Summary
Artificial intelligence (AI) has great potential to transform healthcare by enhancing the workflow and productivity of clinicians, enabling existing staff to serve more patients, improving patient outcomes, and reducing health disparities. In the field of ophthalmology, AI systems have shown performance comparable with or even better than experienced ophthalmologists in tasks such as diabetic retinopathy detection and grading. However, despite these quite good results, very few AI systems have been deployed in real-world clinical settings, challenging the true value of these systems. This review provides an overview of the current main AI applications in ophthalmology, describes the challenges that need to be overcome prior to clinical implementation of the AI systems, and discusses the strategies that may pave the way to the clinical translation of these systems.
Keywords: ophthalmology, eye diseases, artificial intelligence, deep learning, machine learning, clinical translation, real world
Graphical abstract
Li et al. present a comprehensive overview of the primary AI applications in ophthalmology, outline the challenges that should be addressed before the clinical implementation of AI systems, and discuss potential strategies that may facilitate the successful translation of these systems into clinical practice.
Introduction
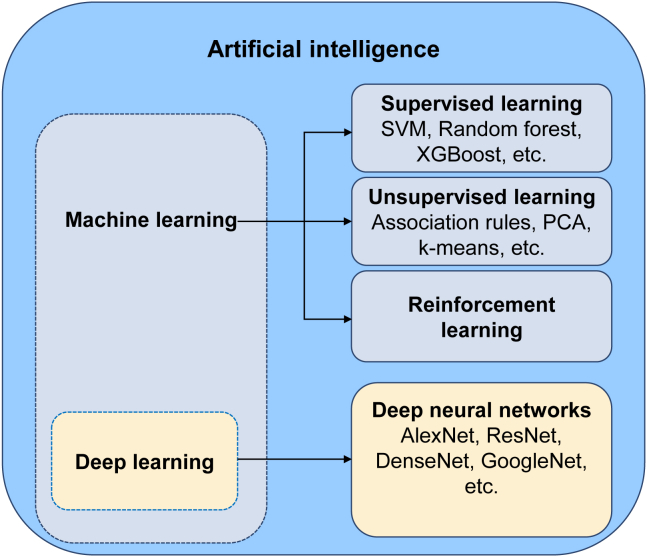
In recent years, artificial intelligence (AI), including machine learning and deep learning (Figure 1), has made a great impact on society worldwide. This is stimulated by the advent of powerful algorithms, exponential data growth, and computing hardware advances.1,2 In the medical and healthcare fields, numerous studies have validated that AI exhibited robust performance in disease diagnoses and treatment response prediction.3,4,5,6,7,8,9 For example, a deep-learning system developed on computed tomography (CT) images, can distinguish patients with COVID-19 pneumonia from patients with other common types of pneumonia and normal controls with an area under the curve (AUC) of 0.971.10 A machine-learning model trained using dual-energy CT radiomics provides a significant additive value for response prediction (AUC = 0.75) in metastatic melanoma patients prior to immunotherapy.11
Figure 1.
Relationship between AI, machine learning, and deep learning
SVM, support vector machine. PCA, principal-component analysis.
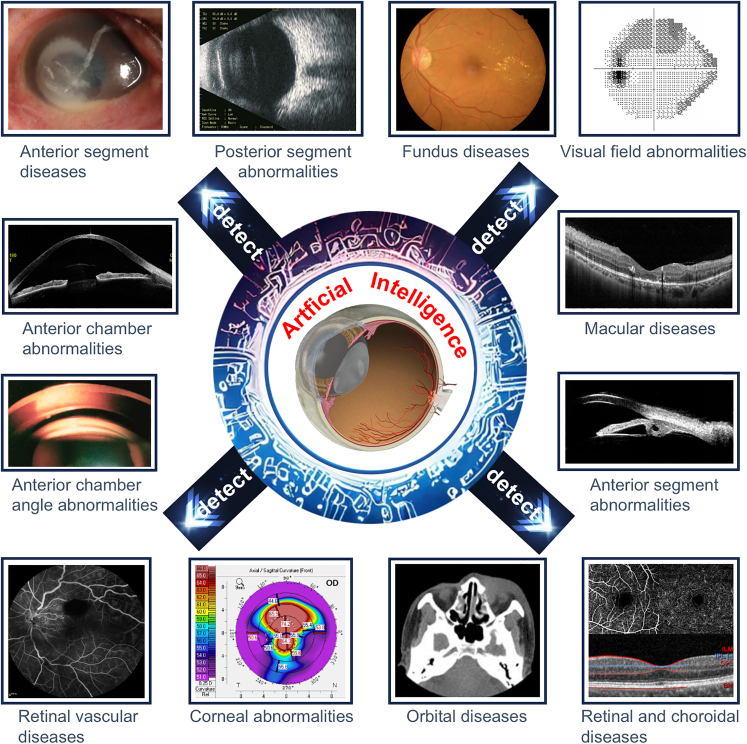
In ophthalmology, the application of AI is very promising, given that the diagnoses and therapeutic monitoring of ocular diseases often rely heavily on image recognition (Figure 2). Based on this technique, diabetic retinopathy (DR), glaucoma, and age-related macular degeneration (AMD) can be accurately detected from fundus images,6,12,13 and keratitis, pterygium, and cataract can be precisely identified from slit-lamp images.14,15,16 Detailed information that describes different imaging types for different purposes in ophthalmology and corresponding AI applications is summarized in Table S1. In addition, AI may support eye doctors in generating individualized views of patients along their care pathways and guide clinical decisions. For instance, the visual prognosis after 12 months in neovascular AMD patients receiving ranibizumab can be predicted by an AI-based model that is developed using their clinical data (e.g., ocular coherence tomography [OCT] and best-corrected visual acuity [BCVA]) collected at baseline and in the first 3 months.17 This method may assist eye doctors in better managing the expectations of patients appropriately during their treatment process.
Figure 2.
Overall schematic diagram describing the practical application of AI in all common ophthalmic imaging modalities
Although there are many reasons to be hopeful for this transformation brought on by AI, hurdles remain to the successful deployment of AI in real-world clinical settings. In this review, we first retrace the current main AI applications in ophthalmology. Second, we describe the major challenges of AI clinical translation. Third, we discuss avenues that could facilitate the real implementation of AI into clinical practice. By stressing issues in the context of present AI applications for clinical ophthalmology, we wish to provide concepts to help promote significative investigations that will finally translate to real-world clinical use.
Application of AI algorithms in posterior-segment eye diseases
DR
The prevalence of diabetes has tripled in the past two decades worldwide. It can cause microvascular damage and retinal dysfunction as a result of chronic exposure to hyperglycemia, and 34.6% of people with diabetes develop DR, which is a leading cause of vision loss in working-age adults (20–74 years).18,19,20 In 2019, approximately 160 million of the population suffered from some form of DR, of whom 47 million suffered from sight-threatening DR.18 By 2045, this number is projected to increase to 242 million for DR and 71 million for sight-threatening DR.18 Early identification and timely treatment of sight-threatening DR can reduce 95% of blindness from this cause.18 Therefore, DR screening programs for patients with diabetes are suggested by the World Health Organization.21 However, conducting these screening programs on a large scale often requires a great deal of manpower and material and financial resources, which is difficult to realize in many low-income and middle-income countries. For this reason, exploring an approach that can reduce costs and increase the efficiency of DR screening programs should be a high priority. The emergence of AI provides potential new solutions, such as applying AI to retinal imaging for automated DR screening and referral (Table 1).
Table 1.
Major studies on the application of AI in DR
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Liu et al.22 | 2022 | retrospective | TFI | 1,171,365 | EfficientNet-B5 | detection of diabetic macular edema | AUC = 0.88–0.96 Sen = 71–100, Spec = 66–88 |
| Dai et al.23 | 2021 | retrospective | TFI | 666,383 | ResNet and Mask R-CNN | DR grading | AUC = 0.92–0.97, Sen = 87.5–93.7, Spec = 74.3–90.0 |
| Lee et al.24 | 2021 | retrospective | TFI | 311,604 | seven algorithms from five companies: OpthAI, AirDoc, Eyenuk, Retina-AI Health, and Retmarker | detection of referable DR | Sen = 51.0–85.9, Spec = 60.4–83.7 |
| Araújo et al.25 | 2020 | retrospective | TFI | 103,062 | A novel Gaussian-sampling method built upon a multiple-instance learning framework | detection of referable DR | Cohen’s quadratic weighted Kappa (κ) = 0.71–0.84 |
| Heydon et al.26 | 2020 | prospective | TFI | 120,000 | machine-learning-enabled software, EyeArt v2.1 | detection of referable DR | Sen = 95.7, Spec = 54.0 |
| Natarajan et al.27 | 2019 | prospective | SFI | 56,986 | Medios AI, a deep-learning-based system | detection of referable DR | Sen = 95.9–100, Spec = 78.7–88.4 |
| Gulshan et al.28 | 2019 | prospective | TFI | 5,762 | Inception-v3 | detection of referable DR | AUC = 0.96–0.98, Sen = 88.9–92.1, Spec = 92.2–95.2 |
| Li et al.29 | 2018 | retrospective | TFI | 177,287 | Inception-v3 | detection of vision-threatening referable DR | AUC = 0.96–0.99, Sen = 92.5–97.0, Spec = 91.4–98.5 |
| Ting et al.6 | 2017 | retrospective | TFI | 189,018 | adapted VGGNet | detection of referable and vision-threatening DR | for referable DR: AUC = 0.89–0.98, Sen = 90.5–100, Spec = 73.3–92.2; for vision-threatening DR: AUC = 0.96, Sen = 100, Spec = 91.1 |
| Gulshan et al.7 | 2016 | retrospective | TFI | 140,426 | Inception-v3 | detection of referable DR | AUC = 0.99, Sen = 87.0–97.5, Spec = 93.9–98.5 |
AUC, area under the curve, representing an aggregate measure of AI performance across all possible classification thresholds; R-CNN, region-based convolutional neural network; Sen, sensitivity, representing the rate of positive samples correctly classified by an AI model; SFI, smartphone-based fundus images; Spec, specificity, representing the rate of negative samples correctly classified by an AI model; TFI, traditional fundus images.
Using deep learning, numerous studies have developed intelligent systems that can accurately detect DR from fundus images. Gulshan et al.7 developed a deep-learning system using 128,175 fundus images (69,573 subjects) and evaluated the system in two external datasets with 11,711 fundus images (5,871 subjects). Their system achieved an AUC over 0.99 in DR screening. Ting et al.6 reported a deep-learning system with an AUC of 0.936, a sensitivity of 90.5%, and a specificity of 91.6% in identifying referable DR, and an AUC of 0.958, a sensitivity of 100%, and a specificity of 91.1% in discerning sight-threatening DR. Tang et al.30 established a deep-learning system for detecting referable DR and sight-threatening DR from ultra-widefield fundus (UWF) images, which had a larger retinal field of view and contained more information about lesions (especially peripheral lesions) compared with traditional fundus images. The AUCs of this system for identifying both referable DR and sight-threatening DR were over 0.9 in the external validation datasets. Justin et al.31 trained a UWF-image-based deep-learning model using ResNet34, and this model had an AUC of 0.915 for DR detection in the test set. Their results indicated that the model developed based on UWF images may be more accurate than that based on traditional fundus images because only the UWF-image-based model could detect peripheral DR lesions without pharmacologic pupil dilation.
In addition to screening for patients with referable DR (i.e., moderate and worse DR) and sight-threatening DR, detecting early-stage DR is also crucial. Evidence indicates that proper intervention to keep glucose, lipid profiles, and blood pressure under control at an early stage can significantly delay the DR progression and even reverse mild non-proliferative DR (NPDR) to the DR-free stage.32 Dai et al.23 reported a deep-learning system named DeepDR with robust performance in detecting early to late stages of DR. Their system was developed based on 466,247 fundus images (121,342 diabetic patients) that were graded for mild NPDR, moderate NPDR, severe NPDR, proliferative DR (PDR), and non-DR by a centered reading group including 133 certified ophthalmologists. The evaluation was performed on 209,322 images collected from three external datasets, China National Diabetic Complications Study (CNDCS), Nicheng Diabetes Screening Project (NDSP), and Eye Picture Archive Communication System (EyePACS), and the average AUCs of the system in these datasets were 0.940, 0.944, and 0.943, respectively. Besides, predicting the onset and progression of DR is essential for the mitigation of the rising threat of DR. Bora et al.33 created a deep-learning system using image data obtained from 369,074 patients to predict the risk of patients with diabetes developing DR within 2 years, and the system achieved an AUC of 0.70 in the external validation set. This automated risk-stratification tool may help optimize DR screening intervals and decrease costs while improving vision-related outcomes. Arcadu et al.34 developed a predictive DR progression algorithm (AUC = 0.79) based on 14,070 stereoscopic seven-field fundus images to provide a timely referral for a fast DR-progressing patient, enabling initiation of treatment prior to irreversible vision loss occurring.
Glaucoma
Glaucoma, characterized by cupping of the optic disc and visual field impairment, is the most frequent cause of irreversible blindness, affecting more than 70 million people worldwide.35,36,37 Due to population growth and aging globally, the number of patients with glaucoma will increase to 112 million by 2040.36 Most vision loss caused by glaucoma can be prevented via early diagnosis and timely treatment.38 However, identifying glaucoma at an early stage, particularly for primary open-angle glaucoma (POAG), normal tension glaucoma (NTG), and chronic primary angle-closure glaucoma (CPACG), is challenging for the following two reasons. First, POAG, NTG, and CPACG are often painless, and visual field defects are inconspicuous at an early stage. Therefore, self-detection of these types of glaucoma by affected people usually occurs at a relatively late stage when central visual acuity is reduced.35,37 Second, the primary approach to detect glaucoma is the examination of the optic disc and retinal nerve fiber layer by a glaucoma specialist through ophthalmoscopy or fundus images.39,40,41 Such manual optic disc assessment is time consuming and labor intensive, which is infeasible to implement in large populations. Accordingly, improvements in screening methods for glaucoma are necessary. AI may pave the road for cost-effective glaucoma screening programs, such as detecting glaucoma from fundus images or OCT images in an automated fashion (Table 2).
Table 2.
