Abstract
Guanylyltransferases are members of the nucleotidyltransferase family and function in mRNA capping by transferring GMP to the phosphate end of nascent RNAs. Although numerous guanylyltransferases have been identified, studies which define the nature of the interaction between the capping enzymes of any origin and their RNA substrates have been limited. Here, we have characterized the RNA-binding activity of VP3, a minor protein component of the core of rotavirions that has been proposed to function as the viral guanylyltransferase and to direct the capping of the 11 transcripts synthesized from the segmented double-stranded RNA (dsRNA) genome of these viruses. Gel shift analysis performed with disrupted (open) virion-derived cores and virus-specific RNA probes showed that VP3 has affinity for single-stranded RNA (ssRNA) but not for dsRNA. While the ssRNA-binding activity of VP3 was found to be sequence independent, the protein does exhibit preferential affinity for uncapped over capped RNA. Like the RNA-binding activity, RNA capping assays performed with open cores indicates that the guanylyltransferase activity of VP3 is nonspecific and is able to cap RNAs initiating with a G or an A residue. These data establish that all three rotavirus core proteins, VP1, the RNA polymerase; VP2, the core capsid protein; and VP3, the guanylyltransferase, have affinity for RNA but that only in the case of the RNA polymerase is the affinity sequence specific.
Rotaviruses, members of the family Reoviridae, are a significant cause of acute gastroenteritis in infants and young children (13). The mature virion is made up of three concentric layers (shells) of protein and contains a segmented double-stranded RNA (dsRNA) genome (9). The innermost shell is a T = 1 icosahedron that is formed by 60 dimers of the RNA-binding protein VP2 (2, 15). One copy each of the minor structural proteins VP1 and VP3 is thought to be associated with each of the pentamers of the VP2 shell. These three proteins along with the viral genome make up the core of the virion (15, 27). Incubation of virion-derived cores in hypotonic buffer causes disruption of the VP2 shell and loss of the dsRNA genome from the particle (4). While VP2 in such core preparations (open cores) remains oligomeric, at least some of the VP1 and VP3 dissociates from the disrupted VP2 shell and becomes soluble (22). In contrast to intact cores, open cores have replicase activity which can catalyze the synthesis of dsRNA from viral mRNA in vitro (4). Double-shelled particles consist of cores surrounded by 260 VP6 trimers and have transcriptase activity which directs the synthesis of the capped, but nonpolyadenylated, mRNAs (6, 12, 19, 26). Reconstitution experiments have indicated that VP6 plays an essential but yet undefined role in viral transcription (1, 29). The outermost shell of the virion consists of the glycoprotein VP7 and the spike protein VP4 (26).
Numerous findings have indicated that the minor core protein, VP1, is the viral RNA-dependent RNA polymerase and functions as both the viral transcriptase and replicase. For instance, sequence analysis has shown that VP1 contains motifs that are shared among RNA-dependent RNA polymerases (7, 10, 21). Also, VP1 is known to bind nucleotides, and cross-linking of the photoreactable nucleotide azido-ATP to VP1 inhibits transcription (33). Additional evidence that VP1 is the viral RNA polymerase comes from gel shift assays which have shown that the protein specifically binds to the 3′ end of viral mRNA (22) and from replicase assays which have shown that this protein is a common component of rotavirus replication intermediates (11) and of baculovirus-expressed virus-like particles that synthesize dsRNA (35). Reconstitution experiments performed with purified recombinant protein have shown that, although VP1 is the candidate RNA polymerase, the protein requires the presence of VP2 for replicase activity (24). This finding confirms earlier studies with rotavirus temperature-sensitive mutants which indicated that VP2 is essential for RNA replication (18).
Of all the core proteins, probably least is known about the properties of VP3. VP3 is a component of early replication intermediates, and it and VP1 seem to be the first two structural proteins to associate with viral mRNA during packaging and RNA replication (11). Recent studies with virus-like particles have provided evidence that VP3 can bind to the N terminus of VP2 (34), a region of the core shell protein to which VP1 and single-stranded RNA (ssRNA) are also known to bind (14) and that is required for RNA replication (24). Studies by Sandino et al. (30) have indicated that VP3 is in direct physical contact with VP6 in the virion and that this interaction may be essential for transcription. Several studies have provided evidence that VP3 is the viral guanylyltransferase and therefore is responsible for capping of viral mRNAs. For example, VP3 contains sequence motifs that are shared among guanylyltransferases (10, 21), and in the presence of GTP, VP3 forms a reversible covalent adduct with GMP (16, 25). Of particular note, experiments by Pizarro et al. (25) indicate that the guanylyltransferase activity of VP3 transfers GMP from the VP3-GMP complex to the pyrophosphate group of GDP, resulting in the formation of a GpppG cap. Unlike the viral mRNA, the minus-strand RNA synthesized by rotaviruses is not capped but instead contains a 5′-pyrophosphate group (12, 19).
Guanylyltransferases are evolutionarily conserved enzymes that have common features with ATP-dependent DNA ligases, RNA ligases, and other members of the nucleotidyltransferase superfamily (8, 31, 32). Because, with a few exceptions, cellular mRNAs and the mRNAs of both DNA and RNA viruses are modified by capping enzymes, the conserved nature of guanylyltransferases suggests that the RNA-binding activity of this enzyme will display common characteristics regardless of origin. However, only the RNA-binding properties of the capping enzyme of vaccinia virus have been investigated to date (17). Here, we have characterized the RNA-binding activity of the rotavirus guanylyltransferase by gel shift assay and have found that the protein has affinity for ssRNA but not for dsRNA. While the ssRNA-binding activity of VP3 is nonspecific, as is also the case for the vaccinia virus guanylyltransferase (17), our results indicate that the protein exhibits preferential affinity for uncapped as opposed to capped ssRNA. Consistent with its nonspecific RNA-binding activity, the 5′ capping activity of VP3 was also shown to be nonspecific and to cap RNAs that initiate with either a G or an A residue.
MATERIALS AND METHODS
Preparation of open cores.
DxRRV is a reassortant rotavirus that was generated by coinfection of MA104 cells with human D and rhesus RRV rotaviruses (20). With the exception of the genome segment encoding VP7, all the other segments of DxRRV originated from the RRV parent. DxRRV virus was propagated in MA104 cells, and the infected cell lysates were treated with 12.5 mM EDTA to remove the outer shell from the triple-shelled virions. The double-shelled virus was pelleted by centrifugation for 2 h in a Beckman type 19 rotor at 18,000 rpm, resuspended in Tris-buffered saline (TBS), and extracted with trichlorotrifluoroethane. After being pelleted again, the virus was resuspended in TBS, purified by twice banding on CsCl gradients, and dialyzed against TBS (23). The concentration of the purified double-shelled virus was approximately 50 A260 U per ml. To prepare (VP6-free) open cores, the double-shelled virus was diluted approximately 10-fold with TBS prior to addition of an equal volume of 2 M CaCl2 (4). To prepare VP6-open cores, the double-shelled virus was treated with CaCl2 without prior dilution. Virus was incubated with CaCl2 for 90 min at 37°C with gentle rocking, and intact cores were recovered from the solution by centrifugation for 2 min at 10,000 × g in an Eppendorf centrifuge. Open cores were prepared by resuspending the intact cores in LSB (2 mM Tris-HCl [pH 7.6], 0.5 mM EDTA, 0.5 mM dithiothreitol) and by dialysis against the same buffer. The protein concentration given for the core preparations was determined by coelectrophoresis with known amounts of bovine serum albumin and is expressed relative to the amount of VP2 that was present. Typically, the concentration of VP2 in the core preparations was 0.1 to 0.3 mg per ml.
35S-labeled open cores and VP6-open cores were prepared from DxRRV-infected cells maintained in 85% methionine-free M199 medium containing 7 μCi of 35S-amino acids (35S-Express; 1,175 Ci/mmol) per ml.
