Abstract
Acute kidney injury (AKI) is characterized by a rapid decrease in renal function with high mortality and risk of progression to chronic kidney disease (CKD). Ischemia and reperfusion injury (IRI) is one of the major causes of AKI. However, the cellular and molecular responses of the kidney to IRI are complex and not fully understood. Herein, we conducted unbiased proteomics and bioinformatics analyses in an IRI mouse model on days 3, 7, and 21, and validated the results using IRI, unilateral ureteral obstruction (UUO), and biopsies from patients with AKI or CKD. The results indicated an obvious temporal expression profile of differentially expressed proteins and highlighted impaired lipid metabolism during the progression of AKI to CKD. Acyl-coenzyme A oxidase 1 (Acox1), the first rate-limiting enzyme of peroxisomal fatty acid beta-oxidation, was then selected, and its disturbed expression in the two murine models validated the proteomic findings. Accordingly, Acox1 expression was significantly downregulated in renal biopsies from patients with AKI or CKD, and its expression was negatively correlated with kidney injury score. Furthermore, in contrast to the decreased Acox1 expression, lipid droplet accumulation was remarkably increased in these renal tissues, suggesting dysregulation of fatty acid oxidation. In conclusion, our results suggest that defective peroxisomal fatty acid oxidation might be a common pathological feature in the transition from AKI to CKD, and that Acox1 is a promising intervention target for kidney injury and repair.
Keywords: Acute kidney injury, Maladaptive repair, Proteome, Fatty acid oxidation, Acox1
Highlights
-
•
Proteomics and bioinformatics analyses reveled that an obvious temporal expression profile of DEPs in IRI mouse models.
-
•
Defective peroxisomal fatty acid oxidation might be a common pathological feature in the transition from AKI to CKD.
-
•
Acox1 is a promising intervention target for kidney injury and repair.
1. Introduction
Acute kidney injury (AKI) is characterized by an abrupt decline in renal function [1], and the in-hospital mortality rate ranges from 50 to 70% depending on comorbidities and specific clinical conditions [2,3]. In addition to its high short-term mortality, AKI is substantially associated with the progression of chronic kidney disease (CKD), with a 9-fold increased risk of progressive CKD compared to patients without AKI [4,5]. Ischemia-reperfusion injury (IRI) is a leading risk factor for AKI. However, the mechanistic features and effective therapeutic strategies for IRI-AKI and its transition to CKD have not been elucidated [6]. Therefore, delineating the pathogenesis of AKI to CKD is required.
Several essential mechanisms have been proposed, especially proximal tubular epithelium maladaptive repair [7,8]. Proximal tubular epithelium cell (RTEC), the major component of kidney, is extremely sensitive to multiple stresses including hypoxia, toxins, and sepsis [9]. Increasing studies have suggested that proximal tubular epithelium plays a vital role in kidney injury and repair processes [7]. Indeed, surviving injured proximal tubular cells with robust dedifferentiation and proliferation capacity contribute to rebuilding damaged epithelial structures and restoring renal function [10]. However, some severe or persistent AKI events can frequently lead to maladaptive or incomplete repair of tubular epithelial cells, ultimately resulting in renal fibrosis and inexorable transition of AKI to CKD [11,12]. Additionally, previous research has suggested that lipid metabolism disorders mediate the pathogenesis of AKI, abnormal kidney repair, and CKD [13]. Calorie restriction or fatty acid binding protein 4 deficiency decreases lipid droplet accumulation, attenuates kidney injury, promotes kidney repair, and restores renal function [13,14]. Peroxisome proliferator-activated receptors (PPARs), the upstream of fatty acid oxidation, might be a promising druggable target to attenuate renal fibrosis via restoring fatty acid oxidation capacity [15,16].
Omics technologies, including genomics, transcriptomics, metabolomics, and proteomics, have remarkably advanced the molecular insight into the understanding of AKI, which might open new avenues for biomarker discovery or identification of drug targets [[17], [18], [19]]. In addition to other omics, proteomics can provide much more information regarding protein production, modification, degradation, and interactions between proteins or other biomolecules [17]. Furthermore, proteomic data are considered more closely related to pathological symptoms and clinical parameters of patients [17]. However, changes in proteins during renal IRI and repair remain largely unclear. In this regard, we conducted an unbiased proteomic study and bioinformatic analysis using a severe bilateral IRI mouse model on days 3, 7, and 21. As a result, we illustrated an obvious temporal expression profile of differentially expressed proteins (DEPs) and highlighted the dysregulated fatty acid oxidation in the progression of AKI to CKD. Proteomic findings were also validated using IRI, unilateral ureteral obstruction (UUO), and renal tissues from patients with AKI or CKD. Together, this study provides new insights into the pathophysiology of AKI progression and may advance therapeutic strategies for kidney injury and repair.
2. Materials and methods
2.1. Mouse ischemic AKI models
All experimental protocols were approved by the ethics committee of the Army Medical University. Male C57BL/6J mice aged 8 weeks and weighing 20–22 g were subjected to bilateral renal ischemia reperfusion injury (IRI) as previously described [20]. In brief, ischemia was induced by the retroperitoneal approach in both kidneys for 35 min at 37 °C (severe IRI). Control-operated mice (control group) underwent the same procedure, but without renal pedicle clamping at the same time (n = 6 per group).
2.2. UUO models
Eight-week-old male C57BL/6J mice were anesthetized and the left proximal ureter was separated and sutured. Next, the ureter was cut between the ligatures. With the exemption of ureter ligation, the control group underwent the same surgery as the model group (n = 6 per group).
2.3. Patients
A total of 52 patients with AKI between January 2018 and December 2020 in Daping Hospital (Chongqing, China) were enrolled in the study. The inclusion criteria were a biopsy-proven AKI (defined by standard morphological criteria), as previous study described [21]. Patients aged < 18 years, with chronic kidney disease (CKD), and hyperlipidemia who were taking lipid-lowering drugs were excluded. Twenty-six control human kidney samples were obtained from the unaffected portion of tumor nephrectomies. The study protocol was approved by the Ethical and Protocol Review Committee of the Army Military Medical University, and written informed consent was obtained from each patient.
2.4. Blood examination
Serum creatinine and urea nitrogen (BUN) levels were measured using an automatic biochemistry analyzer (Unicel DXC 800; Beckman Coulter, Skykesville, MD, USA).
