Abstract
Evidence from human and nonhuman primates suggests that females avoid breeding with close kin and may choose mates based on MHC diversity, which can improve offspring survival. In despotic societies, female mate choice may be hindered by male sexual coercion, but in egalitarian societies, females may be less constrained. Among northern muriquis—an egalitarian, polygynandrous primate with male philopatry—analyses of new data on paternity and variation at microsatellite and MHC loci, combined with behavioural and life-history data, revealed that sires showed higher MHC diversity than expected by chance and were never close kin of dams, consistent with predictions of female mate choice and close inbreeding avoidance. However, females did not differentially reproduce with males who were more distantly related to them or more dissimilar at the MHC than expected by chance, nor with those who had more MHC alleles distinct from their own. The lack of male dominance may permit females to identify and reproduce preferentially with non-offspring males and with males who are more diverse at the MHC. Nonetheless, the absence of disassortative mating at the MHC and neutral loci suggests that female mate choice may be limited by other factors impacting male fertilization success.
Keywords: assortative mating, mate choice, sexual selection, paternity, polygynandry, fitness
1. Introduction
Among mammals, females typically invest considerably greater time and energy in offspring care than do males [1], and this is particularly true among primates, where singleton births and an extended period of juvenile dependency are common. Any reduced fertility or survivorship of offspring is thus expected to impact females' fitness more adversely than males’, suggesting that females should be very selective about potential mates and favour sexual partners with genetic attributes that can enhance offspring's survival [2,3]. Inbreeding avoidance and disassortative mating—the non-random pairing of individuals that are, respectively, non-kin and genetically dissimilar—are two ways in which females may select sexual partners based on genetic characteristics that result in them having offspring who are more genetically diverse [4,5].
A growing body of evidence suggests that the genes of the major histocompatibility complex (MHC) are important in mediating mate choice in vertebrates [6]. Molecules encoded by MHC genes play a key role in the body's immune response to environmental stressors, particularly foreign pathogens, and, theoretically, individuals who are more diverse at MHC loci should be better able to respond to such stressors [7–9]. The complex represents an uncommon example of a balanced polymorphism, where component loci are under strong selective pressure to maintain diversity, thus these loci are potentially an important focus of mate choice selection [8,10–13]. However, analyses of how MHC dissimilarity and/or diversity influences mate choice among mammals, including nonhuman primates and humans, are still rare and are sometimes contradictory. In different human studies, for instance, women have been found to variably prefer either similar or dissimilar male partners—or to show no preference—based on MHC [10]. Moreover, in primate societies with male aggressive behaviour, sexual coercion or intersexual social dominance, females' freedom to choose a mate may be compromised. Indeed, most studies of MHC-mediated mate choice among primates have focused on either despotic or solitary species [10], despite the fact that the effects of female choice on offspring genetics are expected to be more pronounced in species where hierarchical relationships and sexual coercion are rare [14].
Other things being equal, female choice should result in inbreeding avoidance because inbreeding may lead to an increase in homozygosity that can negatively affect offspring fertility, growth, disease susceptibility and survival [5,15]. Selection for mechanisms to avoid inbreeding with close kin should be particularly strong among polygamous primates and other social animals living in mixed-sex groups in which at least one sex is philopatric [16]. Females who avoid mating with close relatives, de facto, also likely avoid having offspring with males who are similar at the MHC, thus these components of female choice are linked.
At the same time, in many species males compete for access to fertile females to maximize their reproductive success, which can result in dominance-related mating and paternity skew among males, thereby undermining female mate choice related to MHC and other inbreeding avoidance behaviour. Male sexual coercion of females can also constrain female choice, overshadowing female inbreeding avoidance behaviour and leading to high paternity skew [17]. Thus, the genetic effects of inbreeding avoidance and mate choice are, theoretically, best examined in taxa where male dominance hierarchies are weak or non-existent, where the opportunity for male sexual coercion of females is minimal, and where female choice is unconstrained.
Northern muriquis (Brachyteles hypoxanthus) are diurnal, egalitarian and polygynandrous primates that live in mixed-sex, patrilocal social groups. The species is characterized by lack of sexual dimorphism, absence of clear dominance hierarchies among males, and minimal evidence of aggression or intersexual dominance among and between males and females [18]. When sexually receptive, females show proceptive behaviour and mate with multiple males without hiding from other group members, and overt male mating competition is minimal [19,20]. Mating is seasonal, with most copulations occurring between September and March (concentrated during the wet season) [21]. Births are likewise seasonal, with most occurring between April and November (concentrated during the dry season) [21,22] after a gestation period of approximately 216 days [23]. Behavioural and demographic data have previously indicated that copulations between mothers and their sexually mature sons are extremely rare or nonexistent, suggesting active inbreeding avoidance [20]. Genetic parentage analysis of one 3-year birth cohort in our study population (n = 22 offspring) confirmed a complete absence of mother-son reproduction [24].
Here, we present new genetic data on these critically endangered platyrrhines, which we combine with rich life-history and demographic data to test predictions about MHC preferences and their relationship to inbreeding avoidance and female sexual selection. After genotyping new samples of offspring, dams, and potential sires from our study population at a panel of neutral autosomal SSR, or ‘microsatellite’, markers, we first conducted parentage analysis for an additional 3-year birth cohort. In doing so, we increased the number of known dam–sire pairs in our longitudinal parentage dataset from 22 to 47, allowing us to expand on a previous assessment of paternity skew in this population and further confirm an absence of mother–son inbreeding [24]. We then generated data on MHC allelic diversity for the entire set of 27 females and 32 potential sires that appear in this parentage dataset and used both the MHC and expanded genotype dataset to evaluate predictions relevant to understanding female mate choice. Specifically, we examined whether muriqui females differentially reproduced with males who are more genetically diverse at MHC loci overall (Prediction 1), are more genetically dissimilar from them at MHC loci, either by sharing fewer alleles or by having a greater number of distinct alleles relative to the female (Prediction 2), or are less closely related to them, on average, at the genome level, as estimated using neutral markers, as expected under inbreeding avoidance (Prediction 3). Inbreeding avoidance is expected to result in dissimilarity at neutral loci, but not necessarily at loci of the MHC, because MHC alleles are presumably not neutral.
2. Methods
(a) . Sampling and laboratory procedures
The study subjects were members of the Matão social group of northern muriquis at the Feliciano Miguel Abdala private natural heritage reserve in the Atlantic Forest of Brazil (19°43'28″ S, 41°48'53″ W). In 2011, the 957 ha forest supported a population of more than 300 individuals living in four mixed-sex groups [24]. The size of these four groups ranged from 37 to 107 animals, and the Matão group has been the subject of a long-term demographic and behavioural study since 1982, during which it grew in size from 22 to over 100 individuals [25,26].
Faecal samples for microsatellite marker and MHC typing were collected from 27 adult females and their offspring born over a six-year period (2005–2010) that covers roughly two consecutive birth cycles for each female, as well as from all potential sires of any of these offspring (i.e. males at least 5 years old who were alive at the time of each conception, n = 32 males total) [20]. Samples were also collected from five additional dams who either did not have any offspring during this time window (n = 2) or had offspring who were not sampled (n = 3). Collectively, this set of animals includes individuals who were genotyped during a previous study of paternity skew in the 2005–2007 cohort, [24], plus new individuals corresponding to offspring born in 2008–2010 and additional potential dams and sires.
Samples were collected by trained personnel who could identify each individual muriqui by its natural facial and body features [18]. Immediately after defecation, roughly 2–4 ml of fresh faeces were transferred to a 8.0 ml vial containing RNAlater or NAP buffer [27] in a minimum 1 : 1 ratio of faeces to preservative and stored at –20°C as soon as possible after collection. Additionally, for two adult males (IV and DI) who died before fresh samples could be collected and were potential sires of at least some infants, we used archival faecal samples stored in desiccating silica gel at an approximate ratio of 1 : 4 of faeces to silica. DNA extraction, PCR amplification and allele scoring at a panel of 15 microsatellite loci (electronic supplementary material, table S1) followed methods described in Strier et al. [24], with some minor adjustments (see electronic supplementary material, Methods for details). For this panel, probability of identity (PI) and probability of identity between full siblings (PIsib) values were 5.0 × 10−13 and 7.8 × 10−6, respectively.