Major studies on the application of AI in glaucoma
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Xiong et al.42 | 2022 | retrospective | VFs and OCT images | 2,463 pairs | multimodal AI algorithm, FusionNet | detection of GON | AUC = 0.87–0.92, Sen = 77.3–81.3, Spec = 84.8–90.6 |
| Li et al.43 | 2022 | retrospective | TFI | 31,040 images and longitudinal data from 7,127 participants | U-Net and PredictNet | prediction of glaucoma incidence and progression | For predicting glaucoma incidence, AUC = 0.88–0.90; for predicting glaucoma progression, AUC = 0.87–0.91 |
| Fan et al.44 | 2022 | retrospective | TFI and VFs | 66,715 | ResNet50 | detection of POAG in patients with ocular hypertension | AUC = 0.74–0.91, Sen = 76–86, Spec = 80–85 |
| Li et al.45 | 2021 | retrospective | anterior-segment OCT images | 1.112 million images of 8694 vol scans | 3D-ResNet-34 and 3D-ResNet-50 | detection of narrow iridocorneal angles (task 1) and peripheral anterior synechiae (task 2) in eyes with suspected PACG | task 1 AUC = 0.943, Sen = 86.7, Spec = 87.8 Task 2 AUC = 0.902, Sen = 90.0, Spec = 89.0 |
| Dixit et al.46 | 2021 | retrospective | a longitudinal dataset of merged VFs and clinical data | 672,123 VF results and 350,437 samples of clinical data | convolutional long short-term memory neural network | assessment of glaucoma progression | AUC = 0.89–0.93 |
| Medeiros et al.47 | 2021 | retrospective | OCT images and TFI | 86,123 pairs | ResNet50 | detection of progressive GON damage | AUC = 0.86–0.96 |
| Li et al.12 | 2020 | retrospective | UWFI | 22,972 | InceptionResNetV2 | detection of GON | AUC = 0.98–1.00, Sen = 97.5–98.2, Spec = 94.3–98.4 |
| Yousefi et al.48 | 2020 | retrospective | VFs | 31,591 | PCA, manifold learning, and unsupervised clustering | monitoring glaucomatous functional loss | Sen = 77, Spec = 94 |
| Ran et al.49 | 2019 | retrospective | OCT images | 6,921 | ResNet-based 3D system | detection of GON | AUC = 0.89–0.97, Sen = 78–90, Spec = 79–96 |
| Martin et al.50 | 2018 | prospective | CLS parameters and initial IOP | 435 subjects | random forest | detection of POAG | AUC = 0.76 |
| Li et al.29 | 2018 | retrospective | TFI | 48,116 | Inception-v3 | detection of referable GON | AUC = 0.99, Sen = 95.6, Spec = 92.0 |
| Asaoka et al.51 | 2016 | retrospective | VFs | 279 | deep feedforward neural network | detection of preperimetric glaucoma | AUC = 0.93, Sen = 77.8, Spec = 90.0 |
CLS, contact lens sensor; GON, glaucomatous optic neuropathy; IOP, intraocular pressure; OCT, optical coherence tomography; PACG, primary angle-closure glaucoma; PCA, principal-component analysis; POAG, primary open-angle glaucoma; UWFI, ultra-widefield fundus images; VF, visual field.
Li et al.29 reported a deep-learning system with excellent performance in detecting referable glaucomatous optic neuropathy (GON) from fundus images. Specifically, they adopted the Inception-v3 algorithm to train the system and evaluated it in 8,000 images. Their system achieved an AUC of 0.986 with a sensitivity of 95.6% and a specificity of 92.0% for discerning referable GON. As fundus imaging is intrinsically a two-dimensional (2D) imaging modality observing the surface of the optic nerve head but glaucoma is a three-dimensional (3D) disease with depth-resolved structural changes, fundus imaging may not able to reach a level of accuracy that could be acquired through OCT, a 3D imaging modality.52,53 Ran et al.49 trained and tested a 3D deep-learning system using 6,921 spectral-domain OCT volumes of optic disc cubes from 1,384,200 2D cross-sectional scans. Their 3D system reached an AUC of 0.969 in detecting GON, significantly outperforming a 2D deep-learning system trained with fundus images (AUC, 0.921). This 3D system also had performance comparable with two glaucoma specialists with over 10 years of experience. The heatmaps indicated that the features leveraged by the 3D system for GON detection were similar to those leveraged by glaucoma specialists.
Primary angle-closure glaucoma is avoidable if the progress of angle closure can be stopped at the early stages.54 Fu et al. developed a deep-learning system using 4,135 anterior-segment OCT images from 2,113 individuals for the automated angle-closure detection.55 The system achieved an AUC of 0.96 with a sensitivity of 0.90 and a specificity of 0.92, which were better than those of the qualitative feature-based system (AUC, 0.90; sensitivity, 0.79; specificity, 0.87). These results indicate that deep learning may mine a broader range of details of anterior-segment OCT images than the qualitative features (e.g., angle opening distance, angle recess area, and iris area) determined by clinicians.
AI can also be used to predict glaucoma progression. Yousefi et al.56 reported an unsupervised machine-learning method to identify longitudinal glaucoma progression based on visual fields from 2,085 eyes of 1,214 subjects. They found that this machine-learning analysis detected progressing eyes earlier (3.5 years) than other methods such as global mean deviation (5.2 years), region-wise (4.5 years), and point-wise (3.9 years). Wang et al.57 proposed an AI approach, the archetype method, to detect visual field progression in glaucoma with an accuracy of 0.77. Moreover, this AI approach had a significantly higher agreement (kappa, 0.48) with the clinician assessment than other existing methods (e.g., the permutation of point-wise linear regression).
AMD
AMD, a disease that affects the macular area of the retina, often causes progressive loss of central vision.58 Age is the strongest risk factor for AMD and almost all late AMD cases happen in people at ages over 60 years.58 With the aging population, AMD will continue to be a major cause of vision impairment worldwide. The number of AMD patients will reach 288 million in 2040, denoting the substantial global burden of AMD.59 Consequently, screening for patients with AMD (especially neovascular AMD) and providing suitable medical interventions in a timely manner can reduce vision loss and improve patient visual outcomes.60
AI has the potential to facilitate the automated detection of AMD and prediction of AMD progression (Table 3). Peng et al.61 constructed and tested a deep-learning system (DeepSeeNet) using 59,302 fundus images from the longitudinal follow-up of 4,549 subjects from the Age-Related Eye Disease Study (AREDS). DeepSeeNet performed well on patient-based multi-class classification with AUCs of 0.94, 0.93, and 0.97 in detecting large drusen, pigmentary abnormalities, and late AMD, respectively. Burlina et al.62 reported a deep-learning system established by AlexNet based on over 130,000 fundus images from 4,613 patients to screen for referable AMD, and their system achieved an average AUC of 0.95. The referable AMD in their study refers to eyes with one of the following conditions: (1) large drusen (size larger than 125 μm); (2) multiple medium-sized drusen and pigmentation abnormalities; (3) choroidal neovascularization (CNV); (4) geographic atrophy.62
Table 3.
Major studies on the application of AI in AMD
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Potapenko et al.63 | 2022 | prospective | OCT images | 106,840 | temporal deep-learning model | detection of CNV activity in AMD patients | AUC = 0.90–0.98, Sen = 80.5–95.6, Spec = 84.6–95.9 |
| Yellapragada et al.64 | 2022 | retrospective | TFI | 100,848 | self-supervised non-parametric instance discrimination | classification of AMD severity | Acc = 65–87 |
| Rakocz et al.65 | 2021 | retrospective | 3D OCT images | 1,942 | SLIVER-net architecture | detection of AMD progression risk factors | AUC = 0.83–0.99 |
| Yim et al.8 | 2020 | retrospective | 3D OCT images | 130,327 | 3D U-Net, 3D dense convolution blocks, and 3D prediction network | predicting conversion to wet AMD | AUC = 0.75–0.89 |
| Hwang et al.66 | 2019 | retrospective | OCT images | 35,900 | VGG16, InceptionV3, and ResNet50 | differentiating normal macula and three AMD types (dry, inactive wet, and active wet AMD) | AUC = 0.98–0.99, Acc = 90.7–92.7 |
| Keel et al.67 | 2019 | retrospective | TFI | 142,725 | Inception-v3 | detection of neovascular AMD | AUC = 0.97–1.00, Sen = 96.7–100, Spec = 93.4–96.4 |
| Peng et al.61 | 2019 | retrospective | TFI | 59,302 | Inception-v3 | classification of patient-based AMD severity | AUC = 0.93–0.97 |
| Grassmann et al.68 | 2018 | retrospective | TFI | 126,211 | a network ensemble of six different neural net architectures | Predicting the AMD stage | Quadratic weighted κ = 0.92, Acc = 63.3 |
| Kermany et al.69 | 2018 | retrospective | OCT images | 208,130 | Inception-v3 | detection of referable AMD | AUC = 0.99–1.00, Sen = 96.6–97.8, Spec = 94.0–97.4 |
| Burlina et al.62 | 2017 | retrospective | TFI | 133,821 | AlexNet | detection of referable AMD | AUC = 0.94–0.96, Sen = 72.8–88.4, Spec = 91.5–94.1 |
Acc, accuracy; CNV, choroidal neovascularization.
AI also has the potential to predict the possibility of progression to late AMD, guiding high-risk patients to start preventive care early (e.g., eating healthy food, abandoning smoking, and taking supplements) and assisting clinicians to decide the interval of the patient’s follow-up examination. In patients diagnosed with wet AMD in one eye, Yim et al.8 introduced an AI system to predict conversion to wet AMD in the second eye. Their system was constructed by a segmentation network, diagnosis network, and prediction network based on 130,327 3D OCT images and corresponding automatic tissue maps for predicting progression to wet AMD within a clinically actionable time window (6 months). The system achieved 80% sensitivity at 55% specificity and 34% sensitivity at 90% specificity. As both genetic and environmental factors can affect the etiology of AMD, Yan et al.70 developed an AI approach with a modified deep convolutional neural network (CNN) using 52 AMD-associated genetic variants and 31,262 fundus images from 1,351 individuals from the AREDS to predict whether an eye would progress to late AMD. Their results showed that the approach based on both fundus images and genotypes could predict late AMD progression with an AUC of 0.85, whereas the approach based on fundus images alone achieved an AUC of 0.81.
Other retinal diseases
Numerous studies also have found that AI could be applied to promote the automated detection of other retinal diseases from clinical images to provide timely referrals for positive cases, solving the issues caused by the unbalanced distribution of ophthalmic medical resources. Milea et al.71 developed a deep-learning system using 14,341 fundus images to detect papilledema. This system achieved an AUC of 0.96 in the external test dataset consisting of 1,505 images. Brown et al.72 established a deep-learning system based on 5,511 retinal images captured by RetCam to diagnose plus disease in retinopathy of prematurity (ROP), a leading cause of blindness in childhood. The AUC of their system was 0.98 with a sensitivity of 93% and a specificity of 93%. In terms of detecting peripheral retinal diseases, such as lattice degeneration and retinal breaks, Li et al.73 trained models with four different deep-learning algorithms (InceptionResNetV2, ResNet50, InceptionV3, and VGG16) using 5,606 UWF images. They found that InceptionResNetV2 had the best performance, which achieved an AUC of 0.996 with 98.7% sensitivity and 99.2% specificity. In addition, AI has also been employed in the automated identification of retinal detachment,74 pathologic myopia,75 polypoidal choroidal vasculopathy,76 etc.
Application of AI algorithms in anterior-segment eye diseases
Cataract
In the past 20 years, although the prevalence of cataracts has been decreasing due to the increasing rates of cataract surgery because of improved techniques and active surgical initiatives, it still affects 95 million people worldwide.77 Cataract remains the leading cause of blindness (accounting for 50% of blindness), especially in low-income and middle-income countries.77 Therefore, exploring a set of strategies to promote cataract screening and related ophthalmic services is imperative. Recent advancements in AI may help achieve this goal, such as diagnosis and quantitative classification of age-related cataract from slit-lamp images (Table 4).
Table 4.
Major studies on the application of AI in cataract
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Keenan et al.16 | 2022 | retrospective | Slit-lamp images | 18,999 | DeepLensNet | diagnosis and quantitative classification of age-related cataract | Mean squared error = 0.23–16.6 |
| Tham et al.78 | 2022 | retrospective | TFI | 25,742 | ResNet50 and XGBoost classifier | detection of visually significant cataract | AUC = 0.92–0.97, Sen = 88.8–96.0, Spec = 81.1–90.3 |
| Xu et al.79 | 2021 | retrospective | TFI | 9,912 | Global-local attention network | Cataract diagnosis and grading | For cataract diagnosis: Acc = 90.7; for cataract grading: Acc = 83.5 |
| Lu et al.80 | 2021 | prospective | Slit-lamp images | 847 | Faster R-CNN and ResNet50 | Cataract grading | AUC = 0.80–0.98, Sen = 85.7–94.7, Spec = 63.6–93.2 |
| Lin et al.81 | 2020 | prospective | Participants’ demographic variables, birth conditions, family medical history, and environmental factors | 1,738 | Random forest and adaptive boosting methods | detection of congenital cataracts | AUC = 0.82–0.96, Sen = 56-82, Spec = 78-98 |
| Xu et al.82 | 2020 | retrospective | TFI | 8,030 | Hybrid global-local feature representation model | Cataract grading | Acc = 86.2, Sen = 79.8–95.0, Spec = 83.3–88.4 |
| Wu et al.83 | 2019 | retrospective | Slit-lamp images | 37,638 | ResNet50 | diagnosis of cataracts and detection of referable cataracts | For cataract diagnosis: AUC = 0.99–1.00; for detecting referable cataracts: AUC = 0.92–1.00 |
| Lin et al.84 | 2019 | prospective | Slit-lamp images | 700 | AI platform, CC-Cruiser | diagnosis of childhood cataracts and provision of treatment recommendation | For cataract diagnosis: Acc = 87.4; for treatment determination: Acc = 70.8 |
| Zhang et al.85 | 2019 | retrospective | TFI | 1,352 | ResNet18, SVM, and FCNN | Cataract grading | Acc = 92.7, Sen = 82.4–99.4, Spec = 81.3–98.5 |
| Gao et al.86 | 2015 | retrospective | Slit-lamp images | 5,378 | Convolutional-recursive neural network | Cataract grading | MAE = 0.304 |
FCNN, fully connected neural network; SVM, support vector machine.