2.5. Histological staining
Kidney sections (4 μm) were stained with periodic acid–Schiff (PAS) and Masson’s trichrome and analyzed using an Olympus microscope (Tokyo, Japan). Kidney damage scores included acute and chronic kidney injury. The degree of acute kidney injury was scored semi-quantitatively on a 0 to 5 scale, according to the percentage of tubular necrosis, loss of brush border, and tubular dilation (0 = no lesion, 1 ≤ 25%, 2 ≥ 25%–50%, 3 ≥ 50%–75%, 4 ≥ 75% to < 100%, 5 = 100%) [22]. Chronic kidney injury was scored semi-quantitatively on a 0 to 4 scale, based on tubular atrophy and interstitial fibrosis (0 = no lesion; 1 ≤ 25%; 2 = 25%–50%; 3 = 50%–75%; 4 ≥ 75%) according to previous studies [23].
2.6. Proteomics analysis
Renal cortex tissue from each sample (three biological replicates) was lysed (lysis buffer: 8 mol/L urea) and digested with trypsin. The ration of protein and trypsin was 50:1 for the first-time digestion, and 100:1 for another time. The protein concentration of each tissue lysate was measured using a BCA protein assay kit (Beyotime, Beijing, China). Subsequently, 1000 μg of protein from each sample was digested with trypsin. The tryptic peptides were fractionated into fractions by high pH reverse-phase HPLC using a Thermo Betasil C18 column (5 μm particles, 10 mm ID, 250 mm length), dissolved in solvent A (0.1% formic acid, 2% acetonitrile/in water), and then loaded onto a home-made reversed-phase analytical column (25 cm, 100 μm i.d.). The gradient comprised an increase from 6% to 24% solvent B (0.1% formic acid in 90% acetonitrile) over 70 min, 24%–35% over 14 min, and climbing to 80% over 3 min, then holding at 80% for the last 3 min, all at a constant flow rate of 450 nL/min on a nanoElute UHPLC system (Bruker Daltonics). The peptides were subjected to capillary source followed by timsTOF Pro (Bruker Daltonics) mass spectrometry coupled online to UPLC. The precursors and fragments were analyzed using a TOF detector. The applied electrospray voltage was 1.6 kV. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states of 0–5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set at 30 s. Raw MS/MS data were processed using the MaxQuant search engine (v.1.6.6.0). Tandem mass spectra were searched against (mouse SwissProt) (17,045 entries) database concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 2 missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in First search and 20 ppm in Main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification and acetylation on protein N-terminal and oxidation on Met were specified as variable modifications. The FDR was adjusted to <1%, and the minimum score for modified peptides was set at > 40.
2.7. Statistical and bioinformatic analysis of the proteomic data
The label free quantification (LFQ) intensities from the “Protein Groups” txt file were extracted and the proteins with “Reverse” and “Contamination” were filtered. To exclude the bias of different replicates and samples, the relative protein abundance was normalized by the mean abundance across samples (Table S1). Data are presented as mean ± standard deviation (SD) for normally distributed variables and as median (interquartile range) for non-normally distributed variables. Differences between the two groups were determined using a two-tailed Student’s t-test. For the three groups, statistical differences were analyzed using ANOVA followed by Bonferroni and Kruskal-Wallis tests for normally and non-parametrically distributed variables, respectively. Statistical significance was set at p < 0.05. Upregulated or downregulated proteins (fold change ≧1.5 or ≦(1/1.5) 0.667, respectively; t-test p < 0.05) were defined as differentially expressed proteins (DEPs). The proteins were functionally annotated according to their biological processes, molecular functions, and cellular components derived from the UniProt-GOA database. A graph theoretical clustering algorithm, molecular complex detection (MCODE), was used to analyze densely connected regions. Principal component analysis (PCA) and hierarchical clustering were performed using R software (v.3.0.1). Co-expression clusters were identified using the WGCNA R package [24].
2.8. Western blot analyses
Western blotting analysis was performed on renal lysates with RIPA solution containing protease inhibitors (Beyotime Biotechnology, Beijing, China). Protein samples (30 μg) were fractionated by 10–12% SDS-PAGE gels and then transferred to polyvinylidene difluoride membranes (PVDFs, Millipore, Billerica, MA, USA). Following blocking with 5% non-fat milk, these PVDFs were incubated with the following primary antibodies: Acox1 (1:1000, 10957-1-AP; Proteintech group, Wuhan, China), Crot (1:500, ab175450; Abcam, Cambridge, UK), Amacr (1:500, ab268062; Abcam), and anti-GAPDH (1:5000, BM3876; Boster Biotechnology, Wuhan, China) and anti-GAPDH (1:5000, 10494-1-AP; Proteintech group, Wuhan, China). The intensity of each band was analyzed using the Quantity One software (Bio-Rad, Hercules, CA, USA).
2.9. Immunohistochemical and immunofluorescent staining
All the specimens were deparaffinized and rehydrated. Following antigen retrieval, the tissues were incubated with primary anti-Acox1 (1:100, 10957-1-AP; Proteintech group) and α-SMA (1:200, BM0002; Boster Biotechnology) antibodies at 4 °C overnight. At least ten fields were randomly selected to evaluate the percentage of Acox1-positive and α-SMA-positive RTECs. For immunofluorescence, all sections were incubated with anti-Acox1 antibody (1:100, 10957-1-AP; Proteintech group) followed by Alexa-488 conjugated goat anti-rabbit antibody (1:200, ab150077; Abcam), anti-α-SMA antibody (1:200, BM0002; Boster Biotechnology), and anti-collagen I (1:100, ab6308; Abcam) followed by Alexa-555 conjugated goat anti-mouse antibody (1:200, ab150118; Abcam) at 4 °C overnight. The cells were then co-stained with DAPI (1:500, C1006; Beyotime Biotechnology). All samples were analyzed using a fluorescence microscope (Leica, Germany). At least six fields were randomly selected to evaluate the percentage of positive staining with Image J software (version 1.37; National Institutes of Health, Bethesda, MD).
2.10. Oil Red O staining
Lipid accumulation was evaluated by Oil Red O staining. Briefly, samples were fixed with 4% paraformaldehyde and stained with Oil Red O for 30 min. Subsequently, the samples were counterstained with hematoxylin for 5 min. The staining was examined using a light microscope (Leica). The percentages of oil-red positive area were quantified using Image J software (version 1.37).
2.11. Statistical analysis
Data are presented as mean ± standard deviation (SD) for normally distributed variables and as median (interquartile range) for non-normally distributed variables. Differences between the two groups were determined using a two-tailed Student’s t-test. For the three groups, statistical differences were analyzed using ANOVA followed by Bonferroni and Kruskal-Wallis tests for normally and non-parametrically distributed variables, respectively. Statistical significance was set at p < 0.05.
