Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) has been consistently identified in Kaposi’s sarcomas (KS), body cavity-based lymphomas (BCBL), and some forms of Castleman’s disease. Previous serological tests with KS patient sera have detected lytic-cycle polypeptides from KSHV-infected BCBL cells. We have found that these polypeptides are predominantly encoded by the K8.1 open reading frame, which is present in the same genomic position as virion envelope glycoproteins of other gammaherpesviruses. The cDNA of K8.1 from BCBL-1 cells was found to encode a glycosylated protein with an apparent molecular mass of 37 kDa. K8.1 was found to be expressed during lytic KSHV replication in BCBL-1 cells and was localized on the surface of cells and virions. The results of immunofluorescence and immunoelectron microscopy suggest that KSHV acquires K8.1 protein on its virion surface during the process of budding at the plasma cell membrane. When KSHV K8.1 derived from mammalian cells was used as an antigen in immunoblot tests, antibodies to K8.1 were detected in 18 of 20 KS patients and in 0 of 10 KS-negative control subjects. These results demonstrate that the K8.1 gene encodes a KSHV virion-associated glycoprotein and suggest that antibodies to K8.1 may prove useful as contributory serological markers for infection by KSHV.
Herpesviruses express a number of transmembrane glycoproteins which are virion associated and involved in binding to and entry of the virus into cells (27). These proteins are expressed on the surface of infected cells and on the virion and are generally glycosylated on asparagine (N-linked) and serine (O-linked) residues. Functions of the gB, gH, and gL glycoproteins of the alphaherpesviruses have been well characterized (27). Most of the glycoproteins encoded by the gammaherpesviruses appear to be unique to this subfamily (10, 17, 29). Glycoproteins with function in virus entry have been identified for two gammaherpesviruses, Epstein-Barr virus (EBV) gp340/220 (17, 31) and murine gammaherpesvirus 68 (MHV 68) gp150 (28). EBV gp340/220 is found to serve as a ligand for CR2 (CD21), which is the receptor for the C3d component of the complement complexes (17, 31). Soluble gp340/220 has been shown to block virus infection, indicating an essential role for the gp340/220-CD21 interaction in virus entry (17). The gene for gp340/220 has been mapped near the center of the virus genome in the BamHI L fragment (32). gp340/220 is synthesized abundantly late in the replication cycle of EBV. The mRNA for gp340 is not spliced, whereas the mRNA for gp220 is spliced in frame (32). The gp150 (BPRF1) gene of MHV 68 is in the same relative position and orientation as the gene encoding EBV gp340/220 (28). It is expressed as a transmembrane glycoprotein and is found to be a component of the virus particle (28). In addition, antibodies to EBV gp340/220 and MHV 68 gp150 neutralize the virus in the absence of complement (28, 34).
Many lines of epidemiological evidence suggest an infectious etiology for Kaposi’s sarcoma (KS). DNA sequences of a novel member of the herpesvirus group, called Kaposi’s sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8, have been consistently identified in KS tumors from human immunodeficiency virus-positive and -negative patients (3, 5, 35). KSHV has also been consistently identified in body cavity-based lymphomas and some forms of Castleman’s disease (3, 5). Analyses of KSHV genomic sequences indicate that KSHV is a gammaherpesvirus that is closely related to herpesvirus saimiri (HVS) (24) and rhesus monkey rhadinovirus (6).
Although KSHV can be produced from latently infected body cavity-based lymphoma (BCBL-1) cells treated with phorbol esters, little is know about virion components. Lin et al. (14) have shown that KSHV open reading frame 65 (ORF 65) encodes a small viral capsid antigen (sVCA) and exhibits significant homology to gammaherpesvirus ORFs which encode sVCA components, namely, EBV BFRF3 (2), HVS ORF65 (1), and MHV 68 ORF65 (33). O’Neill et al. have also shown that KSHV ORF26 encodes one of the viral tegument proteins (18). Both ORF65 and ORF26 genes have been shown to be expressed in the late, lytic phase of viral replication in body cavity-based lymphoma cells (14, 18). Both HVS (25) and KSHV (21) have a very limited range of cells that will support productive infection, and essentially no information is available on what these primate rhadinoviruses may use as receptor for entry to cells. Recent work has shown that different members of cellular proteins that includes the poliovirus receptor family and tumor necrosis factor receptor family are used by different individual alphaherpesviruses as receptors for entry (9, 13, 16).
A variety of serologic tests for KSHV infection have been developed (4, 7, 8, 11, 12, 15). Most involve the detection of a latency-associated nuclear antigen (LANA) that is present in infected B-cell cultures; other tests have examined reactivity to lytic cycle antigens (12, 20). While the tests for anti-LANA are highly specific, they detect only about 80% of infected KS patients. The use of several different tests for antibodies to lytic antigens has yielded prevalence estimates similar to those revealed by anti-LANA testing (7, 11, 26). Specifically, antibodies to recombinant sVCA have been shown to correlate with KS in high-risk populations (14).
In this report, we identify and characterize a novel transmembrane glycoprotein encoded by K8.1 of KSHV, which resides at the same genetic locus and orientation as EBV gp340/220 and MHV 68 gp150. K8.1 was found to be expressed in lytic KSHV replication in BCBL-1 cells and was present on the surface of infected cells and virion particles. In addition, immunoblot and immunofluorescence tests demonstrate that antibodies to K8.1 can serve as a serological marker for KSHV infection.
MATERIALS AND METHODS
Cell culture and transfection.
