Abstract
Individuals infected with human T-cell lymphotropic virus type 1 (HTLV-1) develop a robust immune response to the surface envelope glycoprotein gp46 that is partially protective. The relative contribution of antibodies to conformation-dependent epitopes, including those mediating virus neutralization as part of the humoral immune response, is not well defined. We assess in this report the relationship between defined linear and conformational epitopes and the antibodies elicited to these domains. First, five monoclonal antibodies to linear epitopes within gp46 were evaluated for their ability to abrogate binding of three human monoclonal antibodies that inhibit HTLV-1-mediated syncytia formation and recognize conformational epitopes. Binding of antibodies to conformational epitopes was unaffected by antibodies to linear epitopes throughout the carboxy-terminal half and central domain of HTLV-1 gp46. Second, an enzyme-linked immunoadsorbent assay was developed and used to measure serum antibodies to native and denatured gp46 from HTLV-1-infected individuals. In sera from infected individuals, reactivity to denatured gp46 had an average of 15% of the reactivity observed to native gp46. Third, serum antibodies from 24 of 25 of HTLV-1-infected individuals inhibited binding of a neutralizing human monoclonal antibody, PRH-7A, to a conformational epitope on gp46 that is common to HTLV-1 and -2. Thus, antibodies to conformational epitopes comprise the majority of the immune response to HTLV-1 gp46, and the epitopes recognized by these antibodies do not appear to involve sequences in previously described immunodominant linear epitopes.
Infection with human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 is a growing medical problem worldwide, with over 20 million estimated infections worldwide (reviewed in reference 6). HTLV-1 is the etiologic agent of adult T-cell leukemia and a progressive neurological disease known as tropical spastic paraparesis or HTLV-1-associated myelopathy (TSP/HAM, reviewed in reference 6). HTLV-2, a closely related retrovirus, was originally isolated from a patient with atypical hairy cell leukemia (16) but has been associated recently with a progressive neuropathy similar to TSP/HAM (13, 14, 32). Although a robust immune response is elicited during infection, infection usually persists. Nonetheless, passive immunization studies with HTLV-1 human immune sera in appropriate animal models demonstrated that specific antibody therapy with virus-neutralizing activity could be protective if administered within 24 h of infection (1, 21, 22). Similarly, passive immunization with HTLV-2 human immune sera protected susceptible rabbits from blood-borne HTLV-2 infection (25). Thus, an effective vaccine for HTLV-1 or HTLV-2 should induce the antibody response that mediates virus neutralization as observed in naturally infected individuals.
Analysis of the humoral immune response to HTLV-1 demonstrated that the surface envelope glycoprotein, gp46, is the primary target of neutralizing antibodies (6). Most studies have focused on antibodies to linear epitopes located on the carboxy-terminal half of gp46 (amino acids 170 to 312 [3, 4, 5, 7, 8, 10, 15, 17, 19, 27rsqb;). These antibodies are found in more than 95% of infected individuals (reviewed in references 11 and 18), but the majority of antibodies to these epitopes do not mediate virus neutralization (4, 7, 10). Linear epitopes located in the middle region of the envelope (amino acids 175 to 199), as defined by monoclonal antibodies, are more likely to have neutralizing activity (4). Much less information is available about the role of antibodies to conformation-dependent epitopes on HTLV-1 gp46 in the mediation of virus neutralization. We recently reported on the production and initial characterization of 10 human monoclonal antibodies (HMAbs) to HTLV-1 gp46 (12). Seven of these antibodies recognized conformational epitopes within HTLV-1 gp46, and all seven of these antibodies exhibited varying levels of virus neutralization activity. Competition analysis indicated that these seven HMAbs are directed at four distinct conformational epitopes within HTLV-1 gp46. Two of these HMAbs, PRH-7A and PRH-7B, recognized an epitope common to both HTLV-1 and HTLV-2 gp46 (12). Studies performed with a vaccinia virus construct expressing HTLV-1 gp46 suggested that three of the HMAbs, PRH-7A, PRH-7B, and PRH-11A, could bind to nonglycosylated gp46 produced in cells treated with tunicamycin (2). It is therefore likely that these antibodies do not bind to the carbohydrate moieties directly; little else is known about the locations of conformational epitopes within HTLV-1 gp46.
To better define the role of antibodies to conformational epitopes during natural infection with HTLV-1, studies were performed to measure the overall contribution of antibodies to conformation-dependent epitopes and to a specific conformational epitope as defined by a selected HMAb in sera from HTLV-1-infected individuals. Antibody competition analysis was used to evaluate whether the binding of antibodies to known linear epitopes throughout the carboxy-terminal and central domain of HTLV-1 gp46 inhibits the binding of HMAbs to conformational epitopes. A murine monoclonal antibody (mMAb) that did not affect the binding of HMAbs to conformational epitopes and exhibited equivalent reactivity to native and denatured HTLV-1 gp46 was employed in an enzyme-linked immunoadsorbent assay (ELISA) to quantify the reactivity of individual sera to native and denatured HTLV-1 gp46. We also used competition analysis to measure the relative abundance of PRH-7A-like antibodies to assess the magnitude of antibody response to a specific conformational epitope that mediates virus neutralization and is common to HTLV-1 and HTLV-2. The results of these analyses indicate that antibodies to conformational epitopes predominate in asymptomatic HTLV-1-infected individuals and that a substantial portion of this response is directed at the PRH-7A epitope. The implications of these results are discussed.
MATERIALS AND METHODS
Cell lines and antibodies.
HTLV-1-infected cell lines, HUT-102 and MT-2, and an uninfected T-cell line, RPMI-8402, were propagated as described previously (12). HMAbs antibodies PRH-1, PRH-7A, PRH-11A, PRH-4, and PRH-8 were produced, purified, and biotinylated also as described previously (12). Human hybridoma 0.5 alpha (20) was obtained from the American Type Culture Collection, and secreted antibody was purified as described previously (12). The mMAbs CLONE 65/6C2.2.34 (6C2), CLONE 67/5.5.13.1 (CLN 67), and CLONE 68/4.11.21 (CLN 68) were obtained from Cellular Products (Buffalo, N.Y.).
Antisera.