3. Results
3.1. Histological changes during the progression of AKI induced by IRI
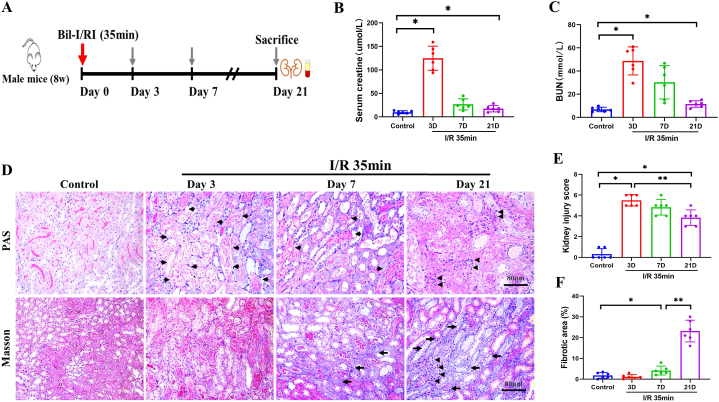
An intact contralateral kidney can compensate for impaired renal function and the kidney owns a powerful reserved function, so we used a bilateral ischemia perfusion injury model (35 min blocking of the renal pedicle) to investigate alterations in protein expression during the initiation and progression of ischemic AKI, as previously described and summarized in Fig. 1A [[25], [26], [27]]. First, we generated a mouse model of I/R-AKI compared to the control group, the ischemic groups, mostly notable on day 3, exhibited significantly increased serum BUN and creatinine levels, confirming the presence of AKI. By 7 and 21 days after I/R, the impaired renal function gradually recovered but did not completely return to baseline, suggesting the persistence of injury (Fig. 1B and C). Further histopathological examinations showed remarkable necrosis of renal tubular epithelial cells and a massive loss of brush borders on day 3 (3D) (Fig. 1D and E). On 7D, these changes seemed to be ameliorated, but inflammatory cell infiltration and tubular atrophy clearly started to appear and increased on day 21, indicating maladaptive repair in progression (Fig. 1D and E). Masson staining showed time-dependent collagen deposition, confirming the progression of renal fibrosis (Fig. 1D, F). Collectively, these results clearly indicate that irreversible kidney injury, maladaptive tubular repair, and advanced interstitial fibrosis contribute to the transition from AKI to CKD following an ischemic insult.
Fig. 1.
Progression of AKI induced by IRI. (A) Schematic diagram of ischemic treatment and sample collection. (B–C) Serum creatinine and BUN levels were determined at different time points (n = 6). (D) PAS (red, glycogen) and Masson staining (blue, collagen) for histological changes after IRI. Long arrows indicate fibrotic area, short arrows represent tubular injury, while arrowheads show the infiltrated inflammatory cells. Scale bar, 80 μm. (E–F) Kidney injury score and fibrotic area were quantified (n = 6). Data are expressed as the means ± SDs for each group. Differences between the two groups were assessed with a two-tailed Student’s t-test. *P < 0.05 versus control; **P < 0.05 versus 3D/7D. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Kidney proteomic remodeling during the progression of AKI induced by IRI
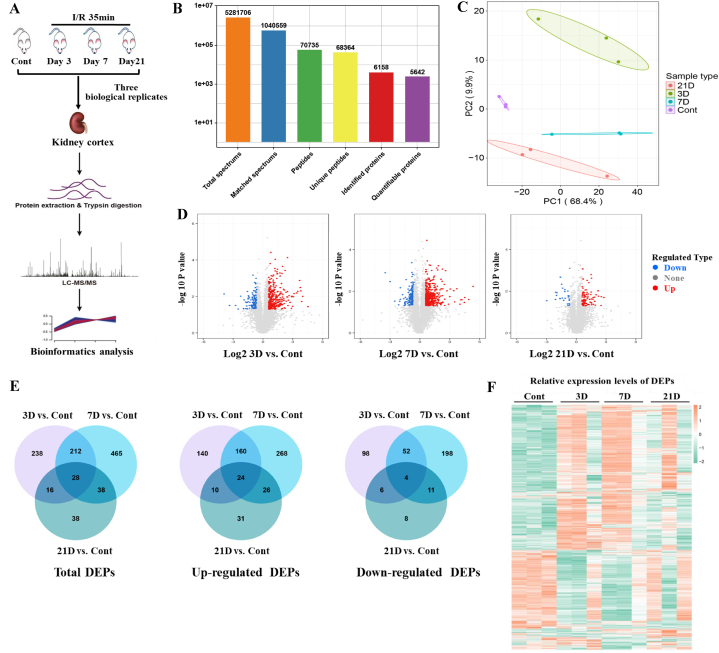
To monitor the underlying longitudinal changes and possible mechanisms that determine the pathogenesis and progression of AKI to CKD, we performed proteomic analysis using LC-MS/MS with a label-free quantitative approach at the indicated time points (Fig. 2A). In total, 2,642,078 MS spectra were detected, and 6158 proteins were quantified from the global kidney proteome, with an estimated FDR of 1% (Fig. 2B and Table S1). As shown in Fig. 2C, principal component analysis (PCA) revealed that proteomics was clearly segregated among the different groups. In contrast to the considerable difference in proteomics on day 3, the proteomics on day 7 seemed to reshape back to normal controls, whereas it still did not completely shift to baseline on day 21, suggesting persistent changes in kidney proteomics. The PCA shows an outlier in each of the AKI groups (3 d, 7 d, and 21 d), which might to be related with the heterogeneity of mice response to IRI surgery. To investigate stage-specific protein expression and molecular characteristics under these conditions, we conducted pairwise comparisons to identify significant different expression proteins (DEPs). Volcano plots, with a log 2 (1.5) FC cutoff value of 0.585 for the differential expressed criteria, provided an overview of the DEPs (Fig. 2D and Fig. S1C). Compared with the hundreds of DEPs in 3/7/21D vs. Cont, the number of DEPs was smaller in 7D vs. 3D, 21D vs. 3D, and 21D vs. 7D, indicating modest changes among these IRI groups (Fig. S1A). Similarly, the common changes in DEPs in 3/7/21D versus Cont. were 28, with 24 upregulated and 4 downregulated, which was more than the DEPs from the ischemic groups (Fig. 2E and Fig. S1B). Finally, heatmaps revealed changes in DEPs at different time points (Fig. 2F). Consistent with the PCA results, some of the remarkably changed proteins on days 3 and 7 seemed to recover on day 21 (Fig. 2F). Collectively, these results revealed that some of the DEPs could recover over time after IRI, but the sustained changes in the expression of certain proteins might contribute to renal fibrosis, suggesting that the proteomes displayed remodeling during the progression of IRI injury.
Fig. 2.
Protein identification in kidney cortex samples from IRI mouse using LC-MS/MS. (A) Experimental scheme. Proteomic sample preparation, mass spectrometry analysis and bioinformatics analysis. (B) Spectrums, peptides and proteins were screened. (C) Principal-component analysis (PCA) of all identified proteins. (D) Volcano plots shows the upregulated and downregulated differentially expressed proteins (DEPs) in different groups. (E) Venn diagram shows the number of DEPs in each group. (F) Heat-map analysis of DEPs in all samples.
