Abstract
Olorofim is a new antifungal in clinical development which has a novel mechanism of action against dihydroorotate dehydrogenase (DHODH). DHODH form a ubiquitous family of enzymes in the de novo pyrimidine biosynthetic pathway and are split into class 1A, class 1B and class 2. Olorofim specifically targets the fungal class 2 DHODH present in a range of pathogenic moulds. The nature and number of DHODH present in many fungal species have not been addressed for large clades of this kingdom. Mucorales species do not respond to olorofim; previous work suggests they have only class 1A DHODH and so lack the class 2 target that olorofim inhibits. The dematiaceous moulds have mixed susceptibility to olorofim, yet previous analyses imply that they have class 2 DHODH. As this is at odds with their intermediate susceptibility to olorofim, we hypothesised that these pathogens may maintain a second class of DHODH, facilitating pyrimidine biosynthesis in the presence of olorofim. The aim of this study was to investigate the DHODH repertoire of clinically relevant species of Mucorales and dematiaceous moulds to further characterise these pathogens and understand variations in olorofim susceptibility. Using bioinformatic analysis, S. cerevisiae complementation and biochemical assays of recombinant protein, we provide the first evidence that two representative members of the Mucorales have only class 1A DHODH, substantiating a lack of olorofim susceptibility. In contrast, bioinformatic analyses initially suggested that seven dematiaceous species appeared to harbour both class 1A-like and class 2-like DHODH genes. However, further experimental investigation of the putative class 1A-like genes through yeast complementation and biochemical assays characterised them as dihydrouracil oxidases rather than DHODHs. These data demonstrate variation in dematiaceous mould olorofim susceptibility is not due to a secondary DHODH and builds on the growing picture of fungal dihydrouracil oxidases as an example of horizontal gene transfer.
Introduction
Invasive fungal infections are a global public health issue: 1.5 million deaths are attributed to fungal disease while over one billion people are affected by it [1]. Market-ready novel antifungal drugs are lacking while the vulnerable population size increases and resistance to current therapies rises due to various geo-ecological and socio-economic factors [2, 3]. Olorofim is a new oral systemic antifungal from the novel class of orotomides [4], currently in phase III clinical development for the treatment of invasive aspergillosis (ClinicalTrials.gov Identifier: NCT05101187) [5, 6]. Due to its novel mechanism of action, olorofim has potent activity against azole-resistant Aspergilli [7] and organisms intrinsically resistant to other antifungals, such as Aspergillus section Terrei [8]. Additionally, it has broad anti-mould activity [6, 9], covering some species that currently have limited or no treatment options, including Lomentospora, Scedosporium, and Scopulariopsis spp. and members of the Rasamsonia agrillacea species complex [10–13]. The spectrum of activity also extends to dimorphic fungi of the genera Coccidioides, Blastomyces, Histoplasma, Talaromyces and Sporothrix but not Candida. [14–16]. Olorofim also lacks activity against Cryptococcus spp. and the Mucorales [17]. Various species of pigmented moulds, termed the dematiaceous moulds, have been subjected to in vitro susceptibility testing with mixed results. Madurella mycetomatis [18] and Phaeoacremonium parasiticum [17] are susceptible to olorofim but Alternaria alternata [9] and the black yeast Exophiala dermatitidis [19] are not. Most of the aforementioned species are recognised as priority fungal pathogens by the World Health Organisation [20].
Olorofim acts through the selective inhibition of fungal dihydroorotate dehydrogenase (DHODH) [4]. DHODH are ubiquitous flavoenzymes that catalyse the fourth step in the de novo pyrimidine biosynthetic pathway. Using a flavin mononucleotide (FMN) cofactor and an electron acceptor, they oxidise dihydroorotate (DHO) to orotate in the production of pyrimidines. DHODH vary in structure, electron acceptor and subcellular localisation: these characteristics dictate their categorisation into one of four classes, summarised in Table 1.
Table 1. Summary of DHODH.
| Class | Form | Electron Acceptor | Localisation | Catalytic base | Species distribution |
|---|---|---|---|---|---|
| 1A | Homodimer | FMN | Cytosol | Cysteine | Gram-positive bacteria, such as Streptococcus mutans [21]; Parasites such as Leishmania major [22] and Trypanosoma spp. [23, 24]; Saccharomyces cerevisiae [25]; Mucorales, predicted [4] |
| 1B | Heterotetramer | NAD+ | Cytosol | Cysteine | Gram-positive bacteria such as Bacillus subtilis [26], Clostridium oroticum [27] and Bifidobacterium bifidum [28] |
| 1S | Heterodimer | Coenzyme Q, O2 | Cytosol | Serine | Sulfolobus solfataricus [29] |
| 2 | Monomer | Quinones | Cell membrane [30] or inner mitochondrial membrane [31] | Serine | Gram-negative bacteria [32]; Plasmodium spp. [33]; Plants [34]; Insects [31]; Mammals [35]; Most fungi, including Aspergillus spp. [36], Candida spp. [37]; Dematiaceous moulds, predicted [4] |
Notably, the bacteria Lactococcus lactis [38, 39] and Enterococcus faecalis [40, 41] harbour both class 1A and class 1B DHODH while the yeasts Lachancea kluyveri [42, 43] and Kluyveromyces marxianus [44] possess both functional class 1A and class 2 DHODH, though K. marxianus has not been experimentally verified. Class 2 DHODH interact with the respiratory chain in some but not all species, such as Anaeromyces robustus [44].
DHODH is a druggable target for therapeutic development in various species. Teriflunomide inhibits human DHODH [45] for rheumatoid arthritis [46] and multiple sclerosis treatment [47]. A novel human DHODH inhibitor MEDS433, based on brequinar, appears to prevent influenza virus replication [48]. Plasmodium DHODH has been validated as a new target for antimalarial development; DSM265 is the first to reach clinical development [49]. Olorofim selectively targets the fungal class 2 DHODH and is predicted to bind in the quinone channel, preventing ubiquinone-mediated reoxidation of FMN and thus inhibiting the oxidation of DHO to orotate [4]. Disruption of de novo pyrimidine biosynthesis has pleiotropic effects, altering cell wall composition, increasing vacuolar size, perturbing cell division and ultimately cell lysis [50, 51]. Variation in class 2 DHODH protein sequence has been shown to account for the differences in susceptibility; Candida spp. vary at residues important for olorofim interaction compared to Aspergillus fumigatus and increased evolutionary distance from susceptible species is concomitant with loss of olorofim susceptibility [4].
While an attractive drug target, the cellular DHODH repertoires of clinically important fungi have not been fully elucidated. Mucorales are predicted to have class 1A DHODH, which would substantiate their lack of olorofim susceptibility [4]. Incidence of mucormycosis has risen due to opportunistic infection caused by COVID-19 and associated treatments [52]. Dematiaceous mould DHODH are not well characterised, but preliminary analyses suggested class 2 enzymes are present in the majority of species [4]. However, olorofim susceptibility is generally lower than Aspergillus spp. with some species responding to the drug in vitro and some not. These genera can infect immunocompetent individuals, causing chromoblastomycosis (eg. Fonsecaea, Phialophora.), phaeohyphomycosis (eg. Alternaria, Cladophialophora, Exophiala, Phialophora, Phaeoacremonium) or eumycetoma (Madurella, Cladophialophora, Phialophora), the latter of which is designated as a Neglected Tropical disease by the World Health Organisation. It is possible that the reduction of olorofim activity in these species is due to the presence of the additional gene products that resemble class 1A DHODH, which may support pyrimidine biosynthesis and cell survival while the class 2 is inhibited. Work published during the course of this study revealed a class 1A DHODH-like gene in A. alternata to be a dihydrouracil oxidase (DHUO) [53], a poorly characterised type of enzyme described in only one other genus of fungi [54]. DHUO have been seen to convert dihydrouracil (DHU) and dihydrothymine to uracil and thymine, respectively, resembling a step that is the reverse of the initial reaction in the reductive pathway of pyrimidine degradation. The biological context and significance of the conversion of DHU to uracil remains to be elucidated. Deeper understanding of the mechanisms by which fungi regulate pyrimidine levels is needed to inform better drug discovery for fungal targets, in addition to broadening basic knowledge on increasingly problematic pathogens.
While tools are evolving to study gene function in some pathogenic moulds, such as Rhizopus microsporus [55] and R. arrhizus [56], Mucor lusitanicus [57] and E. dermatitidis [58], methods are still lacking for the many species in our study. Further, existing methodologies to knock out genes vary greatly in terms of mechanism, reagents and efficiency. To investigate the functionality of metabolic genes from nine different fungal genera, we opted to utilise the genetically tractable model Saccharomyces cerevisiae to obtain reliable and reproducible transformants and results in a time-efficient manner. We set out to understand the DHODH repertoire of clinically important Mucorales and dematiaceous mould species. Bioinformatic analyses of two representative Mucorales species revealed only class 1A DHODH in their proteomes and a yeast complementation study verified these putative proteins could function as DHODH. Biochemical experiments showed these enzymes to be active flavoproteins that use fumarate as electron acceptor, confirming their identity as class 1A DHODH. The same experimental pipeline was applied to seven representative species of dematiaceous fungi. All proteomes assessed appeared to harbour both class 1A-like and class 2-like DHODH; further examination was limited to the putative class 1A-like proteins to understand their importance in the context of olorofim. In contrast to the Mucorales DHODH, complementation analyses and biochemical studies revealed these enzymes to be DHUO, nullifying the hypothesis that these dematiaceous moulds have active class 1A DHODH.
Materials and methods
Protein sequence homology analyses
S. cerevisiae Ura1 and A. fumigatus PyrE protein sequences were used as class 1A and class 2 DHODH sequences, respectively, to query the proteomes of representative clinically important fungi of interest using BLAST. For E. dermatitidis, the DHODH sequence from T. cruzi was used as a query for class 1A orthologs to ensure any class 1A-like sequence was identified in this proteome. Protein sequences were taken from the reference genome for each species. DHODH classification was assigned using Interpro [59]. For phylogenetic analyses, FASTA sequences were submitted to phylogeny.fr (advanced mode) [60]. Sequences were aligned using MUSCLE (full mode) [61] and curated with GBlocks (default settings) [62]. Phylogeny was established by the maximum likelihood method in PhyML, using the approximate likelihood ratio test set to SH-like with WAG substitution [63, 64]. Resultant trees were visualized in TreeDyn [65]. Multiple sequence alignments were performed using MultAlin [66] and visualised graphically using ESPript [67]. All Entrez protein accession numbers used for phylogenetic analysis are listed in S1 Table.
Yeast cloning and plasmid construction
Full-length DHODH genes for heterologous expression in S. cerevisiae were synthesised in plasmids (Eurofins) or gene blocks (IDT) as native (T. cruzi, L. kluyveri, A. fumigatus, C. bantiana, E. dermatitidis, F. pedrosoi, P. americana, A. alternata), E. coli codon optimised (R. arrhizus, M. circinelloides, A. alternata, M. mycetomatis, P. minimum) or S. cerevisiae codon optimised (A. alternata) cDNAs with overhangs for downstream cloning. The A. fumigatus DHODH was cloned from cDNA as described previously [4]. The S. cerevisiae DHODH gene, was amplified from a BG1805 plasmid containing the URA1 ORF (Horizon Discovery Ltd). All PCR was performed using Phusion HSII polymerase (Thermofisher Scientific). A pYCP shuttle plasmid was designed (VectorBuilder) for heterologous DHODH expression in S. cerevisiae and contained the following elements: the pUC origin and an ampicillin resistance gene for replication and maintenance in bacteria, the copper-inducible CUP1 promoter [68], an N-terminal 6xHis tag, the CYC1 terminator [69], LEU2 for yeast selection and the CEN6/ARSH4 sequence for replication in yeast [70]. The negative control construct was included in plasmid design and constitutes amino acids 2–83 of E. coli beta-galactosidase (VectorBuilder). DHODH sequences of interest and controls were inserted behind the N-terminal tag, replacing the negative control sequence, using NEBuilder HiFi (NEB labs). Site-directed mutagenesis was performed either by fusion PCR [71] or using the Phusion Site-Directed Mutagenesis kit (ThermoFisher Scientific).