The 29 HTLV-1-infected individuals whose sera were used in these studies were identified by the testing of samples sent to the Stanford Medical School Blood Center from 1988 to 1993 for HTLV-1 and -2 confirmatory testing. The panel included 24 samples from blood donors that presented at several California blood banks and 5 reference laboratory samples. Blood donation samples were from healthy individuals who would be expected to be asymptomatic for either adult T-cell leukemia or TSP/HAM. Information about the health status of the other five individuals was not available. All sera met standard serological criteria for HTLV-1 infection, including reactivity to p24 gag protein and to recombinant envelope proteins, p21E and MTA-1 (11, 18). Eighteen of these serum samples were from individuals confirmed as HTLV-1 infected by PCR with HTLV-1-specific primers and probes by published procedures (19). For those individuals for whom detailed epidemiological information was available (n = 11 of the potential blood donors), 5 were male and 6 were female; the subjects ranged in age from 28 to 57. Nine of the 11 individuals had an identifiable risk factor for HTLV-1 infection; either they lived in a region where HTLV-1 was endemic or they had a sexual relationship with an individual at increased risk for HTLV-1 infection. Three of the 11 had an antibody response to the hepatitis B virus (HBV) core antigen, but not to the HBV surface antigen. No other antibody or antigen reactivity to other transfusion-transmitted viruses was observed. Control sera were from normal blood donations that were negative for HTLV-1 and -2, human immunodeficiency virus types 1 and 2 (HIV-1 and -2), HBV, and hepatitis C virus by standard screening assays.
Preparation of HTLV-1 and control cell extracts.
MT-2, HUT-102, or RPMI-8402 cells were washed with phosphate-buffered saline (PBS) and resuspended in lysis buffer (150 mM NaCl, 20 mM Tris [pH 7.5], 0.5% deoxycholate, 1.0% Nonidet P-40, 1 mM EDTA). The protease inhibitors, pefabloc (Boehringer Mannheim, Indianapolis, Ind.), aprotinin, leupeptin, and pepstatin, were added to final concentrations of 0.5 mg/ml and 2, 2, and 1 μg/ml, respectively. Fifty microliters of lysis buffer was then added for every 106 cells harvested. Nuclei were pelleted by centrifugation at 18,000 × g at 4°C for 10 min, and the supernatant was either used directly or stored at 4°C for not more than 2 days prior to use. The extracts were diluted 1/5 with BLOTTO (2.5% nonfat dry milk, 2.5% normal goat serum, 0.1% Tween 20 [Sigma, St. Louis, Mo.], 0.02% sodium azide in Tris-buffered saline [TBS]: 150 mM NaCl, 20 mM Tris [pH 7.5]) and added to antibody-coated plates. Denatured extracts were prepared by adding in sodium dodecyl sulfate (SDS) to a final concentration of 0.5%, followed by incubation for 15 min in a 56°C water bath. After heat treatment, the extract was diluted 1/5 in BLOTTO the same way the untreated extracts were. Control experiments indicated that an SDS concentration of 0.1% did not significantly affect the binding of HMAbs recognizing conformational epitopes.
Quantitation of antibody reactivity.
ELISA analysis of antibody reactivity was performed by methods similar to those previously described (23, 24). Briefly, 100 μl of PBS containing 1 μg of the indicated antibody (usually mMAb 6C2) per ml was added to individual wells in 96-well microtiter plates and incubated for 60 min at 37°C. Wells were aspirated, washed once with TBS, and blocked by the addition of 150 μl of BLOTTO at room temperature (RT) for 1 h. The wells were washed once with TBS, and 100 μl of BLOTTO mixed with extracts containing native or heat-denatured env proteins (prepared as described above) was added to each well. After 1 h at RT, the wells were washed three times with TBS, and 100 μl of BLOTTO containing dilutions of HTLV-1 or control sera was added to individual wells and allowed to incubate with bound env protein for 1.5 h at RT. The wells were washed three times with TBS. One hundred microliters of BLOTTO containing 10 ml of a murine immunoglobulin G1 (IgG1) monoclonal antibody (MOPC-21; Sigma) per ml plus either goat anti-human conjugated horseradish peroxidase (HRP; Kirkegaard and Perry, South San Francisco, Calif.) or goat anti-human alkaline phosphatase (Promega, Madison, Wis.) were added. Both secondary antibodies were used at a dilution of 1/5,000. The murine antibody was added to reduce cross-reactivity of the anti-human second antibody with the murine IgG1 antibody (6C2) used to capture env protein. After 1 h at RT, the wells were washed four times with TBS. Bound antibodies were detected by incubation with 100 μl of either a 0.5-mg/ml solution of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) with 0.1% hydrogen peroxide (for HRP) or a 1 mg/ml solution of p-nitrophenyl phosphate (PNPP). Substrate development was allowed to proceed for 15 to 30 min, and the adsorbance of the wells was read at 405 nm with a multiwell plate reader (Du Pont Co., Wilmington, Del.). Results obtained from duplicate wells were averaged.
Antibody competition analysis was performed as described previously (12). Briefly, HMAb PRH-1 was used as the capture antibody, and biotinylated HMAbs PRH-7A, PRH-11A, and PRH-4 were used as detection reagents. Bound biotinylated antibody (Bio HMAb) was detected with alkaline phosphatase-conjugated streptavidin (Amersham, Arlington Heights, Ill.), followed by the addition of 100 μl of 1 mg/ml PNPP per well. Plates were incubated for 30 min at RT, and the adsorbance was read with a multiwell plate reader. Percent inhibition was calculated as follows: [mean optical density (OD) value of Bio HAMb only] − [mean OD value of Bio HMAb plus sera] × 100/[mean OD value of Bio HMAb only]. Statistical analyses were performed with the software programs IN-STAT or PRISIM (Graph Pad Software, San Diego, Calif.).
RESULTS
To investigate the relative contribution of antibodies to conformational and linear epitopes in the immune response to HTLV-1 gp46, we first determined whether known linear epitopes and conformational epitopes, as defined by selected HMAbs, are overlapping. The ability of antibodies to linear epitopes to inhibit the binding of antibodies to conformational epitopes (or vice versa) was measured. A panel of monoclonal antibodies, both human and mouse, to linear epitopes within HTLV-1 gp46 was evaluated to inhibit the binding of selected HMAbs to conformational epitopes. An antibody, HMAb PRH-1, which had been demonstrated to effectively capture HTLV-1 gp46, was used to display the envelope protein to HMAbs recognizing conformational epitopes (12). HMAb PRH-1 recognizes amino acids 260 to 294 of HTLV-1 gp46 both in native and denatured HTLV-1 gp46 (12). The locations of linear epitopes within HTLV-1 gp46 recognized by the antibodies employed in this and other studies are indicated in Fig. 1.