Construction of transcription vectors.
The T7 vectors SP72g8R40 (3), SP65g8R (23), and pMJ5B6.4 (24) were constructed as described previously. Following linearization with SacII and blunt ending with T4 DNA polymerase, SP72g8R40, SP65g8R, and pMJ5B6.4 were transcribed with T7 RNA polymerase to produce SP72-v3′40 RNA, wild-type gene 8 RNA, and wild-type gene 6 RNA, respectively. The vector SP72 was digested with SmaI, XhoI, NdeI, SspI, and PvuII and treated with T4 DNA polymerase to produce nonviral RNAs of 48, 97, 184, 551, and 985 nucleotides (nt), respectively, by runoff transcription with T7 RNA polymerase.
To produce the T7 transcription vector, SP65g8 5′-3′SacII, residues 87 to 1057 in the gene 8 cDNA of SP65g8R were deleted by PCR with the Elongase amplification system (Life Technologies). The reaction mixture contained the negative-sense primer 5′-aataaATTCTCCAAATGAGGATAGCA-3′, the positive-sense primer 5′-AATTTGAGGATGATGATGGCT-3′, and the plasmid SP65g8R and was amplified under the following conditions: 94°C for 1 min, 48°C for 1 min, and 72°C for 2 min (40 cycles). (Virus-specific sequences in primers are shown in uppercase.) The T7 transcription vector, SP65g8 GG44-86/3′SacII, contains the same internal deletion in the gene 8 cDNA that is present in SP65g8 5′-3′SacII (residues 87 to 1057) but also contains a deletion in the gene 8 cDNA extending from residues 2 to 43. The amplification reaction used to generate SP65g8 GG44-86/3′SacII was the same as that used for SP65g8 5′-3′SacII except that the negative-sense primer 5′-AAAAGCAAGCTAGCTCAGCCATGGCCTATAGTGAGTCGTATTA-3′ and positive-sense primer 5′-GCTATCCTCATTTGGAGAAttattAATTTGAGGATGATGATGGCT-3′ were included instead. PCR products were gel purified, self-ligated with T4 DNA ligase, and used to transform competent Escherichia coli DH5α (28) Bacteria containing appropriate plasmids were selected on the basis of antibiotic resistance and digestion with restriction enzymes. The expected nucleotide sequence of the insert in the vectors was confirmed by dideoxynucleotide sequencing with appropriate oligonucleotide primers and a Sequenase version 2.0 kit (Amersham). To produce g8 5′-3′SacII RNA and g8 5′-3′Eco47III RNA, SP65g8 5′-3′SacII was digested with SacII and Eco47III, respectively, and treated with T4 DNA polymerase prior to transcription. To produce g8 GG44-86/3′ RNA, SP65g8 GG44-86/3′SacII was digested with SacII and treated with T4 DNA polymerase prior to transcription.
PCR was also used to produce the T3 transcription vector, pT7B/T3-60/SacII. The amplification reaction contained the positive-sense primer, 5′-caattaaccctcactaaagggATTCGCTATCAATTTGAGGAT-3′; the negative-sense primer, 5′-actcctgcattaggaagcagc-3′; and the plasmid SP65g8R. (Underlined sequences in primers represent the promoter for T3 RNA polymerase.) The PCR product was gel purified and ligated into pT7Blue T vector (Novagen). The resulting construct, pT7B/T3-60/SacII, was linearized with SacII, treated with T4 DNA polymerase, and used in T3 transcription reactions to synthesize the RNA probe, v-3′60. The sequence of the v-3′60 RNA begins with three G residues followed by the 3′-terminal 60 residues of the SA11 gene 8 RNA. To produce the T3 vector, SP72/T3-0/EcoRV, the complementary oligonucleotides 5′-agctcaattaaccctcactaaagggattttgttgca-3′ and 5′-acaaatccctttagtgagggttaattg-3′ were kinase treated and annealed, forming a short DNA hybrid with HindIII and PstI cohesive ends. T4 DNA ligase was used to insert the annealed primers between the HindIII and PstI sites of SP72 (28). The resulting construct, SP72/T3-0/EcoRV, was linearized with EcoRV and used in T3 transcription reactions to produce nv-60, an RNA which initiates with three G residues and is followed by a 60-nt sequence of nonviral origin.
In vitro synthesis of RNAs.
T3 vectors were transcribed by using the Ambion T3 MEGAscript transcription system, and T7 vectors were transcribed with either the Ambion T7 MEGAscript transcription system (SP65g8R and pMJ5B6.4) or the Ambion T7 MEGAshort transcription system (SP72g8R40, SP65g8 5′-3′SacII, SP65g8 GG44-86/3′SacII) (23). To synthesize 32P-labeled RNA probes, the concentration of UTP in the reaction mixture was reduced to one-fourth of the standard level recommended by the manufacturer and 2.5 mCi of [α-32P]UTP (800 Ci/mmol) per ml was included. RNA products were recovered from reaction mixtures by phenol-chloroform extraction and isopropanol precipitation. The quality of the gene 6 and gene 8 RNAs was assessed by electrophoresis on 5% polyacrylamide gels containing 7 M urea (23). 32P-labeled RNA probes were purified by electrophoresis on 8% polyacrylamide gels containing 6 M urea (22). RNA concentrations were calculated from optical densities at 260 nm.
Replicase assays.
Except for minor differences, replication assays were performed as described by Chen et al. (4). Reaction mixtures contained 50 mM Tris-HCl (pH 7.2); 5 mM magnesium acetate; 5 mM dithiothreitol; 200 μM (each) ATP, CTP, and GTP; 20 μM UTP; 10 μCi of [α-32P]UTP (800 Ci/mmol); open cores or VP6-open cores representing 0.5 to 1.0 μg of VP2; and the indicated amount of either gene 6 or gene 8 mRNA. After incubation for 3 h at 32°C, the reaction mixtures were analyzed by electrophoresis on 12% polyacrylamide gels containing sodium dodecyl sulfate (SDS–12% polyacrylamide gel electrophoresis [PAGE]) (23), and the dsRNA products were detected by autoradiography and quantitated with a phosphorimager.
Gel shift assay.
The procedure used for analysis of rotavirus RNA-protein interactions by the gel shift assay was similar to that described earlier (22). Typically, RNA-protein complexes were allowed to form by incubation of open cores or VP6-open cores, containing 3.3 μg of VP2, with 1 to 10 pmol (24 to 250 ng) of 32P-labeled RNA probe for 1 h at 32°C. SA11 dsRNA, rabbit liver tRNA (∼110 nt), and luciferase RNA (2,650 nt; Promega Scientific) were included in some reaction mixtures as cold competitor RNAs. SA11 dsRNA was prepared from virions purified by double banding on CsCl gradients. Reaction mixtures were resolved by electrophoresis for 3 to 4 h at 175 V on nondenaturing 8% polyacrylamide gels containing 50 mM Tris-HCl, pH 9.1 (22). Protein-probe complexes were identified on the gel by autoradiography, and the intensity of radiolabeled bands was quantitated with a phosphorimager.
Preparation of rVP1.
Sf9 cells were mock infected or infected with rBVg1, a recombinant baculovirus containing the gene 1 cDNA of SA11 rotavirus (24). At 5 days postinfection, the cells were scraped into cold phosphate-buffered saline, collected by low-speed centrifugation, washed twice with phosphate-buffered saline, and then lysed by resuspension in LSB-DOC (LSB containing 1% sodium deoxycholate and 500 ng each of aprotinin and leupeptin per ml). The lysates were centrifuged at 2,000 × g for 20 min, the supernatants were removed, and the pellets were resuspended in LSB-DOC and dialyzed exhaustively against LSB. Treatment of the pellets in this manner solubilized the material contained within them. The pellet-containing extracts of the mock-infected and rBVg1-infected cells were used in gel shift assays.
