Abstract
Shark cartilage was created as a cancer-fighting diet because it was believed to have an element that may suppress tumor growth. Due to overfishing, sharks have become endangered recently, making it impossible to harvest natural components from shark cartilage for therapeutic development research. Previously, we identified a peptide SAIF from shark cartilage with an-tiangiogenic and anti-tumor effects, successfully expressed it in Escherichia coli by using genetic engineering techniques. However, we did not elucidate the specific target of SAIF and its antiangiogenic molecular mechanism, which hindered its further drug development. Therefore, in this work, the exact mechanism of action was studied using various techniques, including cellular and in vivo animal models, computer-aided simulation, molecular target capture, and transcriptome sequencing analysis. With VEGF-VEGFR2 interaction and preventing the activation of VEGFR2/ERK signaling pathways, SAIF was discovered to decrease angiogenesis and hence significantly limit tumor development. The findings further demonstrated SAIF's strong safety and pharmaceutically potential. The evidence showed that SAIF, which is expressed by, is a potent and safe angiogenesis inhibitor and might be developed as a candidate peptide drug for the treatment of solid tumors such as hepatocellular carcinoma and other conditions linked with angiogenic overgrowth.
Keywords: Peptides, Shark-derived, Antiangiogenic, Hepatocellular carcinoma
1. Introduction
In recent decades, marine-derived natural compounds have been widely utilized in medication research [[1], [2], [3]]. As an inhibitor of angiogenesis in cancer therapy [[4], [5], [6]], as a joint lubricant in arthritis [7], and as an immunomodulator for psoriasis [8], shark cartilage is believed to have great therapeutic value. Due to recent overfishing and challenges in captive breeding culture, extracting sufficient therapeutic components from shark tissues is no longer viable.
Vascular endothelial growth factor (VEGF) is a significant growth factor that governs angiogenesis, and VEGF exerts its biological effects on angiogenesis predominantly via binding to its receptor, VEGFR2 [9]. Unregulated artery formation promotes several disease processes, including malignancies and intraocular vascular disorders [9,10]. Targeting the VEGF pathway may inhibit ocular angiogenesis [11] and prolong survival in cancer patients [12]. Therefore, VEGF and VEGFR2 are potential targets for angiogenesis in intervention.
Hepatocellular Carcinoma (HCC) [[13], [14], [15], [16]] is common cancer worldwide, fifth in occurrence and fourth in death rates, is the most common cause of cancer-related deaths among Chinese males under 60. The death rate from HCC is higher than any other kind of liver cancer [17,18]. Despite breakthroughs in diagnosis and therapy, most individuals with advanced HCC live less than one year [19]. Currently, there are no clinically defined treatments for HCC [20]. Studies have demonstrated that inhibiting angiogenesis by reducing VEGF/VEGFR2 signaling is one of the most effective treatments for HCC [21,22]. Consequently, developing novel targeted medications for the treatment of HCC must target this signaling pathway.
A shark-derived angiogenesis inhibition factor (SAIF) containing 33 amino acid residues was extracted from shark cartilage and obtained by recombineering technology [23]. It has been proven that SAIF has antiangiogenic and anti-breast cancer properties, but its molecular mechanism of action has not been investigated [23]. This study aimed to elucidate the molecular mechanism behind the antiangiogenic activity of SAIF to assess its pharmaceutically potential.
2. Materials and methods
2.1. Cell lines
Human umbilical vein endothelial cells (HUVEC), normal human liver cells QSG-7701, human hepatoma cells SMMC-7721, Huh7, MHCC97H, MHCC97L, and HepG2 were obtained from Institute of Biomedicine, Jinan University. The HUVEC cells were cultured in ECM medium (ScienCell, USA) with the addition of 5% fetal calf serum, 1% endothelial cell growth supplement (ECGS), and 1% penicillin/streptomycin. The remaining cells were grown in 10% fetal bovine serum-supplemented DMEM medium (Gibco, USA) (Gibco, USA).
2.2. Materials
SAIF peptide (purity >95%) was synthesized by solid phase synthesis by Shanghai Qiang Yao Biotechnology Co. Trypsin (0.25% EDTA+ and Phenol Red) was purchased from Beijing Alltech Biotechnology Co. All recombinant proteins (>98% purity) were purchased from Sino Biological (China). Matrigel was purchased from Corning (USA).
2.3. Cell proliferation assay
Cell proliferation assay was performed as previously described [24]. HUVECs or cancer cells (2.5 × 103 per well/100 μL) were seeded into 96-well plates with ECM medium. The cells were starved for 24 h by being exposed to DMEM media (0.5% FBS) after being incubated for 24 h at 37 °C and 5% CO2. 0.5% starvation media was used to dilute the peptides. The appropriate concentration of peptides was added to a final culture volume of 100 μL of 0.5% starvation medium. The medium was removed by placing back in the incubator at 37 °C for a further 48 h. 100 μL/well of CCK8 working solution (Bimake, China) was added and incubated at 37 °C in a 5% CO2 incubator for 1 h. Finally, the optical density of each well was measured at 450 nm using a multifunctional microplate reader.
2.4. Cell colony formation assay
Cell colony formation assay was similar to the previously described [25]. HUVECs or cancer cells (1 × 103 per well) were incubated for 24 h in ECM medium in 6-well plates. After discarding the previously used medium, 2 mL of various concentrations of made peptide solutions were added, and control received 2 mL fresh medium. The medium was changed every two days, for 14 days. Then 500 μL of 4% paraformaldehyde was added to each well, incubated for 30 min, and then aspirated and air dried. After 30 min, each well received 1 mL of 0.1% crystalline violet solution and PBS washes three times. After wells dry, clonal clusters are photographed and tallied.
2.5. Wound healing experiment
Wound healing experiment was performed as previously described [26]. HUVEC cells (1 × 105 cells per well) were inoculated in 6-well plates and incubated at 37 °C, 5% CO2 for 24 h. After forming a monolayer of cells, they were treated with DMEM (0.5% FBS) for 24 h to starve them. A scratch on the cell monolayer was made by a 200 μL tip. The 6-well plate was carefully shaken with PBS to remove floating or dead cells. Peptides were diluted to different concentrations with DMEM (0.5% FBS), and equal volumes were added to each well for incubation. HUVECs were photographed by microscopy at 0, 16, and 24 h after wounding, and the size of cell migration was quantified using Image J.
