Abstract
Cardiac fibrosis plays a key role in the progression of diabetic cardiomyopathy (DCM). Previous studies demonstrated the cardioprotective effects of natriuretic peptides. However, the effects of natriuretic peptide receptor C (NPRC) on cardiac fibrosis in DCM remains unknown. Here, we observed that myocardial NPRC expression was increased in mice and patients with DCM. NPRC−/− diabetic mice showed alleviated cardiac fibrosis, as well as improved cardiac function and remodeling. NPRC knockdown in both cardiac fibroblasts and cardiomyocytes decreased collagen synthesis and proliferation of cardiac fibroblasts. RNA sequencing identified that NPRC deletion up-regulated the expression of TGF-β–induced factor homeobox 1 (TGIF1), which inhibited the phosphorylation of Smad2/3. Furthermore, TGIF1 up-regulation was mediated by the activation of cAMP/PKA and cGMP/PKG signaling induced by NPRC deletion. These findings suggest that NPRC deletion attenuated cardiac fibrosis and improved cardiac remodeling and function in diabetic mice, providing a promising approach to the treatment of diabetic cardiac fibrosis.
NPRC deletion attenuates cardiac fibrosis in diabetic mice by inhibiting TGF-β1/Smad pathway through the activation of PKA/PKG.
INTRODUCTION
The prevalence of diabetes mellitus (DM) has been continually increasing in the last decade, imposing a substantial burden on global public health. Epidemiologic studies, such as the Framingham Heart Study, have established a strong association between diabetes and an increased risk of heart failure (HF), known as diabetic cardiomyopathy (DCM), which refers to a series of abnormalities in cardiac structure and function independent of diabetic macrovascular complications (1). Despite numerous studies on DCM, there exist few effective therapies, and glycemic control alone often fails to improve cardiac function and prognosis (2). Therefore, further studies on novel therapeutic approaches of DCM are highly warranted.
DCM initially manifests as diastolic dysfunction but gradually progresses to systolic dysfunction. The pathogenesis of DCM is initiated by metabolic disturbances, resulting in mitochondrial dysfunction, oxidative stress, and impaired autophagy. Together, these effects may induce cardiac hypertrophy, fibrosis and inflammation, and cardiomyocyte (CM) death, which inevitably contribute to DCM (3). Ample evidence suggests that patients with diabetes exhibit marked myocardial fibrosis, characterized by the excessive accumulation of extracellular matrix (ECM) in both left and right ventricles (4). Currently, no evidence-based therapies have shown beneficial effects on cardiac fibrosis in diabetic patients (5), indicating that the mechanism of cardiac fibrosis in diabetes should be further explored to identify new therapeutic targets.
The most important progress in the pathogenesis of HF involves neurohumoral activation, which generates many neurohumoral factors that may promote or prevent HF. Among these, natriuretic peptides (NPs) have been recognized as a new class of cardioprotective factors, which consist of three typical members: atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and C-type natriuretic peptide (CNP). ANP and BNP are mainly synthesized and secreted by CMs (6) and less produced by cardiac fibroblasts (CFs) (7, 8). CNP is predominantly secreted by the vascular endothelial cells (9), but CMs and CFs can also produce CNP (10, 11). These NPs bind to three kinds of natriuretic peptide receptors (NPRs): guanylyl cyclase A receptor (GCA), guanylyl cyclase B receptor (GCB), and NPRC. In the heart, NPRC is more abundantly expressed than GCA and GCB, and NPRC expression is more commonly detected in myofibroblasts than CMs (12, 13). GCA and GCB are coupled to guanylyl cyclase (GC), which increases intracellular cyclic guanosine monophosphate (cGMP) and activates protein kinase G (PKG) (14). Unlike GCA or GCB, NPRC is devoid of GC activity, but contains an inhibitory guanine nucleotide regulatory protein (Gi) binding domain in the intracellular region, which inhibits the activity of adenylate cyclase (AC), leading to a reduced level of cyclic adenosine monophosphate (cAMP) and less activated protein kinase A (PKA) (15). In addition to its signaling activity, NPRC also acts as a clearance receptor for NPs to maintain homeostasis (16). ANP, BNP, and CNP, by binding to GCA and GCB, exert multiple cardioprotective roles, such as diuretic (17), anti-hypertension (18), and anti-cardiac fibrosis (19), whereas GCA or GCB deletion worsens cardiac remodeling and dysfunction (20, 21). The clinical application of nesiritide, a recombinant human BNP (22), and sacubitril/valsartan, a drug that inhibits both neprilysin and angiotensin type I receptor (23), has demonstrated a salutary effect of NPs on cardiac remodeling and dysfunction. However, the role of NPRC and its signaling activity in cardiovascular disease remains unclear. NPRC knockout increased atrial fibrosis under angiotensin II treatment, but attenuated atrial fibrosis under transforming growth factor–β1 (TGF-β1) treatment in mice (24, 25). There was evidence that NPRC expression was up-regulated in adipose tissue (26) and skeletal muscles (27) in diabetes, but the expression of NPRC in the diabetic heart has not been observed. Recently, our group conducted a multicenter genome-wide association study in a large Chinese population, which revealed that NPRC gene polymorphisms were associated with increased susceptibility to coronary artery disease (CAD) (28), suggesting the crucial role of NPRC in cardiovascular diseases. Nonetheless, the effect and mechanism of NPRC on DCM have not been reported yet. Here, we hypothesized that NPRC deletion may attenuate cardiac fibrosis in diabetic mice by activating PKA/PKG and inhibiting TGF-β1/Smad pathways, and a series of in vivo and in vitro experiments were designed and performed to test this hypothesis.