COS-1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. BJAB, BC-1, and BCBL-1 cells were grown in RPMI 1640 supplemented with 10% fetal calf serum. BCBL-1 cells were induced with 20 ng of phorbol-12-tetradecanoyl-13-acetate (TPA) per ml. A DEAE transfection was used for transient expression in COS-1 cells.
Cloning of KSHV K8.1 cDNA from BCBL-1 cells.
The KSHV K8.1 cDNA was cloned by reverse transcriptase-mediated PCR. mRNA from BCBL-1 cells was isolated by using an mRNA isolation kit from Qiagen (Santa Clarita, Calif.). Approximately 0.3 μg of mRNA was reverse transcribed by murine leukemia virus reverse transcriptase in a 20-μl reaction mixture with poly(dT) primer for 20 min at 42°C. As a control, cDNA synthesis was performed without reverse transcriptase. One-microliter aliquots of the same cDNA preparation were used for PCR amplification in a 50-μl volume of final reaction mixture with 100 pmol of specific primers [5′ primer (ATGTTCCTGTATGTTGTTTGC; nucleotides 1 to 21) and 3′ poly(dT)] per liter. The primers used for PCR contain a BamHI site at the 5′ end and an EcoRI site at the 3′ end for subsequent cloning. The PCR-amplified DNA was digested with restriction enzymes BamHI and EcoRI and subcloned into vector pSP72 for DNA sequence analysis. KSHV K8.1 cDNA was completely sequenced with an ABI Prism 377 automatic DNA sequence. For transient expression in COS-1 cells, KSHV K8.1 DNA was subcloned into the pFJ vector (30).
Recombinant K8.1 protein and antibodies.
For purification of recombinant K8.1 protein from Escherichia coli, the K8.1 DNA fragment corresponding to amino acid residues 26 to 142 was amplified by PCR using primers containing BamHI and SalI recognition sequences at the ends and subcloned into BamHI and SalI cloning sites of the pQE-40 expression vector (Qiagen, San Diego, Calif.) with the potential of incorporating six histidines at the amino terminus. The lack of unwanted mutations was confirmed by direct DNA sequencing. When E. coli XL-1 Blue containing plasmid pQE40-K8.1 reached an optical density at 600 nm of approximately 0.6, 1 mM isopropyl-β-d-thiogalactopyranoside was added; cells were harvested 3 h after induction and then solubilized with 6 M guanidine hydrochloride. Due to the presence of the affinity tail, His6-K8.1 protein was purified to virtual homogeneity in one step by Ni2+ chelate affinity chromatography. The purified recombinant His6-K8.1 protein was used to generate polyclonal antibody in New Zealand White rabbits. A Ni2+ chelate affinity column containing K8.1 protein was used to purify the antigen-specific antibodies. Antibody specific for K8.1 was eluted with high pH solution (0.1 M triethylamine [pH 11.5]).
Immunoprecipitation and immunoblotting.
Cells were harvested and lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, 50 mM Tris [pH 7.5]) containing 0.1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin). For protein immunoblots, polypeptides in cell lysates corresponding to 105 cells were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane filter. Immunoblot detection was performed with a 1:2,000 dilution of K8.1 antibody or 1:1,000 dilution of KS patient sera, kindly provided by Ellen Feigel (National Cancer Institute human tumor reagent repository).
Construction of recombinant K8.1-GST baculovirus.
The extracellular portion (amino acids 1 to 196) of the K8.1 gene was fused in frame into glutathione S-transferase (GST) gene. KpnI-XhoI fragments containing K8.1-GST gene were inserted into the KpnI and SalI sites of the baculovirus transfer vector pAcSG1 (Pharmingen, San Diego, Calif.). Vector plasmids were cotransfected into Spodoptera frugiperda Sf9 cells with linearized baculovirus DNA. Four days later, virus-containing supernatants were harvested. The recombinant baculovirus was amplified to obtain a high-titer stock solution. Sf9 cells infected with baculovirus were assayed for expression of recombinant protein by immunoblotting. For routine production of recombinant proteins, 106 cells were infected with 0.2 ml of each baculovirus supernatant, and supernatant was harvested at 48 h postinfection. K8.1-GST fusion protein was purified from supernatant on a glutathione-Sepharose column.
Immunofluorescence.
Cells (105) were washed with phosphate-buffered saline (PBS), centrifuged in a Cytospin at 400 rpm for 4 min, and dried overnight. Cells were permeabilized in acetone at −20°C for 15 min, blocked with 10% goat serum for 30 min, and reacted with anti-K8.1 antibody or human serum diluted 1:100 in PBS containing 10% goat serum for 1 h at room temperature. After incubation, cells were washed extensively with PBS, incubated with 1 μg of fluoresceinated goat F(ab′)2 anti-rabbit total immunoglobulin for 30 min at room temperature, and washed three times with PBS. Immunofluorescence was detected with an Olympus immunofluorescence microscope.
Immunoelectron microscopy.
