Summary
Age-related macular degeneration (AMD) is a leading cause of blindness in older adults. Investigating shared genetic components between metabolites and AMD can enhance our understanding of its pathogenesis. We conduct metabolite genome-wide association studies (mGWASs) using multi-ethnic genetic and metabolomic data from up to 28,000 participants. With bidirectional Mendelian randomization analysis involving 16,144 advanced AMD cases and 17,832 controls, we identify 108 putatively causal relationships between plasma metabolites and advanced AMD. These metabolites are enriched in glycerophospholipid metabolism, lysophospholipid, triradylcglycerol, and long chain polyunsaturated fatty acid pathways. Bayesian genetic colocalization analysis and a customized metabolome-wide association approach prioritize putative causal AMD-associated metabolites. We find limited evidence linking urine metabolites to AMD risk. Our study emphasizes the contribution of plasma metabolites, particularly lipid-related pathways and genes, to AMD risk and uncovers numerous putative causal associations between metabolites and AMD risk.
Keywords: age-related macular degeneration, AMD, genome-wide association studies, GWASs, Nurses’ Health Study, NHS, Health Professionals Follow Up Study, HPFS, Canadian Longitudinal Study of Aging, CLSA, Hispanic Community Health Study/Study of Latinos, HCHS/SOL, Mendelian randomization, MR, genomics, metabolomics, UK Biobank
Graphical abstract

Highlights
-
•
Large-scale metabolite genome-wide association studies identify mQTLs in plasma and urine
-
•
Two-sample Mendelian randomization analysis uncovers 108 plasma metabolites associated with AMD
-
•
AMD-associated metabolites are enriched in lipid-related pathways
-
•
Systematic exploration of metabolite connections with AMD subtypes and lipid biomarkers
In this study, Han et al. conduct large-scale integrative genetic and metabolomic analysis and discover 108 plasma metabolites associated with advanced age-related macular degeneration. The identified metabolites are enriched in pathways of glycerophospholipid metabolism, lysophospholipid, triradylcglycerols, and long-chain polyunsaturated fatty acids, providing insight into their potential in AMD pathogenesis.
Introduction
Age-related macular degeneration (AMD) is a leading cause of vision loss among adults aged 50 and above, with a global prevalence of 8.7%.1,2,3,4 Clinically, AMD is characterized by the presence of macular drusen and pigmentary changes in early and intermediate phases when most patients exhibit as minimally asymptomatic. Nevertheless, a subset of patients progress to the late blinding forms—choroidal neovascularization or geographic atrophy. It is currently well established that AMD is a complex disease, involving a combination of environmental and genetic risk factors.3,5 Genome-wide association studies (GWASs) have identified more than 50 single-nucleotide polymorphisms (SNPs) associated with AMD risk, predominantly within the complement pathway (CFH, CFI, C3, and C9) and in lipid-related genes (APOE, CETP, LIPC, and ABCA1).6,7 Recent genetic studies have also identified lipid and inflammatory biomarkers that are associated with AMD susceptibility.8,9,10,11 Despite important advances in elucidating AMD etiology, the underlying pathogenic mechanisms remain incompletely understood. The interplay between genes and environmental risk factors in the development of AMD is largely unknown. Consequently, there is a lack of FDA-approved treatments for non-exudative AMD, which accounts for 90% of cases, and limited therapeutic options to impede the progression of early and intermediate AMD to its severe, vision-impairing forms. A deeper understanding of the biological pathways involved in AMD is essential to inform potential treatment targets.
Metabolites, as small molecules representing intermediate products of a variety of physiological processes, hold promise in addressing the challenges in understanding AMD.12,13,14 Metabolomics, a high-throughput technology, enable the capture of metabolic activity by assaying a large number of metabolites in a given biological sample, such as blood,15 urine,16 and saliva.17 In observational studies, we and others have demonstrated an association between plasma metabolites, predominantly lipids, and AMD risk, as well as a higher likelihood of metabolite association with AMD risk variants located in or near lipid genes.18,19 In urine, amino acid metabolites have been linked to AMD.20 However, existing studies have not determined whether metabolites serve as mediators between risk genes and AMD susceptibility or merely reflect AMD-induced changes. Moreover, the observed associations between metabolites and AMD could be influenced by confounding factors. In such circumstances, traditional observational epidemiological association studies are susceptible to biases stemming from reverse causality and confounding factors.
To partially mitigate against the biases from confounding and reverse association, Mendelian randomization (MR) offers an analytical approach for estimating the putative causal effect of an exposure factor (X; e.g., metabolite) on an outcome (Y; e.g., AMD) using genetic instruments (e.g., lead SNPs associated with the exposure).21 MR can be intuitively understood as analogous to randomized clinical trials in which participants are randomly allocated into treatment and control groups. Similarly, in MR analysis, genetic risk alleles and non-effect alleles are assumed to be randomly assigned at conception.21 In a two-sample MR framework, metabolomic quantitative trait loci (mQTLs) can be identified from a study independent of AMD (e.g., SNP-metabolite and SNP-AMD associations are from two separate studies) to evaluate the potential causal relationships between metabolites and AMD.22,23 In addition, other genetic approaches may reveal novel insights into the shared genetic components between metabolites and AMD. For instance, the Bayesian colocalization approach has identified causal genomic loci shared between complex traits or diseases,24 and a metabolome-wide association study (analogous to transcriptome-wide association study [TWAS]) can uncover new metabolite-disease associations.25,26
Here, we conduct a large-scale integrative genetic and metabolomic analysis across six multi-ancestry and multi-fluid studies to investigate putative causal relationships between metabolites and AMD. The current study comprises five aims: first, we evaluate the discovery and replication of mQTLs in different ancestries and biofluids from up to 28,000 participants; second, metabolites from six multi-ancestry and multi-fluid studies are used to comprehensively investigate the putative causal relationships between metabolites and risk of AMD in a bidirectional two-sample MR framework; third, we identify causal risk variants shared between metabolites and AMD; fourth, we develop a metabolome-wide association study (MWAS) framework to identify metabolite-AMD associations by aggregating genetic effects from multiple mQTLs; and finally, we link the genetic findings to clinical phenotypes, including different AMD subtypes and lipid biomarkers, to interpret the biological mechanisms of the identified metabolites.
Results
Study design
The overall study design is presented in Figure 1. We performed metabolite-based GWASs (mGWASs) using six large-scale datasets (Table S1), comprising plasma data from the Nurses’ Health Study (NHS), NHSII, the Health Professionals Follow Up Study (HPFS), the Canadian Longitudinal Study on Aging (CLSA),27,28 a metabolite study in Finnish (Metabolic Syndrome in Men [METSIM]),29 and the Hispanic Community Health Study/Study of Latinos (HCHS/SOL),30 as well as urinary metabolomic data from Schlosser et al.16 and both plasma and urine measurements from our AMD Biomarkers Study.19 We conducted meta-analysis for plasma and urine separately using the aforementioned studies (Table S2). We obtained AMD GWAS summary statistics from the International AMD Genomics Consortium with 16,144 advanced AMD cases and 17,832 controls (Table S3).7 We performed various genetic analyses to characterize the associations between metabolites and AMD risk.
Figure 1.
Overview of the study design
mGWASs
The mGWASs were performed in each study, and detailed results are described in Table S2. For instance, in the NHS, NHSII, and HPFS cohorts, we observed no evidence of genomic inflation in the mGWAS, with genomic inflation factors (lambda) ranging from 0.99 to 1.03 and linkage disequilibrium (LD) score regression intercepts from 0.977 to 1.01 (Figure S1). Out of the 346 metabolites available in NHS/NHSII/HPFS, the median value of SNP-based heritability was 14.7% (interquartile range [IQR]: 7.2%–21.4%, maximum 70%), with 123 metabolites passing the nominal significance level (SNP-based heritability p < 0.05). Among the 346 metabolites, 243 (70.2%) exhibited at least one genome-wide significant mQTL (p < 5 × 10−8), with a median value of two mQTLs Figure S1). In total, 729 mQTLs were identified from the NHS, NHSII, and HPFS cohorts (Table S2).