Additionally, we amplified a 171 bp fragment of exon 2 of the MHC class II DRB1 gene via PCR with primers JS1 and JS2 [28] and used paired-end ‘next-generation’ sequencing on the Illumina MiSeq platform to characterize MHC diversity among the entire set of dams and possible sires of the offspring in our sample (n = 59 adults total) (see electronic supplementary material, Methods for details). MHC class II molecules are involved in presenting antigens derived, typically, from extracellular pathogens on the surface of infected cells where they can be recognized and dealt with by T cells of the immune system [8,9,29]. We focused on the DRB1 gene, and particularly on exon 2, which contributes to coding the structure of the extracellular antigen-binding region. This gene is exceptionally polymorphic and is the most commonly screened locus in studies of MHC variation in nonhuman primates [30–35]. MHC allele calling was performed using the AmpliSAS pipeline [36]. Briefly, barcoded paired-end reads were demultiplexed by sample, merged, and had primers trimmed, after which the AmpliSAS algorithm was run to cluster, clean, and call the alleles per individual using the parameters outlined in electronic supplementary material, table S3.
(b) . Paternity assignment
Using our 15-locus genotype panel, we assigned paternity for a total of 47 offspring (22 from our previous study and an additional 25 from the 2008–2010 cohort who were newly genotyped) using the maximum-likelihood method implemented in the software CERVUS v.3.0.7 [37]. We did so using a multi-step process that confirmed field assessments of maternity and yielded high confidence (greater than 95%) paternity assignments for all offspring (see electronic supplementary material, Methods for details). For all of our CERVUS analyses, to determine the δ LOD score needed to assign most-likely dams and sires with high confidence, we created datasets of maternity and paternity for 10,000 simulated offspring, where all sampled adult females and males across both cohorts were considered as potential dams or sires for the respective simulations. To be conservative and allow for the possibility that unsampled individuals (e.g. males from an adjacent social group or from the Matão group that were younger than 5 years of age) could have been parents for some offspring, we assumed that our candidate sample included only 90% of potential dams and 80% of potential sires. The simulations were run presuming that 95% of loci were genotyped, which is an underestimate for our dataset. The genotyping error rate and the error rate in the likelihood calculations specified for these simulations were set at 1%.
(c) . Reproductive skew
To test for unequal distribution of paternity among possible sires, we calculated Nonacs' B index of reproductive skew [38] using the {SkewCalc} package (v. 1.0), for the statistical programming software R (v. 4.2.2) [39]. We first determined the number of infants for whom each male was a potential sire, both within each cohort and overall, based on our long-term demographic dataset. A male was considered a potential sire for an infant if he was at least 5 years old and present in the group at the time of the offspring's conception. We then used these determinations, along with information about the number of offspring sired by each male, within each cohort and overall, to calculate skew. The {SkewCalc} function B_index() directly outputs the point estimate for Nonacs' statistic, which usually varies from −1 to 1, and we a used bootstrapping procedure, sampling 20 000 times with replacement, to generate the 95% confidence interval (CI) around the point estimate. We also calculated the minimum B value possible (i.e. if paternities were distributed equally among all potential sires), and the maximum B value possible (i.e. if all paternities were monopolized by a single individual). If the CI around the B index includes zero, then the distribution of paternity among males cannot be concluded to be significantly different from random. When the CI includes the equal sharing value (minimum B), then an equal distribution of paternities among males cannot be excluded, and if it includes the maximum B, then the possibility of complete reproductive skew (i.e. one male being responsible for all paternities) cannot be rejected.
(d) . Relatedness and genetic simulations
We calculated the Queller & Goodnight [40] relatedness index (R) between all pairs of sampled adult individuals in the Matão group using the {related} package (v. 1.0) [41] for the statistical programming software R (v. 4.2.2) [39]. Sexually mature female muriquis within a social group are expected to be less closely related to one another, on average, than are adult males, because females are known to immigrate from other social groups [42,43]. By contrast, adult males, who remain in their natal group for life, are known to belong to patrilines or brotherhoods and are expected to exhibit, on average, higher dyadic relatedness than pairs of random females from within the same group [44–48]. We tested this idea via simulation using 10 000 iterations of a permutation procedure where we randomly shuffled the assigned sex for each individual to break any association between estimated R and adult dyad type (male–male, female–female and mixed sex). We then calculated the mean R for each dyad type for each permutation to generate the null distributions to which we compared observed mean R values.
To further assess how genetic variation was distributed among individuals in our population, we calculated individual heterozygosities among adults for the microsatellite dataset using the Microsoft Excel add-in GenAlEx 6.51b2 [49] and examined whether heterozygosity at microsatellite loci was related to MHC diversity across the set of all individuals for whom MHC sequencing was performed using Kendall's rank correlation, implemented in R (v. 4.2.2) [39].
We then used data on allelic variation at the MHC class II DRB1 locus and microsatellite marker genotypes from dams and possible sires in the Matão population to test our three predictions. We first tested whether females reproduced with males who possessed a greater number of MHC DRB1 alleles than expected by chance (Prediction 1), which would be consistent with female mate choice for males with higher MHC diversity. This idea has been supported in several studies of nonhuman primates, humans and vertebrates in general [50,51]. Second, we tested whether females reproduced with males with whom, on average, they shared fewer MHC DRB1 alleles (Prediction 2a) or who had more ‘distinct’ MHC DRB1 alleles, i.e. alleles different from those they themselves possessed (Prediction 2b), either or both of which would be consistent with female mate choice for MHC dissimilarity. This idea has also been supported in some studies, although not as ubiquitously as Prediction 1. The third prediction we tested was whether females reproduced with males who were less closely related to them, on average, across neutral microsatellite marker loci than expected by chance (Prediction 3a), or who were less closely related than dyads that were likely to be either first- or second-order kin pairs (Prediction 3b), either of which would be consistent with female mate choice to reduce the risk of inbreeding.
To test Predictions 1 to 3a, we used simulations to generate 100 000 sets of 47 dam–random sire dyads and assembled null distributions for the following statistics of interest:
-
—
Prediction 1: average male MHC DRB1 allelic diversity among sires (i.e. the total number of different MHC DRB1 sequences present in a male).
-
—
Prediction 2a: average pairwise MHC ‘dissimilarity’ between parents (i.e. the total number of MHC DRB1 alleles not shared between dams and sires).
-
—
Prediction 2b: average sire MHC ‘distinctiveness’ (i.e. the number of additional MHC DRB1 alleles found in a sire but not in a dam).
-
—
Prediction 3a: average relatedness (R) between dams and sires, operationalized using the Queller & Goodnight [40] relatedness index.
For both maximum likelihood-based parentage analysis and for performing these simulations, we used data on individual long-term life histories, behaviour and parentage to define the set of potential sires for each offspring, based on each candidate male's presence in the group and his age at the most likely conception date for each infant (calculated as the observed birth date minus the muriquis' average gestation length of 216.4 days [43]). For natal Matão males, the minimum age at first observed copulation with ejaculate (which has been considered to represent the earliest onset of sexual maturity) is 5 years [20,24,25]. Potential sires for each offspring were thus limited to only those males who were alive and present in the Matão group and were at least 5 years of age at the time of the offspring's conception. The observed mean value of each of these measures among the set of 47 actual dam–sire pairs confirmed through genetic parentage analysis was then compared to one-tailed critical values determined from these null distributions.
To test Prediction 3b, we compared the average relatedness among genetically confirmed dam–sire pairs to that among sets of genetically confirmed sets of first-order (i.e. parent-offspring and full sibling) and second-order (i.e. half sibling) kin dyads from our full genotype dataset (N = 111 individuals) using permutation procedures implemented in R (v. 4.2.2) [39]. We also generated expected distributions of estimated mean R values for sets of first- and second-order kin dyads and evaluated whether the set of actual dam–sire pairs was, on average, less closely related to one another than were simulated members of these dyad classes, also using permutation. We did this by first simulating 100 dyads of each of parent-offspring, full sibling, half sibling, and unrelated kin classes using the familysim() function from the {related} package [41], with population allele frequencies based on just the set of adult genotypes (n = 64). We then drew 10 000 random samples of 47 dyads from each kinship category, calculated the mean relatedness (R) among these dyads for each sample to generate the relevant null distribution of mean R for dyads of each kin class, and compared the mean R from our set of 47 confirmed dam–sire dyads to these distributions.