Labeling of VP3 of open cores with GTP.
The method used for radiolabeling of VP3 with GTP was a modification of the protocol described by Fukuhara et al. (10). Open cores or VP6-open cores containing 2 to 6.6 μg of VP2 were incubated with 100 μCi of [α-32P]GTP (800 Ci/mmol) and 5 mM MgCl2 in a final volume of 30 μl for 30 min at 37°C. The reaction mixtures were resolved by SDS–12% PAGE. Radiolabeled proteins were detected by autoradiography and quantitated with a phosphorimager.
5′ capping and decapping of RNAs.
The guanylyltransferase activity associated with open cores was used to cap the 5′ ends of RNAs. To prepare capped and [32P]GMP-labeled SP72-v3′40, 20 μg of VP6-open cores in LSB, 5 mM MgCl2, 400 μCi of [α-32P]GTP (800 Ci/mmol), and 2.5 μg of cold SP72-v3′40 were incubated in a final volume of 160 μl for 3 h at 37°C. The sample was deproteinized by phenol-chloroform extraction, and the [32P]GMP-labeled SP72-v3′40 was purified by electrophoresis on and elution from a 7 M urea–8% polyacrylamide gel (22). Capping assay mixtures used for rotavirus gene 8 RNA and influenza virus DI-PA RNA contained 1 μg of open cores, 5 mM MgCl2, 30 μCi of [α-32P]GTP, and 0.3 μg of RNA and were incubated for 1 h at 37°C.
SP72-v3′40 RNAs (10 ng) were treated to remove 5′ cap structures by incubation with 10 U of tobacco acid pyrophosphatase (Epicentre Technologies, Madison, Wis.) in 50 mM sodium acetate (pH 6.0)–1 mM EDTA–0.1% β-mercaptoethanol–0.01% Triton X-100 in a final volume of 20 μl for 1 h at 37°C. The products were analyzed by SDS–14% PAGE.
RESULTS
RNA-protein interactions of open cores.
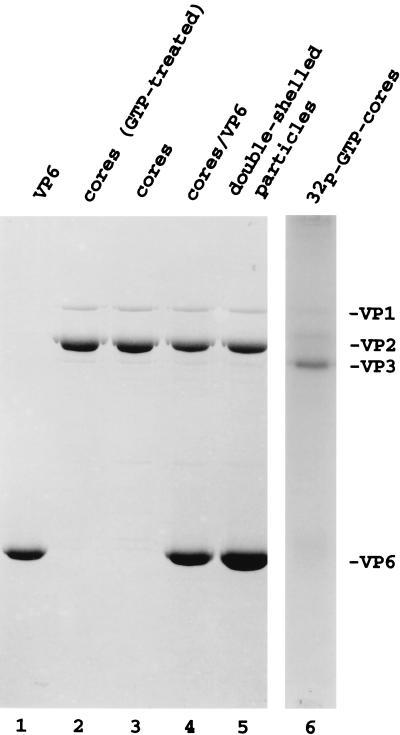
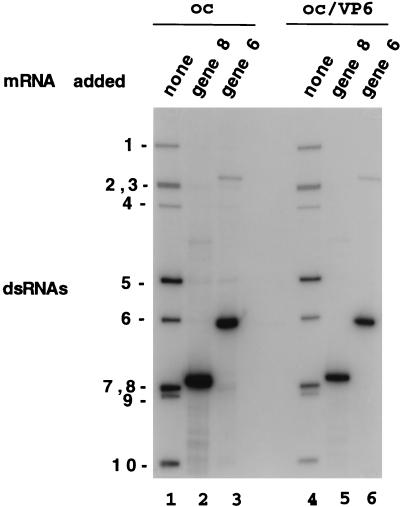
Double-shelled particles were obtained by treatment of purified triple-shelled DxRRV virions with EDTA (Fig. 1, lane 5). Diluted or undiluted preparations of the double-shelled particles were incubated with CaCl2 to disrupt the VP6 layer of protein, and the single-layered core particles were recovered by centrifugation. As shown by gel electrophoresis (Fig. 1, lane 3), exposure of the diluted double-shelled particles to CaCl2 allowed the recovery of cores that were free of VP6. Identification of VP3 in the core preparation was based on the ability of the protein to form a radiolabeled VP3-GMP complex when incubated with [32P]GTP (Fig. 1, lane 6) (25). Consistent with earlier reports (4), dialysis of the intact core particles against hypotonic buffer caused their disruption and generated open cores which had replicase activity that catalyzed the synthesis of dsRNA from viral mRNA in vitro (Fig. 2, lanes 2 and 3). Unlike treatment of the diluted double-shelled particles, treatment of the undiluted double-shelled particles with CaCl2 led to an incomplete disruption of the VP6 protein layer surrounding the cores (Fig. 1, lane 4). Under the conditions used in this study, the efficiency of VP6 removal from the undiluted double-shelled particles was about 50%. Cores containing residual VP6, when disrupted with hypotonic buffer (VP6-open cores), had high levels of replicase activity, albeit levels that were two- to threefold lower than that found for VP6-free open cores (Fig. 2, lanes 5 and 6). Despite the presence of VP6, in vitro analysis of the VP6-open cores showed that they lacked transcriptase activity (data not shown) and therefore did not contain residual intact double-shelled particles.
FIG. 1.
Protein composition of open cores and VP6-open cores. Samples of open cores (lanes 2 and 3), VP6-open cores (lane 4), and double-shelled particles (lane 5), each containing 5 μg of VP2, were analyzed by SDS–12% PAGE followed by staining with Coomassie blue. VP6 (4 μg) recovered from CaCl2-treated double-shelled particles was resolved in lane 1. The cores that were resolved in lane 2 were incubated with [α-32P]GTP prior to electrophoresis. VP3-GMP complexes that formed in this sample were identified by autoradiography (lane 6) and were used to confirm the position of VP3 in the gel.
FIG. 2.
Replicase activity of open cores and VP6-open cores. The products of replicase assays containing open cores (oc) or VP6-open cores (oc/VP6) and no added mRNA (none) or 5 μg of gene 8 or 6 mRNA were resolved by SDS–12% PAGE and detected by autoradiography. Background bands corresponding to the 11 genome segments in core preparations are routinely seen in assays performed with no exogenous RNA and are the products of an undefined activity (4).
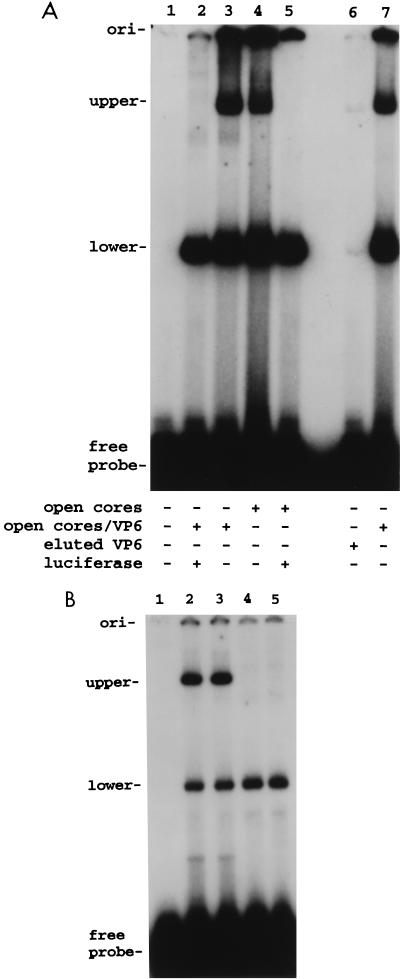
The RNA-binding activity of the proteins in the open core and VP6-open core preparations was evaluated by gel shift assay according to previously published procedures (22). The probe used in the assay, 32P-labeled SP72-v3′40, was 72 nt long, and the last 40 nt of the probe was identical in sequence to the last 40 nt of the SA11 gene 8 mRNA. Electrophoresis of reaction mixtures containing the probe and either open cores or VP6-open cores revealed the presence of two prominent bands representing probe-protein complexes (Fig. 3A, lanes 3 and 4); these complexes were designated as upper and lower based on their migration in the nondenaturing polyacrylamide gel. The fact that the gel shift assay did not reveal any significant differences in the types or levels of probe-protein complexes that were formed by the open cores and the VP6-open cores suggested that VP6 neither bound the probe nor affected the RNA-binding activity of the core proteins. As a direct test of whether VP6 had RNA-binding activity, VP6 which was eluted from double-shelled particles (Fig. 1, lane 1), was incubated with 32P-labeled SP72-v3′40. Analysis of this mixture by electrophoresis on a nondenaturing gel revealed the presence of only background levels of the upper and lower probe-protein complexes (Fig. 3A, lane 6), supporting the idea that VP6 lacks intrinsic RNA-binding activity. The trace levels of probe-protein complexes formed by the VP6 preparation most likely stem from contamination with small amounts of core proteins.
