Abstract
AUG-unrelated translation initiation was found in an insect picorna-like virus, Plautia stali intestine virus (PSIV). The positive-strand RNA genome of the virus contains two nonoverlapping open reading frames (ORFs). The capsid protein gene is located in the 3′-proximal ORF and lacks an AUG initiation codon. We examined the translation mechanism and the initiation codon of the capsid protein gene by using various dicistronic and monocistronic RNAs in vitro. The capsid protein gene was translated cap independently in the presence of the upstream cistron, indicating that the gene is translated by internal ribosome entry. Deletion analysis showed that the internal ribosome entry site (IRES) consisted of approximately 250 bases and that its 3′ boundary extended slightly into the capsid-coding region. The initiation codon for the IRES-mediated translation was identified as the CUU codon, which is located just upstream of the 5′ terminus of the capsid-coding region by site-directed mutagenesis. In vitro translation assays of monocistronic RNAs lacking the 5′ part of the IRES showed that this CUU codon was not recognized by scanning ribosomes. This suggests that the PSIV IRES can effectively direct translation initiation without stable codon-anticodon pairing between the initiation codon and the initiator methionyl-tRNA.
Many insect viruses that are morphologically and biophysically similar to mammalian picornaviruses have been reported (20), and they have been called insect picorna-like viruses. Recently, the complete nucleotide sequences of the genomes of several insect picorna-like viruses have been determined (7, 11, 19, 28, 37). Of these viruses, Drosophila C virus (DCV) (11), Rhopalosiphum padi virus (RhPV) (19) and Plautia stali intestine virus (PSIV) (28) were found to have a novel type of genome organization. The genomes of mammalian picornaviruses consist of positive-strand RNA containing a single large open reading frame (ORF) that codes for the capsid protein precursor in its 5′ part and the nonstructural protein precursor in its 3′ part (27). In contrast, the genomes of DCV, RhPV, and PSIV contain two ORFs that are separated by an intervening region (Fig. 1A). The nonstructural protein precursor is encoded in the 5′-proximal ORF, and the capsid protein precursor is encoded in the 3′-proximal ORF. Previous studies of these three viruses have disclosed two unusual features concerning the translation of capsid proteins: the lack of an in-frame AUG initiation codon and the absence of subgenomic RNA (11, 19, 28).
FIG. 1.
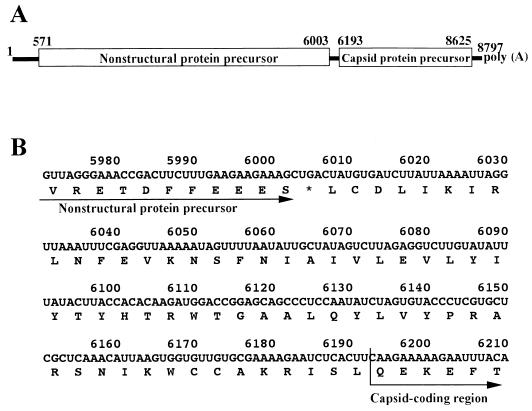
Genome organization of PSIV. (A) Schematic diagram of the PSIV genome. ORFs are shown in open boxes. The numbers indicate nucleotide positions. The first nucleotide of the capsid protein gene represents the 5′-terminal nucleotide of the capsid-coding region. (B) Nucleotide and deduced amino acid sequences of the segment between the nonstructural protein gene and the capsid-coding region. The asterisk indicates the stop codon for the nonstructural protein gene.
Several non-AUG initiation codons are used in the translation of viral and cellular mRNAs; however, their translation efficiency is generally lower than that of the AUG initiation codon (14). Most positive-strand RNA viruses have genome organizations that produce an excess of capsid proteins over nonstructural proteins. When capsid proteins are encoded in the 3′ part of the genome, as in caliciviruses and togaviruses, the viruses produce subgenomic RNA to translate the capsid proteins (31). The excess production of capsid proteins is also observed in DCV in vivo (21), but the virus does not produce subgenomic RNA (11). These observations raise the question of how DCV, RhPV, and PSIV produce capsid proteins effectively.
Previously, we showed that translation of the capsid protein of PSIV occurred independently of the nonstructural protein precursor and that the upstream region of the capsid protein gene was necessary for the translation (28). This observation suggested that the capsid protein of PSIV was translated by internal initiation. Internal initiation of translation was first characterized in mammalian picornavirus RNAs (10, 23). Picornavirus genomic RNAs lack the 5′ cap structure and have long 5′ untranslated regions (5′ UTR). The 5′ UTRs form multiple stem-loop structures, which are in contact with ribosomes. This region is called the internal ribosome entry site (IRES) and conducts cap-independent translation for protein synthesis from the genomic RNA (1, 2, 9, 30). Internal ribosome entry has also been reported for some viral and cellular RNAs such as hepatitis C virus (HCV) RNA (35, 38), cowpea mosaic virus RNA (33), and immunoglobulin heavy-chain binding protein mRNA (18).