Yeast strains, media and genetic methods
The strains used in this study are listed in S2 Table. All parental yeast strains were obtained from Horizon Discovery Ltd. The ura1Δ strain used in this study was derived from a BY4741 ura1Δ strain [72–74] crossed to S288C to make ura1Δ URA3+ HIS3+ strains for transformation with heterologous DHODH plasmids. Genetic crosses were set up on YPD and diploid colonies were selected based on size. Resultant diploids were sporulated in SPO medium (1% KAc, 0.1% Bacto-yeast extract, 0.05% glucose) [75]. Random spore analysis was used to genotype offspring and colony PCR (DreamTaq Green, ThermoScientific) used to confirm the nature of unmarked loci. Yeast strains for transformation were cultured in rich medium with 2% glucose (YPD) and were transformed as described previously [76]. Transformants were selected for on synthetic 2% glucose medium (SD) supplemented with amino acids and vitamins (Formedium) excluding leucine (SD-leu). Serial dilution assays were performed using log-phase cultures grown in SD-leu with 50 μM CuSO4 for primary induction of gene expression. Cells were diluted to 1x105 cells/ml and serially diluted tenfold onto appropriate agar plates. Plates were incubated for three days at 30°C. SD-leu lacking uracil (SD-leu-ura) was used to assess the ability of cells to synthesise pyrimidines. Where appropriate DHU was added to plates at a concentration of 150 mg/L. All pYCP-containing strains were grown, assayed, maintained and stored in the absence of leucine to maintain plasmid selection. Anaerobic growth conditions were generated by sealing agar plates in an airtight bag using the ANAEROGENTM COMPACT system according to the manufacturer’s instructions (Oxoid).
Olorofim susceptibility testing
To determine the sensitivity of class 1A DHODH and DHUO to olorofim, heterologous yeast strains were subjected to antifungal susceptibility testing according to a modified EUCAST microdilution broth methodology for yeast. Olorofim was tested a concentration range of 0.008–8 mg/L. SD-leu-ura with 1 mM CuSO4 broth was used to maintain plasmid selection and ensure pyrimidine synthesis was essential and to ensure robust enzyme expression. For the DHUO strains, 150 mg/L DHU was also added to the broth. Microdilution plates were inoculated with 1x106 cfu/ml and incubated at 35°C for 24 hours. The minimum inhibitory concentration (MIC) was determined visually for each strain. Susceptibility testing was performed in triplicate and results presented as the geometric mean (GM) and range of MICs.
Bacterial cloning and plasmid construction
To generate protein expression plasmids, full-length cDNAs were cloned into the SmaI and HindIII sites (S. cerevisiae, L. kluyveri, M. circinelloides, A. alternata, P. minimum) or SmaI and EcoRI sites (T. cruzi, R. arrhizus) of pET50b (Merck) from gene synthesis plasmids (Eurofins) or gene blocks (IDT). Genes were synthesis as native (T. cruzi, S. cerevisiae, L. kluyveri) or E. coli codon-optimised (all other constructs) cDNAs with flanking restriction sites for cut and paste restriction cloning or overlaps for NEBuilder HiFi (New England Biolabs) assembly into pET50b(+) (Merck). N-terminally truncated A. fumigatus DHODH (residues 89–531) in pET44 vector (previously described in [19]) was used as a template for PCR by Phusion HSII polymerase (ThermoFisher Scientific). The truncated A. fumigatus DHODH PCR product was digested with Cfr9I and HindIII (ThermoFisher Scientific) and subcloned into Cfr9I- and HindIII-digested pET50b.
Recombinant protein expression and purification
Recombinant protein expression was performed as described previously [4]. Briefly, protein expression pET50b plasmids were transformed into E. coli BL21 (DE3) cells. Heterologous protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) in LB broth in the presence of 30 μg/mL kanamycin and 100 μM flavin mononucleotide (FMN) at 18°C for 16–18 hours. Bacterial pellets were lysed with BugBuster (Merck) and the His-tagged fusion proteins were isolated by immobilized metal affinity chromatography using His-Bind nickel resin (Merck) according to the manufacturer’s instructions. Fusion tags were removed by HRV-3C protease (Merck) by on-column digestion (M. circinelloides, A. alternata) or digestion in solution following imidazole elution and desalting (Zeba-spin columns; ThermoFisher Scientific) (all other constructs). Protein concentration was determined by Bradford assay using a Coomassie protein assay kit (ThermoFisher Scientific). 0.75 μg of the final protein fractions were analysed using NuPAGE 4–12% Bis-Tris denaturing gels (Invitrogen) and visualised by staining with GelCode Blue protein stain (ThermoFisher Scientific).
Flavin binding analysis
The amount of flavin bound to the proteins was determined by fluorescence spectroscopy. Protein samples underwent denaturation for 10 minutes at 99°C followed by cooling on ice and centrifugation at 16,000 g for 2 minutes. The resulting supernatants were analysed using an excitation wavelength of 465 nm and an emission wavelength of 518 nm. The signal of the samples was compared to an FMN standard curve for estimation of flavin bound to the enzymes. As Class 1A DHODHs are known to bind FMN and not FAD, and the fluorescence of FMN is approximately 10-fold that of FAD at pH 8, the flavin was assumed to be FMN.
Enzyme assays
All enzyme assays were conducted in the following conditions: room temperature, 150 mM KCl, 50 mM Tris-HCl pH 8 buffer, 50 μl reaction volume, 384-well microplates. Enzyme activity was followed spectrophotometrically. DHODH activity was investigated in three different assays, with 1 mM DHO as a substrate. Firstly, 100 μM 2,6-dichloroindophenol (DCIP) was used as a redox indicator whereby oxidation of DHO was coupled to reduction of DCIP that was measured by a decrease in absorbance at 600 nm. 0.1 μg of each protein fraction was added to start the reaction. Secondly, 1 mM potassium hexacyanoferrate III (ferricyanide, FeCy) was substituted for the DCIP and reduction of FeCy was followed at 420 nm using 0.05 μg of each protein fraction. Thirdly, orotate production was monitored directly by absorbance at 280 nm in a reaction that included 2 mM sodium fumarate and 0.2 μg protein. DHUO activity was investigated using two assays, with 1 mM DHU as a substrate. Firstly, the production of uracil was followed directly by the increase in absorbance at 260 nm. Secondly, the other product of the reaction, hydrogen peroxide (H2O2) production was measured by following the increase in absorbance at 595 nm using the Pierce Quantitative Peroxide Assay Kit (ThermoFisher Scientific). 0.2 μg of protein was added to start the reaction for both DHUO assays. For all assays, absorbance results were converted into nanomoles of product produced by the reaction over time or after 30 minutes.
Olorofim inhibition assays
An adaptation of the DCIP reduction assay described above was used to assess the effect of olorofim and orotate on the activity of DHODH enzymes. The DHO concentration was decreased to 250 μM to allow competition with the product, orotate, which was added at 1 mM where indicated. Olorofim was added at 50 μM where indicated. For the class 2 A. fumigatus DHODH assay, 0.1% Triton X-100 and 50 μM coenzyme Q2 were included. Reactions (200 μl in 96-well microplates) were monitored at 600 nm to measure DCIP reduction during a 20 minute reaction.
For investigation of the effect of inhibitors on DHUO activity, H2O2 production was measured as described above except that the DHU concentration was decreased to 250 μM and the reaction time was 20 minutes. Olorofim was added at 50 μM where indicated. Uracil was added at 1 mM where indicated.
Clustering
DHODH and related sequences from the three domains of life (eukaryotes, bacteria and archaea) were obtained by separate PSI-BLAST searches (three iterations each) using the Tuebingen Toolkit [77]. Redundancy was reduced by using databases non-redundant at 70% sequence identity, and further filtered using MMseqs2 to achieve both minimum sequence identity and minimum alignment coverage of 60%. The results 116 sequences were supplemented with 43 sequences from S4 Table and six bacterial proteins related to Haemophilus paracuniculus DHODH (NCBI Accession WP_078236156.1). These were submitted to CLANS with default settings [78] and clustered to completion (>100,000 rounds in 2D), excluding outliers that have no BLAST hits to the main group stronger than an E-value of 10−30.
Results
Mucorales harbour putative class 1A DHODH genes
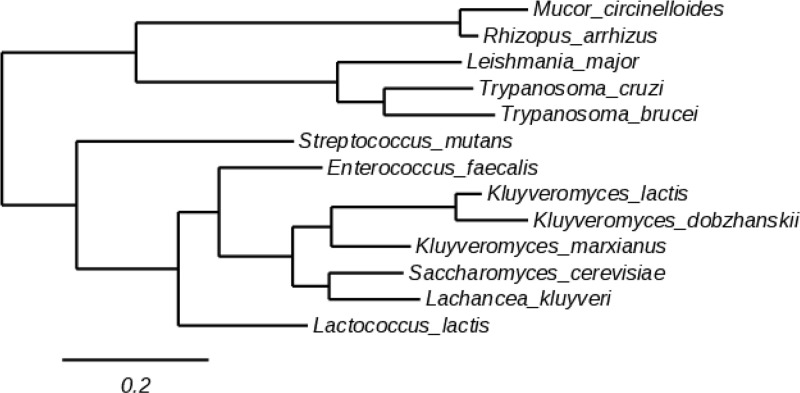
A variety of species are responsible for mucormycosis but Rhizopus and Mucor spp. cause over 50% of cases [79]. Preliminary phylogenetic analyses have previously suggested that the Mucorales fungi Rhizopus arrhizus and Mucor circinelloides possess class 1A DHODH [4]. To confirm this, their proteomes were queried with the S. cerevisiae class 1A (Ura1) and A. fumigatus class 2 (PyrE) protein sequences using BLASTP (NCBI) (S1 Table). Both Mucorales proteomes returned class 1A DHODH proteins with high query coverage and specificity and good identity to the characterised Ura1 protein, indicative of strong homology (S3 Table). Further, analysis of these putative sequences using Interpro classified them as belonging to the class 1A family of DHODH based on conserved domains and important sites [59]. No proteins with class 2 homology were detected for either species. Interestingly, phylogenetic analysis showed these putative class 1A grouped with orthologs from L. major and Trypanosoma species rather than yeasts and bacteria (Fig 1), concordant with the relatively large phylogenetic distance between Mucormycotina and Saccharomycotina within the fungal kingdom [80]. This analysis suggests that Mucorales species possess only a class 1A DHODH and no class 2 counterpart.
Fig 1. Phylogeny of Mucorales class 1A DHODH.
Phylogenetic tree of DHODH sequences from class 1A-containing species. Scale bar is branch length for an expected number of 0.2 substitution per site.