FIG. 1.
Map showing major features of HTLV-1 envelope proteins. HTLV-1 gp21 is indicated in light gray, and HTLV-1 gp46 is indicated in white. CHO indicates glycosylation sites. The dark black boxes indicate the locations of the amino-terminal and central sequences that elicit a neutralizing antibody response (4, 28). The locations of epitopes recognized by various HMAbs and mMAbs employed in these studies have been previously determined (12, 29, 31) and are indicated.
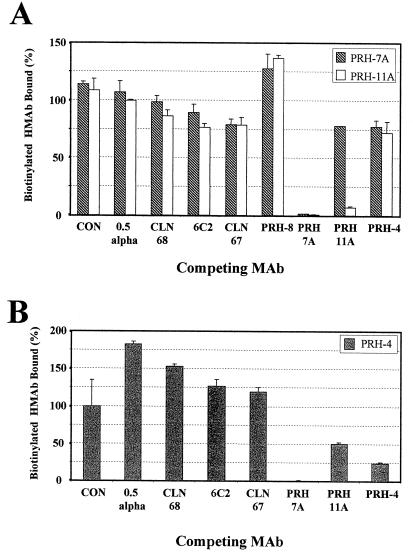
Antibodies to linear epitopes between amino acids 175 and 210 are known to mediate virus neutralization and to be highly prevalent in HTLV-1-infected individuals (4, 5, 11, 15, 18, 27). Two antibodies, HMAb 0.5 alpha and an mMAb, CLN 68, to two overlapping epitopes within the central domain of HTLV-1 gp46 (amino acids 175 to 199) were tested for their ability to inhibit binding of HMAbs PRH-7A, PRH-11A, and PRH-4 to HTLV-1 gp46. HMAb 0.5 alpha recognizes the sequence PPLLPH (amino acids 188 to 193 [31]), and mMAb CLN 68 binds to an epitope within amino acids 190 to 209 (29). Neither 0.5 alpha nor CLN 68 significantly inhibited the binding of HMAbs PRH-7A, PRH-11A, and PRH-4, which recognize separate conformational epitopes (Fig. 2). The apparent enhancement of binding of PRH-4 with HMAb 0.5 alpha (Fig. 2B) was not observed reproducibly. These results suggest that sequences within linear sequences known to encode virus-neutralizing epitopes are not involved in conformational epitopes that also elicit a neutralizing antibody response.
FIG. 2.
Competition analysis with HTLV-1 HMAbs. (A) Extract from 4 × 105 MT-2 cells was captured on microtiter plate wells precoated with 100 ng of HMAb PRH-1. Bound protein was detected with 2 μg of biotinylated PRH-7A (crosshatched bars) per ml or PRH-11A (clear bars) in the presence of 10 μg/ml of the indicated competing antibodies (x axis). HMAb PRH-8 was tested at 6 μg/ml due to the diluted concentration of the antibody. Values are plotted as the percent binding obtained relative to that from samples with no competing antibody. Each bar represents the mean value of two determinations. Error bars indicate 1 standard deviation from the mean. CON is an irrelevant HMAb, R04, that recognizes a cytomegalovirus protein (12). (B) Experiment analogous to that shown in panel A except that 5 μg/ml of biotinylated PRH-4 is the detection antibody. Competing antibodies (x axis) were tested at a concentration of 25 μg/ml. The higher concentration of PRH-4 is required due to a reduced affinity of biotinylated PRH-4 for HTLV-1 gp46 relative to that observed with HMAbs PRH-7A and PRH-11A (12, 12a).
Antibodies to linear epitopes within the carboxy-terminal regions of HTLV-1 gp46 (amino acids 210 to 312), PRH-8, 6C2, and CLN-67 were evaluated in a similar manner. Both CLN-67 and 6C2 recognize denaturation-resistant epitopes common to HTLV-1 and HTLV-2 gp46 (12, 29). As part of this study, we fine mapped the location of the CLN-67 epitope to amino acids 288 to 312 of HTLV-1 gp46 (data not shown), using recombinant proteins and methods as described previously (12). None of the three antibodies in this study significantly inhibited binding of HMAbs PRH-7A and PRH-11A to HTLV-1 gp46. Inhibition of PRH-4 was tested only with antibodies 6C2 and CLN-67 (Fig. 2B) due to the low concentration of HMAb PRH-8. Neither mMAb 6C2 nor CLN-67 had any effect on the binding of HMAb PRH-4. Each conformational HMAb, in contrast, was significantly inhibited by itself. As seen previously (12), HMAbs PRH-11A and PRH-4 were strongly inhibited by HMAb PRH-7A, suggesting that these clearly distinct antibodies may have epitopes that are spatially close. The lack of inhibition of binding by antibodies to sequences within linear epitopes of the carboxy-terminal half of HTLV-1 gp46 suggested a lack of their involvement in the conformational epitopes recognized by HMAbs PRH-4, PRH-7A, and PRH-11A.
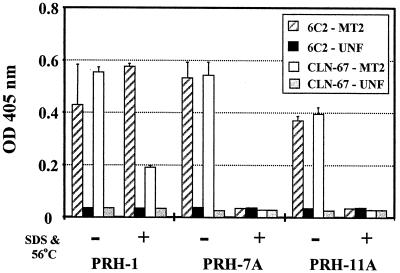
The murine MAbs, CLN-67 and 6C2, were then evaluated as capture antibodies in an ELISA-based detection system to quantify serum antibody binding to native and denatured HTLV-1 env protein. A gentle denaturation treatment of heating HTLV-1 gp46 containing lysate to 56°C for 15 min in the presence of 0.5% SDS was employed. Two HMAbs, PRH-7A and PRH-11A, to different conformational epitopes were used to measure the extent of denaturation obtained with this procedure (Fig. 3). Results obtained with these HMAbs were compared with results obtained with HMAb PRH-1 that recognizes a linear epitope. All three HMAbs exhibited strong binding to native HTLV-1 gp46 captured by either 6C2 or CLN-67. When CLN-67 was the capture antibody, reactivity of HMAb PRH-1 toward denatured HTLV-1 gp46 was reduced by approximately two-thirds relative to native gp46. When 6C2 was employed as the capture antibody, HMAb PRH-1 was equally reactive with either native or denatured HTLV-1 gp46. Consequently, 6C2 was used as the capture antibody in further experiments. No specific binding to denatured HTLV-1 gp46 was observed with either HMAb PRH-7A or PRH-11A when either capture antibody was used. Heating HTLV-1 gp46 to 56°C in the presence of 0.5% SDS was therefore sufficient to denature the tertiary structure of HTLV-1 gp46.