Figure 2 (Figure S2) displays a circle Manhattan plot showing genomic hits from the NHS/NHSII/HPFS, the CLSA, the METSIM, the HCHS/SOL, the plasma meta-analysis, and the urine meta-analysis, exhibiting shared peak signals across different ancestries and biofluids. For plasma data, we meta-analyzed 2,064 plasma metabolites in 26,340 participants and identified 13,065 mQTLs (p < 5 × 10−8 ,Table S2; Figure S3). We meta-analyzed the urine mGWAS from Schlosser et al.16 and the AMD Biomarkers Study for 1,781 urine metabolites in 2,072 participants (Figure S4), identifying 1,443 mQTLs.
Figure 2.
Circular Manhattan plot illustrating metabolite-based genome-wide association studies
The circular Manhattan plot displays the p values from metabolite-based genome-wide association studies (mGWASs). Each circle represents an mGWAS in a specific study. From innermost to outermost circle: NHS/NHSII/HPFS, Canadian Longitudinal Study on Aging (CLSA), Metabolic Syndrome in Men (METSIM) study, HCHS/SOL, plasma meta-analysis, and urine meta-analysis. The circular Manhattan plot is customized to display all genome-wide significant SNPs (p < 5 × 10−8) for available metabolites in each study, with p values truncated at 1 × 10−50.
Multi-ancestry and multi-fluid validation of mQTLs
The mQTLs identified from NHS, NHSII, and HPFS plasma were well replicated in the CLSA, METSIM, and HCHS/SOL studies (Figure 3). For instance, of the 307 SNP-metabolite pairs (mQTLs) with mGWAS summary statistics available in the HCHS/SOL, 111 (36%) achieved genome-wide significance (p < 5 × 10−8 in the HCHS/SOL), and the overall concordance of effect sizes between the discovery (NHS/NHSII/HPFS) and replication cohorts (HCHS/SOL) was remarkably high (Pearson correlation 0.86, p = 6.09 × 10−91; Figure 3A3). We then compared the NHS, NHSII, and HPFS plasma mQTLs with urine mGWAS reported by Schlosser et al.16 (Figure 3A4). Of the 188 mQTLs that were also available from the latter, only 13 (6.9%) passed the genome-wide significance level (p < 5 × 10−8 in Schlosser et al.16). The Pearson correlation of plasma mQTLs and urine was 0.45 (p = 8.25 × 10−11; Figure 3A4), revealing a lower correlation of mQTLs shared between plasma and urine data. Restricting the effect sizes of SNPs to −0.5 to 0.5 showed similar correlations, confirming that the correlations were not solely driven by SNPs with larger effects (Figure S5).
Figure 3.
Multi-ancestry and multi-fluid validation of mQTLs across different studies
These plots show the effect sizes for metabolomic quantitative trait loci (mQTLs) from various datasets, labeled on the x and y axes. mQTLs were selected from the studies labeled on the x axis. Effect sizes are represented as green dots, while the 1.96 standard errors are depicted as gray bars (vertical and horizontal error bars). The red lines indicate the best-fit lines, and the 95% confidence intervals are shown in gray. The plasma meta-analysis included plasma metabolites from NHS/NHSII/HPFS, CLSA, METSIM, HCHS/SOL, and AMD Biomarker Study (Boston and Portugal plasma data). The urine meta-analysis included urine metabolites from Schlosser et al.16 (urine) and AMD Biomarker Study (Boston and Portugal urine data). p = 0 signifies a very small p value in R (smaller than the smallest representable positive double-precision floating point value at 2.225074 × 10−300).
The meta-analysis mQTLs were highly correlated with each input study (Figure S6). In line with the above findings, the mQTLs from plasma meta-analysis exhibited a lower correlation with urine data (Figure 3B). Collectively, these results suggest that effect sizes of metabolites in the same fluid from different ancestries and measurement platforms have a high correlation, whereas the concordance between plasma and urine is lower.
Putative causal associations between metabolites and advanced AMD
We performed two-sample MR analysis using the identified mQTLs and advanced AMD GWAS summary statistics. We discovered a high concordance of MR estimates among 143 available overlapping plasma metabolites in different studies (Figures 4A and S3). We identified 19 metabolites that were associated with advanced AMD risk in at least one study (NHS/NHSII/HPFS, CLSA, METSIM, or HCHS/SOL) and demonstrated a high concordance in effect sizes and directions across different studies, indicating that the high concordance of plasma mQTLs from various platforms and ancestries provided robust MR estimations.
Figure 4.
Replication of MR estimates for the associations between metabolites and AMD from NHS, NHSII, HPFS, CLSA, METSIM, and HCHS/SOL
(A) The replication of 19 overlapping metabolites between NHS/NHSII/HPFS, CLSA, METSIM, and HCHS/SOL. The x axis represents the effect size (beta) of metabolites on advanced AMD risk, while the vertical dashed line corresponds to beta = 0. Four MR methods are depicted with different colors and line types (inverse-variance weighted method, weighted median, weighted mode, and MR-Egger). For metabolites with a single SNP as a genetic instrument, the Wald ratio method is used (grouped into the inverse-variance weighted method).
(B) Comparison of the MR Z scores (MR effect sizes divided by standard errors) from plasma meta-analysis (x axis) and each individual study (y axis). The "MR discovery" indicates whether a metabolite is associated with advanced AMD risk in the plasma meta-analysis. Significant MR p values in each study (MR replication) are denoted by red dots (FDR p < 0.05).
To increase power, we then performed MR analysis using the mQTLs from plasma meta-analysis. In total, we identified 108 plasma metabolites that were associated with advanced AMD after adjusting for multiple testing (false discovery rate [FDR]-corrected p < 0.05; Figure 4B; Table S4). The most common pathways of the associated metabolites were lysophospholipid, glycerophospholipid (i.e., glycerophosphoethanolamine and glycerophosphocholine pathways), triradylcglycerols, long chain polyunsaturated fatty acids (PUFAs), and plasmalogen (Figures 5 and S7). We performed reverse-directional MR analyses and observed limited evidence supporting reverse effects that advanced AMD might alter metabolite levels (Table S4). In the meta-analysis of urine metabolites, we only identified five metabolites associated with advanced AMD (Table S5), reflecting the smaller sample size of urine data or fewer true associations between urine metabolites and advanced AMD. In the urine MR analysis, we found that indolelactate in the tryptophan metabolism pathway was negatively associated with advanced AMD.
Figure 5.
Prioritized metabolite pathways from Mendelian randomization analysis
The x axis displays the names of subpathways (biochemical groups, with “chemical” representing xenobiotics), while the y axis indicates the number of metabolites (with at least two metabolites) associated with advanced AMD risk.
We performed various MR sensitivity analyses to evaluate the MR estimates that may violate MR assumptions. Different MR sensitivity analyses provided consistent estimates and directions (Figure 4A; Table S4). The full descriptions to probe MR assumptions were presented in the discussion section.
Our MR analyses exhibited sufficient power to detect moderate effect sizes for advanced AMD, such as an odds ratio (OR) of 1.2 per standard deviation increase of metabolites (with a mean variance of 3.8% for metabolite genetic instruments). The powers of our analysis for advanced AMD, choroidal neovascularization (CNV), geographic atrophy (GA), mixed AMD, and intermediate AMD are 88%, 77%, 44%, 30%, and 64%, respectively (as shown in Table S6). The genetic instruments used in our analysis are available in Table S7.
Shared common causal variants between metabolites and advanced AMD
From the Bayesian colocalization analysis, we identified 114 potential causal variants for metabolite and advanced AMD pairs (Figure 6; Table S8). The identified colocalization genetic variants between metabolites and advanced AMD were most commonly located in or near APOE, ABCA1, LIPC, and CETP. For example, the SNP rs429358 at the APOE locus was shared between advanced AMD and 47 metabolites, of which 81% were lipid metabolites (Table S8). Overall, we found that 90 (79%) metabolites sharing causal variants with AMD were from lipid pathways, supporting the important role of lipid metabolites in AMD risk (Figure 6).
Figure 6.
