Abstract
Nakayasu et al.’s investigation1 on children from TEDDY study revealed robust predictive value of plasma protein biomarkers in identifying the emergence of persistent autoantibodies and type 1 diabetes. Remarkably, this predictive accuracy was observed six months prior to autoimmunity initiation.
Nakayasu et al.’s investigation on children from TEDDY study revealed robust predictive value of plasma protein biomarkers in identifying the emergence of persistent autoantibodies and type 1 diabetes. Remarkably, this predictive accuracy was observed six months prior to autoimmunity initiation.
Main text
The development of type 1 diabetes (T1D) involves an initial phase of clinically asymptomatic islet autoimmunity (IA) characterized by the appearance of islet autoantibodies, with subsequent progression to dysglycemia and clinical diabetes.2 Understanding the early phase is critical, as the majority of β-cell function has already been lost by the time of T1D onset, making intervention or reversal exceedingly challenging.3 Moreover, children with T1D are prone to suffering from life-threatening complications due to poor metabolic control.4 Therefore, it is imperative to discover biomarkers that can identify early-stage disease in most, if not all, individuals, in order to delay β-cell loss and improve prognostic outcomes. Although islet-specific autoantibodies have proven valuable in predicting future T1D onset, it should be noted that not all autoantibody-positive individuals progress to clinical T1D.5 Nevertheless, it remains unclear whether indicators of future disease development can be identified prior to the autoimmune process. Proteomics analysis has emerged as a promising approach to discover T1D biomarkers,6,7,8 offering valuable insights into the underlying pathogenesis of T1D9,10 and potentially more accurate predictions or diagnoses compared to currently available autoantibody measurements.
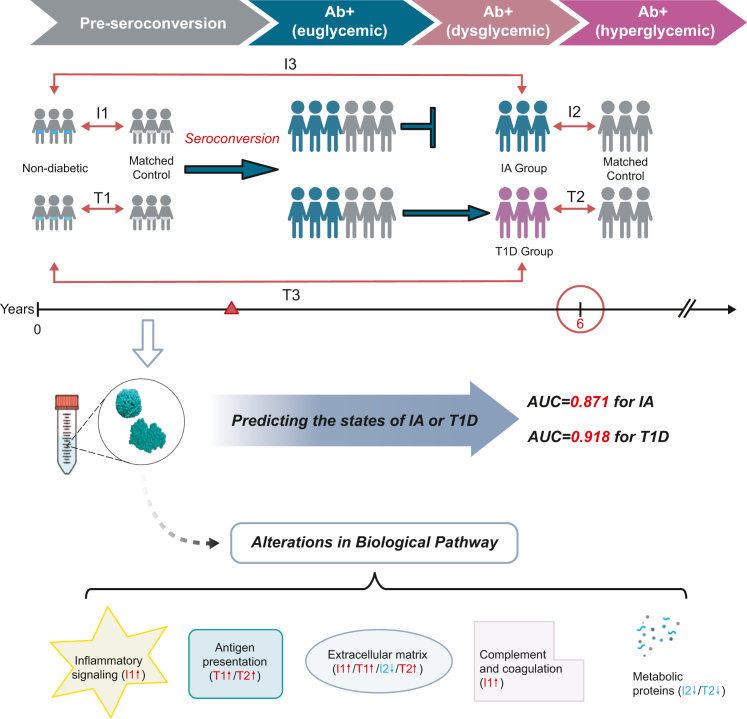
Ernesto S. Nakayasu and colleagues1 conducted a nested case-control study involving children at higher genetic risk of developing T1D from seven different centers in the US and Europe. The study included 94 individuals with developed T1D, 401 individuals with persistent confirmed IA, and their respective matched controls. By using the appearance of autoantibodies against pancreatic islet proteins (known as “seroconversion”) as a turning time point, the researchers conducted six comparisons: I1, IA group vs. controls pre-seroconversion; T1, T1D group vs. controls pre-seroconversion; I2, cases IA group vs. controls post-seroconversion; T2, T1D group vs. controls post-seroconversion; I3, pre- vs. post-seroconversion of IA group; and T3, pre- vs. post-seroconversion of T1D group (Figure 1).
Figure 1.
Proteomic analysis at each stage of type 1 diabetes
Ab, autoantibody; IA, islet autoimmunity; T1D, type 1 diabetes.
These six comparisons span the progression from autoantibody negativity to seroconversion and subsequent diagnosis. In the discovery phase analysis, a total of 376 regulated proteins were identified, revealing alterations in extracellular matrix, antigen presentation, inflammatory signaling, complement and coagulation, and cellular metabolism, of which 83 proteins were validated as biomarkers. Prior to the onset of the autoimmune response, distinct panels of proteins existed between the persistent IA group and T1D group. The IA group exhibited higher levels of complement factors, while the T1D group showed higher levels of antigen-processing proteins compared to their respective controls at pre-seroconversion time point (comparisons I1 and T1). Early changes in protein levels might influence future disease progression. Machine learning analysis demonstrated that panels of peptides prior to seroconversion effectively predicted whether individuals would remain in IA or develop T1D by the age of 6 years (Figure 1). The selected proteins used to construct the two models exhibited some distinct but also overlapping characteristics. Consequently, certain proteins might be regulated throughout all stages of the disease. Notably, most coagulation and complement factors, as well as metabolic proteins in IA group, exhibited similar abundance patterns to those in the T1D group at post-seroconversion time point (comparisons I2 and T2).