FIG. 3.
Effect of mild heat and SDS treatment on HTLV-1 envelope conformation. Cytoplasmic extracts derived from 4 × 105 cells of the T-cell line RPMI-8402 (black and grey bars) or 4 × 105 MT-2 (white and crosshatched bars) cells were either heated to 56°C in the presence of 0.5% SDS (grouped bars labeled +) or not treated (−) and added to microtiter plate wells coated with 100 ng of mMAbs 6C2 (crosshatched and black bars) or CLN-67 (white and grey bars). Bound gp46 was detected with the indicated biotinylated HMAb (x axis) at a concentration of 2 μg/ml. Results are the mean of duplicate determinations, and error bars indicate 1 standard deviation from the mean.
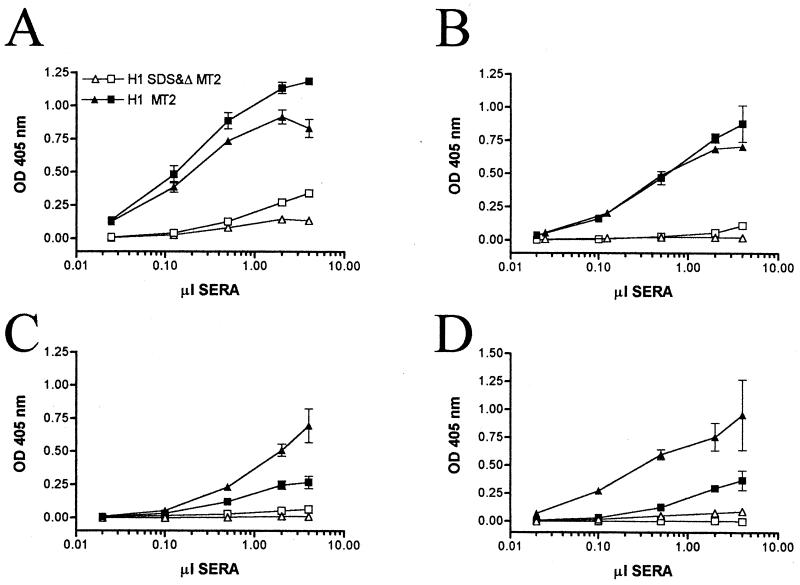
We next evaluated the ability of the capture ELISA to measure antibody reactivity of sera from HTLV-1-infected individuals to both native and denatured HTLV-1 gp46. Titration curves with either representative serum samples (seven from infected blood donors) are presented in Fig. 4. A range of reactivity to native HTLV-1 gp46 was observed. For all sera, a strong signal was obtained with extracts from HTLV-1-infected MT-2 cells, with 2 to 4 μl of serum (equivalent to a dilution of 1/50 to 1/25; Fig. 4). In the same experiments, the specific reactivity observed with 2 μl of sera from uninfected individuals against native or denatured HTLV-1 gp46 ranged from 0 to a maximum OD of 0.018 (data not shown). Denaturation of the HTLV-1 gp46 significantly reduced the reactivity of serum HTLV-1 antibodies to the captured antigen. The two serum samples with the strongest residual reactivity to denatured HTLV-1 gp46 (Fig. 4A) exhibited apparent 50% of maximum binding values towards native gp46 with 0.12 and 0.16 μl of serum (equivalent to dilutions of ∼1/600 and 1/800, respectively). For both these serum samples, an approximately-101.5-greater amount of antibody was required to achieve a given adsorbance value with denatured HTLV-1 gp46 relative to native HTLV-1 gp46 (Fig. 4A). The other six HTLV-1 antiserum samples had even greater differential reactivity to native HTLV-1 gp46. Thus, the antibody response of HTLV-1-infected individuals is directed primarily at denaturation-sensitive or conformational epitopes within HTLV-1 gp46.
FIG. 4.
Titration of sera from HTLV-1-infected individuals against native and denatured HTLV-1 gp46. Native (filled symbols) and denatured (open symbols) extracts from MT-2 cells were added to microtiter plate wells coated with mMAb 6C2. Bound HTLV-1 gp46 was detected with the indicated amounts of sera from HTLV-1-infected individuals in a total volume of 100 μl. Results obtained with sera from two HTLV-1-infected individuals are indicated in each panel (A to D). The individual indicated by (Δ,▴) in panel A was one of five individuals whose health status was undefined. The other seven HTLV-1-infected individuals were potential blood donors and asymptomatic. The values plotted are the mean specific ODs (ODs obtained from MT-2 extract − ODs obtained from uninfected extract). OD values represent two separate determinations, and error bars indicate 1 standard deviation from the mean.
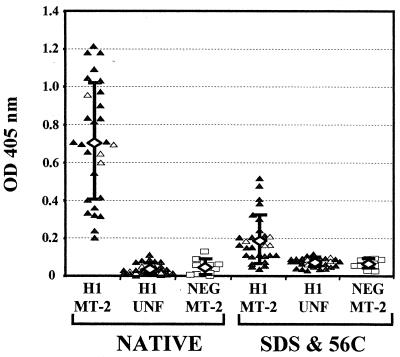
This study was expanded to a larger panel of sera from 28 HTLV-1-infected individuals. The panel included the eight serum samples examined in Fig. 4. Reactivity with 2 μl of serum (dilution of 1/50) was used, since it was at or near the plateau level observed with strongly reactive HTLV-1 antisera (Fig. 4). The mean adsorbance of the 28 serum samples with native gp46 was 0.71, with a range of 0.2 to 1.2 (Fig. 5). This value was significantly different from either the mean reactivity obtained with the same sera against native proteins captured from uninfected extracts (P < 0.0001, repeated measure analysis of variance) or the mean reactivity against HTLV-1 gp46 obtained from 10 serum samples from uninfected individuals (P < 0.0001, unpaired t test with Welch correction). The mean value obtained from the four HTLV-1-infected individuals whose health status was unknown (reference laboratory samples) was essentially equivalent (OD = 0.73) to that obtained with the other 24 serum samples. The mean adsorbance obtained with HTLV-1 sera to denatured HTLV-1 gp46 was 0.196, which was significantly elevated compared to the mean adsorbance obtained with 10 negative serum samples (OD = 0.077; P < 0.0001, unpaired t test with Welch correction). After background reactivity to proteins captured from uninfected extracts was subtracted, the mean serum antibody binding to native HTLV-1 gp46 was 0.674 OD units and the mean binding to denatured HTLV-1 gp46 was 0.119, differences which were also highly significant (P < 0.0001, Table 1). The mean reactivity of individual sera toward denatured gp46 was 14.7% of the value obtained with native HTLV-1 gp46, with values ranging from 1.2 to 43%. The majority of the sera (24 of 28; 86%) had antibody binding with denatured HTLV-1 gp46 that was less than 25% of the values obtained with native HTLV-1 gp46. Overall, the data from both the titration analyses and the larger panel of HTLV-1 sera indicate that the vast majority of the antibody response to HTLV-1 gp46 in an infected individual is directed against conformational epitopes.