Colocalization analysis identifies 114 shared causal variants between metabolite and AMD pairs
The x axis displays the nearest gene name for each shared variant between metabolites and AMD. The y axis represents the number of genes identified in the colocalization analysis. “AMD index SNP” refers to the colocalization analysis for AMD SNPs within a 2 Mb genomic window, and “metabolite index SNP” pertains to the colocalization analysis for metabolite SNPs within a 2 Mb genomic window.
MWAS
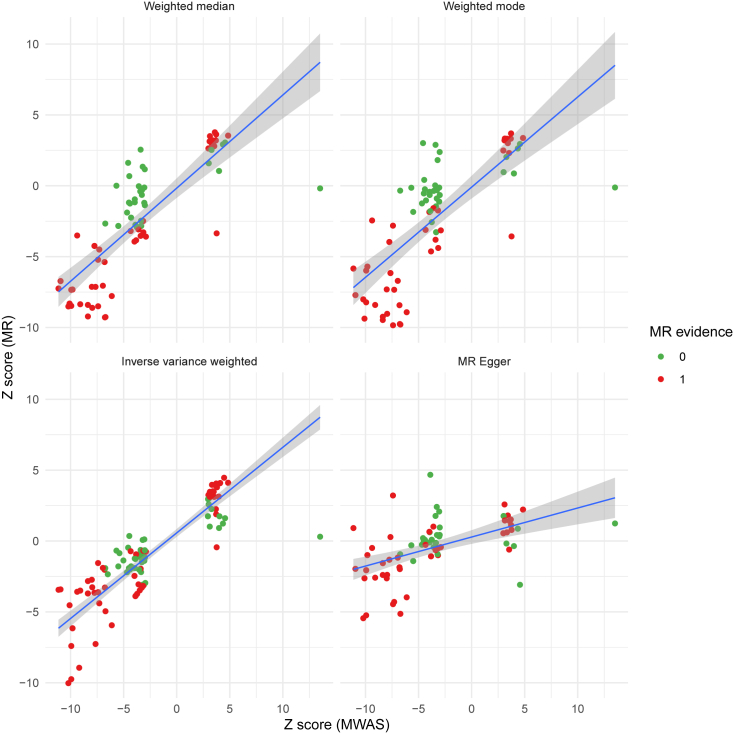
We developed a MWAS pipeline to identify metabolite-AMD associations. From MWAS, we identified 155 plasma metabolites whose combined genetic effects were associated with risk of advanced AMD after adjusting for multiple testing (FDR-corrected p < 0.05; Table S9). Of the 155 metabolites, 96 were also associated with advanced AMD in the MR analysis, and all but one showed a consistent direction of effect sizes between MWAS and the four MR methods (weighted median, weighted mode, and inverse-variance weighted MR methods) (Figure 7). As with the MR analysis, from MWAS, we identified very few urine metabolites that were associated with advanced AMD (Table S10).
Figure 7.
Comparison of results from metabolome-wide association study and Mendelian randomization analysis
The four panels depict the Z scores (association Z statistics) from the MWAS and four MR methods (MR effect sizes divided by standard errors from weighted median, weighted mode, inverse-variance weighted method, and MR-Egger). The x axis shows the Z scores for 155 metabolites that were associated with AMD risk in the MWAS analysis after adjusting for multiple testing. The y axis presents the Z scores from MR analysis. An MR evidence of 1 (displayed in red) indicates that a metabolite is associated with AMD risk from MR analysis after adjusting for multiple testing.
Association of metabolites with AMD subtypes and lipid biomarkers
We systematically evaluated the association of the 108 plasma metabolites identified from the MR analysis in the plasma meta-analysis with four AMD subtypes (CNV, GA, intermediate, and mixed AMD types) and six lipid-related biomarkers (ApoA1, ApoB, CHOL, HDL-C, direct LDL-C, and TG). In the AMD subtype analysis, most identified metabolites for advanced AMD were also significantly associated with CNV AMD, with a high concordance of Z scores (association effect sizes divided by standard errors; Figure S8). For GA, intermediate, and mixed AMD subtypes, the concordances of Z scores with advanced AMD were also high; however, there were substantially fewer significant metabolites, reflecting the much smaller sample size in these AMD subtypes. We also observed that most of the 108 metabolites were associated with the six lipid-related biomarkers (Figure S9), indicating the metabolites are likely to be involved in lipid pathways for the risk of AMD.
Discussion
In this large-scale integrative genetic and metabolomic analysis based on metabolite data from six multi-ancestry studies, including up to 28,000 participants, we identified novel metabolites associated with AMD. Our results demonstrated high concordance of mQTL effect sizes across different ancestries and metabolomic platforms but lower concordance between urine and plasma. This robust mQTL database enabled us to undertake an MR approach to evaluate the causal associations between metabolites and AMD. After accounting for multiple testing, we identified 108 metabolites with potential causal effects on advanced AMD. We further employed colocalization analysis to uncover 114 genetic variants shared between metabolite and advanced AMD pairs, where the most frequent causal variants belong to lipid pathways. The MWAS analysis illustrated that 155 metabolites were associated with advanced AMD, of which 96 metabolites were also significant in the MR analysis. The identified metabolites were enriched in pathways of glycerophospholipid metabolism, lysophospholipid, triradylcglycerols, and long-chain PUFAs. Finally, we systematically evaluated the association of the identified metabolites with different AMD subtypes and lipid biomarkers. To our knowledge, this study presents the first comprehensive large-scale mGWAS in multi-ancestry and multi-fluid datasets to establish putative causal associations of metabolites with risk of AMD.
In the mGWAS, we demonstrated high concordance of genetic effects for metabolites across different ancestries, indicating that the genetic determinants of metabolites in European and Hispanic populations largely overlap. Consistent with a recently published cross-platform mGWAS,15 our mQTLs from different metabolomic platforms based on different instruments (the Broad versus Metabolon) also exhibited high concordance, supporting the use of large-scale data from different studies in this work. Notably, while the correlation between mQTL effect sizes was high from the same fluids, we observed lower concordance between plasma and urine, suggesting that the genetic associations of SNPs with metabolite measurements in plasma and urine are at least partly distinct, yet these results may also be influenced by statistical power.
Despite previous reports of associations between AMD risk SNPs and plasma metabolites, no work has followed a genome-wide approach.18,19,31 For example, we previously assessed AMD risk SNPs and identified 28 significant mQTLs in the LIPC and ASPM genes associated with phosphatidylethanolamine and branched-chain amino acid metabolites.19 Another study from the EYE-RISK Consortium reported 60 metabolites phenotypically associated with AMD, and seven AMD risk SNPs were associated with 34 of the identified metabolites.18 In the current study, we performed a large-scale mGWAS across different ancestries and biofluid samples instead of restricting SNPs to only AMD risk variants (approximately 50 lead SNPs). This approach allowed us to characterize the genetic architecture of metabolites, enabling causal inference based on MR principles and a formal colocalization analysis to provide a comprehensive view of shared genetic effects between metabolites and AMD. For example, SNP rs429358 at the APOE locus is shared between AMD and 47 unique metabolites. As reported, rs429358 is a well-established AMD risk variant (p = 2.39 × 10−42).7 In this study, metabolites with the highest posterior probability of sharing rs429358 with AMD risk were primarily lipid metabolites, such as N-palmitoyl-sphingosine (d18:1/16:0), 1-stearoyl-2-oleoyl-GPE (18:0/18:1), and 1-stearoyl-GPI (18:0).