3.3. Temporal expression profiling of differentially expressed proteins and function analysis
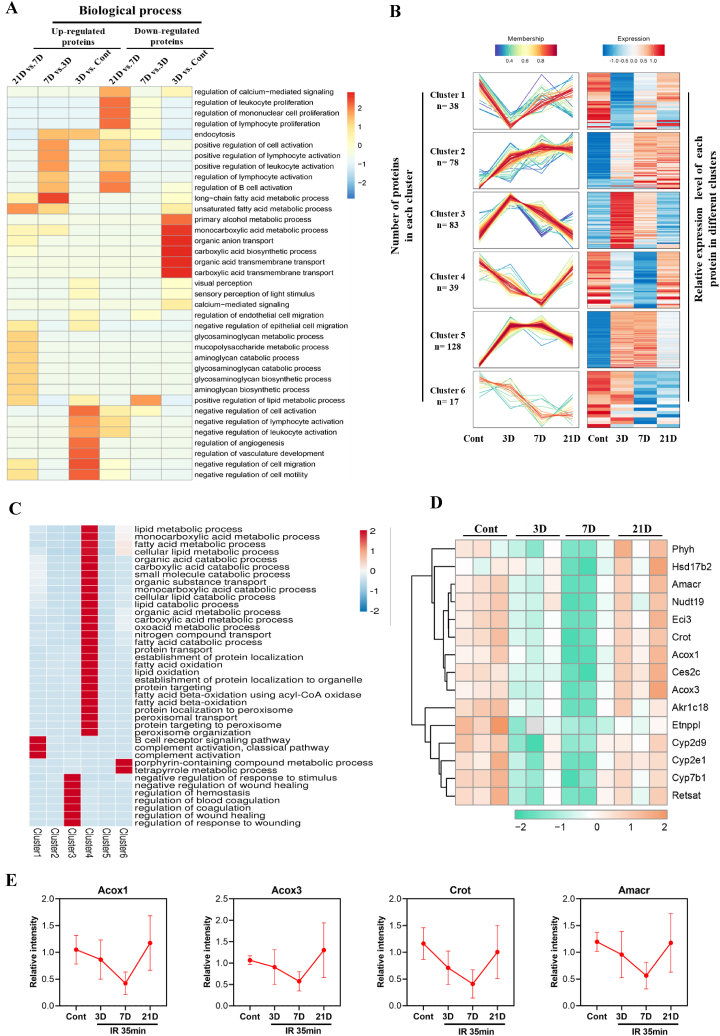
We performed gene ontology (GO) enrichment analysis to gain more insight into DEPs during the progression of IRI injury and repair. GO analysis is divided into three domains: biological processes, molecular functions, and cellular components. As shown in Fig. 3A, the biological processes of upregulated DEPs were enriched in several pathways, including calcium-mediated signaling, endocytosis, immune cell activation and proliferation, cell migration and motility, and angiogenesis, which are associated with inflammation, cell injury and repair, and fibrosis (Fig. 3A) [25]. Similar results were found regarding the molecular function and cellular component analyses (Fig. S2). On the other hand, GO analysis of downregulated DEPs showed enrichment of pathways in the majority of cellular metabolic processes, including fatty acid metabolic process, organic anion and acid transport, and carboxylic acid biosynthetic process (Fig. 3A and S2). All DEPs were further clustered and sorted according to their abundance patterns to investigate molecular changes. They were allotted into six groups by hierarchical clustering analysis. Consistent with the results of the PCA and heatmaps in Fig. 2, sequential variations were found in these clusters, suggesting stage-specific proteome profiles and pathological conditions (Fig. 3B). Additionally, an ingenuity pathway analysis was performed to classify the DEPs based on their functions. In accordance with the GO findings, clusters 1 and 3 revealed variability in complement activation, coagulation, and wound healing in the acute phase, which gradually returned to baseline on day 21 (Fig. 3B). Interestingly, results from clusters 4 and 6 highlighted the dysregulation of cellular metabolic processes, especially fatty acid and lipid metabolism in cluster 4 (Fig. 3C). Evidence from the GO and ingenuity pathway analyses clearly revealed the dysfunctions of lipid metabolism in IRI-AKI progression, which is in agreement with published studies [25,28]. We then analyzed the lipid metabolism-related DEPs in cluster 4 and identified 15 proteins for further study. The heat map showed a remarkable decrease in the levels of certain proteins on day 3, which were further down-regulated on day7, whereas a trend of recovery was clearly observed on day 21 (Fig. 3D). Consistently, four important enzymes of fatty acid oxidation, Acox1, Acox3, Amcar, and Crot, were significantly downregulated on days 3 and 7 but recovered closely to the normal levels on day 21 (Fig. 3E). Together, the temporal expression profiling of DEPs and functional analysis indicated that lipid metabolism, especially fatty acid oxidation, might contribute to the progression of IRI-AKI.
Fig. 3.
