Abstract
PAX5, a member of the paired box gene family of transcription factors, is a B‐cell‐specific activator protein that plays important roles during B lymphopoiesis. Two putative PAX5 binding sites in the human GINS1 promoter region were identified. EMSA, ChIP and luciferase assay showed that PAX5 functions as a positive transcription factor for GINS1 expression. Furthermore, coordinated expression of PAX5 and GINS1 was observed in mice B cells under physiological conditions and LPS stimulation situations. A similar pattern was also observed in human DLBCL cell lines under differentiation‐inducing conditions. In addition, both PAX5 and GINS1 were highly expressed and significantly correlated in DLBCL specimens and cell lines. These findings suggested that dysregulation of PAX5 played an extremely important role in controlling the universal phenomenon of tumor progression through increased expression of GINS1 in DLBCL. In addition, circ1857 that was generated using back splicing of PAX5 pre‐mRNA could further stabilize GINS1 mRNA, modulate GINS1 expression and promote lymphoma progression. To the best of our knowledge, this report is the first to demonstrate the role of GINS1 in DLBCL progression, and the mechanism of GINS1 upregulation using both circ1857 and PAX5 in DLBCL was revealed. Our results suggested that GINS1 may be a possible therapeutic target for DLBCL.
Keywords: DLBCL, GINS1, hsa_circ_0001857, PAX5, PSF1
This report is the first to demonstrate the role of GINS1 in DLBCL progression, and the mechanism of GINS1 upregulation using both circ1857 and PAX5 in DLBCL was revealed. Our results suggest that GINS1 may be a possible therapeutic target for DLBCL.
Abbreviations
- 3′UTR
3′ untranslated regions
- ALL
acute lymphoblastic leukemia
- BSAP
B‐cell‐specific activator protein
- ChIP‐seq
ChIP followed by sequencing
- DLBCL
diffuse large B‐cell lymphoma
- GEPIA
Gene Expression Profiling Interactive Analysis
- GINS1
GINS (Go‐Ichi‐Ni‐San) Complex Subunit 1
- GINS2/3/4
GINS Complex Subunit 2/3/4
- IPTG
isopropyl‐β‐d‐thiogalactoside
- MACS
magnetic‐activated cell sorting
- PAX5
paired box 5
- PRDM1
PR domain zinc finger protein 1
- PSF1
Partner of SLD five 1
- R‐CHOP
rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone
- TCGA
The Cancer Genome Atlas
- TF
transcription factor
1. INTRODUCTION
DLBCL is the most common non‐Hodgkin lymphoma, accounting for 30–40% of new diagnoses. 1 The current R‐CHOP treatment is effective in 60–70% of patients. Approximately 30–40% of patients with DLBCL will relapse after first‐line chemotherapy and the prognosis is poor. 2 Finding novel targets and effective therapies for these DLBCL patients are still urgently needed.
GINS1 (also known as PSF1) is a member of the heterotetrameric GINS complex, which contains GINS1, GINS2, GINS3, and GINS4. 3 , 4 In eukaryotic cells, the GINS complex regulates both the initiation and progression of DNA replication. 5 , 6 Recently, there has been an increasing number of studies on GINS1 in malignant tumors. 7 , 8 , 9 , 10 In addition, GINS1 was highly expressed in several types of leukemias, and knockdown of GINS1 reduced the growth of AML and CML cells, GINS1 was suggested as a possible therapeutic target to enhance the effect of chemotherapy. 11 However, it remains unknown whether GINS1 is involved in DLBCL progression.
PAX5 (also known as BSAP) plays a decisive role in B‐cell proliferation and differentiation. 12 , 13 PAX5 represents a common deregulated molecule in multiple B‐cell malignancies. 14 , 15 , 16 In a significant number of these cancers, PAX5 has been reported to be either upregulated or mutated due to aberrant hypermutation. In lymphoplasmacytic lymphoma and a few cases of DLBCL, elevated expression of PAX5 is the result of chromosomal translocation that juxtapositions PAX5 to the IgH gene promoter. 17 In addition, PAX5 overexpression without translocation was also reported. 18 Moreover, the PAX5 gene has also been revealed to be a regulator in various non‐hematologic tissues and cancers including malignant neuroblastoma and pediatric brain tumors, where PAX5 expression positively correlates with cell proliferation. 19 More interestingly, it has been reported that PAX5 haploinsufficiency could restrain cell proliferation and induce the G0/G1 arrest of lymphoma cells. 20 In addition, a mutation in PAX5 known as PAX5 P80R was reported to mediate a favorable outcome for a subtype of B‐cell precursor ALL prognosis. 21 , 22 All these reports indicated that PAX5 expression might stimulate cells proliferation and contribute to enhanced cell survival.
In this study, a high expression of GINS1 in DLBCL was found and the mechanism of GINS1 upregulation was revealed. Both PAX5 and circ1857 (circBase ID: hsa_circ_0001857, back spliced from PAX5 pre‐mRNA) could promote GINS1 expression in a coordinated way. Our finding indicated that dysregulation of PAX5, the key transcription factor in the B‐cell lineage, plays an extremely important role in controlling the universal phenomenon of tumor progression through increased expression of GINS1 in DLBCL. Moreover, GINS1 regulated by PAX5 was also observed during B‐cell proliferation and differentiation under physiological conditions.
2. MATERIALS AND METHODS
2.1. Cell lines and cell culture
All the cell lines were confirmed using short tandem repeat (STR) profiling. IM‐9 (EBV‐transformed B lymphoblastoid cell line), Farage (a B lymphocyte cell line that was isolated in 1990 from a White, adult female patient with non‐Hodgkin's B‐cell lymphoma, DLBCL GCB subtype), DB (a B lymphoblast cell that was isolated from the ascites of a patient with large cell lymphoma, DLBCL GCB subtype) and SU‐DHL‐2 (a cell line that was isolated in 1974 from the lymph node of a White female with large cell lymphoma, DLBCL ABC subtype) were obtained from ATCC and cultured in RPMI1640 medium supplemented with 10% FBS. HEK293T was cultured in DMEM with 10% FBS.
2.2. Patients and tissues
Fresh human biopsy tissues were obtained from 30 patients with DLBCL collected in Nanjing First Hospital. Lymph node biopsy samples from lymphadenitis individuals (n = 24) were used as negative controls (Table S1).
2.3. RT‐qPCR
RNA isolation was performed using TRIzol reagent. cDNAs were synthesized using oligo(dT) and random hexamers. Real‐time PCR was performed. Relative expression levels of circRNA or mRNA were calculated using the 2−ΔΔCt method. Primer sequences are shown in Table S2.
2.4. Luciferase reporter assay
GINS1 promoter segments (−1500/+50; −550/+50; −400/+50) were obtained using PCR and inserted into pGL‐4.17 vectors to construct luciferase reporter plasmids (Table S3). To prepare mutated promoter plasmids, a mutation was created from a wild‐type promoter plasmid using PCR. Cells were transfected using electroporation and the AMAXA® Cell Line Nucleofector® Kit V (Lonza, Cologne, Germany). The used Nucleofector® programs were G‐016. HEK293T cells were transfected with Lipofectamine 2000. After transfection for 48 h, the luciferase activity of the cells was evaluated using a luciferase assay kit and a β‐galactosidase assay kit.