FIG. 5.
Reactivity of individual sera from HTLV-1-infected (H1) or uninfected (NEG) individuals with native and denatured captured HTLV-1 gp46. Cytoplasmic extracts from 4 × 105 cells of the T-cell line RPMI-8402 (UNF) or MT-2 cells (MT-2) were either heated to 56°C in the presence of 0.5% SDS (SDS & 56C) or not treated (NATIVE) and added to microtiter plates coated with mMAb 6C2. Bound HTLV-1 gp46 was detected with individual sera from HTLV-1-infected potential blood donors (▴, n = 24), HTLV-1-infected individuals with undetermined health status (Δ, n = 4), or uninfected (□, n = 10) individuals. All sera were tested at a dilution of 1/50 (2 μl of sera in 100 μl of BLOTTO). The open diamonds indicate the mean value obtained from all tested individuals (HTLV-1, n = 28) and error bars indicate 1 standard deviation from the mean.
TABLE 1.
Specific reactivity of HTLV-1 sera with native and denatured HTLV-1 gp46
| Protein | Sera | n | Range (OD) | Avg (±1 SD)a |
|---|---|---|---|---|
| Native HTLV-1 gp46 | HTLV-1 | 28 | 0.193–1.135 | 0.674 ± 0.294cd |
| HTLV-1− | 10 | 0.001–0.042 | 0.012 ± 0.013c | |
| Denatured HTLV-1 gp46 | HTLV-1 | 28 | 0.005–0.438 | 0.119 ± 0.122de |
| HTLV-1− | 10 | (−0.061)–0.014 | −0.002 ± 0.022e | |
| % Reactivity remainingb | HTLV-1 | 28 | 1.2–43.7% | 14.7 ± 10.4% |
All values reflect specific reactivity to HTLV-1 envelope proteins, with background reactivity to proteins captured from uninfected extracts subtracted.
% Reactivity remaining is the fraction of the total reactivity for each serum sample that is directed at denatured HTLV-1 gp46 protein. For each serum sample, this value is calculated as follows (OD denatured MT-2 − OD denatured uninfected)/(OD native MT-2 − OD native uninfected) × 100. The average value is the mean % reactivity remaining obtained with all 28 serum samples. This calculation does not produce the same value as dividing the mean OD value for native HTLV-1 gp46 by the mean OD value obtained with denatured HTLV-1 gp46. Significance was calculated by using the unpaired t test with Welch correction.
Significantly different from each other (P < 0.0001).
Significantly different from each other (P < 0.0001).
Significantly different from each other (P < 0.0001).
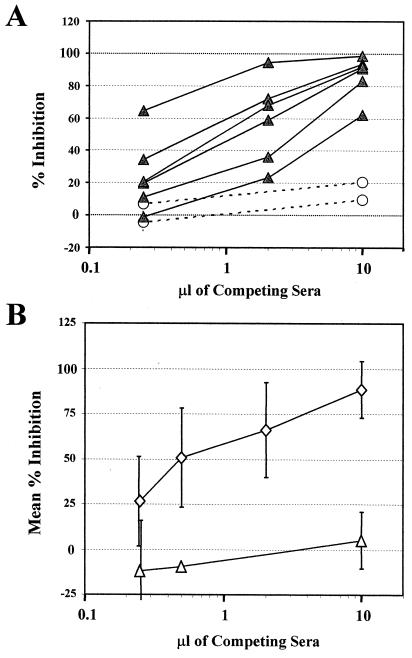
We next measured for serum antibodies to the epitope recognized by HMAb PRH-7A. This epitope was selected because we demonstrated that HMAb PRH-7A mediates virus neutralization and recognizes an epitope common to HTLV-1 and HTLV-2. Furthermore, the lack of inhibition of PRH-7A by any of the other monoclonal antibodies tested and the complete inhibition observed with itself as a blocking antibody suggested that it should be possible to detect the presence of serum antibodies similar to PRH-7A. Antibody competition analysis was used to evaluate sera from HTLV-1-infected and uninfected individuals for the presence of antibodies capable of inhibiting binding of biotinylated PRH-7A to native gp46. Sera from 25 infected individuals were tested. Representative results with six serum samples are presented in Fig. 6A. All six sera exhibited significant inhibition of PRH-7A binding. No significant inhibition was observed with control sera from two uninfected individuals. A broad range in inhibitory activity was observed, with strongly inhibitory sera having approximately 100-fold-higher titers of inhibitory antibodies than those of more weakly inhibitory sera (compare rightmost and leftmost HTLV-1 inhibition curves).
FIG. 6.
HTLV-1 sera contain antibodies similar to HTLV-1 HMAb PRH-7A. (A) Results of a representative experiment evaluating inhibition of PRH-7A binding. Extract from 8 × 105 HUT-102 cells was captured onto microtiter plate wells precoated with 100 ng of HMAb PRH-1. Bound protein was detected with 2 μg/ml solution of biotinylated HMAb PRH-7A in the presence of the indicated amounts (in microliters) of sera from HTLV-1-infected potential blood donors (▴) or uninfected individuals (––○––). The y axis plots the percent inhibition of biotinylated PRH-7A binding relative to that observed in samples not containing any competing sera. The values are the means of two separate determinations. Percent inhibition was calculated as described in Materials and Methods. (B) Results obtained with the entire panel of sera. The mean percentages of inhibitions obtained for sera from HTLV-1-infected (◊) and uninfected individuals (Δ) with the indicated amounts of sera are plotted. Error bars indicate 1 standard deviation from the mean. All assays were done with the indicated amount of sera diluted in 100 μl of BLOTTO containing 2 μg of biotinylated PRH-7A per ml. Twenty-five serum samples from HTLV-1-infected individuals were assayed for PRH-7A inhibition at 10 and 2 μl, 18 samples were assayed at 0.25 μl, and 7 samples were assayed at 0.5 μl. Twelve serum samples from uninfected individuals were tested for PRH-7A inhibition at 10 μl, 8 samples were tested at 0.25 μl, and 2 samples were tested at 0.5 μl.