FIG. 3.
RNA-protein complexes formed by open cores and VP6-open cores. (A) 32P-labeled SP72-v3′40 (7.5 pmol, 0.17 μg) was incubated alone or with open cores or VP6-open cores, each containing 3.3 μg of VP2, or with 2 μg of VP6 eluted from double-shelled particles with CaCl2. Some reaction mixtures also included luciferase RNA (1.7 pmol, 1.0 μg). (B) 32P-labeled SP72-v3′40 (7.5 pmol, 0.17 μg) was incubated alone (lane 1) or with VP6-open cores containing 3.3 μg of VP2 in the absence of competitor RNA for 60 min (lane 2) or 30 min (lane 3) or in the presence of 1.7 μg of luciferase RNA for 60 min (lane 5). Lane 4 shows complexes formed when 32P-labeled SP72-v3′40 was preincubated with the VP6-open cores for 30 min and then, after addition of 1.7 μg of luciferase RNA, the incubation was continued for another 30 min. Probe-protein complexes were detected by electrophoresis on 8% polyacrylamide gels and by autoradiography. ori, origin.
Specificity of RNA-protein interactions.
To examine the specificity of the RNA-protein interactions involved in the formation of the upper and lower complexes, open cores and VP6-open cores were incubated with 32P-labeled SP72-v3′40 and various nonviral competitor RNAs under conditions where the concentration ratio of the competitor RNA to probe was high with respect to mass but low with respect to molarity. When luciferase RNA and the probe were included in the assay at a molar ratio of 0.2:1 and a mass ratio of 6:1, the formation of the upper probe-protein complex was blocked while little effect (<2-fold) was seen on the formation of the lower probe-protein complex (Fig. 3A, lanes 2 and 5). Similar results were seen when Xenopus elongation factor 1a RNA or brome mosaic virus RNA was used in the assay instead of luciferase RNA (data not shown). These results indicated that the formation of the upper complex represented a nonspecific interaction between the probe and protein.
To assess the reversibility of the RNA-protein interaction that led to the formation of the upper probe-protein complex, open cores were incubated with 32P-labeled SP72-v3′40 for 30 min and then luciferase RNA was added and the incubation was continued for 30 more min. Electrophoretic analysis showed that, while upper-probe protein complex was present in the reaction mixture before the addition of competitor RNA, subsequent incubation with luciferase RNA resulted in the loss of the complex (Fig. 3B, lane 4). Thus, the upper complex represents a reversible interaction between the probe and protein.
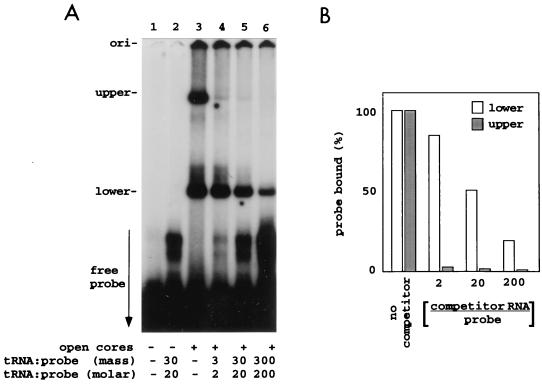
The nature of the RNA-protein interactions occurring between the core proteins and 32P-labeled SP72-v3′40 was further analyzed by including increasing concentrations of rabbit liver tRNA in the gel shift assays. The results showed that even when the concentration of tRNA was in both low molar (twofold) and mass (threefold) excess over probe, the formation of the upper probe-protein complex was nearly completely inhibited (>90%) (Fig. 4). In contrast, the formation of the lower protein complex was only slightly affected (<20%) at this same concentration of tRNA. When the concentration of tRNA in the assay was increased to 20 or 200 molar (or 30 or 300 mass) excess over probe, the level of upper probe-protein complex detected was reduced by 20-fold or more while the level of lower probe-protein complex detected was reduced by two- or fivefold, respectively (Fig. 4). The extreme sensitivity of the upper complex to loss by the addition of competitor RNAs confirms that this complex is generated through a nonspecific interaction between SP72-v3′40 and a core protein. The fact that, even in the presence of 200-fold molar excess competitor tRNA, the formation of lower complex was reduced by only fivefold indicates that this complex represents a specific interaction between SP72-v3′40 and one of the core proteins.
FIG. 4.
Effect of competitor RNA on the interaction of core proteins with a viral 3′-specific RNA probe. 32P-labeled SP72-v3′40 (4.4 pmol, 0.1 μg) was incubated with open cores and with the indicated amount of rabbit liver tRNA. The ratio of tRNA to probe in the reaction mixtures is expressed in terms of mass and moles. (A) Probe-protein complexes were resolved by electrophoresis, and the lower and upper complexes were detected by autoradiography. (B) The intensity of the upper and lower bands was determined with a phosphorimager, and the values were adjusted relative to 100% [probe bound (%)] for the reaction mixture containing no competitor RNA. The percentage of probe bound was plotted versus the molar ratio of tRNA to probe in the reaction mixtures. ori, origin.
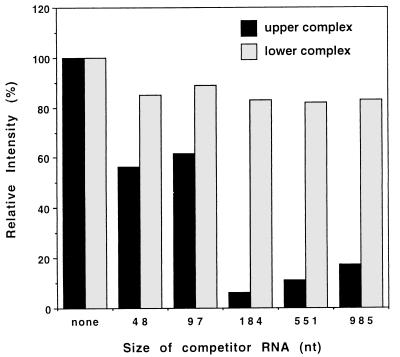
In gel shift assays performed in parallel that contained VP6-open cores; 32P-labeled SP72-v3′40; and equal mass amounts of luciferase RNA (2,650 nt), tRNA (110 nt), or poly(U) (∼3,000 nt), we noted a difference in the efficiency with which these competitor RNAs would interfere with the formation of the upper complex (data not shown). Because this phenomenon raised the possibility that the size of the competitor RNA might affect its ability to interfere with the formation of the upper complex, open cores were incubated with 50 ng of 32P-labeled SP72-v3′40 and 50 ng of one of a set of competitor RNAs ranging in size from 48 to 985 nt. As shown in Fig. 5, the results of the gel shift assay indicated that, on a constant mass basis, longer competitor RNAs (184, 551, and 985 nt) interfered more efficiently with the formation of the upper complex than did shorter competitor RNAs (48 and 97 nt). While the reason for this is unclear, it is noteworthy that the size of the competitor RNA had no influence on the formation of the lower probe-protein complex.
FIG. 5.
Size of the competitor RNA and its impact on formation of the upper protein-probe complex. 32P-labeled SP72-v3′40 (50 ng) was incubated with open cores and no competitor RNA or 50 ng of cold competitor RNAs of the indicated size. The competitor RNAs were produced by runoff transcription of linearized SP72. Upper and lower protein-probe complexes were resolved by electrophoresis, and the band intensities of the complexes detected on the gel were determined with a phosphorimager. The values were adjusted relative to 100% for the assay which contained probe and open cores but no competitor RNA.