2.5. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay was performed as previously described. 23 Cells were fixed in 11% formaldehyde and then re‐suspended in cell lysis buffer. After sonication, the broken chromatin was subsequently incubated with an antibody at 4°C, then protein A+G agarose beads were added and incubated for 2 h. DNA fragments were purified and a PCR was performed. The primers used are shown in Table S4.
2.6. Electrophoretic mobility shift assay
Nuclear extracts were obtained as described above. 23 Synthetic biotin‐labeled oligonucleotides were synthesized and oligonucleotide sequences are shown in Table S4. The probes of annealed oligonucleotides were subsequently incubated with nuclear extract or purified protein. The DNA–protein complex was separated using 6% non‐denaturing polyacrylamide, and detected with HRP‐conjugated streptavidin.
2.7. PAX5 protein purification
Plasmids pET32a‐PAX5 WT or pET32a‐PAX5 P80R were transformed into E. coli BL21 and the expression of Re‐PAX5 protein (recombinant wild‐type PAX5) or Re‐PAX5 P80R protein (recombinant PAX5 P80R) was induced using IPTG. Then, the cultures were centrifuged, and the cell pellet was collected and sonicated. The inclusion body in the precipitate was dissolved in 8 mol/L urea, then recombinant proteins were refolded and purified.
2.8. Generation of stable cell lines
GINS1‐expressing (NM_021067.5) and PAX5‐expressing (NM_016734.3) plasmids were constructed based on pCDH‐CMV‐MCS‐EF1‐copGFP‐T2A‐puro. The circ1857 overexpression vector pLCDH‐ciR‐circ1857 was designed and exons 2–5 of PAX5 was inserted into pLCDH‐ciR that contained an autocyclization sequence (Figure S1). The shRNA expressing vectors were generated by inserting short, double‐stranded DNA oligos encoding a sense–loop–antisense sequence to the targeted gene in pLKO.1‐puro. shRNA target sequences are shown in Table S2. To produce the recombinant viruses, the above recombinant lentiviral vectors were co‐transfected with packaging plasmids into 293 T cells. The lentiviral particles were then used to infect cells in order to generate stable cell lines (Table S5). Briefly, cells were seeded at a density of 5 × 105 cells per well in a six‐well plate, and then infected with virus. Following a 24 h incubation, cells were washed to remove free virus and selection was started using fresh medium containing puromycin; a mixed population of drug‐resistant cells was obtained. To generate stable cell lines, monoclonal cell line screening with 1.5 μg/mL puromycin was performed.
2.9. Western blot
Western blot was conducted with antibodies specific for PAX5, GINS1, PRDM1, a‐tubulin and Flag (Table S6).
2.10. CCK‐8 assays
For the cell proliferation assay, cells were seeded into a 96‐well plate. Cell viability was determined using the Cell Counting Kit‐8 and according to the manufacturer's manual.
2.11. Soft agar colony assay
Cells were seeded into six‐well plates, with a 0.3% top agarose layer and a 0.5% bottom agarose layer in 2× RPMI‐1640 medium containing 20% FBS. Cell colonies were formed following incubation for 10 days. Thereafter, the agarose was fixed with paraformaldehyde, and stained with crystal violet. The number of clones was counted.
2.12. Xenograft lymphoma model
Six‐week‐old male BALB/c‐nude mice were purchased from Gempharmatech Company, China. Each male nude mouse was injected subcutaneously with 8 × 106 cells. Tumor size was monitored. Finally, the mice were anesthetized, and the tumors were obtained and weighed.
2.13. RNA stability assay
Cells were seeded into six‐well plates and incubated with 4.5 μg/mL actinomycin D for the indicated time and then cells were collected.
2.14. B‐cell subpopulation sorting
C57BL/6, 6–8 weeks of age, were obtained from Gempharmatech Company, China. Spleen and bone marrow were isolated and homogenized. Freshly prepared single‐cell suspensions were used for mononuclear cell isolation with Ficoll 400. Subsequent sorting of distinct B‐cell subpopulations was performed using MACS. For spleen CD138+ plasma cell preparation, BSA‐immunized mice were used. Spleen CD138+ plasma cells were isolated using the CD138+ Plasma Cell Isolation Kit (Miltenyi Biotec). 24 , 25 For spleen IgD+ B‐cell isolation, cells were first stained with biotinylated IgD antibody. Subsequently the cells were magnetically labeled with Streptavidin MicroBeads and positively selected (Table S6). For bone marrow IgM+ B cells isolation, anti‐mouse IgM microBeads were used. 26 To increase the purity of each fraction, magnetic cell sorting was conducted twice. The purity of each fraction was determined as ~90% using flow cytometry.
2.15. LPS‐activated mice B220 + splenocytes and IM‐9 cells
Purified spleen B220+ B cells were cultured in RPMI 1640 supplemented with 10% FBS. After 12 h, cells were stimulated with 10 μg/mL LPS. IM‐9 cells were cultured and treated with LPS for the same concentration. After stimulation for the indicated time, cells were harvested and analyzed for western blot.
2.16. CD40‐activated B lymphocytes
SU‐DHL‐2 cells were seeded into six‐well plates and induced to differentiate as previously described. 27 , 28 Briefly, cell culture was supplemented with IL‐21 (50 ng/mL) and anti‐CD40 antibodies (3 μg/mL) and cultured for the indicated time, then cells were collected for analysis.
2.17. GINS1 3′UTR reporter assay
GINS1 3′UTR and GINS1 mutated 3′UTR sequences were amplified and cloned into pmirGLO dual luciferase report vectors. Then constructed pmirGLO GINS1 and pmirGLO GINS1 Mut were co‐transfected with circ1857 overexpression plasmid or circ1857 knockdown plasmid. At 48 h after transfection, the Dual Luciferase Reporter Gene Assay Kit was used to detect luciferase activity.
2.18. Statistical analysis
All experimental data were analyzed using Student's t‐test or one‐way ANOVA and GraphPrism7. All results were presented as the mean ± SD. A p‐value < 0.05 was considered statistically significant.
3. RESULTS
3.1. GINS1 was highly expressed in DLBCL and promoted tumor proliferation in vivo and in vitro
To explore the expression and significance of GINS1 in DLBCL, first, clinical data for DLBCL in TCGA database were used. GEPIA showed that GINS1 was elevated in DLBCL compared with normal. In addition, patients with high GINS1 expression had poorer disease‐free survival than those with low GINS1 expression (Figure 1A1,A2). Next, GINS1 expression was examined in our collected tissue samples using RT‐qPCR and western blotting, and the result was consistent with GEPIA. GINS1 was upregulated in biopsy specimens from DLBCL patients (n = 30) compared with from control lymphadenitis (n = 24) (Figure 1B1,B2). Furthermore, GINS1 expression in three DLBCL cell lines was also investigated, the results showed that GINS1 expression was higher in DLBCL cell lines than in the control IM‐9 cells (Figure 1C1,C2).
FIGURE 1.