As an additional test of Prediction 3b, we also used the software KINGROUP2 [52,53] to identify dyads from our full genotype dataset (n = 111 individuals: 64 adults plus 47 offspring) that were statistically more likely to be either first- or second-order kin than to be unrelated individuals. This involved comparing the likelihood of each dyad being parent-offspring versus unrelated, full sibling versus unrelated, or half sibling versus unrelated to the distributions of the relative likelihoods for those comparisons based on 1,000,000 permutations. We identified as likely ‘close kin’ any dyads where the P value for the likelihood ratio was < 0.05 for any of these comparisons, and then considered whether any genetically confirmed dam–sire pairs were among those identified as potential ‘close kin’.
3. Results
(a) . Paternity assignment
Using the 15-locus dataset, for the newly genotyped cohort of 26 infants conceived during the 2007 to 2009 mating seasons and born in 2008–2010, CERVUS assigned most-likely paternity to a single male with high (greater than 95%) confidence in all cases. Two infants (BLCO and BMB) described as twins from field records were confirmed to be monozygotic twins after finding that their genotypes were identical and were thus considered one sample for statistical purposes. Thus, the effective number of offspring analysed in each 3-year birth cohort was 22 (born in 2005–2007) and 25 (born in 2008–2010). For both cohorts, these numbers represent 76% of all infants born within each period (some infants from each cohort—seven from the first cohort and eight from the second—could not be sampled as they disappeared and presumably died before samples could be collected) (electronic supplementary material, table S4).
The most successful male (NR) sired 6 out of 25 infants born in 2008–2010 (24%), slightly higher than in our previous study, where the most successful male (BE) sired 18% (N = 4) of 22 infants born in 2005–2007 [24]. Still, this is a much lower percentage than has been reported for the most successful male in a sample of other primates living in multimale, mixed-sex groups for which genetic paternity data are available (table 1). Surprisingly, the most successful male in our first study (BE) sired only one infant in the more recent cohort. By contrast, male BLK sired no infant in the first cohort, but was the second most successful sire in the subsequent cohort, where he was assigned paternity for 4 of 25 (16%) infants (electronic supplementary material, table S4).
Table 1.
Selected studies of genetic paternity in wild primates living in multimale-multifemale social groups, expanded from Strier et al. [24].
| taxon | total no. assignments | max % individual paternity success | max (mean) Nonacs’ Ba | mean (range) no. males | no. femalesb | sex ratioc | M–F dom. | reference |
|---|---|---|---|---|---|---|---|
| mountain gorilla 1 | 48 | 85 | 0.432 (0.376) | 3.0 (2–6) | – | – | M > F | [54,55] |
| mountain gorilla 2 | 79 | 83 | – | 5.8 (1–14) | 10.5 (4–17) | 0.55 | M > F | [56,57] |
| yellow baboon | 27 | 81 | – | 5.0 | 11 | 0.45 | M > F | [54,58–60] |
| white-faced capuchin 1 | 41 | 80 | 0.401 (0.237) | 5.8 (2–11) | 7.0 (4–10) | 0.83 | M > F | [61,62] |
| white-faced capuchin 2 | 118 | 85 | – | 4.5 (1–10) | 5.5 (2–11) | 0.82 | M > F | [63] |
| chimpanzee 1 | 38 | 67 | – | 5.2 (1–8) | 3.5 (2–8) † | 1.49 † | M > F | [54,58,64] |
| chimpanzee 2 | 21 | 31 | – | 14.5 (7–17) | 13 | 1.12 | M > F | [65] |
| chimpanzee 3 | 34 | 30 | – | 10.5 (7–12) | 3.9 (1–11) † | 2.69 † | M > F | [66] |
| crested macaque | 63 | 100 | 0.672 (0.330) | 14.7 (7–21) | 20 (17–25) | 0.74 | M > F | [67] |
| Assamese macaque | 43 | 33 | 0.087 | 12 (9–15) | 13.5 (12–15) | 0.89 | M > F | [68] |
| vervets | 94 | 67 | 0.130 (0.045) | 10 (6–14) | 11.3 (6–18) | 0.88 | M > F | [69] |
| bonobo 1 | 10 | 30 | – | 6 | up to 15 | 0.40 | F ≥ M | [70] |
| bonobo 2 | 13 | 62 | 0.220 | 7.4 | 4.9 (1–9) † | 1.51 † | F ≥ M | [71] |
| bonobo 3 | 17 | 82 | 0.510 | 7.7 (5–10) | 9.3 (6–13) | 0.75 | F ≥ M | [72] |
| ring-tailed lemur | 39 | 100 | 0.734 (0.229) | 5.4 (3.7–7.7) | 5.9 (4.7–8.3) | 0.92 | F > M | [73] |
| northern muriqui 1 | 22 | 18 | 0.012 | 24.5 (23–27) | 29.2 (28–31) | 0.84 | F = M | [24] |
| northern muriqui 1 | 22 | 18 | 0.018d | 24.5 (23–27) | 29.2 (28–31) | 0.84 | F = M | This Study |
| northern muriqui 2 | 25 | 24 | 0.055 | 29.9 (29–3) | 33.4 (31–36) | 0.90 | F = M | This Study |
aMaximum and mean values for Nonacs' B index are either taken across groups [55,73], periods of male tenure [62], or a combination of group and birth season [67,69]. Single values indicate results from a single group and time window.
bNumber of males and number of females are based on values presented in original sources or as calculated in [54,58]. In some cases, authors report numbers of individuals of each sex as a range, either across groups or within the same group over time. Extragroup individuals, such as additional candidate sires considered in paternity assessments, are not included. Where authors do not provide an average, we include the midpoint of the range reported or the average, across groups, of such midpoints.
cSex ratio estimates are calculated by dividing the mean no. of males by mean no. of females, as indicated in the preceding column. Because some studies report the census number of females per group and others the number of females inferred to be in estrus and able to conceive, the values included here can be either group composition-based sex ratios (i.e. the ratio of census numbers of either adult or sexually mature males to adult females) or operational sex ratios (i.e. ratio of sexually active males to receptive females) [74], which are often much higher, particularly for species with long interbirth intervals and less reproductive synchrony. Daggers (†) denote those species for which the number of receptive females, rather than the total number of females, is reported and used to calculate the sex ratio estimate.
dIn this study, we recalculated the B index for the same paternity dataset presented in our prior study [24] of Northern muriqui 1 cohort using current software and a slightly refined set of criteria for inclusion of males among the set of candidate sires. Although the recalculated B index is slightly higher than we reported previously, it is nonetheless still quite low compared to other multimale primate species and, as previously reported, is not significantly different from zero.
Of the 31 males who were potential sires for at least one of the 25 infants from the 2008–2010 cohort, 12 were successful (39%)—a lower proportion than that found in our prior study of the 2005–2007 cohort, where 13 of 27 (48%) candidate males sired at least one infant [24]. When the two cohorts are analysed together, 16 out of 32 total potential sires (50%) fathered at least one out of the 47 infants. Of the 27 males that were reproductively active for at least some portion of time during the mating seasons associated with both studies, only nine (33%) sired at least one infant in both birth cohorts. Of the 19 males that were reproductively active across the entire sample period (i.e. for all six mating seasons), only six did not sire any young, and eight (42%) sired at least one infant in both cohorts. For this set of 19 males, paternity success (i.e. siring at least one infant) in one cohort was associated with success in the other (Kendall τ = 0.472, p = 0.045, n = 19), although the number of infants they sired was only weakly correlated between the two cohorts (Kendall τ = 0.333, p = 0.095, n = 19).
Unlike in our previous study, Nonacs' skew index (B) calculated for the 2008–2010 cohort was significantly greater than zero (B = 0.055, n = 25, p < 0.001), although still quite low compared to other primates (table 1). In addition, the 95% confidence interval of B (0.005 to 0.121) did not include either the minimum (−0.039) or maximum B (0.927), suggesting that both a complete sharing scenario and a complete monopoly scenario can be rejected. The combined skew index for both cohorts together was also quite low (B = 0.031, 95% CI = 0.008 to 0.068, n = 47), but, again, was significantly—albeit only slightly—greater than zero (p < 0.01), unlike for the first cohort alone.