Proteins binding to RNA.
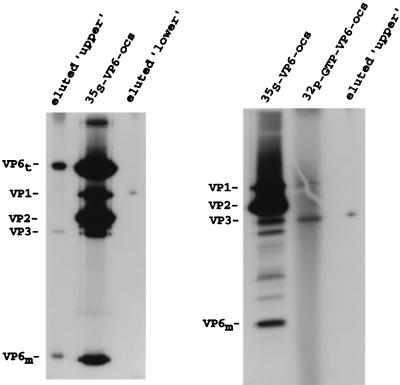
To identify the core proteins involved in the formation of the upper and lower complexes, 35S-labeled open cores and VP6-open cores were prepared and then incubated with 32P-labeled SP72-v3′40. The upper and lower probe-protein complexes in the reaction mixtures were resolved by electrophoresis on a nondenaturing gel, and then the 35S-labeled protein migrating at the position of the complexes was recovered and identified by electrophoresis on an SDS-polyacrylamide gel. For assays performed with 35S-labeled VP6-open cores, VP1 was detected in the lower complex and VP3 and the trimer and monomer forms of VP6 were detected in the upper complex (Fig. 6, left panel). VP3, but not VP6, was present in the upper complex of gel shift assays performed with 35S-labeled open cores (Fig. 6, right panel). These results establish that VP3 has intrinsic RNA-binding activity and furthermore show that, despite evidence for the interaction of VP3 and VP6 in virions (30), VP6 is neither required for nor affects the RNA-binding activity of VP3.
FIG. 6.
Protein composition of the upper and lower probe-protein complexes. 32P-labeled SP72-v3′40 was incubated with 35S-labeled VP6-open cores (ocs) (left) or open cores (right), and the mixtures were resolved by electrophoresis on a nondenaturing 8% polyacrylamide gel. The positions of upper and lower complexes in the gel were identified by autoradiography, and portions of the gel containing the complexes were cut out, soaked in sample buffer, and loaded onto SDS–12% polyacrylamide gels. After electrophoresis, the 35S-labeled proteins were detected by fluorography (shown). The position of proteins in the gel was determined by coelectrophoresis of 35S-labeled VP6-open cores. To confirm the position of VP3, VP3-[32P]GMP complexes were formed by coincubation of cold VP6-open cores and [α-32P]GTP prior to electrophoresis.
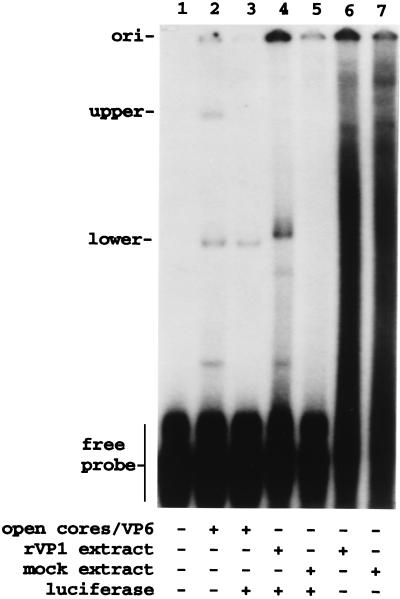
rVP1 produced with a baculovirus expression system was used to verify that VP1 was responsible for the formation of the lower complex (24). As shown in Fig. 7, gel shift assays performed with a cell extract containing rVP1, 32P-labeled SP72-v3′40, and nonviral competitor RNA produced a probe-protein complex that comigrated near the lower complex produced with open cores. When an extract from mock-infected cells was used in the assay instead of the rVP1-containing extract, the lower probe-protein complex was not detected, showing that VP1 was required for its formation. When the rVP1-containing extract was incubated with 32P-labeled SP72-v3′40 but with no competitor RNA, electrophoresis of the mixture produced a smear of probe on the gel (Fig. 7, lane 6). This is probably due to the presence of cellular proteins in the rVP1-containing extract which bound the probe nonspecifically and thereby interfered with the formation or detection of the lower complex.
FIG. 7.
Recombinant VP1 interacts with the viral 3′-specific RNA probe to form the lower complex. 32P-labeled SP72-v3′40 (50 ng) was incubated alone or with either a preparation of open cores, an extract from rBVg1-infected Sf9 cells containing rVP1, or an extract from mock-infected Sf9 cells. Luciferase RNA (0.5 μg) was included in some reaction mixtures. Probe-protein complexes were resolved by electrophoresis and detected by autoradiography. ori, origin.
Specificity of complexes formed with gene 8 5′-terminal sequences.
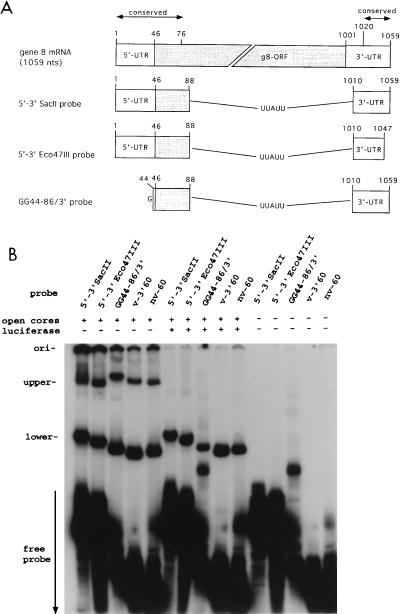
Based on gel shift assays performed with the SP72-v3′40 probe and nonviral competitor RNAs, the possibility that VP3 specifically recognized the 3′-terminal 40 nt of the gene 8 mRNA was ruled out. However, this did not exclude the possibility that, in its role as a guanylyltransferase, VP3 might bind specifically to the 5′ end of the gene 8 mRNA. To address this question, we generated 5′-3′SacII (142 nt), an RNA probe that contains the entire 5′ untranslated region (UTR), the first 40 nt of the open reading frame, and the entire 3′ UTR of the gene 8 mRNA (Fig. 8A). The 5′-3′SacII probe contains all the cis-acting signals of the gene 8 mRNA that are necessary for efficient replication by open cores in vitro (reference 23 and data not shown). Two other probes containing 5′ sequences of the gene 8 mRNA were also produced (Fig. 8A): (i) 5′-3′Eco47III (131 nt), a probe that lacks the last 12 nt of the 5′-3′SacII probe and is therefore replication incompetent (23); and (ii) GG44-86/3′ (100 nt), a probe that lacks the 5′ UTR sequence of the 5′-3′SacII probe.
FIG. 8.
VP3 lacks specificity for the 5′-terminal sequences of viral mRNA. (A) The locations of sequences in RNA probes with respect to the wild-type gene 8 mRNA are indicated. The nonviral sequence UUAUU was used to link the 5′ and 3′ gene 8-specific sequences of the probes. ORF, open reading frame. (B) Five picomoles of the RNA probes, g8 5′-3′SacII, g8 5′-3′Eco47III, g8 GG44-86/3′, v-3′60, and nv-60, was incubated alone or with 3.3 μg of VP6-open cores in the presence or absence of 1.7 pmol (1 μg) of luciferase RNA. The upper and lower probe-protein complexes were resolved by electrophoresis and detected by autoradiography. ori, origin.