Cytospins of TPA-stimulated BCBL-1 cells were fixed with 4% paraformaldehyde–0.1% glutaraldehyde in 0.1 M phosphate buffer, permeabilized, and washed for 15 min in 50 mM glycine-bovine serum albumin with 10% normal goat serum. The slides were incubated overnight in an antigen-specific rabbit polyclonal anti-K8.1 antibody at 4°C and washed in PBS-bovine serum albumin before application of a 10-mm-diameter gold-conjugated goat F(ab′)2 anti-rabbit immunoglobulin G (IgG) antibody overnight at 4°C. After being washed in PBS, the cells were fixed in 1% glutaraldehyde for 10 min and washed in ultrapure water (10 MΩ) before a 20-min silver enhancement (Electron Microscopy Sciences, Fort Washington, Pa.) followed by additional ultrapure water wash. The cells were fixed in 1% osmium tetroxide, dehydrated in graded ethanol, and rinsed with three changes of Eponate 12 resin (Ted Pella, Inc., Redding, Calif.) before placement of inverted microcentrifuge tubes partially filled with resin onto the cells. The resin was cured at 60°C for 24 h, and the tubes with the cells embedded in the resin were separated from the slide by gentle heating of the slides on a hot plate. Sections were cut on a Sorvall Porter-Blum MT-2 ultramicrotome and then stained with methanolic uranyl acetate and SATO’s lead stain. The sections were photographed on a JEOL 1010 electron microscope.
FACS analysis.
Cells (5 × 105) were washed with RPMI medium containing 10% fetal calf serum and incubated with antigen-specific K8.1 antibody followed by fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG for 30 min at 4°C. After washing, each sample was fixed with 1% formalin solution, and fluorescence-activated cell sorting (FACS) analysis was performed with a FACScan (Becton Dickinson Co., Mountain View, Calif.).
RESULTS
Cloning and sequence analysis of K8.1.
Initial DNA sequence analysis of the KSHV genome did not detect an ORF between K8 and ORF52 (24). This ORF was subsequently referred to as K8.1 (19). K8.1 is located at the same genetic locus and orientation as the genes of EBV gp340/220 and MHV 68 gp150 virion-associated proteins. KSHV K8.1 cDNAs were cloned from mRNA of TPA-stimulated BCBL-1 cells by reverse transcriptase-mediated PCR as described in Materials and Methods, using an oligo(dT) primer at the 3′ and a gene-specific primer which overlapped the predicted translational initiational site at the 5′ end of the gene (24). Ten independent cDNA clones were isolated and sequenced; these revealed a splice site (nucleotides 76338 to 76433 of the published KSHV sequence [22]) which removed 94 nucleotides (Fig. 1). The spliced product was predicted to encode an ORF of 228 amino acids for the K8.1 protein. None of 10 clones revealed unspliced mRNA from this region with this orientation. An unspliced mRNA of K8.1 has been detected at low abundance (19).
FIG. 1.
Primary amino acid sequence analysis of K8.1 from BCBL-1 cells. The empty box indicates the signal peptide sequence, asterisks indicate the predicted N-glycosylation sites, and the filled box indicates the hydrophobic region. The underlined sequences indicate the removed nucleotide sequences after splicing.
The putative K8.1 protein encoded by this mRNA has a signal peptide sequence at the amino terminus, an extracellular domain, a transmembrane domain, and a short cytoplasmic tail at the carboxyl terminus (Fig. 1). The position of a potential signal peptide cleavage site is indicated in Fig. 1; cleavage of the signal peptide would yield a mature core protein of 201 residues with a predicted molecular mass of 22 kDa for an unmodified product. The second hydrophobic domain (residues 197 to 217) was long enough to be a membrane-spanning domain. Four potential N-linked glycosylation sites were present in the extracellular domain (Fig. 1). Analysis of the amino acid composition of the protein revealed a high content of serine and threonine residues (21%), which suggests the potential for O-linked glycosylation. These results indicate that K8.1 has structural motifs indicative of a type I membrane-bound glycoprotein.
K8.1 is a glycoprotein.
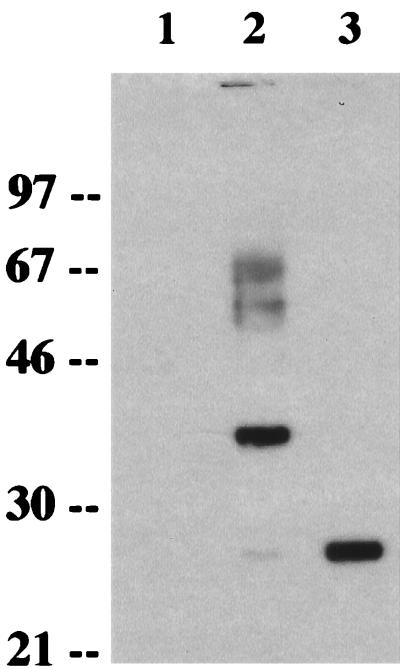
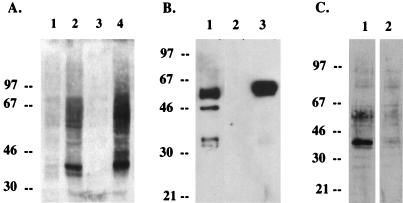
To analyze the K8.1 gene product, we generated a rabbit polyclonal antibody against a purified bacterial His6-K8.1 fusion protein which contained the putative extracellular portion of K8.1 as described in Materials and Methods. When lysates of COS-1 cells prepared 2 days following transfection with expression vector containing K8.1 cDNA were used for an immunoblot assay with the anti-K8.1 antibody, the antibody reacted specifically with a 37-kDa protein as the major species and a 27-kDa protein as a very minor species (Fig. 2, lane 2). Also, 60- to 65-kDa proteins were detected by the anti-K8.1 antibody. In contrast, these proteins were not detected in control COS-1 cells not expressing the K8.1 gene (Fig. 2, lane 1). These results suggested posttranslational modifications of the K8.1 protein. Four potential sites for N-linked glycosylation (NX[S/T]) in the extracellular region likely contributed to the slow migration of the K8.1 protein in SDS-PAGE. To examine this possibility, cells were treated with tunicamycin, which inhibits the addition of N-linked oligosaccharide to glycoprotein. In the presence of tunicamycin, the 37-kDa protein disappeared and the 27-kDa protein was the major band detected (Fig. 2, lane 3). These results indicate that the 27- and 37-kDa proteins are a precursor and an N-glycosylated form of K8.1, respectively.