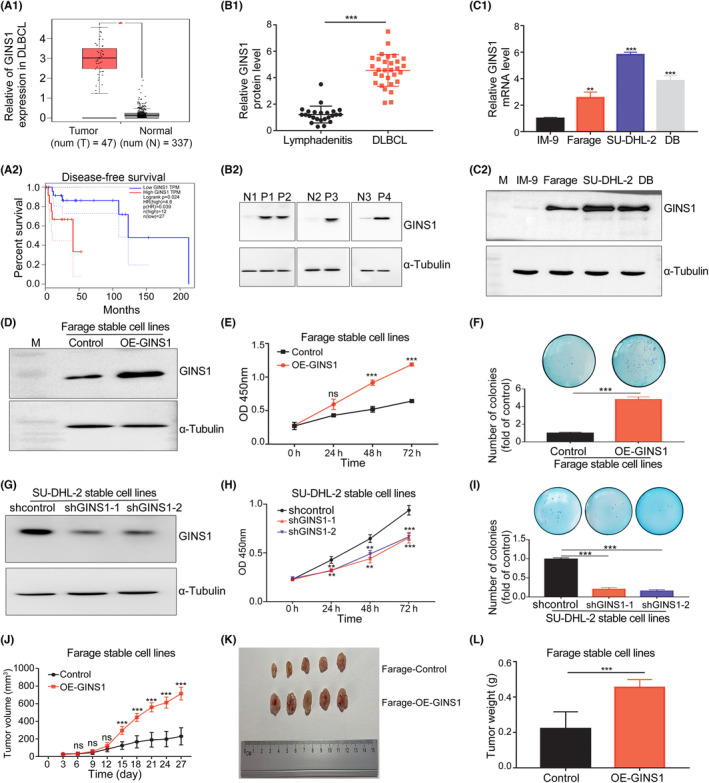
GINS1 was highly expressed and promoted tumor proliferation in DLBCL. (A) GEPIA analysis. A1, GINS1 expression was higher in the DLBCL tumor. A2, Disease‐free survival curves of DLBCL patients. (B) GINS1 protein expression in our collected biopsy specimens from DLBCL (n = 30) and lymphadenitis patients (n = 24). B1, Western blot analysis. B2, Representative western blotting analysis of GINS1 protein expression (P, DLBCL sample; N, non‐tumorous lymphadenitis control sample). (C) GINS1 mRNA and protein expression levels in DLBCL cell lines. C1, RT‐qPCR analysis. C2, Western analysis. (D–F) Overexpression of GINS1 in Farage cells promotes cell proliferation. D, Western blot. E, Cell viability assay. F, Soft agar assay for colony formation. (G–I) Knockdown of GINS1 inhibits the proliferation of SU‐DHL‐2 cells. G, Western blot. H, CCK‐8 assay. I, soft agar colony. (J–L) Overexpression of GINS1 in Farage cells promotes cell proliferation in vivo. J, Tumor growth curve: subcutaneously injecting Farage‐control cells, GINS1 overexpressing stable cells Farage‐OE‐GINS1 into the flank of nude mice, the tumor sizes of two groups were measured (five mice in each group). K, Tumor pictures. L, tumor weight (*p < 0.05, **p < 0.01, ***p < 0.001).
To test the functional significance of GINS1 expression on DLBCL cell proliferation, GINS1 stable expression cell lines Farage‐OE‐GINS1 and GINS1 stable knockdown cell lines SU‐DHL‐2‐shGINS1 were produced (Figure 1D–I). CCK‐8 and clonogenic assays showed that overexpressing GINS1 promoted Farage‐OE‐GINS1 cell proliferation (Figure 1D–F). Knockdown of GINS1 suppressed SU‐DHL‐2‐shGINS1 cell growth (Figure 1G–I). Then, an in vivo experiment further confirmed that the tumors formed using Farage‐OE‐GINS1 cells showed increased rates of proliferation when compared with the control group (Figure 1J–L). These results indicated that GINS1 was critical for DLBCL proliferation.
3.2. PAX5 directly promotes transcription of GINS1
To further explore the mechanisms underlying the upregulation of GINS1 in DLBCL, the promoter region of the human GINS1 gene was analyzed using JASPAR (an open‐access database of curated, non‐redundant TF binding profiles). The presence of two putative PAX5 binding sites was predicted (Figures 2A, S2). More importantly, target genes of PAX5 in public ENCODE ChIP‐seq datasets were analyzed (http://amp.pharm.mssm.edu/Harmonizome/dataset/ENCODE+Transcription+Factor+Targets). 29 ChIP‐seq datasets showed that GINS1 was one of the target genes of PAX5. To further confirm whether PAX5 was involved in GINS1 activation, five promoter reporter plasmids were constructed (Figure 2B). Promoter reporter plasmids and pRSV‐β‐galactosidase control plasmids were co‐transfected into SU‐DHL‐2 cells; the luciferase activity showed that mutation of any one of the predicted PAX5‐binding sites decreased the GINS1 promotor activity. This result indicated that both PAX5‐binding sites on the GINS1 promoter were important.
FIGURE 2.
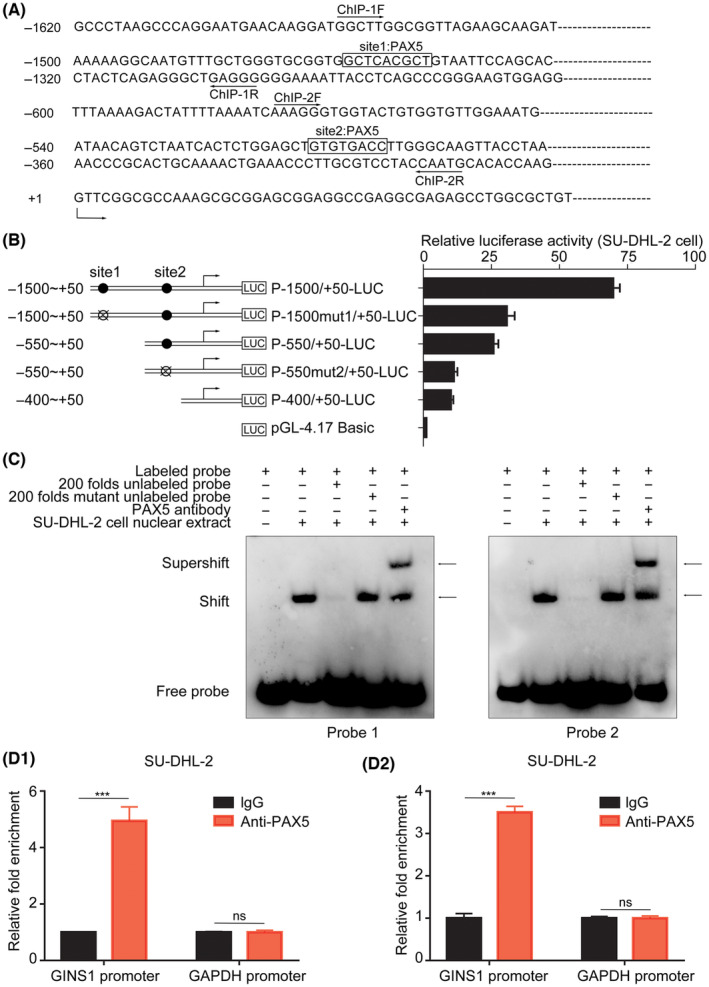
PAX5 is responsible for GINS1 expression. (A) Nucleotide sequence of the promoter region of the GINS1 gene. Two predicted PAX5 binding sites (site1 and site2) are shown. The location of primers used for ChIP‐1 and ChIP‐2 are also shown here. +1 indicates the position of the transcription initiation site of the GINS1 gene. (B) Left: the schematic diagram of the luciferase reporter constructs containing the indicated genomic fragments of the GINS1 gene is shown. Right: the results of the luciferase reporter assay. (C) EMSA analysis. (D) ChIP assay. D1, ChIP‐1. D2, ChIP‐2 (***p < 0.001).