To determine whether VP3 had specific affinity for probes containing sequences found at the 5′ end of viral mRNA, VP6-open cores were incubated with 5 pmol of 32P-labeled 5′-3′SacII, 5′-3′Eco47III, or GG44-86/3′. Analysis of the mixtures by gel shift assay showed that VP1 and VP3 interacted with all three probes to form both the upper and the lower complexes (Fig. 8B). Gel shift assays also showed that the upper and lower complexes were formed with a probe that contained the last 60 nt of the gene 8 mRNA (v-3′60) and another probe of 60 nt that was of nonviral origin (nv-60) (Fig. 8B). The fact that the upper complex was formed with the nv-60 probe is consistent with the findings presented above which indicated that VP3 has sequence-independent affinity for RNA. The observation that the lower complex was also generated with the nv-60 probe supports the conclusion of an earlier study which indicated that VP1 not only has specific RNA-binding activity but also has nonspecific RNA-binding activity as well (22).
The specificity of the probe-protein interactions was further assessed by incubating VP6-open cores with 5 pmol (0.1 to 0.2 μg) of the 5′-3′SacII, 5′-3′Eco47III, or GG44-86/3′ probe and 1.7 pmol (1 μg) of luciferase RNA and then analyzing the mixtures by gel shift assay. The results showed that low molar concentrations of luciferase RNA blocked the formation of the upper complex regardless of the probe used (Fig. 8B). Taken together with results obtained with the 32P-labeled SP72-v3′40 probe (Fig. 3A and 4), these data show that VP3 has nonspecific RNA-binding activity and that the 5′ and 3′ ends of rotavirus mRNA do not contain sequences that VP3 specifically recognizes. Because of the lack of any nucleotide homology within the open reading frames of the rotavirus genes, it is also unlikely that there are specific recognition signals for VP3 in the internal sequences of the viral mRNAs.
The presence of luciferase RNA in the gel shift assay had a remarkably different effect on the formation of the VP1-probe complex than it did on the formation of the VP3-probe complex (Fig. 8B). At the same molar (1:3) and mass (10:1 to 2) ratios of luciferase RNA to probe which blocked the formation of the VP3-probe complex, the formation of the VP1-probe complex was minimally affected (<3-fold) even when the probe was of nonviral origin (nv-60). To summarize, the data show that both VP1 and VP3 have nonspecific RNA-binding activity and thus form complexes with the nonviral probe, nv-60. However, the data also indicate that the formation of the upper complex is remarkably more sensitive to inhibition by the presence of long competitor RNAs than is the formation of the lower complex. A possible explanation of this phenomenon is provided in the Discussion.
VP3 lacks affinity for dsRNA.
To test whether VP3 had affinity for dsRNA, open cores and 50 ng of 32P-labeled SP72-v3′40 were incubated alone or with 500 ng of luciferase RNA or with 50, 250, or 1,250 ng of virion-derived dsRNA. The reaction mixtures were then analyzed by electrophoresis to resolve the VP3-probe and VP1-probe complexes (Fig. 9). The results showed that, unlike in a parallel assay performed with luciferase ssRNA, the presence of dsRNA did not interfere with the formation of the upper complex even when on a mass basis the competitor RNA exceeded the concentration of the probe by 25-fold. These results indicate that VP3 does not have affinity for dsRNA, a finding which is consistent with the idea that the substrates for guanylyltransferases are nascent ssRNAs and not dsRNAs. Interestingly, despite the fact that VP1 is the viral polymerase and therefore must interact with the dsRNA template during transcription, the interaction of VP1 with the probe was not reduced by the presence of dsRNA in the reaction mixture (Fig. 9). This indicates that, under these experimental conditions, the viral RNA polymerase also does not have affinity for the dsRNA template for transcription.
FIG. 9.
VP3 lacks affinity for the dsRNA genome. 32P-labeled SP72-v3′40 (50 ng) was incubated alone or with open cores in the absence or presence of either virion-derived dsRNA or luciferase RNA. The reaction mixtures were analyzed by electrophoresis and autoradiography. A phosphorimager was used to quantitate the levels of VP3- and VP1-probe complexes, and the values were adjusted relative to 100% for the reaction mixture containing probe and open cores but no competitor RNA (lane 2). ori, origin.
The capping activity of the guanylyltransferase is nonspecific.
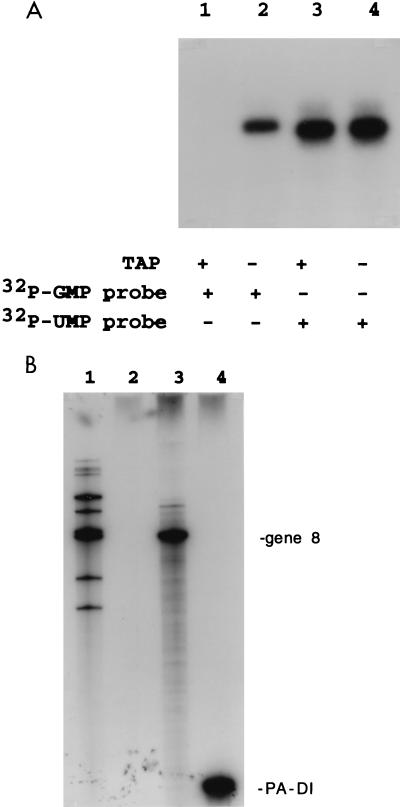
The fact that VP3 was able to bind ssRNA nonspecifically raised the question of whether the activity of the rotavirus guanylyltransferase perhaps was also nonspecific and therefore could cap nonrotaviral RNAs. We were able to address this question because recent studies have shown that open cores contain a guanylyltransferase activity which catalyzes the 5′ capping of exogenous rotavirus mRNAs (3a). The conditions used to assess the specificity of the capping activity were the same that were used to covalently label VP3 with [32P]GTP (Fig. 1, lane 6), except that the reaction mixture also contained added ssRNA. The three different types of RNAs used in the capping assay and their 5′-terminal sequences were as follows: (i) SP72-v3′40, 5′-GGGAGACCGG-3′; (ii) rotavirus gene 8 mRNA, 5′-GGCTTTTAAA-3′; and (iii) influenza virus DI-PA RNA, 5′-AGTAGAAACA-3′. As shown in Fig. 10A (lane 2) and 10B (lanes 3 and 4), all three RNAs became radiolabeled when incubated with [32P]GTP in the capping assay. To verify that it was a guanylyltransferase activity in the open core preparation that was responsible for labeling of the RNAs, an aliquot of the [32P]GMP-labeled SP72-v3′40 was incubated with tobacco alkaline pyrophosphatase, an enzyme known to remove cap structures from RNA by hydrolysis of phosphodiester bonds. As shown in Fig. 10A, lanes 1 and 2, this treatment removed all label from the 32P-labeled RNA ([32P]GMP v3′40 probe), thus establishing that the RNA was modified through the addition of a 32P-guanine cap to its 5′ terminus. In contrast, pyrophosphatase treatment had no effect on SP72-v3′40 ([32P]UMP v3′40 probe) that was labeled internally by synthesis in the presence of [32P]UTP (Fig. 10A, lanes 3 and 4). The fact that the guanylyltransferase capped not only the gene 8 mRNA but also SP72-v3′40 and the DI-PA RNA demonstrated that the capping activity was nonspecific and could cap RNAs that initiate with either of the purine nucleotides, G and A. These data show that there is a direct correlation with the sequence-independent RNA-binding activity observed for the rotavirus guanylyltransferase, VP3, and the suspected capping activity of the protein.
FIG. 10.
The guanylyltransferase activity of VP3 is nonspecific. SP72-v3′40, wild-type gene 8 mRNA, and PA-DI RNA were incubated with [32P]GTP and open cores in a capping assay. (A) SP72-v3′40 RNA ([32P]GMP probe) recovered from the reaction was coelectrophoresed on an SDS–14% polyacrylamide gel (lane 2) with 32P-labeled SP72-v3′40 RNA ([32P]UMP probe) (lane 4) synthesized by runoff transcription in the presence of [32P]UTP (lane 4). Portions of the [32P]GMP and [32P]UMP probes were treated with tobacco acid pyrophosphatase (TAP) prior to electrophoresis (lanes 1 and 3). (B) The gene 8 (lane 3) and PA-DI (lane 4) RNAs were recovered from capping assays by phenol-chloroform extraction, resolved by electrophoresis on a polyacrylamide-urea gel, and detected by autoradiography. 32P-labeled mRNAs made by transcriptionally active double-shelled particles served as markers on the gel (lane 1) (6). Lane 2 shows the products of a capping assay performed in the absence of added RNA.