2.6. HUVEC tube formation assay
The HUVEC tube formation assay was carried out as described previously [27]. The 96-well plate and sterile pipette tips were precooled at −20 °C overnight. The 96-well plates were added with 70 μL Matrigel into each well, and incubated at 37 °C for 30 min. HUVEC cells suspensions (3 × 104 cells/well) containing different doses (0 μM, 25 μM, 50 μM) of peptides were inoculated onto the matrigel and incubated for 4 h at 37 °C. The network of tube lumens was photographed by microscopy.
2.7. The chick chorioallantoic membrane (CAM) assay
The chick embryo chorioallantoic membrane (CAM) assay was carried out as described previously [27]. Freshly fertilized eggs were incubated at 37 °C and 60% humidity for five days (n = 5 eggs/group). The eggs were turned over 180° every 24 h, and the survival rate of the embryos was observed. After 6 days, the seeds were cracked open along the gas chamber, and sterilized rubber rings were placed at the aorta. Saline containing different concentrations of peptides (0, 10, 50, 250μΜ) was added to the center of the rubber ring. The dosing volume was 50 μL, which was then sealed and incubated in the incubator for 48 h. Finally, the area of new blood vessels in the rubber ring was photographed in a stereomicroscope. Analysis was carried out using Angio-Tool software. Vascular density = area of vessels/area of view.
2.8. Effect of peptides on angiogenesis in zebrafish
The detailed protocol was described in previous study [28]. The eggs were collected the next day, and the yolk membrane was broken at 24 h using the membrane breaker pronase (2 mg/mL). The peptides were diluted to different concentrations (0, 10, 50, 250μΜ) using culture water and microinjected under a dissecting microscope. 5 nL of different concentrations of peptides were microinjected into the peritoneal cavity of the zebrafish larvae. Zebrafish larvae were incubated with culture water to dilute PTK787 (VEGFR2 inhibitor) to 0.25 μg/mL, Images of zebrafish angiogenesis were taken at 72hpf using a body fluorescence microscope.
2.9. Transcriptome sequencing and analysis
The detailed protocol was described in previous study [26]. HUVEC cells were lysed on ice by TRIZOL. Cell lysates were transported on dry ice to Aki Biologicals, Guangzhou, for reverse transcription and quality control. RNA with OD260/280 = 1.8–2.0, RIN ≥9.5, 28 S:18 S = 1.6–2.4 were used for sequencing, which were finally sequenced and analyzed using the Illumina HiSeq system. FPKM calculated gene expression. The R statistical package DESeq2 (fold change ≥1, p-value <0.05) was used to identify differentially expressed genes with a false discovery rate (FDR) cut-off <0.05, while GO analysis, KEGG enrichment analysis and GSEA analysis were performed based on differentially expressed genes. More details can be found in Supplementary file 1.
2.10. Surface plasmon resonance (SPR) analysis
The detailed protocol was described in previous study [27]. All the SPR technical services involved in this study were entrusted to Guangzhou Baoser Biotechnology Co. The target protein capture tests made use of a Lumera, USA-manufactured NanoSensor biochip with HUVEC cells lysate as the mobile phase and SAIF molecules as the stationary phase on the chip surface. After testing, the proteins enriched on the surface of the chip were identified by HPLC-MS, and functional analysis was performed using the GEO and KEGG databases. (See Supplementary Document 1 for details).
2.11. In vivo studies
The detailed protocol was described in previous study [25]. All tests on animals were done in accordance with the Medical Laboratory Animal Center of Jinan University's ethical rules (No. 20220612–01). Guangdong Pharmachem Biotechnology provided male BALB/c nude mice. SPF lab mice were fed chow and purified water at room temperature. MHCC97L cells (1 × 107 cells in 0.1 mL saline) were administered subcutaneously into the axillae of five-week-old male BALB/c nude mice. Every two days, tumor volume and body weight were assessed, and tumor volume was estimated as (short diameter)2 × (long diameter)/2. These mice were randomly separated into five groups when tumor volumes reached 50 mm3: model, PTK787 (50 mg/kg), SAIF (10, 20, and 30 mg/kg), and control. SAIF was dissolved in saline and administered by tail vein injection at a volume of 0.1 mL/20 g. The execution was performed 14 days after administration. Blood was collected for blood physiological and blood biochemical analysis, organs were collected for histopathological examination, and tumors were collected for histopathological examination and immunohistochemical experiments.
2.12. Histopathological examination
HE was performed as described previously [25]. Organs or tumors from transplanted mice were fixed with 4% paraformaldehyde overnight at 4 °C. Then, the tissues were processed routinely into paraffin, sectioned at a thickness of 4 μm, and the specimens were stained with Hematoxylin and Eosin (H&E). The samples were observed for histopathological changes using a light microscope.
2.13. Immunohistochemistry experiments
Immunohistochemistry was performed as described previously [25]. The tumor tissue sections were 4 μm in thickness. Microvessel density was identified by immunostaining with CD31 antibody. Tumor cell proliferation was determined by staining with Ki67 antibody.
2.14. Western blotting analysis
The detailed protocol was described in previous study [27]. HUVEC cells were incubated with SAIF for 48 h and then lysed with RIPA buffer. The concentration of the protein samples was quantified using the BCA protein assay kit. Protein samples were separated by electrophoresis on an SDS-PAGE gel and transferred to a PVDF membrane. The membranes were blocked in 5% BSA at room temperature for 2 h. And then, the PVDF membrane were incubated with primary antibodies overnight at 4 °C. After incubation with the secondary antibody for 1 h, signal detection was performed using enhanced chemiluminescence (ECL) reagents. Data was quantified and normalized using Image J. The primary antibodies used were as following: Anti-phospho-ERK1/2 (Abcam, Cat. #ab76299), anti-ERK1/2 (CST, Cat. #9102), anti-VEGFR2 (CST, Cat. #9698), anti-phospho-VEGFR2 (CST, Cat. #3770).
2.15. Molecular docking
Alphafold software [29] was used to model the peptides based on MSA multiple sequence template search or Lenovo redesign, followed by molecular dynamics optimization (Amber force field), ensuring a high quality model. PDB database entry for the structure of the VEGFR2 protein: 3V2A. Using GRAMM-X Docking, the interaction of peptides with VEGFR2 proteins was predicted. The top 10 interaction models with the highest scores were entered into PDBe for free energy calculations. The optimal binding model was determined based on the idea that the critical and competitive binding sites are more stable the lower the free energy value. Finally, analysis and mapping of interaction structure were performed in Discovery Studio 2.1.