Rhizopus and Mucor putative class 1A DHODH genes complement pyrimidine auxotrophy in yeast
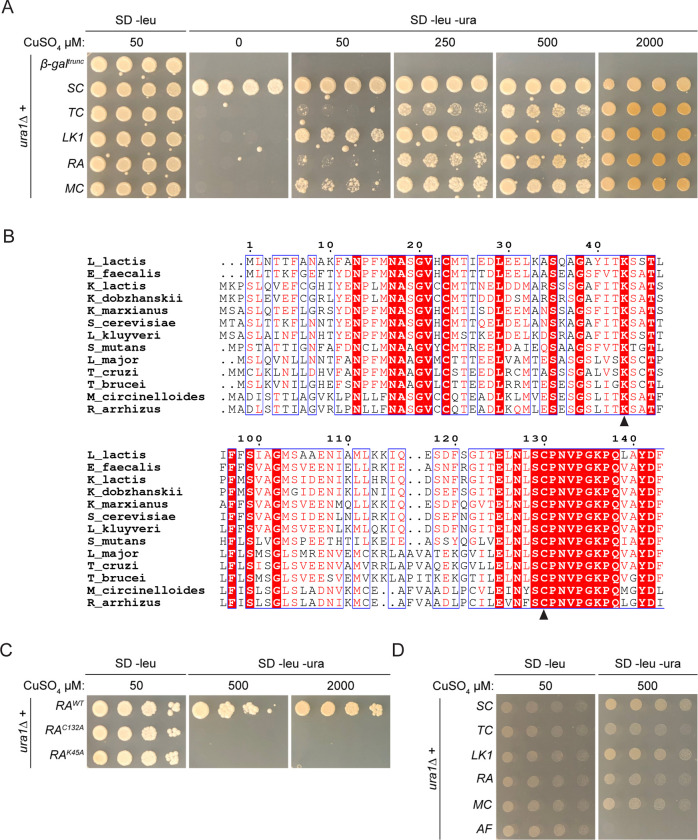
To verify the putative proteins identified bioinformatically are class 1A DHODH, their activity was first assessed using a heterologous system. Previous studies demonstrated that both fungal [43, 44] and non-fungal [81] DHODH can complement ura1Δ S. cerevisiae strains. Therefore, a ura1Δ system was utilised to screen the activity of the putative enzymes through complementation. DHODH sequences and controls were inserted into a vector behind the CUP1 promoter that has leaky expression in regular medium but expression increases with increasing concentrations of Cu2+ [68]. A truncated E. coli gene (beta-galactosidase) and S. cerevisiae URA1+ were included as a negative and positive controls, respectively. Further, characterised class 1A genes from other species were also used to ensure robustness of the system: the yeast L. kluyveri, which also has a functional class 2 DHODH, and the parasite T. cruzi. To ensure an overabundance of copper is not toxic to yeast cells, active class 1A DHODH were induced with a high concentration of copper in pyrimidine-rich media and viability assessed (S1A Fig). Indeed, ura1Δ strains expressing either S. cerevisiae, T. cruzi or L. kluyveri class 1A DHODH (SC, TC, LK1A, respectively) were viable in medium containing 2000 μM copper and uracil, implying no toxic effects of copper accumulation. In line with previous observations, cells were slightly darker in colour due to accumulation of copper sulfide [82, 83]. Transformants were then assessed for their ability to grow in the absence of pyrimidines in various inducing conditions, indicative of DHODH activity (Fig 2A). As expected, the negative control was unable to complement in any condition while SC supported robust growth with just leaky basal expression. LK1A was able to rescue growth in mildly inducing conditions (50 μM CuSO4) and even the non-fungal TC produced robust growth, albeit at very high concentrations of copper (2000 μM CuSO4), reflective of its evolutionary distance. Interestingly, both Mucorales DHODH functioned in yeast but required slightly higher levels of induction for complete pyrimidine prototrophy, compared to the two other fungal enzymes.
Fig 2. Putative Mucorales class 1A DHODH support pyrimidine prototrophy in yeast.
(A) Serial dilution assay of the negative control (β-galtrunc) and confirmed class 1A DHODH from S. cerevisiae (SC), T. cruzi (TC), L. kluveryi (LK1A), R. arrhizus (RA) and M. circinelloides (MC) complementing ura1Δ. (B) Sequence alignment of confirmed class 1A DHODH. Sequences were aligned using MULTALIN [66] and displayed graphically using ESPript [86]. Alignment truncated to show only N-terminus (top) and active site area (bottom). Residue numbers correspond to L. lactis, black arrowheads denote conserved catalytic amino acids mutated. Red highlight, strict identity; red character, similarity within aligned residues; blue frame, similarity across multiple aligned residues. (C) Serial dilution assay comparing the ability of wild-type (RAWT), inactive (RAC132A) and lysine mutant (RAK45A) R. arrhizus DHODH to complement ura1Δ cells. (D) Serial dilution assay of the confirmed class 1A DHODH from S. cerevisiae (SC), T. cruzi (TC), L. kluveryi (LK1A), R. arrhizus (RA), M. circinelloides (MC) and negative control A. fumigatus (AF) complementing ura1Δ in anaerobic conditions.
To confirm that DHODH activity was specifically responsible for the rescue of ura1Δ, predicted key catalytic residues were mutated in the R. arrhizus gene and complementation assessed. The active site cysteine and an N-terminal lysine involved in catalysis were identified in R. arrhizus [84, 85] (Fig 2B) and both were mutated to alanine to negate their function. Both RAC132A and RAK45A were unable to restore growth in the absence of pyrimidines in any concentration of copper (Fig 2C), confirming that production of orotate by class 1A DHODH is responsible for complementation.
Class 1A DHODHs support anaerobic growth through independence of pyrimidine biosynthesis from the respiratory chain and facilitate facultative anaerobiosis in S. cerevisiae and L. kluyveri [25, 42, 43]. To understand if the Mucorales DHODH shared this property, ura1Δ rescue was assessed under anaerobic conditions. Some class 2 DHODH do not support anaerobic pyrimidine prototrophy in this yeast system [43, 44], therefore the DHODH from A. fumigatus was included as a negative control as this enzyme is not toxic and rescues ura1Δ in aerobic conditions (S1B Fig). Growth was slower than that of aerobic conditions but all class 1A DHODH rescued growth under anaerobic conditions, in sharp contrast to the A. fumigatus class 2 (Fig 2D). In this robust system, Mucorales putative class 1A genes appear to have DHODH activity.
Rhizopus and Mucor enzymes have class 1A DHODH activity in vitro
Class 1A DHODH activity for these two Mucorales species was confirmed further using a biochemical approach. Full-length, codon-optimised forms of R. arrhizus and M. circinelloides DHODH cDNAs were synthesised, and recombinant proteins expressed and purified from E. coli (S2 Fig). Initial characterisation of these constructs as DHODH showed they bind flavin in a molar ratio similar to those observed for S. cerevisiae, L. kluyveri 1A and T. cruzi DHODH and to previous reports for class 1A DHODH [87] indicating they are flavoproteins (Table 2). The similarity in flavin fluorescence among the Mucorales and classified class 1A DHODH indicates that it is FMN and not FAD bound to these proteins. Like other characterised DHODHs, both Mucorales enzymes could reduce two artificial electron acceptors: 2,6-dichloroindophenol (DCIP) (Fig 3A) and potassium hexacyanoferrate III (ferricyanide, FeCy) (Fig 3B) in the presence of DHO. Moreover, both enzymes could convert DHO to orotate in the presence of the native class 1A electron acceptor fumarate (Fig 3C). The R. arrhizus enzyme appeared to have higher activity than the M. circinelloides enzyme in all three assays. Taken together, these data suggest that these Mucorales species have active class 1A DHODH enzymes.
Table 2. FMN binding of recombinant class 1A DHODH.
Ratio of FMN to protein of L. kluyveri class 1A (LK1A), T. cruzi (TC), R. arrhizus (RA) and M. circinelloides (MC) DHODH.
| DHODH | SC | LK1A | TC | MC | RA |
|---|---|---|---|---|---|
| μmole FMN/μmole protein | 0.64 | 0.68 | 0.79 | 1.06 | 0.73 |
Fig 3. Mucorales class 1A DHODH are active in vitro.
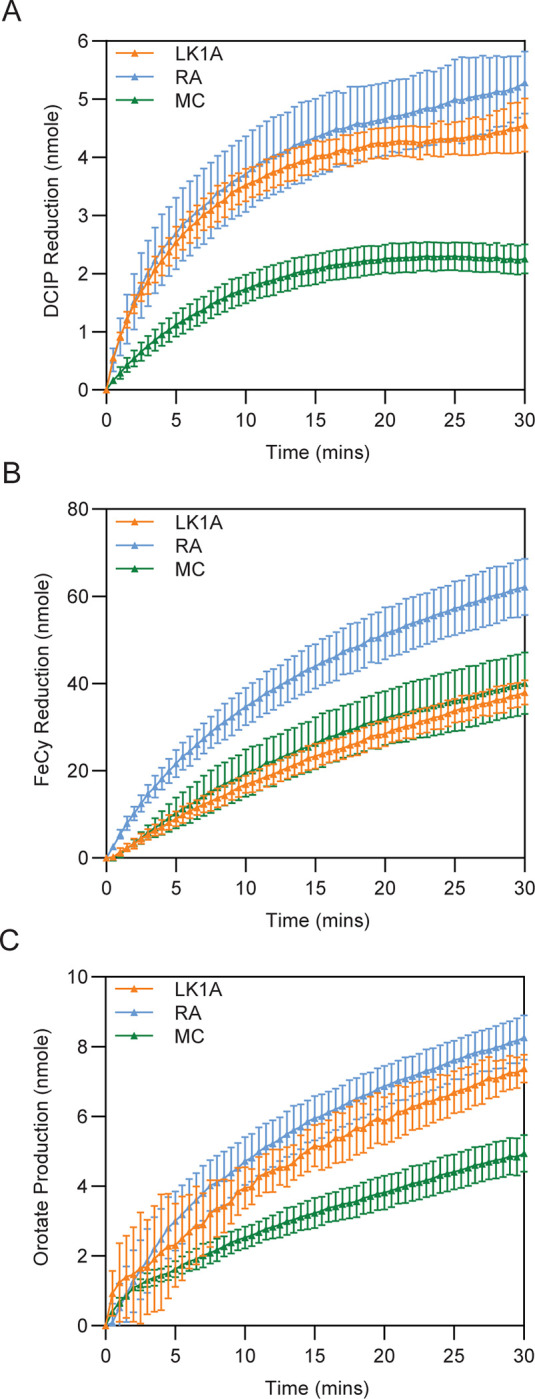
Biochemical analyses of L. kluyveri class 1A (LK1A), R. arrhizus (RA) and M. circinelloides (MC) DHODH. (A) Reduction of DCIP by DHODH over time. (B) Reduction of FeCy by DHODH over time. (C) Production of orotate by DHODH over time.
Class 1A DHODH are not inhibited by olorofim
With the DHODH repertoire of the Mucorales species characterised, we set out to confirm that these enzymes are not inhibited by olorofim and as such are responsible for the resistance of these species. Firstly, the class 1A DHODH heterologous yeast strains were subjected to olorofim susceptibility testing using a broth microdilution method and the minimum inhibitory concentration (MIC) of olorofim determined for each strain. Wild-type was used as a negative control for olorofim activity and the strain expressing the olorofim-susceptible A. fumigatus class 2 DHODH was used as a positive control. As expected, growth of the heterologous A. fumigatus strain was strongly inhibited with a mean MIC of 0.125 mg/L (Table 3), reflective of the potency of olorofim for this DHODH and confirms olorofim could permeate S. cerevisiae cells [4]. The wild-type strain had an MIC of >8 mg/L, suggesting endogenous Ura1 is not inhibited by this antifungal nor are there any off-target effects in this species. All class 1A DHODH strain growth was completely unaffected, suggesting olorofim cannot inhibit class 1A DHODH activity in the yeast system.
Table 3. Olorofim sensitivity of heterologous yeast strains expressing class 1A DHODH.
The geometric mean (GM) MIC and MIC range are presented for a wild-type (WT) strain and heterologous strains expressing either the class 2 DHODH from A. fumigatus (AF) or the class 1A DHODH from S. cerevisiae (SC), T. cruzi (TC), L. kluyveri (LK1A), M. circinelloides (MC) and R. arrhizus (RA).
| ura1Δ + | |||||||
|---|---|---|---|---|---|---|---|
| WT | AF | SC | TC | LK1A | MC | RA | |
| GM (mg/L) | >8 | 0.125 | >8 | >8 | >8 | >8 | >8 |
| Range (mg/L) | >8 | 0.125 | >8 | >8 | >8 | >8 | >8 |
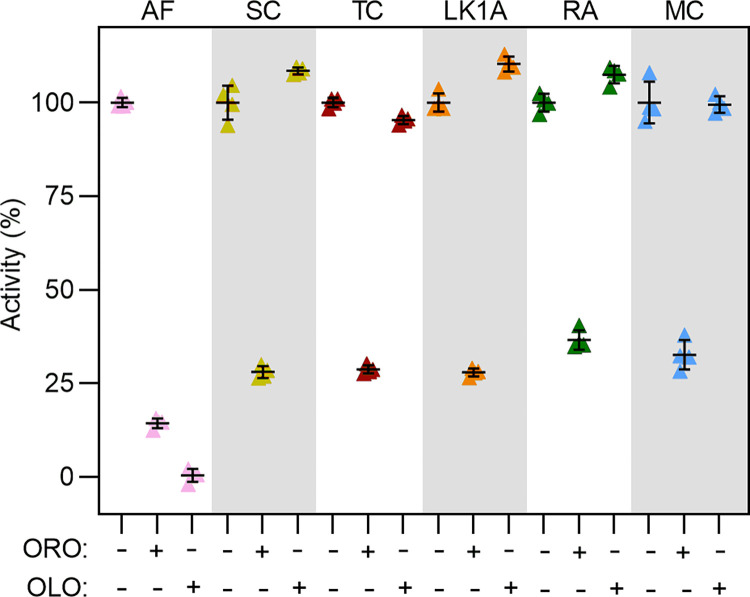
To directly address whether olorofim has any effect on these class 1A DHODH, inhibition assays were performed using the purified proteins and DCIP reduction was monitored. Recombinant class 2 A. fumigatus DHODH was used as a positive control for olorofim activity [4]. The catalytic product orotate was used as a control for general DHODH inhibition, demonstrating >60% inhibition of activity against all enzymes (<50% activity) (Fig 4). While A. fumigatus enzyme activity dropped to almost zero in the presence of olorofim, the antifungal had no inhibitory effect for any of the class 1A DHODH tested, indicative of its selectivity for class 2 enzymes.