In this study, we confirmed that the capsid protein gene of PSIV was translated by internal ribosome entry in vitro by showing that translation of the gene occurs cap independently under dicistronic conditions. The 5′ and 3′ boundaries of the IRES were mapped, and the 3′ boundary was found to slightly overlap the capsid-coding region. We also identified the translation initiation codon of the capsid protein gene by using various site-directed mutants in vitro. The results indicated that translation of the capsid protein gene is initiated at the CUU codon that is located one codon upstream of the 5′ terminus of the capsid-coding region. CUU differs from AUG by two nucleotides, and such an AUG-unrelated initiation codon has not been reported to date. When the 5′ part of the IRES was deleted from a monocistronic RNA carrying the capsid protein gene, scanning ribosomes did not recognize the CUU codon. These data suggest that the PSIV IRES can effectively direct AUG-unrelated initiation.
MATERIALS AND METHODS
Plasmid constructs.
All the RNA templates used for in vitro translation were transcribed from plasmid vectors containing the T7 RNA polymerase promoter sequence.
pCAT3 control vector (Promega) was digested with HindIII and XbaI, and the resultant 0.7-kb fragment, carrying the chloramphenicol acetyltransferase (CAT) gene, was ligated into those sites in pT7Blue (Novagen), generating pT7CAT. A HindIII (blunt-ended)-EcoRI fragment (corresponding to nucleotides [nt] 5375 to 7096) of a cDNA clone of PSIV (28) was ligated into pT7CAT that had been digested with XbaI, blunt ended, and then digested with EcoRI, generating pT7CAT-5375. This plasmid contains two cistrons under the control of the T7 promoter. The first is the CAT gene, and the second is the 5′ part of the capsid protein gene. The upstream sequence of the capsid-coding region is located between the two cistrons.
A series of forward primers (nt 5650 to 5667, 5800 to 5817, 5898 to 5919, 5950 to 5968, 6002 to 6021, and 6100 to 6119) was synthesized to introduce deletions of the 5′ part of the PSIV sequence in pT7CAT-5375. The PSIV sequences with this series of deletions were amplified by PCR with synthesized primers and a 3′ vector-specific primer (5′ GTAAAACGACGGCCAGT), using pT7CAT-5375 as a template. The amplified fragments were blunt ended, digested with EcoRI, and then ligated into pT7CAT that had been digested with XbaI, blunt ended, and digested with EcoRI.
The CAT-IRES-LUC series of constructs have a CAT gene as the first cistron and a luciferase (LUC) gene as the second cistron. PGV-CS2 (Toyo Inki Inc.) was digested with HindIII and EcoRI, and the resultant 1.7-kb fragment, containing a LUC gene, was ligated into those sites in pT7Blue, generating pT7LUC. The LUC gene was amplified by PCR with a synthesized primer and the 3′ vector-specific primer, using pT7LUC as a template. The initiation codon of the LUC gene was deleted and a BamHI site was introduced at the 5′ end by the PCR. The amplified LUC gene was digested with BamHI and EcoRI and then ligated into those sites in pT7CAT, generating pT7CAT-BamLUC. PSIV sequences from nt 5800 to 6192, 6195, 6201, and 6264 were amplified by PCR with a forward primer and reverse primers that contained a BamHI site in their 3′ ends. The amplified fragments were blunt ended, digested with BamHI, and then ligated into pT7CAT-BamLUC that had been digested with SpeI, blunt ended, and digested with BamHI. The constructs generated were named pCAT-IRES6192-LUC, pCAT-IRES6195-LUC, pCAT-IRES6201-LUC, and pCAT-IRES6264-LUC respectively.
Plasmids used to synthesize monocistronic RNAs were generated by inserting PCR products into pT7Blue. Two PSIV cDNA fragments were amplified by PCR with forward primers (corresponding to nt 5800 to 5817 and 6173 to 6193) and the 3′ vector-specific primer, using pT7CAT-5375 as a template. Two PSIV sequences with an AUC-UAA mutation at nt 6184 to 6187 and a CUU-AUG mutation at nt 6190 to 6192 were amplified with forward primers (nt 6173 to 6202) containing the respective mutations and the 3′ vector-specific primer, using p6184TAA or p6190ATG as templates, respectively. These PCR products were blunt ended, digested with EcoRI, and then ligated into pT7Blue that had been digested with HindIII, blunt ended, and digested with EcoRI.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out with a Transformer site-directed mutagenesis kit (Clontech) as described by the manufacturer. Mutations introduced in pT7CAT-5375 were confirmed by nucleotide sequencing with an ABI PRISM Dye Terminator cycle-sequencing kit and a model 377 sequencing system (Perkin-Elmer).
In vitro transcription and in vitro translation.
In vitro transcription of linearized plasmids was carried out with T7 RNA polymerase and an RNA transcription kit (Stratagene). The RNAs were synthesized in the presence or absence of a cap analog, 7mGpppG (Stratagene), as recommended by the manufacturer. After DNase I treatment and phenol-chloroform extraction, transcripts were precipitated with ethanol and ammonium acetate. A portion of the transcripts in each reaction was quantified by agarose gel electrophoresis. Equimolar amounts of RNAs, which corresponded to 10 μg/ml for the transcript of pT7CAT-5375, were used for each in vitro translation reaction. We used an ECL in vitro translation system (Amersham) with 100 mM potassium chloride and 0.5 mM magnesium acetate in a rabbit reticulocyte lysate. The translation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide), blotted onto a polyvinylidine difluoride membrane (Bio-Rad), and then detected by enhanced chemiluminescence.