FIG. 2.
Identification and glycosylation of K8.1. After transfection, COS-1 cells were incubated in the presence of tunicamycin (20 μg/ml) overnight. Cell lysates were used for immunoblot assay with an anti-K8.1 antibody. Lane 1, pFJ; lane 2, pFJ-K8.1; lane 3, pFJ-K8.1 with tunicamycin treatment. Polypeptides from cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane, and reacted with the anti-K8.1 antibody. Sizes are indicated in kilodaltons.
Localization of KSHV K8.1.
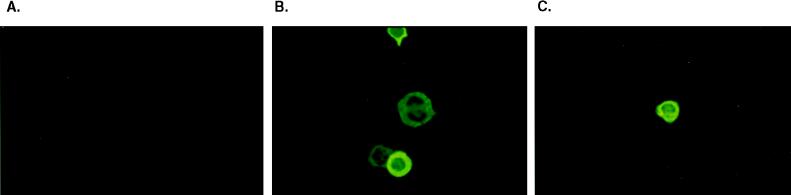
The localization of K1 was determined by indirect immunofluorescence tests. TPA-stimulated BCBL-1 cells were permeabilized with acetone and stained with the anti-K8.1 antibody. The staining pattern suggested that K8.1 was principally associated with the plasma membrane (Fig. 3B). Since the anti-K8.1 antibody was generated against the putative extracellular region of K8.1, nonpermeabilized BCBL-1 cells were also used for immunofluorescence tests. A similar staining pattern with anti-K8.1 antibody was seen regardless of whether the BCBL-1 cells were permeabilized (Fig. 3B and C). These results demonstrate that K8.1 is located principally on the plasma membrane and that the amino terminus of K8.1 was exposed to the extracellular region.
FIG. 3.
Localization of K8.1. After stimulation of TPA, permeabilized (A and B) or nonpermeabilized (C) BCBL-1 cells were treated with preimmune serum (A) or anti-K8.1 antibody (B and C) and then with fluoresceinated goat anti-rabbit total immunoglobulin. Immunofluorescence was detected with an Olympus immunofluorescence microscope. Magnification, ×332.
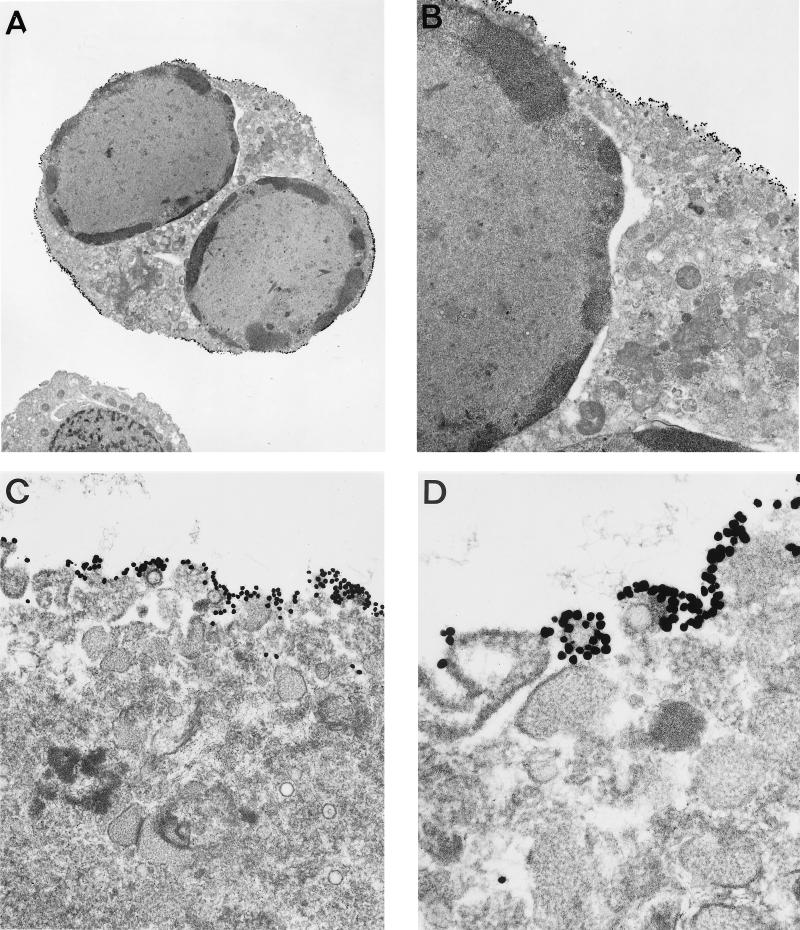
To confirm the localization of K8.1 and examine whether K8.1 was present on the surface of virions, we used immunoelectron microscopic visualization. BCBL-1 cells stimulated with TPA for 92 h were fixed, embedded in resin, and cut into sections in preparation for immunoelectron microscopy. The sections were then reacted with the anti-K8.1 antibody and an anti-rabbit antibody conjugated to gold particles. The K8.1 protein was localized almost exclusively at the periplasmic membrane (Fig. 4A and B). At high magnification, K8.1 was also detected on the surfaces of KSHV virion particles released from the surface of TPA-stimulated BCBL-1 cells (Fig. 4C and D). In contrast, it was not detected on the surface of KSHV virion particles which were in the cytoplasm (Fig. 4C). In addition, the K8.1 protein was detected in cells which produced KSHV virion particles but not detected in cells which did not produced KSHV virion particles (data not shown). The N-terminal domain of K8.1 was localized to the outside of infected BCBL-1 cells and virion membranes, demonstrating that K8.1 is indeed a type I membrane glycoprotein. The distribution of the anti-K8.1 antibody suggested that K8.1 was a component of the surface of infected cells and of virion particles, that the protein protruded from the surface of the particles, and that virions acquired K8.1 protein in the process of budding from the plasma membrane.