2.16. Molecular dynamics
The detailed protocol was described in previous study [28]. The results of molecular docking were utilized to select protein-peptide complexes for molecular dynamics simulation. All simulations of molecular dynamics were conducted using GROMACS 2019 and CHARMM36 m force fields. The complexes were placed in the center of the TIP3P water tank at a filling distance of 1.2 nm from the tank's edge and neutralized with Na+ and Cl−. The system energy was reduced using the quickest descent algorithm and equilibrated under the NVT ensemble for 2 ns. In the NPT system, the ultimate trajectory is calculated at a temperature of 300 K and a pressure of 1 bar.
2.17. Flow cytometry analysis
The detailed protocol was described in previous study [25]. HUVEC (2 × 105 cells/well) were seeded into 6-well plates for 24 h. After 24 h at 37 °C and 5% CO2, the media was withdrawn and the cells were starved with DMEM medium (0.5% FBS). Finally, SAIF were diluted in 0.5% starvation medium, and added at the indicated concentration (0/12.5/25/50 μM) for 48 h. For the cell apoptosis assay, cells were stained using the Annexin V-FITC Apoptosis Analysis Kit (Cas:10-5 CE0 T, DOJINDO) and finally analyzed using the FACSAria™ flow cytometer (BD Biosciences). The cell cycle analysis was carried out according to the protocol in Cell Cycle Analysis Kit (Cas: C1052, Beyotime, Shanghai, China). The percentage of cells in different phases of the cell cycle was examined by measuring the DNA content (propidium iodide intensity) with a flow cytometer (Beckman Coulter, Brea, CA), and populations of G1, S, and G2/M phase cells were determined with the ModFIT software. Each experiment was repeated three independent times.
2.18. Statistical analysis
Data were analyzed using GraphPad Prism 8.0 software, and all results are expressed as mean ± standard deviation (SD). Values for differences between groups were measured using Student's t-test for comparison or one-way ANOVA for multiple comparisons. The setting is not statistically different; p < 0.05 was considered statistically significant. * indicates p < 0.05, ** indicates p < 0.01 and *** indicates p < 0.001.
3. Results
3.1. SAIF inhibits the growth of HUVEC cells
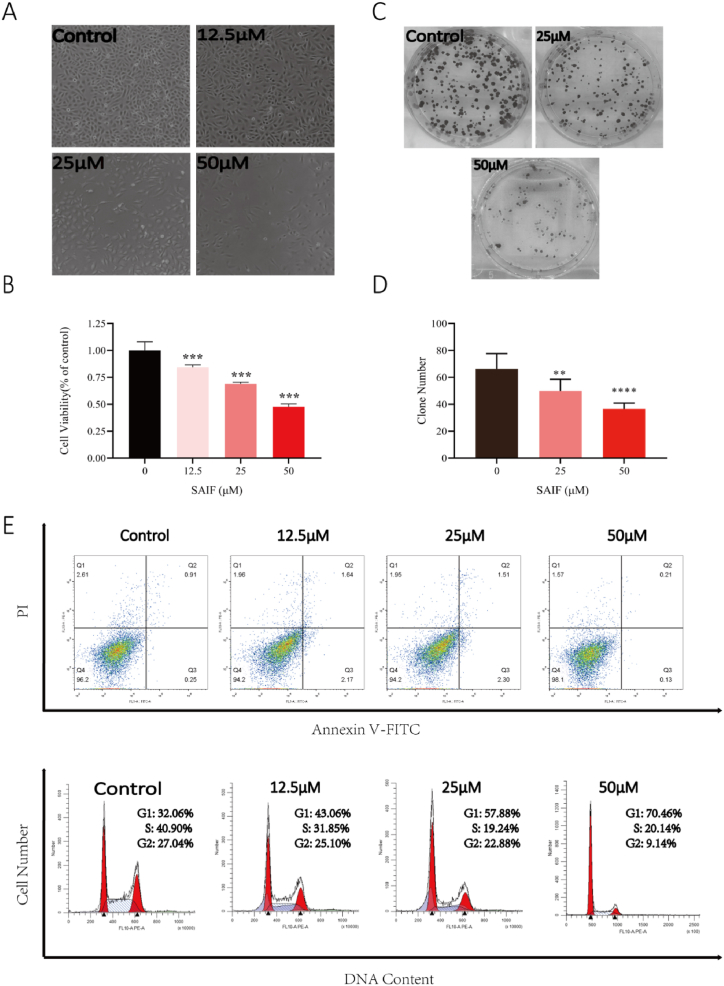
Using HUVEC, the in vitro antiangiogenetic activity of SAIF was evaluated. The results demonstrate that SAIF dramatically inhibited the growth and colony formation of HUVECs in a dose-dependent manner (Fig. 1 A-D). To verify whether SAIF triggers apoptosis or cell cycle arrest, flow cytometry was employed to analyze the SAIF-treated HUVEC cells. Interestingly, SAIF did not result in apoptosis (Fig. 1. E), however, triggers the arrest at G1/S cell cycle checkpoint in HUVEC cells (Fig. 1. F). These results indicate the suppression of SAIF to HUVEC cells in vitro.
Fig. 1.
SAIF inhibited the growth of HUVEC cells. (A) Morphology of HUVEC cells after SAIF (12.5μΜ, 25μΜ, and 50μΜ) treatment. (B) Effect of SAIF (12.5μΜ, 25μΜ, and 50μΜ) on the viability of HUVEC cells. (C) Inhibitory effect of SAIF (25μΜ and 50μΜ) on clone formation of HUVEC cells. (D) Statistical histogram of HUVEC cell clones. (E) The apoptotic effect of SAIF on HUVEC cells was analyzed by Annexin V-FITC/PI staining. (F) SAIF treatment for 48 h altered the cell cycle distribution of HUVEC cells. Data are shown as mean ± SD, n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
3.2. SAIF inhibits HUVECs migration and lumen formation
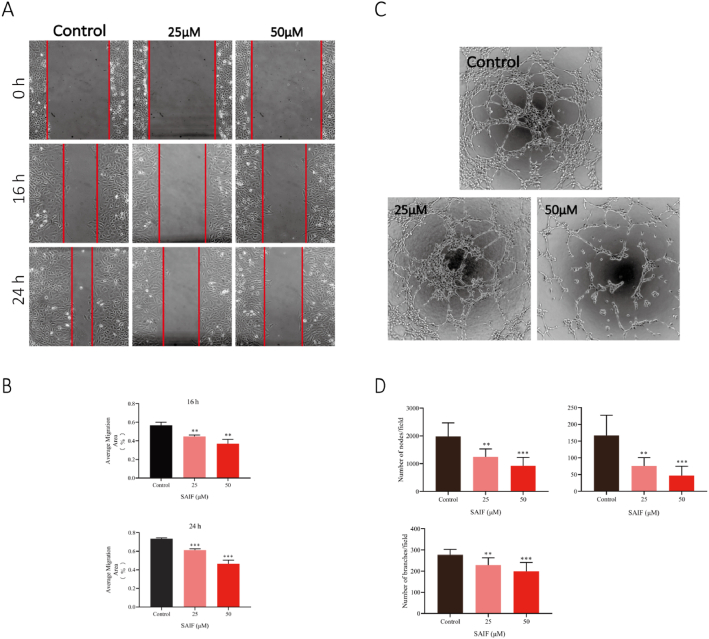
To evaluate the effect of SAIF on cell migration, scratch analysis was employed, and the results demonstrated that migration of HUVEC cells was significantly suppressed by SAIF after 16 and 24 h exposure. Their migratory distance decreases paralleled to the increased peptide concentrations (Fig. 2. A). Moreover, SAIF severely decreased the capacity of HUVEC cells to generate lumens in the lumen formation assay (Fig. 2. C). These results reveal that SAIF inhibits angiogenesis in vitro.