Crucially, previous metabolite-AMD association studies have not included statistical methods designed to address causality. In this work, the large-scale mGWAS enabled a bidirectional two-sample MR approach to investigate putative causal relationships between metabolites and AMD, overcoming limitations of observational association studies prone to confounding factors and reverse causality. From the MR analysis, we identified 108 metabolite-AMD associations enriched in glycerophospholipid metabolism pathways (i.e., glycerophosphoethanolamine and glycerophosphocholine pathways), lysophospholipid, triradylcglycerols, long chain PUFAs, and plasmalogen. Glycerophospholipids play an important role in structural and functional components of biological membranes and serum lipoproteins.32 Most associated metabolites in glycerophospholipid metabolism were negatively associated with AMD risk, suggesting that higher metabolite levels protect against AMD. This observation aligns with our previous work, which demonstrates significant dysregulation of glycerophospholipid metabolism in AMD.33 A recent study in patients treated with anti-VEGF indicated lower glycerophosphocholine levels in the treatment responder group compared with the treatment non-responder group.34 Additionally, we observed that most of these 108 metabolites were associated with six lipid-related biomarkers (such as HDL-C and LDL-C). Notably, metabolites including lysoplasmalogens, 1-(1-enyl-palmitoyl)-GPC (P-16:0), 1-(1-enyl-palmitoyl)-GPE (P-16:0), and 1-(1-enyl-stearoyl)-GPE (P-18:0), transported in lipid particles, are potential putative causal metabolites associated with AMD risk. Altogether, these findings support the important role of glycerophospholipids in AMD risk susceptibility, disease severity, and treatment response. The association of hepatic lipase generation of lysophospholipids with AMD possibly involves complement activation.35 In this study, we also identified PUFAs and plasmalogen (reservoirs of PUFAs), which may exhibit anti-oxidative and anti-inflammatory effects in retinal functions.36 Further functional experiments are warranted to investigate the role of the identified pathways in AMD risk.
A major concern in MR analysis is the possibility of pleiotropic effects of genetic instruments violating MR assumptions.37 For instance, it is possible that a subset of metabolite SNPs might associate with other AMD risk factors through measured or unmeasured confounders, violating one of the MR assumptions.38 Two types of pleiotropy exist in MR analysis: vertical pleiotropy and horizontal pleiotropy.39 In vertical pleiotropy, genetic variants associate with multiple traits on the same pathway, not invalidating the MR assumption, while horizontal pleiotropy indicates genetic variants affecting multiple traits through separate pathways (confounders). We observed that metabolites in the same pathway are more likely to share the same mQTLs, which could be classified as vertical pleiotropy. However, pathways observed through metabolomic platforms do not always correspond to the same biological pathways, leading to the possibility of horizontal pleiotropy effects. To address this concern, we performed various MR sensitivity analyses in the current study. For instance, the MR-Egger method can model directional pleiotropy (non-zero intercept term), and weighted model MR and weighted median MR require only a subset or up to 50% weights from valid variants. We have shown that different MR sensitivity analyses provided consistent estimates and directions (Figure 4A; Table S4).
A key strength of this study is that we performed large-scale mGWASs in different ancestries and biofluid samples, enabling a comprehensive evaluation of the shared genetic components between metabolites and AMD using multiple genetic approaches. We demonstrated that causal metabolite findings were consistent across European and Hispanic ancestral populations. The bidirectional MR approach can delineate association directions and is less likely to be biased by confounding factors and reverse causality.
Limitations of the study
Our results should be interpreted in light of the study limitations. First, in the MR analysis, a concern is the existence of horizontally pleiotropic effects of genetic instruments, where a genetic variant may influence AMD independently of the metabolite exposure. To address this concern, we showed that different MR sensitivity analyses provided consistent estimates and directions (Figure 4A; Table S4). Second, the effect of an exposure is assumed to be stable across a lifetime, meaning the temporary and short-term effects of metabolites on AMD risk would not be reflected. Third, metabolite measurements in this study are from different populations, biological samples, and measurement platforms. Consequently, not all plasma metabolites in the European population were available in the Hispanic population or urine data. However, using overlapping metabolites, the replication of MR findings in different ancestries provided evidence supporting the generalizability of AMD-associated metabolites. In this study, we found limited evidence of associations between urine metabolites and AMD risk, potentially reflecting the smaller sample size of urine mGWASs and the absence of links between urine and ocular tissue. While our study identified a link between lipid-related metabolites and AMD risk using plasma metabolites, it is important to recognize that circulating metabolite concentrations may not accurately reflect the metabolic pathways associated with AMD in the eye. Further research is needed to investigate the influence of metabolites in the retina tissue on AMD risk. Lastly, most study participants were of European descent. To obtain a more comprehensive understanding of the impact of genetic variations on metabolite measurements and AMD risk, it is essential to conduct additional studies that include individuals of Asian and African ancestries.
In conclusion, this study showcases an extensive integrative analysis of multi-omics data pertaining to AMD risk. The metabolites identified with potential causal impact on AMD, along with the shared causal genetic variants between these metabolites and AMD and its subtypes, offer novel insights into the pathogenesis of AMD.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| metabolomicsR | Han et al.40 | https://github.com/XikunHan/metabolomicsR |
| Plink1.9 | Purcell, S. et al.41 | https://www.cog-genomics.org/plink/1.9 |
| REGENIE v2.2.4 | Mbatchou, J. et al.42 | https://rgcgithub.github.io/regenie/ |
| RVTESTS | Zhan et al.43 | http://zhanxw.github.io/rvtests/ |
| TwoSampleMR | Hemani et al.44 | https://mrcieu.github.io/TwoSampleMR/articles/introduction.html |
| coloc | Giambartolomei et al.24 | https://github.com/chr1swallace/coloc |
| METAL | Willer et al.45 | https://genome.sph.umich.edu/wiki/METAL_Documentation |
| mRnd | Brion et al.46 | https://shiny.cnsgenomics.com/mRnd/ |
| FUSION | Gusev et al.26 | http://gusevlab.org/projects/fusion/ |
| MetaboAnalyst 5.0 | Pang et al.47 | https://www.metaboanalyst.ca/MetaboAnalyst/ModuleView.xhtml |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Liming Liang (lliang@hsph.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ethics statement
The study protocol of the Nurses’ Health Study and Nurses’ Health Study II were approved by Institutional Review Boards at both the Harvard T. H. Chan School of Public Health and Brigham and Women’s Hospital; and Health Professionals Follow Up Study was approved by the Harvard T. H. Chan School of Public Health Institutional Review Board. Although these studies were conducted before the requirement for written informed consent, IRBs approved that receipt of the questionnaires and blood samples implied consent.
The CLSA abides by the requirements of the Canadian Institutes of Health Research (CIHR). The protocol of the CLSA has been reviewed and approved by 13 research ethics boards across Canada. A complete and detailed list is available at: https://www.clsa-elcv.ca/participants/privacy/who-ensures-high-ethical-standards/research-ethics -boards.
The Hispanic Community Health Study/Study of Latinos study was approved by institutional review boards at participating institutions. Written informed con-sent was obtained from all participants.
The clinical protocol of AMD Biomarkers Study was conducted in accordance with Health Insurance Portability and Accountability Act requirements and the tenets of the Declaration of Helsinki, and was approved by the Institutional Review Boards at Mass Eye and Ear, Mass General Brigham, FMUC, and the Portuguese National Data Protection Committee. All participants enrolled in the study provided written informed consent.
Method details
In the current study, we included large-scale genetic and metabolomic studies. Detailed information for each study is described below.
The nurses’ health study, nurses’ health study ii, and health professionals follow up study
The NHS, NHSII, and HPFS are three large longitudinal studies comprising approximately 290,000 participants with extensive exposures and outcomes followed for more than 40 years. The detailed descriptions of the study design, data collection, and genotyping have been described previously.48,49,50 Briefly, the NHS was launched in 1976 with 121,701 female nurses aged between 30 and 55 years in the United States; the NHSII started in 1989 involving 116,429 younger female nurses aged between 25 and 42 years at baseline; and the HPFS was established in 1986 and consisted of 51,529 men aged 40 to 75 years at baseline.
Genetic data
In the NHS, NHSII, and HPFS, merging, quality control, and imputation procedures of genetic datasets were previously described.50 Genotype data were available from five genotype platforms after merging genetic data using overlap SNPs. The Haplotype Reference Consortium Release 1.1 data were used as the reference panel for imputation. In this study, we filtered variants with minor allele frequency (MAF) > 0.01 and imputation quality score >0.6, resulting in approximately 8 million SNPs available for metabolite GWAS.