In the in vitro translation assays, a protein band of about 55 kDa were frequently observed in addition to the expected products (see Fig. 2, 3, 4, and 6). We considered that this protein was insufficiently denatured capsid protein, because this band was observed even under monocistronic conditions (see Fig. 6) and was detected by Western blotting with antiserum to PSIV particles (data not shown).
FIG. 2.
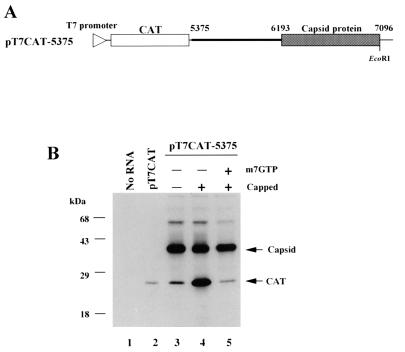
(A) Schematic diagram of pT7CAT-5375. The thin line indicates the vector sequence, and the triangle on the line represents the location of the T7 RNA polymerase promoter. The thick line indicates the PSIV sequence. The open and shaded boxes show the CAT gene and PSIV capsid-coding region, respectively. The nucleotide positions in the PSIV genome are indicated above the line. The EcoRI site used to linearize the plasmid is also shown. (B) Cap influence on in vitro translation of the RNAs transcribed from pT7CAT-5375. Capped and uncapped RNAs were translated in a rabbit reticulocyte lysate with or without a cap analog, m7GTP. To examine the electrophoretic mobility of the CAT protein, uncapped RNA from pT7CAT, which contains only a CAT gene, was also translated. Each product was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide), blotted onto a polyvinylidene difluoride membrane, and then detected by enhanced chemiluminescence. The positions of the translation products (CAT and capsid protein) and molecular mass markers are indicated on the right and left of the panel, respectively. The 55-kDa bands observed in lanes 3 to 5 were thought to be insufficiently denatured proteins translated from the second cistron (see Materials and Methods).
FIG. 3.
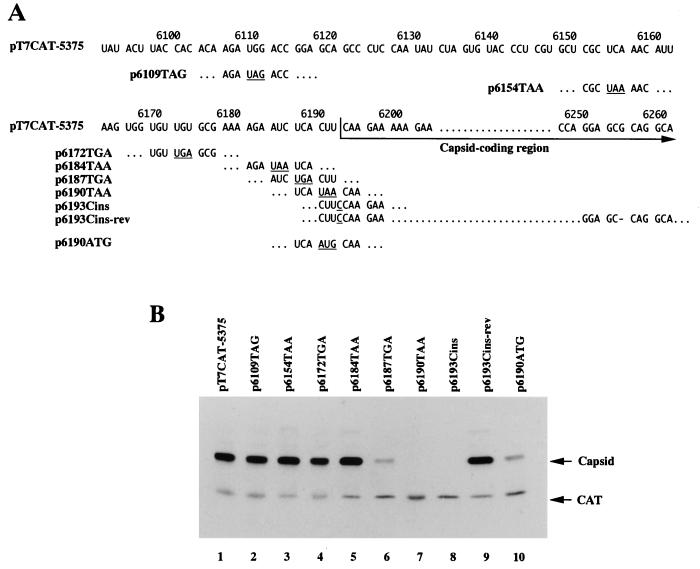
Identification of the initiation codon for capsid protein translation. (A) RNA sequences of mutants derived from pT7CAT-5375. The numbers above the sequence indicate nucleotide positions. Stop codons and inserted nucleotides introduced by site-directed mutagenesis are underlined, and a deleted nucleotide is shown by a dash (p6193Cins-rev). (B) In vitro translation products from uncapped RNAs synthesized from pT7CAT-5375 and the site-directed mutants shown in panel A. The positions of the translation products (CAT and capsid protein) are indicated on the right.
FIG. 4.
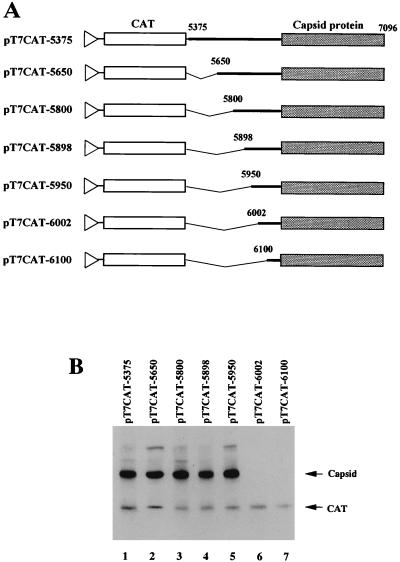
Mapping the 5′ boundary of IRES for PSIV capsid protein translation. (A) Schematic diagrams of pT7CAT-5375 and a series of deletion mutants. The thin lines indicate vector sequences, and the triangles represent the location of the T7 promoter. The thick lines indicate the PSIV sequence. The CAT genes and PSIV capsid-coding regions are shown by open and shaded boxes, respectively. The numbers above the lines indicate nucleotide positions. Deleted regions are shown by angled lines. (B) In vitro translation products of uncapped RNAs synthesized from pT7CAT-5375 and the deletion mutants shown in panel A. The positions of the translation products (CAT and capsid protein) are indicated on the right.