Temporal expression profiling of DEPs and functional analysis. (A) Gene Ontology (GO)-biological process analysis of upregulated and downregulated DEPs in different groups (B) Six co-expression clusters of DEPs. Each line indicates the relative abundance of each protein and is color-coded by the cluster membership. (C) Biological process analysis of DEPs from cluster 4. (D) Heat-map analysis of lipid metabolism related proteins from cluster 4. (E) The relative expression (LFQ intensity) of Acox1, Acox3, Crot and Amacr. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Validation of the proteins involved in fatty acid oxidation in IRI mice
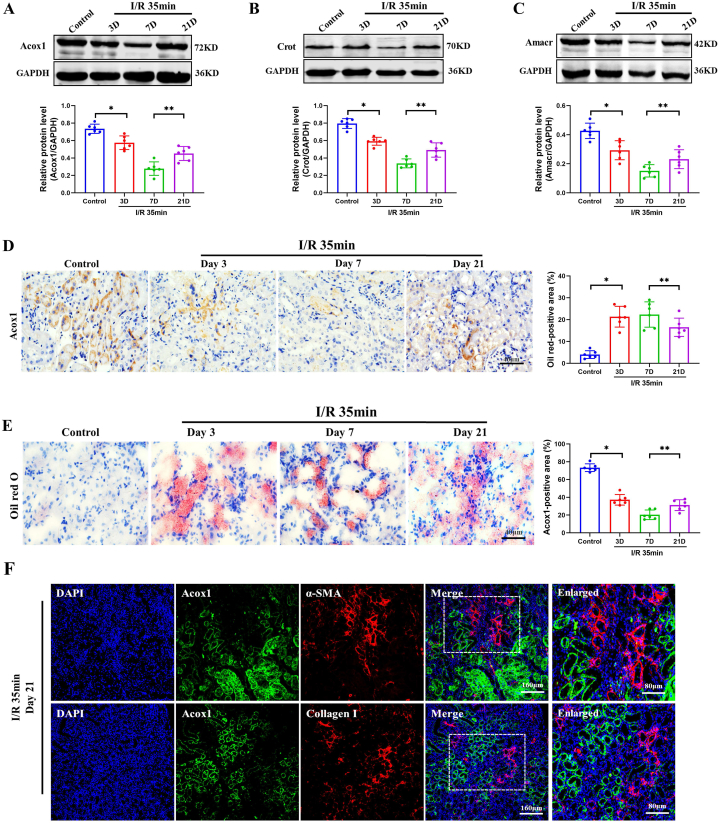
To confirm the above proteomic results, we examined the expression of Acyl-coenzyme A oxidase 1 (Acox1), the first rate-limiting enzyme of peroxisomal fatty acid beta-oxidation, using western blotting and immunohistochemical (IHC) staining. In agreement with the proteomic findings, it was shown that the significantly decreased Acox1 expression on days 3 and 7 was slightly reversed on day 21 after IRI injury (Fig. 4A). Similar results were observed for Crot (Fig. 4B) and Amcar (Fig. 4C) expression. As shown in Fig. 4D, Acox1 was mainly expressed in the tubular cells. Moreover, Acox1 staining was remarkably decreased on day 3 and steadily decreased on day 7, but was slightly elevated on day 21, suggesting recovery at a later stage. Fatty acid oxidation dysfunction leads to lipid droplets accumulation [7,13,29]. Furthermore, we investigated the accumulation of lipid droplets after IRI injury using Oil Red O staining. In contrast to the dramatic accumulation in renal epithelial tubular cells on days 3 and 7, the lipid droplets clearly decreased on day 21 but remained at a higher level than that in the controls (Fig. 4E). Aiming to explore the relationship of peroxisomal fatty acid beta-oxidation dysfunction and renal fibrosis, we co-stained Acox1 with fibrotic markers α-SMA and collagen I. As shown in Fig. 4F, Acox1 did not colocalize with these fibrotic markers, suggesting that dysregulated peroxisomal fatty acid beta-oxidation with downregulated Acox1 in fibrotic regions. Collectively, these results suggest that dysregulated lipid metabolism, especially Acox1-related peroxisomal fatty acid oxidation, is closely associated with IRI-AKI progression and recovery.
Fig. 4.
Validation of the proteins involved in peroxisomal fatty acid oxidation in IRI mice. (A–C) Western blotting was used to determine the expression of Acox1, Crot and Amacr in renal cortex during AKI progression, and their levels are presented relative to those of GAPDH (n = 6). Data are expressed as the means ± SDs for each group. Differences between the two groups were assessed with a two-tailed Student’s t-test. *P < 0.05 versus control. **P < 0.05 versus 7 d. (D) Representative immunostaining micrographs and semi-quantified positive area of Acox1 in IRI mice. (E) Oil red O staining and its semi-quantification of the kidneys after IRI. Scale bar, 40 μm. (F) Colocalization of Acox1 and fibrotic markers (α-SMA and collagen I). Scale bar, 160 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Validation of the proteins involved in peroxisomal fatty acid oxidation in UUO
We further validated the critical role of dysregulated peroxisomal fatty acid oxidation in another model of AKI to CKD using unilateral ureteral obstruction (UUO). Consistent with published data, renal interstitial fibrosis, detected by Masson staining, was slightly increased on day 7 in UUO mice, but was remarkably augmented on days 14 and 21 (Fig. 5A) [30]. Accordingly, relative quantification of Masson’s staining showed a sustained increase compared to the control groups (Fig. 5B). Similarly, the α-SMA-positive area was significantly increased and gradually peaked on day 21 after ureteral obstruction, as shown in Fig. 5A and C. Moreover, western blotting showed that Acox1 was significantly downregulated on day 7, and was further decreased on days 14 and 21, compared with the controls (Fig. 5D–E). Similar results were obtained from IHC staining for Acox1 (Fig. 5F–G). In contrast to the persistently decreased Acox1 expression, lipid droplet accumulation in the renal tubular epithelial areas increased significantly over time (Fig. 5F, H). Overall, these results suggested that dysregulated peroxisomal fatty acid oxidation was a common pathological feature of UUO-induced CKD.
Fig. 5.
Validation of the proteins involved in peroxisomal fatty acid oxidation in the UUO model. (A) Masson staining for histological changes and IHC staining for α-SMA of kidney sections (n = 6). (B–C) Fibrotic and α-SMA positive areas were quantified (n = 6). (D–E) Western blotting was used to determine the expression of Acox1 in UUO, and their levels are expressed relative to those of GAPDH. (F–H) Representative micrographs show renal Acox1 expression and Oil red O staining in UUO mice, and the percentage of positive area was quantified. Scale bar, 40 μm. Data are expressed as the means ± SDs for each group. Differences between the two groups were assessed with Student’s t-test. *P < 0.05 versus control; **P < 0.05 versus 14 d. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
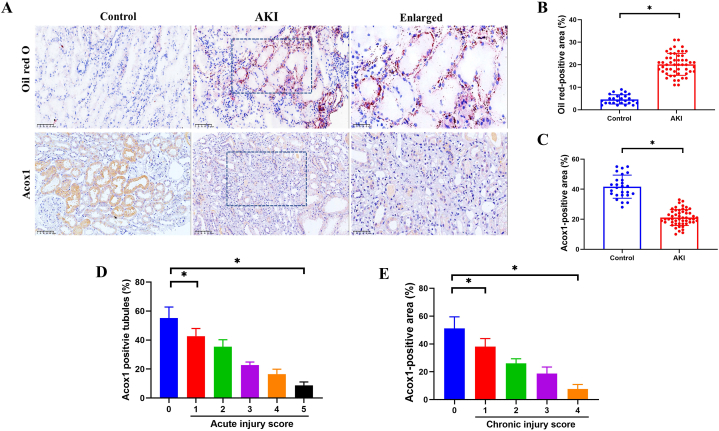
3.6. Validation of the disordered peroxisomal fatty acid oxidation in patients with AKI or CKD
Finally, to validate the findings from the proteomic analysis and murine models, we collected renal biopsies from patients with AKI and performed Oil Red O or IHC staining. The clinical characteristics of the patients (n = 52) and healthy controls (n = 26) are presented in Table S2. In accordance with the murine findings, Oil Red O staining positive areas were markedly increased in renal biopsies from AKI compared with those from controls (Fig. 6A, C). In contrast, Acox1 expression was significantly downregulated (Fig. 6A–B). Furthermore, it was shown that both severe tubular injury with higher acute (4–5) and chronic (3–4) injury scores correlated with lower expression of Acox1, suggesting a negative relationship between renal injury and Acox1 expression (Fig. 6D–E). Similar results were observed in renal tissues of patients with CKD (Fig. S3). Taken together, the results obtained using renal biopsies from patients with AKI or CKD corresponded to the findings in murine models, uncovering the crucial effects of dysregulated peroxisomal fatty acid oxidation in orchestrating AKI progression and indicating the possibility of Acox1 as an intervention target for kidney diseases.