RESULTS
NPRC expression was increased in the hearts of diabetic mice and patients with DM and HF
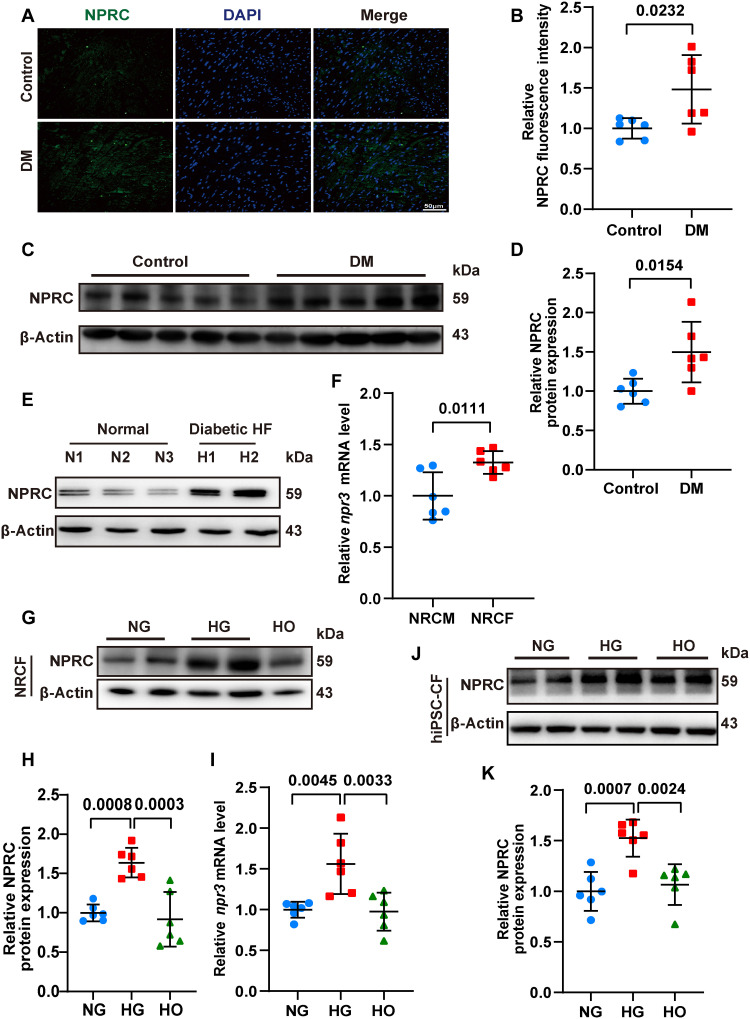
To clarify the role of NPRC in the development of DCM, we constructed a mouse model of DCM by consecutive injections of streptozotocin (STZ), followed by 16 weeks of feeding, as described in the first part of the in vivo experiments. Immunofluorescent staining and Western blot showed increased NPRC expression in the hearts of the DM relative to the control group of mice (Fig. 1). In contrast, no significant difference in GCA and GCB expression was detected between the hearts of the DM and control groups (fig. S3). NPRC expression in hearts from healthy donors and patients with DM and severe HF but without hypertension and coronary artery stenosis were analyzed by Western blot. The results showed that NPRC expression was notably increased in patients with DM and HF compared with that in healthy donors (Fig. 1).