Next, the EMSA was performed. Two sets of probes that contained a putative site1 or site2 were synthesized; DNA–protein complexes were detected when the probes were incubated with the nuclear extracts of SU‐DHL‐2 cells. The results showed that an increased number of unlabeled probes decreased the band for the complexes, and PAX5 antibody also super‐shifted the band for the complexes (Figure 2C). Furthermore, ChIP assays also confirmed that PAX5 could be recruited onto the GINS1 promoters (Figure 2D). These results indicated that PAX5 could bind to the GINS1 promoter.
3.3. PAX5 P80R mutant lacks the ability to bind to the GINS1 promoter
The PAX5 P80R mutation was located in the paired domain for DNA binding (Figure S3A).
It has been reported that patients who have thePAX5 P80R mutation have a significantly higher probability of 5‐year overall survival, 21 and it is suggested that the PAX5 P80R mutation disrupts DNA binding to its target genes. 30 To confirm that GINS1 was regulated by PAX5, recombinant re‐PAX5 and re‐PAX5 P80R protein were expressed in E. coli and purified. The EMSA assay showed that re‐PAX5 P80R had lost its ability for DNA binding to probe1, while re‐PAX5 could form the complex with the probe (Figure S3B). These results confirmed that the PAX5 P80R mutation created the loss of the ability for DNA binding. Consistent with these results, overexpression of PAX5 could activate the promoter activity of the reporter plasmids P‐1500/+50‐LUC significantly in 293T cells, whereas PAX5 P80R failed to stimulate the promoter activity of this reporter plasmid (Figure S3C). Moreover, western blotting demonstrated that PAX5 overexpression enhanced GINS1 expression in 293T cells, whereas PAX5 P80R overexpression did not increase the GINS1 level (Figure S3D). Altogether, these results suggested that GINS1 was the target gene of PAX5.
3.4. PAX5 promotes DLBCL progression through GINS1
Based on the above results, it was proposed that GINS1 upregulation might be caused by the PAX5 abnormality. Therefore, the expression levels of PAX5 and GINS1 in clinical specimens were analyzed, the results showed that both PAX5 and GINS1 were highly expressed and significantly correlated (Figures 1B, 3A1–A3). Moreover, TCGA dataset analysis using GEPIA also showed that the PAX5 level was higher in DLBCL samples than in the control (Figure 3B); a significantly positive correlation between GINS1 and PAX5 expression is shown in Figure S4A. Consistently, compared with the B lymphoblast cell line IM‐9, a higher level of PAX5 expression in DLBCL cell lines was also observed (Figure 3C).
FIGURE 3.
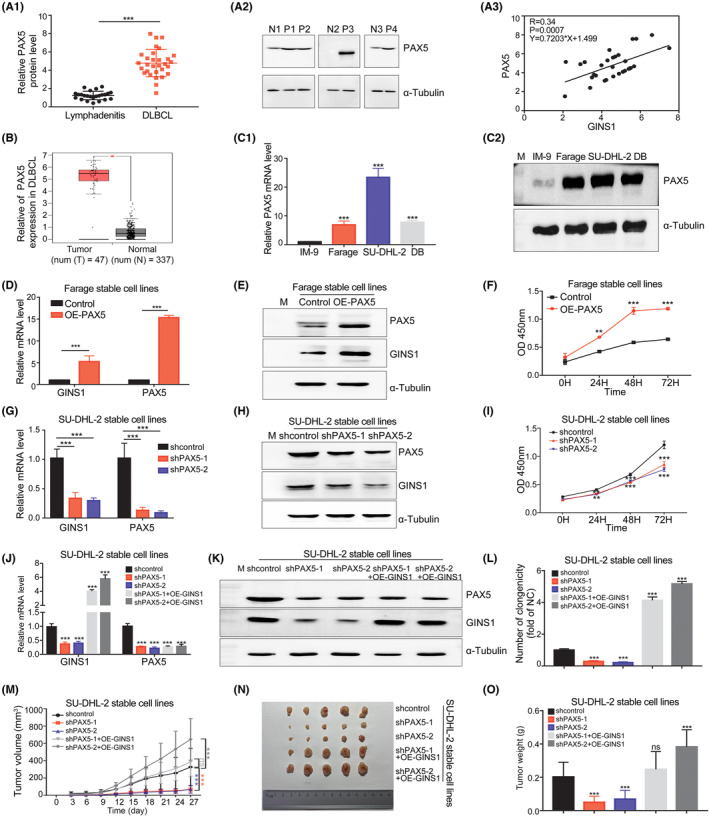
PAX5‐mediated GINS1 expression is critical for DLBCL proliferation. (A) PAX5 protein expression in DLBCL specimens. A1, Western blot analysis of DLBCL (n = 30) and lymphadenitis (n = 24) specimens. A2, Representative western blotting. P, DLBCL patients; N, lymphadenitis control. A3, Correlation analysis. (B) GEPIA analysis. (C) PAX5 level in DLBCL cell lines. C1, RT‐qPCR. C2, Western blot. (D–F) Overexpression of PAX5 in Farage stable cell lines. D, RT‐qPCR. E, Western blot. F, CCK8 assay. (G–I) Stable knockdown of PAX5 in SU‐DHL‐2 cells. G, RT‐qPCR. H, Western blot. I, CCK8 assay. (J–L) GINS1 overexpression reverses the GINS1 expression level decrease caused by PAX5 knockdown in SU‐DHL‐2 stable cell lines. J, RT‐qPCR. K, Western blot. L, Soft agar colony formation assy. (M–O) Xenograft tumor model. M, Tumor growth curve: indicates the groups of stable cell lines subcutaneously injected into the flanks of nude mice; tumor size was measured every 3 days. N, Tumor pictures. O, Tumor weight. *p < 0.05; **p < 0.01; ***p < 0.001.
To further explore the functional significance of PAX5 on GINS1 in DLBCL, stable cell lines were established and evaluated. Overexpressing PAX5 in Farage cell and IM‐9 cells strikingly enhanced the GINS1 level and facilitated cell proliferation (Figures 3D–F, S4B). Knockdown of PAX5 in SU‐DHL‐2 cells and DB cells significantly reduced GINS1 levels and inhibited cell proliferation (Figures 3G–I, S4C). Moreover, GINS1 overexpression reversed the decreased level of GINS1 in PAX5‐knockdown cells and rescued the inhibited proliferation in SU‐DHL‐2 cell and DB cells (Figures 3J–L, S4D). In an in vivo xenograft lymphoma model, the decreased tumor growth in the PAX5‐silencing group was rescued by overexpression of GINS1 in SU‐DHL‐2 stable cells (Figure 3M–O). Taken together, these data suggested that upregulated GINS1 participated in the tumorigenesis of DLBCL.