Fig. 6.
Expression of Acox1 and Oil red O staining of the kidneys in patients with AKI. (A–C) Representative micrographs show Oil red O staining and renal Acox1 expression in healthy controls and patients with AKI, and the percentage of positive area was quantified. (D–E) The positive percentage of Acox1 was quantified according to acute and chronic injury scores. Data are expressed as the means ± SDs for each group. Differences between the two groups were assessed with a two-tailed Student’s t-test. For the three groups, statistical differences were analyzed using ANOVA followed by Bonferroni and Kruskal-Wallis tests for normally and non-parametrically distributed variables, respectively. *P < 0.05 versus 0 group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we investigated the possible mechanisms underlying the transition from AKI to CKD using a bilateral IRI mouse model and an unbiased proteomic study. The results revealed that some of the DEPs recovered over time, but the sustained DEPs were associated to renal fibrosis, suggesting that the proteomes displayed remodeling during the progression of ischemic renal injury and repair. Moreover, temporal expression profiling of DEPs and functional analysis indicated that lipid metabolism, especially dysregulated fatty acid oxidation, might contribute to the progression of IRI-AKI. These findings were further confirmed in UUO-induced CKD and renal biopsies from patients with AKI or CKD, indicating that defective peroxisomal fatty acid oxidation may be a common mechanism in the progression of AKI to CKD.
The kidney has a powerful reserved function, and an intact contralateral kidney can compensate for impaired renal function [27]. Therefore, we used a bilateral ischemia perfusion injury model (35 min blocking of the renal pedicle), which has been proved to be a robust model for acute and chronic kidney injury [26,31]. Consistent with published evidence, we found that IRI-AKI was successfully established with significantly increased BUN and Cre levels and obvious histological changes on day 3 [31]. Moreover, severe IRI-AKI led to the persistence of fibrotic lesions, as evidenced by impaired renal function, increased inflammatory cell infiltration, augmented tubular atrophy, and accumulated collagen deposition on day 7, which was further enhanced on day 21. To comprehensively investigate the proteomic profile during the progression of IRI, we collected renal samples at the early (day 3), middle (day 7), and late (day 21) stages after IRI surgery. To the best of our knowledge, this study is the first to report longitudinal proteomic changes in ischemic kidney injury and repair. Combined with a few proteomic studies, mainly focused on the acute phase of kidney injury, our research could further advance our understanding of the possible underlying mechanism of kidney injury and repair [27,32,33].
Of note, we first showed that the proteomics were clearly segregated among the different time points in this study, indicating the substantially different pathologies over time, which is consistent with published studies, which performed transcriptional analysis of proximal tubular epithelial cells [25]. To uncover stage-specific molecular characteristics, we analyzed the DEPs by pair-wise comparison and GO clarification. The upregulated DEPs were enriched in several pathways associated with inflammation, cell injury and repair, and fibrosis, while the downregulated DEPs showed enrichment in pathways of fatty acid metabolic processes, organic anion and acid transport, and carboxylic acid biosynthetic process, suggesting a dysregulated cellular metabolic condition. Similar results were observed in the Venn diagram (Fig. 2E). These results were consistent with those of previous studies that examined the overall transcriptome patterns related to innate and adaptive immunity, tubular injury/repair, and fibrotic changes [25]. In contrast to elevated fatty acid signaling and increased levels of glycolysis, lipids, and fatty acids at 4 h and 24 h after reperfusion in a proteo-metabolomics study, our results and previous evidence highlight sustained metabolism dysregulation during the progression of AKI to chronic fibrosis [27]. The specific stage of pathophysiology and tubular epithelium response to injuries may account for this discrepancy, which requires further research in the future. We further allotted DEPs into six groups by hierarchical clustering and performed an ingenuity pathway analysis. Sequential variations were consistently found in these clusters, conforming to the stage-specific proteome profiles. Interestingly, results from cluster 4 highlighted the dysregulation of lipid metabolism and fatty acid oxidation (Fig. 3C) during the progression of IRI, which is corroborated by previous studies [13,14,25].
Acox1, the first rate-limiting enzyme of the peroxisomal fatty acid beta-oxidation pathway mediating fatty acid and lipid metabolism, was selected for further study. Previous studies have reported that Acox1 is correlated with multiple disorders, including axon degeneration, severe developmental delay, epilepsy, hepatocarcinogenesis, and adipogenesis of bovine pre-adipocytes [[34], [35], [36]]. In this proteomic study, we found that Acox1 expression was significantly downregulated on days 3 and 7, but gradually restored on day 21, which was also validated by western blotting and IHC in ischemic renal tissues. We further explored whether the disrupted Acox1expression was found in another model of UUO-induced AKI. In accordance with the results at the early and middle stages of repair in IRI, Acox1 expression was clearly decreased on days 7 and 14 following UUO. However, despite the trend of recovery on day 21 in IRI, Acox1 expression was steadily downregulated, suggesting a sustained injury in the UUO model. In accordance with the above murine findings, Acox1 expression was significantly downregulated in renal biopsies from AKI patients, suggesting that Acox1 expression might be negatively related to renal injury. Similar results were observed in patients with CKD. In contrast to the decreased Acox1 expression, lipid droplet accumulation increased significantly in both murine tissues and patient biopsies, which is consistent with previous studies [13,14]. Taken together, these results suggest that dysregulated peroxisomal fatty acid oxidation may be a common pathological feature in the transition from AKI to CKD.
This study has some limitations. First, we performed proteomic analysis to study the possible underlying mechanisms involved in the process of ischemic AKI transitions, but we did not perform transcriptomic or metabolomic studies. However, metabolism-related results were validated in IRI and UUO murine models and in patients with AKI or CKD. Second, proteomic profiling of the very early phase at 4 or 24 h and the terminal stage at 6 or 12 months in ischemic injured kidneys was not included, which needs to be further studied. Even so, this study focused on the mechanism of the transition from AKI to CKD, and the current stage of murine models could nearly represent the entire pathophysiological process. Finally, we did not investigate the mechanism through which abnormal fatty acid oxidation is involved in the progression of AKI to CKD. Activating PPARs, the upstream of fatty acid oxidation proteins, attenuates lipid accumulation and tubulointerstitial fibrosis in the uIRI model [37,38]. Moreover, dysregulated fatty acid oxidation is proved to be related with tubular cell injury (ATP depletion, cell death, dedifferentiation and intracellular lipid deposition), and loss of kidney function through lipotoxicity and oxidative stress in both AKI acute setting and its latter stage [29,39]. Together, PPARs and fatty acid oxidation triggered tubular cell injury might contribute to the progression of AKI to CKD in this study, which needs further studied in the future.
