation and transcriptional activity | 123 |
NSCLC non-small-cell lung cancer, HCC hepatocellular carcinoma, DLBCL diffuse large B-cell lymphoma, NA not applicable.
Histone modifications in immune evasion of CSC
Histone methylation
Several theories have been proposed to explain the unresponsiveness or evasion of ICIs, including intrinsic tumor modifications such as the lack of neoantigens, inactivation of the antigen presentation machinery, blocking of IFN-γ signaling, and extrinsic tumor modifications such as the loss of antigen-presenting cell function through the release of immunosuppressive cytokines, decreased T-cell proliferation and cytokine release, upregulation of additional ICs (e.g., TIM3 and LAG3), and activation of Tregs and MDSCs126,127. Histone methylation is directly involved in immune evasion mechanisms of cancer cells, including breast, prostate, colorectal cancers, and AML6,7, leading to new strategies to regulate the balance of histone methylation for cancer immunotherapy.
G9a and SETDB1 play essential roles in the regulation of pluripotency, stemness, and tumorigenicity128,129. Notably, the expression levels of G9a and SETDB1 correlated with the response to anti-PD-1 immunotherapy. For example, in a mouse bladder cancer model, the dual inhibition of G9a and DNA methyltransferase (DNMT) enhanced the response to anti-PD-L1 therapy, increased the tumor infiltration of NK cells and CD8+ T cells, and facilitated tumor regression130. In addition, inhibition of G9a in neuroblastoma can enhance IFN-γ-stimulated expression of CXCL9 and CXCL10, which are important Th1 chemokines for the recruitment of cytotoxic T cells to the TME of neuroblastoma131. The dual inhibition of histone modification enzymes has also been evaluated. In multiple myeloma cells, simultaneous inhibition of G9a and EZH2 induces the expression of ERV factors and activates the IFN response, causing cell cycle arrest and apoptosis132. In cancer, overexpression of SETDB1 negatively correlates with a characteristic gene signature associated with a positive immune response. Patients with renal cell carcinoma with poor survival response to anti-PD-1 therapy have been shown to have amplification of the SETDB1 gene133. Consistently, SETDB1 knockout in a melanoma mouse model induced the re-expression of ERV antigens presented on the surface of cells by MHC-I to induce a cytotoxic T-cell response. Another study showed that JARID1B (KDM5B), an H3K4 demethylase, can assist SETDB1 by recruiting SETDB1 to the endogenous retrovirus (ERV) element site134, suggesting that SETDB1 is a potential target to restore the immune surveillance of CSCs.
EZH2 has received increased attention as a cause of resistance in tumor immunotherapy because it is overexpressed in various cancer types, including melanoma, and silences tumor-suppressor genes or genes associated with antigen presentation135. The increased expression of EZH2 is associated with the promotion of Treg cell differentiation136, while it is inversely correlated with tumor infiltration of CD8+ T cells137. In a melanoma mouse model, EZH2 was associated with the epigenetic development of resistance against recombinant IL-2 (rIL2) and ICI. Inhibition of EZH2 activity concurrently with rIL2/anti-CTLA-4 immunotherapy downregulates the expression of PD-L1, promotes the expression of IFN-γ, increases the number of intratumoral PD-1lowTIM-3lowLAG-3lowCD8+ T cells, and reverses resistance against anti-CTLA-4 and IL-2 immunotherapies135. EZH2 inhibitors also restore the expression of Th1-type chemokines in ovarian cancer, facilitate tumor infiltration of effector T cells, and enhance the efficacy of anti-PD-L1 treatment137. In contrast to melanoma, which shows high PD-L1 expression, low PD-L1 expression in cancer cells is highly associated with low responsiveness to anti-PD-(L)1 therapy. Xiao et al. showed that EZH2 in hepatocellular carcinoma directly upregulated the level of H3K27me3 at the CD274 promoter to reduce PD-L1 expression138. In addition, EZH2 contributes to the progression of hepatocellular carcinoma by increasing the expression of additional immune checkpoints such as Gal-9 and TIM-3 ligand through H3K27 trimethylation at the miR-22 promoter, with a positive role in the progression of hepatocellular carcinoma139. Goswami et al. observed that peripheral T cells in patients treated with anti-CTLA-4 therapy (ipilimumab) had increased EZH2 expression140. In the same study, the administration of an EZH2 inhibitor (CPI-1205) in bladder cancer and melanoma mouse models increased the cytotoxic activity of CD8+ effector T cells, altered the phenotype and function of Tregs, and increased susceptibility to ipilimumab, providing a rationale for combining immune checkpoint inhibitors and histone modification inhibitors against ICI-resistant cancers (Table 1).
In pancreatic ductal adenocarcinoma, attempts to inhibit PRMT1 to overcome drug resistance against anti-PD-L1 therapy led to a positive outcome141. Unexpectedly, the inhibition of PRMT5 in lung cancer increases CD274 gene expression, eventually activating the PD1/PD-L1 axis and blocking T-cell-mediated antitumor immunity142. Thus, PRMT5 inhibition in combination with anti-PD-L1 therapy could be a breakthrough even though further research is required to investigate its underlying activities.
Elevated LSD1 expression in breast, lung, and other cancer types is not only essential in maintaining CSC stemness and mediating chemoresistance143 but is also inversely proportional to the expression levels of T-cell-attracting chemokines (e.g., CXCL9 and CXCL10) and CD8+ T-cell infiltration, which predicts a poor prognosis in clinical practice144,145. In stem-like cells of sorafenib-resistant hepatocellular carcinoma, LDS1 inhibitors block WNT/β-catenin signaling to overcome drug resistance146. In addition, LSD1 suppresses IFN-mediated antitumor immunity by preventing the activation of ERV factors through demethylation of Argonaute RISC catalytic component 2 (AGO2)144. Hence, LSD1 inhibitors exhibit antitumor effects by derepressing ERV factors, activating type I IFN expression, activating CD8+ T cells, facilitating infiltration in cancer tissues, and promoting DC differentiation in melanoma147. The effect of LSD1 inhibition in overcoming unresponsiveness to ICIs has been verified in various models. In melanoma and TNBC mouse xenograft models, LSD1 inhibitors in combination with anti-PD-1 antibodies increase CD8+ T-cell infiltration and significantly suppress tumor growth and lung metastasis147. The inhibition of LSD1 also enhanced the efficacy of ICI treatment in HNSCC and cervical cancer mouse models148,149. Of note, however, is that the removal of LSD1 in melanoma induced the expression of TGF-β, which suppresses T-cell immunity; as a result, the tumor could not be completely eradicated despite the enhanced efficacy of the combination with ICIs144,147. As a solution, Sheng et al.147 suggested that in ICI treatments, simultaneous inhibition of LSD1 and TGF-β could contribute to the complete eradication of ICI-refractory tumors and prevent tumor recurrence. These findings suggest that the abovementioned triple combination could be applied to the treatment of other types of tumors with low immunogenicity.