3.5. circ1857 upregulated GINS1 expression in DLBCL
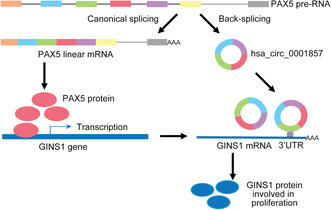
The above results showed that changes in PAX5 could promote GINS1 expression and enhance DLBCL cell proliferation. Accumulating evidence has shown that circRNAs generated by a noncanonical splicing event called back splicing, were involved in human cancer development and progression. 31 It was reported that circ1857 was markedly upregulated in ALL, and could be used for the diagnosis of ALL, 32 but the function of circ1857 was not investigated. Therefore, whether circ1857 was also involved in GINS1 regulation was explored. circ1857 was formed by back splicing exons 2–5 of the linear transcript of the PAX5 gene. In Farage cells, DB cells and the SU‐DHL‐2 cells, the presence of circ1857 was confirmed (Figures 4A, S5A,B). Compared with linear PAX5, circ1857 was more resistant to RNase R digestion, more stable and primarily distributed in the cytoplasm (Figure 4B–D). Subsequently, circ1857 secondary structure was predicted using online RNAfold (http://rna.tbi.univie.ac.at//cgi‐bin/RNAWebSuite/RNAfold.cgi; Figure S5C). More interestingly, the loop in circ1857 was predicted to bind in trans with the 3′UTRs of GINS1 mRNAs (Figure S5C). To determine whether circ1857 acted directly on GINS1 mRNA, luciferase reporter assays were performed, the results showed that circ1857 overexpression promoted GINS1 3′UTR luciferase activity, but had no influence on the GINS1 mutated 3′UTR luciferase activity (Figure 4E). Similarly, circ1857 knockdown inhibited GINS1 3′UTR luciferase activity, but had no effect on the GINS1‐mutated 3′UTR luciferase activity, suggesting that circ1857 directly bound to GINS1 mRNA and promoted its stability. To explore the potential biological function of circ1857 in DLBCL, RNA levels of both GINS1 and circ1857 in biopsy specimens were detected using RT‐qPCR. The results showed that circ1857 and GINS1 were upregulated (Figure 5A,B) and significant correlations between them were found (Figure 5C). Next, the expression of circ1857 in DLBCL cell lines was detected. As shown in Figure 5D, circ1857 levels were also high in DLBCL cell lines. Stable expression of circ1857 in Farage cells resulted in significantly upregulated GINS1 mRNA and protein levels (Figure 5E,F). Furthermore, CCK‐8 assays showed that cell proliferation was significantly increased by overexpressing circ1857 (Figure 5G). Moreover, circ1857 overexpression significantly stabilized the GINS1 mRNA (Figure 5H). These results suggested that circ1857 plays a role in fine tuning the expression of GINS1 at the posttranscriptional level. To further ascertain the role of circ1857 on GINS1, stable knockdown of circ1857 in SU‐DHL‐2 cell lines was established (Figure 5I,J). mRNA and protein levels for GINS1 were markedly decreased in circ1857 stable knockdown cells. There was no significant change for the mRNA and protein levels of PAX5 when circ1857 was overexpressed or knocked down. Lower cell viability was also observed when circ1857 was reduced (Figure 5K). These results suggested that the regulation of circ1857 on the target gene GINS1 was independent of the PAX5 protein. Moreover, the stability of GINS1 decreased in circ1857 knockdown cells (Figure 5L). Altogether, these results indicated that circ1857 could stabilize GINS1 mRNA.
FIGURE 4.
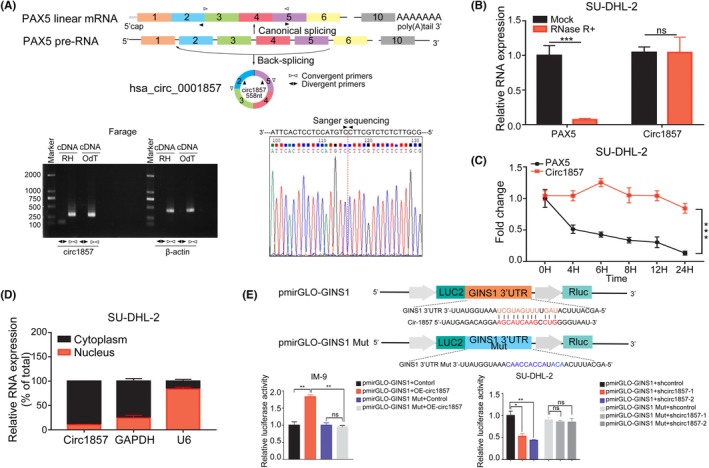
circ1857 in DLBCL. (A) Diagram shows the PAX5 pre‐mRNA canonical splicing and back splicing that produces circ1857. circ1857 in Farage cells was validated using RT‐PCR with convergent and divergent primers and confirmed using Sanger sequencing. PCR analysis for circ1857 and β‐actin in cDNA. RH, random hexamers, OdT, oligo(dT)18 primers. Two independent experiments were carried out with similar results. (B) RT‐qPCR analysis of linear PAX5 mRNA and circ1857 after treatment with RNase R in SU‐DHL‐2. (C) RT‐qPCR analysis of circ1857 and PAX5 mRNA in SU‐DHL‐2 cells after treatment with actinomycin D for the indicated time. (D) Relative expression of circ1857 in the nucleus and cytoplasm, respectively. GAPDH and U6 were used as controls for the cytoplasm and nucleus. (E) Luciferase activity was measured using dual luciferase assays when cells were co‐transfected with a GINS1 3′UTR‐ or a GINS1 mutated 3′UTR‐expressing plasmid with circ1857‐overexpression or ‐knockdown plasmids. The construction of the GINS1 3′UTR or GINS1‐mutated 3′UTR luciferase reporter plasmid is shown. *p < 0.05; **p < 0.01; ***p < 0.001.
FIGURE 5.
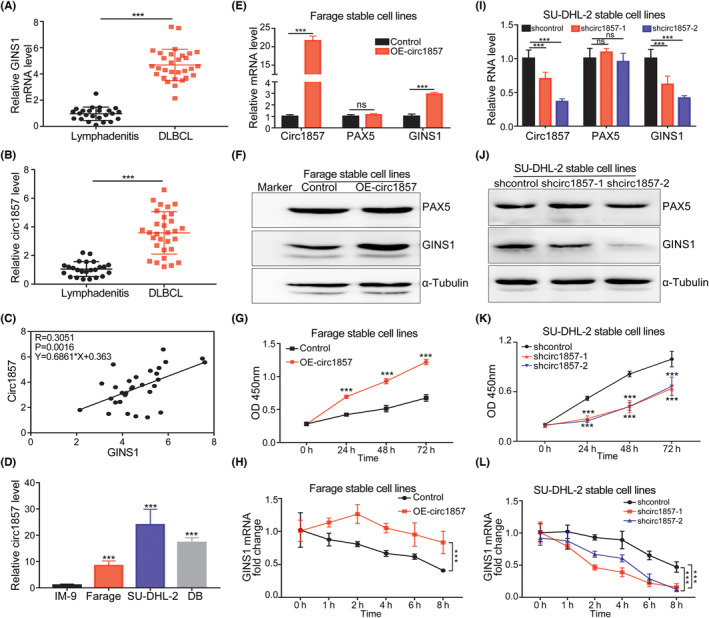
circ1857 promotes GINS1 expression in DLBCL cells. (A, B) GINS1 and circ1857 expression in DLBCL biopsy specimens detected by RT‐qPCR. (C) Correlation between GINS1 and circ1857 in DLBCL specimens. (D) circ1857 levels in DLBCL cell lines analyzed by RT‐qPCR. (E–H) Farage stable cells overexpressing circ1857. E, RT‐qPCR. F, Western blot. G, CCK8. H, RT‐qPCR analysis of GINS1 mRNA in Farage stable cells after treatment with actinomycin D for the indicated time. (I–L) SU‐DHL‐2 stable knockdown of circ1857 cells. I, RT‐qPCR. J, Western blot. K, CCK8. L, After treatment with actinomycin D, RT‐qPCR analysis of expression of GINS1 mRNA in SU‐DHL‐2 stable knockdown circ1857 cells. ***p < 0.001.