In conclusion, we provided an in-depth proteomic characterization of ischemic kidney injury and repair. Our results suggest that dysregulated lipid metabolism, especially peroxisomal fatty acid oxidation, might be a common feature in the progression of AKI to CKD, and that Acox1 is a promising intervention target for kidney injury and repair. These findings contribute to our understanding of AKI progression and provide potential targets for treatment.
5. New and noteworthy
Ischemia and reperfusion injury is one of the major causes of AKI. However, its cellular and molecular responses are not fully understood. This study provided an in-depth proteomic characterization of ischemic kidney injury and repair. Our results suggested that defective peroxisomal fatty acid oxidation might be a common pathological feature in the transition from AKI to CKD, and that Acox1 is a promising intervention target for kidney injury and repair.
Author contribution statement
Jia Chen: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Quan-you Zheng: Analyzed and interpreted the data; Wrote the paper.
Li-ming Wang: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Jia Luo: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Ke-hong Chen: Conceived and designed the experiments.
Ya-Ni He: Conceived and designed the experiments.
Funding statement
Jia Chen was supported by National Natural Science Foundation of China {81800646}.
Quanyou Zheng was supported by National Natural Science Foundation of China {81900628}.
Profr Yani He was supported by National Science and Technology Planning Project {2016YFC1305501}.
Jia Chen was supported by Youth Cultivation Project of Army Military Medical University {2019XQN018}.
Data availability statement
Data included in article/supp. material/referenced in article and available upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18134.
Contributor Information
Ke-hong Chen, Email: chenkehong@foxmail.com.
Ya-ni He, Email: heynmail@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Zuk A., Bonventre J.V. Acute kidney injury. Annu. Rev. Med. 2016;67:293–307. doi: 10.1146/annurev-med-050214-013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoste E.A.J., Kellum J.A., Selby N.M., Zarbock A., Palevsky P.M., Bagshaw S.M., Goldstein S.L., Cerda J., Chawla L.S. Global epidemiology and outcomes of acute kidney injury. Nat. Rev. Nephrol. 2018;14:607–625. doi: 10.1038/s41581-018-0052-0. [DOI] [PubMed] [Google Scholar]
- 3.V.N.A.R.F.T. Network. Palevsky P.M., Zhang J.H., O’Connor T.Z., Chertow G.M., Crowley S.T., Choudhury D., Finkel K., Kellum J.A., Paganini E., Schein R.M., Smith M.W., Swanson K.M., Thompson B.T., Vijayan A., Watnick S., Star R.A., Peduzzi P. Intensity of renal support in critically ill patients with acute kidney injury. N. Engl. J. Med. 2008;359:7–20. doi: 10.1056/NEJMoa0802639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coca S.G., Singanamala S., Parikh C.R. Chronic kidney disease after acute kidney injury: a systematic review and meta-analysis. Kidney Int. 2012;81:442–448. doi: 10.1038/ki.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk A., Bonventre J.V. Recent advances in acute kidney injury and its consequences and impact on chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2019;28:397–405. doi: 10.1097/MNH.0000000000000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rewa O., Bagshaw S.M. Acute kidney injury-epidemiology, outcomes and economics. Nat. Rev. Nephrol. 2014;10:193–207. doi: 10.1038/nrneph.2013.282. [DOI] [PubMed] [Google Scholar]
- 7.Ferenbach D.A., Bonventre J.V. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat. Rev. Nephrol. 2015;11:264–276. doi: 10.1038/nrneph.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato Y., Takahashi M., Yanagita M. Pathophysiology of AKI to CKD progression. Semin. Nephrol. 2020;40:206–215. doi: 10.1016/j.semnephrol.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Grgic I., Campanholle G., Bijol V., Wang C., Sabbisetti V.S., Ichimura T., Humphreys B.D., Bonventre J.V. Targeted proximal tubule injury triggers interstitial fibrosis and glomerulosclerosis. Kidney Int. 2012;82:172–183. doi: 10.1038/ki.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang-Panesso M., Kadyrov F.F., Lalli M., Wu H., Ikeda S., Kefaloyianni E., Abdelmageed M.M., Herrlich A., Kobayashi A., Humphreys B.D. FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury. J. Clin. Investig. 2019;129:5501–5517. doi: 10.1172/JCI125519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusaba T., Lalli M., Kramann R., Kobayashi A., Humphreys B.D. Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B.C., Tang T.T., Lv L.L., Lan H.Y. Renal tubule injury: a driving force toward chronic kidney disease. Kidney Int. 2018;93:568–579. doi: 10.1016/j.kint.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 13.Chung K.W., Lee E.K., Lee M.K., Oh G.T., Yu B.P., Chung H.Y. Impairment of PPARalpha and the fatty acid oxidation pathway aggravates renal fibrosis during aging. J. Am. Soc. Nephrol. : JASN (J. Am. Soc. Nephrol.) 2018;29:1223–1237. doi: 10.1681/ASN.2017070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao Y., Liu L., Yin L., Xu L., Tang Z., Qi Y., Mao Z., Zhao Y., Ma X., Peng J. FABP4 contributes to renal interstitial fibrosis via mediating inflammation and lipid metabolism. Cell Death Dis. 2019;10:382. doi: 10.1038/s41419-019-1610-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Chung K.W., Ha S., Kim S.M., Kim D.H., An H.J., Lee E.K., Moon H.R., Chung H.Y. PPARalpha/beta activation alleviates age-associated renal fibrosis in sprague Dawley rats. J. Gerontol. A Biol. Sci. Med. Sci. 2020;75:452–458. doi: 10.1093/gerona/glz083. [DOI] [PubMed] [Google Scholar]
- 16.Nemeth A., Mozes M.M., Calvier L., Hansmann G., Kokeny G. The PPARgamma agonist pioglitazone prevents TGF-beta induced renal fibrosis by repressing EGR-1 and STAT3. BMC Nephrol. 2019;20:245. doi: 10.1186/s12882-019-1431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinschen M.M., Saez-Rodriguez J. The tissue proteome in the multi-omic landscape of kidney disease. Nat. Rev. Nephrol. 2021;17:205–219. doi: 10.1038/s41581-020-00348-5. [DOI] [PubMed] [Google Scholar]