Overall, 24 of 25 serum samples (96%) inhibited the binding of PRH-7A to native HTLV-1 gp46. The mean percent inhibition of PRH-7A binding observed with 10 μl of competing sera for all HTLV-1-infected individuals was 89% (Fig. 6B). This value was significantly elevated relative to the mean percent inhibition of 5% obtained with 12 serum samples from uninfected individuals (P < 0.0001, unpaired t test with Welch correction). For the entire panel, 50% inhibition of PRH-7A binding was observed with ∼0.5 μl of competing sera (dilution = 1/200). The one serum sample that did not exhibit significant inhibition of HMAb PRH-7A had a relatively weak antibody response to native HTLV-1 gp46 (highest OD value, 0.370) and essentially no reactivity to denatured HTLV-1 gp46 (Fig. 4D, squares). The largest amount of this serum tested (10 μl) inhibited PRH-7A binding by 27%. The majority of HTLV-1 sera tested, 21 of 25 serum samples (84%), exhibited greater than 80% inhibition of PRH-7A binding with 10 μl of sera regardless of whether overall reactivity to native HTLV-1 gp46 was strong or weak. These findings strongly argue that antibodies analogous to an HTLV-1 HMAb recognizing a conformational epitope and having strong neutralization activity (12) are highly prevalent among HTLV-1-infected individuals.
DISCUSSION
In this study, we determined the relative amounts of antibodies to conformational versus linear epitopes of HTLV-1 gp46 present during natural infection in a panel of asymptomatic individuals. Titration analysis and evaluation of a larger panel of sera at a low dilution indicated that the majority of the immune response to HTLV-1 gp46 in asymptomatic infected individuals is directed to conformational epitopes within HTLV-1 gp46. One implication of this finding is that capture assays employing native HTLV-1 gp46 may have some diagnostic utility. In this small panel, 28 of the 28 HTLV-1-infected individuals tested would have been identified as HTLV-1 positive with reasonable cutoffs (Fig. 3A). Use of the capture assay may be relevant for the testing of individuals with indeterminate serology or as an alternative to current confirmatory antibody assays. There is currently no information on whether the antibody response to conformational epitopes precedes or follows development of antibody response to linear epitopes within the HTLV-1 envelope proteins or in other antigens. Such studies, with either appropriate animal models or seroconversion panels would be informative.
The locations of amino acids involved in conformational epitopes are unknown. However, the observation that antibodies to linear epitopes within the carboxy-terminal region did not affect the binding of HMAbs PRH-7A, PRH-11A, and PRH-4 to gp46 suggests that the epitopes recognized by these HMAbs reside primarily in the amino-terminal half of HTLV-1 gp46. One sequence located in the amino-terminal half of HTLV-1 gp46 has been shown to induce HTLV-1-specific neutralizing antibodies when it is inoculated into goats (amino acids 90 to 98 [28]). However, in a recent study mMAbs generated to a similar peptide (amino acids 89 to 110) did not immunoprecipitate native HTLV-I gp46 or recognize HTLV-I gp46 on the surfaces of infected cells (10). Consequently, this particular sequence may not be accessible in native gp46. More focused studies aimed at mapping the locations of conformational epitopes within HTLV-1 gp46 will be required to evaluate the exposure and immunogenicity of the amino-terminal region of HTLV-1 gp46.
We also determined that 96% of 25 serum samples from HTLV-1-infected individuals possessed antibodies that strongly inhibited the binding of an antibody, PRH-7A, to a specific conformational epitope. Control experiments performed with monoclonal antibodies to known linear and conformational epitopes within HTLV-1 gp46 indicated that the inhibition assay was highly specific for antibodies analogous to PRH-7A. PRH-7A has been shown to strongly inhibit HTLV-1-mediated syncytium formation and recognizes a conformational epitope common to HTLV-1 and -2 (12). Thus, antibodies similar to PRH-7A have the potential to play a significant role in containing HTLV-1 infection in an infected individual. Although it would be desirable to evaluate HTLV-1 sera for the presence of other conformational antibodies, the high prevalence of antibodies to PRH-7A and the inhibitory effect of PRH-7A on binding of either PRH-4 or PRH-11A precluded these experiments. As the epitopes recognized by these other HMAbs are mapped, it may be possible to develop assays for the frequency of these antibodies by using epitope (−) mutants.
The one serum sample that did not inhibit PRH-7A binding was unremarkable except that it had a relatively low reactivity with native HTLV-1 gp46 (OD value of 0.297 with 2 μl of sera; mean for panel of 0.674, as summarized in Table 1). The other four serum samples with the lowest overall reactivity to native HTLV-1 gp46 (specific OD range of 0.192 to 0.386) exhibited between 82 and 96% inhibition of PRH-7A binding with 10 μl of sera. Thus, sera with relatively weak reactivity to native HTLV-1 gp46 can still contain substantial amounts of antibodies similar to PRH-7A. Other potential explanations for the failure of a particular HTLV-1 serum to contain antibodies analogous to PRH-7A include infection with an HTLV-1 isolate that has a mutation in one or more of the amino acids comprising the epitope recognized by PRH-7A. Alternatively, an individual may be recently infected and present with an incomplete antibody response. A third explanation is that the immune response to other conformational epitopes (such as that recognized by HMAb PRH-4) may predominate in select individuals.
The observation that the majority of the antibody response to the HTLV-1 envelope is directed at conformational epitopes is in agreement with similar studies performed with sera of HIV-infected individuals to native and denatured HIV envelope proteins (23, 24). One implication of these studies is that an effective HTLV-1 vaccine needs to contain antigens that are able to induce antibodies to conformational epitopes mediating virus neutralization similar to those observed during natural infection. In contrast to HIV, evidence for a protective humoral immune response to both HTLV-1 and HTLV-2 is extensive as cited in references 1, 9, 21, 22, and 25. These include passive immunization studies with human gammaglobulin obtained from HTLV-1-infected donors, protecting susceptible rabbits from milk-borne and blood-borne HTLV-1 infection (22). Another, perhaps more subtle, implication of these studies concerns the relationship of antibody titer to HTLV-1 proviral load and disease status. Several studies have noted an increase in reactivity to denatured HTLV-1 viral proteins or to HTLV-1 envelope synthetic peptides in individuals with TSP/HAM (3, 26). Higher antibody titers have also been observed in individuals with high proviral loads or individuals whose lymphocytes exhibit spontaneous lymphoproliferation (30, 33). However, the assays employed in these studies did not measure antibody response to conformational epitopes within HTLV-1 gp46, and the conclusions drawn were based on the assessment of a minority of the total antibody response to HTLV-1 gp46.