Keenan et al.16 trained deep-learning models, named DeepLensNet, to detect and quantify nuclear sclerosis (NS) from 45° slit-lamp images and cortical lens opacity (CLO) and posterior subcapsular cataract (PSC) from retroillumination images. NS grading was considered on 0.9–7.1 scale. CLO and PSC grading were both considered as percentages. In the full test set, mean squared error values for DeepLensNet were 0.23 for NS, 13.1 for CLO, and 16.6 for PSC. The results indicate that this framework can perform automated and quantitative classification of cataract severity with high accuracy, which has the potential to increase the accessibility of cataract evaluation globally. Except for slit-lamp images, Tham et al.78 found that fundus images could also be used to develop an AI system for cataract screening. Based on 25,742 fundus images, they constructed a framework with ResNet50 and XGBoost classifier for the automated detection of visually significant cataracts (BCVA < 20/60), achieving AUCs of 0.916–0.965 in three external test sets. One merit of this system is that it can screen for cataracts with a single imaging modality, which is different from the traditional method that requires slit-lamp and retroillumination images alongside BCVA measurement. The other merit is that this system can be readily integrated into existing fundus-image-based AI systems, allowing simultaneous screening for other posterior-segment diseases.
Other than cataract screening, AI can also offer real-time guidance for phacoemulsification cataract surgery (PCS). Nespolo et al.87 invented a computer vision-based platform using a region-based CNN (Faster R-CNN) built on ResNet50, a k-means clustering technique, and an optical-flow-tracking technology to enhance the surgeon experience during the PCS. Specifically, this platform can be used to receive frames from the video source, locate the pupil, discern the surgical phase being performed, and provide visual feedback to the surgeon in real time. The results showed that the platform achieved AUCs of 0.996, 0.972, 0.997, and 0.880 for capsulorhexis, phacoemulsification, cortex removal, and idle-phase recognition, respectively, with a dice score of 90.23% for pupil segmentation and a mean processing speed of 97 frames per second. A usability survey suggested that most surgeons would be willing to perform PCS for complex cataracts with this platform and thought it was accurate and helpful.
Keratitis
Keratitis is a major global cause of corneal blindness, often affecting marginalized populations.88 The burden of corneal blindness on patients and the wider community can be huge, particularly as it tends to occur in people at a younger age than other blinding eye diseases such as AMD and cataracts.89 Keratitis can get worse quickly with time, which may lead to permanent visual impairment and even corneal perforation.90 Early detection and timely management of keratitis can halt the disease progression, resulting in a favorable prognosis.91
Li et al.15 found that AI had high accuracy in screening for keratitis and other corneal abnormalities from slit-lamp images. In terms of the deep-learning algorithms, they used Inception-v3, DenseNet121, and ResNet50, with DensNet121 performing best. To be specific, the optimal algorithm DenseNet121 reached AUCs of 0.988–0.997, 0.982–0.990, and 0.988–0.998 for the classification of keratitis, other corneal abnormalities (e.g., corneal dystrophies, corneal degeneration, corneal tumors), and normal cornea, respectively, in three external test datasets. Interestingly, their system also performed well on cornea images captured by smartphone under the super-macro mode, with an AUC of 0.967, a sensitivity of 91.9%, and a specificity of 96.9% in keratitis detection. This smartphone-based approach will be extremely cost-effective and convenient for proactive keratitis screening by high-risk people (e.g., farmers and contact lens wearers) if it can be applied to clinical practice.
To give prompt and precise treatment to patients with infectious keratitis, Xu et al.92 proposed a sequential-level deep-learning system that could effectively discriminate among bacterial keratitis, fungal keratitis, herpes simplex virus stromal keratitis, and other corneal diseases (e.g., phlyctenular keratoconjunctivitis, acanthamoeba keratitis, corneal papilloma), with an overall diagnostic accuracy of 80%, outperforming the mean diagnostic accuracy (49.27%) achieved by 421 ophthalmologists. The strength of this system was that it could extract the detailed patterns of the cornea region and assign local features to an ordered set to conform to the spatial structure and thereby learn the global features of the corneal image to perform diagnosis, which achieved better performance than conventional CNNs. Major AI applications in keratitis diagnosis are described in Table 5.
Table 5.
Major studies on the application of AI in keratitis
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Redd et al.93 | 2022 | retrospective | corneal images | 980 | MobileNet | differentiation of BK and FK | AUC = 0.83–0.86, For detecting BK, Acc = 75; for detecting FK, Acc = 81 |
| Ren et al.94 | 2022 | prospective | conjunctival swabs | 149 | random forest | differentiation of BK, FK, and VK using the conjunctival bacterial microbiome characteristics | for referring microbiota composition, Acc = 96.3; for referring gene functional composition, Acc = 93.8 |
| Wu et al.95 | 2022 | prospective | tear Raman spectroscopy | 75 | CNN and RNN | detection of keratitis | Acc = 92.7–95.4, Sen = 93.8–95.6, Spec = 94.8–97.4 |
| Tiwari et al.96 | 2022 | retrospective | corneal images | 2,746 | VGG-16 | differentiation of active corneal ulcers from healed scars | AUC = 0.95–0.97, Sen = 78.2–93.5, Spec = 84.4–91.3 |
| Li et al.15 | 2021 | retrospective | slit-lamp and smartphone images | 13,557 | DenseNet121 | detection of keratitis | AUC = 0.97–1.00, Sen = 91.9–97.7, Spec = 96.9–98.2 |
| Ghosh et al.97 | 2021 | retrospective | slit-lamp images | 2,167 | ensemble learning | discrimination between BK and FK | AUC = 0.90, Sen = 77, F1 score = 0.83 |
| Wang et al.98 | 2021 | retrospective | slit-lamp and smartphone images | 6,073 | Inception-v3 | differentiation of BK, FK, and VK | AUC = 0.85–0.96, QWK = 0.54–0.91 |
| Xu et al.92 | 2020 | retrospective | slit-lamp images | 115,408 | deep sequential feature learning | differentiation of BK, FK, VK, and other types of infectious keratitis | Acc = 80.0 |
| Lv et al.99 | 2020 | retrospective | IVCM images | 2,088 | ResNet101 | diagnosis of FK | AUC = 0.98–0.99, Sen = 82.6–91.9 Spec = 98.3–98.9 |
| Gu et al.100 | 2020 | retrospective | slit-lamp images | 5,325 | Inception-v3 | detection of infectious and non-infectious keratitis | AUC = 0.88–0.95 |
BK, bacterial keratitis; CNN, convolutional neural network; FK, fungal keratitis; IVCM, in vivo confocal microscopy; QWK, quadratic weighted kappa; RNN, recurrent neural network; VK, viral keratitis.
Keratoconus
Keratoconus is a progressive corneal ectasia with central or paracentral stroma thinning and corneal protrusion, resulting in irreversible visual impairment due to irregular corneal astigmatism or the loss of corneal transparency.101 Early identification of keratoconus, especially subclinical keratoconus, and subsequent treatment (e.g., corneal crosslinking and intrastromal corneal ring segments) are crucial to stabilize the disease and improve the visual prognosis.101 Advanced keratoconus can be detected by classic clinical signs (e.g., Vogt’s striae, Munson’s sign, Fleischer ring) through slit-lamp examination or by corneal topographical characteristics such as increased corneal refractive power, steeper radial axis tilt, and inferior-superior (I-S) corneal refractive asymmetry from corneal topographical maps. However, the detection of subclinical keratoconus remains challenging.102
AI may accurately diagnose subclinical keratoconus and keratoconus and predict their progress trends (Table 6). Luna et al.103 reported machine-learning techniques, decision tree, and random forest for the diagnosis of subclinical keratoconus based on Pentacam topographic and Corvis biomechanical metrics, such as the flattest keratometry curvature, steepest keratometry curvature, stiffness parameter at the first flattening, and corneal biomechanical index. The optimal model achieved an accuracy of 89% with a sensitivity of 93% and a specificity of 86%. Meanwhile, they found that the stiffness parameter at the first flattening was the most important determinant in identifying subclinical keratoconus. Timemy et al.104 introduced a hybrid deep-learning construct for the detection of keratoconus. This model was developed using corneal topographic maps from 204 normal eyes, 215 keratoconus eyes, and 123 subclinical keratoconus eyes and was tested in an independent dataset including 50 normal eyes, 50 keratoconus eyes, and 50 subclinical keratoconus eyes. The proposed model reached an accuracy of 98.8% with an AUC of 0.99 and F1 score of 0.99 for the two-class task (normal vs. keratoconus) and an accuracy of 81.5% with an AUC of 0.93 and F1 score of 0.81 for the three-class task (normal vs. keratoconus vs. subclinical keratoconus).
Table 6.
Major studies on the application of AI in keratoconus
| Study | Year | Study design | Data type | Data size | AI type | Task | Performance |
|---|---|---|---|---|---|---|---|
| Junior et al.105 | 2022 | retrospective | corneal tomographic images | 2,893 | BESTi | detection of subclinical KC | AUC = 0.99, Sen = 87.0–97.5, Spec = 93.9–98.5 |
| Timemy et al.104 | 2021 | retrospective | corneal tomographic images | 4,844 | EfficientNet-b0 and SVM | detection of suspected KC and KC | AUC = 0.93–0.99, Acc = 81.5–98.8 |
| García106 | 2021 | retrospective | KC patients measured with Pentacam | 743 | TDNN | detection of KC progression | Sen = 70.8, Spec = 80.6 |
| Luna et al.103 | 2021 | retrospective | subjects with Pentacam topographic and Corvis biomechanical metrics | 81 | decision tree and random forest | diagnosis of Subclinical KC | Acc = 89, Sen = 93, Spec = 86 |
| Xie et al.107 | 2020 | retrospective | corneal tomographic images | 6,465 | InceptionResNetV2 | detection of suspected irregular cornea and KC | AUC = 0.99, Sen = 91.9, Spec = 98.7 |
| Zéboulon et al.108 | 2020 | retrospective | subjects with corneal topography raw data | 3000 | ResNet | detection of KC | Acc = 99.3, Sen = 100, Spec = 100 |
| Shi et al.109 | 2020 | retrospective | eyes with both corneal tomographic and UHR-OCT images | 121 | machine-learning-derived classifier | detection of subclinical KC | AUC = 0.93, Sen = 98.5, Spec = 94.7 |
| Cao et al.110 | 2020 | retrospective | eyes with complete Pentacam parameters | 267 | random forest | detection of subclinical KC | Acc = 98, Sen = 97, Spec = 98 |
| Issarti et al.111 | 2019 | retrospective | subjects with corneal elevation and thickness data | 851 | FNN and Grossberg-Runge Kutta architecture | detection of suspected KC | Acc = 96.6, Sen = 97.8, Spec = 95.6 |
| Hidalgo et al.112 | 2016 | retrospective | eyes with Pentacam parameters | 860 | SVM | detection of KC | Acc = 98.9, Sen = 99.1, Spec = 98.5 |
BESTi, boosted ectasia susceptibility tomography index; FNN, feedforward neural network; KC, keratoconus; TDNN, time-delay neural network; UHR-OCT, ultra-high-resolution ocular coherence tomography.
Early and accurate prediction of progress trends in keratoconus is critical for the prudent and cost-effective use of corneal crosslinking and the determination of timing of follow-up visits. García et al.106 reported a time-delay neural network to predict keratoconus progression using two prior tomography measurements from Pentacam. This network received six characteristics as input (e.g., average keratometry, the steepest radius of the front surface, and the average radius of the back surface), evaluated in two consecutive examinations, forecasted the future values, and obtained the result (stable or suspect progressive) leveraging the significance of the variation from the baseline. The average positive and negative predictive values of the network were 71.4% and 80.2%, indicating it had the potential to assist clinicians to make a personalized management plan for patients with keratoconus.
Other anterior-segment diseases
A large number of studies have also proved the possibility of using AI to detect other anterior-segment diseases. For example, Chase et al.113 demonstrated that the deep-learning system, developed by a VGG19 network based on 27,180 anterior-segment OCT images, was able to identify dry eye diseases, with 84.62% accuracy, 86.36% sensitivity, and 82.35% specificity. The performance of this system was significantly better than some clinical dry eye tests, such as Schirmer’s test and corneal staining, and was comparable with that of tear break-up time and Ocular Surface Disease Index. In addition, Zhang et al.114 developed a deep-learning system for detecting obstructive meibomian gland dysfunction (MGD) and atrophic MGD using 4,985 in vivo laser confocal microscope images and validated the system on 1,663 images. The accuracy, sensitivity, and specificity of the system for obstructive MGD were 97.3%, 88.8%, and 95.4%, respectively; for atrophic MGD, 98.6%, 89.4%, and 98.4%, respectively; and for healthy controls, 98.0%, 94.5%, and 92.6%, respectively. Moreover, Li et al.3 introduced an AI system based on Faster R-CNN and DenseNet121 to detect malignant eyelid tumors from photographic images captured by ordinary digital cameras. In an external test set, the average precision score of the system was 0.762 for locating eyelid tumors and the AUC was 0.899 for discerning malignant eyelid tumors.
Application of AI algorithms in predicting systemic diseases based on retinal images
AI has the potential to detect hidden information that clinicians are normally unable to perceive from digital health data. In ophthalmology, with the continuous advancement of AI technologies, the application of AI based on retinal images has extended from the detection of multiple fundus diseases to the screening for systemic diseases. These breakthroughs can be attributed to the following three reasons: (1) the unique anatomy of the eye offers an accessible “window” for the in vivo visualization of microvasculature and cerebral neurons; (2) the retina manifestations can be signs of many systemic diseases, such as diabetes and heart disease; (3) retinal changes can be recorded through non-invasive digital fundus imaging, which is low cost and widely available in different levels of medical institutions.
Cardiovascular disease
Cardiovascular disease (CVD) is a leading cause of death globally, taking an estimated 17.9 million lives annually.115 Overt retinal vascular damage (such as retinal hemorrhages) and subtle changes (such as retinal arteriolar narrowing) are markers of CVD.116 To improve present risk-stratification approaches for CVD events, Rim et al.117 developed and validated a deep-learning-based cardiovascular risk-stratification system using 216,152 retinal images from five datasets from Singapore, South Korea, and the United Kingdom. This system achieved an AUC of 0.742 in predicting the presence of coronary artery calcium (a preclinical marker of atherosclerosis and strongly associated with the risk of CVD). Poplin et al.118 reported that deep-learning models trained on data from 284,355 patients could extract new information from retinal images to predict cardiovascular risk factors, such as age (mean absolute error [MAE] within 3.26 years), gender (AUC = 0.97), systolic blood pressure (MAE within 11.23 mm Hg), smoking status (AUC = 0.71), and major adverse cardiac events (AUC = 0.70). Meanwhile, they demonstrated that the deep-learning models generated each prediction using anatomical features, such as the retinal vessels or the optic disc.