Fig 4. Class 1A DHODH are not inhibited by olorofim.
DCIP reduction assays of A. fumigatus (AF), S. cerevisiae (SC), T. cruzi (TC), L. kluveryi (LK1A), R. arrhizus (RA) and M. circinelloides (MC) in the absence (-) or presence (+) of either orotate (ORO) or olorofim (OLO). Activity in the presence of inhibitors was expressed as a percentage of control enzyme activity in absence of inhibitors after 20 minutes (100%).
Dematiaceous moulds appear to harbour two classes of putative DHODH genes
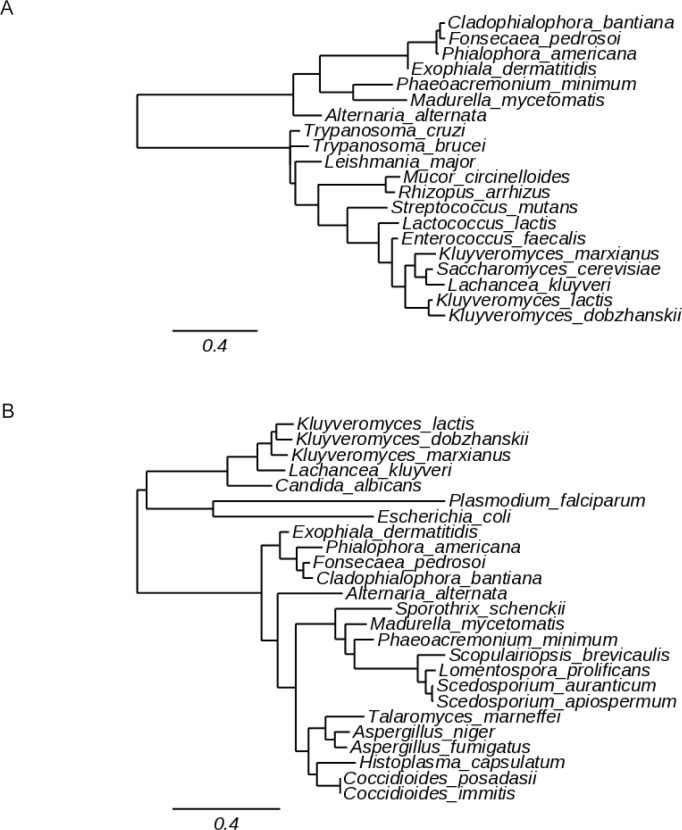
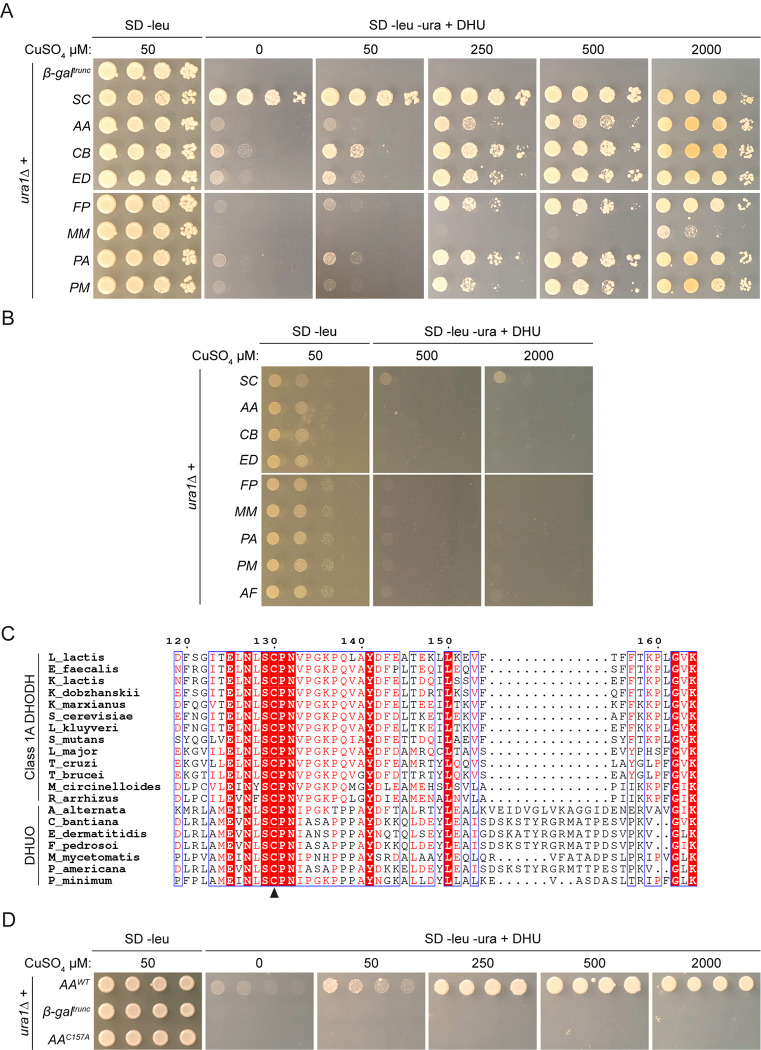
To determine the class of DHODH present in the dematiaceous moulds, their proteomes were queried for class 1A and class 2 orthologs, again using Ura1 and PyrE, respectively. All seven species returned both putative class 1A DHODH- and class 2 DHODH-like proteins with varying degrees of sequence identity to the queries, however sequence identity and E-values were consistently better for PyrE (>54% identity) than for Ura1 (24–31%) in all species (S4 Table). However, Interpro classified all Ura1 orthologs as belonging to the class 1A DHODH protein family. Remarkably, class 1A DHODH sequences from a range of species revealed the dematiaceous moulds form a distinctly separate clade to the fungi proven to have the enzyme (Fig 5A) The dematiaceous class 2 sequences are slightly more integrated with other species containing this type of enzyme, suggesting these enzymes are more closely related than the putative class 1A are (Fig 5B). This analysis suggests dematiaceous moulds may have both class 1A and class 2 DHODH. We investigated these phylogenetically distinct putative class 1A proteins further to determine whether they had DHODH activity.
Fig 5. Phylogeny of dematiaceous mould DHODH.
Phylogenetic trees of (A) class 1A and (B) class 2 DHODH sequences. Scale bars represent branch length for an expected number of 0.4 substitution per site.
Dematiaceous mould putative class 1A DHODH genes do not complement pyrimidine auxotrophy in yeast
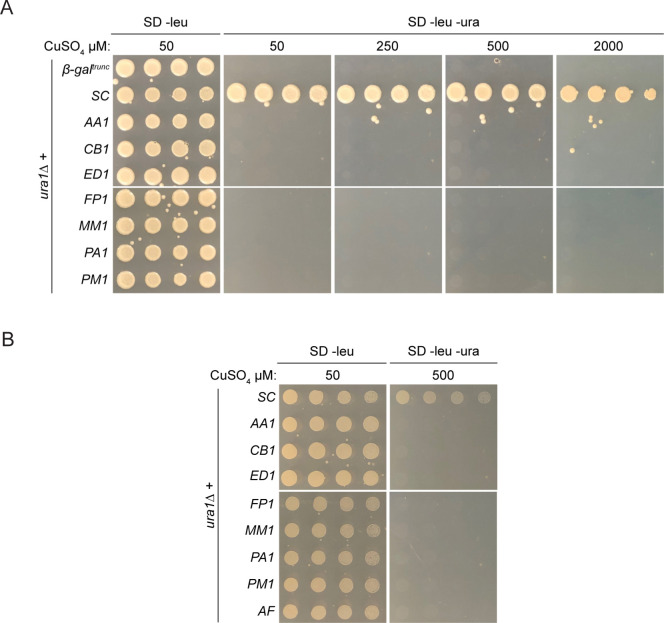
With evidence that heterologous class 1A activity from species with multiple DHODH genes produces sufficient pyrimidines for ura1Δ growth, the activity of the putative class 1A DHODH genes from dematiaceous moulds was assessed using the same system. In sharp contrast to SC, class 1A constructs from all seven species evaluated were unable to complement ura1Δ, even under strong inducing conditions (2000 μM CuSO4) (Fig 6A).
Fig 6. Dematiaceous mould genes do not support pyrimidine prototrophy in yeast.
(A) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and putative genes from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells (B) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and putative class 1A DHODH from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells under anaerobic conditions.
To ensure that codon usage was not a limiting factor in the ability of these exogenous genes to rescue ura1Δ, three variations of the A. alternata class 1A were assessed for complementation. An E. coli codon-optimised construct (AA1AEc), a native CDS construct (AA1ANat) and a yeast-codon optimised construct (AA1ASc) all behaved similarly in complementation testing; none rescued the ura1Δ strain (S3 Fig). Therefore, any potential translation issues are not responsible for lack of DHODH activity.
In attempts to uncover any potential DHODH functionality of these putative genes, their activity was investigated under anaerobic conditions. Only ura1Δ strains expressing confirmed class 1A DHODH were able to grow anaerobically. Like the A. fumigatus DHODH, the putative enzymes were unable to support anaerobic growth (Fig 6B). Taken together, these results suggest that these putative class 1A genes do not harbour DHODH activity.
Dematiaceous mould putative genes use DHU to support yeast pyrimidine prototrophy
During the course of this study, another group demonstrated that yeast cell lysates containing the A. alternata putative class 1A DHODH gene could oxidise DHU to uracil and as such the enzyme has been classified as a DHUO [53]. In the absence of DHODH activity and considering the A. alternata findings, the other dematiaceous moulds were assessed for DHUO activity in the yeast complementation system. Transformants were again assayed for their ability to grow in the absence of pyrimidines in various inducing conditions but in the presence of DHU [53], indicative of their ability to oxidise the substrate to uracil and rescue the ura1Δ pyrimidine auxotrophy. As expected, the negative control was unable to complement in any condition and the SC growth control grew well in all conditions (Fig 7A). The previously characterised A. alternata DHUO [53] rescued the auxotrophy well at 250 μM copper and above. Intriguingly, all dematiaceous enzymes complemented ura1Δ growth using DHU, implicating them as DHUO, but pyrimidine prototrophy was not identical for each DHUO at equivalent copper levels, suggestive of variation in oxidase activity between enzymes of different species. In line with previous findings [53], none of the dematiaceous mould enzymes could use DHU to rescue pyrimidine auxotrophy in anaerobic conditions (Fig 7B). These data show that these putative enzymes use DHU and oxygen to produce uracil, implicating them as DHUO rather than DHODH. Again, codon usage had no effect on ability of a construct to rescue ura1Δ the in the presence of DHU (S4 Fig).
Fig 7. Dematiaceous mould putative genes have DHUO activity in the yeast model.
(A) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and putative genes from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells in the presence of DHU. (B) Serial dilution assay of the confirmed class 1A DHODH from S. cerevisiae (SC) and putative genes from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells under anaerobic conditions in the presence of DHU. (C) Sequence alignment of confirmed class 1A DHODH and dematiaceous DHUO. Sequences were aligned using MULTALIN [66] and displayed graphically using ESPript [86]. Alignment truncated to show only active site area. Residue numbers correspond to L. lactis, black arrowhead denotes conserved catalytic cysteine mutated. Red highlight, strict identity; red character, similarity within aligned residues; blue frame, similarity across multiple aligned residues. (D) Serial dilution assay comparing the ability of wild-type (AAWT), negative control (β-galtrunc) and cysteine mutant (AAC157A) A. alternata DHUO to complement ura1Δ cells.