When comparing this study to previous investigations on T1D biomarkers, certain similarities can be observed. Previous studies,6,7 similar to the comparison made in this study (T2), have also demonstrated changes in serum proteomic following the onset of IA or disease. The altered abundance of these proteins likely reflects a secondary or systemic immune response to the pathology of T1D. Notably, both Finnish DIPP study and the present work detected lower levels of APOC2 in children who later developed T1D compared to their relative controls, even before the appearance of autoantibodies.8 However, the current study goes beyond previous efforts by conducting a comprehensive analysis of protein profiles at each stage of T1D development, from early infancy to the diagnosis of IA or T1D. Through proteomics analysis, they identified proteins and biological processes associated with disease cause and progression. Importantly, they discovered proteins biomarkers that can accurately predict persistent IA or developing T1D by the age of 6 years, even as early as 6 months prior to the onset of autoimmunity. This early detection of proteins biomarkers provides an opportunity for physicians to implement intervention strategies to prevent or delay the development of IA or progression to T1D in children who are at risk. Although widespread screening may be challenging due to the relatively low prevalence of T1D and substantial testing costs, it is valuable to apply these approaches to children with a genetic predisposition to diabetes.
In summary, this study offers insights into early diagnosis and the preservation of pancreatic β cell in T1D and opens avenues for further research and therapeutic interventions targeting the identified pathways. Looking ahead, further research is needed to explore the biological and genetic mechanisms underlying the identified protein biomarkers. Understanding the intricate pathways and molecular interactions involved in T1D development will facilitate the development of targeted interventions and personalized treatment strategies. Additionally, efforts should be made to translate these biomarkers into clinical practice, enabling early detection and intervention in individuals at high risk for T1D.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
References
- 1.Nakayasu E.S., Bramer L.M., Ansong C., Schepmoes A.A., Fillmore T.L., Gritsenko M.A., Claus T.R., Gao Y., Piehowski P.D., Stanfill B.A., et al. Plasma protein biomarkers predict the development of persistent autoantibodies and type 1 diabetes 6 months prior to the onset of autoimmunity. Cell Reports Medicine. 2023;4 doi: 10.1016/j.xcrm.2023.101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dayan C.M., Korah M., Tatovic D., Bundy B.N., Herold K.C. Changing the landscape for type 1 diabetes: the first step to prevention. Lancet. 2019;394:1286–1296. doi: 10.1016/S0140-6736(19)32127-0. [DOI] [PubMed] [Google Scholar]
- 3.Dayan C.M., Besser R.E.J., Oram R.A., Hagopian W., Vatish M., Bendor-Samuel O., Snape M.D., Todd J.A. Preventing type 1 diabetes in childhood. Science. 2021;373:506–510. doi: 10.1126/science.abi4742. [DOI] [PubMed] [Google Scholar]
- 4.Varkevisser R.D.M., Birnie E., Mul D., van Dijk P.R., Aanstoot H.J., Wolffenbuttel B.H.R., van der Klauw M.M. Type 1 diabetes management: Room for improvement. J. Diabetes. 2023;15:255–263. doi: 10.1111/1753-0407.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler A.G., Rewers M., Simell O., Simell T., Lempainen J., Steck A., Winkler C., Ilonen J., Veijola R., Knip M., et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q., Fillmore T.L., Schepmoes A.A., Clauss T.R.W., Gritsenko M.A., Mueller P.W., Rewers M., Atkinson M.A., Smith R.D., Metz T.O. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J. Exp. Med. 2013;210:191–203. doi: 10.1084/jem.20111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhi W., Sharma A., Purohit S., Miller E., Bode B., Anderson S.W., Reed J.C., Steed R.D., Steed L., Hopkins D., She J.X. Discovery and validation of serum protein changes in type 1 diabetes patients using high throughput two dimensional liquid chromatography-mass spectrometry and immunoassays. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moulder R., Bhosale S.D., Erkkilä T., Laajala E., Salmi J., Nguyen E.V., Kallionpää H., Mykkänen J., Vähä-Mäkilä M., Hyöty H., et al. Serum proteomes distinguish children developing type 1 diabetes in a cohort with HLA-conferred susceptibility. Diabetes. 2015;64:2265–2278. doi: 10.2337/db14-0983. [DOI] [PubMed] [Google Scholar]
- 9.Caseiro A., Barros A., Ferreira R., Padrão A., Aroso M., Quintaneiro C., Pereira A., Marinheiro R., Vitorino R., Amado F. Pursuing type 1 diabetes mellitus and related complications through urinary proteomics. Transl. Res. 2014;163:188–199. doi: 10.1016/j.trsl.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Singh H., Yu Y., Suh M.J., Torralba M.G., Stenzel R.D., Tovchigrechko A., Thovarai V., Harkins D.M., Rajagopala S.V., Osborne W., et al. Type 1 Diabetes: Urinary Proteomics and Protein Network Analysis Support Perturbation of Lysosomal Function. Theranostics. 2017;7:2704–2717. doi: 10.7150/thno.19679. [DOI] [PMC free article] [PubMed] [Google Scholar]