Metabolomic data
The measurements of 346 named plasma metabolites were retrieved from previous sub-studies based on the NHS, NHSII, and HPFS as reported elsewhere.51 Metabolites were measured at the Broad Institute of Harvard University and MIT (Cambridge, MA) using high-throughput liquid chromatography-tandem mass spectrometry (LC-MS) technique. Quality control and standardization procedures were previously described.51 Briefly, we removed low-quality measurements (individuals or metabolites with <75% detection rate, or metabolites with no between-person variations or with an intraclass correlation coefficient <0.4) and imputing missing values for each metabolite with the half minimum values. In the present study, participants with both genetic and metabolomic data were included in the subsequent association analysis to identify genetic variants associated with metabolites. The median sample size of GWAS for the 346 metabolites was 6,610 participants (interquartile range [IQR]: 3,829 - 6,961).
Metabolite-based genome-wide association studies (mGWAS)
The mGWAS were performed in the NHS, NHSII, and HPFS within each genotype platform using the RVTESTS software (version 20190205).43 For each metabolite, after adjusting for age, fasting status, cohort (NHS, NHSII, and HPFS), indicators of subcohort outcome for both the original genetic and metabolomic studies, batch effects in metabolomics, and the first four genetic principal components, the residual phenotypes were inversely normal transformed to obtain score statistics (--inverseNormal and --useResidualAsPhenotype in RVTESTS).43 The mGWAS summary statistics were meta-analyzed across different genotype platforms based on the inverse variance-weighted method (METAL software).45
The canadian longitudinal study on aging (CLSA)
The Canadian Longitudinal Study on Aging (CLSA) is a national, longitudinal study encompassing 51,338 participants aged between 45 and 85 years from 10 Canadian provinces.27,28
Genetic data
In the CLSA, genetic data were available for 26,622 participants (version 3) genotyped for 794,409 markers on the Affymetrix Axiom array and were further imputed to approximately 308 million SNPs based on the TOPMed reference panel.52 The genetic quality assessment and imputation procedures were described elsewhere.52 In the current study, we removed non-European ancestry participants based on genetic principal components, and 9,021 participants with both genetic and metabolomic data were included in the genetic association analysis.53 SNPs with MAF >0.01 and imputation quality score >0.6 were retained for association tests.
Metabolomic data
In the CLSA, the metabolomic data were profiled by Metabolon using non-targeted LC-MS. In total, 1,458 metabolites were measured in 10,204 participants. In this study, no sample was removed for missing rate >50%, and no samples had outliers >6 standard deviation in the first two metabolite principal components. We observed that 36 metabolites have a missing rate >90%. With a missing rate at 10%, the effective sample is still approximately 1,000, therefore, these metabolites were included in the following GWAS. 21 metabolites were removed with no variance. In this study, 144 samples were measured in each batch. A batch-normalization method was applied to adjust for batch effects. The missing values were imputed using the minimum value in each batch. The measurements of metabolites were transformed using the rank-based inverse normal method to interpret the metabolites in one standard deviation unit. We included 1,437 metabolites from 9,021 European ancestry participants based on the batch-normalized data in the genetic association tests.
mGWAS
The association tests were performed in REGENIE (v2.2.4)42 to run GWAS for 50 metabolites in each job. In the regression models, we adjusted for age, gender, fasting time, genetic batch, and the first ten genetic principal components.
Plasma metabolite study in finnish
Genetic associations for 1391 plasma metabolites in 6136 Finnish men were recently performed from the METabolic Syndrome In Men (METSIM) study.29 Briefly, The genotype data (OmniExpress platform) were imputed using the METSIM integrative panel genome sequence data. 1,544 plasma metabolites were assayed by Metabolon using non-targeted LC-MS/MS, and 1,391 metabolites quantified were included in the mGWAS. For each metabolite, inverse normalized metabolite residual values were used in association tests by linear mixed models.29 The detailed genetic data, metabolite data, and association analysis were described elsewhere.29
The hispanic community health study/study of latinos
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is a community-based cohort study comprising 16,415 Hispanic adults aged between 18 and 74 years in the United States.54 The study design, metabolite measurements, and genetic data have been described elsewhere.30,54 Briefly, the metabolite measurements were quantified using untargeted LC-MS at Metabolon (Durham, NC). We included only known metabolites with missing rates ≤25%. The participants of HCHS/SOL were genotyped on a customized Illumina array and were further imputed based on the 1000 Genomes Project phase III reference panel. SNPs were filtered with MAF ≥1% and imputation quality ≥0.3 for the genetic association analyses. In the current study, we included mGWAS summary statistics for 640 plasma metabolites in a random subset of 3,926 participants.30
Urine metabolite GWAS from schlosser et al.
A previous study reported urinary mGWAS for 1,172 metabolites among 1,627 patients with reduced kidney function.16 The metabolites were quantified using non-targeted MS analysis performed by Metabolon using ultra-high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) method.16 In this study, we obtained urinary mGWAS summary statistics.
AMD biomarkers study
We developed a cross-sectional observational study including AMD cases and controls at two sites: the Department of Ophthalmology at Massachusetts Eye and Ear (Mass Eye and Ear) and Harvard Medical School (Boston, MA), and the Faculty of Medicine of the University of Coimbra (FMUC) and the “Centro Hospitalar e Universitário de Coimbra” (Coimbra, Portugal). The clinical protocol was conducted in accordance with Health Insurance Portability and Accountability Act requirements and the tenets of the Declaration of Helsinki, and was approved by the Institutional Review Boards at Mass Eye and Ear, Mass General Brigham, FMUC, and the Portuguese National Data Protection Committee. All participants enrolled in the study provided written informed consent.
Sample collection
The detailed study protocol for blood and urine samples have been described elsewhere.19,55 In summary, patients diagnosed with AMD, as well as participants with no evidence of AMD, aged more than 50 years were included at both study sites (Boston and Portugal) and underwent a complete ophthalmological exam for phenotypic characterization. For all participants, overnight fasting samples were collected the next morning. Two blood samples were collected: one into a sodium-heparin tube that was centrifuged within 30 min to obtain plasma for metabolomic analysis and the other into an ethylenediaminetetraacetic acid tube that was used for DNA extraction.19 We also collected urine samples into sterile cups and then stored into sterile cryovials of 1.5 mL (MEE) and 5 mL (FMUC/AIBILI), which were stored at −80°.55
Metabolomic data
The non-targeted MS analysis (UPLC-MS/MS) was used for metabolomic profiling by Metabolon.19,55 The standard quality control and data processing pipeline were used for metabolite measurements, with 907 plasma metabolites and 1,417 urine metabolites included in the current analysis after batch normalization. Briefly, we removed metabolites with missing rate >50% or no variance. Measurement outliers were defined as values >5 standard deviation and were winsorized. Datasets were merged after batch normalization and missing values were imputed with half of the minimum value in each batch. The rank-based inverse normal transformation algorithm was applied to the metabolite measurements.