To confirm that DHUO activity was specifically responsible for the rescue of ura1Δ, the predicted key catalytic residue was identified and mutated in the A. alternata gene and complementation assessed. The potential active was identified in the dematiaceous fungi through sequence alignment with confirmed class 1A DHODH, where a clear SCPNV motif was conserved across both enzyme classes (Fig 7C). On the hypothesis that class 1A DHODH and DHUO catalytic mechanisms may share an important cysteine, this residue was mutated to alanine in A. alternata to negate activity. AAC157A was unable to restore growth using DHU in the absence of pyrimidines in any concentration of copper (Fig 7D). These results not only confirm that production of uracil is responsible for complementation in this system but provides the first insights into the sequence and catalytic mechanism of DHUO enzymes.
Dematiaceous mould enzymes display DHUO activity in vitro
Enzymatic identity and activity for representative dematiaceous proteins were further assessed biochemically. Full-length, codon-optimised A. alternata and P. minimum cDNAs were synthesised and recombinant protein expressed and purified from E. coli (S5A Fig). At 37.5 and 35.5 kDa, respectively, these proteins are slightly bigger than the five class 1A DHODHs studied above, ranging from 33.6 to 35.1 kDa. Again, characterisation of these enzymes as flavoproteins revealed them to bind flavin with a molar ratio similar to that observed for the known FMN-binding class 1A DHODHs, indicating that it is FMN and not FAD bound (Table 4). The observed lack of DHODH activity observed in the complementation study was confirmed by following the reduction of DCIP and FeCy and production of orotate in the presence of DHO; neither dematiaceous enzyme was active in any of these in vitro assays compared to class 1A DHODHs (S5B–S5D Fig), supporting the notion that these enzymes are not class 1A DHODH. Subsequently, based on findings from Bouwknegt and colleagues, DHUO activity for these enzymes was assessed in vitro, following production of uracil and hydrogen peroxide levels as the two products of DHU oxidation [54, 88]. Concordant with previous observations, the A. alternata enzyme as well as that from P. minimum converted DHU to uracil while the M. circinelloides class 1A DHODH could not (Fig 8A). Furthermore, both dematiaceous enzymes produced hydrogen peroxide while the proven class 1A had no such activity (Fig 8B). The enzyme from P. minimum appeared to be more active than the A. alternata counterpart in both DHUO assays. Taken together, these data suggest that these species of dematiaceous mould have active DHUOs.
Table 4. FMN binding of dematiaceous mould enzymes.
Ratio of FMN to protein of A. alternata (AA) and P. minimum (PM) DHUO.
| DHODH | AA | PM |
|---|---|---|
| μmole FMN/μmole protein | 0.50 | 0.91 |
Fig 8. Dematiaceous mould enzymes have DHUO activity in vitro.
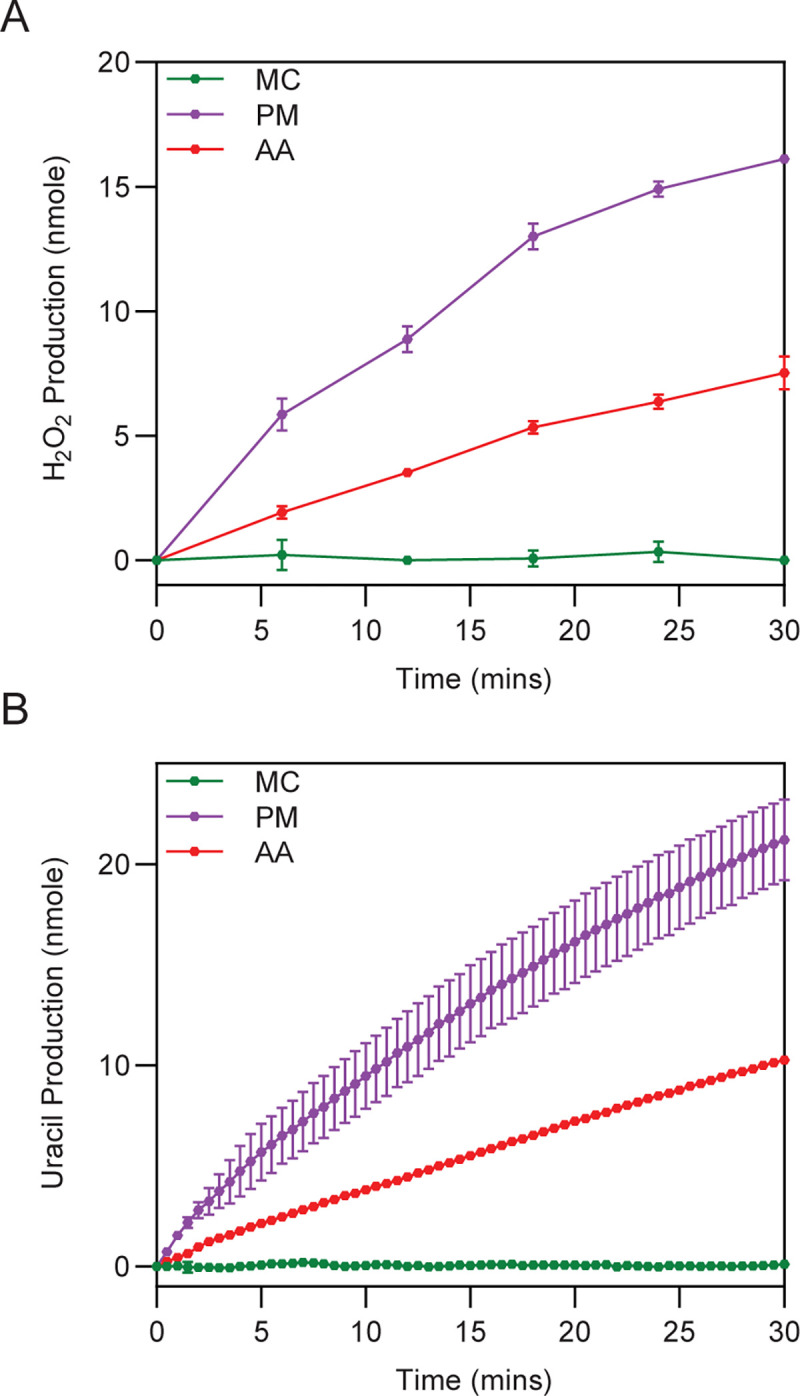
Biochemical analyses of M. circinelloides class 1A DHODH (MC) and A. alternata (AA) and P. minimum (PM) DHUO. (A) Production of uracil over time. (B) Production of H2O2 over time.
DHUO are not inhibited by olorofim
With the knowledge that these dematiaceous genes are not DHODH, we set out to understand whether olorofim has any effect on this class of enzymes. Like the wild-type strain, all DHUO strain growth was completely unaffected, implying olorofim does not inhibit heterologous DHUO activity (Table 5).
Table 5. Olorofim sensitivity of heterologous yeast strains expressing DHUO.
The geometric mean (GM) MIC and MIC range are presented for a wild-type (WT) strain and heterologous strains expressing either the class 2 DHODH from A. fumigatus (AF) or the DHUO from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PA) and P. minimum (PM).
| ura1Δ + | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | AF | AA | CB | ED | FP | MM | PA | PM | |
| GM (mg/L) | >8 | 0.125 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Range (mg/L) | >8 | 0.125 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
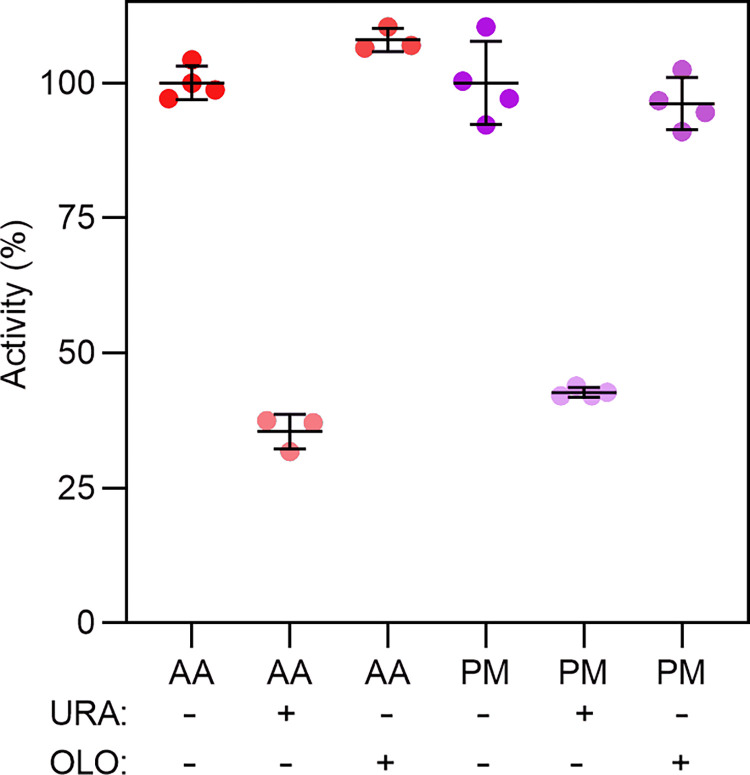
Building on the yeast system results, the direct effects of olorofim on DHUO were investigated biochemically using the hydrogen peroxide assay. The catalytic product uracil was used as a positive control for DHUO inhibition [88], reducing both A. alternata and P. minimum enzyme activity to less than 50% (Fig 9). However, olorofim showed no substantial inhibitory effect for either DHUO, confirming that this antifungal does not target this class of enzymes.
Fig 9. DHUO are not inhibited by olorofim.
H2O2 production assays of A. alternata (AA) and P. minimum (PM) DHUO in the absence (-) or presence (+) of either uracil (URA) or olorofim (OLO). Activity in the presence of inhibitors was expressed as a percentage of control enzyme activity in absence of inhibitors after 20 minutes (100%).
Discussion
DHODH are ubiquitous enzymes important to cellular metabolism, yet their diversity between and within classes makes them attractive therapeutic targets. In this study, we have shed light on the DHODH repertoire of two different types of clinically important fungi. Firstly, we have validated the prior suggestion that the Mucorales species M. circinelloides and R. arrhizus harbour active class 1A DHODH; these enzymes utilise fumarate to produce orotate and are able to functionally replace the S. cerevisiae class 1A enzyme. Secondly, we have disproved our hypothesis that certain dematiaceous moulds may contain class 1A DHODH: these putative enzymes behave as DHUO that catalyse the oxidation of DHU to uracil in vitro and in the yeast system tested.
Preliminary bioinformatics from our previous study suggested Mucorales species harbour class 1A DHODH due to their sequence similarity to known enzymes of this class [4]. Building on this work, proteomes of the Mucorales were found to harbour only orthologs of class 1A DHODH; we pursued characterisation of enzymes from two representative species that are the most common etiological agents of mucormycosis. As with all characterised class 1A DHODHs, the M. circinelloides and R. arrhizus enzymes were seen to be flavoproteins that bound FMN with a molar ratio of FMN to protein of 1.06 and 0.73, respectively, values that support the presence of one FMN molecule per subunit in active enzyme. Both enzymes were able to utilise fumarate as a terminal electron acceptor typical of class 1A DHODH for oxidation of DHO to orotate. It is interesting to note that the R. arrhizus DHODH was more active than the other Mucorales enzyme and even the L. kluyveri class 1A, perhaps reflective of different nutritional requirements in different species. Moreover, their ability to support anaerobic pyrimidine prototrophy in yeast provides further evidence that these enzymes belong to the cytosolic class that do not interact with the respiratory chain, allowing oxygen-independent growth in some species [43]. Mucor spp. can grow anaerobically and exhibit morphological dimorphism between aerobic and anaerobic conditions [89, 90], in line with an oxygen-independent method of synthesising pyrimidines using class 1A DHODHs [25]. We also confirmed that the class 2 DHODH of A. fumigatus was unable to support anaerobic pyrimidine prototrophy which is consensus for most class 2 DHODHs, as they donate electrons to the respiratory chain [25], but not for all [44]. Further, we expanded on structural observations that an N-terminal lysine is vital for catalysis by making a hydrogen bond to the FMN cofactor [85, 91]: mutagenesis of this residue in the R. arrhizus enzyme replicated the loss of activity seen in mutation of the active site cysteine responsible for class 1A catalysis. While general sequence similarity is low among class 1A DHODHs, conservation of important residues across kingdoms is useful for future therapeutic drug design and so further mutagenesis studies should be conducted in a variety of species. Characterisation of these Mucorales DHODH as class 1A enzymes substantiates the lack of olorofim activity seen for this group of moulds. In fact, the drug had no indirect or direct activity on any class 1A DHODH tested in yeast or biochemically, respectively. Olorofim susceptibility and the identity and activity of DHODH enzymes in other species of the Mucorales order of fungi, though predictable from this study, remain to be confirmed but would enrich the current understanding of these opportunistic pathogens.