The age of the youngest sire at the time of conception of offspring born in the 2008–2010 cohort (male FS) was 7.5 years, which is slightly younger than the age of the youngest sire found in our previous study (8.4 years). Similar to the prior cohort, there was at least a two-year delay between the age at which males are usually first seen copulating (approximately 5 years) and their age at first successful conception [24]. Also, consistent with our finding for the first cohort, older males (i.e. those in the upper half of the age distribution of candidate males) sired more infants (n = 16) than younger males (n = 9), although, as in the first cohort, this was not a significant difference (binomial test, one-tailed p = 0.115).
(b) . MHC variation
MHC DRB1 PCR amplicons of dams and potential sires were sequenced to a median depth of 293 622 reads per sample and had a median depth of 156 477 reads per sample after AmpliSAS removed those that did not fit the inclusion criteria. AmpliSAS initially scored 39 MHC variants in the total sample of 59 adult individuals. Seventeen of these putative alleles were removed prior to statistical analyses. Fifteen of these 17 were scored in only one individual and had a very low number of total reads (less than 4000). These are likely to be PCR or sequencing artefacts, rather than true alleles. The other two of these 17 variants were scored in only 3 and 8 individuals, respectively, but with low total read count across individuals (less than 3400 and less than 12 500, respectively) and low read counts per individual (all less than 3000) (electronic supplementary material, table S6). The remaining 22 alleles were found in an average of 14.7 individuals (range = 1 to 59, median = 7) and represent a reasonably high MHC diversity for a population that expanded from only 50 individuals since 1983 [75] (electronic supplementary material, figure S2, and tables S5 and S6). By comparison, only three haplotypes were found to be present in hypervariable region 1 of the mitochondrial DNA in this same population [76].
The median number of distinct MHC DRB1 alleles found in the set of 32 potential sires was 5 (range = 3 to 7) and in the set of 27 dams was 6 (range = 3 to 8), while the median number of dissimilar alleles between all pairs of such males and females was 6 (range = 0 to 12). There was no correlation between microsatellite heterozygosity and the number of MHC alleles per sample among either potential sires (Kendall's τ = −0.188, p = 0.201, n = 32), dams (Kendall's τ = −0.080, p = 0.610, n = 27), or among the total set of 59 individuals for whom we characterized MHC diversity (Kendall's τ = −0.119, p = 0.259).
(c) . Within-group relatedness
As expected for primates such as muriquis with male philopatry and strongly female-biased dispersal, adult male dyads in the Matão group were, on average, more closely related to one another than expected by chance (mean R among male–male dyads = 0.027, p = 0.006, one-tailed test), while adult females were somewhat less closely related than expected by chance (mean R among female–female dyads = −0.038, p = 0.054, one-tailed test). Mixed-sex dyads were also somewhat less closely related than expected by chance (mean R among mixed-sex dyads = −0.024, p = 0.053, two-tailed test). Given these results, not surprisingly, the average R among adult male dyads was significantly greater than that among both adult female and mixed-sex dyads (p = 0.013 and p = 0.010, respectively) (see also electronic supplementary material, figure S3).
(d) . Female sexual selection and inbreeding avoidance
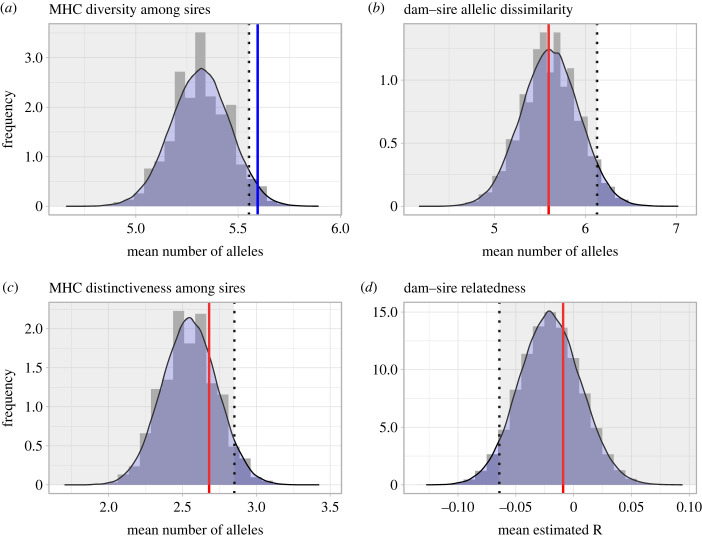
Consistent with Prediction 1, female muriquis produced infants with males who were more diverse at the MHC than expected by chance (mean MHC diversity of sires = 5.60, p = 0.021; figure 1a). However, there was no evidence that females produced offspring with males who were more dissimilar to them at the MHC than expected by chance, either when this was operationalized as the total number of MHC DRB1 alleles that dams and sires do not have in common (Prediction 2a: mean MHC ‘dissimilarity’ between dams and sires = 5.60, p = 0.502; figure 1b) or as the number of alleles found in the sire that are different from those seen in the dam (Prediction 2b: mean MHC ‘distinctiveness’ of sires = 2.68, p = 0.219; figure 1c).
Figure 1.
Null distributions of (a) average MHC diversity among sires, (b) average MHC ‘dissimilarity’ between dams and sires, (c) average MHC ‘distinctiveness’ among sires, and (d) average estimated R [40] between dams and sires at microsatellite loci based on 100 000 simulations of sets of dam-random sire pairs from the Matão group. For each simulation, a random male was selected as the sire for each of 47 offspring with a confirmed dam from the set of males who were present in the group and older than 5 years at the time of the offspring's conception. In each panel, the dotted line marks the critical value for testing the relevant prediction (i.e. either the 5% or 95% quantile of the simulation distribution, depending on whether the relevant test is upper- or lower-tailed). Solid lines mark the mean observed value for the relevant statistic calculated from 47 actual dam–sire pairs. The blue solid line reflects a significant test (p < 0.05) while the red solid lines reflect non-significant tests.
Based on estimates of dyadic genetic relatedness (R) using the 15-locus microsatellite dataset for adults only, there was also no support for the idea that dyads of males and females who successfully produced offspring had a lower mean R, on average, than would be expected by chance (Prediction 3a: mean R of actual dam–sire pairs across 47 offspring = −0.009, p = 0.663, figure 1d). However, consistent with our prior results from the 2005–2007 birth cohort, none of the 25 infants born in 2008–2010 resulted from a mother-son liaison, and the same was true for an additional eight successfully assigned paternities for natal adult males residing within the Matão group (electronic supplementary material, table S7).
Additionally, confirmed dam–sire dyads were, on average, neither more nor less closely related to one another as adult female–female (p = 0.427, two-tailed test), male–male (p = 0.146, two-tailed test), or mixed-sex dyads (p = 0.786, two-tailed test). However, confirmed dam–sire dyads were significantly less closely related to one another, on average, than either observed dyads of first- and second-order kin (p < 0.001 for all comparisons, based on the full genotype dataset) or simulated sets of such kin (p < 0.001 for all comparisons, using only adult genotypes as the basis for simulation; Prediction 3b; see also electronic supplementary material, figure S4).
4. Discussion
The results of this study contribute to our understanding of mating systems and the processes of female sexual selection and inbreeding avoidance in a group-living primate characterized by egalitarian social relationships.
Based on a total of 33 new parentage assignments (25 offspring born in the 2008–2010 cohort plus eight additional assignments for natal adult males in the Matão group), this study confirms the total avoidance of inbreeding between mothers and their adult sons reported in prior behavioural studies [20,24,77] and in our previous genetic analysis of parentage in an earlier cohort of 22 offspring born in this population [24]. It also lowers the minimum age for successful paternity in wild muriquis from 8.4 to 7.5 years and increases the degree of male reproductive skew seen in wild muriquis from 18% to 24% of paternities for the most successful sire within each cohort. However, whether these differences reflect cohort-specific variation (e.g. number of potential male competitors excluding close kin, female preferences) or are the result of the increased sample size is not clear. It is also notable that the reproductive success of individual males varied dramatically between cohorts, as females tended to have successive offspring with different sires. Parentage data from additional cohorts are needed to gain a fuller understanding of the causes and true range of variation in male age at first paternity and male reproductive skew.