FIG. 4.
Immunoelectron microscopy of TPA-stimulated BCBL-1 cells with anti-K8.1 antibody. TPA-stimulated BCBL-1 cells were fixed, embedded in low-temperature resin, sectioned, and labeled with a combination of antigen-specific anti-K8.1 antibody and 5-nm-diameter colloidal gold. The sections were analyzed with a JEOL 1010 electron microscope. Magnifications: (A) ×3,600; (B) ×12,000; (C) ×36,000; (D) ×72,000.
Lytic expression of K8.1 protein.
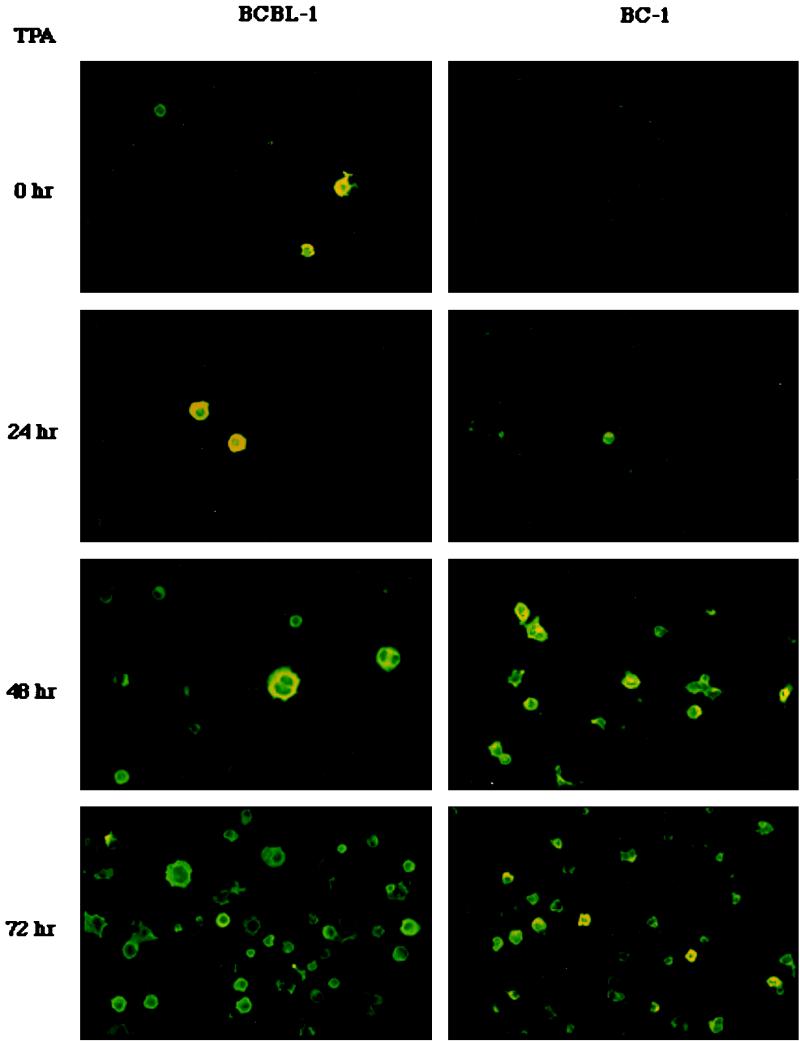
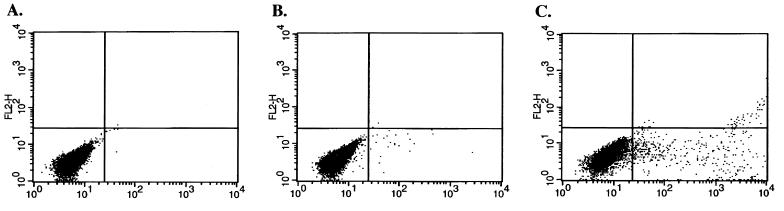
To examine the level of K8.1 expression, BCBL-1 and BC-1 cells were stimulated with TPA for 24, 48, and 72 h and then subjected to immunofluorescence staining with anti-K8.1 antibody as described above (Fig. 3). Unstimulated BCBL-1 and BC-1 cells were used as controls. K8.1 was not detected in unstimulated BC-1 cells. In contrast, the level of K8.1 gradually increased in BC-1 cells during the time course of TPA stimulation: 1 to 2% at 24 h, 10 to 15% at 48 h, and 30% at 72 h (Fig. 5). Since BCBL-1 cells exhibit approximately 1% spontaneous lytic viral replication (22), K8.1 expression was detected at a low level before the TPA stimulation in these cells (Fig. 5). The level of K8.1 also gradually increased during TPA stimulation of the BCBL-1 cells: 2 to 5% at 24 h, 15 to 20% at 48 h, and 30% at 72 h (Fig. 5). A sustained or slightly increased level of K8.1 expression was detected after 96 h of TPA stimulation (data not shown). Since K8.1 was expressed on the cell surface, the level of expression of K8.1 was further examined by FACS analysis with antigen-specific anti-K8.1 antibody. Less than 5% of unstimulated BCBL-1 cells exhibited surface expression of K8.1, whereas over 30% of BCBL-1 cells exhibited surface expression of K8.1 after TPA stimulation (Fig. 6). These results indicate that K8.1 is a lytic gene product whose expression is greatly stimulated by TPA treatment.