VP3 preferentially binds uncapped RNA.
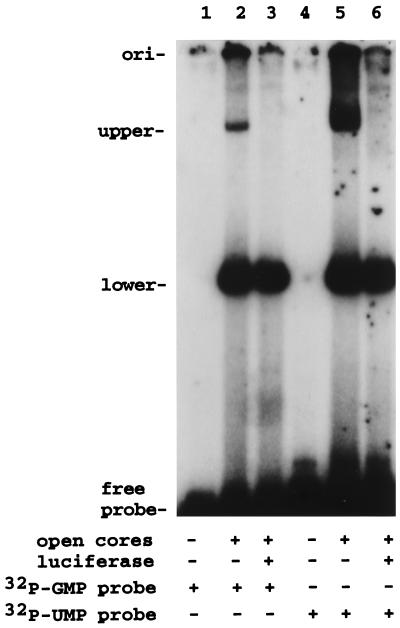
Because uncapped nascent RNAs are the substrates for guanylyltransferases, we tested whether VP3 differed in its binding activity for capped and uncapped ssRNA. To perform this analysis, two different pools of 32P-labeled SP72-v3′40 were prepared. One pool, [32P]UMP v3′40, was not capped and was labeled internally by synthesis in the presence of [32P]UTP (Fig. 10A, lane 4). The other pool, [32P]GMP v3′40, was capped and labeled by incubating cold SP72-v3′40 with open cores and [32P]GTP (Fig. 10A, lane 2). The probes were incubated with VP6-open cores, and the probe-protein complexes were resolved by electrophoresis and detected by autoradiography (Fig. 11). The results showed that the upper and lower complexes were formed with both the [32P]GMP v3′40 and [32P]UMP v3′40 probes. Thus, VP3 and VP1 have RNA-binding activity for both capped and uncapped RNAs. However, based on quantitation of the amount of probe in the upper and lower complexes, VP3 was calculated to bind three to four times less capped probe than uncapped probe per constant amount of VP1. Hence, while VP3 has affinity for both capped and uncapped RNAs, VP3 seems to have an affinity that is severalfold higher for uncapped RNAs than for capped RNAs.
FIG. 11.
VP3 preferentially binds uncapped RNA. Capped radiolabeled probe was made by incubating SP72-v3′40 with [32P]GTP and open cores ([32P]GMP probe). Noncapped radiolabeled probe was made by runoff transcription of SP72-v3′40 with [32P]UTP ([32P]UMP probe). Purified probes (0.12 μg) were incubated with 3.3 μg of VP6-open cores in the presence or absence of 1 μg of luciferase RNA. Probe-protein complexes were detected by electrophoresis and autoradiography. ori, origin.
DISCUSSION
Gel shift assays were used in this study to characterize the RNA-binding activity of proteins that are contained within the core of the virion and that function as the viral RNA polymerase (VP1) and the viral guanylyltransferase (VP3). Assays performed with the virus-specific probe SP72-v3′40 and nonviral competitor RNAs showed that VP1 of open cores (Fig. 4) and baculovirus-expressed rVP1 (Fig. 7) specifically recognize the 3′ end of the gene 8 mRNA. It remains to be determined if the recognition signal for VP1 includes the last 7 nt of the mRNA, a sequence that is highly conserved and is required for synthesis of minus-strand RNA (30). Gel shift assays performed with the nonviral probe, nv-60, showed that VP1 also has nonspecific affinity for RNA (Fig. 8B). The fact that VP1 has both nonspecific and specific RNA-binding activity was previously noted for this protein (22) and is an expected feature of RNA polymerases, since they not only have to recognize a promoter but also must move along a DNA or an RNA template during RNA synthesis. In this study, we also found that viral dsRNA does not competitively interfere with binding of VP1 to the gene 8 3′-specific probe, SP72-v3′40, and thus VP1 does not have affinity for dsRNA under the same buffer conditions (i.e., LSB) in which the protein binds to and replicates viral RNA (Fig. 9) (23). The inability of VP1 to bind to dsRNA suggests that, for the RNA polymerase to initiate transcription, the 5′ end of the dsRNA template for mRNA synthesis must first undergo denaturation by a helicase or unwindase.
We provide in this work the first direct evidence that VP3 is an RNA-binding protein. Gel shift assays performed with open cores showed that VP3 was able to bind to virus-specific probes containing the 3′ end or the 5′ and 3′ ends of the gene 8 mRNA (Fig. 4 and 8B). But the affinity of VP3 for these probes was nonspecific, as the presence of even low amounts of nonviral competitor RNA completely suppressed the formation of VP3-probe complexes. The fact that VP3 also interacted with nonviral probes to form VP3-probe complexes verifies that this protein has nonspecific RNA binding activity (Fig. 8B). The presence of viral dsRNA in gel shift assays did not interfere with binding of VP3 to ssRNA probes (Fig. 9); thus, VP3 is like VP1 in that it too apparently lacks affinity for the dsRNA template for transcription. Perhaps this is not surprising, however, given that the substrate for the capping activities proposed for VP3 is ssRNA and not dsRNA, and thus, enzymatically, there is no known need for the protein to bind to dsRNA.
Since only nonspecific RNA-binding activity was observed for VP3, we tested whether the guanylyltransferase activity of VP3 also lacked specificity. Indeed, this is the case, as guanyl-yltransferase assays performed with open cores showed that the enzyme could cap both rotaviral and influenza virus RNAs and could cap RNAs initiating with a G or an A residue. Thus, the RNA-binding activity observed for VP3 parallels the capping activity of the protein, in that both are nonspecific. The nonspecific nature of the capping activity of VP3 is characteristic of capping enzymes in general (5). Gel shift assays comparing capped and uncapped RNAs indicated that VP3 preferentially binds uncapped RNA (Fig. 11). This implies that, while VP3 may not recognize the 5′-terminal sequence of the RNA, it may recognize features of the 5′ end that are associated with the presence or the absence of a cap structure, e.g., a 5′-terminal γ- or β-phosphate group.
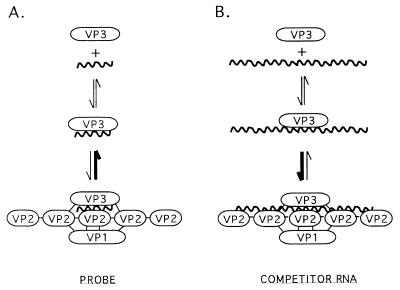
In analyzing the nonspecific RNA-binding activity of VP3, we unexpectedly found that longer competitor RNAs (≥184 nt) interfered much more efficiently with the formation of VP3-probe complexes than did shorter RNAs (≤97 nt) (Fig. 5). We also found that if VP3-probe complexes were allowed to form by incubation of open cores with 50 ng of 32P-labeled SP72-v3′40, and then 50 ng of a long competitor RNA (luciferase RNA, 2,650 nt) was added, 30 min later it was not possible to detect any VP3–SP72-v3′40 complexes. This is remarkable because the final mixture contained equal amounts of the SP72-v3′40 probe and the luciferase competitor RNA, and thus, the level of VP3-probe complex detected after addition of the competitor would be predicted to be reduced by 50% and not by the observed 100%. Both of these findings suggest that the interaction of VP3 with longer RNAs generates a VP3-RNA complex that is more “stable” than the complex generated by the interaction of VP3 with shorter RNAs. However, the fact that the RNA-binding activity of VP3 is nonspecific makes it seem unlikely that the actual affinity of VP3 for RNA would differ based on the length of the RNA. Instead, we believe that the factor that accounts for the effect that RNA length has on the stability of the VP3-RNA complex is VP2, the major protein component present in the open core preparations.