- 18.Gerhardt L.M.S., McMahon A.P. Multi-omic approaches to acute kidney injury and repair. Curr. Opin. Biomed. Eng. 2021;20 doi: 10.1016/j.cobme.2021.100344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govender M.A., Brandenburg J.T., Fabian J., Ramsay M. The use of 'omics for diagnosing and predicting progression of chronic kidney disease: a scoping review. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.682929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canaud G., Brooks C.R., Kishi S., Taguchi K., Nishimura K., Magassa S., Scott A., Hsiao L.L., Ichimura T., Terzi F., Yang L., Bonventre J.V. Cyclin G1 and TASCC regulate kidney epithelial cell G2-M arrest and fibrotic maladaptive repair. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Section 2: AKI definition. Kidney Int. Suppl. 2012;2011(2):19–36. doi: 10.1038/kisup.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L., Brooks C.R., Xiao S., Sabbisetti V., Yeung M.Y., Hsiao L.L., Ichimura T., Kuchroo V., Bonventre J.V. KIM-1-mediated phagocytosis reduces acute injury to the kidney. J. Clin. Investig. 2015;125:1620–1636. doi: 10.1172/JCI75417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L., Besschetnova T.Y., Brooks C.R., Shah J.V., Bonventre J.V. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 2010;16:535–543. doi: 10.1038/nm.2144. 531p following 143. 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan H., Yang K., Li Y., Shaw T.I., Wang Y., Blanco D.B., Wang X., Cho J.H., Wang H., Rankin S., Guy C., Peng J., Chi H. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity. 2017;46:488–503. doi: 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Xing Y., Li W., Yang F., Lang Y., Yang J., Liu Z. Renal tubules transcriptome reveals metabolic maladaption during the progression of ischemia-induced acute kidney injury. Biochem. Biophys. Res. Commun. 2018;505:432–438. doi: 10.1016/j.bbrc.2018.08.111. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y., Tang C., Cai J., Chen G., Zhang D., Dong Z. Rodent models of AKI-CKD transition. Am. J. Physiol. Ren. Physiol. 2018;315:F1098–F1106. doi: 10.1152/ajprenal.00199.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H., van Dullemen L.F.A., Akhtar M.Z., Faro M.L., Yu Z., Valli A., Dona A., Thezenas M.L., Charles P.D., Fischer R., Kaisar M., Leuvenink H.G.D., Ploeg R.J., Kessler B.M. Proteo-metabolomics reveals compensation between ischemic and non-injured contralateral kidneys after reperfusion. Sci. Rep. 2018;8:8539. doi: 10.1038/s41598-018-26804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malagrino P.A., Venturini G., Yogi P.S., Dariolli R., Padilha K., Kiers B., Gois T.C., Cardozo K.H., Carvalho V.M., Salgueiro J.S., Girardi A.C., Titan S.M., Krieger J.E., Pereira A.C. Proteome analysis of acute kidney injury - discovery of new predominantly renal candidates for biomarker of kidney disease. J. Proteonomics. 2017;151:66–73. doi: 10.1016/j.jprot.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Kang H.M., Ahn S.H., Choi P., Ko Y.A., Han S.H., Chinga F., Park A.S., Tao J., Sharma K., Pullman J., Bottinger E.P., Goldberg I.J., Susztak K. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M., Cao X., Zhang K., Li Y., Zheng Q.Y., Li G.Q., He Q.H., Li S.J., Xu G.L., Zhang K.Q. Celastrol alleviates renal fibrosis by upregulating cannabinoid receptor 2 expression. Cell Death Dis. 2018;9:601. doi: 10.1038/s41419-018-0666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilflingseder J., Willi M., Lee H.K., Olauson H., Jankowski J., Ichimura T., Erben R., Valerius M.T., Hennighausen L., Bonventre J.V. Enhancer and super-enhancer dynamics in repair after ischemic acute kidney injury. Nat. Commun. 2020;11:3383. doi: 10.1038/s41467-020-17205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spath M.R., Bartram M.P., Palacio-Escat N., Hoyer K.J.R., Debes C., Demir F., Schroeter C.B., Mandel A.M., Grundmann F., Ciarimboli G., Beyer A., Kizhakkedathu J.N., Brodesser S., Gobel H., Becker J.U., Benzing T., Schermer B., Hohne M., Burst V., Saez-Rodriguez J., Huesgen P.F., Muller R.U., Rinschen M.M. The proteome microenvironment determines the protective effect of preconditioning in cisplatin-induced acute kidney injury. Kidney Int. 2019;95:333–349. doi: 10.1016/j.kint.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y.H., Platt M.P., Fu H., Gui Y., Wang Y., Gonzalez-Juarbe N., Zhou D., Yu Y. Global proteome and phosphoproteome characterization of sepsis-induced kidney injury. Mol. Cell. Proteomics : MCP. 2020;19:2030–2047. doi: 10.1074/mcp.RA120.002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin E.N., Ackerman S.L. Lipid metabolism and axon degeneration: an ACOX1 balancing act. Neuron. 2020;106:551–553. doi: 10.1016/j.neuron.2020.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Chen X.F., Tian M.X., Sun R.Q., Zhang M.L., Zhou L.S., Jin L., Chen L.L., Zhou W.J., Duan K.L., Chen Y.J., Gao C., Cheng Z.L., Wang F., Zhang J.Y., Sun Y.P., Yu H.X., Zhao Y.Z., Yang Y., Liu W.R., Shi Y.H., Xiong Y., Guan K.L., Ye D. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018;19 doi: 10.15252/embr.201745124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F., Xiong Q., Tao H., Liu Y., Zhang N., Li X.F., Suo X.J., Yang Q.P., Chen M.X. ACOX1, regulated by C/EBPalpha and miR-25-3p, promotes bovine preadipocyte adipogenesis. J. Mol. Endocrinol. 2021;66:195–205. doi: 10.1530/JME-20-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jao T.M., Nangaku M., Wu C.H., Sugahara M., Saito H., Maekawa H., Ishimoto Y., Aoe M., Inoue T., Tanaka T., Staels B., Mori K., Inagi R. ATF6alpha downregulation of PPARalpha promotes lipotoxicity-induced tubulointerstitial fibrosis. Kidney Int. 2019;95:577–589. doi: 10.1016/j.kint.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Q., Yu X., Yu X., Liu S., Jiang J., Cheng Y., Lin H., Wang Y., Zhang X., Ye X., Xiang Z. An integrated network pharmacology and cell metabolomics approach to reveal the role of rhein, a novel PPARalpha agonist, against renal fibrosis by activating the PPARalpha-CPT1A axis. Phytomedicine. 2022;102 doi: 10.1016/j.phymed.2022.154147. [DOI] [PubMed] [Google Scholar]
- 39.Gomez H. Reprogramming metabolism to enhance kidney tolerance during sepsis: the role of fatty acid oxidation, aerobic glycolysis, and epithelial de-differentiation. Nephron. 2022:1–4. doi: 10.1159/000527392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article and available upon reasonable request.