Histone acetylation
In various tumor mouse models, treatment with HDAC inhibitors has shown interesting effects in reversing drug resistance against ICIs through varying mechanisms150. Inhibition of HDAC using romidepsin (a class I HDACi) in a melanoma mouse model increased the expression of genes related to MHC I antigen processing and presentation, including transporter 1, ATP binding cassette subfamily B member (TAP1), proteasome 20S subunit beta 9 (LMP2), and β-2-microglobulin (B2M), and improved the cytotoxic activity of CD8+ T cells151. The use of romidepsin in a lung tumor model increased the response to PD-1 blockade immunotherapy and improved T-cell infiltration152. Additionally, Woods et al. showed that panobinostat (pan-HDACi) in melanoma increased the expression of MHC-I in cancer cells and increased the production of IL-2 and IFN-γ from CD4+ T cells, subsequently increasing the sensitivity to anti-PD-(L)1 therapy153,154. Panobinostat regulates the levels of various cytokines associated with T-cell activation in patients with Hodgkin’s lymphoma155. Vorinostat (a pan-HDACi) increases sensitivity to anti-PD-L1 therapy by increasing PD-L1 expression and downregulating CD4+ FOXP3+ Tregs in TNBC156. Similarly, Briere et al. reported that in a syngeneic tumor model, a class I/IV HDACi (mocetinostat) increased PD-L1 expression and exhibited a significantly improved antitumor immune response in combination with anti-PD-L1 therapy157. In addition, givinostat, a class I/II HDAC inhibitor, blocked tumor regression and metastasis when used with DNMTi (azacitidine), anti-CTLA-4 mAb, or anti-PD-1 mAb in an NSCLC model158. Entinostat (a class I HDACi) inhibited the function of MDSCs and increased the antitumor effect of anti-PD-1 therapy in metastatic BC159. In other studies, entinostat inhibited MDSCs in lung cancer and renal cell carcinoma mouse models to reinforce the effects of anti-PD1 therapy160 and induce the expression of genes related to antigen presentation in BC161. In addition, Knox et al. reported that treatment with a selective HDAC6i (nexturastat A) increased IFN-γ and IL-2 expression in the SM1 murine melanoma model and improved the TME to maximize the effects of anti-PD-1 therapy162. ACY241, another HDAC6 inhibitor, upregulated the expression of costimulatory (CD28 and CD40L) and activating (CD38, CD69, and CD137) molecules in bone marrow cells in multiple myeloma and increased T-cell activity163. To date, clinical studies on the combined use of HDAC inhibitors and immune checkpoint inhibitors have been positive. However, some HDAC inhibitors also have negative effects on immune cell viability and function that possibly limit the effect of immune therapy, such as inhibiting the activation and proliferation of lymphocytes, thereby requiring further investigation164,165.
Unlike HDAC inhibitors, the positive effects of HAT inhibitors have not yet been reported. Nevertheless, HAT also has the potential to become an important target molecule for immunotherapy. Fan et al. reported that an upregulated level of HAT1 (histone acetyltransferase 1, KAT1) was correlated with a poor prognosis of pancreatic cancer166. The study also showed that HAT1 regulated the expression of PD-L1 at the transcriptional level in both in vitro and in vivo models and that HAT1 knockdown reduced pancreatic tumor cell proliferation. In addition, PD-L1 expression was positively correlated with HAT1 expression in pancreatic tumor tissues. These results indicate that in patients with acquired resistance due to abnormal expression of HAT1 and PD-L1, targeting HAT1 activity could maximize the antitumor immune response to ICIs by blocking the PD-1/PD-L1 axis and increasing sensitivity to ICIs.
These results collectively suggest that histone modification enzymes regulate the adaptive resistance mechanisms against tumor immunotherapy to act as epigenetic checkpoints to suppress immune responses and that these histone modifiers could serve as potential therapeutic targets for the improvement of immunotherapy (Fig. 3).
Fig. 3. Overcoming cancer resistance to immune checkpoint inhibitors using histone modification inhibitors.
The expression of PD-L1 and tumor mutational burden (TMB) in tumors are critical factors for responsiveness to immunotherapy. TMB increases the probability of neo-antigen expression and eventually leads to an effective immune response. Histone-modifying enzymes (e.g., HDACs and EZH2), which are overexpressed in CSCs, enhance stemness and decrease immunogenicity through epigenetic modulation in CSCs, making them resistant to immunotherapy. This tumor immune evasion is acquired through the decreased expression of neoantigens, secretion of immunosuppressive cytokines, decreased antigen processing and presentation, and attenuation of IFN signaling. Inhibitors of histone-modifying enzymes promote the expression of neoantigens, activate the antigen presentation pathway, increase the secretion of immune-enhancing cytokines (e.g., IFNs) in resistant tumors, and downregulate immunosuppressive factors in the TME, leading to tumor rerecognition by the immune system and increasing the efficacy of anti-PD-(L)1 therapy.
Inhibitors for histone modificatons
As previously mentioned, epigenetic mechanisms exert a significant impact on both host immune cells and tumor cells, and epigenetic drugs enhance the efficacy of cancer immunotherapy through various mechanisms. In line with this, the combination of conventional anticancer chemotherapy drugs or ICIs with histone modification inhibitors is highly attractive, with dramatic activation effects, as shown in numerous studies using cellular and animal models. Combined treatments with histone modification inhibitors and other anticancer drugs are currently being tested in studies targeting various cancer types.