(a) . Paternity, inbreeding avoidance and relatedness
Our confirmation that adult females are not reproducing with their adult sons or other close kin implies the existence of mechanisms of kin recognition (e.g. familiarity during development) to avoid incestuous matings. Considering the demographic history of this small, isolated population [78], it is not surprising that some dam–sire pairs had high estimated dyadic relatedness (as high as 0.369) and that a few pairs were statistically more likely to be ‘close kin’ than unrelated (see electronic supplementary material, figure S3). However, parentage tests excluded all such pairs as likely parent–offspring dyads, in agreement with observational data collected over multiple field seasons [20,77].
Overall, the distribution of paternities among adult male muriquis in both cohorts contrasts with patterns of male parentage and reproductive skew seen in other multimale–multifemale primate species where male social relationships are more hierarchical (table 1). This result is consistent with the non-aggressive and promiscuous behaviour of muriquis and with a scramble rather than contest model of male reproductive competition [79]. Indeed, in contrast to chimpanzees, where alpha males sire a smaller share of infants as the number of male competitors increases [29,32], we found the opposite pattern in muriquis, with reproductive skew slightly higher for the cohort with more potential male competitors (31 versus 27). Instead, patterns of female mate choice and the social influence of adult males' mothers seem to have a bigger impact on the variation we observe in muriquis, with matriarchs and their adult sons possibly collaborating to mutually increase each other's fitness [24]. Such kin-selected advantage also seems evident in bonobos and chimpanzees, in which, respectively, transfer of social status [80,81] and postweaning maternal care [82] are argued to increase adult sons' reproductive success. Indeed, grandmaternal success in muriquis tended to be associated with the presence of and social proximity between mothers and their sons [24,83].
Muriquis also have marked reproductive seasonality, with the mating season occurring from September through March [20], although this can vary annually depending on rainfall and, presumably, resource availability [84]. Temporal concentration of reproductive opportunities, if associated with female reproductive synchrony, should reduce any given male's ability to monopolize mating access to multiple females. Coupled with female polyandry, this may act to spread paternities among males in multimale–multifemale groups, thereby reducing paternity skew [58]. However, reproductive seasonality and female reproductive synchrony are only weakly correlated in nonhuman primates, and reproductive seasonality alone has been shown to be a poor predictor of male reproductive skew [54]. In addition, female muriquis lack conspicuous signs of ovulation (e.g. sexual swellings) [23,85], although males do seem to obtain pheromonal cues of females' status by inspecting their genitalia [86]. Moreover, males have large testis size, which suggests that sperm competition is an important component of male muriquis’ reproductive strategy [87]. Reproductive synchrony, concealed ovulation, sperm competition and behavioural inbreeding avoidance all probably contribute collectively to explaining the low paternity skew seen in muriquis. Yet, given that some males—older ones, in particular—showed a tendency to sire a higher number of infants [24], it is still possible that some level of proficiency in reading female signs of ovulation or other attributes that can enhance a male's efficiency at fertilizing the females during their conceptive cycles (e.g. female preference, differential sperm viability, etc.) plays a role in male muriquis' reproductive tactics. Indeed, behavioural observations show that older male muriquis are more likely to complete a copulation bout with ejaculation during the conception season than are younger males [88].
(b) . MHC diversity
Our finding that female muriquis tended to have offspring with males that carried higher allelic diversity at the MHC locus than expected by chance may also contribute to the persistence of this small, isolated population. In fact, all 47 infants were sired by males that had at least five MHC alleles. This may be in part explained by the availability of many males in the group with moderate to high levels of diversity (mode = 5 alleles). Only three potential sires had fewer than five alleles at the DRB1 exon 2 locus, and all individuals in our sample, regardless of sex, had at least three alleles. This low variance in the level of MHC diversity seen across individuals has elsewhere been referred to as the ‘lek paradox’, in which sexual selection tends to purge lower diversity males from the population [89,90]. This is particularly striking given the fact that our study population of muriquis, which was over 300 individuals in 2011 [24], had expanded from a considerably smaller population of 50 individuals in the early 1980s [75]. The high level of diversity seen at the MHC may be one of the reasons why this isolated population has been able to grow so steadily in the last three and a half decades [78]. Our results concur with the meta-analysis of Winternitz et al. [10], which found a significant signal for primate choice for MHC diversity in a comparison of six studies. However, these authors also suggest that there is a tendency for relative testis size to be negatively associated with choice for MHC diversity, which does not seem to be the case among muriquis.
Contrary to our expectation, we did not find evidence of females preferring more dissimilar males at the MHC, which also contrasts with the overall trend that has been reported for primates [10], nor did we find evidence that breeders were less closely related, on average, than expected using estimates of relatedness based on the microsatellite markers. Two main hypotheses may explain how female reproductive choice increases offspring fitness. The ‘good genes’ hypothesis predicts that specific cues signal a male's genetic quality (e.g. diversity in his MHC alleles), while the ‘compatibility’ hypothesis suggests that genetic dissimilarity (e.g. having different MHC alleles) between mates increases the genetic diversity and, hence, the fitness of offspring [91]. Evolving a mechanism to discriminate whether another individual is genetically dissimilar may be more complex than being able to discriminate diversity. The former requires integrating information on one's own and another's genetic makeup, while the latter may require only reading the same kinds of cues (e.g. olfactory or visual) [51,92] across the set of possible partners.
Another possibility is that the power of the tests we could employ with this dataset may be limited due to the number of dam–sire pairs (n = 47) compared to some suggested theoretical sample sizes of hundreds or even thousands of dyads [10,93] and the short length of DRB1 exon 2 relative to the full MHC gene family. Information on additional paternities and exons may be needed in order to better test disassortative mating in muriquis, especially in a scenario where allelic variance may be high and the effect weak.
Acknowledgements
We appreciate the work and support of all of the people and institutions that contributed in different stages of this research, including Todd Disotell, Clifford Jolly, Terry Harrison, Sérgio Mendes, Valéria Fagundes, Carla Possamai, Fernanda Tabacow, Rogério Ribeiro, Simone Lóss, Juliana Justino, the New York Consortium in Evolutionary Primatology (NYCEP), the Molecular Anthropology Laboratory at New York University, and the Primate Molecular Ecology and Evolution Laboratory and the Genome Sequencing and Analysis Facility at the University of Texas at Austin. We also thank Alvaro Sebastian for assistance in using AmpliSAS, as well as two anonymous reviewers whose thoughtful feedback improved the manuscript.
Contributor Information
Karen B. Strier, Email: kbstrier@wisc.edu.
Anthony Di Fiore, Email: anthony.difiore@austin.utexas.edu.
Ethics
This study complied with research protocols and sample collection and transfer approvals issued by the University of Wisconsin-Madison (no. L00104-0-07-08), the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (Sisbio permit no. 37854-1), the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA/CITES) (export permit no. 09BR003125/DF), and the US Centers for Disease Control and Prevention (CDC) (import permit no. 2008-11-096). The work fully adhered to the legal requirements of Brazil and to the American Society of Primatologists' Principles for the Ethical Treatment of Primates.
Data accessibility
MHC DRB1 exon 2 allele sequences (171 bp) are provided in electronic supplementary material, table S5, and are deposited in GenBank (accession numbers OR340584–OR340605; GenBank sequences are 216 bp and include primer-binding regions). Additional datasets (e.g., multilocus genotypes) and information supporting this article have also been provided as part of electronic supplementary material [94]. R scripts and associated datasets for key analyses are available at https://github.com/difiore/muriqui-MHC.
Authors' contributions
P.B.C.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; K.B.S.: conceptualization, data curation, formal analysis, funding acquisition, methodology, project administration, resources, writing—review and editing; A.D.: conceptualization, data curation, formal analysis, funding acquisition, methodology, project administration, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, BEX-1872-07-9), New York University, the New York Consortium in Evolutionary Primatology (NYCEP), The University of Texas at Austin, and the University of Wisconsin-Madison.