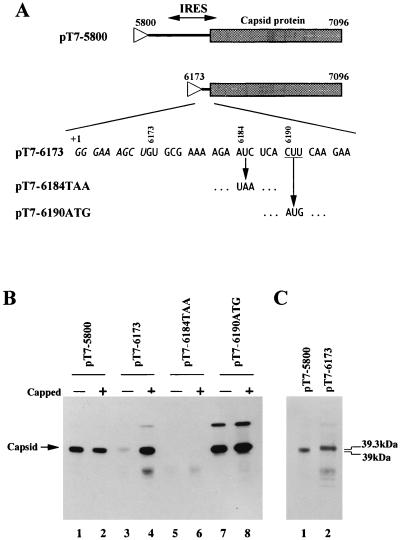
FIG. 6.
(A) Schematic diagrams of plasmids used to synthesize monocistronic RNAs. The lines indicate PSIV sequences and the capsid-coding regions are shown as shaded boxes. Triangles represent the location of the T7 promoter. The 5′-terminal nucleotide sequence of the RNA transcribed from the plasmid pT7-6173 is shown. Italic and roman letters indicate the vector and PSIV sequences, respectively. +1 represents the transcription start site for T7 RNA polymerase. The CUU initiation codon is underlined. For pT7-6184TAA and pT7-6190ATG, only mutated codons are shown. (B) In vitro translation products of uncapped and capped RNAs synthesized from pT7-5800, pT7-6173, pT7-6184TAA, and pT7-6190ATG. The position of the translation products (capsid protein) is indicated on the left of the panel. (C) Comparison of the molecular masses of the translation products from pT7-5800 and pT7-6173. To show the difference in mobility, the same amounts of products were electrophoresed.
RESULTS
The PSIV capsid protein precursor is translated cap independently by IRES-mediated initiation.
We previously indicated that the PSIV capsid protein gene is translated independently of the first ORF and that the upstream sequence of the capsid-coding region is necessary for translation (28). The mechanisms by which a downstream ORF in a eukaryotic polycistronic mRNA is translated are known to include leaky scanning, stop codon readthrough, reinitiation (13), and internal initiation (8, 16). Of these mechanisms, translation by internal initiation is cap independent while translation by the other mechanisms is cap dependent. To confirm that translation of the PSIV capsid protein occurs by internal initiation, the effect of cap structure on translation of the capsid protein gene was investigated in vitro. The dicistronic vector, pT7CAT-5375, contains a CAT gene as the first cistron and the 5′ part of the PSIV capsid protein gene with its upstream sequence as the second cistron (Fig. 2A). The uncapped RNA transcribed from pT7CAT-5375 produced 39- and 25-kDa proteins; the former was much more efficiently translated than the latter (Fig. 2B, lane 3). Antiserum directed against purified PSIV particles confirmed that this 39-kDa protein was the second cistron product (28), while the 25-kDa protein was believed to be the CAT gene product, based on the molecular mass of the product from an RNA containing only the CAT gene (pT7CAT; lane 2). When the capped RNA transcribed from pT7CAT-5375 was translated, the yield of the first cistron product increased significantly whereas the yield of the second cistron product was not affected (lane 4). It is known that cap analogs in the translation reaction mixture compete with the 5′ cap structure of mRNA. Therefore, cap-dependent translation should be inhibited by cap analogs. When the capped RNA was translated in the presence of a cap analog, 7mGpppG (0.8 mM), translation of the first cistron was apparently inhibited but that of the second cistron was not affected (lane 5). These results indicate that translation of the second cistron occurs cap independently. Taking our previous result into consideration, we concluded that the PSIV capsid protein is produced by IRES-mediated translation.
Identification of the capsid protein translation initiation codon.
The N terminus of the PSIV capsid protein precursor was mapped at the CAA codon at nt 6193 to 6195 (28). The nucleotide sequence between the stop codon of the nonstructural protein gene and the CAA codon had no in-frame AUG initiation codon or stop codon (Fig. 1B). To identify the initiation codon for capsid protein translation, stop codons were introduced into the upstream sequence of the capsid-coding region in pT7CAT-5375 by site-directed mutagenesis and transcripts from the mutants were translated in rabbit reticulocyte lysate (Fig. 3). The mutation of the codons at nt 6109 to 6111, 6154 to 6156, 6172 to 6174, and 6184 to 6186 to stop codons (p6109TAG, p6154TAA, p6172TGA and p6184TAA, respectively) did not affect translation of the second cistron (Fig. 3B, lanes 2 to 5). When the UCA codon at nt 6187 to 6189 was mutated to a UGA stop codon (p6187TGA), expression of the second cistron was reduced but not eliminated (lane 6). These results mean that translation of the capsid proteins is initiated at either the CUU codon at nt 6190 to 6192 or the CAA codon at nt 6193 to 6195. When the CUU codon at nt 6190 to 6192 was converted to a UAA stop codon (p6190TAA), there was no translation of the second cistron (lane 7). This suggests that the CUU codon at nt 6190 to 6192 was the initiation codon or that initiation at the CAA codon at nt 6193 to 6195 was affected by this mutation.