Genetic data
Genetic profiling was performed at the Broad Genomics Institute using the OmniExpress array. Before imputation, we performed genetic quality control in both variant and sample levels. Briefly, we excluded variants with MAF less than 1%, p values less than 1 × 10−5 from Hardy-Weinberg equilibrium tests, and missing rates greater than 5%. Non-European participants were removed based on self-reported ancestry information and genetic principal components. The Michigan Imputation Server was used for imputation with the 1,000 Genomes Project Phase III as the reference panel. Approximately 8.6 million SNPs with MAF >0.01 and imputation quality score >0.6 were retained for association analysis.
mGWAS
In the AMD Biomarkers Study, after quality control of metabolomics and genetic data, for plasma mGWAS, 183 participants from Boston, MA and 286 from Coimbra, Portugal were included (total sample size 647). For urine mGWAS, 183 participants from Boston, MA and 286 from Coimbra, Portugal were included (total sample size 445). We performed mGWAS separately for Boston, MA and Coimbra, Portugal separately using the PLINK software (version 20190205).41 Gender, age, and the first ten genetic principal components were adjusted for in the association tests. We then meta-analyzed the mGWAS summary statistics from the two sub-cohorts using the inverse variance-weighted method (METAL software).45
The International AMD Genomics Consortium
The International AMD Genomics Consortium (IAMDGC) has reported a large AMD GWAS collected from 26 studies, including 16,144 advanced AMD cases and 17,832 controls of European ancestry.7 Cases of advanced AMD included both geographic atrophy and/or choroidal neovascularization, and the controls were without any known advanced or intermediate AMD. In the present study, AMD GWAS summary statistics of 12,023,117 SNPs were available in our analysis. We also included different AMD subtype GWAS, including 5,336 intermediate AMD cases, 8,544 CNV cases, 2,656 GA cases, and 1,511 mixed AMD cases with both CNV and GA (Supplementary Table 3). The detailed diagnosis criteria of advanced AMD and different AMD subtypes were described elsewhere.7,10,11 Briefly, advanced AMD included choroidal neovascularization and/or geographic atrophy in at least one eye and age at first diagnosis more than 50 years old; intermediate AMD cases had pigmentary changes in the retinal pigment epithelium or more than five macular drusen greater than 63 μm in diameter and age at first diag-nosis more than 50 years old.7,10,11
UK biobank lipid biomarker GWAS
To evaluate the effects of metabolites on lipid related biomarkers, we included 6 lipid related biomarkers: apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), total cholesterol (CHOL), high-density lipoprotein cholesterol (HDL-C), direct low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG).11 The GWAS were performed on approximately 400,000 participants in the UK Biobank. The detailed descriptions of UK Biobank, the lipid data, and GWAS analyses were described elsewhere.11
Quantification and statistical analysis
Metabolite-based genome-wide association studies and meta-analysis
The mGWAS for each study was described above. mGWAS summary statistics were meta-analyzed based on the inverse variance-weighted method (METAL software).45 For plasma meta-analysis, we included plasma metabolites from NHS/NHSII/HPFS, CLSA, METSIM, HCHS/SOL, and AMD Biomarker Study (Boston and Portugal plasma data). For urine meta-analysis, we included urine metabolites from Schlosser et al. (urine), AMD Biomarker Study (Boston and Portugal urine data). The detailed information of sample size and number of metabolites were available in the Supplementary Table 2. Genome-wide significant SNPs were obtained using the clumping method in PLINK 1.9 software (MAF >0.01, p value <5 × 10−8, r2 < 0.01, and a window of 1Mb).41 The European ancestry participants from 1000 Genomes Project phase III were used as the reference panel.
Bidirectional mendelian randomization analysis
We performed bidirectional MR to evaluate the associations between metabolites and risk of AMD (R package TwoSampleMR, version 0.5.6).44 For metabolites, top independent SNPs were selected using the PLINK software (MAF >0.01, p value <5 × 10−8, r2 < 0.01, and a window of 1Mb).41 We utilized the mRnd method (http://cnsgenomics.com/shiny/mRnd/) to evaluate the power of our MR analyzes across various AMD subtypes.46 We conducted a series of MR analyses with different assumptions and strengths to investigate the putative causal associations between metabolites and AMD in the two-sample MR framework, including inverse-variance weighted method (MR-IVW), weighted median, weighted mode, and MR-Egger.37,56,57 Some MR methods (MR-Egger weighted mode, weighted median) requiring at least three genome-wide significant SNPs as genetic instruments.58 For metabolites with only one SNP as the genetic instrument, the Wald ratio method was used in the MR analysis. To assess the potential reverse causality, we selected top genetic risk variants for AMD and calculated the effects on each metabolite to perform reverse-directional MR analysis (AMD precedes and leads to changes in metabolites instead of vice versa). To adjust for multiple testing, we used false discovery rate (FDR, Benjamini & Hochberg method) < 5% as the significant threshold to account for multiple MR tests and metabolites.59
Colocalization analysis
A Bayesian colocalization approach by the coloc package (R package, version 5.1.0) was used to identify shared genetic variants for metabolites and AMD.24 In the Bayesian colocalization analysis, the posterior probabilities were calculated for the following five hypotheses: H0, no association with either trait; H1 and H2, association with trait 1 or trait 2; H3, association with both traits but two independent SNPs; and H4: association with both traits with a shared variant. A posterior probability for H4 (PP4) more than 0.8 supports a shared causal variant affecting both metabolite and AMD. To identify the shared variants in the colocalization analysis, we tested independent metabolite SNPs (p < 5 × 10−8) and AMD SNPs (p < 1 × 10−6) with a genomic window of 2 Mb of the independent SNPs.
Metabolome-wide association study
The metabolome-wide association studies (MWAS) pipeline was adapted from the transcriptome-wide association analysis (FUSION) to leverage the metabolites and AMD GWAS summary statistics identifying metabolite-AMD associations.26 In the TWAS approach, the individual level data of genotypes and gene expression measurements in a training sample were used to model genetically regulated gene expression.25,26 With a reference panel of genotype, TWAS can impute gene expression in an outcome dataset with only GWAS summary statistics, allowing to evaluate the association between gene expressions and the outcome. In our MWAS analysis, the individual level data were not available for all studies, therefore, we applied a clump method to select independent genome-wide significant SNPs (r2 = 0.01, p value <5 × 10−8) for prediction (a polygenic risk score approach), imputed the metabolite levels into the AMD GWAS summary statistics, and performed MWAS to identify significant metabolite-AMD associations.25,26,60 In our sensitivity analysis, we modeled two other approaches: the single most significantly associated SNP (top1) and all independent SNPs with p values <1 × 10−5 when there were no genome-wide significant metabolite SNPs available. The FDR (Benjamini & Hochberg method) corrected p value <5% was used to control for multiple testing.
Pathway analysis
We counted the number of significant metabolites from MR analysis in each pathway based on available pathway annotation information. Then we performed chemical classification enrichment statistical analysis using the Kolmogorov-Smirnov test. Enrichment significance was calculated based on the chemical similarity of these metabolites (ChemRICH method).47,61
Acknowledgments
We would like to thank the research participants in the NHS, NHSII, and HPFS, the CLSA, the METSIM, the HCHS/SOL, the urinary metabolomic data from Schlosser et al.,16 and the AMD Biomarkers Study for making this work possible.
The opinions expressed in this manuscript are the authors’ own and do not reflect the views of the CLSA or any affiliated institution. This research was made possible using the data/biospecimens collected by the CLSA. Funding for the CLSA is provided by the government of Canada through the Canadian Institutes of Health Research (CIHR) under grant reference LSA 94473 and the Canada Foundation for Innovation, as well as the following provinces: Newfoundland, Nova Scotia, Quebec, Ontario, Manitoba, Alberta, and British Columbia. This research has been conducted using a CLSA dataset (Baseline Comprehensive Dataset v.6.0, Follow-up 1 Comprehensive Dataset v.3.0, Genome-wide Genetic Data v.3.0, and Metabolomics v.1.0) under application number 2006016. The CLSA is led by Drs. Parminder Raina, Christina Wolfson, and Susan Kirkland.
The HCHS/SOL is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I/N01-HC-65233), the University of Miami (HHSN268201300004I/N01-HC-65234), the Albert Einstein College of Medicine (HHSN268201300002I/N01-HC-65235), the University of Illinois at Chicago (HHSN268201300003I/N01- HC-65236 Northwestern University), and San Diego State University (HHSN268201300005I/N01-HC-65237). The following institutes/centers/offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: the National Institute on Minority Health and Health Disparities, the National Institute on Deafness and Other Communication Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Support for metabolomics data was graciously provided by the JLH Foundation (Houston, TX, USA).
The authors thank all of the participants who took part in the UK Biobank and the International AMD Genomics Consortium and support staff who made this study possible. This lipid GWAS work was conducted using the UK Biobank Resource (application number 25331). For the AMD datasets, all contributing sites and additional funding information are acknowledged in this publication: Fritsche et al.,7 The International AMD Genomics consortium’s web page is http://eaglep.case.edu/iamdgc_web/, and additional information is available at http://csg.sph.umich.edu/abecasis/public/amd2015/. The AMD case-control datasets used for the analyses described in this manuscript were obtained from the NEI Study of Age-Related Macular Degeneration (NEI-AMD) Database found at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001039.v1.p1 through dbGaP accession number 20740. Funding support for NEI-AMD was provided by the National Eye Institute. We would like to thank NEI-AMD participants and the NEI-AMD Research Group for their valuable contribution to this research.