The efforts of the past two decades have been successful, and several histone deacetylation inhibitors (HDACis) and histone methylation inhibitors (EZH2is) have been clinically approved. Vorinostat was the first HDAC inhibitor approved by the FDA in 2006 for the treatment of cutaneous T-cell lymphoma (CTCL). Romidepsin is an HDAC inhibitor approved in 2009 for the treatment of CTCL and peripheral T-cell lymphoma. Belinostat is the third HDAC inhibitor approved for the treatment of peripheral T-cell lymphoma. Subsequently, panobinostat was approved for the treatment of multiple myeloma in 2015. In 2020, tazemetostat, an EZH2 inhibitor, was approved for the treatment of epithelioid sarcoma, making it the first approved histone “writer” inhibitor and the first used to treat solid tumors167. Most recently, the EZH1/2 dual inhibitor valemetostat was approved for the first time in Japan for the treatment of adult T-cell leukemia/lymphoma, for which there is no suitable therapeutic option168. The recent results of clinical trials of histone modification inhibitors alone or in combination with chemotherapy or immunotherapy for the treatment of drug-resistant cancer are summarized in Table 2.
Table 2.
Recent results of clinical trials using histone modification inhibitors alone or in combination with chemo- or immunotherapy for the treatment of drug-resistant cancers.
| Inhibitor | Target | Combination therapy | Cancer type | Phase | Endpoint outcome | Clinical trial ID |
|---|---|---|---|---|---|---|
| Vorinostat | HDACs | Hydroxychloroquine, Regorafenib | Chemotherapy-refractory metastatic colorectal cancer | II | Not achieved | NCT02316340 |
| NA | Recurrent/metastatic transitional cell carcinoma of the urothelium | II | Not achieved | NCT00363883 | ||
| Bortezomib, dexamethasone | Relapsed myeloma | II | Achieved | ISRCTN08577602 | ||
| Pembrolizumab | Recurrent/metastatic HNSCC and salivary gland cancer | II | Achieved | NCT02538510. | ||
| Bevacizumab | Recurrent glioblastoma | II | Not achieved | NCT01266031 | ||
| Pembrolizumab | Advanced/metastatic non-small-cell lung cancer | I/IIb | Achieved | NCT02638090 | ||
| Bortezomib | Mantle Cell Lymphoma | II | Achieved | NCT00703664 | ||
| Rituximab-CHOP | Advanced diffuse large B-cell lymphoma | I/II | Not achieved | NCT00972478 | ||
| Bevacizumab | Grade 4 malignant glioma | II | Not achieved | NCT01738646 | ||
| Romidepsin | Class I HDACs | 5-Azacytidine | Peripheral T-cell lymphoma | II | Achieved | NCT01998035 |
| Lenalidomide | Relapsed/refractory lymphoma | I | Achieved | NCT01755975, 02341014 | ||
| Alisertib | Relapsed/refractory aggressive B-cell and T-cell lymphoma | I | Achieved | NCT01897012 | ||
| Belinostat | HDACs | Followed by zevalin | Relapsed aggressive high-risk lymphoma | II | Not achieved | NCT01686165 |
| Bortezomib | Released/refractory acute leukemia, myelodysplastic syndrome | I | Not achieved | NCT01075425 | ||
| Cisplatin and etoposide | Small-cell lung cancer | I | Achieved | NCT00926640 | ||
| Panobinostat | HDACs | Everolimus | Relapsed/Refractory Diffuse Large B-Cell Lymphoma | II | Not achieved | NCT00978432 |
| Bortezomib and dexamethasone | Relapsed/Refractory Multiple Myeloma | II | Achieved | NCT02290431 | ||
| NA | Graft-versus-host disease | II | Achieved | NCT02588339 | ||
| NA | Multiple myeloma (a complete response failed after transplantation) | II | Achieved | ACTRN12613000219785 | ||
| Bortezomib and dexamethasone | Relapsed/refractory multiple myeloma | II | Achieved | NCT02654990 | ||
| Carfilzomib | Relapsed/refractory multiple myeloma | I/II | Achieved | NCT01496118 | ||
| Tazemetostat | EZH2 | NA | Relapsed/refractory, BAP1-inactivated malignant pleural mesothelioma | II | Not achieved | NCT02860286 |
| Atezolizumab | Relapsed/refractory diffuse large B-cell lymphoma | Ib | Achieved | NCT02220842 | ||
| NA | Relapsed/refractory B-cell non-Hodgkin lymphoma with EZH2 mutation | II | Achieved | NCT03456726 | ||
| NA | Relapsed/refractory follicular lymphoma | II | Achieved | NCT01897571 |
CHOP cyclophosphamide, doxorubicin, vincristine, and prednisone, HNSCC head and neck squamous cell carcinoma, NA not applicable.
