Abstract
Transcriptional bursting is a prevalent feature of gene expression. The transient assembly of transcription factor clusters at regulatory DNAs is critical to control bursting dynamics.
In September 1957, Francis Crick proposed the “central dogma” of molecular biology (1). According to this dogma, once genetic information is transferred into a protein, it cannot be reversed back into the DNA sequence. This initiated a paradigm shift in the logic of biology. The initial step in the transfer of genetic information is a process known as “transcription,” where an enzyme called RNA polymerase synthesizes an RNA copy of the DNA sequence. Through past biochemical, structural, and whole-genome sequencing studies, the molecular mechanisms of transcription by RNA polymerase have been elucidated in great detail. However, the temporal dynamics of this process still remains an outstanding question in modern biology.
From bacteria to humans, emerging evidence suggests that the prevalent feature of gene expression conserved across species is intermittent bursts of de novo transcription or transcriptional bursting. Transcriptional bursting regulates gene expression during cell fate determination and disease processes in mammals. It has also been shown that transcriptional bursting plays a critical role in dosage compensation mechanisms of X-linked genes during sex specification, as well as in establishing the temporal and spatial patterns of gene expression during Drosophila embryogenesis. Transcriptional bursting is also reported to serve as a major source of random effects in gene expression and contribute to cellular variability within a population of cells. The first evidence for transcriptional bursting was obtained from electron microscopy analysis of chromatin spreads prepared from early Drosophila embryos more than 40 years ago (2). Recent advances in high-resolution imaging methods such as MS2/PP7 RNA labeling and single-molecule fluorescence in situ hybridization (FISH) methods enable scientists to directly visualize transcriptional bursting at single-cell resolution in a highly quantitative manner. However, the molecular mechanism underlying the control of transcriptional bursting still remains a central question in the field.
ROLES OF ENHANCERS IN THE CONTROL OF TRANSCRIPTIONAL BURSTING
Enhancers are short segments of regulatory DNA that consist of a cluster of binding sites for sequence-specific transcription factors (TFs) and coactivators. They play a fundamentally important role in determining when and in which cell types genes should be transcribed in response to intrinsic and extrinsic signals. Recent whole-genome studies by the ENCODE consortium suggested that the human genome contains ~900,000 putative enhancers. Quantitative MS2/PP7 live-imaging and single-molecule FISH studies revealed that enhancers regulate the level of gene activities by modulating the kinetics of transcriptional bursting from linked genes in Drosophila and mammalian cells (3, 4). Single-cell RNA-seq analysis demonstrated that regulation of transcriptional bursting by enhancers is a widespread mechanism throughout the mammalian genome.
According to a textbook model, enhancers are thought to activate transcription through stable association with target gene promoters by looping out intervening sequences. However, dynamic modulation of bursting kinetics by enhancers appears to be incompatible with this traditional static “looping” model. Importantly, MS2/PP7 live-imaging analysis demonstrated that a single enhancer can co-activate two linked genes simultaneously (4), giving rise to the possibility that enhancers activate transcription by producing a nuclear microenvironment where transcription machineries are locally concentrated at specific genomic locations, thereby acting as a “hub” for driving transcriptional bursting.
In line with these findings, increasing evidence suggests that many TFs and coactivators contain intrinsically disordered regions (IDRs) to facilitate dynamic condensate formation within a nucleus. However, there is a critical lack of understanding in the functional significance of these TF/coactivator condensates in the control of gene expression, and, therefore, it remains to be determined whether enhancers actually form “hubs” during induction of transcriptional bursting in living cells. A recent super-resolution live visualization of TFs together with transcriptional bursting provided direct evidence that the dynamic assembly and disassembly of TF clusters at enhancers are a major source of transcriptional bursting in developing Drosophila embryos (5). Fusion of IDRs with TFs facilitated both the formation of TF clusters and the rate of burst induction, suggesting that multivalent protein-protein interactions mediated by IDRs serve as a key regulatory step to control transcriptional bursting. Recent whole-genome studies have reported that the majority of enhancer-promoter contacts, as well as the level of nascent RNA synthesis, are not affected by acute depletion of architectural proteins such as CTCF and cohesin in mammalian cells (6) or massive rearrangements of genome organization in Drosophila (i.e., balancer chromosomes). This suggests that TF clusters assembled at enhancers can exert their burst-inducing activity independently of the formation of topologically associating domains (TADs).