S.M. is supported by a research fellowship and program grant (1150144) from the Australian National Healthand Medical Research Council (NHMRC). The Richards research group is supported by the Canadian Institutes of Health Research (CIHR: 365825, 409511, and 100558), the Lady Davis Institute of the Jewish General Hospital, the Canadian Foundation for Innovation, the NIH Foundation, Cancer Research UK, Genome Québec, the Public Health Agency of Canada, Genome Québec, McGill University, and the Fonds de Recherche Québec Santé (FRQS). J.B.R. is supported by an FRQS Clinical Research Scholarship. Support from Calcul Québec and Compute Canada is acknowledged. This work was supported by Cancer Research UK (grant number C18281/A29019). TwinsUK is funded by the Wellcome Trust, the Medical Research Council, the European Union, the National Institute for Health Research (NIHR)-funded BioResource, and the Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. This work is supported by MSL Renewed Hope Foundation and Ines and Fredrick Yeatts Foundation Award. These funding agencies had no role in the design, implementation, or interpretation of this study.
The NHS study is funded by UM1 CA186107 and R01 CA49449, the NHSII is funded by U01 CA176726 and R01 CA67262, and the HPFS study is funded by U01 CA167552.
This study was supported by RO1EY030088 (D.H.), the Miller Retina Research Fund (Mass. Eye and Ear), the Champalimaud Vision Award (Joan Miller), the Portuguese Foundation for Science and Technology/Harvard Medical School Portugal Program (HMSP-ICJ/006/2013) (I.L.), and R01 AR049880 (K.C.).
The opinions expressed in this manuscript are the author’s own and do not reflect the views of the CLSA.
Author contributions
L.L., X.H., and D.H. conceived the study. X.H. conducted the data analysis and drafted the initial version of the manuscript. X.H., I.L., Jun Li, B.Y., J.B.R., D.H., and L.L. contributed to data collection. Jun Li prepared the metabolite phenotypic data for the NHS/NHSII/HPFS cohorts. X.H., I.L., J.B.R., S.M., D.H., and L.L. interpreted the data and provided critical revisions to the manuscript. All authors contributed to and critically reviewed the manuscript.
Declaration of interests
J.B.R.’s institution has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline, and Biogen for projects unrelated to this research. He is the CEO of 5 Prime Sciences (www.5primesciences.com), which provides research services for biotech, pharma, and venture capital companies for projects unrelated to this research. J.L.-S. is a scientific advisor to Precion Inc. D.G.V. is a consultant for Sumitomo/Sunovion, Inhibikase, OLix Pharma, Twenty/Twenty, and Valitor.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 21, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101085.
Contributor Information
Xikun Han, Email: xikun_han@hsph.harvard.edu.
Deeba Husain, Email: deeba_husain@meei.harvard.edu.
Liming Liang, Email: lliang@hsph.harvard.edu.
Supplemental information
Data and code availability
-
•
UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. Data are available from the Canadian Longitudinal Study on Aging (www.clsa-elcv.ca) for researchers who meet the criteria for access to de-identified CLSA data (https://www.clsa-elcv.ca/researchers/data-support-documentation). The data generated from NHS/NHSII are not publicly available due to participant confidentiality and privacy concerns but are available upon request. Further information including the procedures to obtain and access data from the Nurses’ Health Studies is described at https://www.nurseshealthstudy.org/researchers. The leading SNPs of metabolites were available in Supplementary Table 7.
-
•
The following software packages were used for data analysis: metabolomicsR: https://github.com/XikunHan/metabolomicsR.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Friedman D.S., O’Colmain B.J., Muñoz B., Tomany S.C., McCarty C., de Jong P.T.V.M., Nemesure B., Mitchell P., Kempen J., Eye Diseases Prevalence Research Group Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 2.Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.-Y., Wong T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 4.Fleckenstein M., Keenan T.D.L., Guymer R.H., Chakravarthy U., Schmitz-Valckenberg S., Klaver C.C., Wong W.T., Chew E.Y. Age-related macular degeneration. Nat. Rev. Dis. Prim. 2021;7 doi: 10.1038/s41572-021-00265-2. 31-25. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarthy U., Wong T.Y., Fletcher A., Piault E., Evans C., Zlateva G., Buggage R., Pleil A., Mitchell P. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X., Gharahkhani P., Mitchell P., Liew G., Hewitt A.W., MacGregor S. Genome-wide meta-analysis identifies novel loci associated with age-related macular degeneration. J. Hum. Genet. 2020;65:657–665. doi: 10.1038/s10038-020-0750-x. [DOI] [PubMed] [Google Scholar]
- 7.Fritsche L.G., Igl W., Bailey J.N.C., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Q., Maranville J.C., Fritsche L., Sim X., Cheung C.M.G., Chen L.J., Gorski M., Yamashiro K., Ahn J., Laude A., et al. HDL-cholesterol levels and risk of age-related macular degeneration: a multiethnic genetic study using Mendelian randomization. Int. J. Epidemiol. 2017;46:1891–1902. doi: 10.1093/ije/dyx189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess S., Davey Smith G. Mendelian randomization implicates high-density lipoprotein cholesterol-associated mechanisms in etiology of age-related macular degeneration. Ophthalmology. 2017;124:1165–1174. doi: 10.1016/j.ophtha.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X., An Y., Zhou Y., Liu C., Yin W., Xia X. Using Mendelian randomization to evaluate the causal relationship between serum C-reactive protein levels and age-related macular degeneration. Eur. J. Epidemiol. 2020;13:139–146. doi: 10.1007/s10654-019-00598-z. [DOI] [PubMed] [Google Scholar]
- 11.Han X., Ong J.-S., Hewitt A.W., Gharahkhani P., MacGregor S. The effects of eight serum lipid biomarkers on age-related macular degeneration risk: a Mendelian randomization study. Int. J. Epidemiol. 2021;50:325–336. doi: 10.1093/ije/dyaa178. [DOI] [PubMed] [Google Scholar]
- 12.Patti G.J., Yanes O., Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laíns I., Gantner M., Murinello S., Lasky-Su J.A., Miller J.W., Friedlander M., Husain D. Metabolomics in the study of retinal health and disease. Prog. Retin. Eye Res. 2019;69:57–79. doi: 10.1016/j.preteyeres.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Lotta L.A., Pietzner M., Stewart I.D., Wittemans L.B.L., Li C., Bonelli R., Raffler J., Biggs E.K., Oliver-Williams C., Auyeung V.P.W., et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat. Genet. 2021;53:54–64. doi: 10.1038/s41588-020-00751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlosser P., Li Y., Sekula P., Raffler J., Grundner-Culemann F., Pietzner M., Cheng Y., Wuttke M., Steinbrenner I., Schultheiss U.T., et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat. Genet. 2020;52:167–176. doi: 10.1038/s41588-019-0567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nag A., Kurushima Y., Bowyer R.C.E., Wells P.M., Weiss S., Pietzner M., Kocher T., Raffler J., Völker U., Mangino M., et al. Genome-wide scan identifies novel genetic loci regulating salivary metabolite levels. Hum. Mol. Genet. 2020;29:864–875. doi: 10.1093/hmg/ddz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acar İ.E., Lores-Motta L., Colijn J.M., Meester-Smoor M.A., Verzijden T., Cougnard-Gregoire A., Ajana S., Merle B.M.J., de Breuk A., Heesterbeek T.J., et al. Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration: the EYE-RISK consortium. Ophthalmology. 2020;127:1693–1709. doi: 10.1016/j.ophtha.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Lains I., Zhu S., Han X., Chung W., Yuan Q., Kelly R.S., Gil J.Q., Katz R., Nigalye A., Kim I.K., et al. Genomic-metabolomic associations support the role of LIPC and glycerophospholipids in age-related macular degeneration. Ophthalmol. Sci. 2021;1:100017. doi: 10.1016/j.xops.2021.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laíns I., Duarte D., Barros A.S., Martins A.S., Carneiro T.J., Gil J.Q., Miller J.B., Marques M., Mesquita T.S., Barreto P., et al. Urine nuclear magnetic resonance (NMR) metabolomics in age-related macular degeneration. J. Proteome Res. 2019;18:1278–1288. doi: 10.1021/acs.jproteome.8b00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skrivankova V.W., Richmond R.C., Woolf B.A.R., Yarmolinsky J., Davies N.M., Swanson S.A., VanderWeele T.J., Higgins J.P.T., Timpson N.J., Dimou N., et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA. 2021;326:1614–1621. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- 23.Skrivankova V.W., Richmond R.C., Woolf B.A.R., Davies N.M., Swanson S.A., VanderWeele T.J., Timpson N.J., Higgins J.P.T., Dimou N., Langenberg C., et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. doi: 10.1136/bmj.n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., GTEx Consortium. Nicolae D.L., et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raina P.S., Wolfson C., Kirkland S.A., Griffith L.E., Oremus M., Patterson C., Tuokko H., Penning M., Balion C.M., Hogan D., et al. The Canadian longitudinal study on aging (CLSA) Can. J. Aging. 2009;28:221–229. doi: 10.1017/S0714980809990055. [DOI] [PubMed] [Google Scholar]