Conclusion
Epigenetic alterations influence normal gene expression and consequently play a critical role in the onset and progression of diverse cancer types. CSCs are the main cause of cancer recurrence, metastasis, and treatment failure. The mechanisms that enable the characteristic drug resistance and immune evasion of CSCs are influenced by epigenetic regulation. These epigenetic modifications can be altered or reversed, thus ensuring new promising strategies in the therapeutic approach to CSCs. While advancements over the past several decades have yielded information to overcome drug-resistant cancer through the control of histone modification, several questions remain unanswered. Histone-modifying enzymes extensively remodel the TME to enable cancer cells to resist various anticancer therapies. Thus, the role of histone modification inhibitors should include not only blocking the intrinsic function of CSCs, which attenuates the antitumor response but also interfering with CSC crosstalk with immune cells to restore immune cell activity, ultimately converting aggressive CSCs into controllable “naïve” cells through complete reorganization of the epigenetic landscape. Despite the positive results of the combination of histone-modifying enzyme inhibitors for synergistic activity of tumor immune therapies, the diversity of genes targeted by these inhibitors makes it difficult to fully elucidate the mechanisms underlying complementary or synergistic actions with ICIs in the TME. In addition, approaches to cancer types in which the effects of histone modification inhibitors are relatively poor must be addressed. Because the combination effect with other anticancer drugs is maximized through the formation of suitable CSCs or immunological conditions via epigenetic priming by histone modification inhibitors, suitable biomarkers need to be identified to achieve a satisfactory combination effect. Nonetheless, modulation of histone modifications involved in CSC drug resistance and immune evasion will maximize clinical benefits through appropriate chemotherapeutic and immunotherapeutic combinatorial approaches, paving the way for personalized precision medicine.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2020R1A6A1A03043708, 2021R1A2C1011132, and 2020R1I1A1A01051942).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vasan N, et al. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 4.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Saleh R, Elkord E. Acquired resistance to cancer immunotherapy: Role of tumor-mediated immunosuppression. Semin. Cancer Biol. 2020;65:13–27. doi: 10.1016/j.semcancer.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Sasidharan Nair V, et al. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin. Epigenet. 2018;10:78. doi: 10.1186/s13148-018-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Oto A, et al. Histone demethylases and their roles in cancer epigenetics. J. Med. Oncol. Ther. 2016;1:34–40. [PMC free article] [PubMed] [Google Scholar]
- 8.Bayik D, Lathia JD. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer. 2021;21:526–536. doi: 10.1038/s41568-021-00366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Wang Y. Role of epigenetic regulation in plasticity of tumor immune microenvironment. Front. Immunol. 2021;12:640369. doi: 10.3389/fimmu.2021.640369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valkenburg KC, et al. Wnt/β-catenin signaling in normal and cancer stem cells. Cancers. 2011;3:2050–2079. doi: 10.3390/cancers3022050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheinbay E, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho S, et al. Notch1 regulates the expression of the multidrug resistance gene ABCC1/MRP1 in cultured cancer cells. Proc Natl Acad Sci USA. 2011;108:20778–20783. doi: 10.1073/pnas.1019452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res. 2014;74:6364–6374. doi: 10.1158/0008-5472.CAN-14-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, et al. STRAP promotes stemness of human colorectal cancer via epigenetic regulation of the NOTCH pathway. Cancer Res. 2017;77:5464–5478. doi: 10.1158/0008-5472.CAN-17-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghoshal P, et al. Loss of the SMRT/NCoR2 corepressor correlates with JAG2 overexpression in multiple myeloma. Cancer Res. 2009;69:4380–4387. doi: 10.1158/0008-5472.CAN-08-3467. [DOI] [PubMed] [Google Scholar]
- 17.Cochrane CR, et al. Hedgehog signaling in the maintenance of cancer stem cells. Cancers. 2015;7:1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canettieri G, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 19.To KK, et al. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol. Cancer Res. 2008;6:151–164. doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial–mesenchymal transition. Cells. 2020;9:217. doi: 10.3390/cells9010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan ZL, et al. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 22.Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am. J Cancer Res. 2015;5:1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 23.Spangle JM, et al. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:123–131. doi: 10.1016/j.bbcan.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 25.Toh TB, et al. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao P, et al. Histone demethylase PHF8 promotes epithelial to mesenchymal transition and breast tumorigenesis. Nucleic Acids Res. 2017;45:1687–1702. doi: 10.1093/nar/gkw1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Gamero, C. et al. LSD1: expanding functions in stem cells and differentiation. Cells10, 3252 (2021). [DOI] [PMC free article] [PubMed]
- 29.Miller SA, et al. Lysine-specific demethylase 1 mediates AKT activity and promotes epithelial-to-mesenchymal transition in PIK3CA-mutant colorectal cancer. Mol. Cancer Res. 2020;18:264–277. doi: 10.1158/1541-7786.MCR-19-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang HS, et al. TSPAN8 promotes colorectal cancer cell growth and migration in LSD1-dependent manner. Life Sci. 2020;241:117114. doi: 10.1016/j.lfs.2019.117114. [DOI] [PubMed] [Google Scholar]