To examine these possibilities, we constructed p6193Cins (Fig. 3A). This plasmid has a cytosine inserted immediately downstream of the CUU codon to change the reading frame of the capsid protein gene. We assumed that if CAA was the initiation codon, this insertion should not severely affect translation of the capsid protein. However, a transcript from this plasmid did not produce the second cistron product (Fig. 3B, lane 8). This result strongly suggests that the CUU codon is the initiation codon. To rule out the possibility that this insertion affects initiation at the CAA, p6193Cins-rev was constructed (Fig. 3A). In this plasmid, a guanosine at nt 6255 was deleted compared with p6193Cins. Therefore, the reading frame of the capsid-coding region was consistent with the CUU codon. When the RNA transcribed from p6193Cins-rev was translated, the second cistron product reappeared (Fig. 3B, lane 9). These results indicate that the CUU codon at nt 6190 to 6192 is the translation initiation codon of the PSIV capsid protein gene.
Mapping the 5′ boundary of the IRES for translation initiation of the capsid protein gene.
To map the 5′ boundary of the IRES for PSIV capsid protein translation, we introduced a series of deletions into the intercistronic region of pT7CAT-5375 (Fig. 4A). When the nucleotide sequence from nt 5375 to 5949 was deleted (pTCAT-5950), the second cistron was translated as efficiently as in pT7CAT-5375 (Fig. 4B). However, translation of the second cistron was not observed when the nucleotide sequence was deleted up to nt 6001. These results indicate that the 5′ boundary of the IRES is located between nt 5950 and 6001.
Mapping the 3′ boundary of the IRES for initiation of translation of the capsid protein gene.
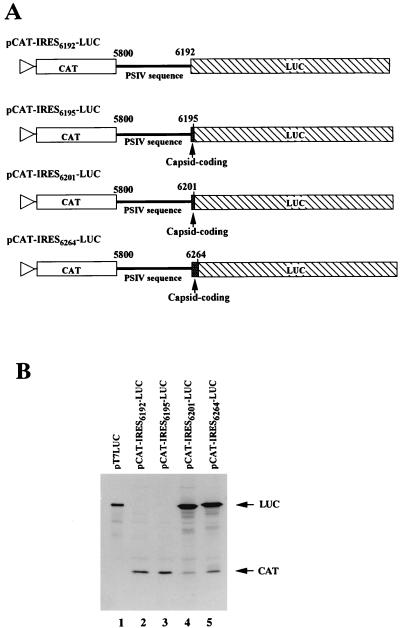
To map the 3′ boundary of the IRES, a LUC gene was used as the second cistron. In the CAT-IRES-LUC series of constructs, LUC genes without an AUG initiation codon were ligated to the PSIV sequences so that they were in frame (Fig. 5A). When the LUC gene was fused just downstream of the CUU initiation codon (pCAT-IRES6192-LUC), translation of the LUC gene was not observed (Fig. 5B, lane 2). An RNA synthesized from pCAT-IRES6195-LUC also failed to produce the second cistron product (lane 3). The LUC gene was efficiently translated when fused downstream of nt 6201 (pCAT-IRES6201-LUC) and nt 6264 (pCAT-IRES6264-LUC) (lanes 4 and 5). These results indicate that the 3′ boundary of the IRES is located between nt 6196 and 6201. This means that the IRES extends into the capsid-coding region.
FIG. 5.
Mapping the 3′ boundary of IRES for PSIV capsid protein translation. (A) Schematic diagrams of the pCAT-IRES-LUC series of constructs. The triangles represent the location of the T7 promoter. The CAT and LUC genes are shown by open and hatched boxes, respectively. The thick lines indicate the PSIV sequences, and the shaded boxes show the PSIV capsid-coding regions. The numbers above the lines indicate the positions of the 5′- and 3′-terminal nucleotides of PSIV sequences. (B) In vitro translation products of uncapped RNAs synthesized from the pCAT-IRES-LUC series of constructs. To examine the electrophoretic mobility of the LUC protein, uncapped RNA synthesized from pT7LUC, which contains only a LUC gene, was also analyzed. The positions of the translation products (CAT and LUC) are indicated on the right of the panel.
The CUU initiation codon does not function in scanning-dependent translation.
The initiator methionyl-tRNA (Met-tRNAi) is the only initiator tRNA (25), and methionine is the initiating residue even when translation is initiated at non-AUG triplets (22). Since previously reported non-AUG initiation codons, such as CUG, GUG, ACG, and AUU, differ from AUG by only one nucleotide, it is thought that at least two base pairs should be required between the initiation codon and the anticodon of Met-tRNAi to initiate translation and that a codon that is completely unrelated to AUG cannot function as an initiation codon (14). The initiation codon for translating the PSIV capsid protein gene, CUU, differs from AUG by two nucleotides. This raised the question whether base pairing between the CUU initiation codon and the anticodon of Met-tRNAi is important for translation initiation of the PSIV capsid protein. If it is important, a mutation of the CUU initiation codon to AUG is assumed to increase the yield of the capsid protein. To examine this, we first introduced a CUU-AUG mutation into pT7CAT-5375 to construct p6190ATG (Fig. 3A). The in vitro translation assay of the RNA transcribed from p6190ATG showed that the CUU-AUG mutation reduced the yield of the second cistron product (Fig. 3B; lane 10). This was an unexpected result because the translation efficiency of most of the previously reported non-AUG codons was improved by mutation to AUG (14, 32, 36).