Chronic kidney disease and type 2 diabetes
Chronic kidney disease (CKD) is a progressive disease with high morbidity and mortality that occurs in the general adult population, particularly in people with diabetes and hypertension.119 Type 2 diabetes is another common chronic disease that accounts for nearly 90% of the 537 million cases of diabetes worldwide.120 Early diagnosis and proactive management of CKD and diabetes are critical in reducing microvascular and macrovascular complications and mortality burden. As CKD and diabetes have manifestations in the retina, retinal images can be used to detect and monitor these diseases. Zhang et al.121 reported that deep-learning models developed based on 115,344 retinal images from 56,672 patients were able to detect CKD and type 2 diabetes solely from retinal images or in combination with clinical metadata (e.g., age, sex, body mass index, and blood pressure) with AUCs of 0.85–0.93. The models can also be utilized to predict estimated glomerular filtration rates and blood-glucose levels, with MAEs of 11.1–13.4 mL min−1 per 1.73 m2 and 0.65–1.1 mmol L−1, respectively.121 Sabanayagam et al.122 established a deep-learning algorithm using 12,790 retinal images to screen for CKD. In this study, the model trained solely by retinal images achieved AUCs of 0.733–0.911 in validation and testing datasets, indicating the feasibility of employing retinal photography as an adjunctive screening tool for CKD in community and primary care settings.122
Alzheimer’s disease
Alzheimer’s disease (AD), a progressive neurodegenerative disease, is the most common type of dementia in the elderly worldwide and is becoming one of the most lethal, expensive, and burdening diseases of this century.123 Diagnosis of AD is complex and normally involves expensive and sometimes invasive tests (such as amyloid positron emission tomography [PET] imaging and cerebrospinal fluid assays), which are not usually available outside of highly specialized clinical institutions. The retina is an extension of the central nervous system and offers a distinctively accessible insight into brain pathology. Research has found potentially measurable structural, vascular, and metabolic changes in the retina at the early stages of AD.124 Therefore, using noninvasive and low-cost retinal photography to detect AD is feasible. Cheung et al.125 demonstrated that a deep-learning model had the capability to identify AD from retinal images alone. They trained, validated, and tested the model using 12,949 retinal images from 648 AD patients and 3,240 individuals without the disease.125 The model had accuracies ranging from 79.6% to 92.1% and AUCs ranging from 0.73 to 0.91 for detecting AD in testing datasets. In the datasets with PET information, the model can also distinguish between participants who were β-amyloid positive and those who were β-amyloid negative, with accuracies ranging from 80.6% to 89.3% and AUCs ranging from 0.68 to 0.86. This study showed that a retinal-image-based deep-learning algorithm had high accuracy in detecting AD and this approach could be used to screen for AD in a community setting.
Challenges in the AI clinical translation
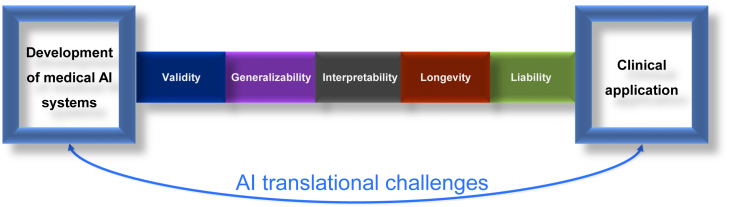
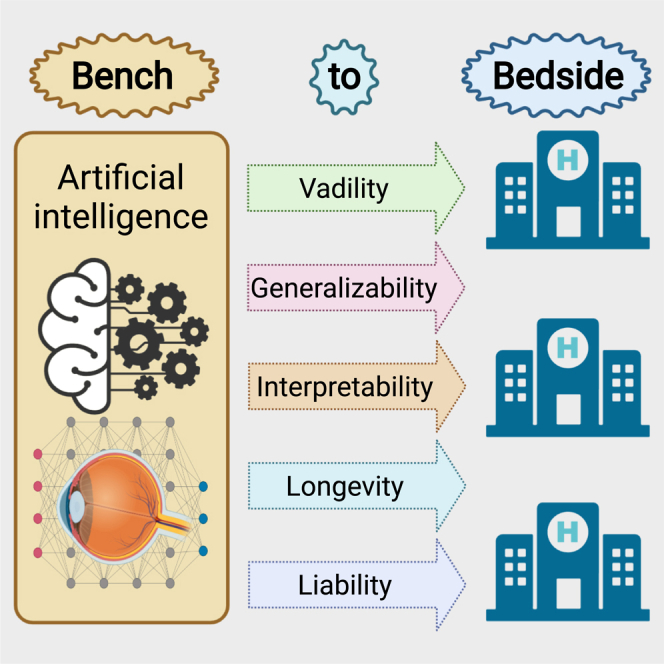
Although AI systems have shown great performance in a wide variety of retrospective studies, relatively few of them have been translated into clinical practice. Many challenges, such as the generalizability of AI systems, still exist and stand in the path of true clinical adoption of AI tools (Figure 3). In this section, we highlight some critical challenges and the research that has already been conducted to tackle these issues.
Figure 3.
Medical AI translational challenges between system development and routine clinical application
There are five major challenges in the path of AI clinical translation: validity, generalizability, interpretability, longevity, and liability.
Validity of AI systems
Data issues in developing robust AI systems
Data sharing
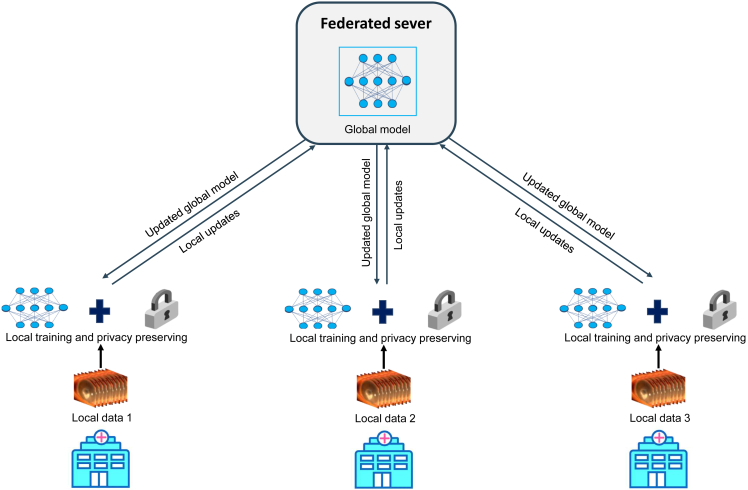
Large datasets are required to facilitate the development of a robust AI system. The lack of high-quality public datasets that are truly representative of real-world clinical practice stands in the path of clinical translation of AI systems. Data sharing might be a good solution but it generates ethical and legal challenges in general. Even if data are obtained in an anonymized manner, it can potentially put patient privacy at risk.126 Protecting patient privacy and acquiring approval for data use are important rules to comply with. Unfortunately, these rules may hinder data sharing among different medical research groups, not to mention making the data publicly available. The adoption of federated learning is a good alternative to training AI models with diverse data from multiple clinical institutions without the centralization of data.127 This strategy can address the issue that data reside in different institutions, removing barriers to data sharing and circumventing the problem of patient privacy (Figure 4). Meanwhile, federated learning can facilitate rapid data science collaboration and thus improve the robustness of AI systems.128 In addition, controlled-access data sharing, an approach that requires a request for access to datasets to be approved, is another alternative solution for researchers to acquire data to solve relevant research issues while protecting participant privacy.
Figure 4.
Framework of federated learning
Local hospitals are given a copy of a current global model from a federated server to train on their own datasets. After a certain number of iterations, the local hospitals send model updates back to the federated server and keep their datasets in their own secure infrastructure. The federated server aggerates the contributions from these hospitals. Then the updated global model is shared with the local hospitals and they can continue local training. The main advantage of federated learning is that it establishes a global model without directly sharing datasets, preserving patient privacy across sites.
Data annotation
Accurate clinical data annotations are crucial for the development of reliable AI systems,2,129 as annotation inconsistencies may lead to unpredictable clinical consequences, such as erroneous classifications.130 Several approaches have been leveraged to resolve disagreements between graders and obtain ground truth. One method consists of adopting the majority decision from a panel of three or more professional graders.7 Another consists of recruiting two or more professional graders to label data independently and then employing another senior grader to arbitrate disagreements, with the senior grader’s decision used as the ground truth.131 Third, some data annotations can be conducted using the recognized gold standard. For example, Li et al.3 annotated images of benign and malignant eyelid tumors based on unequivocal histopathological diagnoses.
Normally, annotations can be divided into two categories: the annotation of interest regions in images (e.g., retinal hemorrhages, exudates, and drusen) and clinical annotations (e.g., disease classification, treatment response, vision prognosis). Conducting manual annotations for large-scale datasets before the model training is a considerably time-consuming and labor-intensive task that needs a lot of professional graders or ophthalmologists, hindering the construction of robust AI systems.132,133,134,135 Therefore, exploring techniques to promote the efficient production of annotations is important, although manual annotations are still necessary. Training models using weakly supervised learning may be a good approach to reduce the workload of manual annotations.133,134 For image segmentation, weak supervision requires sparse manual annotations of small interest regions using dots via experts, whereas full supervision needs dense annotations, in which all pixels of images are manually labeled.132,134 Playout et al.132 have reported that weak supervision in combination with advanced learning in model training can achieve performance comparable with fully supervised models for retinal lesion segmentation in fundus images.
Standardization of clinical data collection
In the past two decades, health systems have heavily invested in the digitalization of every aspect of their operation. This transformation has resulted in unprecedented growth in the volume of medical electronic data, facilitating the development of AI-based medical devices.136 Although the size of datasets has increased, data collection is not done in a standardized manner, affecting the ready utilization of these data for AI model training and testing. This issue leads to a growing number of multicentric efforts to deal with the large variability in examination items, the timing of laboratory tests, image quality, etc. To improve the usability of data, standardization of clinical data collection should be implemented to generate high-quality data with complete and consistent information to support the development of robust medical AI products.137,138 For example, medical text data collection should include basic information such as age, gender, and examination date, and health examination records should have all examination items and complete results. Besides, as low-quality image data often result in a loss of diagnostic information and affect AI-based image analyses,139 image quality assessment is necessary at the stage of data allocation to filter out low-quality images, which could improve the performance of AI-based diagnostic models in real-world settings.140 To address this issue, Liu et al.141 developed a deep-learning-based flow-cytometry-like image quality classifier for the automated, high-throughput, and multidimensional classification of fundus image quality, which can detect low-quality images in real time and then guide a photographer to acquire high-quality images immediately. Li et al.142 reported a deep-learning-based image-quality-control system that could discern low-quality slit-lamp images. This system can be used as a prescreening tool to filter out low-quality images and ensure that only high-quality images will be transferred to the subsequent AI-based diagnostic systems. Shen et al.143 established a new multi-task domain adaptation framework for the automated fundus image quality assessment. The proposed framework can offer interpretable quality assessment with both quantitative scores and quality visualization, which outperforms different state-of-the-art methods. Dai et al.23 demonstrated that the AI-based image quality assessment could reduce the proportion of poor-quality images and significantly improve the accuracy of an AI model for DR diagnosis.
Real-world performance of AI systems
Recently, several reports showed that AI systems in practice were less helpful than retrospective studies described.144,84 For example, Kanagasingam et al.144 evaluated their deep-learning-based system for DR screening based on retinal images in a real-world primary care clinic. They found that the system had a high false-positive rate (7.9%) and a low positive predictive value (12%). The possible reason for this unsatisfactory performance of the system is that the incidence of DR in the primary care clinic was 1%, whereas their AI system was developed using retrospective data in which the incidence of DR was much higher (33.3%). Long et al.145 developed an AI platform that had 98.25% accuracy in childhood cataract diagnosis and 92.86% accuracy in treatment suggestions in external datasets retrospectively collected from three hospitals. However, when they applied the platform to unselected real-world datasets prospectively obtained from five hospitals, accuracies decreased to 87.4% in cataract diagnosis and 70.8% in treatment determination.84 The possible explanation for this phenomenon is that the retrospective datasets often undergo extensive filtering and cleaning, which makes them less representative of real-world clinical practice. Randomized controlled trials (RCTs) and prospective research can bridge such gaps between theory and practice, showing the true performance of AI systems in real healthcare settings and demonstrating how useful the systems are for the clinic.146
Generalizability of AI systems to clinical practice
Although numerous studies reported that their AI systems showed robust performance in detecting eye diseases and had the potential to be applied in clinics, most AI-based medical devices had not yet been authorized for market distribution for clinical management of diseases such as AMD, glaucoma, and cataracts. One of the most important reasons for this is that the generalizability of AI systems to populations of different ethnicities and different countries, different clinical application scenarios, and images captured using different types of cameras remains uncertain. Lots of AI studies only evaluated their systems in data from a single source, hence the systems often performed poorly in real-world datasets that had more sources of variation than the datasets utilized in research papers.84 To improve the generalizability of AI systems, first, we need to build large, multicenter, and multiethnic datasets for system development and evaluation. Milea et al.71 developed a deep-learning system for papilledema detection using data collected from 19 sites in 11 countries and evaluated the system in data obtained from five other sites in five countries. The AUCs of their system in internal and external test datasets were 0.99 and 0.96, verifying that the system had broad generalizability. In addition, transfer learning, a technique that aims to transfer knowledge from one task to a different but related task, can help decrease generalization errors of AI systems via reusing the weights of a pretrained model, particularly when faced with tasks with limited data.147 Kermany et al.69 demonstrated that AI systems trained with a transfer-learning algorithm had good performance and generalizability in the diagnosis of common diseases from different types of images, such as detecting diabetic macular edema from OCT images (accuracy = 98.2%) and pediatric pneumonia from chest X-ray images (accuracy = 92.8%). Third, the generalizability of AI networks can be improved by utilizing a data-augmentation (DA) strategy that creates more training samples for increasing the diversity of the training data.148 Zhou et al.149 proposed an approach named DA-based feature alignment that could consistently and significantly improve the out-of-distribution generalizability (up to +16.3% mean of clean AUC) of AI algorithms in glaucoma detection from fundus images. Fourth, an AI algorithm trained based on lesion labels can broaden its generalizability in disease detection. Li et al.150 reported that the algorithm trained with the image-level classification labels and the anatomical and pathological labels displayed better performance and generalizability than that trained with only the image-level classification labels in diagnosing ophthalmic disorders from slit-lamp images (accuracies, 99.22%–79.47% versus 90.14%–47.19%).