- 31.Hsu JM, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meder L, et al. Combined VEGF and PD-L1 blockade displays synergistic treatment effects in an autochthonous mouse model of small cell lung cancer. Cancer Res. 2018;78:4270–4281. doi: 10.1158/0008-5472.CAN-17-2176. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Advancing to the era of cancer immunotherapy. Cancer Commun. (Lond) 2021;41:803–829. doi: 10.1002/cac2.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spranger S, et al. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31:711–723.e714. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuchiya H, Shiota G. Immune evasion by cancer stem cells. Regen. Ther. 2021;17:20–33. doi: 10.1016/j.reth.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krempski J, et al. Tumor-infiltrating programmed death receptor-1+ dendritic cells mediate immune suppression in ovarian cancer. J. Immunol. 2011;186:6905–6913. doi: 10.4049/jimmunol.1100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki BT, Farrar WL. Cancer stem cells, CD200 and immunoevasion. Trends Immunol. 2008;29:464–468. doi: 10.1016/j.it.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Tan Y, et al. Tumor-associated macrophages: a potential target for cancer therapy. Front. Oncol. 2021;11:693517. doi: 10.3389/fonc.2021.693517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dianat-Moghadam H, et al. Immune evader cancer stem cells direct the perspective approaches to cancer immunotherapy. Stem Cell Res. Ther. 2022;13:150. doi: 10.1186/s13287-022-02829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinushi M, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl Acad. Sci. USA. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan QM, et al. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014;352:160–168. doi: 10.1016/j.canlet.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Lei MML, Lee TKW. Cancer stem cells: emerging key players in immune evasion of cancers. Front. Cell Dev. Biol. 2021;9:692940. doi: 10.3389/fcell.2021.692940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veglia F, et al. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021;21:485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welte T, et al. Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation. Nat. Cell Biol. 2016;18:632–644. doi: 10.1038/ncb3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ai L, et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer. 2019;18:88. doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee CR, et al. Myeloid-derived suppressor cells are controlled by regulatory T cells via TGF-beta during murine colitis. Cell Rep. 2016;17:3219–3232. doi: 10.1016/j.celrep.2016.11.062. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, et al. Regulatory T cells promote glioma cell stemness through TGF-beta-NF-kappaB-IL6-STAT3 signaling. Cancer Immunol. Immunother. 2021;70:2601–2616. doi: 10.1007/s00262-021-02872-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, et al. Sox2 communicates with Tregs through CCL1 to promote the stemness property of breast cancer cells. Stem Cells. 2017;35:2351–2365. doi: 10.1002/stem.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezalotfi A, et al. Gastric cancer stem cells effect on Th17/Treg balance; a bench to beside perspective. Front. Oncol. 2019;9:226. doi: 10.3389/fonc.2019.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh, E. et al. Regulatory T cells induce metastasis by increasing Tgf-beta and enhancing the epithelial-mesenchymal transition. Cells8, 1387 (2019). [DOI] [PMC free article] [PubMed]
- 51.Beck B, et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- 52.Castriconi R, et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009;182:3530–3539. doi: 10.4049/jimmunol.0802845. [DOI] [PubMed] [Google Scholar]
- 53.Tallerico R, et al. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 2013;190:2381–2390. doi: 10.4049/jimmunol.1201542. [DOI] [PubMed] [Google Scholar]
- 54.Jewett A, et al. Natural killer cells preferentially target cancer stem cells; role of monocytes in protection against NK cell mediated lysis of cancer stem cells. Curr. Drug Deliv. 2012;9:5–16. doi: 10.2174/156720112798375989. [DOI] [PubMed] [Google Scholar]
- 55.Akhter MZ, et al. Aggressive serous epithelial ovarian cancer is potentially propagated by EpCAM(+)CD45(+) phenotype. Oncogene. 2018;37:2089–2103. doi: 10.1038/s41388-017-0106-y. [DOI] [PubMed] [Google Scholar]
- 56.Zhong Y, et al. Spheres derived from the human SK-RC-42 renal cell carcinoma cell line are enriched in cancer stem cells. Cancer Lett. 2010;299:150–160. doi: 10.1016/j.canlet.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 57.Malladi S, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sainz B, Jr., et al. ISG15 is a critical microenvironmental factor for pancreatic cancer stem cells. Cancer Res. 2014;74:7309–7320. doi: 10.1158/0008-5472.CAN-14-1354. [DOI] [PubMed] [Google Scholar]
- 59.Martin-Hijano L, Sainz B., Jr. The interactions between cancer stem cells and the innate interferon signaling pathway. Front. Immunol. 2020;11:526. doi: 10.3389/fimmu.2020.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Y, et al. Influence of interferon-alpha on the expression of the cancer stem cell markers in pancreatic carcinoma cells. Exp. Cell Res. 2014;324:146–156. doi: 10.1016/j.yexcr.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 61.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Musella M, et al. Type I IFNs promote cancer cell stemness by triggering the epigenetic regulator KDM1B. Nat. Immunol. 2022;23:1379–1392. doi: 10.1038/s41590-022-01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song M, et al. Low-dose IFNgamma induces tumor cell stemness in tumor microenvironment of non-small cell lung cancer. Cancer Res. 2019;79:3737–3748. doi: 10.1158/0008-5472.CAN-19-0596. [DOI] [PubMed] [Google Scholar]
- 65.Yang, L. et al. Histone H3K4 methyltransferases as targets for drug-resistant cancers. Biology10, 581 (2021). [DOI] [PMC free article] [PubMed]
- 66.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu. Rev. Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 68.Zeisig BB, et al. Reconstructing the disease model and epigenetic networks for MLL-AF4 leukemia. Cancer Cell. 2008;14:345–347. doi: 10.1016/j.ccr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Fiskus, W. et al. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J.12, 5 (2022). [DOI] [PMC free article] [PubMed]
- 70.Malik R, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grinat J, et al. The epigenetic regulator Mll1 is required for Wnt-driven intestinal tumorigenesis and cancer stemness. Nat. Commun. 2020;11:6422. doi: 10.1038/s41467-020-20222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen R, et al. Kmt2c mutations enhance HSC self-renewal capacity and convey a selective advantage after chemotherapy. Cell Rep. 2021;34:108751. doi: 10.1016/j.celrep.2021.108751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, et al. Histone methyltransferase KMT2D cooperates with MEF2A to promote the stem-like properties of oral squamous cell carcinoma. Cell Biosci. 2022;12:49. doi: 10.1186/s13578-022-00785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X, et al. MLL5 (KMT2E): structure, function, and clinical relevance. Cell. Mol. Life Sci. 2017;74:2333–2344. doi: 10.1007/s00018-017-2470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin ML, et al. SETD1A-SOX2 axis is involved in tamoxifen resistance in estrogen receptor α-positive breast cancer cells. Theranostics. 2022;12:5761–5775. doi: 10.7150/thno.72599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, L. et al. SETD1A promotes proliferation of castration-resistant prostate cancer cells via FOXM1 transcription. Cancers (Basel)12, 1736 (2020). [DOI] [PMC free article] [PubMed]
- 77.Wang R, et al. An SETD1A/Wnt/β-catenin feedback loop promotes NSCLC development. J. Exp. Clin. Cancer Res. 2021;40:318. doi: 10.1186/s13046-021-02119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 79.Hou Z, et al. Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. Cancer Lett. 2020;487:63–73. doi: 10.1016/j.canlet.2020.05.029. [DOI] [PubMed] [Google Scholar]