FUNCTIONAL INTERPLAY BETWEEN TETHERING ELEMENTS AND TF CLUSTERS
As discussed above, one of the notable features of enhancers is their ability to co-activate multiple genes simultaneously (4). Visualization of this co-activation process with TFs in living Drosophila embryos showed that two co-activated genes physically share a common TF cluster during the induction of transcriptional bursting (5), further supporting the idea that enhancers act as a hub for gene activation. A key question arises: Does co-regulation of multiple genes by shared enhancers occur in the context of endogenous eukaryotic genomes? Recent high-resolution Micro-C analysis revealed the existence of focal promoter-promoter contacts between two neighboring genes, both in Drosophila and mammals (6, 7). In Drosophila, such focal promoter-promoter contacts are often found to connect two distant paralogous genes that exhibit very similar expression patterns during early embryogenesis (7). CRISPR-Cas9–mediated genome-editing analysis demonstrated that the expression of a pair of connected paralogs is driven by shared enhancers. MS2/PP7 live-imaging analysis further revealed that these genes exhibit highly coordinated bursting kinetics although they are separated by very large linear genomic distances (7).
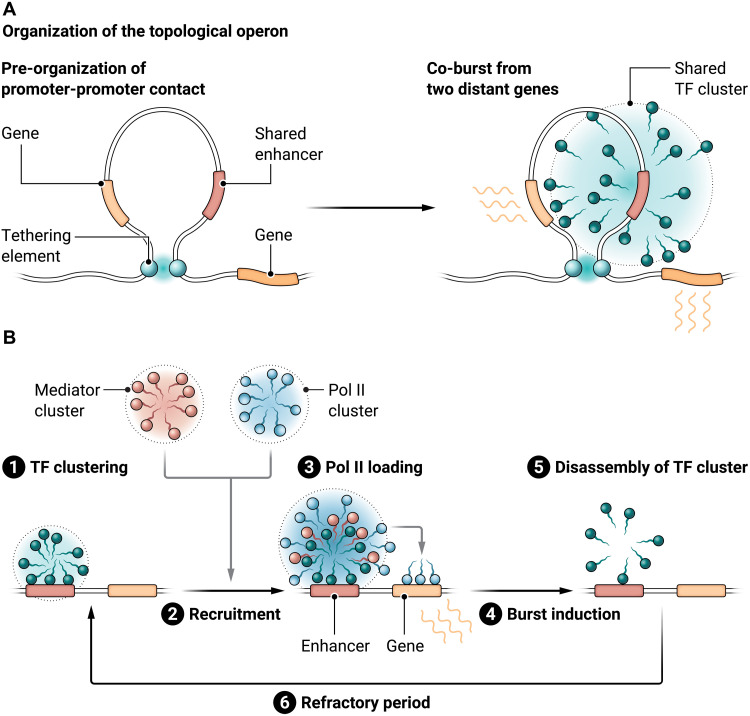
If this is the case, then how do two distant paralogous genes come into the physical proximity of common TF clusters formed at shared enhancers? Promoter-promoter contacts mediated by a previously uncharacterized class of regulatory DNAs called “tethering elements” appear to underlie long-range regulatory connectivity between two paralogs by bringing the genes close together in 3D genomic space. Tethering elements have a unique capability of mediating long-range focal interaction independently of TADs but do not possess enhancer activity by themselves (8). Tethering elements can even mediate focal interaction between two distant genomic loci bypassing several TADs. When a tethering element is lost from one of a pair of paralogous genes, shared enhancers are no longer able to equally access two promoters at the same time, resulting in biased burst induction from the enhancer-proximal gene (7). Together, these observations suggest that promoter-promoter contacts mediated by tethering elements enable common TF clusters formed at shared enhancers to drive coordinated transcriptional bursting from the two genes (Fig. 1A). Analogous to the control of multiple genes by a single ON/OFF regulatory switch in the bacterial operon system (e.g., the lac operon), a term “topological operon” was recently coined to describe the co-regulation of multiple genes by shared enhancers in eukaryotic genomes (7). It is reasonable to assume that the dynamic assembly and disassembly of common TF clusters within a topological operon facilitate coordinated regulation of functionally related genes at the single-cell level. In addition to Drosophila, Arabidopsis was recently suggested as having a topological operon at the thalianol biosynthetic gene cluster.