Interpretability of AI systems
AI systems are often described as black boxes due to the nature of these systems (being trained instead of being explicitly programmed).151 It is difficult for clinicians to understand the precise underlying functioning of the systems. As a result, correcting some erroneous behaviors might be difficult, and acceptance by clinicians as well as regulatory approval might be hampered. Decoding AI for clinicians can mitigate such uncertainty. This challenge has provided a stimulus for research groups and industries to focus on explainable AI. Techniques that enable a good understanding of the working principle of AI systems are developed. For instance, Niu et al.152 reported a method that could enhance the interpretability of an AI system in detecting DR. To be specific, they first define novel pathological descriptors leveraging activated neurons of the DR detector to encode both the appearance and spatial information of lesions. Then, they proposed a novel generative adversarial network (GAN), Patho-GAN, to visualize the signs that the DR detector identified as evidence to make a prediction. Xu et al.153 developed an explainable AI system for diagnosing fungal keratitis from in vivo confocal microscopy images based on gradient-weighted class activation mapping (Grad-CAM) and guided Grad-CAM techniques. They found that the assistance from the explainable AI system could boost ophthalmologists’ performance beyond what was achievable by the ophthalmologist alone or with the black-box AI assistance. Overall, these interpretation frameworks may facilitate AI acceptance for clinical usage.
Longevity of AI systems
The performance of AI systems has the potential to degrade over time as the characteristics of the world, such as disease distribution, population characteristics, health infrastructure, and cyber technologies, are changing all the time. This requires that AI systems should have the ability of lifelong continuous learning to keep and even improve their performance over time. The continuous learning technique, meta-learning, which aims to improve the AI algorithm itself, is a potential approach to address this issue.154
Liability
Although medical AI systems can help physicians in clinics, such as disease diagnosis, recommendations for treatment, and prognosis prediction, it is still unclear whether healthcare providers, developers, sellers, or regulators should be held accountable if an AI system makes mistakes in real-world clinical practice even after being thoroughly clinically validated. For example, AI systems may miss a retinal disease in a fundus image or recommend an incorrect treatment strategy. As a result, patients may be injured. In this case, we have to determine who is responsible for this incident. The allocation of liability makes clear not only whether and from whom patients acquire redress but also whether, potentially, AI systems will make their way into clinical practice.155 At present, the suggested solution is to treat medical AI systems as a confirmatory tool rather than as a source of ways to improve care.156 In other words, a physician should check every output from the medical AI systems to ensure the results that meet and follow the standard of care. Therefore, the physician would be held liable if malpractice occurs due to using these systems. This strategy may minimize the potential value of medical AI systems as some systems may perform better than even the best physicians but the physicians would choose to ignore the AI recommendation when it conflicts with standard practice. Consequently, the approach that can balance the safety and innovation of medical AI needs to be further explored.
Future directions
Generally, AI models were directly trained using existing open-sourced machine-learning packages frequently utilized by others to address the issue of interest without additional customization or refinement. This approach may limit the optimal performance of AI applications as no generalized solution exists in most cases. To improve the performance of AI tools, in-depth knowledge of clinical problems as well as the features of AI algorithms is indispensable. Therefore, applicable customization of the algorithms should be conducted according to the specific challenges of each problem, which usually needs interdisciplinary collaboration among ophthalmologists, computer scientists (e.g., AI experts), policymakers, and others.
Although AI studies have seen enormous progress in the past decade, they are predominantly based on fixed datasets and stationary environments. The performance of AI systems is often fixed by the time they are developed. However, the world is not stationary, which requires that AI systems should have the ability as clinicians to improve themselves constantly and evolve to thrive in dynamic learning settings. Continual learning techniques, such as gradient-based learning, modular neural network, and meta-learning, may enable AI models to obtain specialized solutions without forgetting previous ones, namely learning over a lifetime, as a clinician does.157 These techniques may take AI to a higher level by improving learning efficiency and enabling knowledge transfer between related tasks.
In addition to current diagnostic and predictive tasks, AI methods can also be employed to support ophthalmologists with additional information impossible to obtain by sole visual inspection. For instance, the objective quantification of the area of corneal ulcer via a combination of segmentation and detection techniques can assist ophthalmologists in precisely evaluating whether the treatment is effective on patients in follow-up visits. If the area becomes smaller, it indicates that the condition has improved. Otherwise, it denotes that the condition has worsened and treatment strategies may need to change.
To date, AI is not immune to the garbage-in, garbage-out weakness, even with big data. Appropriate data preprocessing to acquire high-quality training sets is critical to the success of AI systems.7,158 While AI systems have good performance (e.g., detecting corneal diseases) in high-quality images, they often perform poorly in low-quality images.159,160 Nevertheless, the performance of human doctors in low-quality images is better than that of an AI system, exposing a vulnerability of the AI system.159 As low-quality images are inevitable in real-world settings,142,161 exploring approaches that can improve the performance of AI systems in low-quality images is needed to enhance the robustness of AI-based products in clinical practice.
A lot of studies have drawn overly optimistic conclusions based on AI systems’ good performance on external validation datasets. However, such results are not evidence of the clinical usefulness of AI systems.162 Well-conducted and well-reported prospective studies are essential to provide proof to truly demonstrate the added value of AI systems in ophthalmology and pave the way to clinical implementation. Recent guidelines, such as the Standard Protocol Items: Recommendations for Interventional Trials (SPIRIT)-AI extension, Consolidated Standards of Reporting Trials (CONSORT)-AI extension, and Standards for Reporting of Diagnostic Accuracy Study (STARD)-AI, may improve the design, transparency, reporting, and nuanced conclusions of AI studies, rigorously validating the usefulness of medical AI, and ultimately improving the quality of patient care.163,164,165 In addition, an international team established an evaluation framework termed Translational Evaluation of Healthcare AI (TEHAI) focusing on the assessment of translational aspects of AI systems in medicine.166 The evaluation components (e.g., capability, utility, and adoption) of TEHAI can be used at any stage of the development and deployment of medical AI systems.166
Patient privacy and data security are major concerns in medical AI development and application. Several approaches may help address these issues. First, sensitive data should be obtained and used in research with patient consent, and anonymization and aggregation strategies should be adopted to obscure personal details. Any clinical institution should handle patient data responsibly, for example, by utilizing appropriate security protocols. Second, differential privacy (DP), a data-perturbation-based privacy approach that is able to retain the global information of a dataset while reducing information about a single individual, can be employed to reduce privacy risks and protect data security.167 Based on this approach, an outside observer cannot infer whether a specific individual was utilized for acquiring a result from the dataset. Third, homomorphic encryption, an encryption scheme that allows computation on encrypted data, is widely treated as a gold standard for data security. This approach has successfully been applied to AI algorithms and to the data that allow secure and joint computation.168
Recently, regulatory agencies, such as the Food and Drug Administration (FDA), have proposed a regulatory framework to evaluate the safety and effectiveness of AI-based medical devices during the initial premarket review.169 Specifically, manufacturers have to illustrate what aspects they intend to achieve through AI devices, and how the devices will learn and change while remaining effective and safe, as well as strategies to reduce performance loss. This regulatory framework is good guidance for research groups to better develop and report their AI-based medical products.
Conclusions
AI in ophthalmology has made huge strides over the past decade. Plenty of studies have shown that the performance of AI is equal to and even superior to that of ophthalmologists in many diagnostic and predictive tasks. However, much work remains to be done before deploying AI products from bench to bedside. Issues such as real-world performance, generalizability, and interpretability of AI systems are still insufficiently investigated and will require more attention in future studies. The solution of data sharing, data annotation, and other related problems will facilitate the development of more robust AI products. Strategies such as customization of AI algorithms for a specific clinical task and utilization of continual learning techniques may further improve AI’s ability to serve patients. RCTs and prospective studies following special guidelines (e.g., SPIRIT-AI extension, STARD-AI, and FDA’s guidance) can rigorously demonstrate whether AI devices would bring a positive impact to real healthcare settings, contributing to the clinical translation of these devices. Although this field is not completely mature yet, we hope AI will play an important role in the future of ophthalmology, making healthcare more efficient, accurate, and accessible, especially in regions lacking ophthalmologists.
Acknowledgments
This study received funding from the National Natural Science Foundation of China (grant nos. 82201148, 62276210), the Natural Science Foundation of Zhejiang Province (grant no. LQ22H120002), the Medical and Health Science and Technology Project of Zhejiang Province (grant nos. 2022RC069, 2023KY1140), the Natural Science Foundation of Ningbo (grant no. 2023J390), the Natural Science Basic Research Program of Shaanxi (grant no. 2022JM-380), and Ningbo Science & Technology Program (grant no. 2021S118). The funding organization played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conception and design, Z.L., Y.S., and W.C.; identification of relevant literature, Z.L., L.W., X.W., J.J., W.Q., H.X., H.Z., S.W., Y.S., and W.C.; manuscript writing, all authors; final approval of the manuscript, all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101095.
Contributor Information
Zhongwen Li, Email: li.zhw@qq.com.
Yi Shao, Email: freebee99@163.com.
Wei Chen, Email: chenwei@eye.ac.cn.
Supplemental information
References
- 1.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 2.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 3.Li Z., Qiang W., Chen H., Pei M., Yu X., Wang L., Li Z., Xie W., Wu X., Jiang J., Wu G. Artificial intelligence to detect malignant eyelid tumors from photographic images. NPJ Digit. Med. 2022;5:23. doi: 10.1038/s41746-022-00571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lotter W., Diab A.R., Haslam B., Kim J.G., Grisot G., Wu E., Wu K., Onieva J.O., Boyer Y., Boxerman J.L., et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat. Med. 2021;27:244–249. doi: 10.1038/s41591-020-01174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landhuis E. Deep learning takes on tumours. Nature. 2020;580:551–553. doi: 10.1038/d41586-020-01128-8. [DOI] [PubMed] [Google Scholar]
- 6.Ting D.S.W., Cheung C.Y.L., Lim G., Tan G.S.W., Quang N.D., Gan A., Hamzah H., Garcia-Franco R., San Yeo I.Y., Lee S.Y., et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 8.Yim J., Chopra R., Spitz T., Winkens J., Obika A., Kelly C., Askham H., Lukic M., Huemer J., Fasler K., et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat. Med. 2020;26:892–899. doi: 10.1038/s41591-020-0867-7. [DOI] [PubMed] [Google Scholar]
- 9.Li Z., Guo C., Nie D., Lin D., Cui T., Zhu Y., Chen C., Zhao L., Zhang X., Dongye M., et al. Automated detection of retinal exudates and drusen in ultra-widefield fundus images based on deep learning. Eye. 2022;36:1681–1686. doi: 10.1038/s41433-021-01715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;182:1360. doi: 10.1016/j.cell.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brendlin A.S., Peisen F., Almansour H., Afat S., Eigentler T., Amaral T., Faby S., Calvarons A.F., Nikolaou K., Othman A.E. A Machine learning model trained on dual-energy CT radiomics significantly improves immunotherapy response prediction for patients with stage IV melanoma. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Guo C., Lin D., Nie D., Zhu Y., Chen C., Zhao L., Wang J., Zhang X., Dongye M., et al. Deep learning for automated glaucomatous optic neuropathy detection from ultra-widefield fundus images. Br. J. Ophthalmol. 2021;105:1548–1554. doi: 10.1136/bjophthalmol-2020-317327. [DOI] [PubMed] [Google Scholar]