Whether the antibody response to conformational epitopes is correlated with either disease status or HTLV-1 proviral load remains an open question. In this panel, up to a 100-fold range in the overall titer to native HTLV-1 gp46 and the amount of PRH-7A-like antibodies was observed in sera from primarily healthy infected blood donors. Although we cannot exclude the possibility that one or more individuals included in this panel progressed to disease, the results obtained reflect the antibody profile of infected individuals with disease containment. It is possible that the wide range in antibody response is due to infection with different HTLV-1 genotypes. Other studies, however, of similar populations would predict that 90% or more of the individuals in this study would be infected with the cosmopolitan genotype (34). Additional studies evaluating the full range of antibody response in relation to HTLV-1 disease progression and other predictive parameters, such as spontaneous lymphocyte proliferation, proviral load, and the overall immune status of the infected individual, are clearly warranted. The assays described in our study provide a means of addressing this issue. Furthermore, evaluation of the antibody response to native HTLV-1 gp46 in concert with studies directed at better defining the locations of conformational epitopes mediating virus neutralization should provide important insights into the structure and function of the HTLV-1 envelope protein.
ACKNOWLEDGMENTS
We acknowledge Susan Perkins for assistance with the culture and purification of many of the antibodies employed in these studies. We also acknowledge Wanda Washington for help with administrative matters.
This work was supported in part by Public Health Service grant DA-06596 to S.K.H.F.
REFERENCES
- 1.Akari H, Suzuki T, Ikeda K, Hoshinio H, Tsugikazu T, Murotsuka T, Terao K, Ito H, Yoshikawa Y. Prophylaxis of experimental HTLV-1 infection in cynomologus monkeys by passive immunization. Vaccine. 1997;15:1391–1395. doi: 10.1016/s0264-410x(97)00055-8. [DOI] [PubMed] [Google Scholar]
- 2.Arp J, Levatte M, Rowe J, Perkins S, King E, Leystra-Lantz C, Foung S K H, Dekaban G A. A source of glycosylated human T-cell lymphotropic virus type 1 envelope protein: expression of gp46 by the vaccinia virus-T7 polymerase system. J Virol. 1996;70:7349–7359. doi: 10.1128/jvi.70.11.7349-7359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astier-Gin T, Portail J P, Londos-Gagliardi D, Moynet D, Blanchard S, Dalibart R, Pouliquen J F, Georget-Courbot M C, Hajjar C, Sainte-Foie S, Guillermain B. Neutralizing activity and antibody reactivity toward immunogenic regions of the human T cell leukemia virus type I surface glycoprotein in sera of infected patients with different clinical states. J Infect Dis. 1997;175:716–719. doi: 10.1093/infdis/175.3.716. [DOI] [PubMed] [Google Scholar]
- 4.Baba E, Nakamura M, Tanaka Y, Kuroki M, Itoyama Y, Nakano S, Niho Y. Multiple neutralizing B-cell epitopes of human T-cell leukemia virus type 1 (HTLV-1) identified by human monoclonal antibodies. J Immunol. 1993;151:1013–1024. [PubMed] [Google Scholar]
- 5.Buckner C, Roberts C R, Foung S K H, Lipka J, Reyes G R, Hadlock K, Chan L, Gongora-Biachi R A, Hjelle B, Lal R B. Immune responsiveness to the immunodominant recombinant envelope epitopes of human T lymphotropic virus types I and II in diverse geographic populations. J Infect Dis. 1992;166:1160–1163. doi: 10.1093/infdis/166.5.1160. [DOI] [PubMed] [Google Scholar]
- 6.Cann A J, Chen I S Y. Human T-cell leukemia virus types I and II. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1501–1527. [Google Scholar]
- 7.Carrington C V F, Paul N, Cordell J, Schulz T F. Probing the conformation of the human T-lymphotropic virus I envelope protein with monoclonal antibodies. J Gen Virol. 1996;77:2025–2029. doi: 10.1099/0022-1317-77-9-2025. [DOI] [PubMed] [Google Scholar]
- 8.Desgranges C, Souche S, Vernant J C, Smadja D, Vahlne A, Horal P. Identification of novel neutralization-inducing regions of the human T-cell lymphotropic virus type I envelope glycoproteins with human HTLV-1-seropositive sera. AIDS Res Hum Retroviruses. 1994;10:163–173. doi: 10.1089/aid.1994.10.163. [DOI] [PubMed] [Google Scholar]
- 9.De The, G., and M. Kazanji. 1996. An HTLV-1/II vaccine: from animal models to clinical trials? J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1):S191–S198. [DOI] [PubMed]
- 10.Grange M P, Rosenberg A R, Horal P, Desgranges C. Identification of exposed epitopes on the envelope glycoproteins of human T cell lymphotropic virus type I. (HTLV-I) Int J Cancer. 1998;75:804–813. doi: 10.1002/(sici)1097-0215(19980302)75:5<804::aid-ijc22>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Hadlock K G. Recent advances in the diagnosis of HTLV-I and HTLV-II infection. Expert Opin Therapeutic Pat. 1995;5:923–943. [Google Scholar]
- 12.Hadlock K G, Rowe J, Perkins S, Bradshaw P, Song G-Y, Cheng C, Yang J, Gascon R, Halmos J, Rehman M, McGrath M S, Foung S K H. Neutralizing human monoclonal antibodies to conformational epitopes of HTLV-1 and HTLV-2 gp46. J Virol. 1997;71:5828–5840. doi: 10.1128/jvi.71.8.5828-5840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Hadlock, K. G. Unpublished observations.