Therefore, we carried out an experiment to examine whether the CUU codon is recognized by Met-tRNAi. Monocistronic RNAs that are designed to be translated by the scanning mechanism were synthesized and assayed (pT7-6173; Fig. 6A), because codon-anticodon pairing is essential for scanning-dependent translation initiation. Capping of the RNA transcribed from pT7-6173 increased the translational efficiency (Fig. 6B, lanes 3 and 4), indicating that this translation occurred by the scanning mechanism. Transcripts from pT7-6173 generated a product with a slightly larger molecular mass (39.3 kDa) than the 39-kDa product observed in the IRES-mediated translation (pT7-5800) (Fig. 6B and C). This larger protein was thought to be the product of translation initiated at AUC at nt 6184 to 6186, since the codon was the only in-frame potential initiation codon upstream of the CUU codon in the RNA synthesized from pT7-6173 (Fig. 6A). Indeed, a transcript with an AUC-AUG mutation at nt 6184 to 6186 produced a polypeptide with the same size as the product from pT7-6173 (data not shown). A mutation of the AUC codon to UAA (pT7-6184TAA) abolished production of the larger protein (Fig. 6B, lanes 5 and 6), and the RNAs with a CUU-AUG mutation at nt 6190 to 6192 (pT7-6190ATG) produced a major protein with the same molecular mass as the product in the IRES-mediated translation (lanes 7 and 8). These results show that the CUU codon is not recognized by Met-tRNAi in scanning-dependent translation. This suggests that translation mediated by the PSIV IRES is initiated without stable codon-anticodon pairing between the CUU initiation codon and Met-tRNAi.
DISCUSSION
The PSIV capsid proteins are encoded in the 3′ part of the genome. Since PSIV produces no subgenomic RNA and the capsid protein gene lacks an AUG initiation codon (28), the unusual mechanism must be involved in capsid protein translation. In this study, we have provided data explaining this unusual mechanism: initiation of translation of the PSIV capsid protein gene occurs at a CUU codon by internal ribosome entry in vitro.
Previously, we indicated that the PSIV capsid protein gene is translated independently of the first ORF encoding nonstructural proteins and that the upstream sequence of the capsid-coding region is necessary for translation of the capsid protein precursor (28). Here, we showed that translation of the capsid protein occurs cap independently in the presence of the upstream cistron (Fig. 2), confirming IRES-mediated translation of the capsid protein precursor in vitro. The 5′ boundary of the IRES was mapped between nt 5950 and 6001, and the 3′ boundary was mapped between nt 6196 and 6201. It is noteworthy that the 3′ boundary of the IRES lies 6 to 11 nt downstream of the initiation codon (Fig. 5). It has been reported that the activity of the IRESs of a flavivirus, HCV (17, 26) and a picornavirus, hepatitis A virus (HAV) (5), depends on the coding sequences downstream of the initiation codon. The IRESs of picornaviruses and HCV consist of approximately 450 and 300 nt, respectively (2, 15). The PSIV IRES, which consists of approximately 250 nt, is shorter than those of picornaviruses and HCV.
The most noteworthy result described here is that the CUU codon at nt 6190 to 6192 is the initiation codon for capsid protein translation. Non-AUG codons are known to act as the initiation codon for protein synthesis in eukaryotes, such as CUG (6, 24), GUG (3, 32), ACG (4), and AUU (29). These differ from AUG by only one nucleotide, while the CUU codon differs by two nucleotides. The in vitro translation assay with monocistronic RNAs showed that scanning ribosomes did not recognize the CUU codon (Fig. 6). This result suggests that the codon-anticodon pairing between the CUU initiation codon and Met-tRNAi is not essential for translation initiation mediated by the PSIV IRES. In the HCV IRES, mutation of the authentic AUG initiation codon to CUG or AUU only slightly reduced the efficiency of translation initiation (26). However, when the AUG codon was mutated to GAG or GCG, the initiation site changed to another downstream codon. This means that codon-anticodon pairing plays an important role in translation initiation mediated by the HCV IRES. Therefore, the PSIV IRES has different mechanism for translation initiation from the HCV IRES.
Picornavirus IRESs are divided into two classes based on their location relative to the authentic AUG initiation codon. In the enteroviruses and rhinoviruses, the 3′ end of the IRES is located ∼150 bases upstream of the initiation codon and ribosomes that enter the IRES appear to scan until the initiation codon. On the other hand, in cardiovirus and aphthovirus IRESs, ribosomes contact the initiation codon directly (2). In our in vitro translation assay of monocistronic RNAs, the CUU codon did not function as the initiation codon in scanning-dependent translation. Instead, the AUC codon at nt 6184 to 6186, which is located 2 codons upstream of the CUU codon, was recognized as the initiation site (Fig. 6). However, under the dicistronic conditions, a mutation at the AUC codon (p6184TAA; Fig. 3) did not affect translation of the second cistron. Considering these results, it is probable that the 40S ribosomal subunit contacts the CUU initiation codon directly without scanning. If the tertiary structure of the IRES makes the CUU initiation codon come into contact with the anticodon-loop of Met-tRNAi within the 40S subunit, translation initiation may occur efficiently without stable codon-anticodon pairing.