- 13.Dow E.R., Keenan T.D.L., Lad E.M., Lee A.Y., Lee C.S., Loewenstein A., Eydelman M.B., Chew E.Y., Keane P.A., Lim J.I., Collaborative Community for Ophthalmic Imaging Executive Committee and the Working Group for Artificial Intelligence in Age-Related Macular Degeneration From data to deployment: the collaborative community on ophthalmic imaging roadmap for artificial intelligence in Age-Related macular degeneration. Ophthalmology. 2022;129:e43–e59. doi: 10.1016/j.ophtha.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ting D.S.J., Foo V.H., Yang L.W.Y., Sia J.T., Ang M., Lin H., Chodosh J., Mehta J.S., Ting D.S.W. Artificial intelligence for anterior segment diseases: emerging applications in ophthalmology. Br. J. Ophthalmol. 2021;105:158–168. doi: 10.1136/bjophthalmol-2019-315651. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Jiang J., Chen K., Chen Q., Zheng Q., Liu X., Weng H., Wu S., Chen W. Preventing corneal blindness caused by keratitis using artificial intelligence. Nat. Commun. 2021;12:3738. doi: 10.1038/s41467-021-24116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keenan T.D.L., Chen Q., Agrón E., Tham Y.C., Goh J.H.L., Lei X., Ng Y.P., Liu Y., Xu X., Cheng C.Y., et al. DeepLensNet: deep learning automated diagnosis and quantitative classification of cataract type and severity. Ophthalmology. 2022;129:571–584. doi: 10.1016/j.ophtha.2021.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt-Erfurth U., Bogunovic H., Sadeghipour A., Schlegl T., Langs G., Gerendas B.S., Osborne A., Waldstein S.M. Machine learning to analyze the prognostic value of current imaging biomarkers in neovascular Age-Related macular degeneration. Ophthalmol. Retina. 2018;2:24–30. doi: 10.1016/j.oret.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Burton M.J., Ramke J., Marques A.P., Bourne R.R.A., Congdon N., Jones I., Ah Tong B.A.M., Arunga S., Bachani D., Bascaran C., et al. The lancet global health commission on global eye health: vision beyond 2020. Lancet Global Health. 2021;9:e489–e551. doi: 10.1016/S2214-109X(20)30488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung N., Mitchell P., Wong T.Y. Diabetic retinopathy. Lancet. 2010;376:124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 20.Vujosevic S., Aldington S.J., Silva P., Hernández C., Scanlon P., Peto T., Simó R. Screening for diabetic retinopathy: new perspectives and challenges. Lancet Diabetes Endocrinol. 2020;8:337–347. doi: 10.1016/S2213-8587(19)30411-5. [DOI] [PubMed] [Google Scholar]
- 21.WHO . 2020. Diabetic Retinopathy Screening: A Short Guide: Increase Effectiveness, Maximize Benefits and Minimize Harm.https://apps.who.int/iris/handle/10665/336660 [Google Scholar]
- 22.Liu X., Ali T.K., Singh P., Shah A., McKinney S.M., Ruamviboonsuk P., Turner A.W., Keane P.A., Chotcomwongse P., Nganthavee V., et al. Deep learning to detect OCT-derived diabetic macular edema from color retinal photographs: a multicenter validation study. Ophthalmol. Retina. 2022;6:398–410. doi: 10.1016/j.oret.2021.12.021. [DOI] [PubMed] [Google Scholar]
- 23.Dai L., Wu L., Li H., Cai C., Wu Q., Kong H., Liu R., Wang X., Hou X., Liu Y., et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat. Commun. 2021;12:3242. doi: 10.1038/s41467-021-23458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee A.Y., Yanagihara R.T., Lee C.S., Blazes M., Jung H.C., Chee Y.E., Gencarella M.D., Gee H., Maa A.Y., Cockerham G.C., et al. Multicenter, Head-to-Head, Real-World validation study of seven automated artificial intelligence diabetic retinopathy screening systems. Diabetes Care. 2021;44:1168–1175. doi: 10.2337/dc20-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araújo T., Aresta G., Mendonça L., Penas S., Maia C., Carneiro Â., Mendonça A.M., Campilho A. DR|GRADUATE: uncertainty-aware deep learning-based diabetic retinopathy grading in eye fundus images. Med. Image Anal. 2020;63 doi: 10.1016/j.media.2020.101715. [DOI] [PubMed] [Google Scholar]
- 26.Heydon P., Egan C., Bolter L., Chambers R., Anderson J., Aldington S., Stratton I.M., Scanlon P.H., Webster L., Mann S., et al. Prospective evaluation of an artificial intelligence-enabled algorithm for automated diabetic retinopathy screening of 30 000 patients. Br. J. Ophthalmol. 2021;105:723–728. doi: 10.1136/bjophthalmol-2020-316594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natarajan S., Jain A., Krishnan R., Rogye A., Sivaprasad S. Diagnostic accuracy of Community-Based diabetic retinopathy screening with an offline artificial intelligence system on a smartphone. JAMA Ophthalmol. 2019;137:1182–1188. doi: 10.1001/jamaophthalmol.2019.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulshan V., Rajan R.P., Widner K., Wu D., Wubbels P., Rhodes T., Whitehouse K., Coram M., Corrado G., Ramasamy K., et al. Performance of a Deep-Learning algorithm vs manual grading for detecting diabetic retinopathy in India. JAMA Ophthalmol. 2019;137:987–993. doi: 10.1001/jamaophthalmol.2019.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Keel S., Liu C., He Y., Meng W., Scheetz J., Lee P.Y., Shaw J., Ting D., Wong T.Y., et al. An automated grading system for detection of Vision-Threatening referable diabetic retinopathy on the basis of color fundus photographs. Diabetes Care. 2018;41:2509–2516. doi: 10.2337/dc18-0147. [DOI] [PubMed] [Google Scholar]
- 30.Tang F., Luenam P., Ran A.R., Quadeer A.A., Raman R., Sen P., Khan R., Giridhar A., Haridas S., Iglicki M., et al. Detection of diabetic retinopathy from Ultra-Widefield scanning laser ophthalmoscope images: a multicenter deep learning analysis. Ophthalmol. Retina. 2021;5:1097–1106. doi: 10.1016/j.oret.2021.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Engelmann J., McTrusty A.D., MacCormick I.J.C., Pead E., Storkey A., Bernabeu M.O. Detecting multiple retinal diseases in ultra-widefield fundus imaging and data-driven identification of informative regions with deep learning. Nat. Mach. Intell. 2022;4:1143–1154. [Google Scholar]
- 32.Liu Y., Wang M., Morris A.D., Doney A.S.F., Leese G.P., Pearson E.R., Palmer C.N.A. Glycemic exposure and blood pressure influencing progression and remission of diabetic retinopathy: a longitudinal cohort study in GoDARTS. Diabetes Care. 2013;36:3979–3984. doi: 10.2337/dc12-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bora A., Balasubramanian S., Babenko B., Virmani S., Venugopalan S., Mitani A., de Oliveira Marinho G., Cuadros J., Ruamviboonsuk P., Corrado G.S., et al. Predicting the risk of developing diabetic retinopathy using deep learning. Lancet. Digit. Health. 2021;3:e10–e19. doi: 10.1016/S2589-7500(20)30250-8. [DOI] [PubMed] [Google Scholar]
- 34.Arcadu F., Benmansour F., Maunz A., Willis J., Haskova Z., Prunotto M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit. Med. 2019;2:92. doi: 10.1038/s41746-019-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonas J.B., Aung T., Bourne R.R., Bron A.M., Ritch R., Panda-Jonas S. Glaucoma. Lancet. 2017;390:2183–2193. doi: 10.1016/S0140-6736(17)31469-1. [DOI] [PubMed] [Google Scholar]
- 36.Tham Y.C., Li X., Wong T.Y., Quigley H.A., Aung T., Cheng C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GBD 2019 Blindness and Vision Impairment Collaborators. Vision Loss Expert Group of the Global Burden of Disease Study. Briant P.S., Flaxman S.R., Taylor H.R.B., Jonas J.B., Abdoli A.A., Abrha W.A., Abualhasan A., Abu-Gharbieh E.G., et al. Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Global Health. 2021;9:e144–e160. doi: 10.1016/S2214-109X(20)30489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M., Lin C., Weinreb R.N., Lai G., Chiu V., Leung C.K.S. Risk of visual field progression in glaucoma patients with progressive retinal nerve fiber layer thinning: a 5-Year prospective study. Ophthalmology. 2016;123:1201–1210. doi: 10.1016/j.ophtha.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 40.King A., Azuara-Blanco A., Tuulonen A. Glaucoma. BMJ. 2013;346:f3518. doi: 10.1136/bmj.f3518. [DOI] [PubMed] [Google Scholar]
- 41.Hollands H., Johnson D., Hollands S., Simel D.L., Jinapriya D., Sharma S. Do findings on routine examination identify patients at risk for primary open-angle glaucoma? The rational clinical examination systematic review. JAMA. 2013;309:2035–2042. doi: 10.1001/jama.2013.5099. [DOI] [PubMed] [Google Scholar]
- 42.Xiong J., Li F., Song D., Tang G., He J., Gao K., Zhang H., Cheng W., Song Y., Lin F., et al. Multimodal machine learning using visual fields and peripapillary circular OCT scans in detection of glaucomatous optic neuropathy. Ophthalmology. 2022;129:171–180. doi: 10.1016/j.ophtha.2021.07.032. [DOI] [PubMed] [Google Scholar]
- 43.Li F., Su Y., Lin F., Li Z., Song Y., Nie S., Xu J., Chen L., Chen S., Li H., et al. A deep-learning system predicts glaucoma incidence and progression using retinal photographs. J. Clin. Invest. 2022;132 doi: 10.1172/JCI157968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan R., Bowd C., Christopher M., Brye N., Proudfoot J.A., Rezapour J., Belghith A., Goldbaum M.H., Chuter B., Girkin C.A., et al. Detecting glaucoma in the ocular hypertension study using deep learning. JAMA Ophthalmol. 2022;140:383–391. doi: 10.1001/jamaophthalmol.2022.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F., Yang Y., Sun X., Qiu Z., Zhang S., Tun T.A., Mani B., Nongpiur M.E., Chansangpetch S., Ratanawongphaibul K., et al. Digital gonioscopy based on three-dimensional Anterior-Segment OCT: an international multicenter study. Ophthalmology. 2022;129:45–53. doi: 10.1016/j.ophtha.2021.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Dixit A., Yohannan J., Boland M.V. Assessing glaucoma progression using machine learning trained on longitudinal visual field and clinical data. Ophthalmology. 2021;128:1016–1026. doi: 10.1016/j.ophtha.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medeiros F.A., Jammal A.A., Mariottoni E.B. Detection of progressive glaucomatous optic nerve damage on fundus photographs with deep learning. Ophthalmology. 2021;128:383–392. doi: 10.1016/j.ophtha.2020.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yousefi S., Elze T., Pasquale L.R., Saeedi O., Wang M., Shen L.Q., Wellik S.R., De Moraes C.G., Myers J.S., Boland M.V. Monitoring glaucomatous functional loss using an artificial Intelligence-Enabled dashboard. Ophthalmology. 2020;127:1170–1178. doi: 10.1016/j.ophtha.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran A.R., Cheung C.Y., Wang X., Chen H., Luo L.Y., Chan P.P., Wong M.O.M., Chang R.T., Mannil S.S., Young A.L., et al. Detection of glaucomatous optic neuropathy with spectral-domain optical coherence tomography: a retrospective training and validation deep-learning analysis. Lancet. Digit. Health. 2019;1:e172–e182. doi: 10.1016/S2589-7500(19)30085-8. [DOI] [PubMed] [Google Scholar]
- 50.Martin K.R., Mansouri K., Weinreb R.N., Wasilewicz R., Gisler C., Hennebert J., Genoud D., Research Consortium Use of machine learning on contact lens Sensor-Derived parameters for the diagnosis of primary open-angle glaucoma. Am. J. Ophthalmol. 2018;194:46–53. doi: 10.1016/j.ajo.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Asaoka R., Murata H., Iwase A., Araie M. Detecting preperimetric glaucoma with standard automated perimetry using a deep learning classifier. Ophthalmology. 2016;123:1974–1980. doi: 10.1016/j.ophtha.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 52.Girard M.J.A., Schmetterer L. Artificial intelligence and deep learning in glaucoma: current state and future prospects. Prog. Brain Res. 2020;257:37–64. doi: 10.1016/bs.pbr.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Hood D.C., La Bruna S., Tsamis E., Thakoor K.A., Rai A., Leshno A., de Moraes C.G.V., Cioffi G.A., Liebmann J.M. Detecting glaucoma with only OCT: implications for the clinic, research, screening, and AI development. Prog. Retin. Eye Res. 2022;90 doi: 10.1016/j.preteyeres.2022.101052. [DOI] [PubMed] [Google Scholar]
- 54.Ran A.R., Tham C.C., Chan P.P., Cheng C.Y., Tham Y.C., Rim T.H., Cheung C.Y. Deep learning in glaucoma with optical coherence tomography: a review. Eye. 2021;35:188–201. doi: 10.1038/s41433-020-01191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu H., Baskaran M., Xu Y., Lin S., Wong D.W.K., Liu J., Tun T.A., Mahesh M., Perera S.A., Aung T. A deep learning system for automated Angle-Closure detection in anterior segment optical coherence tomography images. Am. J. Ophthalmol. 2019;203:37–45. doi: 10.1016/j.ajo.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 56.Yousefi S., Kiwaki T., Zheng Y., Sugiura H., Asaoka R., Murata H., Lemij H., Yamanishi K. Detection of longitudinal visual field progression in glaucoma using machine learning. Am. J. Ophthalmol. 2018;193:71–79. doi: 10.1016/j.ajo.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Wang M., Shen L.Q., Pasquale L.R., Petrakos P., Formica S., Boland M.V., Wellik S.R., De Moraes C.G., Myers J.S., Saeedi O., et al. An artificial intelligence approach to detect visual field progression in glaucoma based on spatial pattern analysis. Invest. Ophthalmol. Vis. Sci. 2019;60:365–375. doi: 10.1167/iovs.18-25568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 59.Wong W.L., Su X., Li X., Cheung C.M.G., Klein R., Cheng C.Y., Wong T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 60.Heier J.S., Khanani A.M., Quezada Ruiz C., Basu K., Ferrone P.J., Brittain C., Figueroa M.S., Lin H., Holz F.G., Patel V., et al. Efficacy, durability, and safety of intravitreal faricimab up to every 16 weeks for neovascular age-related macular degeneration (TENAYA and LUCERNE): two randomised, double-masked, phase 3, non-inferiority trials. Lancet. 2022;399:729–740. doi: 10.1016/S0140-6736(22)00010-1. [DOI] [PubMed] [Google Scholar]