FIG. 5.
Lytic expression of K8.1. BC-1 and BCBL-1 cells were stimulated with TPA for 0, 24, 48, and 72 h, permeabilized with methanol-ethanol, reacted with rabbit anti-K8.1 antibody, and then incubated with 1 μg of fluoresceinated goat F(ab′)2 anti-mouse total immunoglobulin. Immunofluorescence was detected with an Olympus immunofluorescence microscope.
FIG. 6.
FACS analysis of surface expression of K8.1. Unstimulated (B) or TPA-stimulated (C) BCBL-1 cells were reacted with purified antigen-specific K8.1 antibody followed by FITC-conjugated anti-rabbit IgG. As a negative control, unstimulated BCBL-1 cells (A) were stained with FITC conjugated anti-rabbit IgG without incubation with the anti-K8.1 antibody.
Detection of K8.1 by KS-positive human sera.
Human KS-positive sera reacted predominantly with a 37-kDa protein as well as other proteins on immunoblots with lysates from TPA-stimulated BCBL-1 cells (Fig. 7A, lane 2). This 37-kDa protein comigrated with K8.1 protein produced by transfection of cloned cDNA in COS-1 cells (Fig. 7A, lane 4). To determine whether the 37-kDa protein detected by KS-positive human sera was encoded by the K8.1 gene, we attempted to deplete K8.1-specific antibodies from the KS-positive human sera using purified glycosylated K1 fusion protein. The amino-terminal region from residues 1 to 196 of K8.1, which corresponds to the putative extracellular region, was fused in frame to GST protein, and the K8.1-GST fusion gene was expressed in insect cells, using a baculovirus expression system. Purified glycosylated K8.1-GST protein from the supernatant migrated with an apparent molecular mass of 50 kDa in SDS-PAGE (Fig. 7B). KS-positive human serum was depleted of K8.1-specific antibodies by mixing with K8.1-GST conjugated with Sepharose beads or mixing with GST conjugated with Sepharose beads. The treated sera were then used for immunoblot analysis with TPA-stimulated BCBL-1 cell lysates. While the 37-kDa protein was readily detected by KS-positive serum preincubated with GST protein, the reactivity of human KS-positive sera with the 37-kDa protein was specifically and dramatically reduced by preincubation with K8.1-GST fusion protein (Fig. 7C). These results demonstrate that the 37-kDa K8.1 protein in BCBL-1 cell lysates is a major protein reactive to human KS-positive sera by immunoblotting.
FIG. 7.
Identification of K8.1 as an immunogen detected by KS-positive human sera. (A) Immunoblot analysis with KS-positive human sera. Lane 1, BCBL-1 cell lysates without treatment; lane 2, BCBL-1 cell lysates with treatment of TPA for 72 h; lane 3, COS-1 cells transfected with pFJ vector; lane 4, COS-1 cells transfected with pFJ-K8.1. KS-positive human sera were used at 1:1,000 dilution. Enhanced chemiluminescence was used to detect proteins. (B) Expression and purification of K8.1-GST fusion protein from S. frugiperda Sf9 cells, using recombinant baculovirus. Sf9 cells were infected with recombinant baculovirus K8.1-GST. After 72 h, supernatant was used for purification of K8.1-GST protein on a glutathione-Sepharose column. Whole-cell lysates (lane 1), purified proteins from mock-infected insect cells (lane 2), and purified protein from recombinant K8.1-GST baculovirus-infected insect cells (lane 3) was subjected to immunoblot analysis with anti-K8.1 antibody. (C) Depletion of K8.1-specific antibody from KS-positive human sera. Lysates from TPA-stimulated BCBL-1 cells were used for immunoblot analysis with KS-positive human serum mixed with GST beads (lane 1) or with KS-positive human serum mixed with K8.1-GST beads (lane 2). Sizes are indicated in kilodaltons.
Prevalence of antibodies to KSHV K8.1.