3.6. Coordinated expression of PAX5 and GINS1 in B cells
PAX5 was physiologically expressed in normal B cells and silenced in plasma cells. Next PAX5 and GINS1 protein expression patterns at different B‐cell stages were evaluated. For PAX5 and the plasma cell regulator PRDM1 were mutually exclusive and cross‐antagonized each other, therefore the PRDM1 level was also analyzed here. PAX5 and GINS1 were coordinately expressed in sorted bone marrow IgM+ B cells and splenic IgD+ B cells but both declined in spleen CD138+ plasma cells (Figure 6A,B). LPS is a mitogen that stimulates B cells, causing proliferation and then differentiation into plasma cells. 33 The expression of PAX5 and the GINS1 expression pattern in LPS‐treated splenocytes were analyzed next. In sorted spleen B220+ cells, expression of both PAX5 and GINS1 in splenocytes increased after LPS treatment for 12 h, during which time cells entered an active proliferation stage. Following this increase period, the expression of both proteins began to decline at 24 h, which could be explained using terminal cell differentiation (Figure 6C). In IM‐9 cells after LPS stimulation, PAX5 and GINS1 levels also increased at 12 h, and then both decreased at 48 h (Figure 6D). More interestingly, circ1857 expression showed a similar trend as GINS1 mRNA in LPS‐stimulated IM‐9 cells (Figure S6A).
FIGURE 6.
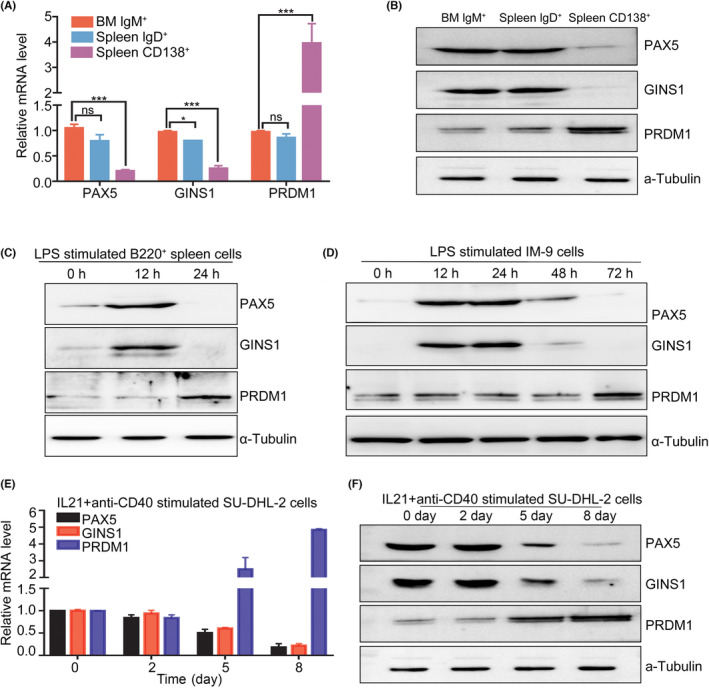
GINS1 is regulated by PAX5 in B‐cell proliferation and differentiation. (A, B) Expression of PAX5 and GINS1 during the mouse B‐cell proliferation and differentiation. (C) After LPS stimulation, the expression of GINS1, PAX5 and PRDM1 in mouse spleen cells was analyzed. Cells were incubated with 10 mg/mL LPS for the indicated time and harvested for western blot analysis. (D) After LPS stimulation, the expression of GINS1, PAX5 and PRDM1 in IM‐9 cells was analyzed using western blot. (E, F) After anti‐CD40 and IL21 stimulation for the indicated time, cells were collected, the expression levels of PAX5, GINS1 and PRDM1 were analyzed using RT‐qPCR and western blot. ***p < 0.001.
It was noted that PAX5 was dysregulated in B‐cell tumors ranging from ALL to many mature B‐cell lymphoma subtypes. 14 , 17 , 19 , 27 At the normal physical late stage of B‐cell differentiation, PAX5 expression was downregulated. High‐level PAX5 at this stage of differentiation would perturb the plasma cell differentiation program initiated by PAX5 repression, thereby contributing to the development of a fraction of the DLBCL. Therefore, DLBCL cells were induced to differentiate into plasma cells. SU‐DHL‐2 was induced to differentiate under differentiation‐inducing conditions. Through stimulation with interleukin 21 and anti‐CD40 (CD40L), when PAX5 was observed to decrease at day 5, GINS1 was also decreased simultaneously. In addition the plasma cell‐specific gene PRDM1 was induced (Figure 6E,F). The correlation of circ1857 and GINS1 expression in SU‐DHL‐2 cells under induced differentiation conditions was also evaluated (Figure S6B). All these results indicated that GINS1 was one of the PAX5 targeted genes. Tight control of PAX5 is crucial not only for normal B lymphopoiesis, but also for preventing tumor formation.
4. DISCUSSION
In this work, aberrant expression levels of PAX5, circ1857 and GINS1 in DLBCL were observed. We further demonstrated that knocking down one of them suppressed cell proliferation. Mechanistically, PAX5 is a transactivator for GINS1 gene transcription, and circ1857 stabilized GINS1 mRNA. circ1857 and PAX5 coordinately promoted DLBCL progression by upregulating GINS1 expression (Figure 7).
FIGURE 7.
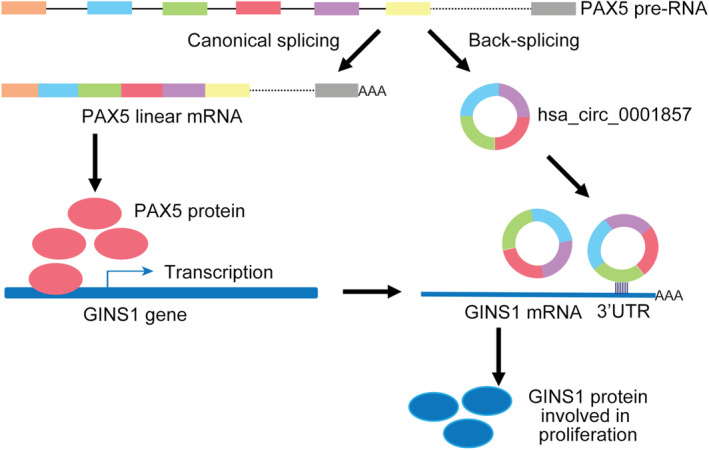
Schematic diagram of the regulation mechanism of GINS1 expression using PAX5 and circ1857 in normal B cells and DLBCL.
GINS1 is essential for the initiation and elongation of DNA replication by recruiting DNA polymerase. 6 , 34 Initially, GINS1 was found to be predominantly expressed in highly proliferating cells such as stem cells and progenitor cells instead of mature cells. 35 , 36 GINS1 homozygous null mutations in mice were embryonic lethal and heterozygous mutations of GINS1 in humans displayed intrauterine growth retardation, chronic neutropenia, and NK cell deficiency. 37 , 38 Later, it was found that GINS1 was involved in the tumorigenesis of various cancers. 11 , 39 , 40 However, GINS1 in DLBCL was never investigated.