References
- 1.Trivers RL. 1972. Parental investment and sexual selection. In Sexual selection and the descent of Man: 1871–1971 (ed. Campbell B), pp. 136-179. Chicago, IL: Aldine de Gruyter. [Google Scholar]
- 2.Kappeler PM. 2012. Mate choice. In The evolution of primate societies (eds Mitani JC, Call J, Kappeler PM, Palombit RA, Silk JB), pp. 367-386. Chicago, IL: University of Chicago Press. [Google Scholar]
- 3.Tregenza T, Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage. Mol. Ecol. 9, 1013-1027. ( 10.1046/j.1365-294x.2000.00964.x) [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y, Bolnick DI, Kirkpatrick M. 2013. Assortative mating in animals. Am. Nat. 181, E125-E138. ( 10.1086/670160) [DOI] [PubMed] [Google Scholar]
- 5.Pusey A, Wolf M. 1996. Inbreeding avoidance in animals. Trends Ecol. Evol. 11, 201-206. ( 10.1016/0169-5347(96)10028-8) [DOI] [PubMed] [Google Scholar]
- 6.Kamiya T, O'Dwyer K, Westerdahl H, Senior A, Nakagawa S. 2014. A quantitative review of MHC-based mating preference: the role of diversity and dissimilarity. Mol. Ecol. 23, 5151-5163. ( 10.1111/mec.12934) [DOI] [PubMed] [Google Scholar]
- 7.Hughes AL, Yeager M. 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32, 415-435. ( 10.1146/annurev.genet.32.1.415) [DOI] [PubMed] [Google Scholar]
- 8.Hughes AL, Hughes MK, Howell CY, Nei M, Howard JC, Higgs P. 1994. Natural selection at the class II major histocompatibility complex loci of mammals. Phil. Trans. R. Soc. B 346, 350-367. [DOI] [PubMed] [Google Scholar]
- 9.Klein J. 1986. Natural history of the major histocompatibility complex. New York, NY: John Wiley. [Google Scholar]
- 10.Winternitz J, Abbate JL, Huchard E, Havlíček J, Garamszegi LZ. 2017. Patterns of MHC-dependent mate selection in humans and nonhuman primates: a meta-analysis. Mol. Ecol. 26, 668-688. ( 10.1111/mec.13920) [DOI] [PubMed] [Google Scholar]
- 11.Setchell JM, Huchard E. 2010. The hidden benefits of sex: evidence for MHC-associated mate choice in primate societies. Bioessays 32, 940-948. ( 10.1002/bies.201000066) [DOI] [PubMed] [Google Scholar]
- 12.Piertney SB, Oliver MK. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7-21. ( 10.1038/sj.hdy.6800724) [DOI] [PubMed] [Google Scholar]
- 13.Edwards SV, Hedrick PW. 1998. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol. Evol. 13, 305-311. ( 10.1016/S0169-5347(98)01416-5) [DOI] [PubMed] [Google Scholar]
- 14.Alberts SC. 2012. Magnitude and sources of variation in male reproductive performance. In The evolution of primate societies (eds Mitani JC, Call J, Kappeler PM, Palombit RA, Silk JB), pp. 412-431. Chicago: University of Chicago Press. [Google Scholar]
- 15.Charlesworth B, Charlesworth D. 1999. The genetic basis of inbreeding depression. Genet. Res. 74, 329-340. ( 10.1017/S0016672399004152) [DOI] [PubMed] [Google Scholar]
- 16.Charpentier MJE, Widdig A, Alberts SC. 2007. Inbreeding depression in non-human primates: a historical review of methods used and empirical data. Am. J. Primatol. 69, 1370-1386. ( 10.1002/ajp.20445) [DOI] [PubMed] [Google Scholar]
- 17.Nonacs P, Hager R. 2011. The past, present and future of reproductive skew theory and experiments. Biol. Rev. 86, 271-298. ( 10.1111/j.1469-185X.2010.00144.x) [DOI] [PubMed] [Google Scholar]
- 18.Strier KB. 1999. Faces in the forest: The endangered muriqui monkeys of Brazil, 2nd edition. Cambridge, MA: Harvard University Press. [Google Scholar]
- 19.Strier KB. 1990. New World primates, new frontiers: Insights from the woolly spider monkey, or muriqui (Brachyteles arachnoides). Int. J. Primatol. 11, 7-19. ( 10.1007/BF02193693) [DOI] [Google Scholar]
- 20.Strier KB. 1997. Mate preferences of wild muriqui monkeys (Brachyteles arachnoides): reproductive and social correlates. Folia Primatol. 68, 120-133. ( 10.1159/000157242) [DOI] [Google Scholar]
- 21.Strier K. 1996. Reproductive ecology of female muriquis (Brachyteles arachnoides). In Adaptive radiations of neotropical primates (eds Norconk M, Rosenberger A, Garber P), pp. 511-532. New York: Plenum Press. [Google Scholar]
- 22.Strier KB, Mendes SL, Santos RR. 2001. Timing of births in sympatric brown howler monkeys (Alouatta fusca clamitans) and northern muriquis (Brachyteles arachnoides hypoxanthus). Am. J. Primatol. 55, 87-100. ( 10.1002/ajp.1042) [DOI] [PubMed] [Google Scholar]
- 23.Strier KB, Ziegler TE. 1997. Behavioral and endocrine characteristics of the reproductive cycle in wild muriqui monkeys, Brachyteles arachnoides. Am. J. Primatol. 42, 299-310. () [DOI] [PubMed] [Google Scholar]
- 24.Strier KB, Chaves PB, Mendes SL, Fagundes V, Di Fiore A. 2011. Low paternity skew and the influence of maternal kin in an egalitarian, patrilocal primate. Proc. Natl Acad. Sci. USA 108, 18 915-18 919. ( 10.1073/pnas.1116737108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strier KB, Boubli JP, Possamai CB, Mendes SL. 2006. Population demography of northern muriquis (Brachyteles hypoxanthus) at the Estação Biológica de Caratinga/Reserva particular do Patrimônio Natural-Felìciano Miguel Abdala, Minas Gerais, Brazil. Am. J. Phys. Anthropol. 130, 227-237. ( 10.1002/ajpa.20366) [DOI] [PubMed] [Google Scholar]
- 26.Strier KB, Mendes SL. 2012. The northern muriqui (Brachyteles hypoxanthus): lessons on behavioral plasticity and population dynamics from a critically endangered species. In Long-Term field studies of primates (eds Kappeler PM, Watts DP), pp. 125-140. Berlin, Germany: Springer. [Google Scholar]
- 27.Camacho-Sanchez M, Burraco P, Gomez-Mestre I, Leonard JA. 2013. Preservation of RNA and DNA from mammal samples under field conditions. Mol. Ecol. Resour. 13, 663-673. ( 10.1111/1755-0998.12108) [DOI] [PubMed] [Google Scholar]
- 28.Schad J, Sommer S, Ganzhorn JU. 2004. MHC variability of a small lemur in the littoral forest fragments of southeastern Madagascar. Conserv. Genet. 5, 299-309. ( 10.1023/B:COGE.0000031137.50239.d3) [DOI] [Google Scholar]
- 29.Knapp LA. 2005. The ABCs of MHC. Evol. Anthropol. 14, 28-37. ( 10.1002/evan.20038) [DOI] [Google Scholar]
- 30.Schwensow N, Eberle M, Sommer S. 2008. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc. R. Soc. B 275, 555-564. ( 10.1098/rspb.2007.1433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huchard E, Knapp LA, Wang J, Raymond M, Cowlishaw G. 2010. MHC, mate choice and heterozygote advantage in a wild social primate. Mol. Ecol. 19, 2545-2561. ( 10.1111/j.1365-294X.2010.04644.x) [DOI] [PubMed] [Google Scholar]
- 32.Arguello–Sánchez LE, Arguello JR, García-Feria LM, García-Sepúlveda CA, Santiago-Alarcon D, Espinosa De Los Monteros A. 2018. MHC class II DRB variability in wild black howler monkeys (Alouatta pigra), an endangered New World primate. Anim. Biodivers. Conserv. 41, 389-404. ( 10.32800/abc.2018.41.0389) [DOI] [Google Scholar]
- 33.Huchard E, Baniel A, Schliehe-Diecks S, Kappeler PM. 2013. MHC-disassortative mate choice and inbreeding avoidance in a solitary primate. Mol. Ecol. 22, 4071-4086. ( 10.1111/mec.12349) [DOI] [PubMed] [Google Scholar]
- 34.Setchell JM, Charpentier MJE, Abbott KM, Wickings EJ, Knapp LA. 2009. Opposites attract: MHC-associated mate choice in a polygynous primate. J. Evol. Biol. 23, 136-148. ( 10.1111/j.1420-9101.2009.01880.x) [DOI] [PubMed] [Google Scholar]
- 35.Petersen RM, Bergey CM, Roos C, Higham JP. 2022. Relationship between genome-wide and MHC class I and II genetic diversity and complementarity in a nonhuman primate. Ecol. Evol. 12. ( 10.1002/ece3.9346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebastian A, Herdegen M, Migalska M, Radwan J. 2016. AMPLISAS: a web server for multilocus genotyping using next-generation amplicon sequencing data. Mol. Ecol. Resour. 16, 498-510. ( 10.1111/1755-0998.12453) [DOI] [PubMed] [Google Scholar]
- 37.Kalinowski S, Taper M, Marshall T. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099-1106. ( 10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 38.Nonacs P. 2000. Measuring and using skew in the study of social behavior and evolution. Am. Nat. 156, 577-579. ( 10.1086/316995) [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. 2022. R: a language and environment for statistical computing. See https://www.r-project.org/.