The same prior phylogenetic analyses also found class 2 DHODH in a limited number of dematiaceous moulds, including Alternaria spp. and Exophiala, although sequences grouped away from those of genera against which olorofim is most potent, such as Aspergillus spp., Coccidioides, Histoplasma and Blastomyces [4]. We posited that the lack of olorofim activity in some of these species that appear to contain the drug target may be due to the presence of a secondary DHODH of a different class, specifically a class 1A as is the repertoire of L. kluyveri [42, 43]. Indeed, we found DHODH-like proteins belonging to both class 1A and class 2 families in this selection of dematiaceous moulds, though overall the class 2 orthologs had stronger identity to PyrE than the class 1As did to Ura1. Nevertheless, the putative Ura1 orthologs had enough conservation of important residues for them to be classified as class 1A DHODH proteins. The presence of putative class 1A in all species queried was unexpected as some show sensitivity to olorofim, such as M. mycetomatis and Phaeoacremonium spp. [17, 18], while others have lowered susceptibility, including A. alternata and E. dermatitidis [9, 19]. With the view to understand the effects of a secondary DHODH in species containing the olorofim target DHODH, we focused on characterising the putative class 1A DHODH genes in a selection of dematiaceous moulds, with remarkable results.
Surprisingly, no putative class 1A DHODH gene from any of the pathogenic species was able to support aerobic or anaerobic pyrimidine prototrophy in S. cerevisiae. Further, purified recombinant A. alternata and P. minimum proteins did not exhibit classical class 1A biochemistry; neither could produce orotate or use any of the electron acceptors tested. However, both enzymes bound FMN, indicating they are flavoproteins but lack apparent DHODH activity. These data were unexpected as annotation of most of these genes and protein products assigned or predicted them to be class 1A DHODH.
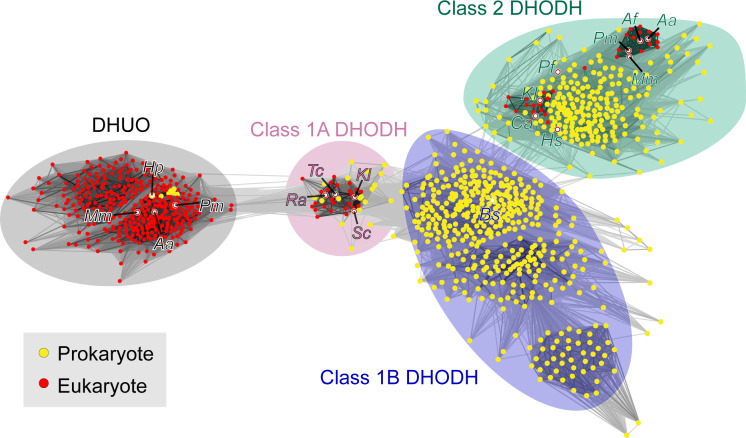
While our work was underway, a study used yeast complementation and lysate assays to demonstrate that the Ura1-like gene from A. alternata encodes a DHUO: a redox enzyme that utilises oxygen to convert DHU or dihydrothymine into uracil or thymine, plus hydrogen peroxide [53]. In line with this work, we subsequently found that the putative dematiaceous enzymes facilitated pyrimidine prototrophy in yeast when supplemented with DHU but only under aerobic conditions. Our biochemical data provide direct evidence that A. alternata possesses a DHUO and that P. minimum can be added to the list of organisms with this class of metabolic enzyme. Limited work on DHUO has demonstrated that they are flavoproteins that utilise only oxygen as an electron acceptor for oxidase activity [88]. Our observations corroborate this; both dematiaceous enzymes bind FMN and cannot rescue yeast growth in anaerobic conditions even in the presence of DHU. Expanding on preliminary sequence analysis [53], all class 1A DHODH and DHUO proteins assessed in this study shared a highly conserved active site that is likely a large contribution to the in silico assignment of the latter as the former in protein databases. Further, we provide the first insight into the mechanism of DHU oxidation through the identification of a catalytic cysteine in the A. alternata enzyme. Conservation of this cysteine across DHUO and class 1A DHODH is remarkable yet unsurprising as sequence or structure is frequently seen to be similar among enzymes that catalyse different reactions as an example of divergent evolution [92], providing an insight into a possible relationship between these two classes of metabolic enzyme. Assessing sequence similarity between enzymes from this study and across all of evolution defines the DHUOs as a separate, large family of enzymes that group together away from the three well-established classes of DHODH (Fig 10). Sequences from several bacteria in the Pasteurellaceae family cluster within this group, suggesting for the first time that bacteria, such as Haemophilus paracuniculus, may have enzymes with DHUO activity. This also suggests that horizontal gene transfer may be the source of fungal DHUOs in a manner parallel to the appearance of class 1A DHODH in fungi [53].
Fig 10. DHUOs are a separate enzyme family in fungi and bacteria.
Cluster map of >1000 DHODH sequences obtained from all domains of organisms, indicating the classes of DHODH (1A, 1B and 2) and the origin in eukaryotes (red) or prokaryotes (yellow). A small number of species are highlighted by names: A. alternata (Aa), A. fumigatus (Af), B. subtilis (Bs), C. albicans (Ca), H. paracuniculus (Hp), H. sapiens (Hs), K. lactis (Kl), M. mycetomatis (Mm), P. falciparum (Pf), P. minimum (Pm), R. arrhizus (Ra), S. cerevisiae (Sc) and T. cruzi (Tc).
Interestingly, DHUO activity seemed to vary significantly between species; enzymes from C. bantiana and E. dermatitidis supported the most robust growth of yeast while the M. mycetomatis counterpart did not reach comparable levels of growth. Further experiments are required to validate these differences in DHUO activity directly in vitro. Moreover, the P. minimum DHUO appeared more enzymatically active than that from A. alternata. Differences in DHUO activity may be caused by species requiring a different balance of pyrimidine metabolism and catabolism influenced by variation in activity of input enzymes. Previously, DHUOs were hypothesised to contribute to dihydropyrimidine detoxification [53] but it may be they also play a role in recycling of pyrimidines. Regardless of their role, these enzymes are not targeted by olorofim and as such, variations in dematiaceous mould susceptibility to the orotomide cannot be attributed to their DHUO.
While the phenomenon of having two different classes of DHODH does exist in select yeasts [42, 43], it appears that dematiaceous moulds are excluded from this. Further work is required to understand why certain species respond to olorofim and others appear more resistant as there are multiple possibilities for why this might be. Some species could have a stronger arsenal of efflux pumps that render them hardier in the face of xenobiotic compounds, as is seen for certain fungal species [93]. Perhaps local concentrations of olorofim never reach a threshold for target inhibition due to active export, rendering species less susceptible to the drug. Conversely, it may be differences in the target class 2 DHODH protein sequence that are responsible for reduced olorofim sensitivity. Indeed, this was shown to be the case for Candida albicans [4] and for instances of olorofim resistance where substitution mutation is the root cause [94]. Future work will address differences between class 2 DHODH proteins from susceptible and non-susceptible species. While the true cause for variation of olorofim sensitivity in dematiaceous moulds remains to be elucidated, it can be concluded that these species do not possess class 1A DHODH but do harbour DHUO in their repertoire of pyrimidine network enzymes.
Supporting information
(A) Serial dilution assay of the negative control (β-galtrunc) and confirmed class 1A DHODH from S. cerevisiae (SC), T. cruzi (TC), L. kluveryi (LK1A) in ura1Δ cells in the presence of high copper concentration and pyrimidines. (B) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and confirmed class 2 DHODH from A. fumigatus (AF) in ura1Δ cells in the presence of high copper concentration and pyrimidines.
(TIF)
NuPAGE analysis of purified recombinant proteins. T. cruzi (TC), L. kluveryi (LK1A), S. cerevisiae (SC), M. circinelloides (MC) and R. arrhizus (RA).
(TIF)
Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and three putative A. alternata genes in ura1Δ cells. AAEc, E. coli codon-optimised construct; AANat, native A. alternata sequence construct; AASc, S. cerevisiae codon-optimised construct.
(TIF)
Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and three putative A. alternata genes in ura1Δ cells. AAEc, E. coli codon-optimised construct; AANat, native A. alternata sequence construct; AASc, S. cerevisiae codon-optimised construct in the presence of DHU.
(TIF)
(A) NuPAGE analysis of purified proteins. A. alternata (AA), P. minimum (PM). (B) Amount of DCIP reduced by proven and putative DHODH after 30 minutes. (C) Amount of FeCy reduced by proven and putative DHODH after 30 minutes. (D) Amount of orotate produced by proven and putative DHODH after 30 minutes.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by F2G Ltd. https://www.f2g.com/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bongomin F, Gago S, Oladele RO, Denning DW. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J Fungi (Basel). 2017; 3(4). Epub 2018/01/27. doi: 10.3390/jof3040057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gow NAR, Johnson C, Berman J, Coste AT, Cuomo CA, Perlin DS, et al. The importance of antimicrobial resistance in medical mycology. Nat Commun. 2022; 13(1): 5352. Epub 2022/09/13. doi: 10.1038/s41467-022-32249-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol. 2022; 20(9): 557–71. Epub 2022/03/31. doi: 10.1038/s41579-022-00720-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver JD, Sibley GEM, Beckmann N, Dobb KS, Slater MJ, McEntee L, et al. F901318 represents a novel class of antifungal drug that inhibits dihydroorotate dehydrogenase. Proceedings of the National Academy of Sciences of the United States of America. 2016; 113(45): 12809–14. doi: 10.1073/pnas.1608304113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seyedmousavi S, Chang Y, Law D, Birch M, Rex JH, Kwon-Chung KJ. Efficacy of Olorofim (F901318) against Aspergillus fumigatus, A. nidulans, and A. tanneri in Murine Models of Profound Neutropenia and Chronic Granulomatous Disease. Antimcrobial Agents and Chemotherapy. 2019; 63(6): 13. doi: 10.1128/AAC.00129-19CE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiederhold NP. Review of the Novel Investigational Antifungal Olorofim. Journal of Fungi. 2020; 6(3). doi: 10.3390/jof6030122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buil JB, Rijs AJMM, Meis JF, Birch M, Law D, Melchers WJG, et al. In vitro activity of the novel antifungal compound F901318 against difficult-to-treat Aspergillus isolates. The Journal of antimicrobial chemotherapy. 2017; 72(9): 2548–52. doi: 10.1093/jac/dkx177 [DOI] [PubMed] [Google Scholar]
- 8.Lackner M, Birch M, Naschberger V, Grässle D, Beckmann N, Warn P, et al. Dihydroorotate dehydrogenase inhibitor olorofim exhibits promising activity against all clinically relevant species within Aspergillus section Terrei. Journal of Antimicrobial Chemotherapy. 2018; 73(11): 3068–73. doi: 10.1093/jac/dky329 [DOI] [PubMed] [Google Scholar]
- 9.Singh A, Singh P, Meis JF, Chowdhary A. In vitro activity of the novel antifungal olorofim against dermatophytes and opportunistic moulds including Penicillium and Talaromyces species. J Antimicrob Chemother. 2021. Epub 2021/01/10. doi: 10.1093/jac/dkaa562 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiederhold NP, Law D, Birch M. Dihydroorotate dehydrogenase inhibitor F901318 has potent in vitro activity against Scedosporium species and Lomentospora prolificans. Journal of Antimicrobial Chemotherapy. 2017; 72(7): 1977–80. doi: 10.1093/jac/dkx065 [DOI] [PubMed] [Google Scholar]
- 11.Biswas C, Law D, Birch M, Halliday C, Sorrell TC, Rex J, et al. In vitro activity of the novel antifungal compound F901318 against Australian Scedosporium and Lomentospora fungi. Medical mycology. 2018; 56(8): 1050–4. doi: 10.1093/mmy/myx161 [DOI] [PubMed] [Google Scholar]
- 12.Kirchhoff L, Dittmer S, Weisner A-K, Buer J, Rath P-M, Steinmann J. Antibiofilm activity of antifungal drugs, including the novel drug olorofim, against Lomentospora prolificans. Journal of Antimicrobial Chemotherapy. 2020: 1–8. doi: 10.1093/jac/dkaa157 [DOI] [PubMed] [Google Scholar]
- 13.Wiederhold NP, Patterson HP, Sanders CJ, Canete-Gibas C. Dihydroorotate dehydrogenase inhibitor olorofim has potent in vitro activity against Microascus/Scopulariopsis, Rasamsonia, Penicillium and Talaromyces species. Mycoses. 2022. Epub 2022/11/28. doi: 10.1111/myc.13548 . [DOI] [PubMed] [Google Scholar]
- 14.Wiederhold NP, Najvar LK, Jaramillo R, Olivo M, Birch M, Law D, et al. The orotomide olorofim is efficacious in an experimental model of central nervous system coccidioidomycosis. Antimicrobial Agents and Chemotherapy. 2018; 62(9): 1–8. doi: 10.1128/AAC.00999-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borba-Santos LP, Rollin-Pinheiro R, da Silva Fontes Y, Dos Santos GMP, de Sousa Araujo GR, Rodrigues AM, et al. Screening of Pandemic Response Box Library Reveals the High Activity of Olorofim against Pathogenic Sporothrix Species. J Fungi (Basel). 2022; 8(10). Epub 2022/10/28. doi: 10.3390/jof8101004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fothergill A, Wiederhold N, Sibley G, Kennedy A, Oliver J, Birch M, et al. Spectrum of Activity of F901318, the First Agent from the New Orotomide Class of Antifungals. 2008. [Google Scholar]
- 17.Georgacopoulos O, Nunnally NS, Ransom EM, Law D, Birch M, Lockhart SR, et al. In Vitro Activity of Novel Antifungal Olorofim against Filamentous Fungi and Comparison to Eight Other Antifungal Agents. J Fungi (Basel). 2021; 7(5). Epub 2021/06/03. doi: 10.3390/jof7050378 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim W, Eadie K, Konings M, Rijnders B, Fahal AH, Oliver JD, et al. Madurella mycetomatis, the main causative agent of eumycetoma, is highly susceptible to olorofim. Journal of Antimicrobial Chemotherapy. 2020; 75(4): 936–41. doi: 10.1093/jac/dkz529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchhoff L, Dittmer S, Buer J, Rath PM, Steinmann J. In vitro activity of olorofim (F901318) against fungi of the genus, Scedosporium and Rasamsonia as well as against Lomentospora prolificans, Exophiala dermatitidis and azole-resistant Aspergillus fumigatus. Int J Antimicrob Agents. 2020; 56(3): 106105. Epub 2020/07/30. doi: 10.1016/j.ijantimicag.2020.106105 . [DOI] [PubMed] [Google Scholar]
- 20.Organization GWH. WHO fungal priority pathogens list to guide research development and public health action report 2022. 2022. [Google Scholar]
- 21.Liu Y, Gao ZQ, Liu CP, Xu JH, Li LF, Ji CN, et al. Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011; 67(Pt 2): 182–7. Epub 2011/02/09. doi: 10.1107/S1744309110048414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feliciano PR, Cordeiro AT, Costa-Filho AJ, Nonato MC. Cloning, expression, purification, and characterization of Leishmania major dihydroorotate dehydrogenase. Protein Expr Purif. 2006; 48(1): 98–103. Epub 2006/04/08. doi: 10.1016/j.pep.2006.02.010 . [DOI] [PubMed] [Google Scholar]
- 23.Arakaki TL, Buckner FS, Gillespie JR, Malmquist NA, Phillips MA, Kalyuzhniy O, et al. Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies. Mol Microbiol. 2008; 68(1): 37–50. Epub 2008/03/04. doi: 10.1111/j.1365-2958.2008.06131.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takashima E, Inaoka DK, Osanai A, Nara T, Odaka M, Aoki T, et al. Characterization of the dihydroorotate dehydrogenase as a soluble fumarate reductase in Trypanosoma cruzi. Mol Biochem Parasitol. 2002; 122(2): 189–200. Epub 2002/07/11. doi: 10.1016/s0166-6851(02)00100-7 . [DOI] [PubMed] [Google Scholar]
- 25.Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci U S A. 1992; 89(19): 8966–70. Epub 1992/10/01. doi: 10.1073/pnas.89.19.8966 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahler AE, Nielsen FS, Switzer RL. Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its isolated subunits. Arch Biochem Biophys. 1999; 371(2): 191–201. Epub 1999/11/05. doi: 10.1006/abbi.1999.1455 . [DOI] [PubMed] [Google Scholar]
- 27.Argyrou A, Washabaugh MW, Pickart CM. Dihydroorotate dehydrogenase from Clostridium oroticum is a class 1B enzyme and utilizes a concerted mechanism of catalysis. Biochemistry. 2000; 39(34): 10373–84. Epub 2000/08/24. doi: 10.1021/bi001111d . [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki S, Satoh T, Todoroki M, Niimura Y. b-type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum. Appl Environ Microbiol. 2009; 75(3): 629–36. Epub 2008/12/09. doi: 10.1128/AEM.02111-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorensen G, Dandanell G. A new type of dihydroorotate dehydrogenase, type 1S, from the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles. 2002; 6(3): 245–51. Epub 2002/06/20. doi: 10.1007/s00792-001-0249-0 . [DOI] [PubMed] [Google Scholar]
- 30.Couto SG, Cristina Nonato M, Costa-Filho AJ. Site directed spin labeling studies of Escherichia coli dihydroorotate dehydrogenase N-terminal extension. Biochem Biophys Res Commun. 2011; 414(3): 487–92. Epub 2011/10/12. doi: 10.1016/j.bbrc.2011.09.092 . [DOI] [PubMed] [Google Scholar]
- 31.Loffler M, Knecht W, Rawls J, Ullrich A, Dietz C. Drosophila melanogaster dihydroorotate dehydrogenase: the N-terminus is important for biological function in vivo but not for catalytic properties in vitro. Insect Biochem Mol Biol. 2002; 32(9): 1159–69. Epub 2002/09/06. doi: 10.1016/s0965-1748(02)00052-8 . [DOI] [PubMed] [Google Scholar]
- 32.Larsen JN, Jensen KF. Nucleotide sequence of the pyrD gene of Escherichia coli and characterization of the flavoprotein dihydroorotate dehydrogenase. Eur J Biochem. 1985; 151(1): 59–65. Epub 1985/08/15. doi: 10.1111/j.1432-1033.1985.tb09068.x . [DOI] [PubMed] [Google Scholar]
- 33.Krungkrai J. Purification, characterization and localization of mitochondrial dihydroorotate dehydrogenase in Plasmodium falciparum, human malaria parasite. Biochim Biophys Acta. 1995; 1243(3): 351–60. Epub 1995/04/13. doi: 10.1016/0304-4165(94)00158-t . [DOI] [PubMed] [Google Scholar]
- 34.Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992; 2(3): 417–22. Epub 1992/05/01. doi: 10.1111/j.1365-313x.1992.00417.x . [DOI] [PubMed] [Google Scholar]
- 35.Bader B, Knecht W, Fries M, Loffler M. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr Purif. 1998; 13(3): 414–22. Epub 1998/08/07. doi: 10.1006/prep.1998.0925 . [DOI] [PubMed] [Google Scholar]
- 36.Gustafson G, Davis G, Waldron C, Smith A, Henry M. Identification of a new antifungal target site through a dual biochemical and molecular-genetics approach. Current Genetics. 1996. doi: 10.1007/s002940050115 [DOI] [PubMed] [Google Scholar]
- 37.Zameitat E, Gojkovic Z, Knecht W, Piskur J, Loffler M. Biochemical characterization of recombinant dihydroorotate dehydrogenase from the opportunistic pathogenic yeast Candida albicans. FEBS J. 2006; 273(14): 3183–91. Epub 2006/06/16. doi: 10.1111/j.1742-4658.2006.05327.x . [DOI] [PubMed] [Google Scholar]
- 38.Nielsen FS, Rowland P, Larsen S, Jensen KF. Purification and characterization of dihydroorotate dehydrogenase A from Lactococcus lactis, crystallization and preliminary X-ray diffraction studies of the enzyme. Protein Sci. 1996; 5(5): 852–6. Epub 1996/05/01. doi: 10.1002/pro.5560050506 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen FS, Andersen PS, Jensen KF. The B form of dihydroorotate dehydrogenase from Lactococcus lactis consists of two different subunits, encoded by the pyrDb and pyrK genes, and contains FMN, FAD, and [FeS] redox centers. J Biol Chem. 1996; 271(46): 29359–65. Epub 1996/11/15. doi: 10.1074/jbc.271.46.29359 . [DOI] [PubMed] [Google Scholar]
- 40.Marcinkeviciene J, Tinney LM, Wang KH, Rogers MJ, Copeland RA. Dihydroorotate dehydrogenase B of Enterococcus faecalis. Characterization and insights into chemical mechanism. Biochemistry. 1999; 38(40): 13129–37. Epub 1999/10/21. doi: 10.1021/bi990674q . [DOI] [PubMed] [Google Scholar]
- 41.Marcinkeviciene J, Jiang W, Locke G, Kopcho LM, Rogers MJ, Copeland RA. A second dihydroorotate dehydrogenase (Type A) of the human pathogen Enterococcus faecalis: expression, purification, and steady-state kinetic mechanism. Arch Biochem Biophys. 2000; 377(1): 178–86. Epub 2000/04/25. doi: 10.1006/abbi.2000.1769 . [DOI] [PubMed] [Google Scholar]
- 42.Zameitat E, Knecht W, Piškur J, Löffler M. Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri. FEBS Letters. 2004; 568(1–3): 129–34. doi: 10.1016/j.febslet.2004.05.017 [DOI] [PubMed] [Google Scholar]
- 43.Gojković Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, Pynyaha Y, et al. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Molecular Genetics and Genomics. 2004; 271(4): 387–93. doi: 10.1007/s00438-004-0995-7 [DOI] [PubMed] [Google Scholar]
- 44.Bouwknegt J, Koster CC, Vos AM, Ortiz-Merino RA, Wassink M, Luttik MAH, et al. Class-II dihydroorotate dehydrogenases from three phylogenetically distant fungi support anaerobic pyrimidine biosynthesis. Fungal Biol Biotechnol. 2021; 8(1): 10. Epub 2021/10/18. doi: 10.1186/s40694-021-00117-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene S, Watanabe K, Braatz-Trulson J, Lou L. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochem Pharmacol. 1995; 50(6): 861–7. Epub 1995/09/07. doi: 10.1016/0006-2952(95)00255-x . [DOI] [PubMed] [Google Scholar]
- 46.Fox RI, Herrmann ML, Frangou CG, Wahl GM, Morris RE, Strand V, et al. Mechanism of action for leflunomide in rheumatoid arthritis. Clin Immunol. 1999; 93(3): 198–208. Epub 1999/12/22. doi: 10.1006/clim.1999.4777 . [DOI] [PubMed] [Google Scholar]
- 47.O’Connor PW, Li D, Freedman MS, Bar-Or A, Rice GP, Confavreux C, et al. A Phase II study of the safety and efficacy of teriflunomide in multiple sclerosis with relapses. Neurology. 2006; 66(6): 894–900. Epub 2006/03/29. doi: 10.1212/01.wnl.0000203121.04509.31 . [DOI] [PubMed] [Google Scholar]
- 48.Sibille G, Luganini A, Sainas S, Boschi D, Lolli ML, Gribaudo G. The Novel hDHODH Inhibitor MEDS433 Prevents Influenza Virus Replication by Blocking Pyrimidine Biosynthesis. Viruses. 2022; 14(10). Epub 2022/10/28. doi: 10.3390/v14102281 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips MA, Lotharius J, Marsh K, White J, Dayan A, White KL, et al. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci Transl Med. 2015; 7(296): 296ra111. Epub 2015/07/17. doi: 10.1126/scitranslmed.aaa6645 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.du Pré S, Beckmann N, Almeida C, Sibley GEM, Law D, Brand AC, et al. Effect of the Novel Antifungal Drug F901318 (Olorofim) on Growth and Viability of Aspergillus fumigatus. 2018; 901318: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.du Pré S, Birch M, Law D, Beckmann N, Sibley GEM, Bromley MJ, et al. The dynamic influence of olorofim (F901318) on the cell morphology and organization of living cells of Aspergillus fumigatus. Journal of Fungi. 2020; 6(2). doi: 10.3390/jof6020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoenigl M, Seidel D, Carvalho A, Rudramurthy SM, Arastehfar A, Gangneux JP, et al. The emergence of COVID-19 associated mucormycosis: a review of cases from 18 countries. Lancet Microbe. 2022; 3(7): e543–e52. Epub 2022/02/01. doi: 10.1016/S2666-5247(21)00237-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouwknegt J, Vos AM, Ortiz Merino RA, van Cuylenburg DC, Luttik MAH, Pronk JT. Identification of fungal dihydrouracil-oxidase genes by expression in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 2022; 115(11): 1363–78. Epub 2022/10/15. doi: 10.1007/s10482-022-01779-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis CH, Putnam MD, Thwaites WM. Metabolism of dihydrouracil in Rhodosporidium toruloides. J Bacteriol. 1984; 158(1): 347–50. Epub 1984/04/01. doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lax C, Navarro-Mendoza MI, Perez-Arques C, Navarro E, Nicolas FE, Garre V. Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus. Cell Rep Methods. 2021; 1(8): 100124. Epub 2022/04/28. doi: 10.1016/j.crmeth.2021.100124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuboi Y, Sakuma T, Yamamoto T, Horiuchi H, Takahashi F, Igarashi K, et al. Gene manipulation in the Mucorales fungus Rhizopus oryzae using TALENs with exonuclease overexpression. FEMS Microbiol Lett. 2022; 369(1). Epub 2022/02/10. doi: 10.1093/femsle/fnac010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lax C, Canovas-Marquez JT, Tahiri G, Navarro E, Garre V, Nicolas FE. Genetic Manipulation in Mucorales and New Developments to Study Mucormycosis. Int J Mol Sci. 2022; 23(7). Epub 2022/04/13. doi: 10.3390/ijms23073454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng M, Cooper CR Jr., Szaniszlo PJ. Genetic transformation of the pathogenic fungus Wangiella dermatitidis. Appl Microbiol Biotechnol. 1995; 44(3–4): 444–50. Epub 1995/12/01. doi: 10.1007/BF00169942 . [DOI] [PubMed] [Google Scholar]
- 59.Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021; 49(D1): D344–D54. Epub 2020/11/07. doi: 10.1093/nar/gkaa977 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36(Web Server issue): W465–9. Epub 2008/04/22. doi: 10.1093/nar/gkn180 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32(5): 1792–7. Epub 2004/03/23. doi: 10.1093/nar/gkh340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000; 17(4): 540–52. Epub 2000/03/31. doi: 10.1093/oxfordjournals.molbev.a026334 . [DOI] [PubMed] [Google Scholar]
- 63.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003; 52(5): 696–704. Epub 2003/10/08. doi: 10.1080/10635150390235520 . [DOI] [PubMed] [Google Scholar]
- 64.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006; 55(4): 539–52. Epub 2006/06/21. doi: 10.1080/10635150600755453 . [DOI] [PubMed] [Google Scholar]
- 65.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006; 7: 439. Epub 2006/10/13. doi: 10.1186/1471-2105-7-439 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988; 16(22): 10881–90. Epub 1988/11/25. doi: 10.1093/nar/16.22.10881 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999; 15(4): 305–8. Epub 1999/05/13. doi: 10.1093/bioinformatics/15.4.305 . [DOI] [PubMed] [Google Scholar]
- 68.Koller A, Valesco J, Subramani S. The CUP1 promoter of Saccharomyces cerevisiae is inducible by copper in Pichia pastoris. Yeast. 2000; 16: 651–656. doi: . [DOI] [PubMed] [Google Scholar]
- 69.Montgomery DL, Hall BD, Gillam S, Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978; 14(3): 673–80. Epub 1978/07/01. doi: 10.1016/0092-8674(78)90250-7 [DOI] [PubMed] [Google Scholar]
- 70.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989; 122(1): 19–27. Epub 1989/05/01. doi: 10.1093/genetics/122.1.19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007; 2(4): 924–32. Epub 2007/04/21. doi: 10.1038/nprot.2007.132 . [DOI] [PubMed] [Google Scholar]
- 72.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994; 10(13): 1793–808. Epub 1994/12/01. doi: 10.1002/yea.320101310 . [DOI] [PubMed] [Google Scholar]
- 73.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999; 285(5429): 901–6. Epub 1999/08/07. doi: 10.1126/science.285.5429.901 . [DOI] [PubMed] [Google Scholar]
- 74.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002; 418(6896): 387–91. Epub 2002/07/26. doi: 10.1038/nature00935 . [DOI] [PubMed] [Google Scholar]
- 75.Ohkuni K, Hayashi M, Yamashita I. Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae. Yeast. 1998; 14(7): 623–31. Epub 1998/06/25. doi: . [DOI] [PubMed] [Google Scholar]
- 76.Gietz RD, Schiestl RH. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007; 2(1): 1–4. Epub 2007/04/03. doi: 10.1038/nprot.2007.17 . [DOI] [PubMed] [Google Scholar]
- 77.Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Soding J, et al. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics. 2020; 72(1): e108. Epub 2020/12/15. doi: 10.1002/cpbi.108 . [DOI] [PubMed] [Google Scholar]
- 78.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004; 20(18): 3702–4. Epub 2004/07/31. doi: 10.1093/bioinformatics/bth444 . [DOI] [PubMed] [Google Scholar]
- 79.Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis. 2005; 41(5): 634–53. Epub 2005/08/05. doi: 10.1086/432579 . [DOI] [PubMed] [Google Scholar]
- 80.Li Y, Steenwyk JL, Chang Y, Wang Y, James TY, Stajich JE, et al. A genome-scale phylogeny of the kingdom Fungi. Curr Biol. 2021; 31(8): 1653–65 e5. Epub 2021/02/20. doi: 10.1016/j.cub.2021.01.074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zameitat E, Freymark G, Dietz CD, Loffler M, Bolker M. Functional expression of human dihydroorotate dehydrogenase (DHODH) in pyr4 mutants of Ustilago maydis allows target validation of DHODH inhibitors in vivo. Appl Environ Microbiol. 2007; 73(10): 3371–9. Epub 2007/03/21. doi: 10.1128/AEM.02569-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu W, Farrell RA, Stillman DJ, Winge DR. Identification of SLF1 as a new copper homeostasis gene involved in copper sulfide mineralization in Saccharomyces cerevisiae. Mol Cell Biol. 1996; 16(5): 2464–72. Epub 1996/05/01. doi: 10.1128/MCB.16.5.2464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruta LL, Farcasanu IC. Saccharomyces cerevisiae Concentrates Subtoxic Copper onto Cell Wall from Solid Media Containing Reducing Sugars as Carbon Source. Bioengineering (Basel). 2021; 8(3). Epub 2021/04/04. doi: 10.3390/bioengineering8030036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bjornberg O, Rowland P, Larsen S, Jensen KF. Active site of dihydroorotate dehydrogenase A from Lactococcus lactis investigated by chemical modification and mutagenesis. Biochemistry. 1997; 36(51): 16197–205. Epub 1998/01/24. doi: 10.1021/bi971628y . [DOI] [PubMed] [Google Scholar]
- 85.Rowland P, Nielsen FS, Jensen KF, Larsen S. The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis. Structure. 1997; 5(2): 239–52. Epub 1997/02/15. doi: 10.1016/s0969-2126(97)00182-2 . [DOI] [PubMed] [Google Scholar]
- 86.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014; 42(Web Server issue): W320–4. Epub 2014/04/23. doi: 10.1093/nar/gku316 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jordan DB, Bisaha JJ, Picollelli MA. Catalytic properties of dihydroorotate dehydrogenase from Saccharomyces cerevisiae: studies on pH, alternate substrates, and inhibitors. Arch Biochem Biophys. 2000; 378(1): 84–92. Epub 2000/06/28. doi: 10.1006/abbi.2000.1823 . [DOI] [PubMed] [Google Scholar]
- 88.Owaki J, Uzura K, Minami Z, Kusai K. Partial Purification and Characterization of Dihydrouracil Oxidase, a Flavoprotein from Rhodotorula glutinis. J Ferment Technol. 1986; 64(3): 205–10. [Google Scholar]
- 89.Valle-Maldonado MI, Jacome-Galarza IE, Gutierrez-Corona F, Ramirez-Diaz MI, Campos-Garcia J, Meza-Carmen V. Selection of reference genes for quantitative real time RT-PCR during dimorphism in the zygomycete Mucor circinelloides. Mol Biol Rep. 2015; 42(3): 705–11. Epub 2014/11/14. doi: 10.1007/s11033-014-3818-x . [DOI] [PubMed] [Google Scholar]
- 90.Homa M, Ibragimova S, Szebenyi C, Nagy G, Zsindely N, Bodai L, et al. Differential Gene Expression of Mucor lusitanicus under Aerobic and Anaerobic Conditions. J Fungi (Basel). 2022; 8(4). Epub 2022/04/22. doi: 10.3390/jof8040404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinheiro MP, Iulek J, Cristina Nonato M. Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase from Y strain. Biochem Biophys Res Commun. 2008; 369(3): 812–7. Epub 2008/02/28. doi: 10.1016/j.bbrc.2008.02.074 . [DOI] [PubMed] [Google Scholar]
- 92.Ribeiro AJM, Riziotis IG, Tyzack JD, Borkakoti N, Thornton JM. Using mechanism similarity to understand enzyme evolution. Biophys Rev. 2022; 14(6): 1273–80. Epub 2023/01/21. doi: 10.1007/s12551-022-01022-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abou Ammar G, Tryono R, Doll K, Karlovsky P, Deising HB, Wirsel SG. Identification of ABC transporter genes of Fusarium graminearum with roles in azole tolerance and/or virulence. PLoS One. 2013; 8(11): e79042. Epub 2013/11/19. doi: 10.1371/journal.pone.0079042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buil JB, Oliver JD, Law D, Baltussen T, Zoll J, Hokken MWJ, et al. Resistance profiling of Aspergillus fumigatus to olorofim indicates absence of intrinsic resistance and unveils the molecular mechanisms of acquired olorofim resistance. Emerg Microbes Infect. 2022; 11(1): 703–14. Epub 2022/02/04. doi: 10.1080/22221751.2022.2034485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Serial dilution assay of the negative control (β-galtrunc) and confirmed class 1A DHODH from S. cerevisiae (SC), T. cruzi (TC), L. kluveryi (LK1A) in ura1Δ cells in the presence of high copper concentration and pyrimidines. (B) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and confirmed class 2 DHODH from A. fumigatus (AF) in ura1Δ cells in the presence of high copper concentration and pyrimidines.
(TIF)
NuPAGE analysis of purified recombinant proteins. T. cruzi (TC), L. kluveryi (LK1A), S. cerevisiae (SC), M. circinelloides (MC) and R. arrhizus (RA).
(TIF)
Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and three putative A. alternata genes in ura1Δ cells. AAEc, E. coli codon-optimised construct; AANat, native A. alternata sequence construct; AASc, S. cerevisiae codon-optimised construct.
(TIF)
Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and three putative A. alternata genes in ura1Δ cells. AAEc, E. coli codon-optimised construct; AANat, native A. alternata sequence construct; AASc, S. cerevisiae codon-optimised construct in the presence of DHU.
(TIF)
(A) NuPAGE analysis of purified proteins. A. alternata (AA), P. minimum (PM). (B) Amount of DCIP reduced by proven and putative DHODH after 30 minutes. (C) Amount of FeCy reduced by proven and putative DHODH after 30 minutes. (D) Amount of orotate produced by proven and putative DHODH after 30 minutes.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.