In DLBCL, frequent alterations of the PAX5 gene were reported. 17 IgH‐PAX5 translocation resulted in the intact coding sequences for PAX5 under the control of strong enhancers or promoters of the IgH. 41 Consequently, the expression of translocated PAX5 was strongly increased. 42 , 43 Other mechanisms also contributed to the increased PAX5. 18 , 44
circRNAs are a group of covalently closed loop‐like RNAs. 45 Growing numbers of studies have shown that circRNAs are important regulator molecules in cancer progression, but they have been poorly studied in DLBCL. circ1857 was previously found in ALL. 32 Here, circ1857 and its host gene product PAX5 protein were found both upregulated in DLBCL. Although some antisense circular RNA was found to suppress parental gene splicing and translation, 46 actually more circular RNA was found positively correlated with that of linear parental mRNA. 47 Here both circ1857 and PAX5 upregulated the same target gene GINS1 in DLBCL in a coordinated manner. To the best of our knowledge, this kind of coordinated function pattern of circular RNA and its parental mRNA product has not been reported previously.
In conclusion, we showed that proper control of GINS1 expression was critical for B‐cell proliferation and differentiation. From an oncological view, aberrant high expression of PAX5 and GINS1 blocked B‐cell differentiation. In addition, GINS1 recently was reported to have anti‐apoptotic functions, 48 , 49 all these findings suggested that deregulated PAX5 and GINS1 are an oncogene for a fraction of DLBCL. Furthermore more cell lines, more tissue samples and further studies are needed to establish the therapeutic potential of GINS1 in DLBCL.
AUTHOR CONTRIBUTIONS
Ting Wang performed experiments and analyzed the data. Zhenfa Chen, Cui Li, Wei Zhang and Wenbin Huang provided suggestions and performed the analysis. Jun Xue and Jundong Wang provided patient samples. Shufeng Li designed experiments, reviewed the data, and wrote the manuscript. All authors read and approved the final submitted manuscript.
FUNDING INFORMATION
This work was supported by a grant from the Chinese National Nature Science Foundation (31070706) and by the funds from the Applied Basic Research Programs of Science and Technology Commission Foundation of Jiangsu Province (BK20161416).
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Review Board: This study was approved by the ethics committee of the Nanjing First Hospital Medical University.
Informed Consent: Written informed consent was obtained from each individual.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: Research protocols were approved by the Animal Experimentation Ethics Committee of Southeast University.
Supporting information
Table S1:
Wang T, Chen Z, Li C, et al. PAX5 and circ1857 affected DLBCL progression and B‐cell proliferation through regulating GINS1 . Cancer Sci. 2023;114:3203‐3215. doi: 10.1111/cas.15856
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2. Hertzberg M. R‐CHOP in DLBCL: priming for success. Blood. 2022;139(8):1121‐1122. doi: 10.1182/blood.2021013620 [DOI] [PubMed] [Google Scholar]
- 3. MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489‐500. doi: 10.1042/Bj20091531 [DOI] [PubMed] [Google Scholar]
- 4. Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17(9):1153‐1165. doi: 10.1101/gad.1065903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamada K. The GINS complex: structure and function. Subcell Biochem. 2012;62:135‐156. doi: 10.1007/978-94-007-4572-8_8 [DOI] [PubMed] [Google Scholar]
- 6. Jedrychowska M, Denkiewicz‐Kruk M, Alabrudzinska M, et al. Defects in the GINS complex increase the instability of repetitive sequences via a recombination‐dependent mechanism. PLoS Genet. 2019;15(12):e1008494. doi: 10.1371/journal.pgen.1008494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahmad M, Hameed Y, Khan M, et al. Up‐regulation of GINS1 highlighted a good diagnostic and prognostic potential of survival in three different subtypes of human cancer. Braz J Biol. 2021;84:e250575. doi: 10.1590/1519-6984.250575 [DOI] [PubMed] [Google Scholar]
- 8. Bu F, Zhu X, Zhu J, et al. Bioinformatics analysis identifies a novel role of GINS1 gene in colorectal cancer. Cancer Manag Res. 2020;12:11677‐11687. doi: 10.2147/CMAR.S279165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S, Wu L, Zhang H, et al. GINS1 induced Sorafenib resistance by promoting cancer stem properties in human hepatocellular cancer cells. Front Cell Dev Biol. 2021;9:711894. doi: 10.3389/fcell.2021.711894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li M, Shi M, Hu C, Chen B, Li S. MALAT1 modulated FOXP3 ubiquitination then affected GINS1 transcription and drived NSCLC proliferation. Oncogene. 2021;40(22):3870‐3884. doi: 10.1038/s41388-021-01816-3 [DOI] [PubMed] [Google Scholar]
- 11. Hsieh HY, Jia WZ, Jin ZC, Kidoya H, Takakura N. High expression of PSF1 promotes drug resistance and cell cycle transit in leukemia cells. Cancer Sci. 2020;111(7):2400‐2412. doi: 10.1111/cas.14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xue K, Song JZ, Yang Y, et al. PAX5 promotes pre‐B cell proliferation by regulating the expression of pre‐B cell receptor and its downstream signaling. Mol Immunol. 2016;73:1‐9. doi: 10.1016/j.molimm.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 13. Liu GJ, Jaritz M, Wohner M, et al. Repression of the B cell identity factor Pax5 is not required for plasma cell development. J Exp Med. 2020;217(11):e20200147. doi: 10.1084/jem.20200147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamada T, Yonetani N, Ueda C, et al. Expression of the PAX5/BSAP transcription factor in haematological tumour cells and further molecular characterization of the t(9;14)(g13;q32) translocation in B‐cell non‐Hodgkin's lymphoma. Brit J Haematol. 1998;102(3):691‐700. [DOI] [PubMed] [Google Scholar]
- 15. Montesinos‐Rongen M, Van Roost D, Schaller C, Wiestler OD, Deckert M. Primary diffuse large B‐cell lymphomas of the central nervous system are targeted by aberrant somatic hypermutation. Blood. 2004;103(5):1869‐1875. doi: 10.1182/blood-2003-05-1465 [DOI] [PubMed] [Google Scholar]
- 16. Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto‐oncogenes in B‐cell diffuse large‐cell lymphomas. Nature. 2001;412(6844):341‐346. doi: 10.1038/35085588 [DOI] [PubMed] [Google Scholar]
- 17. Ohno H, Nakagawa M, Kishimori C, et al. Diffuse large B‐cell lymphoma carrying t(9;14)(p13;q32)/PAX5‐immunoglobulin heavy chain gene is characterized by nuclear positivity of MUM1 and PAX5 by immunohistochemistry. Hematol Oncol. 2020;38(2):171‐180. doi: 10.1002/hon.2716 [DOI] [PubMed] [Google Scholar]
- 18. Balasenthil S, Gururaj AE, Talukder AH, et al. Identification of Pax5 as a target of MTA1 in B‐cell lymphomas. Cancer Res. 2007;67(15):7132‐7138. doi: 10.1158/0008-5472.Can-07-0750 [DOI] [PubMed] [Google Scholar]