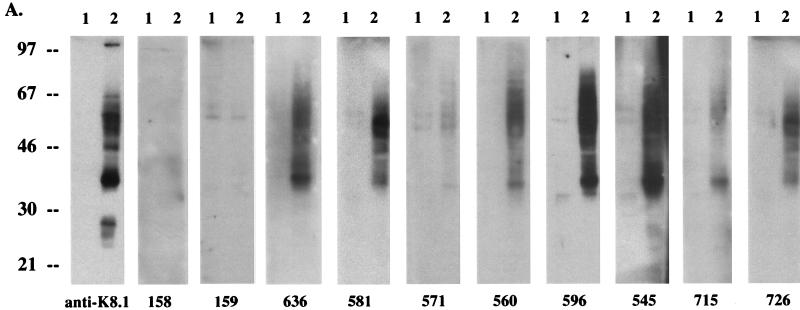
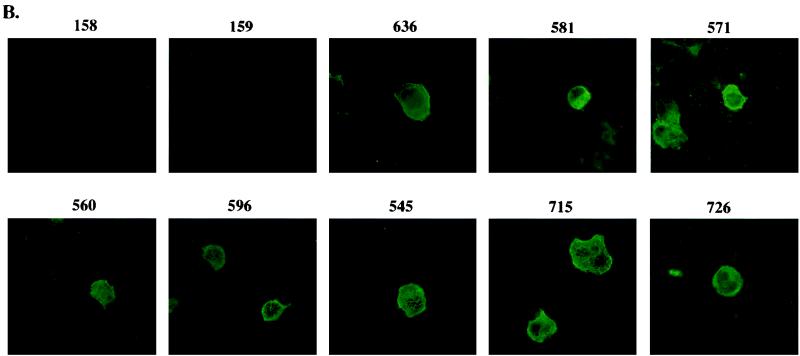
We next examined the ability of K8.1 to serve as a serological marker to detect KSHV infection. K8.1 expressed in COS-1 cells was used as an antigen in immunoblotting and immunofluorescence tests with KS-positive human sera. A rabbit anti-K8.1 antibody was used as a positive control for both assays. Immunoblot screening with 20 KS-positive human sera and 10 KS-negative control human sera showed that antibodies to K8.1 were detected in 90% of 20 KS patients and in none of 10 KS-negative control subjects (Fig. 8A and Table 1). No such protein was detected in control cells lacking the K8.1 gene in COS-1 cells with KS-positive and KS-negative human sera. In addition, different levels of reactivity against the K8.1 protein were detected among KS-positive sera (Fig. 8A and Table 1). To further investigate the ability of K8.1 to serve as an antigen to detect KSHV infection, COS-1 cells transfected with a K8.1 expression vector were used for immunofluorescence tests. Transfected COS-1 cells were permeabilized with acetone and reacted with human sera at a 1:100 dilution. The rabbit anti-K8.1 antibody was used as a positive control. Consistent with the results obtained from immunoblot screening, 80% of KS-positive human sera showed reactivity to the K8.1 protein in immunofluorescence tests (Fig. 8B and Table 1). The staining pattern of K8.1 with KS-positive sera was essentially the same as that obtained with the rabbit anti-K8.1 antibody (data not shown).
FIG. 8.
Evaluation of anti-K8.1 antibody as a serological marker for KSHV infection. (A) Immunoblot screening. COS-1 cells were transfected with pFJ vector (lane 1) or with pFJ-K8.1 (lane 2). Polypeptides from these cells were resolved by SDS-PAGE in reducing conditions and reacted with 1:1,000-diluted KS-negative control human sera (158 and 159) or KS-positive human sera (636, 581, 571, 560, 596, 545, 715, and 726). Rabbit anti-K8.1 antibody was used as a positive control. (B) Immunofluorescence tests. COS-1 cells transfected with pFJ-K8.1 were permeabilized with methanol-ethanol, reacted with 1:100-diluted human sera as described above, and then incubated with 1 μg of fluoresceinated goat F(ab′)2 anti-human total immunoglobulin. Immunofluorescence was detected with an Olympus immunofluorescence microscope.
TABLE 1.
Serological screen of anti-K8.1 antibody in KS patients
| Source of sera | ||||||||
|---|---|---|---|---|---|---|---|---|
| Immunoblot assay
|
Immunofluoresence assay
|
|||||||
| No. of patients with intensitya of:
|
No. of patients with antibody to K8.1 | No. of patients with intensity of:
|
No. of patients with antibody to K8.1 | |||||
| − | + | ++ | +++ | − | + | |||
| KS-positive patients (n = 20) | 2 | 3 | 4 | 10 | 18 | 4 | 16 | 16 |
| KS-negative controls (n = 10) | 10 | 0 | 0 | 0 | 0 | 9 | 1 | 1 |
−, negative; +, very weak; ++, weak; +++, strong.
DISCUSSION
In this report, we have demonstrated that viral polypeptides detected by KS-positive human sera are encoded by the K8.1 ORF which is present in the same genomic position and orientation as ORFs for virion envelope glycoproteins of other gammaherpesviruses. The glycosylated K8.1 protein is expressed during lytic KSHV replication in BCBL-1 cells and is localized on the surface of infected cells and virion particles. Antibodies to K8.1 were detected by immunoblotting in 18 of 20 KS patients and in 0 of 10 KS-negative control subjects. Our results demonstrate that the K8.1 gene encodes a KSHV virion-associated glycoprotein and suggest that antibodies to K8.1 can serve as a serological marker for infection with KSHV.
In gammaherpesviruses for which sequence data are available, a potential glycoprotein gene has been found at the same locus as the gene for K8.1: gp340/220 of EBV (17), gp150 of MHV 68 (28), and ORF51 of HVS (1). In EBV, gp340/220 has been shown to be responsible for the binding of EBV to B lymphocytes via the CD21 surface protein and mediating the initial step of virus infection (17, 31). In addition to virus absorption, this interaction leads to the activation of a signal transduction pathway, resulting in an increased CD19 tyrosine phosphorylation level, which ultimately facilitates efficient expression of viral genes following infection (17). The motif on gp340/220 that is responsible for CD21 interaction is EDPGFFNVE, which is located near the amino terminus (17). In MHV 68, gp150 has been identified as a virion envelope protein and antibodies to gp150 neutralize virus infectivity (28). This finding suggests that gp150 of MHV 68 is involved in the binding of the virus to a cellular receptor (28). However, K8.1 of KSHV and gp150 of MHV 68 do not contain sequence homology with the CD21-interacting motif of gp340/220 of EBV. While KSHV and MHV 68, like EBV, are tropic for B cells, they may employ the different cellular molecules for their entry.