Fig. 2.
SAIF inhibited HUVEC migration and lumen formation in vitro. (A) SAIF (25μΜ and 50μΜ) inhibited the migration of HUVECs. (B) Quantitative analysis of the migrated area was performed by Image J. (C) SAIF (25 μM, 50 μM) inhibited the tube formation of HUVECs. (D) The number of tubes, branches, and fulcrum was analyzed by angiogenesis analysis of Image J. Data are shown as mean ± SD, n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
3.3. SAIF inhibits angiogenesis chick chorioallantoic membrane (CAM) and zebrafish models
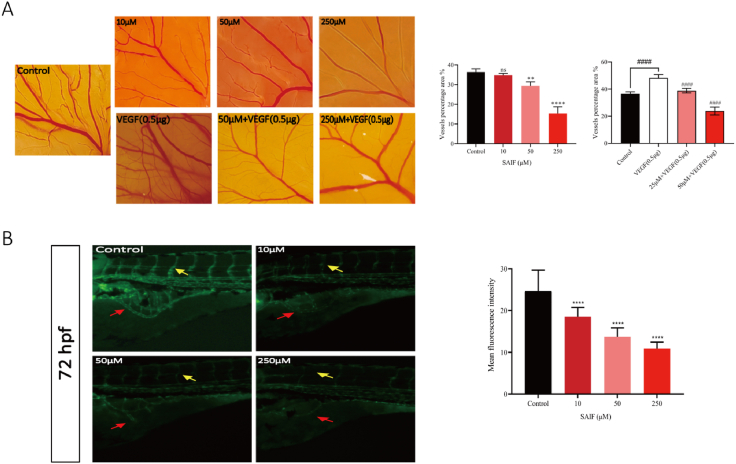
The chick chorioallantoic membrane experiment validated SAIF's antiangiogenic effect (Fig. 3. A). SAIF (50 μM, 250 μM) reduced capillaries and branching arteries compared to saline. VEGF (0.5 μg) significantly increased neoangiogenesis, and SAIF reduced this effect (50 μM, 250 μM) (Fig. 3. A).
Fig. 3.
SAIF inhibited angiogenesis in vivo. (A) SAIF inhibits angiogenesis in CAM model. Blood vessels formed on the chick chorioallantoic membrane (CAM) after 48 h of treatment with SAIF (10, 50, and 250 μM) and VEGF (0.5 μg) for 48 h. Angio-Tool was used to analyzing vessel density. Data are shown as mean ± SD, n = 5 (B) The effect of SAIF on zebrafish angiogenesis. Zebrafish larvae (24hpf) were microinjected with SAIF (0, 10, 50, and 250 μM) in a 5 nL. After incubation for 48 h, intersegmental vessels (ISV, yellow arrows) and subintestinal vessels (SIV, red arrows) of zebrafish larvae (72hpf) were photographed fluorescently. The SIV fluorescence intensity was quantified using Image J. Data are shown as mean ± SD, n = 30. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
In vivo, SAIF's antiangiogenic impact was tested in the zebrafish juvenile angiogenesis model (fli1:EGFP) (Fig. 3. B). After 72 h, microinjection of the peptide solution into juvenile zebrafish (24hpf) suppressed subintestinal vascular (SIV) development. Zebrafish larvae formed less intersegmental vessels (ISV) as peptide concentrations increased.
3.4. SAIF suppresses the activation of VEGFR2 and ERK through interfering with the binding of VEGF to VEGFR2
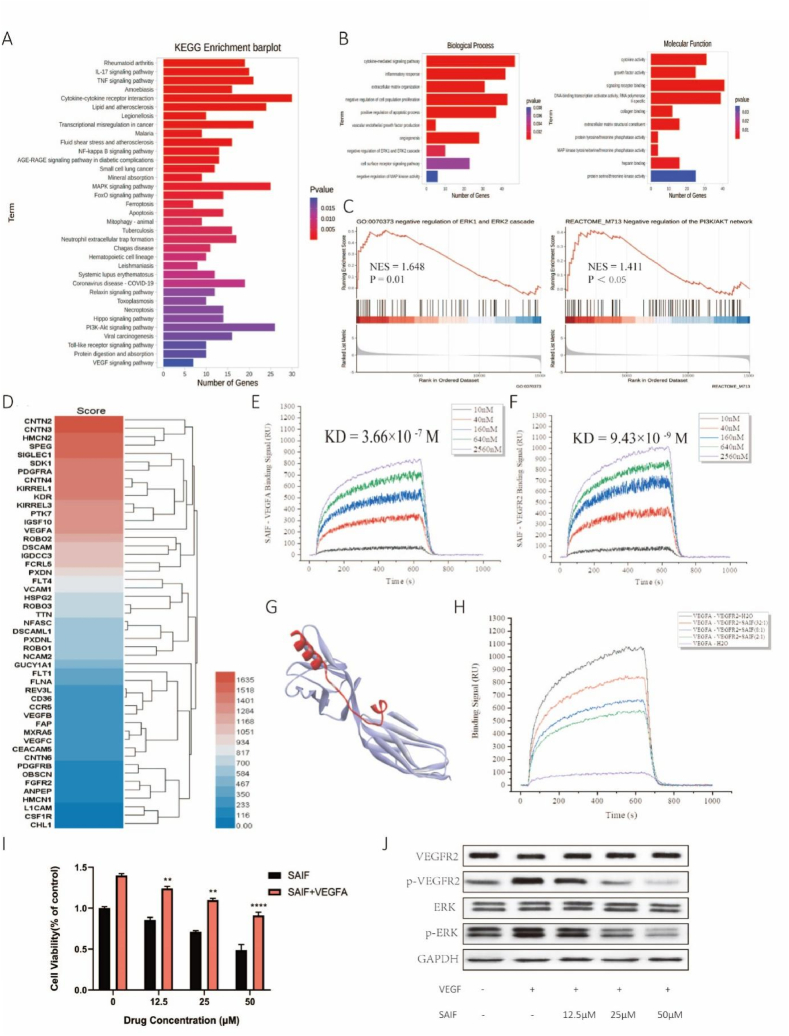
SAIF-treated HUVEC cell RNA was used for transcriptome analysis to examine SAIF's antiangiogenic mechanisms. Transcriptome data revealed the top 35 SAIF-regulated KEGG pathways (Fig. 4. A). SAIF dramatically affected HUVECs' VEGF, MAPK, and PI3K-AKT signaling pathways. Angiogenesis involves several signaling pathways, suggesting that SAIF inhibits them to decrease cell development.