- 80.Bergin CJ, et al. G9a controls pluripotent-like identity and tumor-initiating function in human colorectal cancer. Oncogene. 2021;40:1191–1202. doi: 10.1038/s41388-020-01591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu S, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6:6887–6901. doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L, et al. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. 2018;9:129. doi: 10.1038/s41419-017-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciechomska IA, et al. BIX01294, an inhibitor of histone methyltransferase, induces autophagy-dependent differentiation of glioma stem-like cells. Sci. Rep. 2016;6:38723. doi: 10.1038/srep38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu H, et al. Combined tazemetostat and MAPKi enhances differentiation of papillary thyroid cancer cells harbouring BRAF(V600E) by synergistically decreasing global trimethylation of H3K27. J. Cell. Mol. Med. 2020;24:3336–3345. doi: 10.1111/jcmm.15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adelaiye-Ogala R, et al. EZH2 modifies sunitinib resistance in renal cell carcinoma by kinome reprogramming. Cancer Res. 2017;77:6651–6666. doi: 10.1158/0008-5472.CAN-17-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang L, et al. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget. 2014;5:10665–10677. doi: 10.18632/oncotarget.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nassa G, et al. Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells. Sci. Adv. 2019;5:eaav5590. doi: 10.1126/sciadv.aav5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kurani H, et al. DOT1L is a novel cancer stem cell target for triple-negative breast cancer. Clin. Cancer Res. 2022;28:1948–1965. doi: 10.1158/1078-0432.CCR-21-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hwang JW, et al. Protein arginine methyltransferases: promising targets for cancer therapy. Exp. Mol. Med. 2021;53:788–808. doi: 10.1038/s12276-021-00613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao Y, et al. The dual function of PRMT1 in modulating epithelial-mesenchymal transition and cellular senescence in breast cancer cells through regulation of ZEB1. Sci. Rep. 2016;6:19874. doi: 10.1038/srep19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Katsuno Y, et al. Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J. Biol. Chem. 2018;293:13059–13072. doi: 10.1074/jbc.RA118.002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiang K, et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21:3498–3513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng BN, et al. Targeting PRMT5 activity inhibits the malignancy of hepatocellular carcinoma by promoting the transcription of HNF4α. Theranostics. 2019;9:2606–2617. doi: 10.7150/thno.32344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, et al. Arginine methyltransferase PRMT5 methylates and stabilizes KLF5 via decreasing its phosphorylation and ubiquitination to promote basal-like breast cancer. Cell Death Differ. 2021;28:2931–2945. doi: 10.1038/s41418-021-00793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sachamitr P, et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021;12:979. doi: 10.1038/s41467-021-21204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Almeida-Rios D, et al. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget. 2016;7:53018–53028. doi: 10.18632/oncotarget.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakakido M, et al. PRMT6 increases cytoplasmic localization of p21CDKN1A in cancer cells through arginine methylation and makes more resistant to cytotoxic agents. Oncotarget. 2015;6:30957–30967. doi: 10.18632/oncotarget.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang T, et al. PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol. Cell. 2021;81:1276–1291 e1279. doi: 10.1016/j.molcel.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.AlAbdi L, et al. Oct4-mediated inhibition of Lsd1 activity promotes the active and primed state of pluripotency enhancers. Cell Rep. 2020;30:1478–1490.e1476. doi: 10.1016/j.celrep.2019.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu C, et al. LSD1 stimulates cancer-associated fibroblasts to drive Notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 2018;78:938–949. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 102.Yin F, et al. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol. Cell. Biol. 2014;34:158–179. doi: 10.1128/MCB.00631-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Verigos, J. et al. The histone demethylase LSD1/ΚDM1A mediates chemoresistance in breast cancer via regulation of a stem cell program. Cancers (Basel)11, 1585 (2019). [DOI] [PMC free article] [PubMed]
- 104.Boulding T, et al. LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer. Sci. Rep. 2018;8:73. doi: 10.1038/s41598-017-17913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cuyàs E, et al. The LSD1 inhibitor iadademstat (ORY-1001) targets SOX2-driven breast cancer stem cells: a potential epigenetic therapy in luminal-B and HER2-positive breast cancer subtypes. Aging. 2020;12:4794–4814. doi: 10.18632/aging.102887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao LJ, et al. Lysine demethylase LSD1 delivered via small extracellular vesicles promotes gastric cancer cell stemness. EMBO Rep. 2021;22:e50922. doi: 10.15252/embr.202050922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramadoss S, et al. Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene. 2017;36:6508. doi: 10.1038/onc.2017.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.MacPherson L, et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature. 2020;577:266–270. doi: 10.1038/s41586-019-1835-6. [DOI] [PubMed] [Google Scholar]
- 109.Di Martile M, et al. Histone acetyltransferase inhibitor CPTH6 preferentially targets lung cancer stem-like cells. Oncotarget. 2016;7:11332–11348. doi: 10.18632/oncotarget.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baell JB, et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 2018;560:253–257. doi: 10.1038/s41586-018-0387-5. [DOI] [PubMed] [Google Scholar]
- 111.Yu B, et al. KAT6A acetylation of SMAD3 regulates myeloid-derived suppressor cell recruitment, metastasis, and immunotherapy in triple-negative breast cancer. Adv. Sci. 2021;8:e2100014. doi: 10.1002/advs.202100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu W, et al. KAT6A, a novel regulator of beta-catenin, promotes tumorigenicity and chemoresistance in ovarian cancer by acetylating COP1. Theranostics. 2021;11:6278–6292. doi: 10.7150/thno.57455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simonsson M, et al. The balance between acetylation and deacetylation controls Smad7 stability. J. Biol. Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 114.Chiu CC, et al. Grp78 as a therapeutic target for refractory head-neck cancer with CD24(-)CD44(+) stemness phenotype. Cancer Gene Ther. 2013;20:606–615. doi: 10.1038/cgt.2013.64. [DOI] [PubMed] [Google Scholar]