While our studies isolated a single spliced form of K8.1, an alternatively spliced form of the K8.1 gene has been detected as a minor population in TPA-stimulated BCBL-1 cells (19). These results suggest that multiple forms of K8.1 gene products may exist, similar to what is observed for the EBV gp340/220 (32). However, whether the alternatively spliced form of K8.1 indeed encodes a protein has not yet been studied.
Herpesviruses have been typically found to bud from the nuclear membrane (23). However, it has been suggested that herpes simplex virus may often lose this envelope coat by fusion with cytoplasmic membranes (23). Our data quite clearly indicate that KSHV acquires K8.1 by a process of budding from the plasma membrane. Further studies are required to elucidate the detailed mechanisms of KSHV envelopment and release.
Of sera from humans with KS, 80 to 90% reacted with mammalian-expressed K8.1 protein by immunoblot or immunofluorescence assays. In contrast, all KS-negative control sera used in this study did not detect K8.1 in the immunoblot screening, and 90% of KS-negative control sera did not detect K8.1 in the immunofluorescence tests. These observations suggest that antibodies to K8.1 may be useful serological marker for KSHV infection. However, as found in serological tests with lytic antigens and LANA by other investigators (4, 7, 8, 11, 12, 15), a significant proportion (10 to 20%) of sera from human with KS did not detect the K8.1 protein in our immunoblot and immunofluorescence tests. Such incomplete sensitivity may be accounted by the insensitivity of the test systems, the requirement of conformational epitopes on K8.1, the large genetic complexity of KSHV, or genetic polymorphism of K8.1. Any of these possibilities would require the use of multiple antigens or testing formats in order to generate a highly sensitive assay system to screen human sera for KSHV infection. However, it is also possible that some human infected with KSHV scant or no antibodies to the virus because of immune deficiency, antigen excess, or other factors.
ACKNOWLEDGMENTS
We thank E. Feigel for providing KS-positive sera and L. Alexander and R. Means for critical reading of the manuscript. We also thank J. Newton for manuscript preparation, Kristen Toohey for photography support, and Maryann DeMaria for FACS analysis.
This work was supported by Public Health Service grants CA31363, AI38131, and RR00168.
ADDENDUM IN PROOF
After the manuscript was submitted, Chandran et al. (B. Chandran, C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat, Virology 249:140–149, 1998) published similar results.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M, Coleman H, Fleckenstein B, Honness R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Séguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Chandran B, Smith M S, Koelle D M, Corey L, Horvat R, Goldstein E. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology. 1998;243:208–217. doi: 10.1006/viro.1998.9055. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 6.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to human Kaposi sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 8.Gao S-J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herepesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 9.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman G J, Lazarowitz S G, Hayward S D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralising antigen. Proc Natl Acad Sci USA. 1980;77:2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedes D, Operaiski E, Busch M, Kohn R, Flood J, Ganem D E. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 12.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Hereps simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S-F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi’s sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S-J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 16.Montgomery R I, Warner M S, Lum B J L, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 17.Nemerow G R, Houghton R A, Moore M D, Cooper N R. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2) Cell. 1989;56:369–377. doi: 10.1016/0092-8674(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 18.O’Neill E, Dougland J L, Chien M-L, Garcia J V. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 82-kilodalton protein expressed in a body cavity-based lymphoma cell line. J Virol. 1997;71:4791–4797. doi: 10.1128/jvi.71.6.4791-4797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raab M-S, Albrecht J-C, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renne R, Blcckbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Ganem D. Lytic growth of Kaposi’s sarcoma-associate herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 23.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 24.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi’s sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmer B, Alt M, Buckreus I, Berthold S, Fleckenstein B, Platzer E, Grassmann R. Persistence of selectable herpesvirus saimiri in various human haematopoietic and epithelial cell lines. J Gen Virol. 1991;72:1953–1958. doi: 10.1099/0022-1317-72-8-1953. [DOI] [PubMed] [Google Scholar]
- 26.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, deRuiter A, Hatzakis A, Tedder S, Weller I V D, Weiss R A, Moore P S. Prevalence of Kaposi’s sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 27.Spear P G. The herpesviruses. Vol. 3. New York, N.Y: Plenum Publishing Corp.; 1985. [Google Scholar]
- 28.Stewart J P, Janjua N J, Pepper S D V, Bennion G, Mackett M, Allen T, Nash A A, Arrand J R. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J Virol. 1996;70:3528–3535. doi: 10.1093/benz/9780199773787.article.b00034574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunil-Chandra N P, Efstathiou S, Nash A A. Interactions of murine gammaherpesvirus 68 with B and T cell lines. Virology. 1993;193:825–833. doi: 10.1006/viro.1993.1191. [DOI] [PubMed] [Google Scholar]
- 30.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 32.van Grunsven W M, van Heerde E C, de Haard H J, Spaan W J, Middeldorp J M. Gene mapping and expression of two immunodominant Epstein-Barr virus capsid proteins. J Virol. 1993;67:3908–3916. doi: 10.1128/jvi.67.7.3908-3916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virgin H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whang Y, Silberkland M, Morgan A, Munshi S, Lenny A, Kieff E. Expression of the Epstein-Barr virus gp350/220 gene in rodent and primate cells. J Virol. 1987;61:1796–1807. doi: 10.1128/jvi.61.6.1796-1807.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]