Fig. 1. Dynamic assembly of TF cluster at enhancers is critical to control transcriptional bursting.
(A) Tethering elements pre-organize promoter-promoter contacts between two distant paralogous genes (left). Formation of common TF clusters at shared enhancers drives co-bursts from two genes (right). (B) Enhancers serve as hubs for the transient assembly of TF clusters and Pol II/Mediator during induction of transcriptional bursting. Credit: Austin Fisher/Science Advances
THE MOLECULAR FUNCTION OF TF CLUSTERS AT ENHANCERS
The next question is how the dynamic assembly and disassembly process of TF clusters at enhancers leads to transcriptional bursting. Previous super-resolution analysis demonstrated that RNA polymerase II (Pol II) and Mediator also undergo dynamic clustering, frequently overlapping with each other, in living mouse ES cells (9). Given that TFs exert their regulatory functions through recruitment of coactivators such as Mediator, it is tempting to speculate that TF clusters formed at enhancers drive transcriptional bursting by transiently increasing the local concentration of Mediator and Pol II at the site of transcription through molecular blending with these clusters (Fig. 1B). After the consumption of locally concentrated Pol II through its consecutive loading onto associated genes for burst induction, TF clusters at enhancers disappear very rapidly (5), suggesting that Pol II serves as a sort of “molecular glue” stabilizing TF clustering at enhancers. Importantly, purification of optogenetically induced TF clusters revealed that they form a macromolecular activator complex containing Mediator and Pol II constituents (10).
A recent live-imaging study suggested that TF clusters are typically formed ~40 s before the induction of transcriptional bursting, and this time lag becomes much less pronounced (~8 s) when Mediator clusters were visualized instead of TFs (5), suggesting that there is a temporal hierarchy between the clustering of TFs, Mediator, and Pol II at enhancers (Fig. 1B). Overall, the recent literature is consistent with the idea that enhancers serve as hubs for the transient assembly of TF clusters and Pol II/Mediator during induction of transcriptional bursting. It is likely that simultaneous live visualization of TF clustering together with Pol II/Mediator dynamics in living cells will provide further mechanistic insights into the regulatory process of transcriptional bursting.
REFERENCES
- 1.Cobb M., 60 years ago, Francis Crick changed the logic of biology. PLOS Biol. 15, e2003243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKnight S. L., Miller O. L. Jr., Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell 17, 551–563 (1979). [DOI] [PubMed] [Google Scholar]
- 3.Bartman C. R., Hsu S. C., Hsiung C. C., Raj A., Blobel G. A., Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 62, 237–247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawasaki K., Fukaya T., Functional coordination between transcription factor clustering and gene activity. Mol. Cell 83, 1605–1622.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Hsieh T. S., et al. , Enhancer-promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat. Genet. 54, 1919–1932 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levo M., Raimundo J., Bing X. Y., Sisco Z., Batut P. J., Ryabichko S., Gregor T., Levine M. S., Transcriptional coupling of distant regulatory genes in living embryos. Nature 605, 754–760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batut P. J., Bing X. Y., Sisco Z., Raimundo J., Levo M., Levine M. S., Genome organization controls transcriptional dynamics during development. Science 375, 566–570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cisse I. I., Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y. J., Lee M. Jr., Lee Y.-T., Jing J., Sanders J. T., Botten G. A., He L., Lyu J., Zhang Y., Mettlen M., Ly P., Zhou Y., Xu J., Light-activated macromolecular phase separation modulates transcription by reconfiguring chromatin interactions. Sci. Adv. 9, eadg1123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]