Fig. 4.
SAIF interferes with the binding of VEGF to VEGFR2 and inhibits the activation of VEGFR2 and ERK. (A)KEGG pathway enrichment analysis, including the top 35 most significantly enriched pathways. (B)GO enrichment analysis. (C) Effects of SAIF treatment on ERK pathway and PI3K/AKT pathway. (D) The target protein of SAIF. (E) The binding affinity of SAIF to VEGF. (F) The binding affinity of SAIF to VEGFR2. (G) The predicted binding pattern of SAIF to VEGFR2. (H) VEGF binds VEGFR2 competitively with SAIF. (I) SAIF inhibition inhibits the VEGF-induced proliferation of HUVEC cells. (J) Western blot-ting analysis of VEGFR2 and ERK phosphorylation following treatment of HUVEC cells with SAIF (12.5 μM, 25 μM, 50 μM) for 48 h. Both the total and phosphorylated forms of VEGFR2 and ERK were detected. GAPDH was used as a loading control. Data are shown as mean ± SD, n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
Since SAIF inhibits neoangiogenesis and transcriptome sequencing data shows that SAIF significantly affects the VEGF signaling cascade pathway, we wondered if SAIF affected VEGF-VEGFR2 binding. SPR was used to discover SAIF's target proteins. SAIF captured 49 proteins (Fig. 4D). PDGFRA, VEGF, and VEGFR2 were high-affinity angiogenesis targets. SAIF affinity for VEGF and VEGFR2 was then assessed. SAIF binds VEGFR2 more strongly and persistently than VEGFA (Supplementary file 2), with an affinity parameter KD of 3.66 × 10-7 M for SAIF-VEGF (Fig. 4. E) and 9.43 × 10-9 M for SAIF-VEGFR2. We examined SAIF binding to VEGFR2 (Fig. 4. G) and performed molecular dynamics simulations (Fig. S1). SAIF maintained VEGFR2 hydrogen bonding throughout the simulation.
SAIF's high-affinity interaction with VEGFR2 may interfere with VEGF's binding. SPR was used to test SAIF's competitive inhibition. VEGF's VEGFR2 affinity was initially determined. VEGF-VEGFR2 had a KD of 1.21 × 10-9 M. VEGF's affinity for VEGFR2 decreased significantly with varied SAIF concentration ratios. SAIF may reduce VEGF-VEGFR2 competition (Fig. 4. H).
Cell proliferation assay was used to confirm SAIF's capacity to reduce VEGF-induced HUVEC cell growth (Fig. 4. I). We then studied VEGFR2 activation and its MAPK signaling pathway. p-VEGFR2 activation decreased whereas VEGFR2 expression did not. ERK phosphorylation also decreased (Fig. 4. J). These findings imply that SAIF inhibits VEGF/VEGFR2/ERK signaling to inhibit angiogenesis.
3.5. SAIF reduces the proliferation of HCC cells obtained from humans in vitro and transplanted malignancies in vivo
Given that SAIF can affect VEGF/VEGFR2 signaling and inhibit angiogenesis, and that HCC development is typically characterized by an expanded vascular system [10], resulting in an abnormal tumor vascular system and immunosuppressive environment, we believe that SAIF inhibits HCC development by disrupting angiogenesis in vivo.
After 48 h of treatment with various doses of SAIF in five HCC cell lines (7721, HepG2, Huh7, MHCC97H, MHCC97L), SAIF inhibited the proliferation of human HCC cells in a concentration-dependent manner (Fig. 5. A). SAIF inhibits HCC cell growth in long-term colony formation tests (Fig. 5. B & C). SAIF toxicity was also tested in the human normal regular hepatocyte cell line 7701.7701 cells were unaffected by long-term culture (Fig. 5. B) despite a little decrease in cellular activity in the cell proliferation experiment (Fig. 5. A).
Fig. 5.
SAIF inhibited human HCC cell growth. (A) The effect of SAIF on the viability of human hepatocyte 7701 and human hepatoma cell lines (7721, MHCC97H, HepG2, Huh7, MHCC97L). (B) Inhibition of SAIF on clone formation in human hepatocytes 7701 and human hepatoma cells (7721, MHCC97H, HepG2, Huh7, MHCC97L). (C) Statistical histogram of clonal cell clusters. Data are shown as mean ± SD, n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
We created a subcutaneous tumor model in BALB/c nude mice using MHCC97L from the evaluated HCC cell lines to assess SAIF's anticancer effects. Nude mice were weighed and tumor-developed every two days for fourteen days during treatment. SAIF and PTK787 (VEGFR2 inhibitor) decreased MHCC97L transplanted tumor growth and significantly reduced mean tumor volume (Fig. 6A&B) and weight (Fig. 6C) compared to the control group. Table 1 shows that SAIF (10, 20, and 30 mg/kg) inhibited tumors at 14.13, 51.15, and 72.65%, respectively. SAIF-treated naked mice had similar body weight alterations to the control group (Fig. 6D). The treatment and control groups had similar liver and kidney organ indicators (Fig. 6. E). H&E staining of the liver and kidney in the SAIF-treated group (10, 20, and 30 mg/kg) showed no significant histological changes compared to the control group (Fig. 6. F). These findings showed SAIF does not affect mice's metabolic organs. SAIF's ability to suppress tumor cell growth was confirmed by ki67 staining (Fig. 6. G).
Fig. 6.