Since Met-tRNAi is the only initiator tRNA (25), methionine would also be the initiating amino acid when the capsid protein precursor of PSIV is translated by internal ribosome entry. The N-terminal amino acid of the capsid protein precursor of PSIV is glutamine, which is encoded by the CAA codon at nt 6193 to 6195 (Fig. 1B) (28). Therefore, the initiating residue, methionine, should be removed from the capsid protein precursor. In enteroviruses and rhinoviruses of the family Picornaviridae, the N-terminal methionine of the capsid protein precursor (P1) is removed and then the penultimate amino acid, glycine, is myristoylated (27). The N-terminal methionine of nascent polypeptide chains is known to be removed cotranslationally by a methionine aminopeptidase. This processing depends on the penultimate residue (12). Since the Met-Gln pair is poorly cleaved by methionine aminopeptidase (12), removal of the methionine in PSIV may occur during capsid formation or by another host protease or viral protease.
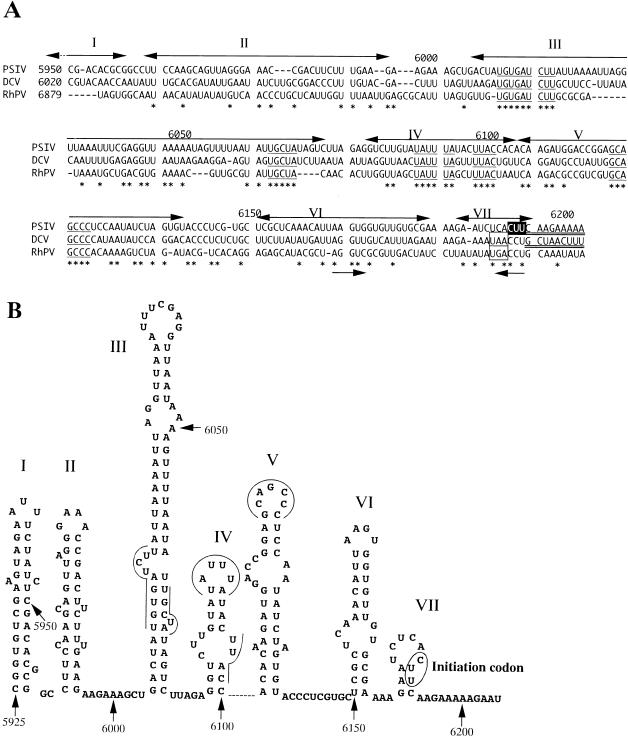
The other two insect picorna-like viruses, DCV (11) and RhPV (19), have similar genome organizations to that of PSIV. The PSIV RNA sequence containing the IRES was aligned with the sequences upstream of the capsid-coding regions of DCV and RhPV by using CLUSTAL W (34) (Fig. 7A), and the secondary structure of the PSIV RNA of this region was predicted with the program MFOLD (Fig. 7B). In the multiple alignment, PSIV, DCV, and RhPV shared several conserved short RNA segments. These conserved segments were located at similar positions when the secondary structures of the DCV and RhPV RNAs were predicted (data not shown). It is likely that these short conserved RNA segments play important roles in IRES activity. In addition, we found another characteristic sequence, an inverted repeat at nt 6163 to 6167 and 6188 to 6192 (Fig. 7A). In the DCV and RhPV sequences, the inverted repeat in this region was also conserved. When the UCA codon at nt 6187 to 6189 and the CUU codon at nt 6190 to 6192 were mutated to other codons, the translational efficiency of the capsid protein gene by internal ribosome entry was reduced (Fig. 3). This result may suggest the importance of this inverted-repeat sequence on IRES-mediated translation. Since this inverted repeat is located in the loop segments of stem-loop structures VI and VII (Fig. 7B), it is possible that the segment from nt 6163 to 6167 interacts with that from nt 6188 to 6192.
FIG. 7.
(A) Multiple alignment of nucleotide sequences upstream of PSIV (28), DCV (11), and RhPV (19) (accession no. AB006531, AF014388, and AF022937, respectively) capsid-coding regions. The numbers on the left indicate the starting nucleotide positions of the aligned sequences, and the numbers above the sequences represent the nucleotide positions in the PSIV sequence. The initiation codon for the PSIV capsid protein translation is shown in reverse type. In the PSIV and DCV sequences, the capsid-coding regions are doubly underlined. The DCV sequence has a stop codon (boxed) 2 codons upstream of the 5′ terminus of the capsid-coding region, and the RhPV sequence also has a stop codon in the same position. Asterisks indicate nucleotides conserved in all three viruses, and the conserved short RNA segments are underlined. Two arrows below the sequences show an inverted repeat. The double-headed arrows above the sequences represent stem-loop segments in the secondary structure predicted for the PSIV RNA, and the roman numerals correspond to those shown in panel. (B) Computer-predicted secondary structure of the PSIV RNA sequence containing the IRES for the capsid protein translation. The numbers indicate nucleotide positions. The initiation codon is circled. The lines and curves indicate the conserved short RNA segments. Stem-loop structures are numbered from I to VII.