- 40.Queller D, Goodnight K. 1989. Estimating relatedness using genetic markers. Evolution 43, 258-275. ( 10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- 41.Pew J, Muir PH, Wang J, Frasier TR. 2015. related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 15, 557-561. ( 10.1111/1755-0998.12323) [DOI] [PubMed] [Google Scholar]
- 42.Printes RC, Strier KB. 1999. Behavioral correlates of dispersal in female muriquis (Brachyteles arachnoides). Int. J. Primatol. 20, 941-960. ( 10.1023/A:1020882719850) [DOI] [Google Scholar]
- 43.Strier KB, Ziegler TE. 2000. Lack of pubertal influences on female dispersal in muriqui monkeys, Brachyteles arachnoides. Anim. Behav. 59, 849-860. ( 10.1006/anbe.1999.1387) [DOI] [PubMed] [Google Scholar]
- 44.Di Fiore A. 2003. Molecular genetic approaches to the study of primate behavior, social organization, and reproduction. Yearbook of Physical Anthropology 46, 62-99. ( 10.1002/ajpa.10382) [DOI] [PubMed] [Google Scholar]
- 45.Di Fiore A. 2012. Genetic consequences of primate social organization. In The evolution of primate societies (eds Mitani JC, Call J, Kappeler PM, Palombit RA, Silk JB), pp. 269-292. Chicago, IL: University of Chicago Press. [Google Scholar]
- 46.Lawson Handley LJ, Perrin N. 2007. Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 16, 1559-1578. ( 10.1111/j.1365-294X.2006.03152.x) [DOI] [PubMed] [Google Scholar]
- 47.Langergraber KE, Mitani JC, Vigilant L. 2007. The limited impact of kinship on cooperation in wild chimpanzees. Proc. Natl Acad. Sci. USA 104, 7786-7790. ( 10.1073/pnas.0611449104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strier KB. 1994. Brotherhoods among atelins: kinship, affiliation, and competition. Behaviour 130, 151-167. ( 10.1163/156853994X00505) [DOI] [Google Scholar]
- 49.Peakall R, Smouse PE. 2012. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537-2539. ( 10.1093/bioinformatics/bts460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, et al. 2020. MHC-based mate choice in wild golden snub-nosed monkeys. Front. Genet. 11, article 609414. ( 10.3389/fgene.2020.609414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winternitz J, Abbate JL. 2015. Examining the evidence for major histocompatibility complex-dependent mate selection in humans and nonhuman primates. Res. Rep. Biol. 6, 73-88. ( 10.2147/RRB.S58514) [DOI] [Google Scholar]
- 52.Konovalov DA, Heg D. 2008. A maximum-likelihood relatedness estimator allowing for negative relatedness values. Mol. Ecol. Resour. 8, 256-263. ( 10.1111/j.1471-8286.2007.01940.x) [DOI] [PubMed] [Google Scholar]
- 53.Konovalov DA, Manning C, Henshaw MT. 2004. KINGROUP: A program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol. Ecol. Notes 4, 779-782. ( 10.1111/j.1471-8286.2004.00796.x) [DOI] [Google Scholar]
- 54.Gogarten JF, Koenig A. 2013. Reproductive seasonality is a poor predictor of receptive synchrony and male reproductive skew among nonhuman primates. Behav. Ecol. Sociobiol. 67, 123-134. ( 10.1007/s00265-012-1432-2) [DOI] [Google Scholar]
- 55.Bradley BJ, Robbins MM, Williamson EA, Steklis HD, Steklis NG, Eckhardt N, Boesch C, Vigilant L. 2005. Mountain gorilla tug-of-war: silverbacks have limited control over reproduction in multimale groups. Proc. Natl Acad. Sci USA 102, 9418-9423. ( 10.1073/pnas.0502019102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vigilant L, Roy J, Bradley BJ, Stoneking CJ, Robbins MM, Stoinski TS. 2015. Reproductive competition and inbreeding avoidance in a primate species with habitual female dispersal. Behav. Ecol. Sociobiol. 69, 1163-1172. ( 10.1007/s00265-015-1930-0) [DOI] [Google Scholar]
- 57.Rosenbaum S, Hirwa JP, Silk JB, Vigilant L, Stoinski TS. 2015. Male rank, not paternity, predicts male–immature relationships in mountain gorillas, Gorilla beringei beringei. Anim. Behav. 104, 13-24. ( 10.1016/j.anbehav.2015.02.025) [DOI] [Google Scholar]
- 58.Ostner J, Nunn CL, Schülke O. 2008. Female reproductive synchrony predicts skewed paternity across primates. Behav. Ecol. 19, 1150-1158. ( 10.1093/beheco/arn093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altmann J, et al. 1996. Behavior predicts genetic structure in a wild primate group. Proc. Natl Acad. Sci USA 93, 5797-5801. ( 10.1073/pnas.93.12.5797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muruthi P, Altmann J, Altmann S. 1991. Resource base, parity, and reproductive condition affect females' feeding time and nutrient intake within and between groups of a baboon population. Oecologia 87, 467-472. ( 10.1007/BF00320408) [DOI] [PubMed] [Google Scholar]
- 61.Muniz L, Perry S, Manson J, Gilkenson H, Groslouis J, Vigilant L. 2006. Father–daughter inbreeding avoidance in a wild primate population. Curr. Biol. 16, R156-R157. ( 10.1016/j.cub.2006.02.055) [DOI] [PubMed] [Google Scholar]
- 62.Muniz L, Perry S, Manson JH, Gilkenson H, Gros-Louis J, Vigilant L. 2010. Male dominance and reproductive success in wild white-faced capuchins (Cebus capucinus) at Lomas Barbudal, Costa Rica. Am. J. Primatol. 72, 1118-1130. ( 10.1002/ajp.20876) [DOI] [PubMed] [Google Scholar]
- 63.Wikberg EC, Jack KM, Fedigan LM, Campos FA, Yashima AS, Bergstrom ML, Hiwatashi T, Kawamura S. 2017. Inbreeding avoidance and female mate choice shape reproductive skew in capuchin monkeys (Cebus capucinus imitator). Mol. Ecol. 26, 653-667. ( 10.1111/mec.13898) [DOI] [PubMed] [Google Scholar]
- 64.Boesch C, Kohou G, Néné H, Vigilant L. 2006. Male competition and paternity in wild chimpanzees of the Taï forest. Am. J. Phys. Anthropol. 130, 103-115. ( 10.1002/ajpa.20341) [DOI] [PubMed] [Google Scholar]
- 65.Newton-Fisher NE, Thompson ME, Reynolds V, Boesch C, Vigilant L. 2010. Paternity and social rank in wild chimpanzees (Pan troglodytes) from the Budongo Forest, Uganda. Am. J. Phys. Anthropol. 142, 417-428. ( 10.1002/ajpa.21241) [DOI] [PubMed] [Google Scholar]
- 66.Wroblewski EE, Murray CM, Keele BF, Schumacher-Stankey JC, Hahn BH, Pusey AE. 2009. Male dominance rank and reproductive success in chimpanzees, Pan troglodytes schweinfurthii. Anim. Behav. 77, 873-885. ( 10.1016/j.anbehav.2008.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engelhardt A, Muniz L, Perwitasari-Farajallah D, Widdig A. 2017. Highly polymorphic microsatellite markers for the assessment of male reproductive skew and genetic variation in critically endangered crested macaques (Macaca nigra). International Journal of Primatology 38, 672-691. ( 10.1007/s10764-017-9973-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sukmak M, Wajjwalku W, Ostner J, Schülke O. 2014. Dominance rank, female reproductive synchrony, and male reproductive skew in wild Assamese macaques. Behav. Ecol. Sociobiol. 68, 1097-1108. ( 10.1007/s00265-014-1721-z) [DOI] [Google Scholar]