SAIF inhibits the growth of MHCC97L xenografts. (A) Visual graph of tumor size. (B) Tumor volume curve. (C) Tumor weight after 14 days of SAIF administration. (D) Body weight change of nude mice during drug treatment. (E) Organ coefficients of the liver and kidney of nude mice. (F) Histological analysis of the liver and kidney of tumor-bearing mice by H&E staining. (G) H&E staining and immunohistochemistry of tumor tissues. Data are shown as mean ± SD, n = 5. *p < 0.05, **p < 0.01 and ***p < 0.001; one-way ANOVA followed by Dunnett's multiple comparison test.
Table 1.
Effect of SAIF on tumor inhibition rate (%) of MHCC97L xenograft mice.
| Group | Dose | Tumor Weight (g) | Tumor Inhibition Rate (%) |
|---|---|---|---|
| Saline (Control) | 0.1 mL/20 g | 1.302 ± 0.117 | / |
| SAIF (low dose) | 10 mg/kg | 1.118 ± 0.273 * | 14.13% |
| SAIF (middle dose) | 20 mg/kg | 0.636 ± 0.068 *** | 51.15% |
| SAIF (high dose) | 30 mg/kg | 0.356 ± 0.092 *** | 72.65% |
| PTK787 | 50 mg/kg | 0.146 ± 0.049 *** | 88.78% |
Values represent the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001, compared with the Saline group (n = 5).
We examined tumor tissue histologically for CD31 expression to determine if SAIF inhibited angiogenesis in the transplanted tumor (Fig. 6. G). Neovascularisations were lower in the treated group. SAIF may reduce tumor angiogenesis since fewer cells expressed CD31.
4. Discussion
Antibodies and small compound drugs that target tyrosine kinases [9,10] are the primary antiangiogenesis agents used clinically at present. These agents reduce tumor formation by inhibiting VEGF/VEGFR2 signaling, which in turn inhibits angiogenesis. While monoclonal antibodies that hinder VEGF/VEGFR2 pathway can successfully kill cancer cells, they can also harm normal cells and tissues leading to immune-related side effects [[30], [31], [32], [33]]. Although small molecule compounds such as sunitinib, sorafenib, and lenvatinib, which target VEGFR2 angiogenesis inhibitors, are effective in treating hepatocellular carcinoma [[34], [35], [36], [37]], however, they tend to cause severe toxic side effects due to low specificity, easy drug resistance, and high toxicity [9,10,38]. As a result of their superior safety profile, minimal immunogenicity, and significantly greater selectivity [39,40], peptides have become a promising area of focus for novel medication development.
More than 60 peptide drugs have been approved by the FDA for marketing, while more than 600 peptides are undergoing clinical and preclinical testing, demonstrating the promise of peptide drug development [[41], [42], [43]]. Active ingredients derived from marine organisms are a valuable therapeutic resource. This work acquired the SAIF sequence from a shark [23]. Through a series of experiments, we have proven that SAIF exerts considerable antiangiogenic and antitumor effects and uncovers its molecular mode of action. In addition, we have accomplished large scale SAIF synthesis via recombinant expression, thereby laying the foundation for its therapeutic development.
In addition to SAIF's demonstrated capacity to block the VEGF-VEGFR2 signaling axis, transcriptome sequencing results indicate that SAIF also activates the TNF signaling pathway. Activation of the TNF signaling system has increased found to increase the permeability of tumor arteries, allowing therapeutic medicines to reach tumor tissue [44,45] more effectively. Besides, long-term TNFαtreatment triggered endothelial cell death and inhibited angiogenesis [46,47]. Moreover, based on target capture and affinity analysis tests, we discovered that SAIF could interact with VEGF-B, VEGFR1, and VEGFR3, as well as block VEGF and VEGFR2 binding [10]. In contrast to VEGFR2, VEGFR1 is solely engaged in pathological angiogenesis; consequently, inhibiting VEGFR1 signaling can benefit cancer therapy [48,49]. In addition, the activation of VEGFR3 can encourage the development of lymphatic capillaries within and surrounding tumors, facilitating the spread of malignancies in a coordinated fashion. Moreover, according to certain research, the activated VEGFC/VEGFR3 axis helps tumors withstand the apoptotic effects of chemotherapeutic drugs [50]. Given propensity of SAIF to interact with several members of the VEGF or VEGFR families, we hypothesize that SAIF may exert its antitumor effects via multiple direct or indirect modes of action in addition to blocking VEGF/VEGFR2 signaling, as described in this study. In addition, we discovered that SAIF has a remarkable binding affinity for PDGFA, a known tumor therapeutic target [51]. The FDA has licensed several PDGFR inhibitors for the treatment of several oncological disorders, including gastrointestinal mesenchymal tumors (GIST), leukemia, and glioma [52]. In addition to predominantly targeting the VEGF-VEGFR2 signaling axis, we hypothesize that SAIF may be connected with activating the TNF signaling pathway and inhibiting activation of the TNF signaling pathway and inhibiting the other related receptors of other receptors related to angiogenesis. This phenomenon shows that SAIF has enormous therapeutic potential for treating disorders characterized by the paths above pathways and receptors.
Data availability statement
Sequencing data are available in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA893416. All other data supporting the findings of this work are available within the paper and its supplementary information files.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki and ap-proved by the Ethics Committee of Animal Experiments of Jinan University, Guangzhou, PR China, Ji'nan University approved all experiments conducted in this study (approval No.: 20,220,612–01).
Author contribution statement
Junye Xie, Fu Li: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yuling Cai, Yibo Zhang: Performed the experiments.
Jinting Zhang, Zijian Su, Rongzhan Liu, Weicai Li, Dianlong Kang: Contributed reagents, materials, analysis tools or data.
Zhaodong Zhai, Xue Chen, Minghua Lei: Analyzed and interpreted the data.
Xiaojia Chen: Conceived and designed the experiments; Wrote the paper.
An Hong: Conceived and designed the experiments.
Funding statement
Miss Xiaojia Chen was supported by National Natural Science Foundation of China {82273833}, Guangdong Provincial Key R&D Program {2022B1111070007}.
An Hong was supported by National Natural Science Foundation of China {82173729}, Program of Department of Natural Resources of Guangdong Province {GDNRC [2021] 50}, Science & Technology Program of Guangzhou City {20212210007}.
Xue Chen was supported by Special Funds for the Cultivation of Guangdong College Students' Scientific and Technological Innovation {pdjh2021a0051}.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We want to thank Ms. Guo Shujun and Ms. Zhou Yuying from the Biomedical Research Institute of School of Life Science and Technology of Jinan University for help in the technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18240.