The capsid protein genes of DCV and RhPV also lack an in-frame AUG initiation codon. The capsid protein gene of RhPV has an AUG codon in a different reading frame, and its translation is suggested to occur through a −1 frameshift (19), while the translation initiation site of the DCV capsid protein gene is not clearly defined (11). The alignment data showed that the RNA segments upstream of the capsid-coding regions of PSIV, DCV, and RhPV have similar primary and secondary structures. In addition, the DCV and RhPV sequences have stop codons that precede the CCU codons in the position corresponding to the CUU initiation codon of PSIV (Fig. 7A). These data suggest that translation of the capsid proteins of PSIV, DCV, and RhPV is initiated by a similar mechanism, namely, IRES-mediated translation initiation at an AUG-unrelated codon.
ACKNOWLEDGMENTS
We thank Hiroaki Noda for critical reading of the manuscript.
This work was supported by Enhancement of Centers of Excellence, Special Coordination Funds for Promoting Science and Technology, Science and Technology Agency, Japan.
REFERENCES
- 1.Agol V I. The 5′-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeck R, Curran J, Matsuoka Y, Compans R, Kolakofsky D. The parainfluenza virus type 1 P/C gene uses a very efficient GUG codon to start its C′ protein. J Virol. 1992;66:1765–1768. doi: 10.1128/jvi.66.3.1765-1768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graff J, Ehrenfeld E. Coding sequences enhance internal initiation of translation by hepatitis A virus RNA in vitro. J Virol. 1998;72:3571–3577. doi: 10.1128/jvi.72.5.3571-3577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hann S R, King M W, Bentley D L, Anderson C W, Eisenman R N. A non-AUG translation initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 7.Isawa H, Asano S, Sahara K, Iizuka T, Bando H. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch Virol. 1998;143:127–143. doi: 10.1007/s007050050273. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov P A, Karpova O V, Skulachev M V, Tomashevskaya O L, Rodionova N P, Dorokhov Y L, Atabekov J G. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- 9.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 10.Jang S K, Kräusslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson K N, Christian P D. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol. 1998;79:191–203. doi: 10.1099/0022-1317-79-1-191. [DOI] [PubMed] [Google Scholar]
- 12.Kendall R L, Yamada R, Bradshaw R A. Cotranslational amino-terminal processing. Methods Enzymol. 1990;185:398–407. doi: 10.1016/0076-6879(90)85035-m. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 15.Lemon S M, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 16.Liu D X, Inglis S C. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J Virol. 1992;66:6143–6154. doi: 10.1128/jvi.66.10.6143-6154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H-H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosome entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 19.Moon J S, Domier L L, McCoppin N K, D’Arcy C J, Jin H. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology. 1998;243:54–65. doi: 10.1006/viro.1998.9043. [DOI] [PubMed] [Google Scholar]
- 20.Moore N F, Reavy B, King L A. General characteristics, gene organization and expression of small RNA viruses of insects. J Gen Virol. 1985;66:647–659. [Google Scholar]
- 21.Moore N F, Reavy B, Pullin J S K, Plus N. The polypeptides induced in Drosophila cells by Drosophila C virus (strain Ouarzazate) Virology. 1981;112:411–416. doi: 10.1016/0042-6822(81)90288-9. [DOI] [PubMed] [Google Scholar]
- 22.Peabody D S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 23.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 24.Prats H, Kaghad M, Prats A C, Klagsburn M, Lelias J M, Liauzun P, Chalon P, Tauber J P, Amalric F, Smith J A, Caput D. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.RajBhandary U L, Chow C M. Initiator tRNAs and initiation of protein synthesis. In: Söll D, RajBhandary U L, editors. Transfer RNA: structure, biosynthesis, and function. Washington, D.C: American Society for Microbiology; 1995. pp. 511–528. [Google Scholar]
- 26.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carrol A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 609–654. [Google Scholar]
- 28.Sasaki J, Nakashima N, Saito H, Noda H. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology. 1998;244:50–58. doi: 10.1006/viro.1998.9094. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz J, Prüfer D, Rohde W, Tacke E. Non-canonical translation mechanisms in plants: efficient in vitro and in planta initiation at AUU codons of the tobacco mosaic virus enhancer sequence. Nucleic Acids Res. 1996;24:257–263. doi: 10.1093/nar/24.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart S R, Semler B L. RNA determinants of picornavirus cap-independent translation initiation. Semin Virol. 1997;8:242–255. [Google Scholar]
- 31.Strauss E G, Strauss J H, Levine A J. Virus evolution. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 153–171. [Google Scholar]
- 32.Sugihara H, Andrisani V, Salvaterra P M. Drosophila choline acetyltransferase uses a non-AUG initiation codon and full length RNA is inefficiently translated. J Biol Chem. 1990;265:21714–21719. [PubMed] [Google Scholar]
- 33.Thomas A M, ter Haar E, Wellink K, Voorma H O. Cowpea mosaic virus middle component RNA contains sequence that allows internal binding of ribosomes and that requires eukaryotic initiation factor 4F for optimal translation. J Virol. 1991;65:2953–2959. doi: 10.1128/jvi.65.6.2953-2959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J D, Higgins G D, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Wilk F, Dullemans A M, Verbeek M, Van den Heuvel J F J M. Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology. 1997;238:353–362. doi: 10.1006/viro.1997.8835. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]