- 61.Peng Y., Dharssi S., Chen Q., Keenan T.D., Agrón E., Wong W.T., Chew E.Y., Lu Z. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126:565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burlina P.M., Joshi N., Pekala M., Pacheco K.D., Freund D.E., Bressler N.M. Automated grading of Age-Related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135:1170–1176. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Potapenko I., Thiesson B., Kristensen M., Hajari J.N., Ilginis T., Fuchs J., Hamann S., la Cour M. Automated artificial intelligence-based system for clinical follow-up of patients with age-related macular degeneration. Acta Ophthalmol. 2022;100:927–936. doi: 10.1111/aos.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yellapragada B., Hornauer S., Snyder K., Yu S., Yiu G. Self-Supervised feature learning and phenotyping for assessing Age-Related macular degeneration using retinal fundus images. Ophthalmol. Retina. 2022;6:116–129. doi: 10.1016/j.oret.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rakocz N., Chiang J.N., Nittala M.G., Corradetti G., Tiosano L., Velaga S., Thompson M., Hill B.L., Sankararaman S., Haines J.L., et al. Automated identification of clinical features from sparsely annotated 3-dimensional medical imaging. NPJ Digit. Med. 2021;4:44. doi: 10.1038/s41746-021-00411-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hwang D.K., Hsu C.C., Chang K.J., Chao D., Sun C.H., Jheng Y.C., Yarmishyn A.A., Wu J.C., Tsai C.Y., Wang M.L., et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics. 2019;9:232–245. doi: 10.7150/thno.28447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keel S., Li Z., Scheetz J., Robman L., Phung J., Makeyeva G., Aung K., Liu C., Yan X., Meng W., et al. Development and validation of a deep-learning algorithm for the detection of neovascular age-related macular degeneration from colour fundus photographs. Clin. Exp. Ophthalmol. 2019;47:1009–1018. doi: 10.1111/ceo.13575. [DOI] [PubMed] [Google Scholar]
- 68.Grassmann F., Mengelkamp J., Brandl C., Harsch S., Zimmermann M.E., Linkohr B., Peters A., Heid I.M., Palm C., Weber B.H.F. A deep learning algorithm for prediction of Age-Related eye disease study severity scale for Age-Related macular degeneration from color fundus photography. Ophthalmology. 2018;125:1410–1420. doi: 10.1016/j.ophtha.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 69.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., et al. Identifying medical diagnoses and treatable diseases by Image-Based deep learning. Cell. 2018;172:1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 70.Yan Q., Weeks D.E., Xin H., Swaroop A., Chew E.Y., Huang H., Ding Y., Chen W. Deep-learning-based prediction of late Age-Related macular degeneration progression. Nat. Mach. Intell. 2020;2:141–150. doi: 10.1038/s42256-020-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milea D., Najjar R.P., Zhubo J., Ting D., Vasseneix C., Xu X., Aghsaei Fard M., Fonseca P., Vanikieti K., Lagrèze W.A., et al. Artificial intelligence to detect papilledema from ocular fundus photographs. N. Engl. J. Med. 2020;382:1687–1695. doi: 10.1056/NEJMoa1917130. [DOI] [PubMed] [Google Scholar]
- 72.Brown J.M., Campbell J.P., Beers A., Chang K., Ostmo S., Chan R.V.P., Dy J., Erdogmus D., Ioannidis S., Kalpathy-Cramer J., et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Zhang L., Xu F., Jin C., Zhang X., et al. A deep learning system for identifying lattice degeneration and retinal breaks using ultra-widefield fundus images. Ann. Transl. Med. 2019;7:618. doi: 10.21037/atm.2019.11.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Wu X., Xu F., Jin C., Zhang X., et al. Deep learning for detecting retinal detachment and discerning macular status using ultra-widefield fundus images. Commun. Biol. 2020;3:15. doi: 10.1038/s42003-019-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y., Foo L.L., Wong C.W., Li J., Hoang Q.V., Schmetterer L., Ting D.S.W., Ang M. Pathologic myopia: advances in imaging and the potential role of artificial intelligence. Br. J. Ophthalmol. 2023;107:600–606. doi: 10.1136/bjophthalmol-2021-320926. [DOI] [PubMed] [Google Scholar]
- 76.Tsai Y.Y., Lin W.Y., Chen S.J., Ruamviboonsuk P., King C.H., Tsai C.L. Diagnosis of polypoidal choroidal vasculopathy from fluorescein angiography using deep learning. Transl. Vis. Sci. Technol. 2022;11:6. doi: 10.1167/tvst.11.2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y.C., Wilkins M., Kim T., Malyugin B., Mehta J.S. Lancet. 2017;390:600–612. doi: 10.1016/S0140-6736(17)30544-5. [DOI] [PubMed] [Google Scholar]
- 78.Tham Y.C., Goh J.H.L., Anees A., Lei X., Rim T.H., Chee M.L., Wang Y.X., Jonas J.B., Thakur S., Teo Z.L., et al. Detecting visually significant cataract using retinal photograph-based deep learning. Nat. Aging. 2022;2:264–271. doi: 10.1038/s43587-022-00171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu X., Li J., Guan Y., Zhao L., Zhao Q., Zhang L., Li L. GLA-Net: a global-local attention network for automatic cataract classification. J. Biomed. Inf. 2021;124 doi: 10.1016/j.jbi.2021.103939. [DOI] [PubMed] [Google Scholar]
- 80.Lu Q., Wei L., He W., Zhang K., Wang J., Zhang Y., Rong X., Zhao Z., Cai L., He X., et al. Lens Opacities Classification System III-based artificial intelligence program for automatic cataract grading. J. Cataract Refract. Surg. 2022;48:528–534. doi: 10.1097/j.jcrs.0000000000000790. [DOI] [PubMed] [Google Scholar]
- 81.Lin D., Chen J., Lin Z., Li X., Zhang K., Wu X., Liu Z., Huang J., Li J., Zhu Y., et al. A practical model for the identification of congenital cataracts using machine learning. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.102621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu X., Zhang L., Li J., Guan Y., Zhang L. A hybrid Global-Local representation CNN model for automatic cataract grading. IEEE J. Biomed. Health Inform. 2020;24:556–567. doi: 10.1109/JBHI.2019.2914690. [DOI] [PubMed] [Google Scholar]
- 83.Wu X., Huang Y., Liu Z., Lai W., Long E., Zhang K., Jiang J., Lin D., Chen K., Yu T., et al. Universal artificial intelligence platform for collaborative management of cataracts. Br. J. Ophthalmol. 2019;103:1553–1560. doi: 10.1136/bjophthalmol-2019-314729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin H., Li R., Liu Z., Chen J., Yang Y., Chen H., Lin Z., Lai W., Long E., Wu X., et al. Diagnostic efficacy and therapeutic decision-making capacity of an artificial intelligence platform for childhood cataracts in eye clinics: a multicentre randomized controlled trial. EClinicalMedicine. 2019;9:52–59. doi: 10.1016/j.eclinm.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang H., Niu K., Xiong Y., Yang W., He Z., Song H. Automatic cataract grading methods based on deep learning. Comput. Methods Progr. Biomed. 2019;182 doi: 10.1016/j.cmpb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 86.Gao X., Lin S., Wong T.Y. Automatic feature learning to grade nuclear cataracts based on deep learning. IEEE Trans. Biomed. Eng. 2015;62:2693–2701. doi: 10.1109/TBME.2015.2444389. [DOI] [PubMed] [Google Scholar]
- 87.Garcia Nespolo R., Yi D., Cole E., Valikodath N., Luciano C., Leiderman Y.I. Evaluation of artificial Intelligence-Based intraoperative guidance tools for phacoemulsification cataract surgery. JAMA Ophthalmol. 2022;140:170–177. doi: 10.1001/jamaophthalmol.2021.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flaxman S.R., Bourne R.R.A., Resnikoff S., Ackland P., Braithwaite T., Cicinelli M.V., Das A., Jonas J.B., Keeffe J., Kempen J.H., et al. Global causes of blindness and distance vision impairment 1990-2020: a systematic review and meta-analysis. Lancet Global Health. 2017;5:e1221–e1234. doi: 10.1016/S2214-109X(17)30393-5. [DOI] [PubMed] [Google Scholar]
- 89.Burton M.J. Prevention, treatment and rehabilitation. Community Eye Health. 2009;22:33–35. [PMC free article] [PubMed] [Google Scholar]
- 90.Singh P., Gupta A., Tripathy K. Keratitis. StatPearls. 2020 [Google Scholar]
- 91.Lin A., Rhee M.K., Akpek E.K., Amescua G., Farid M., Garcia-Ferrer F.J., Varu D.M., Musch D.C., Dunn S.P., Mah F.S., American Academy of Ophthalmology Preferred Practice Pattern Cornea and External Disease Panel Bacterial keratitis preferred practice Pattern(R) Ophthalmology. 2019;126:P1–P55. doi: 10.1016/j.ophtha.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 92.Xu Y., Kong M., Xie W., Duan R., Fang Z., Lin Y., Zhu Q., Tang S., Wu F., Yao Y.F. Deep sequential feature learning in clinical image classification of infectious keratitis. Engineering. 2021;7:1002–1010. [Google Scholar]
- 93.Redd T.K., Prajna N.V., Srinivasan M., Lalitha P., Krishnan T., Rajaraman R., Venugopal A., Acharya N., Seitzman G.D., Lietman T.M., et al. Image-Based differentiation of bacterial and fungal keratitis using deep convolutional neural networks. Ophthalmol. Sci. 2022;2 doi: 10.1016/j.xops.2022.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ren Z., Li W., Liu Q., Dong Y., Huang Y. Profiling of the conjunctival bacterial microbiota reveals the feasibility of utilizing a Microbiome-Based machine learning model to differentially diagnose microbial keratitis and the core components of the conjunctival bacterial interaction network. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.860370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu W., Huang S., Xie X., Chen C., Yan Z., Lv X., Fan Y., Chen C., Yue F., Yang B. Raman spectroscopy may allow rapid noninvasive screening of keratitis and conjunctivitis. Photodiagnosis Photodyn. Ther. 2022;37 doi: 10.1016/j.pdpdt.2021.102689. [DOI] [PubMed] [Google Scholar]
- 96.Tiwari M., Piech C., Baitemirova M., Prajna N.V., Srinivasan M., Lalitha P., Villegas N., Balachandar N., Chua J.T., Redd T., et al. Differentiation of active corneal infections from healed scars using deep learning. Ophthalmology. 2022;129:139–146. doi: 10.1016/j.ophtha.2021.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ghosh A.K., Thammasudjarit R., Jongkhajornpong P., Attia J., Thakkinstian A. Deep learning for discrimination between fungal keratitis and bacterial keratitis: DeepKeratitis. Cornea. 2022;41:616–622. doi: 10.1097/ICO.0000000000002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L., Chen K., Wen H., Zheng Q., Chen Y., Pu J., Chen W. Feasibility assessment of infectious keratitis depicted on slit-lamp and smartphone photographs using deep learning. Int. J. Med. Inf. 2021;155 doi: 10.1016/j.ijmedinf.2021.104583. [DOI] [PubMed] [Google Scholar]
- 99.Lv J., Zhang K., Chen Q., Chen Q., Huang W., Cui L., Li M., Li J., Chen L., Shen C., et al. Deep learning-based automated diagnosis of fungal keratitis with in vivo confocal microscopy images. Ann. Transl. Med. 2020;8:706. doi: 10.21037/atm.2020.03.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu H., Guo Y., Gu L., Wei A., Xie S., Ye Z., Xu J., Zhou X., Lu Y., Liu X., Hong J. Deep learning for identifying corneal diseases from ocular surface slit-lamp photographs. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-75027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferdi A.C., Nguyen V., Gore D.M., Allan B.D., Rozema J.J., Watson S.L. Keratoconus natural progression: a systematic review and meta-analysis of 11 529 eyes. Ophthalmology. 2019;126:935–945. doi: 10.1016/j.ophtha.2019.02.029. [DOI] [PubMed] [Google Scholar]
- 102.de Sanctis U., Loiacono C., Richiardi L., Turco D., Mutani B., Grignolo F.M. Sensitivity and specificity of posterior corneal elevation measured by Pentacam in discriminating keratoconus/subclinical keratoconus. Ophthalmology. 2008;115:1534–1539. doi: 10.1016/j.ophtha.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 103.Castro-Luna G., Jiménez-Rodríguez D., Castaño-Fernández A.B., Pérez-Rueda A. Diagnosis of subclinical keratoconus based on machine learning techniques. J. Clin. Med. 2021;10 doi: 10.3390/jcm10184281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Al-Timemy A.H., Mosa Z.M., Alyasseri Z., Lavric A., Lui M.M., Hazarbassanov R.M., Yousefi S. A hybrid deep learning construct for detecting keratoconus from corneal maps. Transl. Vis. Sci. Technol. 2021;10:16. doi: 10.1167/tvst.10.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Almeida J.G., Guido R.C., Balarin S.H., Brandao C.C., Carlos D.M.L., Lopes B.T., Machado A.P., Ambrosio R.J. Novel artificial intelligence index based on Scheimpflug corneal tomography to distinguish subclinical keratoconus from healthy corneas. J. Cataract Refract. Surg. 2022 doi: 10.1097/j.jcrs.0000000000000946. [DOI] [PubMed] [Google Scholar]
- 106.Jiménez-García M., Issarti I., Kreps E.O., Ní Dhubhghaill S., Koppen C., Varssano D., Rozema J.J., The REDCAKE Study Group Forecasting progressive trends in keratoconus by means of a time delay neural network. J. Clin. Med. 2021;10 doi: 10.3390/jcm10153238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Y., Zhao L., Yang X., Wu X., Yang Y., Huang X., Liu F., Xu J., Lin L., Lin H., et al. Screening candidates for refractive surgery with corneal Tomographic-Based deep learning. JAMA Ophthalmol. 2020;138:519–526. doi: 10.1001/jamaophthalmol.2020.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zéboulon P., Debellemanière G., Bouvet M., Gatinel D. Corneal topography raw data classification using a convolutional neural network. Am. J. Ophthalmol. 2020;219:33–39. doi: 10.1016/j.ajo.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 109.Shi C., Wang M., Zhu T., Zhang Y., Ye Y., Jiang J., Chen S., Lu F., Shen M. Machine learning helps improve diagnostic ability of subclinical keratoconus using Scheimpflug and OCT imaging modalities. Eye Vis. 2020;7:48. doi: 10.1186/s40662-020-00213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cao K., Verspoor K., Chan E., Daniell M., Sahebjada S., Baird P.N. Machine learning with a reduced dimensionality representation of comprehensive Pentacam tomography parameters to identify subclinical keratoconus. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104884. [DOI] [PubMed] [Google Scholar]
- 111.Issarti I., Consejo A., Jiménez-García M., Hershko S., Koppen C., Rozema J.J. Computer aided diagnosis for suspect keratoconus detection. Comput. Biol. Med. 2019;109:33–42. doi: 10.1016/j.compbiomed.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 112.Ruiz Hidalgo I., Rodriguez P., Rozema J.J., Ní Dhubhghaill S., Zakaria N., Tassignon M.J., Koppen C. Evaluation of a Machine-Learning classifier for keratoconus detection based on scheimpflug tomography. Cornea. 2016;35:827–832. doi: 10.1097/ICO.0000000000000834. [DOI] [PubMed] [Google Scholar]