- 115.Witt AE, et al. Identification of a cancer stem cell-specific function for the histone deacetylases, HDAC1 and HDAC7, in breast and ovarian cancer. Oncogene. 2017;36:1707–1720. doi: 10.1038/onc.2016.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu MY, et al. MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer Lett. 2014;354:311–319. doi: 10.1016/j.canlet.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 117.Yano M, et al. Association of histone deacetylase expression with histology and prognosis of ovarian cancer. Oncol. Lett. 2018;15:3524–3531. doi: 10.3892/ol.2018.7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta M, et al. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012;26:1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bora-Singhal N, et al. Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci. Rep. 2020;10:4722. doi: 10.1038/s41598-020-61295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen X, et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci. Rep. 2014;4:7481. doi: 10.1038/srep07481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim. Biophys. Acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kumar B, et al. Suberoylanilide hydroxamic acid (SAHA) reverses chemoresistance in head and neck cancer cells by targeting cancer stem cells via the downregulation of nanog. Genes Cancer. 2015;6:169–181. doi: 10.18632/genesandcancer.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nalls D, et al. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen PL, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 128.Haebe JR, et al. Emerging role of G9a in cancer stemness and promises as a therapeutic target. Oncogenesis. 2021;10:76. doi: 10.1038/s41389-021-00370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee S, et al. Network inference analysis identifies SETDB1 as a key regulator for reverting colorectal cancer cells into differentiated normal-like cells. Mol. Cancer Res. 2020;18:118–129. doi: 10.1158/1541-7786.MCR-19-0450. [DOI] [PubMed] [Google Scholar]
- 130.Segovia C, et al. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat. Med. 2019;25:1073–1081. doi: 10.1038/s41591-019-0499-y. [DOI] [PubMed] [Google Scholar]
- 131.Seier, J. A. et al. Druggable epigenetic suppression of interferon-induced chemokine expression linked to MYCN amplification in neuroblastoma. J. Immunother. Cancer9, e001335 (2021). [DOI] [PMC free article] [PubMed]
- 132.Ishiguro K, et al. Dual EZH2 and G9a inhibition suppresses multiple myeloma cell proliferation by regulating the interferon signal and IRF4-MYC axis. Cell Death Discov. 2021;7:7. doi: 10.1038/s41420-020-00400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griffin GK, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–314. doi: 10.1038/s41586-021-03520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang SM, et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature. 2021;598:682–687. doi: 10.1038/s41586-021-03994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zingg D, et al. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 2017;20:854–867. doi: 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 136.Wang D, et al. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 2018;23:3262–3274. doi: 10.1016/j.celrep.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Peng D, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiao G, et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer. 2019;7:300. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen S, et al. EZH2 promotes hepatocellular carcinoma progression through modulating miR-22/galectin-9 axis. J. Exp. Clin. Cancer Res. 2018;37:3. doi: 10.1186/s13046-017-0670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goswami S, et al. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 2018;128:3813–3818. doi: 10.1172/JCI99760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zheng NN, et al. Combining protein arginine methyltransferase inhibitor and anti-programmed death-ligand-1 inhibits pancreatic cancer progression. World J. Gastroenterol. 2020;26:3737–3749. doi: 10.3748/wjg.v26.i26.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hu R, et al. PRMT5 inhibition promotes PD-L1 expression and immuno-resistance in lung cancer. Front. Immunol. 2021;12:722188. doi: 10.3389/fimmu.2021.722188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Verigos, J. et al. The histone demethylase LSD1/KappaDM1A mediates chemoresistance in breast cancer via regulation of a stem cell program. Cancers11, 1585 (2019). [DOI] [PMC free article] [PubMed]
- 144.Sheng W, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;174:549–563.e519. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qin Y, et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene. 2019;38:390–405. doi: 10.1038/s41388-018-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang M, et al. Targeting KDM1A attenuates Wnt/beta-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett. 2017;398:12–21. doi: 10.1016/j.canlet.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 147.Sheng W, et al. Simultaneous inhibition of LSD1 and TGFbeta enables eradication of poorly immunogenic tumors with anti-PD-1 treatment. Cancer Discov. 2021;11:1970–1981. doi: 10.1158/2159-8290.CD-20-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu S, et al. LSD1 silencing contributes to enhanced efficacy of anti-CD47/PD-L1 immunotherapy in cervical cancer. Cell Death Dis. 2021;12:282. doi: 10.1038/s41419-021-03556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Han Y, et al. Targeting LSD1 suppresses stem cell-like properties and sensitizes head and neck squamous cell carcinoma to PD-1 blockade. Cell Death Dis. 2021;12:993. doi: 10.1038/s41419-021-04297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hogg SJ, et al. Targeting the epigenetic regulation of antitumour immunity. Nat. Rev. Drug Discov. 2020;19:776–800. doi: 10.1038/s41573-020-0077-5. [DOI] [PubMed] [Google Scholar]
- 151.Khan AN, et al. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol. Immunother. 2008;57:647–654. doi: 10.1007/s00262-007-0402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zheng H, et al. HDAC inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2016;22:4119–4132. doi: 10.1158/1078-0432.CCR-15-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Woods DM, et al. The antimelanoma activity of the histone deacetylase inhibitor panobinostat (LBH589) is mediated by direct tumor cytotoxicity and increased tumor immunogenicity. Melanoma Res. 2013;23:341–348. doi: 10.1097/CMR.0b013e328364c0ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Woods DM, et al. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol. Res. 2015;3:1375–1385. doi: 10.1158/2326-6066.CIR-15-0077-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Oki Y, et al. Immune regulatory effects of panobinostat in patients with Hodgkin lymphoma through modulation of serum cytokine levels and T-cell PD1 expression. Blood Cancer J. 2014;4:e236. doi: 10.1038/bcj.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Terranova-Barberio M, et al. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. 2017;8:114156–114172. doi: 10.18632/oncotarget.23169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Briere D, et al. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol. Immunother. 2018;67:381–392. doi: 10.1007/s00262-017-2091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Topper MJ, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell. 2017;171:1284–1300.e1221. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim K, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc. Natl Acad. Sci. USA. 2014;111:11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Orillion A, et al. Entinostat neutralizes myeloid-derived suppressor cells and enhances the antitumor effect of PD-1 inhibition in murine models of lung and renal cell carcinoma. Clin Cancer Res. 2017;23:5187–5201. doi: 10.1158/1078-0432.CCR-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Truong, A. S. et al. Entinostat induces antitumor immune responses through immune editing of tumor neoantigens. J. Clin. Invest.131, e138560 (2021). [DOI] [PMC free article] [PubMed]