- 19. Nasrabadi PN, Martin D, Gharib E, Robichaud GA. The pleiotropy of PAX5 gene products and function. Int J Mol Sci. 2022;23(17):10095. doi: 10.3390/ijms231710095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liang X, Gu J, Li TJ, et al. PAX5 haploinsufficiency induce cancer cell dormancy in Raji cells. Exp Cell Res. 2018;367(1):30‐36. doi: 10.1016/j.yexcr.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 21. Passet M, Boissel N, Sigaux F. PAX5 P80R mutation identifies a novel subtype of B‐cell precursor acute lymphoblastic leukemia with favorable outcome (vol 133, pg 280, 2019). Blood. 2020;135(22):2011. doi: 10.1182/blood.2020006141 [DOI] [PubMed] [Google Scholar]
- 22. Bastian L, Schroeder MP, Eckert C, et al. PAX5 biallelic genomic alterations define a novel subgroup of B‐cell precursor acute lymphoblastic leukemia. Leukemia. 2019;33(8):1895‐1909. doi: 10.1038/s41375-019-0430-z [DOI] [PubMed] [Google Scholar]
- 23. Wang T, Zhang W, Huang WB, Hua ZC, Li SF. LncRNA MALAT1 was regulated by HPV16 E7 independently of pRB in cervical cancer cells. J Cancer. 2021;12(21):6344‐6355. doi: 10.7150/jca.61194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Forster M, Farrington K, Petrov JC, et al. MYSM1‐dependent checkpoints in B cell lineage differentiation and B cell‐mediated immune response. J Leukoc Biol. 2017;101(3):643‐654. doi: 10.1189/jlb.1AB0415-177RR [DOI] [PubMed] [Google Scholar]
- 25. Kawano Y, Fujiwara S, Wada N, et al. Multiple myeloma cells expressing low levels of CD138 have an immature phenotype and reduced sensitivity to lenalidomide. Int J Oncol. 2012;41(3):876‐884. doi: 10.3892/ijo.2012.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y, Yuan X, Mu P, Li Q, Ao J, Chen X. Development of monoclonal antibody against IgM of large yellow croaker (Larimichthys crocea) and characterization of IgM(+) B cells. Fish Shellfish Immunol. 2019;91:216‐222. doi: 10.1016/j.fsi.2019.05.035 [DOI] [PubMed] [Google Scholar]
- 27. van Keimpema M, Gruneberg LJ, Mokry M, et al. The forkhead transcription factor FOXP1 represses human plasma cell differentiation. Blood. 2015;126(18):2098‐2109. doi: 10.1182/blood-2015-02-626176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andorsky DJ, Timmerman JM. Interleukin‐21: biology and application to cancer therapy. Expert Opin Biol Ther. 2008;8(9):1295‐1307. doi: 10.1517/14712598.8.9.1295 [DOI] [PubMed] [Google Scholar]
- 29. Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database‐Oxford. 2016;2016:baw100. doi: 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre‐B cell acute lymphoblastic leukemia. Nat Genet. 2013;45(10):1226‐1231. doi: 10.1038/ng.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Decruyenaere P, Offner F, Vandesompele J. Circulating RNA biomarkers in diffuse large B‐cell lymphoma: a systematic review. Exp Hematol Oncol. 2021;10(1):13. doi: 10.1186/s40164-021-00208-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo S, Li B, Chen Y, et al. Hsa_circ_0012152 and Hsa_circ_0001857 accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Front Oncol. 2020;10:1655. doi: 10.3389/fonc.2020.01655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee IS, Choi WH, Kim JY, et al. Transcriptional regulation of the murine 1‐cys peroxiredoxin gene by the B cell‐specific activator protein, Pax5. J Cell Biochem. 2008;104(2):465‐476. doi: 10.1002/jcb.21638 [DOI] [PubMed] [Google Scholar]
- 34. Dmowski M, Makiela‐Dzbenska K, Jedrychowska M, Denkiewicz‐Kruk M, Fijalkowska IJ. Mutation spectrum data for Saccharomyces cerevisiae psf1‐1 pol2‐M644G mutants. Data Brief. 2022;42:108223. doi: 10.1016/j.dib.2022.108223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagahama Y, Ueno M, Miyamoto S, et al. PSF1, a DNA replication factor expressed widely in stem and progenitor cells, drives tumorigenic and metastatic properties. Cancer Res. 2010;70(3):1215‐1224. doi: 10.1158/0008-5472.CAN-09-3662 [DOI] [PubMed] [Google Scholar]
- 36. Kimura T, Cui D, Kawano H, Yoshitomi‐Sakamoto C, Takakura N, Ikeda E. Induced expression of GINS complex is an essential step for reactivation of quiescent stem‐like tumor cells within the peri‐necrotic niche in human glioblastoma. J Cancer Res Clin. 2019;145(2):363‐371. doi: 10.1007/s00432-018-2797-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cottineau J, Kottemann MC, Lach FP, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127(5):1991‐2006. doi: 10.1172/JCI90727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ueno M, Itoh M, Kong L, Sugihara K, Asano M, Takakura N. PSF1 is essential for early embryogenesis in mice. Mol Cell Biol. 2005;25(23):10528‐10532. doi: 10.1128/MCB.25.23.10528-10532.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou L, Sun XJ, Liu C, et al. Overexpression of PSF1 is correlated with poor prognosis in hepatocellular carcinoma patients. Int J Biol Markers. 2015;30(1):e56‐e64. doi: 10.5301/jbm.5000105 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J, Wu Q, Wang Z, et al. Knockdown of PSF1 expression inhibits cell proliferation in lung cancer cells in vitro. Tumour Biol. 2015;36(3):2163‐2168. doi: 10.1007/s13277-014-2826-8 [DOI] [PubMed] [Google Scholar]
- 41. Busslinger M, Klix N, Pfeffer P, Graninger PG, Kozmik Z. Deregulation of PAX‐5 by translocation of the emu enhancer of the IgH locus adjacent to two alternative PAX‐5 promoters in a diffuse large‐cell lymphoma. Proc Natl Acad Sci U S A. 1996;93(12):6129‐6134. doi: 10.1073/pnas.93.12.6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463‐470. doi: 10.1038/ni1454 [DOI] [PubMed] [Google Scholar]
- 43. Poppe B, De Paepe P, Michaux L, et al. PAX5/IGH rearrangement is a recurrent finding in a subset of aggressive B‐NHL with complex chromosomal rearrangements. Genes Chromosomes Cancer. 2005;44(2):218‐223. doi: 10.1002/gcc.20214 [DOI] [PubMed] [Google Scholar]
- 44. Li W, He Y, Cheng Z. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA‐338‐3p/PAX5 axis. Eur J Cancer Prev. 2021;30(2):132‐142. doi: 10.1097/CEJ.0000000000000596 [DOI] [PubMed] [Google Scholar]
- 45. Kristensen LS, Okholm TLH, Veno MT, Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15(2):280‐291. doi: 10.1080/15476286.2017.1409931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma J, Du WW, Zeng K, et al. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther. 2021;29(9):2754‐2768. doi: 10.1016/j.ymthe.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang L, Yu W, Wang Y, Li H, Shen Z. Anlotinib inhibits synovial sarcoma by targeting GINS1: a novel downstream target oncogene in progression of synovial sarcoma. Clin Transl Oncol. 2019;21(12):1624‐1633. doi: 10.1007/s12094-019-02090-2 [DOI] [PubMed] [Google Scholar]
- 49. Varga M, Csalyi K, Bertyak I, et al. Tissue‐specific requirement for the GINS complex during zebrafish development. Front Cell Dev Biol. 2020;8:373. doi: 10.3389/fcell.2020.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1:


