Abstract
Marek’s disease is a herpesvirus (Marek’s disease virus [MDV])-induced pathology of chickens characterized by paralysis and the rapid appearance of T-cell lymphomas. Lymphoblastoid cell lines (LBCLs) derived from MDV-induced tumors have served as models of MDV latency and transformation. We have recently reported the construction of mutant MDVs having a deletion (M. S. Parcells et al., J. Virol. 69:7888–7898, 1995) and an insertion (A. S. Anderson et al., J. Virol. 72:2548–2553, 1998) within the unique short region of the virus genome. These mutant MDVs retained oncogenicity, and LBCLs have been established from the mutant-induced tumors. We report the characterization of these cell lines with respect to (i) virus structure within and reactivated from the cell lines, (ii) surface antigen expression, (iii) kinetics of MDV and marker gene induction, (iv) localization and colocalization of induced MDV antigens and β-galactosidase (β-Gal), and (v) methylation status of the region of lacZ insertion in recombinant- and non-recombinant-derived cell lines. Our results indicate that (i) recombinant-derived cell lines contain no parental virus, (ii) the established cell lines are predominantly CD4+ CD8−, (iii) the percentage of Lac-expressing cells is low (1 to 3%) but increases dramatically upon 5′-iododeoxyuridine (IUdR) treatment, (iv) lacZ expression is induced with the same kinetics as several MDV lytic-phase genes (pp38, US1, gB, gI, and US10), and (v) the regulation of lacZ expression is not mediated by methylation. Furthermore, the MDV-encoded oncoprotein, Meq, could be detected in cells expressing β-Gal and various lytic antigens but did not appear to be induced by IUdR treatment. Our results indicate that regulation of the lacZ marker gene can serve as sensitive measure of virus lytic-phase induction and the reactivation from latency.
Marek’s disease is a pathology of chickens characterized by paralysis, peripheral nerve demyelination, and, most commonly, the rapid formation of lymphomas (reviewed in references 21 and 86). The etiologic agent, Marek’s disease virus (MDV), is an acute-transforming, cell-associated alphaherpesvirus that is ubiquitous in commercial poultry production. In chickens, MDV undergoes lytic, albeit productive/restrictive, replication in B and T cells at early times postinfection (17, 19). A strictly cell associated viremia ensues in the peripheral blood, with an accompanying immunosuppression and a switch from lytic to latent infection (12). The early immunosuppression induced by MDV has been attributed to bursal and thymic atrophy and virus-induced host factors. The switch to latency appears to be mediated by host immune factors including interferon (10, 11, 45) and may be biphasic in onset (88). MDV latent infection occurs primarily in CD4+ T cells (T-helper [TH] cells) that express major histocompatibility complex (MHC) class II (MHC-II; Ia) (13, 17, 20). The T-cell expression of MHC-II has led to the conclusion that latently infected T cells are activated (13, 18, 19) and to the hypothesis that T-cell activation precedes and is necessary for T-cell infection.
Following primary immunosuppression, a secondary cytolytic infection occurs at peripheral sites, including the feather follicle epithelium, where infectious virus is shed in the dander (15). In susceptible chickens, the secondary cytolytic infection is followed by a profound immunosuppression and the appearance of lymphomas. This immunosuppression appears to be caused by tumor cell-associated factors (45, 85), apoptosis of CD4+ T cells, and the down regulation of CD8 expression among cytotoxic T cells in the peripheral blood (54).
Lymphoblastoid cell lines (LBCLs) established from MDV-induced tumors are primarily CD4+, suggesting that transformed cells arise from the pool of latently infected T cells (56, 57, 67, 75, 76). These LBCLs retain some responsiveness to lymphokines (42) and interferon (88, 89). Moreover, several LBCLs were shown to be immunosuppressive to proliferating chicken spleen cells even after glutaraldehyde fixation, suggesting that receptors present on the surface of LBCLs mediated this effect (70). MDV can be rescued from many LBCLs via cocultivation with chicken embryo fibroblasts (CEF) or chicken kidney cells, and the level of spontaneous virus reactivation appears to vary among cell lines and with length of time in culture (20, 58a). The state of the MDV genome in LBCLs has been the subject of some controversy (84), but recent studies (28, 29) have shown that integration of the MDV genome is a common feature of LBCLs as well as MDV primary lymphomas. Reactivation of MDV from LBCLs is therefore believed to entail either an excision or replication from the latent, integrated genome copies (29). Consequently, LBCLs have served as the model for MDV latency as well as for transformation, although the relationship between these events remains unclear.
To study the function of MDV-encoded gene products in the context of infection, we constructed mutant MDVs with insertions or deletions in the virus genome and characterized these viruses in cell culture and in chickens (22, 59, 60). More recently, we constructed mutant MDVs derived from a highly pathogenic/oncogenic strain of MDV, RB1B (4, 53, 61, 62). During the characterization of these mutant MDVs in vivo, we were able to establish LBCLs. These cell lines are T-lymphoblastoid cells and are predominantly CD4+, i.e., having a TH immunophenotype. We have found that in the recombinant-MDV-induced tumors, as well as in the established cell lines, the lacZ marker gene used in construction of these mutants is repressed, despite constitutive expression of lacZ during the lytic infection of CEF with these viruses (61). We have hypothesized that this gene can serve as a marker for virus reactivation from latency. To support this hypothesis, we now show that (i) the lacZ gene is present in the genome of the MDVs within the recombinant cell lines, and no parental virus is present or can be reactivated from these lines; (ii) treatment of these cell lines with 5′-iodo-2′-deoxyuridine (IUdR) induces the expression of β-galactosidase (β-Gal) as well as MDV-encoded lytic gene products with indistinguishable kinetics, (iii) recombinant-derived LBCLs express both MDV antigens and β-Gal, suggesting that lacZ induction correlates with MDV gene induction. We have also determined that the observed regulation does not appear to involve methylation, as the segment of the unique short (US) region into which the lacZ cassette has been inserted in the recombinant-derived as well as parent virus-derived LBCLs is largely nonmethylated. Given the observed regulation of a heterologous promoter within the context of the MDV genome, these mutant-MDV-derived cell lines promise to provide a novel method for the study of MDV gene regulation in LBCLs.
MATERIALS AND METHODS
Cells and viruses.
The construction of mutant MDVs from strain RB1B, a strain of exceptionally high virulence (74), has been described previously (61). In the deletion mutant MDV, RB1BΔ4.5lac, six genes are removed from the US region of the virus genome and replaced with a lacZ expression cassette. The structures of RB1BΔ4.5lac and RB1BΔ4.5lac reisolated from infected chickens have been reported previously (61). The insertion mutant MDV, RB1BUS6lacgpt, contains an insertion of a lacZ-gpt cassette within the US6 (glycoprotein D [gD] homolog) open reading frame (ORF). The structures of RB1BUS6lacgpt, RB1BUS6lacgpt in the derived cell lines (MDCC-UD22 to MDCC-UD29), and RB1BUS6lacgpt reactivated from these cell lines are reported elsewhere (4).
LBCLs were established from lymphomas induced by the mutant MDVs as described elsewhere (4, 61). Essentially, lymphomas were removed aseptically from experimentally infected specific-pathogen-free single-comb White Leghorn chickens exhibiting signs of Marek’s disease (paralysis and cachexia). Excised tumors were homogenized on ice in M199 medium supplemented with antibiotics (penicillin G [50 μg/ml], streptomycin sulfate [50 μg/ml], and neomycin sulfate [100 μg/ml]; Gibco-BRL Life Technologies, Bethesda, Md. [Gibco]), using Tenbroeck homogenizers. The homogenate was crudely filtered through sterile cheesecloth into 50-ml polypropylene tubes. Lymphocytes were purified from the homogenate by density gradient centrifugation using Histopaque 1119 (Sigma Chemical Corp., St. Louis, Mo. [Sigma]), washed twice with M199 supplemented with 3% calf serum and antibiotic (Gibco), counted, and resuspended to 2 × 107 cells/ml in modified LM Hahn medium (76) without tryptose phosphate broth. The lymphocytes/blasts were plated at 0.1 × 107, 0.5 × 107, and 1 × 107 cells/ml in 12-well tissue culture plates (Corning Glassworks, Corning, N.Y.), monitored daily, and split 1:3 to 1:5 every 4 to 7 days until established. Most cell lines were established by five to eight passages, and stocks were frozen in 10% dimethyl sulfoxide (Sigma) in fetal bovine serum (Gibco). Unless otherwise stated, all cell lines used for analysis were maintained in modified LM Hahn medium.
The MDV-transformed cell line MDCC-MSB1 (3) was obtained from Sandra Cloud, Department Animal and Food Sciences, University of Delaware. Non-MDV-transformed chicken B- and T-LBCL controls consisting of reticuloendotheliosis virus (REV)-transformed B- and T-cell lines RECC-CU60 and RECC-CU91, respectively, were obtained from Karel A. Schat, Department of Microbiology and Immunology, Veterinary Medical Center, Cornell University.
Antibodies.
Monoclonal antibodies to chicken CD3, CD4, CD8α, CD8β, and CD28 antigens and to T-cell receptor class 1 (TCR1; γδ), TCR2 (α-Vβ1), and TCR3 (α-Vβ2) were obtained commercially (Southern Biotechnology Associates, Inc., Birmingham, Ala. [SBT]). Monoclonal antibodies to chicken MHC-II, P2M11 and CIa-1, were obtained from Hyun Lillehoj (Infectious Diseases Research Laboratory, U.S. Department of Agriculture [USDA], Beltsville, Md.) and Donald Ewert (Wistar Institute, Philadelphia, Pa.), respectively. Monoclonal antibodies to chicken immunoglobulin M (IgM) and IgD and a pan-B-cell antigen (Bu-1a) were also obtained from Hyun Lillehoj.
Monoclonal antibodies to MDV pp38 (H19.47), Meq (23B46), and gB (1AN86.17) were obtained from Lucy Lee, USDA-ADOL, East Lansing, Mich. Rabbit polyclonal antiserum to MDV US7 (gI)-, US1 (ICP22 homolog)-, and US10-encoded proteins were obtained from Peter Brunovskis, Department of Microbiology and Molecular Biology, Case Western Reserve University Cleveland, Ohio, and Leland Velicer, Department of Microbiology, Michigan State University, East Lansing. Rabbit polyclonal antiserum to β-Gal was obtained commercially (5 Prime→3 Prime, Inc., Boulder, Colo.).
Indirect immunofluorescence analysis of fixed cells (dual staining).
For indirect immunofluorescence analysis, 50 μl of paraformaldehyde-fixed (see below) lymphoblastoid cells was pipetted onto 10-well Teflon-coated slides (Cel-Line, Newfield, N.J.). After approximately 5 min, the liquid was removed by pipetting and the remaining meniscus of liquid was allowed to air dry for 5 to 10 min. Dried slides were rehydrated in 1× phosphate-buffered saline, pH 7.4 (PBS), and blocked with 3% goat serum in PBS for 1 h at 37°C. The fixed cells were then stained with monoclonal (anti-pp38, -Meq, or -gB) and polyclonal (anti-US1, -US7, -US10, or anti-β-Gal) antibodies which had been preadsorbed with fixed RECC-CU91 cells (see below) and diluted in the blocking solution (1:50) for 1 to 2 h at 37°C in a humidified chamber. The slides were washed three times in PBS and stained with goat anti-mouse Ig–Alexa 546 conjugate (Molecular Probes, Eugene, Oreg.) and goat anti-rabbit Ig–fluorescein isothiocyanate (FITC) conjugate (Sigma) diluted 1:100 in blocking buffer. The slides were incubated for 1 h at 37°C in a humidified chamber, washed three times with PBS, and covered with VectaShield containing 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml; Vector Laboratories Inc., Burlingame, Calif.) and a coverslip. Fluorescence-labeled cells were observed in an Olympus BX50 fluorescence microscope (Olympus, Lake Success, N.Y.) using a triple-cube (MF) filter (Omega Optical, Brattleboro, Vt.) and 60× or 100× oil immersion lenses. Fluorescent images were captured with ImagePro Plus (Media Cybernetics, Silver Spring, Md.), and color and contrast levels were adjusted by using Adobe Photoshop 4.0 (Adobe Systems Inc., Salinas, Calif.). Composite figures were assembled with QuarkXpress 3.31 software (Quark Inc., Denver, Colo.). For each antibody set, CU91 cells were also stained and observed as a negative control in addition to secondary-antibody-only controls.
Flow cytometric analysis. (i) Immunophenotypic analysis of cell lines.
For immunophenotypic analysis, cell lines were stained with anti-chicken T-lymphocyte monoclonal antibodies directly conjugated to FITC. In addition, analyses were repeated using indirect immunofluorescence to detect low-level receptor expression. Standard direct and indirect immunostaining methods were used on living cells according to the recommendations of the manufacturer (SBT).
For flow cytometric analysis, FACSort (Becton Dickenson, San Jose, Calif.) flow cytometers were used with Hewlett-Packard (University of Delaware) and Macintosh (University of Arkansas) computer hardware. For each sample, 104 ungated cells were acquired. During analysis, live cells were gated according to their scatter profiles after identification by propidium iodide (PI; 10 μg/ml in PBS–1% bovine serum albumin [BSA]–1% sodium azide; Sigma) exclusion. For data analysis, Lysis II and CellQuest (Becton Dickinson) software packages were used. Immunophenotypic analysis was performed on each cell line two to four times.
(ii) Flow cytometry of fixed, solubilized LBCLs.
For flow cytometric analysis of cytoplasmic antigen expression during reactivation, we used two methods of fixation and cellular permeabilization. Initially, we used an ethanol (EtOH) fixation method which worked well with anti-pp38, anti-US1, anti-US7, anti-gB, and anti-β-Gal antibodies. Since the anti-Meq antibody did not react with EtOH-fixed cells, we then used a paraformaldehyde fixation method. In any event, the results obtained with either fixation method were essentially identical for the pp38, US1, US7, gB, and β-Gal antibodies.
EtOH fixation method.
The method for EtOH fixation and permeabilization was used as detailed in the appendix of the PharMingen (San Diego, Calif. catalog for the quantitation of cyclin expression. Essentially, 2 × 107 lymphoblastoid cells were pelleted by centrifugation, the supernatant medium was decanted, and the pellet was loosened by agitation. Cells were fixed via the dropwise addition of cold 75% EtOH to a total of 10 ml while the cells were agitated with a vortex mixer. Cells were stored at −20°C overnight, after which time they were recovered by centrifugation, the EtOH was decanted, and the pellet was loosened by agitation. The cells were rehydrated in PBS (Sigma) and washed successively with 10 ml of additional PBS and 10 ml wash buffer (PBS containing 1% BSA [fraction V; Fisher Biotechnology, St. Louis, Mo.], 1% fetal bovine serum [Gibco], 1% goat serum [Sigma], 0.1% saponin [Sigma], and 0.1% sodium azide [Sigma]). The fixed, rehydrated, and permeabilized cells were then blocked in 1 ml of wash buffer containing 5% goat serum (diluent) for 1 h at room temperature with agitation. After blocking, 50 μl (106) cells were added to primary antibody diluted 1:50 in permeabilizing diluent.
To remove nonspecific anti-chicken lymphocyte antibodies, all primary antibodies were preabsorbed overnight at a 1:25 dilution, using EtOH-fixed RECC-CU91 cells (107/ml). The cells were then removed by centrifugation, and the preadsorbed antisera were diluted with an equal volume of diluent and filtered through 0.45-μm-pore-size filters (Acrodisc; Gelman Sciences, Fisher). Likewise, secondary staining reagents goat anti-mouse–FITC conjugate (Sigma) and goat anti-rabbit–FITC conjugate (Sigma) were preadsorbed by using EtOH-fixed RECC-CU91 cells and filtered as described above. Cells were incubated with primary and the appropriate secondary antisera for 30 min at room temperature (rt) with agitation. Between incubations and after secondary staining, cells were washed twice with 3 ml of wash buffer. For the final wash, cells were resuspended in wash buffer containing 10 μg of PI/ml, incubated for 10 min, pelleted as described above, and resuspended in 0.3 ml of PBS–1% BSA–0.1% sodium azide.
Paraformaldehyde fixation method.
The method used for paraformaldehyde fixation was similar to that of Baigent et al. (5). Briefly, cells were harvested by centrifugation, washed once with 10 ml of PBS, and resuspended to a concentration of 107/ml in 0.4% paraformaldehyde (Sigma) in PBS. The cells were fixed on ice with agitation for 30 min, washed once with 10 ml of PBS, and then permeabilized as described above. The remaining blocking and staining steps were the same as for the EtOH fixation method.
During analysis, cell permeabilization and antibody staining were monitored by FL-2 (orange/red, PI) and FL-1 (green, FITC) fluorescence, respectively. Samples consisting of RECC-CU91 cells and secondary reagent only were included as controls for the specificity of antibody staining. In addition, since MDCC-UD01 cells were transformed by RB1BΔ4.5lac, a virus having a deletion of six genes including US1, these cells served as an MDV-derived cell line control for the anti-US1 protein staining of cell lines MDCC-UD14 and MDCC-UD24. Likewise, MDCC-UD14 cells were transformed by the parent virus, RB1B, and therefore served as an MDV-derived cell line control for the anti-β-Gal staining of cell lines MDCC-UD01 and MDCC-UD24. MSB-1 cells were not included as controls because this line has undergone extensive passage in culture and was found to have altered surface antigen expression since its original characterization (76) (see Table 2). For each sample, 104 fixed and permeabilized cells were acquired for analysis. To confirm results obtained by fluorescence-activated cell sorting (FACS), samples of cells were taken before and after fixation as well as after staining and placed onto 10-well Teflon-coated slides. The prefixation samples (50 μl from each culture) were placed on slides, air dried, and stored at −20°C. Micrographs of cells poststaining shown in Fig. 4A are from the samples used for flow cytometric analysis.
TABLE 2.
Immunophenotypic characterization of cell lines
| Cell line | Relative fluorescence intensitya
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3b | CD4 | CD8α | CD8β | CD28 | TCR1 | TCR2 | TCR3 | MHC-IIcd | IgMc | Buc | |
| MDCC-UD01 | ++ | +++ | − | − | ++ | − | ++ | − | ++++ | − | − |
| MDCC-UD02 | ++ | +++ | − | − | + | − | + | − | + | − | − |
| MDCC-UD03 | +/− | +++ | − | − | + | − | +/− | − | ++ | − | − |
| MDCC-UD13 | +/− | − | ++ | − | ++ | − | +/− | − | ++++ | − | − |
| MDCC-UD14 | ++ | +++ | − | − | ++ | − | ++ | + | ++++ | − | − |
| MDCC-UD22 | + | +++ | − | ND | ND | − | ++ | − | + | − | − |
| MDCC-UD24 | + | +++ | − | − | − | − | + | − | +++ | − | − |
| MDCC-UD25 | + | +++ | − | ND | ND | − | + | − | ++++ | − | − |
| MDCC-UD27 | + | +++ | − | ND | ND | − | + | − | ++ | − | − |
| MDCC-UD28 | ++ | +++ | − | − | +++ | − | ++ | + | +++ | − | − |
| MDCC-UD29 | + | +++ | − | ND | ND | − | + | − | ++ | − | − |
| MDCC-MSB1e | ++ | +++ | − | − | − | − | ++ | − | +/− | − | − |
| RECC-CU60f | − | − | − | ND | ND | − | − | − | − | ++ | ++++ |
| RECC-CU91 | + | − | − | − | + | − | − | − | ++++ | − | − |
Number of plus signs indicates relative fluorescence intensity determined by direct staining methods; −, no detectable expression by direct or indirect staining methods; +/−, surface expression detectable only via indirect staining methods; ND, not determined.
Anti-chicken T-cell antibody obtained from SBT.
Anti-chicken MHC-II (P2M11), IgM/IgD, and Bu-1a monoclonal antibodies obtained from H. Lillehoj.
Anti-chicken MHC-II (CIa-1) antibody obtained from D. Ewert.
Originally described in reference 3; obtained from S. Cloud.
Originally described in reference 77; obtained from K. A. Schat.
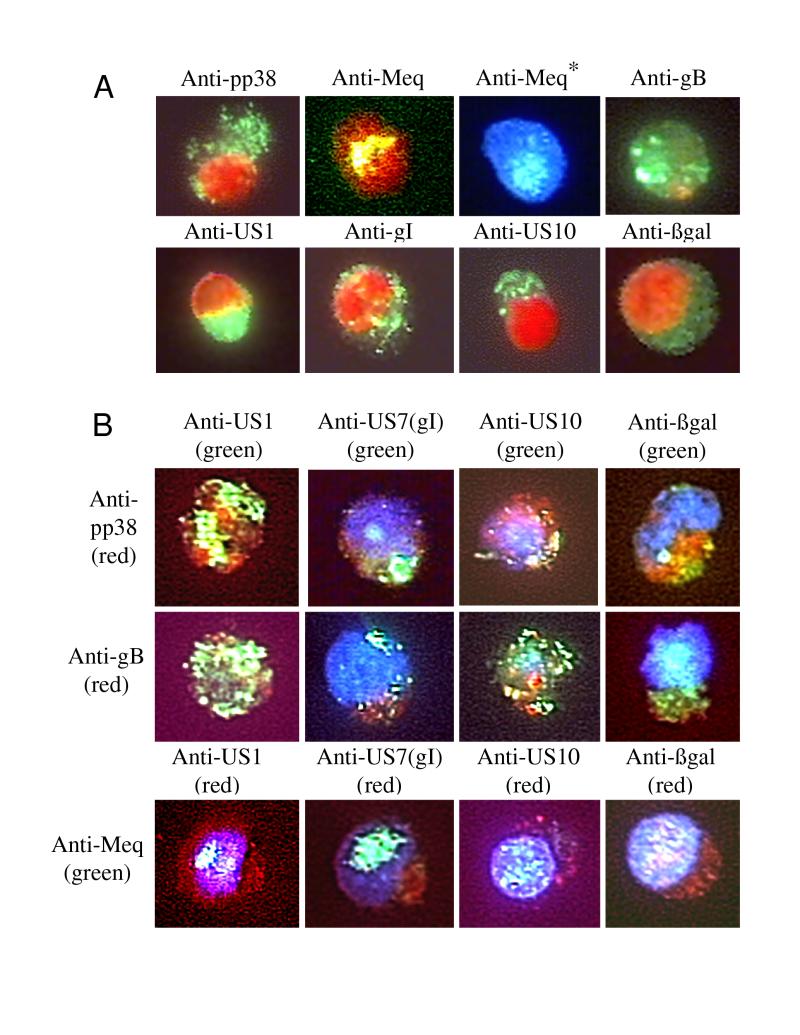
FIG. 4.
(A) Single-antibody immunofluorescence analysis of induced antigen expression. Samples from cells stained for FACS analysis were examined under oil immersion (total magnification, ×600). The cells were stained as described below for FACS analysis of internal antigens. Nuclei were stained with PI (10 μg/ml) or DAPI (VectaShield; Vector Laboratories). For each antibody, RECC-CU91 cells were stained and observed for any background (data not shown). The cell lines shown are MDCC-UD14 (anti-pp38, anti-Meq, anti-gB, anti-US1, anti-gI, and anti-US10) and MDCC-UD01 (anti-βgal). ∗, Meq in the nuclei was difficult to observe in PI-stained cells; therefore the staining observed with DAPI-stained nuclei is also presented. (B) Dual-antibody immunofluorescence analysis of induced antigen expression. For the upper two rows, monoclonal antibodies (anti-pp38 and anti-gB) were detected with anti-mouse Ig Alexa 546 conjugate (Molecular Probes), and polyclonal antibodies (anti-US1, anti-US7, anti-US10, and anti-βgal) were detected with anti-rabbit Ig–FITC conjugate (Sigma). For the bottom row, the anti-Meq monoclonal antibody was detected with anti-mouse Ig–FITC and the polyclonal antibodies were detected with anti-rabbit Alexa 546 conjugate. This color scheme was used because the Alexa 546 dye was less stable than FITC under the high-power illumination. The cells were observed with an oil immersion lens (total magnification, ×1,000). For anti-β-Gal staining, MDCC-UD01 cells were used; in all other cases, MDCC-UD14 cells were used.
IUdR treatment.
To induce MDV antigen expression in the mutant and RB1B-derived cell lines, cells were treated with IUdR (Sigma) as originally described (30). Cells were harvested by centrifugation and resuspended at a concentration of 106/ml in medium containing IUdR (25 μg/ml). Cells were then incubated for 0, 24, and 48 h prior to harvest for flow cytometric and immunofluorescence analyses.
Southern blot hybridizations.
To examine the genomic structure of MDVs in the recombinant- and parental-MDV-derived cell lines, 10-μg aliquots of DNA from cell lines MDCC-UD01, -UD02, -UD03, -UD13, and -UD14, and from CEF infected with virus reactivated from each cell line, were digested with EcoRI. The DNA fragments were separated on a 0.8% agarose gel in 1× 90 mM Tris-borate–2 mM EDTA and transferred to Hybond reinforced nitrocellulose (Amersham Life Sciences Inc., Arlington Heights, Ill. [Amersham]), using standard methods (72). The immobilized DNA fragments were hybridized with a random-primed [α-32P]dCTP-labeled (Rediprime; Amersham) 12-kbp HindIII-BamHI subfragment of the BamHI-A clone of an MDV genomic library (31). This subfragment contains most of the US region of MDV and spans the region deleted in RB1BΔ4.5lac (Fig. 1A). The blot was hybridized for 2 h at 65°C with agitation. The probe was removed, and the blot was washed with increasing stringency (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate to 0.1× SSC–0.1% sodium dodecyl sulfate). The washed blot was exposed to X-ray film (XAR; Kodak, Rochester, N.Y.) for exposures ranging from 30 min to 2 h. For study of the methylation status of the MDV genome in cell lines MDCC-MSB-1, MDCC-UD01, and MDCC-UD14, and the viruses reactivated from these lines (BC-1, RB1BΔ4.5lac, and RB1B, respectively), 30-μg aliquots of cell line and virus DNAs were digested with EcoRI overnight at 37°C and then divided into three tubes. One tube (10 μg) was not digested further, one (10 μg) was digested with high-concentration MspI (methylation insensitive; New England Biolabs), and the third was digested with high-concentration HpaII (methylation sensitive; New England Biolabs). The EcoRI-, EcoRI-, and MspI-digested and EcoRI and HpaII-digested samples were separated on a 1% agarose gel, transferred to nitrocellulose, probed, washed, and exposed to film (as described above). The probe used was a 2.4-kbp SacI fragment from plasmid pMD190 (59). This fragment spans the 5′ end of the lacZ cassette and the promoter region of the US2 and US3 genes (Fig. 5A).
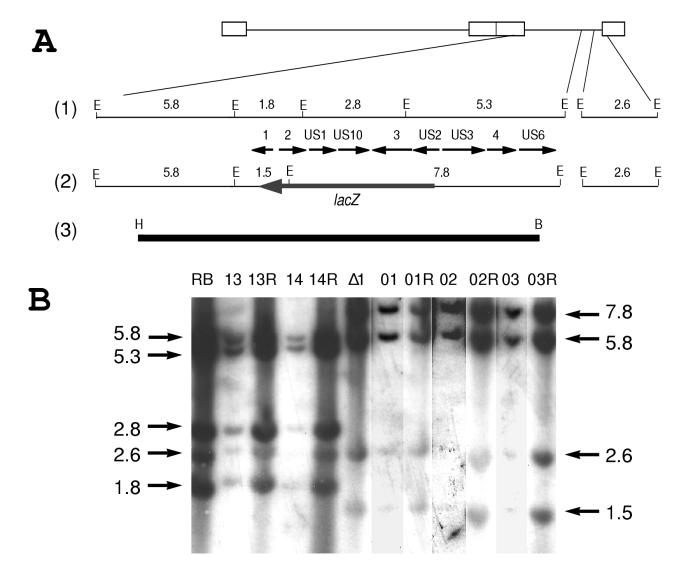
FIG. 1.
(A) Diagrams of the MDV genome: 1, the unique-short (US) region of MDV including the ORFs encoded; 2, the US region of the deletion mutant MDV, RB1BΔ4.5lac; 3, a 12-kbp HindIII (H)-BamHI (B) subfragment of the BamHI-A fragment of the MDV genome (31) spanning the internal-repeat short (IRs)/US junction and containing most of the US region. This probe spans the region deleted in RB1BΔ4.5lac. The EcoRI restriction sites (E) are shown for the region, with their approximate sizes given in kilobase pairs. The ORFs are named according to their homology to herpes simplex virus, and numbers 1 through 4 denote MDV-specific ORFs according to the nomenclature of Brunovskis and Velicer (9). (B) Southern blot hybridization of DNAs purified from cell lines and from CEF infected with MDV reactivated from the RB1B- or RB1BΔ4.5lac-derived cell lines. Lanes: RB, RB1B-infected CEF DNA (parental virus); 13, MDCC-UD13 DNA; 13R, UD13 reactivated onto CEF DNA; 14, MDCC-UD14 DNA; 14R, UD14 reactivated onto CEF DNA; Δ1, RB1BΔ4.5lac-infected CEF DNA (mutant virus); 01, MDCC-UD01 DNA; 01R, UD01 reactivated onto CEF DNA; 02, MDCC-UD02 DNA; 02R, UD02 reactivated onto CEF DNA; 03, MDCC-UD03 DNA; 03R, UD03 reactivated onto CEF DNA. Fragment sizes are given in kilobase pairs.
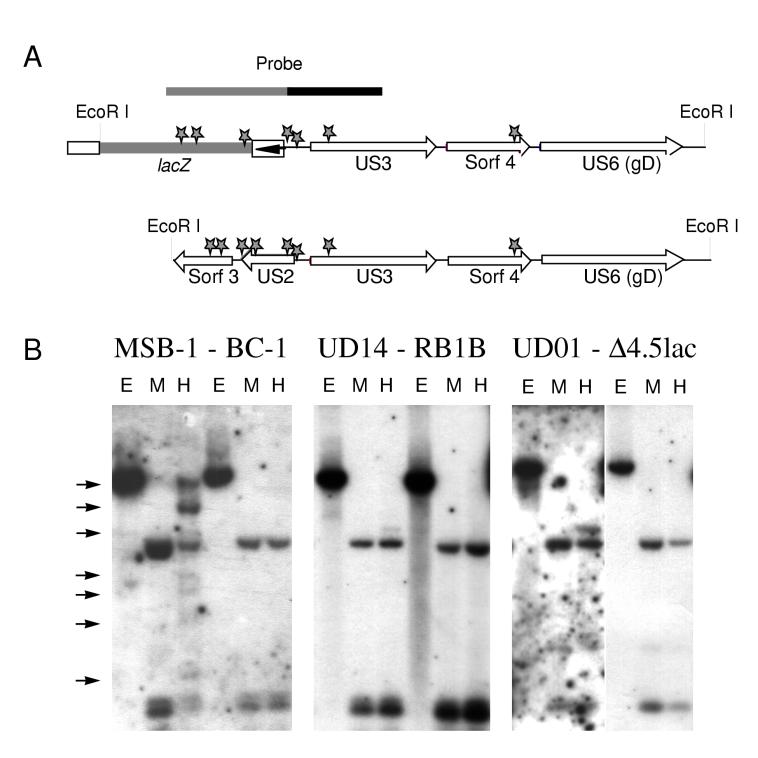
FIG. 5.
Examination of the methylation status of the US region of BC-1, RB1B, and RB1BΔ4.5lac genomes. (A) The 7.8-kbp EcoRI fragment of the US region of RB1BΔ4.5lac and the corresponding 5.3-kbp EcoRI fragment from the parental RB1B strain of MDV (see the genomic map in Fig. 1A). Stars, MspI/HpaII restriction sites (5′-CCGG-3′); hatched line and arrow, lacZ sequence and the SV40 early promoter, respectively. The hatched line above the RB1BΔ4.5lac genome denotes the probe used for panel B, a 2.4-kbp SacI fragment spanning the SV40 promoter as well as the US2 and US3 promoter regions. This probe was obtained from the plasmid used to generate RB1BΔ4.5lac, pMD190 (59, 61). (B) Southern blot hybridization analysis of cell line and reactivated virus-infected CEF DNAs. Each panel consists of three lanes corresponding to a cell line DNA followed by three lanes of the reactivated virus-infected CEF DNA. Each set of DNAs was digested with EcoRI (E lanes), EcoRI plus MspI (M lanes), or EcoRI plus HpaII (H lanes). The DNAs used were from cell lines MDCC-MSB-1 (3), MDCC-UD14 and MDCC-UD01 (this report), and the corresponding reactivated virus-infected CEF DNAs BC-1, RB1B, and RB1BΔ4.5lac, respectively. The arrows denote fragments containing CpG methylation as seen as HpaII-resistant bands.
RESULTS
Recombinant-MDV-derived cell lines.
We have established a number of cell lines from the highly oncogenic RB1B strain of MDV, an RB1B-derived deletion mutant, and an RB1B-derived mutant having an insertion into the US6 (gD homolog) gene (Table 1). These cell lines were established from primary lymphomas found in gonads, spleen, liver, and kidney tissues. Each cell line was established within 5 to 10 serial passages from explant. Cell lines MDCC-UD22/UD23, -UD24/UD25, and -UD27/UD28 were established from lymphomas at separate sites within a single chicken; i.e., MDCC-UD22 and -UD23 were established from kidney and ovarian lymphomas, respectively, from the same chicken, etc.
TABLE 1.
Cell lines, viruses, and tissue sources
| Cell linea | Virusb | Lymphomac | Inductiond | Spontaneous reactione |
|---|---|---|---|---|
| MDCC-UD01 | RB1BΔ4.5lac | Ovary | Inoculate | 200/104 |
| MDCC-UD02 | RB1BΔ4.5lac | Spleen | Inoculate | 5/104 |
| MDCC-UD03 | RB1BΔ4.5lac | Liver | Inoculate | 50/104 |
| MDCC-UD13 | RB1B | Visceralf | Inoculate | <1/106 |
| MDCC-UD14 | RB1B | Kidney | Inoculate | 200/104 |
| MDCC-UD22 | RB1BUS6lacgpt | Kidney | Inoculate | NDg |
| MDCC-UD23 | RB1BUS6gptlac | Ovary | Inoculate | 40/104 |
| MDCC-UD24 | RB1BUS6lacgpt | Liver | Inoculate | 400/104 |
| MDCC-UD25 | RB1BUS6lacgpt | Testes | Inoculate | ND |
| MDCC-UD27 | RB1BUS6lacgpt | Kidney | Contact | ND |
| MDCC-UD28 | RB1BUS6lacgpt | Testes | Contact | ND |
| MDCC-UD29 | RB1BUS6lacgpt | Visceralf | Contact | ND |
Cell lines are named according to agreed nomenclature: MDCC (Marek’s Disease cell culture), followed by two-letter designation of institution (UD [University of Delaware]) and unique number.
Viruses used to induce tumors: RB1BΔ4.5lac (61), RB1B (originally described in reference 74), and RB1BUS6lacgpt (4).
Lymphoma site used to establish cell line.
Inoculate, tumor induced by inoculated virus; contact, tumor induced by horizontally transmitted virus.
Approximate number of PFU induced on secondary CEF after cocultivation with given number of cells from each cell line. Each number represents an average, rounded to the nearest 5 PFU obtained in at least three independent trials.
Site of lymphoma ambiguous; more than one tumor taken; information lost on final establishment of line.
ND, not determined.
Our characterization of these cell lines has included the evaluation of their spontaneous reactivation frequency, that is, the number of MDV-induced plaques obtained via cocultivation of the lymphoblastoid cells with CEF. This reactivation frequency was characteristic for each cell line and ranged over several orders of magnitude (from 400 PFU/104 [1 PFU/25 cells] for MDCC-UD24 cells to <1 PFU/106 for MDCC-UD13 cells). To obtain virus from MDCC-UD13 cells, these cells were cocultivated with CEF and 7 days later the monolayers were blind passaged. After this passage, a few virus plaques were observed. These were passaged until heavily infected monolayers were obtained.
Analyses of RB1BΔ4.5lac and RB1B genomes in the derived cell lines.
Southern blot hybridization analysis using cell line and reactivated virus-infected CEF DNAs (Fig. 1) verified that RB1BΔ4.5lac was the transforming virus for cell lines MDCC-UD01, -UD02, and -UD03 and that no detectable parental virus was present. In addition, we examined the US region of RB1B-transformed cell lines MDCC-UD13 and -UD14, using cell line and reactivated virus-infected CEF DNAs.
After EcoRI digestion, the separated DNAs were transferred to nitrocellulose and hybridized with a radiolabeled 12.2-kbp HindIII-BamHI probe which spans the region of deletion in the mutant viruses. In RB1B-infected CEF and RB1B-derived cell lines (UD13 and UD14) and the viruses reactivated from these cell lines, the probe detected 5.8-, 5.3-, 2.8-, 2.6-, and 1.8-kbp fragments. In RB1BΔ4.5lac-infected CEF and RB1BΔ4.5lac-derived cell lines (UD01, UD02, and UD03) and the viruses reactivated from these lines, the probe detected 7.8-, 5.8-, 2.6-, and 1.5-kbp bands (Fig. 1). Deletion of 4.5 kbp of the US region from the genome of RB1BΔ4.5lac, and the insertion of a 3.9-kbp fragment encoding the lacZ gene at the site of the deletion, resulted in the following changes in hybridization patterns. First, the 2.8-kbp fragment present in the parent virus and derived cell lines is absent from the mutant virus and derived cell lines. Second, the 1.8-kbp parental fragment is truncated to 1.5 kbp in the mutant. Third, the 5.3-kbp parental fragment has 0.9 kbp deleted and 3.4 kbp added from the lacZ cassette, resulting in a fragment of 7.8 kbp.
The 5.8- and 2.6-kbp parental and mutant bands lie outside the region of deletion and are therefore detected in both parent and mutant DNAs. The 2.6-kbp band actually shows sequence identity with the 5.8-kbp band, as these fragments span the junctions of the US region with the terminal and internal repeats, respectively. These results are consistent with the changes in genome structure described for the initial RB1BΔ4.5lac mutant virus (Fig. 1 and references 59 and 61).
Immunophenotypes of mutant- and RB1B-derived cell lines.
Flow cytometric analysis of mutant- and RB1B-derived cell lines (Table 2) revealed the following: (i) most of the cell lines were CD4+ CD8−, consistent with a TH cell immunophenotype, (ii) one cell line, MDCC-UD13, was CD4− CD8+ CD3low TCR2low, and (iii) one cell line, MDCC-UD03, was CD4+ CD3low TCR2low. The immunophenotypes of the latter two cell lines are consistent with those of immature T cells found in the thymic cortex (33). All of the MDV-transformed cell lines expressed TCR2 (αβ) to various levels. Cell lines UD14 and UD28 also expressed a low level of TCR3 (α-Vβ2). Further analysis of UD14 cells revealed that they were indeed TCR2+ TCR3+ and were not a mixed population of TCR2+ and TCR3+ cells (data not shown).
All cells expressed MHC-II (Ia) to various levels, supporting the hypothesis that MDV-transformed T cells are activated. Our MSB-1 cells expressed very low levels of MHC-II, contrary to previous reports (76), but this cell line has been distributed widely, has been highly passaged, and has been shown to undergo changes in other laboratories (28). All of the T-cell lines analyzed, except for MSB-1s, expressed the accessory antigen CD28, a receptor involved in coactivation and present on T cells in the thymic cortex and medulla and in the periphery (33). None of the MDV-transformed cell lines described here expressed detectable Bu-1a (pan-B-cell antigen) or surface Ig (IgM/IgD), indicating that all of the established lines were not B cells.
The controls used for B- and T-cell markers were REV-transformed cell lines RECC-CU60 and RECC-CU91, respectively (77). RECC-CU60 cells expressed both Bu-1a and surface Ig (IgM/IgD) but no detectable levels of MHC-II (Table 2); RECC-CU91 cells expressed low levels of CD3, CD28, and TCR3 and high levels of MHC-II but expressed no CD4 and/or CD8 (Table 2).
Flow cytometric analysis of IUdR-treated cell lines.
From our construction of recombinant MDVs (2, 4, 59–61), we found that a lacZ gene driven by the simian virus 40 (SV40) early promoter was constitutively expressed during the lytic infection of CEF. We recently reported, however, that lacZ expression is repressed in tumors induced by recombinant MDVs and cell lines derived from these tumors (4, 61). Upon reactivation of the recombinant virus from these cell lines onto CEF, lacZ expression again became constitutive (61).
To obtain a representative measure of the induction of MDV antigens and β-Gal in recombinant- and parent-MDV-derived cell lines, we performed flow cytometric analysis on fixed, IUdR-treated cell lines (Fig. 3 and 4). Representative cell lines were chosen for analysis: MDCC-UD01 (RB1BΔ4.5lac transformed), MDCC-UD13 (RB1B transformed, nonproducer), MDCC-UD14 (RB1B transformed, producer), MDCC-UD24 (RB1BUS6lacgpt-derived), and RECC-CU91 (non-MDV-transformed T-lymphoblastoid cells) (included as a control). Cell lines UD01, UD14, and UD24 were selected according to their similarity of immunophenotype (CD3+ CD4+ TCR2+ MHC-II+ CD28+) and spontaneous MDV reactivation frequencies (200 to 400 PFU/104 cells, [Table 1]). Cell line UD13 was included to examine induction of MDV antigen expression in a CD8+, nonproducer cell line (<1 PFU/106 cells plated).
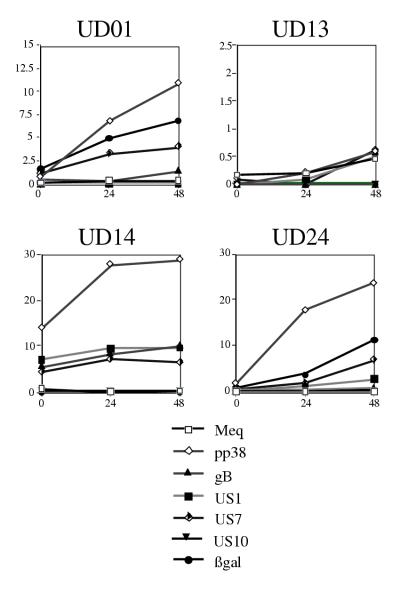
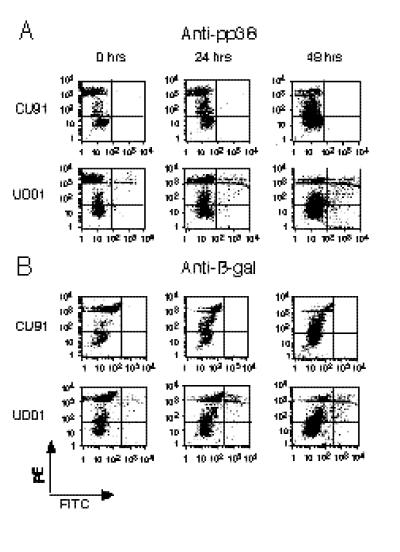
FIG. 3.
Comparative analysis of internal antigen induction. Axes are x axis, hours after IUdR treatment; y axis, percentage of cells having FL-1 (FITC) fluorescence above controls. The controls for mouse anti-Meq (Meq), mouse anti-pp38 (pp38), mouse anti-gB (gB), rabbit anti-US1 protein (US1), rabbit anti-glycoprotein I (US7), and rabbit anti-US10 protein (US10) were RECC-CU91 cells stained with each antibody, and each cell line stained with secondary antibody alone. The controls for rabbit anti-β-Gal (βgal) were RECC-CU91, MDCC-UD13, and MDCC-UD14 cells stained with the antibody as well as secondary antibody alone. Induction and antibody staining were performed twice with anti-Meq, five times with anti-pp38, three times with anti-gB, four times with anti-US1, four times with anti-US7, twice with anti-US10, and five times with anti-β-Gal. Note differences in y axes: MDCC-UD01 (maximum, 15%), MDCC-UD13 (maximum, 2.5%); MDCC-UD14 and MDCC-UD24 (maximum, 30%).
Our flow cytometric results (Fig. 2 and 3) confirmed our initial findings, namely, that the marker gene (lacZ) was induced in the recombinant-derived cell lines (UD01 and UD24) with the same kinetics as MDV gene products pp38, and US1-, US7- and US10-encoded proteins (Fig. 4). The induction of these genes was also highly specific for MDV-transformed cell lines and for the appropriate MDV cell lines (recombinant or parental). These results can be summarized as follows: (i) a REV-transformed chicken T-cell line (RECC-CU91) did not show induction of any of the MDV gene products or β-Gal (Fig. 2 and data not shown); (ii) induction of the MDV US1-encoded protein was not detected for UD01 cells, the region encoding US1 having been deleted from the genome of RB1BΔ4.5lac (Fig. 1 and reference 61); and (iii) induction of the lacZ gene product (β-Gal) was specific for cell lines UD01 and UD24, no expression being detected in cell line UD13, UD14 (parental), or CU91 (non-MDV) (Fig. 2 and 3).
FIG. 2.
Flow cytometric analysis of MDV internal antigen induction. Shown are dot blot analyses of RECC-CU91 and MDCC-UD01 cell lines treated with IUdR for 0, 24, and 48 h. FL-1 (FITC) fluorescence (x axis) and FL-2 (phycoerythrin-PI) fluorescence (y axis) are shown in fluorescence intensity units (log scale). All cells were fixed with paraformaldehyde, stained with antibodies to anti-pp38 (L. Lee) (A) or anti-β-Gal (5 Prime→3 Prime) (B), and then stained with goat anti-mouse–FITC (Sigma) (A) or goat anti-rabbit FITC (Sigma) (B). Finally, all samples were stained with PI (Sigma) at 10 μg/ml for 10 min. Gates were set by using RECC-CU91 (non-MDV-derived cell line) stained with each anti-MDV antibody, MDCC-UD14 cells (non-recombinant-MDV-derived cell line) stained with anti-β-Gal, as well as cell lines stained with secondary antibody reagents only.
For cell lines UD01, UD14, and UD24, pp38 expression was induced to a greater extent than expression of any of the other gene products examined (Fig. 2 to 4). Monoclonal antibody H19.47 was developed against MDV serotype 1-specific phosphoproteins and reacted with pp38 and pp24 antigens (4, 78). Genes encoding pp38 and pp24 span the repeats flanking the unique long (UL) region and, as a result, share promoters and 62 amino acid residues at their N termini (6, 49, 91). Since this antibody (H19.47) detects two gene products, the expression of which is driven by identical promoters, the level of pp38 induction actually reflects the sum total of pp38 and pp24 induction.
In contrast to the producer cell lines, UD13 showed very limited induction after IUdR treatment (<1% of cells expressed any of the antigens examined). This level of induction for pp38 expression was more than 10-fold less than observed for UD01 cells, 20-fold less than observed for UD24 cells, and 30-fold less than observed for UD14 cells (Fig. 3). Despite the high percentage of cells that could be induced to express pp38, the UD14 cells showed only twofold induction over time and very little increase in expression between 24 and 48 h (Fig. 3). This pattern of expression is also seen for the other lytic antigens examined, i.e., US1, US7, US10, and gB, and is consistent with the pattern of induction of some expressor MDV cell lines described previously (20).
Immunostaining of MDV-derived cell lines.
To gain insight into the localization of MDV antigens and β-Gal within LBCLs and to determine if the expression of antigens was mutually exclusive in the induced cell lines, we performed immunostaining in combination with PI (Fig. 4A) and DAPI (Fig. 4B) DNA staining. The monoclonal antibody to pp38 detects this antigen throughout the cytoplasm in a mesh-like pattern (Fig. 4A, upper left). Expression of pp38 was also detected throughout the cytoplasm of cells also expressing the US1-encoded protein, gI, the US10-encoded protein, and β-Gal (Fig. 4B, top row). Costaining for pp38 and the US1-encoded protein (ICP22 homolog, p24/27) showed distinct regions of pp38 surrounded by p24/27. This pattern was seen repeatedly. Both proteins are phosphorylated (9, 25–27) and appear to be associated with distinct sites in the cytoplasm. In contrast, pp38 did not appear to associate with gI, the US10-encoded protein, or β-Gal (Fig. 4B, top row).
The monoclonal antibody to the gB homolog of MDV detected distinct patches of gB on the surface of the IUdR-treated cells (Fig. 4A, top right; Fig. 4B, middle row). These patches of gB may denote sights of virus particle envelopment, as gI was also found to colocalize to patches on the cell surface. The US10 protein of MDV localized to cytoplasmic patches having a very distinct pattern of expression (Fig. 4A, second from right; Fig. 4B, column 3). These US10 cytoplasmic patches seemed to localize near patches of gB on the cell surface (Fig. 4B, middle row, column 3), suggesting that this protein may be associated with virion assembly. The functions of alphaherpesvirus US10-encoded proteins are unknown, but these genes are dispensable for MDV infection (59, 61).
The monoclonal antibody to Meq detected a nuclear antigen that is found throughout the nucleus and in patches in some cells (Fig. 4A, top row, columns 2 and 3; Fig. 4B, bottom row). This patch-like pattern of nuclear expression is particularly notable in the US1-encoded protein- and gI-expressing cells (Fig. 4B, columns 1 and 2). It has been postulated that the Meq protein may play a role in virus replication as well as in transactivation (69), and since the MDV US1-encoded protein and gI have been reported to be late antigens (8, 9), the observed patch-like pattern may indicate sights of viral DNA replication. Meq was difficult to visualize in PI-stained cells (Fig. 4A, column 2) but was more easily detected in nuclei stained with DAPI (Fig. 4A, column 3; Fig. 4B, bottom row).
The polyclonal antibody to β-Gal detects diffuse cytoplasmic expression in IUdR-treated UD01 and UD24 cells (Fig. 4A, bottom right; Fig. 4B, column 4). There did not appear to be any association between β-Gal and any of the other antigens examined.
DISCUSSION
MDV infection, like other herpesvirus infections, is separable into lytic (productive) and latent (nonproductive) phases. In the case of MDV, however, the latent infection does not seem readily separable from a transforming infection. Studies on the progression of MDV infection suggest that latency is established in T cells within the first weeks of infection followed by secondary productive cytolytic infection and transformation of a subset of the latently infected T cells (13, 17, 19). Although CD4+ T cells (TH cells) have been predominantly associated with MDV latent/transforming infection, MDV can transform T cells of various immunophenotypes (14, 56, 67, 75). All MDV T-LBCLs to date, however, are TCR2 (αβ+), TCR3 (α-Vβ2+), or TCR null+ (reference 75 and this report), and no TCR1 (γδ+) MDV cell lines have been isolated despite the susceptibility of this lineage to transformation by REV (51).
We now report the characterization of LBCLs transformed by mutant MDVs. Like most MDV-transformed cell lines reported, these cell lines have the predominant activated TH cell immunophenotype (CD3+ CD4+ TCR2+ MHC-II+ CD28+), with a few notable exceptions.
Cell lines UD14 and UD28 express both TCR2 and TCR3 (Table 2). We studied this dual expression further in UD14 cells and found that these cells do indeed express both TCR2 and a lower level of TCR3 and are not a mix of TCR2 and TCR3 single-positive cells (data not shown). The functional significance of this subpopulation of T cells is unknown, but such aberrant receptor expression has been noted for other MDV cell lines (75). Our current understanding of TCR gene rearrangement and expression during T-cell ontogeny does not explain such a lineage (33). This aberrant TCR expression may represent a novel T-cell lineage or may simply be a result of transforming events induced by MDV.
Cell lines UD13 (CD8+ CD4−) and UD03 (CD4+ CD8−) are interesting in that they express very low (i.e., undetectable by direct antibody staining) levels of TCR and CD3 (Table 2). Consequently, these cells appear to be of an immature T-cell lineage that have increased single accessory molecule expression without increases in TCR and CD3 surface expression (33). The establishment of these lines from visceral lymphomas suggests that MDV can transform such immature lineages. T-cell activation requires stimulation of an adequate number of TCRs in the presence of other costimulatory events as well as a fully mature T cell (87). Since we and others (14, 75) have identified MDV cell lines of apparent immature T-cell lineages, it appears that activation of a T cell, in the true sense, is not an absolute requirement for MDV infection. As mentioned above, the aberrant expression of surface antigens may be a result of MDV transformation and passage in culture, but the lines described here were all analyzed within 30 passages in culture. Also, by using the local-lesion method for the generation of MDV cell lines, cell lines with similar immunophenotypes have been established (14, 75). Recently, a lineage of cytotoxic T cells that are CD4− CD8− has been identified (82); therefore, other nonclassical T-cell lineages may also have a function in the chicken.
In LBCLs, few MDV gene products are produced (73, 78, 79, 83), and these are encoded by the repeat regions of the viral genome. Despite this paucity of genome expression, fixed LBCLs have been used to confer immunity to MDV tumor formation (66), indicating that MDV-specific gene products are expressed. Due to their expression in transformed/latently infected LBCLs, these MDV gene products have been inferentially associated with MDV oncogenicity (7, 25, 27, 37, 38, 40, 50, 63–65, 73, 79, 80, 83).
Probably the most compelling of the MDV putative oncogenes is meq (MDV EcoRI-Q-encoded gene). The encoded gene product, Meq, was identified in cell lines and has since been identified in MDV tumors (40, 71). Meq is a transcriptional activator of the bZIP class having basic and leucine zipper domains (40, 68, 69). Meq forms heterodimers with c-Jun and transactivates its own promoter through an AP-1-like binding motif. Recently, Meq-Meq homodimers were found to bind two DNA consensus sequences (Meq-responsive elements); one consensus contains tetradecanoyl phorbol acetate and cyclic AMP response sequences, and the other contains a CACA motif present near the MDV origin of lytic replication (69).
Moreover, Meq has recently been shown to morphologically transform Rat-2 cells, as well as induce serum-independent growth and prevent tumor necrosis factor alpha-, C2-ceramide-, and UV irradiation-induced apoptosis (48). The mechanism of apoptotic block appears to be due to the induction of bcl-2 expression and a repression of bax expression, again relating the oncogenic potential of Meq to its ability to transactivate.
Meq has been localized to the nucleus and nucleolus of Rat-1 and Cos-1 cells transfected with a Meq expression vector (47). We have observed two types of Meq nuclear staining similar to that described in the localization studies of Liu et al. (47). One type of staining was described as discrete points within the nucleus (coiled bodies) and is similar to some of our observations (Fig. 4B, with anti-US10 and with anti-β-Gal). The other type of staining, and what we found to be more common, was patches of concentrated staining with anti-Meq (Fig. 4A, anti-Meq panels; Fig. 4B, with anti-US1 and anti-US7). This concentrated, patch-like staining appears very similar to the nucleolar staining previously found in Rat-2 and Cos-1 cells transfected with a Meq expression vector (47). Our results extend the work of Liu et al., as we show similar patterns of anti-Meq staining in the context of the MDV-transformed LBCLs.
Another putative MDV oncogene product is pp38, a phosphorylated, cytoplasmically localized immediate-early/early protein of putative regulatory significance (25, 26, 39, 55). This protein shows identity with pp24 (62 amino-terminal residues), the genes encoding these proteins initiating within the repeats flanking the UL region (6, 49, 91).
The association of pp38 and pp24 with oncogenicity has been questioned since these gene products are expressed by oncogenic and attenuated strains of MDV, and homologs have been identified in the nononcogenic serotypes of MDV, MDV serotype 2 (58), and herpesvirus of turkeys (81). In fact, some researchers regard the expression of pp38 and pp24 as a hallmark of cytolytic infection (5, 71), despite evidence that low level pp38 expression may be required for the maintenance of LBCL proliferation (43, 90) and the identification of pp38 in LBCLs in the absence of IUdR treatment (25).
A problem inherent in the functional analysis of Meq and pp38 expression in LBCLs has been a tendency to examine their expression collectively through Northern or Western blot analyses (25–27, 40). This is particularly evident in the case of pp38 expression, in which relatively few cells in a population may be expressing the protein. Those cells which do express pp38, however, express large amounts of the protein (Fig. 4). Likewise, relatively few cells appear to express the Meq protein (Fig. 3), yet both the transcript encoding Meq and the protein are readily detectable by blotting methods (40, 48). To provide a context for the expression of these proteins, we examined the percentages of cells expressing each protein (Fig. 3), the visual localization of each protein (Fig. 4A), and the ability of LBCLs to coexpress these proteins (Fig. 4B). It should be noted that the immunostaining method used may be less sensitive than blotting methods, since in situ, proteins may be less accessible when complexed to other proteins or cellular structures. Our work here does give an indication, however, of the percentages of cells expressing readily detectable levels of the specific antigens and further demonstrate that their expression or induction of expression is not uniform throughout the cell population.
The results presented here and previous work indicate a marked increase in pp38 and pp24 expression with IUdR treatment of cells (Fig. 2 and 3; references 26, 27, and 39) and are consistent with the hypothesis that pp38-pp24 expression is associated with cytolytic infection. Moreover, we demonstrate that other MDV gene products having true late kinetics (US1, US10, and gI) are also induced to various degrees, consistent with the induction of a cytolytic infection. The higher level of pp38 induction, its identification as an immediate-early/early protein (26), and the coexpression of pp38 with other lytic infection-associated antigens which are induced in lower percentages of cells at the times examined suggest that pp38 may regulate the expression of these antigens. These results extend other findings of other studies in which single antigen or nondefined antiserum was used to examine antigen induction (20, 25, 30, 39). Upon cocultivation of our IUdR-treated cell lines, however, we saw a variable increase in the reactivation frequency of virus from the cell lines (data not shown). Since IUdR treatment does affect the viability of the cells, and the production of MDV appears to be closely associated with cell viability, the induction elicited by IUdR may also result in the death of cells prior to the production or spread of infectious virions.
Our initial interest in the study of MDV gene induction during reactivation came from the observation that the lacZ expression cassette, driven by the SV40 early promoter and inserted into the MDV genome, was constitutively expressed during the lytic infection of CEF (2, 59–61). In tumors induced by recombinant MDVs and the cell lines derived from these tumors, however, expression of the lacZ cassette was highly repressed (4, 61). Upon reactivation of these viruses from cell lines and infection of CEF, lacZ expression was again constitutive (61). When the cell lines were fixed and stained with antibodies to different MDV gene products as well as anti-β-Gal, in all cases only a very small percentage of cells were stained. The observed repression of lacZ expression does not appear to be due to a decreased activity of the SV40 early promoter in chicken T-LBCLs for the following reasons: (i) this promoter has been used to drive expression of neomycin phosphotransferase in chicken T-LBCLs for the generation of stable expression cell lines (58a, 77); and (ii) upon treatment of the recombinant-derived cell lines with IUdR in this study, the activity of this promoter is clearly demonstrable (Fig. 2 to 4). Therefore, the repression of the lacZ cassette appears to be associated with its context within the MDV genome.
The regulation of the MDV genome in LBCLs, like the regulation of gene expression during the latency of other herpesviruses, is poorly understood. As mentioned previously, the MDV genome, or at least portions of it, is integrated into the host chromosomes (28), although no common sites of insertion have been identified. MDV integration does appear to be telomeric, an interesting finding given the identification of telomere-like repeats at the junction between the inverted repeats (44). These findings, however, do not explain the silencing of the MDV genome in these cells. Studies into the mechanism of gene regulation in MDV cell lines have implicated methylation (34, 36, 41), antisense transcripts to the ICP4 gene (23, 24, 46, 52), and higher-order genomic structure (35). Our results examining the SV40 promoter and flanking MDV genomic region in MDCC-UD01 cells, as well as the homologous region from a parental-strain-derived LBCL (Fig. 5), clearly demonstrate that this region is not methylated in most of the cells. A minority of the cells have one or two sites methylated in the US3 gene (Fig. 5). Our MSB-1 cells, however, contain populations that are methylated to various degrees within this region (Fig. 5). We have also examined the regions encoding pp24 and pp38 and found that these regions remain largely nonmethylated even in MSB-1 cells which have been passaged extensively in cell culture. During productive infection of CEF with the viruses reactivated from each cell line (MSB-1→BC-1, UD14→RB1B, and UD01→RB1BΔ4.5lac), no methylation is noted in the US- or pp24/pp38-encoding region (Fig. 5 and data not shown).
Studies on the mechanisms of genome repression in MDV-transformed LBCLs could have enormous implications regarding the study of herpesvirus latency in general and may hold some keys to MDV transformation. It is notable that the MDV genome is also repressed when introduced into avian leukosis virus-transformed B cells (32), REV-transformed T cells (77), and chemically transformed CEF (1, 2). Like MDV-transformed LBCLs, these other transformed cells showed very limited expression of the MDV genome, although the state of the MDV genome (integrated versus episomal) in these systems has not been reported. Introduction of the MDV genome does increase the proliferation status of non-MDV-transformed LBCLs (16), despite limited expression of the MDV genome. The expression of latency-associated RNAs (22, 23, 46, 52) has been recently reported for the MDV genome inserted into a chemically transformed chicken fibroblast line, OU2.2 (2), suggesting that these transcripts may be necessary for the repression of the MDV genome. Transcripts antisense to ICP4-, pp38-, and Meq-encoding genes were recently shown to decrease proliferation of MSB-1 cells (43, 90), suggesting that these gene products are somehow involved in the maintenance of latency and transformation, but the mechanism eliciting this effect is unclear.
In summary, we report the establishment and characterization of recombinant-MDV-derived cell lines. The immunophenotypes of these cell lines are consistent with those of LBCLs derived by other groups using nonrecombinant MDV strains (75). Moreover, it is clear that the mutations engineered in these viruses have not affected their ability to transform chicken T cells. Examination of the virus structure within, and reactivated from, these cell lines has shown that these lines are free of parental virus (this report and reference 4) and that the original mutations are stable within the cell lines. We also report that the marker gene inserted within the genomes of these recombinant cell lines is regulated in the same manner as MDV lytic genes. Treatment of these cell lines with IUdR induces the expression of this marker gene with the same kinetics as for several MDV genes. Consequently, expression of this marker gene serves as a means for the measurement of lytic-phase induction. The dual-staining studies have demonstrated that pp38 and Meq are expressed at the same time as other lytic-phase genes and, in the case of the recombinant lines, lacZ. These results demonstrate that expression of pp38 and Meq and that of lytic-phase genes are not mutually exclusive.
In cell lines transformed by RB1BUS6gptlac, the marker cassette also contains a gene for positive selection of virus-infected cells, the guanine phosphoribosyltransferase gene of Escherichia coli (4). Since this cassette can be used for the selection of cells resistant to mycophenolic acid, we plan on examining the expression of this cassette within and outside the context of the MDV genome in LBCLs.
Moreover, we have recently established a recombinant MDV-derived cell line having a green fluorescent protein expression cassette driven by the cytomegalovirus immediate-early promoter inserted into the US2 gene of MDV (58a). We have observed that in tumors induced by this virus (RB1BUS2gfp), and the resulting cell line established (MDCC-UA04), the green fluorescent protein expression cassette is also repressed (58a). This cell line and the recombinant cell lines described here promise to provide much needed insight into the mechanisms of MDV latency and reactivation.
ACKNOWLEDGMENTS
We thank Peter Brunovskis, Donald Ewert, Lucy Lee, Hyun Lillehoj, Ton Schat, and Leland Velicer for gifts of antibodies and cell lines. We also thank Gisela Erf and John Kirby for critical reading of the manuscript and Tina Bersi for technical assistance.
This work was, in part, funded by USDA-NRI grant 95-37204-2640.
Footnotes
Publication no. 98060 of the Arkansas Agricultural Experiment Station.
REFERENCES
- 1.Abujoub A, Coussens P M. Development of a sustainable chick cell line infected with Marek’s disease virus. Virology. 1995;214:541–549. doi: 10.1006/viro.1995.0065. [DOI] [PubMed] [Google Scholar]
- 2.Abujoub A, Coussens P M. Evidence that Marek’s disease virus exists in a latent state in a sustainable fibroblast cell line. Virology. 1996;229:309–321. doi: 10.1006/viro.1997.8444. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama Y, Kato S. Two cell lines from lymphomas of Marek’s disease. Biken J. 1974;17:105–116. [PubMed] [Google Scholar]
- 4.Anderson A S, Parcells M S, Morgan R W. The glycoprotein D (US6) homolog is not essential for oncogenicity or horizontal transmission of Marek’s disease virus. J Virol. 1997;72:2548–2553. doi: 10.1128/jvi.72.3.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baigent S J, Ross L J N, Davison T F. A flow cytometric method for identifying Marek’s disease virus pp38 expression in lymphocyte subpopulations. Avian Pathol. 1996;25:255–267. doi: 10.1080/03079459608419140. [DOI] [PubMed] [Google Scholar]
- 6.Becker Y, Asher Y, Tabor E, Davidson I, Malkinson M. Open reading frames in a 4556 nucleotide sequence within MDV-1 BamHI-D DNA fragment: evidence for splicing of mRNA from a new viral glycoprotein gene. Virus Genes. 1994;8:55–69. doi: 10.1007/BF01703602. [DOI] [PubMed] [Google Scholar]
- 7.Bradley G, Hayashi M, Lancz G, Tanaka A, Nonoyama M. Structure of the Marek’s disease virus BamHI-H gene family: genes of putative importance for tumor induction. J Virol. 1989;63:2534–2542. doi: 10.1128/jvi.63.6.2534-2542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunovskis P, Chen X, Velicer L. Proceedings of the 4th International Symposium on Marek’s disease. Wageningen, The Netherlands: Ponsen & Looijen; 1992. Analysis of Marek’s disease virus glycoproteins D, I, and E; pp. 118–122. [Google Scholar]
- 9.Brunovskis P, Velicer L F. Proceedings of the 4th International Symposium on Marek’s disease. Wageningen, The Netherlands: Ponsen & Looijen; 1992. Genetic organization of the Marek’s disease virus unique short region and identification of US-encoded polypeptides; pp. 74–78. [Google Scholar]
- 10.Buscaglia C, Calnek B W, Schat K A. Effect of immune competence on the establishment and maintenance of latency with Marek’s disease herpesvirus. J Gen Virol. 1988;69:1067–1077. doi: 10.1099/0022-1317-69-5-1067. [DOI] [PubMed] [Google Scholar]
- 11.Buscaglia C, Calnek B W. Maintenance of Marek’s disease herpesvirus latency in vitro by a factor found in conditioned medium. J Gen Virol. 1988;69:2809–2818. doi: 10.1099/0022-1317-69-11-2809. [DOI] [PubMed] [Google Scholar]
- 12.Calnek B W, Spencer J L. Marek’s disease virus and lymphoma. In: Rapp F, editor. Oncogenic herpesviruses. Vol. 1. Boca Raton, Fla: CRC Press; 1985. pp. 103–143. [Google Scholar]
- 13.Calnek B W. Proceedings of the International Symposium on Marek’s disease. Kennet Square, Pa: AAAP Press; 1985. Update on Marek’s disease research; pp. 1–4. [Google Scholar]
- 14.Calnek B W, Lucio B, Schat K A, Lillehoj H S. Pathogenesis of Marek’s disease virus-induced local lesions. 1. Lesion characterization and cell line establishment. Avian Dis. 1989;33:291–302. [PubMed] [Google Scholar]
- 15.Calnek B W, Adldinger H K, Kahn D E. Feather follicle epithelium: a source of enveloped and infectious cell-free herpesvirus from Marek’s disease. Avian Dis. 1970;14:219–233. [PubMed] [Google Scholar]
- 16.Calnek B W, Schat K A. Proliferation of chicken lymphoblastoid cells after in vitro infection with Marek’s disease virus. Avian Dis. 1991;35:728–737. [PubMed] [Google Scholar]
- 17.Calnek B W, Schat K A, Ross L J N, Shek W R, Chen C-L H. Further characterization of Marek’s disease virus infected lymphocytes. I. In vivo infection. Int J Cancer. 1984;33:389–398. doi: 10.1002/ijc.2910330318. [DOI] [PubMed] [Google Scholar]
- 18.Calnek B W, Schat K A, Ross L J N, Chen C-L H. Further characterization of Marek’s disease virus-infected lymphocytes. II. In vitro infection. Int J Cancer. 1984;33:399–406. doi: 10.1002/ijc.2910330319. [DOI] [PubMed] [Google Scholar]
- 19.Calnek B W, Schat K A, Shek W R, Chen C-L H. In vitro infection of lymphocytes with Marek’s disease virus. J Natl Cancer Inst. 1982;69:709–713. [PubMed] [Google Scholar]
- 20.Calnek B W, Shek W R, Schat K A. Spontaneous and induced herpesvirus genome expression in Marek’s disease tumor cell lines. Infect Immun. 1981;34:483–492. doi: 10.1128/iai.34.2.483-491.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calnek B W, Witter R L. Marek’s disease. In: Calnek B W, editor. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 367–413. [Google Scholar]
- 22.Cantello J L, Anderson A S, Francesconi A, Morgan R W. Isolation of a Marek’s disease virus (MDV) recombinant containing the lacZ gene of Escherichia coli stably inserted within the MDV US2 gene. J Virol. 1991;65:1585–1588. doi: 10.1128/jvi.65.3.1584-1588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantello J L, Anderson A S, Morgan R W. Identification of latency associated transcripts that map antisense to the ICP4 homolog gene of Marek’s disease virus. J Virol. 1994;68:6280–6290. doi: 10.1128/jvi.68.10.6280-6290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantello J L, Parcells M S, Anderson A S, Morgan R W. Marek’s disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J Virol. 1997;71:1353–1361. doi: 10.1128/jvi.71.2.1353-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Sondermeijer P J A, Velicer L F. Identification of a unique Marek’s disease virus gene which encodes a 38-kilodalton phosphoprotein and is expressed in both lytically infected cells and latently infected lymphoblastoid tumor cells. J Virol. 1992;66:85–94. doi: 10.1128/jvi.66.1.85-94.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Z, Lee L F, Liu J-L, Kung H-J. Structural analysis and transcriptional mapping of the Marek’s disease virus gene encoding pp38, an antigen associated with transformed cells. J Virol. 1991;65:6509–6515. doi: 10.1128/jvi.65.12.6509-6515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui Z, Yan D, Lee L F. Marek’s disease virus gene clones encoding virus-specific phosphorylated polypeptides and serological characterization of fusion proteins. Virus Genes. 1990;3:309–322. doi: 10.1007/BF00569038. [DOI] [PubMed] [Google Scholar]
- 28.Delecluse H-J, Hammerschmidt W. Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common. J Virol. 1993;67:82–92. doi: 10.1128/jvi.67.1.82-92.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delecluse H-J, Schuller S, Hammerschmidt W. Latent Marek’s disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn K, Nazerian K. Induction of Marek’s disease virus antigens by IdUrd in a chicken lymphoblastoid cell lines. J Gen Virol. 1977;34:413–419. doi: 10.1099/0022-1317-34-3-413. [DOI] [PubMed] [Google Scholar]
- 31.Fukuchi K, Sudo M, Lee Y-S, Tanaka M, Nonoyama M. Structure of the Marek’s disease virus DNA: detailed restriction enzyme map. J Virol. 1984;51:102–109. doi: 10.1128/jvi.51.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fynan E F, Ewert D, Block T M. Latency and reactivation of Marek’s disease virus in B lymphocytes transformed by avian leukosis virus. J Gen Virol. 1993;74:2163–2170. doi: 10.1099/0022-1317-74-10-2163. [DOI] [PubMed] [Google Scholar]
- 33.Göbel T W F. The T-dependent immune system. In: Davison T F, Morris T R, Payne L N, editors. Poultry immunology. Oxfordshire, England: Carfax Publishing Co.; 1996. pp. 31–45. [Google Scholar]
- 34.Hayashi M, Furuichi S, Ren S, Isogai E, Nonoyama M, Namioka S. Enhancement of mRNA synthesis from Marek’s disease virus genome in the lymphoblastoid cell line, MDCC-MSB1, by 5-azacytidine. J Vet Med Sci. 1994;56:287–291. doi: 10.1292/jvms.56.287. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi M, Nakamura K, Isogai E, Namioka S. Nucleosomal structure of Marek’s disease virus genome in transformed lymphoblastoid cell lines, MDCC-MSB1 and MDCC-RP1. Microbiol Immunol. 1991;8:643–653. doi: 10.1111/j.1348-0421.1991.tb01596.x. [DOI] [PubMed] [Google Scholar]
- 36.Hirai K, Ikuta K, Kitamoto N, Kato S. Latency of herpesvirus of turkey and Marek’s disease virus genomes in a chicken T-lymphoblastoid cell line. J Gen Virol. 1981;53:133–143. doi: 10.1099/0022-1317-53-1-133. [DOI] [PubMed] [Google Scholar]
- 37.Hong Y, Coussens P M. Identification of an immediate-early gene in the Marek’s disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide. J Virol. 1994;68:3593–3603. doi: 10.1128/jvi.68.6.3593-3603.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong Y, Frame M, Coussens P M. A 14-kDa immediate-early phosphoprotein is specifically expressed in cells infected with oncogenic Marek’s disease virus strains and their attenuated derivatives. Virology. 1995;206:695–700. doi: 10.1016/s0042-6822(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 39.Ikuta K, Nakajima K, Naito M, Ann S H, Ueda S, Kato S, Hirai K. Identification of Marek’s disease virus-specific antigens in Marek’s disease lymphoblastoid cell lines using monoclonal antibody against virus-specific phosphorylated polypeptides. Int J Cancer. 1985;35:257–264. doi: 10.1002/ijc.2910350219. [DOI] [PubMed] [Google Scholar]
- 40.Jones D, Lee L, Liu J-L, Kung H-J, Tillotson J K. Marek’s disease encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanamori A, Ikuta K, Ueda S, Kato S, Hirai K. Methylation of Marek’s disease virus DNA in chicken T-lymphoblastoid cell lines. J Gen Virol. 1987;68:1485–1490. doi: 10.1099/0022-1317-68-5-1485. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan M, Dhar A, Brown T R, Sundick R S. Marek’s disease virus-transformed chicken T-cell lines respond to lymphokines. Vet Immunol Immunopathol. 1992;34:63–79. doi: 10.1016/0165-2427(92)90152-g. [DOI] [PubMed] [Google Scholar]
- 43.Kawamura M, Hayashi M, Furuichi T, Nonoyama M, Isogai E, Namioka S. The inhibitory effects of oligonucleotides complementary to Marek’s disease virus mRNA transcribed from the BamHI-H region, on the proliferation of transformed lymphoblastoid cells, MDCC-MSB1. J Gen Virol. 1991;72:1105–1111. doi: 10.1099/0022-1317-72-5-1105. [DOI] [PubMed] [Google Scholar]
- 44.Kishi M, Bradley G, Jessip J, Tanaka A, Nonoyama M. Inverted repeat regions of Marek’s disease virus DNA possess a structure similar to that of the a sequence of herpes simplex virus DNA and contain host cell telomere sequences. J Virol. 1991;65:2791–2797. doi: 10.1128/jvi.65.6.2791-2797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee L F, Sharma J M, Nazerian K, Witter R L. Suppression of mitogen-induced proliferation of normal spleen cells by macrophages from chickens inoculated with Marek’s disease virus. J Immunol. 1978;120:1554–1559. [PubMed] [Google Scholar]
- 46.Li D-S, Pastorek J, Zelnik V, Smith G D, Ross L J N. Identification of novel transcripts complementary to the Marek’s disease virus homologue of the ICP4 gene of herpes simplex virus. J Gen Virol. 1994;75:1713–1722. doi: 10.1099/0022-1317-75-7-1713. [DOI] [PubMed] [Google Scholar]
- 47.Liu J-L, Lee L F, Ye Y, Qian Z, Kung H-J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J-L, Ye Y, Lee L F, Kung H-J. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J Virol. 1998;72:388–395. doi: 10.1128/jvi.72.1.388-395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makimura K, Peng F-Y, Tsuji M, Hasegawa S, Kamai Y, Nonoyama M, Tanaka A. Mapping of Marek’s disease virus genome: identification of junction sequences between unique long and inverted repeat regions. Virus Genes. 1994;8:15–24. doi: 10.1007/BF01703598. [DOI] [PubMed] [Google Scholar]
- 50.Maray T, Malkinson M, Becker Y. RNA transcripts of Marek’s disease virus (MDV) serotype-1 infected and transformed cells. Virus Genes. 1988;2:49–68. doi: 10.1007/BF00569736. [DOI] [PubMed] [Google Scholar]
- 51.Marmor M D, Benatar T, Ratcliffe M J H. Retroviral transformation in vitro of chicken T cells expressing either α/β or γ/δ T cell receptors by reticuloendotheliosis virus strain T. J Exp Med. 1993;177:647–656. doi: 10.1084/jem.177.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKie E A, Ubukata E, Hasegawa S, Zhang S, Nonoyama M, Tanaka A. The transcripts from the sequences flanking the short component of Marek’s disease virus during latent infection form a unique family of 3′-coterminal RNAs. J Virol. 1995;69:1310–1314. doi: 10.1128/jvi.69.2.1310-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan R W, Anderson A, Kent J, Parcells M. Characterization of Marek’s disease virus RB1B-based mutants having disrupted glycoprotein C or glycoprotein D homolog genes. In: Silva R F, et al., editors. Proceedings of the 5th International Symposium on Marek’s disease. Tallahassee, Fla: Rose Printing Co., Inc.; 1996. pp. 207–212. [Google Scholar]
- 54.Morimura T, Hattori M, Ohashi K, Sugimoto C, Onuma M. Immunomodulation of peripheral T cells in chickens infected with Marek’s disease virus: involvement in immunosuppression. J Gen Virol. 1995;76:2979–2985. doi: 10.1099/0022-1317-76-12-2979. [DOI] [PubMed] [Google Scholar]
- 55.Nakajima K, Ikuta K, Naito M, Ueda S, Kato S, Hirai K. Analysis of Marek’s disease virus serotype 1-specific phosphorylated polypeptides in virus-infected cells and Marek’s disease lymphoblastoid cells. J Gen Virol. 1987;68:1379–1389. doi: 10.1099/0022-1317-68-5-1379. [DOI] [PubMed] [Google Scholar]
- 56.Nazerian K J, Sharma J M. Detection of T-cell surface antigens in a Marek’s disease lymphoblastoid cell line. J Natl Cancer Inst. 1975;54:277–279. doi: 10.1093/jnci/54.1.277. [DOI] [PubMed] [Google Scholar]
- 57.Nazerian K J, Witter R L. Properties of a chicken lymphoblastoid cell line from Marek’s disease tumor. J Natl Cancer Inst. 1975;54:453–458. [PubMed] [Google Scholar]
- 58.Ono M, Kawaguchi Y, Maeda K, Kamiya N, Tohya Y, Kai C, Niikura M, Mikami T. Nucleotide sequence analysis of Marek’s disease virus (MDV) serotype 2 homolog of MDV serotype 1 pp38, an antigen associated with transformed cells. Virology. 1994;201:142–146. doi: 10.1006/viro.1994.1275. [DOI] [PubMed] [Google Scholar]
- 58a.Parcells, M. S. Unpublished observation.
- 59.Parcells M S, Anderson A S, Cantello J L, Morgan R W. Characterization of Marek’s disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short component open reading frames. J Virol. 1994;68:8239–8253. doi: 10.1128/jvi.68.12.8239-8253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parcells M S, Anderson A S, Morgan R W. Characterization of a Marek’s disease virus mutant containing a lacZ insertion in the US6 (gD) homologue gene. Virus Genes. 1994;9:5–13. doi: 10.1007/BF01703430. [DOI] [PubMed] [Google Scholar]
- 61.Parcells M S, Anderson A S, Morgan R W. Retention of oncogenicity by a Marek’s disease virus mutant lacking six unique short region genes. J Virol. 1995;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parcells M S, Anderson A S, Morgan R W. Rapid construction of mutant Marek’s disease viruses using a green fluorescent protein (GFP) expression cassette. In: Silva R F, et al., editors. Proceedings of the 5th International Symposium on Marek’s disease. Tallahassee, Fla: Rose Printing Co., Inc.; 1996. pp. 284–289. [Google Scholar]
- 63.Peng F, Bradley G, Tanaka A, Lancz G, Nonoyama M. Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek’s disease virus. J Virol. 1992;66:7389–7396. doi: 10.1128/jvi.66.12.7389-7396.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng F, Specter S, Tanaka A, Nonoyama M. A 7 kDa protein encoded by the BamHI-H gene family of Marek’s disease virus is produced in lytically and latently infected cells. Int J Oncol. 1994;4:799–802. doi: 10.3892/ijo.4.4.799. [DOI] [PubMed] [Google Scholar]
- 65.Peng Q, Zeng M, Bhuiyan Z A, Ubukata E, Tanaka A, Nonoyama M, Shirazi Y. Isolation and characterization of Marek’s disease virus (MDV) cDNAs mapping to the BamHI-I2, BamHI-Q2, and BamHI-L fragments of the MDV genome from lymphoblastoid cells transformed and persistently infected with MDV. Virology. 1995;213:590–599. doi: 10.1006/viro.1995.0031. [DOI] [PubMed] [Google Scholar]
- 66.Powell P C. Immunity to Marek’s disease induced by glutaraldehyde-treated cells of Marek’s disease lymphoblastoid cell lines. Nature. 1975;257:684–685. doi: 10.1038/257684a0. [DOI] [PubMed] [Google Scholar]
- 67.Powell P C, Payne L N, Frazier J A, Rennie M. T lymphoblastoid cell lines from Marek’s disease lymphomas. Nature. 1974;251:79–80. doi: 10.1038/251079a0. [DOI] [PubMed] [Google Scholar]
- 68.Qian Z, Brunovskis P, Rauscher III F, Lee L, Kung H-J. Transactivation activity of Meq, a Marek’s disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J Virol. 1995;69:4037–4044. doi: 10.1128/jvi.69.7.4037-4044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian Z, Brunovskis P, Lee L, Vogt P K, Kung H-J. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J Virol. 1996;70:7161–7170. doi: 10.1128/jvi.70.10.7161-7170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quere P. Suppression mediated in vitro by Marek’s disease virus-transformed T-lymphoblastoid cell lines: effect on lymphoproliferation. Vet Immunol Immunopathol. 1992;32:149–164. doi: 10.1016/0165-2427(92)90076-3. [DOI] [PubMed] [Google Scholar]
- 71.Ross N, O’Sullivan G, Smith G, Davison F, Rennie M. Expression of MDV genes in lymphomas and their roles in oncogenesis. In: Silva R F, et al., editors. Proceedings of the 5th International Symposium on Marek’s disease. Tallahassee, Fla: Rose Printing Co., Inc.; 1996. pp. 40–46. [Google Scholar]
- 72.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 73.Schat K A, Buckmaster A, Ross L J N. Partial transcription map of Marek’s disease herpesvirus in lytically infected cells and lymphoblastoid cell lines. Int J Cancer. 1989;44:101–109. doi: 10.1002/ijc.2910440119. [DOI] [PubMed] [Google Scholar]
- 74.Schat K A, Calnek B W, Fabricant J. Characterization of two highly oncogenic strains of Marek’s disease virus. Avian Pathol. 1982;11:593–605. doi: 10.1080/03079458208436134. [DOI] [PubMed] [Google Scholar]
- 75.Schat K A, Chen C-L H, Calnek B W, Char D. Transformation of T lymphocyte subsets by Marek’s disease herpesvirus. J Virol. 1991;65:1408–1413. doi: 10.1128/jvi.65.3.1408-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schat K A, Chen C-L, Shek W R, Calnek B W. Surface antigens on Marek’s disease lymphoblastoid cell lines. J Natl Cancer Inst. 1982;69:715–720. [PubMed] [Google Scholar]
- 77.Schat K A, Pratt W D, Morgan R, Weinstock D, Calnek B W. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 1992;36:432–439. [PubMed] [Google Scholar]
- 78.Silva R F, Lee L F. Monoclonal antibody-mediated immunoprecipitation of proteins from cells infected with Marek’s disease virus or turkey herpesvirus. Virology. 1984;136:307–320. doi: 10.1016/0042-6822(84)90167-3. [DOI] [PubMed] [Google Scholar]
- 79.Silver S, Tanaka A, Nonoyama M. Transcription of the Marek’s disease virus genome in a nonproductive chicken lymphoblastoid cell line. Virology. 1979;93:127–133. doi: 10.1016/0042-6822(79)90281-2. [DOI] [PubMed] [Google Scholar]
- 80.Silver S, Smith M, Nonoyama M. Transcription of the Marek’s disease virus genome in virus-induced tumors. J Virol. 1979;30:84–89. doi: 10.1128/jvi.30.1.84-89.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith G D, Zelnik V, Ross L J N. Gene organization in herpesvirus of turkeys: identification of a novel open reading frame in the long unique region and a truncated homologue of pp38 in the internal repeat. Virology. 1995;207:205–216. doi: 10.1006/viro.1995.1067. [DOI] [PubMed] [Google Scholar]
- 82.Stenger S, Mazzaccaro R J, Uyemura K, Cho S, Barnes P F, Rosat J-P, Sette A, Brenner M B, Porcelli S A, Bloom B R, Modlin R L. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 83.Sugaya K, Bradley G, Nonoyama M, Tanaka A. Latent transcripts of Marek’s disease virus are clustered in the short and long repeat regions. J Virol. 1990;64:5773–5782. doi: 10.1128/jvi.64.12.5773-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka A, Silver S, Nonoyama M. Biochemical evidence of the non-integrated status of Marek’s disease virus DNA in virus transformed lymphoblastoid cells of chicken. Virology. 1978;88:19–24. doi: 10.1016/0042-6822(78)90105-8. [DOI] [PubMed] [Google Scholar]
- 85.Theis G A. Effects of lymphocytes from Marek’s disease-infected chickens on mitogen responses of syngeneic normal chicken spleen cells. J Immunol. 1977;118:887–894. [PubMed] [Google Scholar]
- 86.Venugopal K, Payne L N. Molecular pathogenesis of Marek’s disease-recent developments. Avian Pathol. 1995;24:597–609. doi: 10.1080/03079459508419100. [DOI] [PubMed] [Google Scholar]
- 87.Viola A, Lanzavecchia A. T cell activation determined by T cell receptor number and tunable thresholds. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 88.Volpini L M, Calnek B W, Sekellick M J, Marcus P I. Stages of Marek’s disease virus latency defined by variable sensitivity to interferon modulation of viral antigen expression. Vet Microbiol. 1995;47:99–109. doi: 10.1016/0378-1135(95)00056-g. [DOI] [PubMed] [Google Scholar]
- 89.von Bülow V, Klasen A. Growth inhibition of Marek’s disease T-lymphoblastoid cell lines by chicken bone-marrow-derived macrophages activated in vitro. Avian Pathol. 1983;12:161–178. doi: 10.1080/03079458308436161. [DOI] [PubMed] [Google Scholar]
- 90.Xie Q, Anderson A S, Morgan R W. Marek’s disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J Virol. 1996;70:1125–1131. doi: 10.1128/jvi.70.2.1125-1131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu G S, Iwata A, Gong M, Ueda S, Hirai K. Marek’s disease virus type 1-specific phosphorylated proteins pp38 and pp24 with common amino acid termini are encoded from the opposite junction regions between the long unique and inverted repeat sequences of viral genome. Virology. 1994;200:816–820. doi: 10.1006/viro.1994.1249. [DOI] [PubMed] [Google Scholar]