VP2 is an RNA-binding protein with nonspecific affinity for both ssRNA and dsRNA (2, 14). The RNA-binding domain of VP2 is located near the N terminus of the protein (14), as are the binding domains for VP1 and VP3 (34). Since open core preparations contain VP1, VP2, and VP3 but much of the VP1 and VP3 is soluble and not associated with the oligomeric VP2, the affinity of VP1 and VP3 for VP2 would not seem to be particularly strong (22). The model presented in Fig. 12 explains how the RNA-binding activities of VP2 and the affinity of VP2 for VP3 would cause VP3 to more stably interact with longer RNAs than with shorter RNAs. The model predicts that when a short RNA (probe) is added to open cores, the short RNA will interact with VP3 to form a VP3-short RNA complex. The VP3-short RNA complex may interact with VP2 oligomers but probably not stably because the presence of VP3 on the small RNA restricts the ability of VP2 to fully bind to the RNA and/or the small size of the RNA allows only a very limited number of molecules of the VP2 oligomer to bind to the VP3-small RNA complex (Fig. 12A). In contrast, the model predicts that, in the presence of long RNAs, a VP3-long RNA complex will form and this complex will interact more stably with the VP2 oligomer because the length of the RNA allows it to interact simultaneously with a greater number of RNA-binding sites on the VP2 oligomer (Fig. 12B). The stability of the VP3-long RNA-VP2 complex may be further enhanced by the affinity, albeit possibly weak, that VP3 has for VP2. In summary, because the interaction of short probes with VP3 is reversible (Fig. 3B), the addition of long competitive RNAs to gel shift assays would have the effect of driving VP3 out of the VP3-probe complexes and into VP3-competitive RNA-VP2 complexes.
FIG. 12.
Model for the effect that RNA size has on the formation of VP3-probe complexes.
The effect of long competitor RNAs on the formation of VP1-probe complexes in gel shift assays was quite different than that observed for the VP3-probe complexes. In assays where the 3′-terminal gene 8 probe, SP72-v3′40, was used (Fig. 5), the reason for the lack of any significant effect by long competitor RNAs is because VP1 has a specific and therefore, by default, a higher affinity for the probe than for the competitor RNA. In assays where a nonviral probe was used, the presence of low amounts of long competitive RNAs had little effect on the formation of VP1-probe complexes but completely blocked the formation of VP3-probe complexes (Fig. 8B). This suggests that, in contrast to VP3, the interaction of VP1 with the short RNA leads to a more stable complex than does the interaction of VP1 with a long RNA. A part of the explanation for this phenomenon may be that, for the VP1-short RNA complex, the limited size of the RNA does not allow VP2 to bind to the RNA at the same time as VP1 and thus it is not possible to form a VP1-short RNA-VP2 complex. But this does not explain why, as proposed to occur for VP3, VP1 of the VP1-short RNA complex would not be chased into a VP1-long RNA-VP2 complex and, therefore, result in the loss of VP1-probe complexes in gel shift assays containing long competitor RNAs. Instead, the experimental data raise the possibility that the affinity of VP2 oligomers for the RNA component of the VP1-long RNA complex would cause the release of VP1. That is, the stable interaction of an RNA with a VP2 oligomer may preclude the nonspecific binding of the RNA polymerase to the VP2-RNA complex.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Melinda Jones on this project. We also thank Robert Chanock, Kim Green, and Albert Kapikian for reviews of the manuscript.
REFERENCES
- 1.Bican P, Cohen J, Charpilienne A, Scherrer R. Purification and characterization of bovine rotavirus cores. J Virol. 1982;43:1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle J F, Holmes K V. RNA-binding proteins of bovine rotavirus. J Virol. 1986;51:561–568. doi: 10.1128/jvi.58.2.561-568.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Patton J T. Rotavirus RNA replication requires a single-stranded 3′ terminus for efficient minus-strand synthesis. J Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Chen, D., and J. T. Patton. Unpublished data.
- 4.Chen D Y, Zeng C Q-Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho E-J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J. Ribonucleic acid polymerase activity associated with purified calf rotavirus. J Gen Virol. 1977;36:395–402. doi: 10.1099/0022-1317-36-3-395. [DOI] [PubMed] [Google Scholar]
- 7.Cohen J, Charpilienne A, Chilmonczyk S, Estes M K. Nucleotide sequence of bovine rotavirus gene 1 and expression of the gene product in baculovirus. J Virol. 1989;170:131–140. doi: 10.1016/0042-6822(89)90519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong P, Shuman S. Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J Biol Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 9.Desselberger U, McCrae M A. The rotavirus genome. Curr Top Microbiol Immunol. 1994;68:5945–5952. doi: 10.1007/978-3-642-78256-5_3. [DOI] [PubMed] [Google Scholar]
- 10.Fukuhara N, Nishikawa K, Gorziglia M, Kapikian A Z. Nucleotide sequence of gene segment 1 of a porcine rotavirus strain. Virology. 1989;173:743–749. doi: 10.1016/0042-6822(89)90590-4. [DOI] [PubMed] [Google Scholar]
- 11.Gallegos C O, Patton J T. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 12.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus double-stranded RNA segments. J Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapikian A Z. Overview of viral gastroenteritis. Arch Virol. 1996;12:7–19. doi: 10.1007/978-3-7091-6553-9_2. [DOI] [PubMed] [Google Scholar]
- 14.Labbe M, Baudoux P, Charpilienne A, Poncet D, Cohen J. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J Gen Virol. 1994;75:3423–3430. doi: 10.1099/0022-1317-75-12-3423. [DOI] [PubMed] [Google Scholar]
- 15.Lawton J A, Zeng C Q-Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Mattion N M, Estes M K. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Shuman S. RNA binding properties of vaccinia virus capping enzyme. J Biol Chem. 1993;268:21253–21262. [PubMed] [Google Scholar]
- 18.Mansell E A, Patton J T. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J Virol. 1990;64:4988–4996. doi: 10.1128/jvi.64.10.4988-4996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCrae M A, McCorquodale J G. Molecular biology of rotaviruses. V. Terminal structure of viral RNA species. Virology. 1983;126:204–212. doi: 10.1016/0042-6822(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 20.Midthun K, Greenberg H B, Hoshino Y, Kapikian A Z, Wyatt R G, Chanock R M. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J Virol. 1985;53:949–954. doi: 10.1128/jvi.53.3.949-954.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell D B, Both G W. Completion of the genomic sequence of the simian rotavirus SA11: nucleotide sequences of segments 1, 2, and 3. Virology. 1990;177:324–331. doi: 10.1016/0042-6822(90)90487-c. [DOI] [PubMed] [Google Scholar]
- 22.Patton J T. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNAs. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton J T, Jones M T, Kalbach A N, He Y-W, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizarro J L, Sandino A M, Pizarro J M, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 26.Prasad B V V, Wang G J, Clerx J P M, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 27.Prasad B V V, Rothnagel R, Zeng C Q-Y, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sandino A M, Jashes M, Faundez G, Spencer E. Role of the inner protein capsid on in vitro human rotavirus transcription. J Virol. 1986;60:797–802. doi: 10.1128/jvi.60.2.797-802.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandino A M, Fernandez J, Pizarro J, Vasquez M, Spencer E. Structure of rotavirus particle: interaction of the inner capsid protein VP6 with the core polypeptide VP3. Biol Res. 1994;27:39–48. [PubMed] [Google Scholar]
- 31.Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotide transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 33.Valenzuela S, Pizarro J, Sandino A M, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J Virol. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng C Q, Estes M K, Charpilienne A, Cohen J. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J Virol. 1998;72:201–208. doi: 10.1128/jvi.72.1.201-208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Q-Y, Wentz M J, Estes M K, Ramig R F. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]