- 69.Minkner MMI, Young C, Amici F, McFarland R, Barrett L, Grobler JP, Henzi SP, Widdig A. 2018. Assessment of male reproductive skew via highly polymorphic STR markers in wild vervet monkeys, Chlorocebus pygerythrus. J. Hered. 109, 780-790. ( 10.1093/jhered/esy048) [DOI] [PubMed] [Google Scholar]
- 70.Gerloff U, Hartung B, Fruth B, Hohmann G, Tautz D. 1999. Intracommunity relationships, dispersal pattern, and paternity success in a wild living community of bonobos (Pan paniscus) determined from DNA analysis of faecal samples. Proc. R. Soc. B 266, 1189-1195. ( 10.1098/rspb.1999.0762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Surbeck M, Langergraber KE, Fruth B, Vigilant L, Hohmann G. 2017. Male reproductive skew is higher in bonobos than chimpanzees. Curr. Biol. 27, R640-R641. ( 10.1016/j.cub.2017.05.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishizuka S, Kawamoto Y, Sakamaki T, Tokuyama N, Toda K, Okamura H, Furuichi T. 2018. Paternity and kin structure among neighbouring groups in wild bonobos at Wamba. R. Soc. Open Sci. 5, 171006. ( 10.1098/rsos.171006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parga JA, Sauther ML, Cuozzo FP, Youssouf Jacky IA, Lawler RR, Sussman RW, Gould L, Pastorini J. 2016. Paternity in wild ring-tailed lemurs (Lemur catta): Implications for male mating strategies. Am. J. Primatol. 78, 1316-1325. ( 10.1002/ajp.22584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jennions MD, Fromhage L. 2017. Not all sex ratios are equal: the Fisher condition, parental care and sexual selection. Phil. Trans. R. Soc. B 372, 20160312. ( 10.1098/rstb.2016.0312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strier KB, Ives AR. 2012. Unexpected demography in the recovery of an endangered primate population. PLoS ONE 7, e44407. ( 10.1371/journal.pone.0044407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaves PB, Alvarenga CS, Possamai CD, Dias LG, Boubli JP, Strier KB, Mendes SL, Fagundes V. 2011. Genetic diversity and population history of a critically endangered primate, the northern muriqui (Brachyteles hypoxanthus). PLoS ONE 6, e20722. ( 10.1371/journal.pone.0020722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Possamai CB, Young RJ, Mendes SL, Strier KB. 2007. Socio-sexual behavior of female northern muriquis (Brachyteles hypoxanthus). Am. J. Primatol. 69, 766-776. ( 10.1002/ajp.20399) [DOI] [PubMed] [Google Scholar]
- 78.Strier KB. 2021. The limits of resilience. Primates 62, 861-868. ( 10.1007/s10329-021-00953-3) [DOI] [PubMed] [Google Scholar]
- 79.Strier KB, Dib LT, Figueira JEC. 2002. Social dynamics of male muriquis (Brachyteles arachnoides hypoxanthus). Behaviour 139, 315-342. ( 10.1163/156853902760102690) [DOI] [Google Scholar]
- 80.Furuichi T. 2011. Female contributions to the peaceful nature of bonobo society. Evol. Anthropol. 20, 131-142. ( 10.1002/evan.20308) [DOI] [PubMed] [Google Scholar]
- 81.Surbeck M, Mundry R, Hohmann G. 2011. Mothers matter! Maternal support, dominance status and mating success in male bonobos (Pan paniscus). Proc. R. Soc. B 278, 590-598. ( 10.1098/rspb.2010.1572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crockford C, Samuni L, Vigilant L, Wittig RM. 2020. Postweaning maternal care increases male chimpanzee reproductive success. Sci. Adv. 6, eaaz5746. ( 10.1126/sciadv.aaz5746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tolentino K, Roper JJ, Passos FC, Strier KB. 2008. Mother–offspring associations in northern muriquis, Brachyteles hypoxanthus. Am. J. Primatol. 70, 301-305. ( 10.1002/ajp.20488) [DOI] [PubMed] [Google Scholar]
- 84.Strier K, Ziegler T, Wittwer D. 1999. Seasonal and social correlates of fecal testosterone and cortisol levels in wild male muriquis (Brachyteles arachnoides). Horm. Behav. 35, 125-134. ( 10.1006/hbeh.1998.1505) [DOI] [PubMed] [Google Scholar]
- 85.Strier KB, Ziegler TE. 1994. Insights into ovarian function in wild muriqui monkeys (Brachyteles arachnoides). Am. J. Primatol. 32, 31-40. ( 10.1002/ajp.1350320104) [DOI] [PubMed] [Google Scholar]
- 86.Strier KB. 1992. Causes and consequences of nonaggression in the woolly spider monkey, or muriqui (Brachyteles arachnoides). In Aggression and peacefulness in humans and other primates (eds Silverberg J, Gray JP), pp. 100-116. New York: Oxford University Press. [Google Scholar]
- 87.Milton K. 1985. Mating patterns of woolly spider monkeys, Brachyteles arachnoides: implications for female choice. Behav. Ecol. Sociobiol. 17, 53-59. ( 10.1007/BF00299429) [DOI] [Google Scholar]
- 88.Possamai CB, Young RJ, de Oliveira RCR, Mendes SL, Strier KB. 2005. Age-related variation in copulations of male northern muriquis (Brachyteles hypoxanthus). Folia Primatol. 76, 33-36. ( 10.1159/000082453) [DOI] [PubMed] [Google Scholar]
- 89.Kotiaho JS, Simmons LW, Tomkins JL. 2001. Towards a resolution of the lek paradox. Nature 410, 684-686. ( 10.1038/35070557) [DOI] [PubMed] [Google Scholar]
- 90.Kotiaho JS, LeBas NR, Puurtinen M, Tomkins JL. 2008. On female choice, heterozygosity and the lek paradox. Anim. Behav. 75, e1-e3. ( 10.1016/j.anbehav.2007.08.011) [DOI] [Google Scholar]
- 91.Mays HL, Hill GE. 2004. Choosing mates: Good genes versus genes that are a good fit. Trends Ecol. Evol. 19, 554-559. ( 10.1016/j.tree.2004.07.018) [DOI] [PubMed] [Google Scholar]
- 92.Charpentier MJE, Boulet M, Drea CM. 2008. Smelling right: The scent of male lemurs advertises genetic quality and relatedness. Mol. Ecol. 17, 3225-3233. ( 10.1111/j.1365-294X.2008.03831.x) [DOI] [PubMed] [Google Scholar]
- 93.Hoover B, Nevitt G. 2016. Modeling the importance of sample size in relation to error in MHC-based mate-choice studies on natural populations. Integr. Comp. Biol. 56, 925-933. ( 10.1093/icb/icw105) [DOI] [PubMed] [Google Scholar]
- 94.Chaves PB, Strier KB, Di Fiore A. 2023. Paternity data reveal high MHC diversity among sires in a polygynandrous, egalitarian primate. Figshare. ( 10.6084/m9.figshare.c.6743143) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Chaves PB, Strier KB, Di Fiore A. 2023. Paternity data reveal high MHC diversity among sires in a polygynandrous, egalitarian primate. Figshare. ( 10.6084/m9.figshare.c.6743143) [DOI] [PMC free article] [PubMed]
Data Availability Statement
MHC DRB1 exon 2 allele sequences (171 bp) are provided in electronic supplementary material, table S5, and are deposited in GenBank (accession numbers OR340584–OR340605; GenBank sequences are 216 bp and include primer-binding regions). Additional datasets (e.g., multilocus genotypes) and information supporting this article have also been provided as part of electronic supplementary material [94]. R scripts and associated datasets for key analyses are available at https://github.com/difiore/muriqui-MHC.