- 28.Raina P., Wolfson C., Kirkland S., Griffith L.E., Balion C., Cossette B., Dionne I., Hofer S., Hogan D., van den Heuvel E.R., et al. Cohort profile: the Canadian longitudinal study on aging (CLSA) Int. J. Epidemiol. 2019;48:1752–1753j. doi: 10.1093/ije/dyz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X., Chan L.S., Bose D., Jackson A.U., VandeHaar P., Locke A.E., Fuchsberger C., Stringham H.M., Welch R., Yu K., et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 2022;13:1644. doi: 10.1038/s41467-022-29143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feofanova E.V., Chen H., Dai Y., Jia P., Grove M.L., Morrison A.C., Qi Q., Daviglus M., Cai J., North K.E., et al. A genome-wide association study discovers 46 loci of the human metabolome in the hispanic community health study/study of Latinos. Am. J. Hum. Genet. 2020;107:849–863. doi: 10.1016/j.ajhg.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laíns I., Chung W., Kelly R.S., Gil J., Marques M., Barreto P., Murta J.N., Kim I.K., Vavvas D.G., Miller J.B., et al. Human plasma metabolomics in age-related macular degeneration: meta-analysis of two cohorts. Metabolites. 2019;9:127. doi: 10.3390/metabo9070127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hishikawa D., Hashidate T., Shimizu T., Shindou H. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 2014;55:799–807. doi: 10.1194/jlr.R046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laíns I., Kelly R.S., Miller J.B., Silva R., Vavvas D.G., Kim I.K., Murta J.N., Lasky-Su J., Miller J.W., Husain D. Human plasma metabolomics study across all stages of age-related macular degeneration identifies potential lipid biomarkers. Ophthalmology. 2018;125:245–254. doi: 10.1016/j.ophtha.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y., Teo Y.C.K., Beuerman R.W., Wong T.Y., Zhou L., Cheung C.M.G. A serum metabolomics study of patients with nAMD in response to anti-VEGF therapy. Sci. Rep. 2020;10:1341. doi: 10.1038/s41598-020-58346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma W., Paik D.C., Barile G.R. Bioactive lysophospholipids generated by hepatic lipase degradation of lipoproteins lead to complement activation via the classical pathway. Invest. Ophthalmol. Vis. Sci. 2014;55:6187–6193. doi: 10.1167/iovs.14-14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Leeuwen E.M., Emri E., Merle B.M.J., Colijn J.M., Kersten E., Cougnard-Gregoire A., Dammeier S., Meester-Smoor M., Pool F.M., de Jong E.K., et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye Res. 2018;67:56–86. doi: 10.1016/j.preteyeres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Burgess S., Bowden J., Fall T., Ingelsson E., Thompson S.G. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani G., Bowden J., Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 2018;27:R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X., Liang L. metabolomicsR: a streamlined workflow to analyze metabolomic data in R. Bioinform. Adv. 2022;2:vbac067. doi: 10.1093/bioadv/vbac067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mbatchou J., Barnard L., Backman J., Marcketta A., Kosmicki J.A., Ziyatdinov A., Benner C., O’Dushlaine C., Barber M., Boutkov B., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhan X., Hu Y., Li B., Abecasis G.R., Liu D.J. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brion M.-J.A., Shakhbazov K., Visscher P.M. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.É., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu F.B., Manson J.E., Stampfer M.J., Colditz G., Liu S., Solomon C.G., Willett W.C. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N. Engl. J. Med. 2001;345:790–797. doi: 10.1056/NEJMoa010492. [DOI] [PubMed] [Google Scholar]
- 49.Grobbee D.E., Rimm E.B., Giovannucci E., Colditz G., Stampfer M., Willett W. Coffee, caffeine, and cardiovascular disease in men. N. Engl. J. Med. 1990;323:1026–1032. doi: 10.1056/nejm199010113231504. [DOI] [PubMed] [Google Scholar]
- 50.Lindström S., Loomis S., Turman C., Huang H., Huang J., Aschard H., Chan A.T., Choi H., Cornelis M., Curhan G., et al. A comprehensive survey of genetic variation in 20,691 subjects from four large cohorts. PLoS One. 2017;12:e0173997. doi: 10.1371/journal.pone.0173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Guasch-Ferré M., Chung W., Ruiz-Canela M., Toledo E., Corella D., Bhupathiraju S.N., Tobias D.K., Tabung F.K., Hu J., et al. The Mediterranean diet, plasma metabolome, and cardiovascular disease risk. Eur. Heart J. 2020;41:2645–2656. doi: 10.1093/eurheartj/ehaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forgetta V., Li R., Darmond-Zwaig C., Belisle A., Balion C., Roshandel D., Wolfson C., Lettre G., Pare G., Paterson A.D., et al. Cohort profile: genomic data for 26 622 individuals from the Canadian Longitudinal Study on Aging (CLSA) BMJ Open. 2022;12:e059021. doi: 10.1136/bmjopen-2021-059021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han X., Steven K., Qassim A., Marshall H.N., Bean C., Tremeer M., An J., Siggs O.M., Gharahkhani P., Craig J.E., et al. Automated AI labeling of optic nerve head enables insights into cross-ancestry glaucoma risk and genetic discovery in >280,000 images from UKB and CLSA. Am. J. Hum. Genet. 2021;108:1204–1216. doi: 10.1016/j.ajhg.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaVange L.M., Kalsbeek W.D., Sorlie P.D., Avilés-Santa L.M., Kaplan R.C., Barnhart J., Liu K., Giachello A., Lee D.J., Ryan J., et al. Sample design and cohort selection in the hispanic community health study/study of Latinos. Ann. Epidemiol. 2010;20:642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lains I., Mendez K.M., Gil J.Q., Miller J.B., Kelly R.S., Barreto P., Kim I.K., Vavvas D.G., Murta J.N., Liang L., et al. Urinary mass spectrometry profiles in age-related macular degeneration. J. Clin. Med. 2022;11:940. doi: 10.3390/jcm11040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 60.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barupal D.K., Fiehn O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017;7:14567. doi: 10.1038/s41598-017-15231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. Data are available from the Canadian Longitudinal Study on Aging (www.clsa-elcv.ca) for researchers who meet the criteria for access to de-identified CLSA data (https://www.clsa-elcv.ca/researchers/data-support-documentation). The data generated from NHS/NHSII are not publicly available due to participant confidentiality and privacy concerns but are available upon request. Further information including the procedures to obtain and access data from the Nurses’ Health Studies is described at https://www.nurseshealthstudy.org/researchers. The leading SNPs of metabolites were available in Supplementary Table 7.
-
•
The following software packages were used for data analysis: metabolomicsR: https://github.com/XikunHan/metabolomicsR.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.