- 162.Knox T, et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci. Rep. 2019;9:6136. doi: 10.1038/s41598-019-42237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bae J, et al. Histone deacetylase (HDAC) inhibitor ACY241 enhances anti-tumor activities of antigen-specific central memory cytotoxic T lymphocytes against multiple myeloma and solid tumors. Leukemia. 2018;32:1932–1947. doi: 10.1038/s41375-018-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wong DJ, et al. Exposure to a histone deacetylase inhibitor has detrimental effects on human lymphocyte viability and function. Cancer Immunol. Res. 2014;2:459–468. doi: 10.1158/2326-6066.CIR-13-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kroesen M, et al. HDAC inhibitors and immunotherapy; a double edged sword? Oncotarget. 2014;5:6558–6572. doi: 10.18632/oncotarget.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fan P, et al. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J. Exp. Clin. Cancer Res. 2019;38:47. doi: 10.1186/s13046-019-1044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gounder M, et al. Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet. Oncol. 2020;21:1423–1432. doi: 10.1016/S1470-2045(20)30451-4. [DOI] [PubMed] [Google Scholar]
- 168.Izutsu K. An open-label, single-arm, phase 2 trial of valemetostat in relapsed or refractory adult T-cell leukemia/lymphoma. Blood. 2022;141:1159–1168. doi: 10.1182/blood.2022016862. [DOI] [PubMed] [Google Scholar]
- 169.Xue K, et al. Vorinostat, a histone deacetylase (HDAC) inhibitor, promotes cell cycle arrest and re-sensitizes rituximab- and chemo-resistant lymphoma cells to chemotherapy agents. J. Cancer Res. Clin. Oncol. 2016;142:379–387. doi: 10.1007/s00432-015-2026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nakajima W, et al. Combination with vorinostat overcomes ABT-263 (navitoclax) resistance of small cell lung cancer. Cancer Biol. Ther. 2016;17:27–35. doi: 10.1080/15384047.2015.1108485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Booth L, et al. Prior exposure of pancreatic tumors to [sorafenib + vorinostat] enhances the efficacy of an anti-PD-1 antibody. Cancer Biol. Ther. 2019;20:109–121. doi: 10.1080/15384047.2018.1507258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Piro G, et al. Vorinostat potentiates 5-fluorouracil/cisplatin combination by inhibiting chemotherapy-induced EGFR nuclear translocation and increasing cisplatin uptake. Mol. Cancer Ther. 2019;18:1405–1417. doi: 10.1158/1535-7163.MCT-18-1117. [DOI] [PubMed] [Google Scholar]
- 173.Lin CY, et al. Vorinostat combined with brigatinib overcomes acquired resistance in EGFR-C797S-mutated lung cancer. Cancer Lett. 2021;508:76–91. doi: 10.1016/j.canlet.2021.03.022. [DOI] [PubMed] [Google Scholar]
- 174.Park SE, et al. Combination treatment with docetaxel and histone deacetylase inhibitors downregulates androgen receptor signaling in castration-resistant prostate cancer. Invest. New Drugs. 2018;36:195–205. doi: 10.1007/s10637-017-0529-x. [DOI] [PubMed] [Google Scholar]
- 175.To KK, et al. Reversal of platinum drug resistance by the histone deacetylase inhibitor belinostat. Lung Cancer. 2017;103:58–65. doi: 10.1016/j.lungcan.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 176.Llopiz D, et al. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol. Immunother. 2019;68:379–393. doi: 10.1007/s00262-018-2283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Savickiene J, et al. Epigenetic and molecular mechanisms underlying the antileukemic activity of the histone deacetylase inhibitor belinostat in human acute promyelocytic leukemia cells. Anticancer Drugs. 2014;25:938–949. doi: 10.1097/CAD.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 178.Mondello P, et al. Panobinostat acts synergistically with ibrutinib in diffuse large B cell lymphoma cells with MyD88 L265P mutations. JCI Insight. 2018;3:e125568. doi: 10.1172/jci.insight.125568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jiang XJ, et al. Synergistic effect of panobinostat and bortezomib on chemoresistant acute myelogenous leukemia cells via AKT and NF-kappaB pathways. Cancer Lett. 2012;326:135–142. doi: 10.1016/j.canlet.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 180.Tate CR, et al. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 2012;14:R79. doi: 10.1186/bcr3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shahbazi J, et al. The bromodomain inhibitor JQ1 and the histone deacetylase inhibitor panobinostat synergistically reduce N-Myc expression and induce anticancer effects. Clin. Cancer Res. 2016;22:2534–2544. doi: 10.1158/1078-0432.CCR-15-1666. [DOI] [PubMed] [Google Scholar]
- 182.Medon M, et al. HDAC inhibitor panobinostat engages host innate immune defenses to promote the tumoricidal effects of trastuzumab in HER2(+) tumors. Cancer Res. 2017;77:2594–2606. doi: 10.1158/0008-5472.CAN-16-2247. [DOI] [PubMed] [Google Scholar]
- 183.Lee WY, et al. Panobinostat sensitizes KRAS-mutant non-small-cell lung cancer to gefitinib by targeting TAZ. Int. J. Cancer. 2017;141:1921–1931. doi: 10.1002/ijc.30888. [DOI] [PubMed] [Google Scholar]
- 184.Ferrari AC, et al. Epigenetic therapy with panobinostat combined with bicalutamide rechallenge in castration-resistant prostate cancer. Clin. Cancer Res. 2019;25:52–63. doi: 10.1158/1078-0432.CCR-18-1589. [DOI] [PubMed] [Google Scholar]
- 185.Zang H, et al. Overcoming acquired resistance of epidermal growth factor receptor-mutant non-small cell lung cancer cells to osimertinib by combining osimertinib with the histone deacetylase inhibitor panobinostat (LBH589) Cancer. 2020;126:2024–2033. doi: 10.1002/cncr.32744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wanitpongpun C, et al. Tamoxifen enhances romidepsin-induced apoptosis in T-cell malignant cells via activation of FOXO1 signaling pathway. Leukemia Lymphoma. 2021;62:1585–1596. doi: 10.1080/10428194.2021.1876857. [DOI] [PubMed] [Google Scholar]
- 187.Cooper SJ, et al. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol. Cancer Ther. 2012;11:2105–2115. doi: 10.1158/1535-7163.MCT-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]