- 13.Harrington W J, Jr, Sheremata W, Hjelle B, Dube D K, Bradshaw P, Foung S K H, Snodgrass S, Toedter G, Cabral L, Poiesz B. Spastic ataxia associated with human T-cell lymphotropic virus type II infection. Ann Neurol. 1993;33:411–414. doi: 10.1002/ana.410330416. [DOI] [PubMed] [Google Scholar]
- 14.Hjelle B, Appelzeller O, Mills R, Alexander S, Torrez-Martinez N, Jahnke R, Ross G. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet. 1992;339:645–646. doi: 10.1016/0140-6736(92)90797-7. [DOI] [PubMed] [Google Scholar]
- 15.Horal P, Hall W W, Svennerholm B, Lycke J, Jeansson S, Rymo L, Kaplan M H, Vahlne A. Identification of type-specific linear epitopes in the glycoproteins gp46 and gp21 of human T-cell leukemia viruses type I and type II using synthetic peptides. Proc Natl Acad Sci USA. 1991;88:5754–5758. doi: 10.1073/pnas.88.13.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalyanaraman V S, Sarangadharan M D, Miyoshi I, Blayney D, Golde D, Gallo R C. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 17.Kuroki M, Nakamura M, Itoyama Y, Tanaka Y, Shiraki H, Baba E, Esaki T, Tatsumoto T, Nagafuchi S, Nakano S, Niho Y. Identification of new epitopes recognized by human monoclonal antibodies with neutralizing and antibody-dependent cellular cytotoxicity activities specific for human T cell leukemia virus type 1. J Immunol. 1992;149:940–948. [PubMed] [Google Scholar]
- 18.Lal, R. B. 1996. Delineation of immunodominant epitopes of human T-lymphotropic virus types I and II and their usefulness in developing serologic assays for the detection of antibodies to HTLV-I and HTLV-II. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1):170–178. [DOI] [PubMed]
- 19.Lipka J J, Bui K, Reyes G R, Moeckli R, Wiktor S Z, Blattner W A, Murphy E L, Shaw G M, Hanson C V, Sninsky J J, Foung S K H. Determination of a unique and immunodominant epitope of human T cell lymphotropic virus type I. J Infect Dis. 1990;162:353–357. doi: 10.1093/infdis/162.2.353. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita S, Robert-Guroff M, Trepel J, Cossman J, Mitsuya H, Broder S. Human monoclonal antibody directed against an envelope glycoprotein of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1986;83:2672–2676. doi: 10.1073/pnas.83.8.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi I, Yoshimoto S, Kubonishi I, Fujishita M, Yamato K, Hirose S, Taguchi H, Niiya K, Kobayashi M. Infectious transmission of human T-cell leukemia virus-I to rabbits. Int J Cancer. 1985;35:81–85. doi: 10.1002/ijc.2910350113. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi, I., N. Takehara, T. Sawada, Y. Iwahara, R. Kataoka, D. Yang, and H. Hoshino. 1992. Immunoglobulin prophylaxis against HTLV-1 in a rabbit model. Leukemia 6(Suppl. 1):24–26. [PubMed]
- 23.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore J P, Ho D D. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morishita N, Ishii K, Tanaka Y, Sawada T, Hoshino H, Foung S K H, Miyoshi I. Immunoglobulin prophylaxis against human T cell lymphotropic virus type II in rabbits. J Infect Dis. 1994;169:620–623. doi: 10.1093/infdis/169.3.620. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura M, Kuroki M, Kira J I, Itoyama Y, Shiraki H, Kuroda N, Washitani Y, Nakano S, Nagafuchi S, Anazai K, Tasumoto T, Esaki T, Maeda Y, Niho Y. Elevated antibodies to synthetic peptides of HTLV-1 envelope transmembrane glycoproteins in patients with HAM/TSP. J Neuroimmunol. 1991;35:167–178. doi: 10.1016/0165-5728(91)90171-3. [DOI] [PubMed] [Google Scholar]
- 27.Palker T J, Tanner M E, Scearce R M, Streilein R D, Clark M E, Haynes B F. Mapping of immunogenic regions of human T cell leukemia virus type I (HTLV-1) gp46 and gp21 envelope glycoproteins with env-encoded synthetic peptides and a monoclonal antibody to gp46. J Immunol. 1989;142:971–978. [PubMed] [Google Scholar]
- 28.Palker T J, Riggs E R, Spragion D E, Muir A J, Scearce R M, Randall R R, McAdams M W, McKnight A, Clapham P R, Weiss R A, Haynes B F. Mapping of homologous, amino-terminal neutralizing regions of human T-cell lymphotropic virus type I and II gp46 envelope glycoproteins. J Virol. 1992;66:5879–5889. doi: 10.1128/jvi.66.10.5879-5889.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papsidero L D, Dittmer R P, Vaickus L, Poiesz B J. Monoclonal antibodies and chemiluminescence immunoassay for detection of the surface protein of human T-cell lymphotropic virus. J Clin Microbiol. 1992;30:351–358. doi: 10.1128/jcm.30.2.351-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince H E, Lee H, Jensen E R, Swanson P, Weber D, Fitzpatrick L, Doyle M, Kleinman S. Immunologic correlates of spontaneous lymphocyte proliferation in human T-lymphotropic virus infection. Blood. 1991;78:169–174. [PubMed] [Google Scholar]
- 31.Ralston S, Hoeprich P, Akita R. Identification and synthesis of the epitope for a human monoclonal antibody which can neutralize human T-cell leukemia/lymphotropic virus type I. J Biol Chem. 1989;264:16343–16346. [PubMed] [Google Scholar]
- 32.Sheremata W A, Harrington W J, Jr, Bradshaw P A, Foung S K H, Raffanti S P, Berger J R, Snodgrass S, Resnick L, Poiesz B J. Association of ‘(tropical) ataxic neuropathy’ with HTLV-II. Virus Res. 1993;29:71–77. doi: 10.1016/0168-1702(93)90126-8. [DOI] [PubMed] [Google Scholar]
- 33.Shinzato O, Kamihira S, Ikeda S, Kondo H, Kanda T, Nagata Y, Nakayama E, Shiku H. Relationship between the anti-HTLV-1 antibody level, the number of abnormal lymphocytes and the viral-genome dose in HTLV-1 infected individuals. Int J Cancer. 1993;54:208–212. doi: 10.1002/ijc.2910540208. [DOI] [PubMed] [Google Scholar]
- 34.Vidal A U, Gessain A, Yoshida M, Tekaia F, Garin B, Guillemain B, Schulz T, Farid R, De The G. Phylogenetic classification of human T cell leukemia/lymphoma virus type I genotypes in five major molecular and geographical subtypes. J Gen Virol. 1994;75:3655–3666. doi: 10.1099/0022-1317-75-12-3655. [DOI] [PubMed] [Google Scholar]