Contributor Information
Xiaojia Chen, Email: tchenxj@jnu.edu.cn.
An Hong, Email: tha@jnu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Skropeta D., Wei L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014;31(8):999–1025. doi: 10.1039/c3np70118b. [DOI] [PubMed] [Google Scholar]
- 2.Lyu P., Kwok H.F. High-throughput strategy accelerates the progress of marine anticancer peptide drug development. Recent Pat. Anti-Cancer Drug Discov. 2019;14(1):2–4. doi: 10.2174/1574892813999181114152127. [DOI] [PubMed] [Google Scholar]
- 3.Zuo W., Kwok H.F. Development of marine-derived compounds for cancer therapy. Mar. Drugs. 2021;19(6):342. doi: 10.3390/md19060342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patra D., Sandell L.J. Antiangiogenic and anticancer molecules in cartilage. Expet Rev. Mol. Med. 2012;14:e10. doi: 10.1017/erm.2012.3. [DOI] [PubMed] [Google Scholar]
- 5.Lu C., Lee J.J., Komaki R., Herbst R.S., Feng L., Evans W.K., Choy H., Desjardins P., Esparaz B.T., Truong M.T. Chemoradiotherapy with or without Ae-941 in Stage iii Non–Small cell lung cancer: a randomized Phase iii trial. J. Natl. Cancer Inst.: J. Natl. Cancer Inst. 2010;102(12):859–865. doi: 10.1093/jnci/djq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabbani-Chadegani A., Abdossamadi S., Bargahi A., Yousef-Masboogh M. Identification of low-molecular-weight protein (SCP1) from shark cartilage with anti-angiogenesis activity and sequence similarity to parvalbumin. J. Pharmaceut. Biomed. Anal. 2008;46(3):563–567. doi: 10.1016/j.jpba.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Chen L., Bao B., Wang N., Xie J., Wu W. Oral administration of shark type II collagen suppresses complete Freund's adjuvant-induced rheumatoid arthritis in rats. Pharmaceuticals. 2012;5(4):339–352. doi: 10.3390/ph5040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauder D.N., DeKoven J., Champagne P., Croteau D., Dupontb É. Neovastat (AE-941), an inhibitor of angiogenesis: randomized phase I/II clinical trial results in patients with plaque psoriasis. J. Am. Acad. Dermatol. 2002;47(4):535–541. doi: 10.1067/mjd.2002.124702. [DOI] [PubMed] [Google Scholar]
- 9.Apte R.S., Chen D.S., Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176(6):1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayson G.C., Kerbel R., Ellis L.M., Harris A.L. Antiangiogenic therapy in oncology: current status and future directions. Lancet. 2016;388(10043):518–529. doi: 10.1016/S0140-6736(15)01088-0. [DOI] [PubMed] [Google Scholar]
- 11.Formica M.L., Awde Alfonso H.G., Palma S.D. Biological drug therapy for ocular angiogenesis: anti‐VEGF agents and novel strategies based on nanotechnology. Pharmacology Research & Perspectives. 2021;9(2) doi: 10.1002/prp2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Qu X., Wang Y., Dong W., Shen H., Zhang T., Ni Y., Liu Q., Du J. The impact of ramucirumab on survival in patients with advanced solid tumors: a systematic review and meta-analysis of randomized II/III controlled trials. Clin. Drug Invest. 2016;36(1):27–39. doi: 10.1007/s40261-015-0355-x. [DOI] [PubMed] [Google Scholar]
- 13.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 14.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C., Al-Raddadi R., Alvis-Guzman N., Amoako Y., Artaman A. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R., Cheng Q., Owusu-Ansah K.G., Song G., Jiang D., Zhou L., Xu X., Wu J., Zheng S. NKILA, a prognostic indicator, inhibits tumor metastasis by suppressing NF-κB/Slug mediated epithelial-mesenchymal transition in hepatocellular carcinoma. Int. J. Biol. Sci. 2020;16(3):495. doi: 10.7150/ijbs.39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elserag H.B. 2011. Hepatocellular Carcinoma — NEJM. [Google Scholar]
- 18.El–Serag H.B., Rudolph K.L. 2007. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. [DOI] [PubMed] [Google Scholar]
- 19.Ryerson A.B., Eheman C.R., Altekruse S.F., Ward J.W., Jemal A., Sherman R.L., Henley S.J., Holtzman D., Lake A., Noone A.-M. Annual Report to the Nation on the Status of Cancer, 1975‐2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristofanilli M., Charnsangavej C., Hortobagyi G.N. Angiogenesis modulation in cancer research: novel clinical approaches. Nat. Rev. Drug Discov. 2002;1(6):415–426. doi: 10.1038/nrd819. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez–Gea V., Toffanin S., Friedman S.L., Llovet J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144(3):512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu A.X., Sahani D.V., Duda D.G., Di Tomaso E., Ancukiewicz M., Catalano O.A., Sindhwani V., Blaszkowsky L.S., Yoon S.S., Lahdenranta J. Efficacy, safety, and potential biomarkers of sunitinib monotherapy in advanced hepatocellular carcinoma: a phase II study. J. Clin. Oncol. 2009;27(18):3027. doi: 10.1200/JCO.2008.20.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q., Yao S., Chen X., Xu L., Peng W., Zhang L., Zhang Q., Liang X.-F., Hong A. A polypeptide from shark troponin I can inhibit angiogenesis and tumor growth. Mol. Biol. Rep. 2012;39(2):1493–1501. doi: 10.1007/s11033-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 24.Su Z., Zhang Y., Cao J., Sun Y., Cai Y., Zhang B., He L., Zhang Z., Xie J., Meng Q. Hyaluronic acid-FGF2-derived peptide bioconjugates for suppression of FGFR2 and AR simultaneously as an acne antagonist. J. Nanobiotechnol. 2023;21(1):55. doi: 10.1186/s12951-023-01812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Zhang Y., Cao J., Su Z., Li F., Zhang P., Zhang B., Liu R., Zhang L., Xie J. FGFR4 and EZH2 inhibitors synergistically induce hepatocellular carcinoma apoptosis via repressing YAP signaling. J. Exp. Clin. Cancer Res. 2023;42(1):1–16. doi: 10.1186/s13046-023-02659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao J., Zhang Y., Yang Y., Xie J., Su Z., Li F., Li J., Zhang B., Wang Z., Zhang P. Turning gray selenium and sublimed sulfur into a nanocomposite to accelerate tissue regeneration by isothermal recrystallization. J. Nanobiotechnol. 2023;21(1):57. doi: 10.1186/s12951-023-01796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H., Cong Q., Du Z., Liao W., Zhang L., Yao Y., Ding K. Sulfated fucoidan FP08S2 inhibits lung cancer cell growth in vivo by disrupting angiogenesis via targeting VEGFR2/VEGF and blocking VEGFR2/Erk/VEGF signaling. Cancer Lett. 2016;382(1):44–52. doi: 10.1016/j.canlet.