- 113.Chase C., Elsawy A., Eleiwa T., Ozcan E., Tolba M., Abou Shousha M. Comparison of autonomous AS-OCT deep learning algorithm and clinical dry eye tests in diagnosis of dry eye disease. Clin. Ophthalmol. 2021;15:4281–4289. doi: 10.2147/OPTH.S321764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y.Y., Zhao H., Lin J.Y., Wu S.N., Liu X.W., Zhang H.D., Shao Y., Yang W.F. Artificial intelligence to detect meibomian gland dysfunction from in-vivo laser confocal microscopy. Front. Med. 2021;8 doi: 10.3389/fmed.2021.774344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.GBD 2013 Mortality and Causes of Death Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Seidelmann S.B., Claggett B., Bravo P.E., Gupta A., Farhad H., Klein B.E., Klein R., Di Carli M., Solomon S.D. Retinal vessel calibers in predicting Long-Term cardiovascular outcomes: the atherosclerosis risk in communities study. Circulation. 2016;134:1328–1338. doi: 10.1161/CIRCULATIONAHA.116.023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rim T.H., Lee C.J., Tham Y.C., Cheung N., Yu M., Lee G., Kim Y., Ting D.S.W., Chong C.C.Y., Choi Y.S., et al. Deep-learning-based cardiovascular risk stratification using coronary artery calcium scores predicted from retinal photographs. Lancet. Digit. Health. 2021;3:e306–e316. doi: 10.1016/S2589-7500(21)00043-1. [DOI] [PubMed] [Google Scholar]
- 118.Poplin R., Varadarajan A.V., Blumer K., Liu Y., McConnell M.V., Corrado G.S., Peng L., Webster D.R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018;2:158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 119.Kalantar-Zadeh K., Jafar T.H., Nitsch D., Neuen B.L., Perkovic V. Chronic kidney disease. Lancet. 2021;398:786–802. doi: 10.1016/S0140-6736(21)00519-5. [DOI] [PubMed] [Google Scholar]
- 120.Ahmad E., Lim S., Lamptey R., Webb D.R., Davies M.J. Type 2 diabetes. Lancet. 2022;400:1803–1820. doi: 10.1016/S0140-6736(22)01655-5. [DOI] [PubMed] [Google Scholar]
- 121.Zhang K., Liu X., Xu J., Yuan J., Cai W., Chen T., Wang K., Gao Y., Nie S., Xu X., et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat. Biomed. Eng. 2021;5:533–545. doi: 10.1038/s41551-021-00745-6. [DOI] [PubMed] [Google Scholar]
- 122.Sabanayagam C., Xu D., Ting D.S.W., Nusinovici S., Banu R., Hamzah H., Lim C., Tham Y.C., Cheung C.Y., Tai E.S., et al. A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet. Digit. Health. 2020;2:e295–e302. doi: 10.1016/S2589-7500(20)30063-7. [DOI] [PubMed] [Google Scholar]
- 123.Scheltens P., De Strooper B., Kivipelto M., Holstege H., Chételat G., Teunissen C.E., Cummings J., van der Flier W.M. Alzheimer's disease. Lancet. 2021;397:1577–1590. doi: 10.1016/S0140-6736(20)32205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gupta V.B., Chitranshi N., den Haan J., Mirzaei M., You Y., Lim J.K., Basavarajappa D., Godinez A., Di Angelantonio S., Sachdev P., et al. Retinal changes in Alzheimer's disease- integrated prospects of imaging, functional and molecular advances. Prog. Retin. Eye Res. 2021;82 doi: 10.1016/j.preteyeres.2020.100899. [DOI] [PubMed] [Google Scholar]
- 125.Cheung C.Y., Ran A.R., Wang S., Chan V.T.T., Sham K., Hilal S., Venketasubramanian N., Cheng C.Y., Sabanayagam C., Tham Y.C., et al. A deep learning model for detection of Alzheimer's disease based on retinal photographs: a retrospective, multicentre case-control study. Lancet. Digit. Health. 2022;4:e806–e815. doi: 10.1016/S2589-7500(22)00169-8. [DOI] [PubMed] [Google Scholar]
- 126.Price W.N., Cohen I.G. Privacy in the age of medical big data. Nat. Med. 2019;25:37–43. doi: 10.1038/s41591-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rieke N., Hancox J., Li W., Milletarì F., Roth H.R., Albarqouni S., Bakas S., Galtier M.N., Landman B.A., Maier-Hein K., et al. The future of digital health with federated learning. NPJ Digit. Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dayan I., Roth H.R., Zhong A., Harouni A., Gentili A., Abidin A.Z., Liu A., Costa A.B., Wood B.J., Tsai C.S., et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021;27:1735–1743. doi: 10.1038/s41591-021-01506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krause J., Gulshan V., Rahimy E., Karth P., Widner K., Corrado G.S., Peng L., Webster D.R. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125:1264–1272. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 130.Sylolypavan A., Sleeman D., Wu H., Sim M. The impact of inconsistent human annotations on AI driven clinical decision making. NPJ Digit. Med. 2023;6:26. doi: 10.1038/s41746-023-00773-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin D., Xiong J., Liu C., Zhao L., Li Z., Yu S., Wu X., Ge Z., Hu X., Wang B., et al. Application of Comprehensive Artificial intelligence Retinal Expert (CARE) system: a national real-world evidence study. Lancet. Digit. Health. 2021;3:e486–e495. doi: 10.1016/S2589-7500(21)00086-8. [DOI] [PubMed] [Google Scholar]
- 132.Playout C., Duval R., Cheriet F. A novel weakly supervised multitask architecture for retinal lesions segmentation on fundus images. IEEE Trans. Med. Imag. 2019;38:2434–2444. doi: 10.1109/TMI.2019.2906319. [DOI] [PubMed] [Google Scholar]
- 133.Wang J., Li W., Chen Y., Fang W., Kong W., He Y., Shi G. Weakly supervised anomaly segmentation in retinal OCT images using an adversarial learning approach. Biomed. Opt Express. 2021;12:4713–4729. doi: 10.1364/BOE.426803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xing R., Niu S., Gao X., Liu T., Fan W., Chen Y. Weakly supervised serous retinal detachment segmentation in SD-OCT images by two-stage learning. Biomed. Opt Express. 2021;12:2312–2327. doi: 10.1364/BOE.416167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cai W., Xu J., Wang K., Liu X., Xu W., Cai H., Gao Y., Su Y., Zhang M., Zhu J., et al. EyeHealer: a large-scale anterior eye segment dataset with eye structure and lesion annotations. Precis. Clin. Med. 2021;4:85–92. doi: 10.1093/pcmedi/pbab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen D., Liu S., Kingsbury P., Sohn S., Storlie C.B., Habermann E.B., Naessens J.M., Larson D.W., Liu H. Deep learning and alternative learning strategies for retrospective real-world clinical data. NPJ Digit. Med. 2019;2:43. doi: 10.1038/s41746-019-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang Y., Li R., Xiang Y., Lin D., Yan A., Chen W., Li Z., Lai W., Wu X., Wan C., et al. Standardization of collection, storage, annotation, and management of data related to medical artificial intelligence. Intelligent Medicine. 2021 [Google Scholar]
- 138.Li X.T., Huang R.Y. Standardization of imaging methods for machine learning in neuro-oncology. Neurooncol. Adv. 2020;2:v49–v55. doi: 10.1093/noajnl/vdaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Z., Chen W. Solving data quality issues of fundus images in real-world settings by ophthalmic AI. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.100951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu R., Wang X., Wu Q., Dai L., Fang X., Yan T., Son J., Tang S., Li J., Gao Z., et al. DeepDRiD: diabetic Retinopathy-Grading and image quality estimation challenge. Patterns (N Y) 2022;3 doi: 10.1016/j.patter.2022.100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu L., Wu X., Lin D., Zhao L., Li M., Yun D., Lin Z., Pang J., Li L., Wu Y., et al. DeepFundus: a flow-cytometry-like image quality classifier for boosting the whole life cycle of medical artificial intelligence. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2022.100912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Z., Jiang J., Chen K., Zheng Q., Liu X., Weng H., Wu S., Chen W. Development of a deep learning-based image quality control system to detect and filter out ineligible slit-lamp images: a multicenter study. Comput. Methods Progr. Biomed. 2021;203 doi: 10.1016/j.cmpb.2021.106048. [DOI] [PubMed] [Google Scholar]
- 143.Shen Y., Sheng B., Fang R., Li H., Dai L., Stolte S., Qin J., Jia W., Shen D. Domain-invariant interpretable fundus image quality assessment. Med. Image Anal. 2020;61 doi: 10.1016/j.media.2020.101654. [DOI] [PubMed] [Google Scholar]
- 144.Kanagasingam Y., Xiao D., Vignarajan J., Preetham A., Tay-Kearney M.L., Mehrotra A. Evaluation of artificial Intelligence-Based grading of diabetic retinopathy in primary care. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Long E., Lin H., Liu Z., Wu X., Wang L., Jiang J., An Y., Lin Z., Li X., Chen J., et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat. Biomed. Eng. 2017;1 [Google Scholar]
- 146.Rajpurkar P., Chen E., Banerjee O., Topol E.J. AI in health and medicine. Nat. Med. 2022;28:31–38. doi: 10.1038/s41591-021-01614-0. [DOI] [PubMed] [Google Scholar]
- 147.Wang N., Cheng M., Ning K. Overcoming regional limitations: transfer learning for cross-regional microbial-based diagnosis of diseases. Gut. 2022 doi: 10.1136/gutjnl-2022-328216. 2022-328216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6 doi: 10.1186/s40537-021-00492-0. 60-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou C., Ye J., Wang J., Zhou Z., Wang L., Jin K., Wen Y., Zhang C., Qian D. Improving the generalization of glaucoma detection on fundus images via feature alignment between augmented views. Biomed. Opt Express. 2022;13:2018–2034. doi: 10.1364/BOE.450543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li W., Yang Y., Zhang K., Long E., He L., Zhang L., Zhu Y., Chen C., Liu Z., Wu X., et al. Dense anatomical annotation of slit-lamp images improves the performance of deep learning for the diagnosis of ophthalmic disorders. Nat. Biomed. Eng. 2020;4:767–777. doi: 10.1038/s41551-020-0577-y. [DOI] [PubMed] [Google Scholar]
- 151.Duran J.M., Jongsma K.R. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J. Med. Ethics. 2021 doi: 10.1136/medethics-2020-106820. [DOI] [PubMed] [Google Scholar]
- 152.Niu Y., Gu L., Zhao Y., Lu F. Explainable diabetic retinopathy detection and retinal image generation. IEEE J. Biomed. Health Inform. 2022;26:44–55. doi: 10.1109/JBHI.2021.3110593. [DOI] [PubMed] [Google Scholar]
- 153.Xu F., Jiang L., He W., Huang G., Hong Y., Tang F., Lv J., Lin Y., Qin Y., Lan R., et al. The clinical value of explainable deep learning for diagnosing fungal keratitis using in vivo confocal microscopy images. Front. Med. 2021;8 doi: 10.3389/fmed.2021.797616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hospedales T.M., Antoniou A., Micaelli P., Storkey A.J. IEEE Trans Pattern Anal Mach Intell; 2021. Meta-Learning in Neural Networks: A Survey. [DOI] [PubMed] [Google Scholar]
- 155.Maliha G., Gerke S., Cohen I.G., Parikh R.B. Artificial intelligence and liability in medicine: balancing safety and innovation. Milbank Q. 2021;99:629–647. doi: 10.1111/1468-0009.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Price W.N., Gerke S., Cohen I.G. Potential liability for physicians using artificial intelligence. JAMA. 2019;322:1765–1766. doi: 10.1001/jama.2019.15064. [DOI] [PubMed] [Google Scholar]
- 157.Hadsell R., Rao D., Rusu A.A., Pascanu R. Embracing change: continual learning in deep neural networks. Trends Cognit. Sci. 2020;24:1028–1040. doi: 10.1016/j.tics.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 158.Li Z., Jiang J., Zhou H., Zheng Q., Liu X., Chen K., Weng H., Chen W. Development of a deep learning-based image eligibility verification system for detecting and filtering out ineligible fundus images: a multicentre study. Int. J. Med. Inf. 2021;147 doi: 10.1016/j.ijmedinf.2020.104363. [DOI] [PubMed] [Google Scholar]
- 159.Li Z., Jiang J., Qiang W., Guo L., Liu X., Weng H., Wu S., Zheng Q., Chen W. Comparison of deep learning systems and cornea specialists in detecting corneal diseases from low-quality images. iScience. 2021;24 doi: 10.1016/j.isci.2021.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Z., Li M., Wang D., Hou P., Chen X., Chu S., Chai D., Zheng J., Bai J., Xu F., et al. Deep learning from “passive feeding” to “selective eating” of real-world data. Cell Biosci. 2020;10:143. doi: 10.1038/s41746-020-00350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Trucco E., Ruggeri A., Karnowski T., Giancardo L., Chaum E., Hubschman J.P., Al-Diri B., Cheung C.Y., Wong D., Abràmoff M., et al. Validating retinal fundus image analysis algorithms: issues and a proposal. Invest. Ophthalmol. Vis. Sci. 2013;54:3546–3559. doi: 10.1167/iovs.12-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nagendran M., Chen Y., Lovejoy C.A., Gordon A.C., Komorowski M., Harvey H., Topol E.J., Ioannidis J.P.A., Collins G.S., Maruthappu M. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368:m689. doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cruz Rivera S., Liu X., Chan A.W., Denniston A.K., Calvert M.J., SPIRIT-AI and CONSORT-AI Working Group Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Lancet. Digit. Health. 2020;2:e549–e560. doi: 10.1016/S2589-7500(20)30219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu X., Cruz Rivera S., Moher D., Calvert M.J., Denniston A.K., SPIRIT-AI and CONSORT-AI Working Group Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 2020;26:1364–1374. doi: 10.1038/s41591-020-1034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sounderajah V., Ashrafian H., Aggarwal R., De Fauw J., Denniston A.K., Greaves F., Karthikesalingam A., King D., Liu X., Markar S.R., et al. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: the STARD-AI Steering Group. Nat. Med. 2020;26:807–808. doi: 10.1038/s41591-020-0941-1. [DOI] [PubMed] [Google Scholar]
- 166.Reddy S., Rogers W., Makinen V.P., Coiera E., Brown P., Wenzel M., Weicken E., Ansari S., Mathur P., Casey A., Kelly B. Evaluation framework to guide implementation of AI systems into healthcare settings. BMJ Health Care Inform. 2021;28 doi: 10.1136/bmjhci-2021-100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhu T., Ye D., Wang W., Zhou W., Yu P.S. More than privacy: applying differential privacy in key areas of artificial intelligence. IEEE Trans. Knowl. Data Eng. 2022;34:2824–2843. [Google Scholar]
- 168.Hesamifard E., Takabi H., Ghasemi M. 2019. Deep Neural Networks Classification over Encrypted Data,ACM; pp. 97–108. [Google Scholar]
- 169.US Food and Drug Administration (FDA) Proposed Regulatory Framework for Modifcations to Artifcial Intelligence/Machine Learning (AI/ML)-Based Sofware as a Medical Device (SaMD)—Discussion Paper and Request for Feedback. https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.