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Wei R., Wu Q., Ai N., Wang L., Zhou M., Shaw C., Chen T., Ye R.D., Ge W., Siu S.W. A novel bioengineered fragment peptide of Vasostatin-1 exerts smooth muscle pharmacological activities and anti-angiogenic effects via blocking VEGFR signalling pathway. Comput. Struct. Biotechnol. J. 2021;19:2664–2675. doi: 10.1016/j.csbj.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burger R.A., Brady M.F., Bookman M.A., Fleming G.F., Monk B.J., Huang H., Mannel R.S., Homesley H.D., Fowler J., Greer B.E. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N. Engl. J. Med. 2011;365(26):2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 31.Hurwitz H., Fehrenbacher L., Novotny W., Cartwright T., Hainsworth J., Heim W., Berlin J., Baron A., Griffing S., Holmgren E. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 32.Sandler A., Gray R., Perry M.C., Brahmer J., Schiller J.H., Dowlati A., Lilenbaum R., Johnson D.H. Paclitaxel–carboplatin alone or with bevacizumab for non–small-cell lung cancer. N. Engl. J. Med. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 33.Simpkins F., Belinson J.L., Rose P.G. Avoiding bevacizumab related gastrointestinal toxicity for recurrent ovarian cancer by careful patient screening. Gynecol. Oncol. 2007;107(1):118–123. doi: 10.1016/j.ygyno.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y., Lin H., Meng N., Lu W., Li G., Han Y., Dai X., Xia Y., Song X., Yang S. YL529, a novel, orally available multikinase inhibitor, potently inhibits angiogenesis and tumour growth in preclinical models. Br. J. Pharmacol. 2013;169(8):1766–1780. doi: 10.1111/bph.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z., Wang L., Liu X., Chen C., Wang B., Wang W., Hu C., Yu K., Qi Z., Liu Q. Discovery of a highly selective VEGFR2 kinase inhibitor CHMFL-VEGFR2-002 as a novel anti-angiogenesis agent. Acta Pharm. Sin. B. 2020;10(3):488–497. doi: 10.1016/j.apsb.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudo M., Finn R.S., Qin S., Han K.-H., Ikeda K., Piscaglia F., Baron A., Park J.-W., Han G., Jassem J. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 37.Llovet J., Shepard K.V., Finn R.S., Ikeda M., Sung M., Baron A.D., Kudo M., Okusaka T., Kobayashi M., Kumada H. A phase Ib trial of lenvatinib (LEN) plus pembrolizumab (PEMBRO) in unresectable hepatocellular carcinoma (uHCC): updated results. Ann. Oncol. 2019;30:v286–v287. [Google Scholar]
- 38.Fosgerau K., Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov. Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Li C.M., Haratipour P., Lingeman R.G., Perry J.J.P., Gu L., Hickey R.J., Malkas L.H. Novel peptide therapeutic approaches for cancer treatment. Cells. 2021;10(11):2908. doi: 10.3390/cells10112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erak M., Bellmann-Sickert K., Els-Heindl S., Beck-Sickinger A.G. Peptide chemistry toolbox–Transforming natural peptides into peptide therapeutics. Bioorg. Med. Chem. 2018;26(10):2759–2765. doi: 10.1016/j.bmc.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Craik D.J., Fairlie D.P., Liras S., Price D. The future of peptide‐based drugs. Chem. Biol. Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 42.Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov. Today. 2010;15(1–2):40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Kaspar A.A., Reichert J.M. Future directions for peptide therapeutics development. Drug Discov. Today. 2013;18(17–18):807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 44.van Horssen R., Ten Hagen T.L., Eggermont A.M. TNF-α in cancer treatment: molecular insights, antitumor effects, and clinical utility. Oncol. 2006;11(4):397–408. doi: 10.1634/theoncologist.11-4-397. [DOI] [PubMed] [Google Scholar]
- 45.Newman G., Gonzalez-Perez R.R. Leptin–cytokine crosstalk in breast cancer. Mol. Cell. Endocrinol. 2014;382(1):570–582. doi: 10.1016/j.mce.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crosby L., Casey W., Morgan K., Ni H., Yoon L., Easton M., Misukonis M., Burleson G., Ghosh D.K. Murine J774 macrophages recognize LPS/IFN-g, non-CpG DNA or two-CpG DNA-containing sequences as immunologically distinct. Nitric Oxide. 2010;22(3):242–257. doi: 10.1016/j.niox.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., Bian K., Vallabhaneni S., Zhang B., Wu R.C., O'Malley B.W., Long W. ERK3 promotes endothelial cell functions by upregulating SRC‐3/SP1‐mediated VEGFR2 expression. J. Cell. Physiol. 2014;229(10):1529–1537. doi: 10.1002/jcp.24596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lacal P.M., Graziani G. Therapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacol. Res. 2018;136:97–107. doi: 10.1016/j.phrs.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 49.Uemura A., Fruttiger M., D'Amore P.A., De Falco S., Joussen A.M., Sennlaub F., Brunck L.R., Johnson K.T., Lambrou G.N., Rittenhouse K.D. VEGFR1 signaling in retinal angiogenesis and microinflammation. Prog. Retin. Eye Res. 2021;84 doi: 10.1016/j.preteyeres.2021.100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dias S., Choy M., Alitalo K., Rafii S. Vascular endothelial growth factor (VEGF)–C signaling through FLT-4 (VEGFR-3) mediates leukemic cell proliferation, survival, and resistance to chemotherapy. Blood, The Journal of the American Society of Hematology. 2002;99(6):2179–2184. doi: 10.1182/blood.v99.6.2179. [DOI] [PubMed] [Google Scholar]
- 51.Roskoski R., Jr. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol. Res. 2018;129:65–83. doi: 10.1016/j.phrs.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 52.Heldin C.-H., Lennartsson J., Westermark B. Involvement of platelet‐derived growth factor ligands and receptors in tumorigenesis. J. Intern. Med. 2018;283(1):16–44. doi: 10.1111/joim.12690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available in the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA893416. All other data supporting the findings of this work are available within the paper and its supplementary information files.