Fig. 1. NPRC expression was increased in the hearts of diabetic patients, diabetic mice, and glucose-treated CFs.
(A) Representative immunofluorescence staining of NPRC (green) in tissue sections from the left ventricle of control and diabetic mice. The nuclei were counterstained with DAPI (blue). Bar = 50 μm. (B) Quantification of fluorescence intensity of NPRC in (A). n = 6 per group. (C) Representative Western blot images of NPRC expression in the left ventricular samples from control and diabetic mice. (D) Quantification of the protein expression of NPRC in (C). n = 6 per group. (E) Western blot images of NPRC expression in human heart samples from healthy donors and patients with DM and HF. (F) Relative mRNA levels of npr3 in primary NRCMs and NRCFs. n = 6 per group. (G) Representative Western blot images of NPRC expression in NRCFs treated with normal concentration of glucose (NG), high concentration of glucose (HG), and high osmotic medium (HO). (H) Quantification of the protein expression of NPRC in (G). n = 6 per group. (I) Relative mRNA levels of npr3 in NRCFs treated with NG, HG, or HO. n = 6 per group. (J) Representative Western blot images of NPRC expression in hiPSC-CFs treated with NG, HG, or HO. (K) Quantification of the protein expression of NPRC in (J). n = 6 per group. Normal distributions were tested by Shapiro-Wilk method. Unpaired two-tailed Student’s t tests were applied in (B), (D), and (F). One-way ANOVA was applied in (H), (I), and (K).
As the heart is composed of approximately 30% CMs and 70% non-CMs, predominantly CFs (29), we compared the expression of NPRC in primary neonatal rat CMs (NRCMs) and CFs (NRCFs). Polymerase chain reaction (PCR) analysis showed a higher mRNA level of npr3 in NRCFs than in NRCMs (Fig. 1). The distribution of the three NPRs in NRCFs was determined by immunofluorescent staining, which showed that under the same exposure conditions, the intensity of NPRC was much higher than that of GCA or GCB. PCR analysis also showed a higher mRNA level of npr3 than that of npr1 and npr2 (fig. S3). These results suggested that NPRC accounted for most of the NPRs in cultured NRCFs, which was consistent with previous reports (12, 13). Then, primary NRCFs were treated with different concentrations of glucose. Compared with the normal glucose group (NG; 5.5 mM), the mRNA and protein expression of NPRC in NRCFs in the high glucose group (HG; 33.3 mM) increased significantly, as detected by Western blot and PCR analyses, while no significant difference in NPRC expression was observed between the high osmotic (HO) and NG groups, thus excluding the impact of elevated osmotic pressure on NPRC expression. Moreover, human CFs differentiated from human induced pluripotent stem cells (hiPSC-CFs) were also treated with NG, HG, and HO. Western blot showed that NPRC increased in the HG group compared with the NG and HO groups in hiPSC-CFs (Fig. 1). These results indicated that NPRC may be involved in the function of CFs and the development of DCM. Additionally, we treated NRCMs with NG, HG, and HO and found that the expression of NPRC was also higher in HG-treated NRCMs than in NG- or HO-treated NRCMs (fig. S3), suggesting that NPRC in CMs may likely participate in the development of DCM.
NPRC deletion improved cardiac function and remodeling in diabetic mice
To investigate the role of NPRC in DCM progression, we generated NPRC−/− mice and the deletion of NPRC was confirmed by Western blot analysis of the heart tissues from wild-type (WT) and NPRC−/− mice (fig. S4). In the second part of the in vivo experiments, NPRC−/− nondiabetic and diabetic mice exhibited skeletal abnormalities, including hunched backs, elongated tails, and increased body length, compared with the littermate WT mice (fig. S4). For the next 16 weeks, blood glucose levels and body weight of the mice in the DM group differed significantly from those of the control group. However, neither body weight nor blood glucose levels showed significant differences between WT and NPRC−/− mice (WT + control versus NPRC−/− + control; WT + DM versus NPRC−/− + DM). Systolic blood pressure (SBP) of NPRC−/− mice was lower than that of WT mice, whereas no significant difference was observed in diastolic blood pressure (DBP) and heart rate (WT + control versus NPRC−/− + control; WT + DM versus NPRC−/− + DM). There were no significant differences in the serum levels of total cholesterol (TC), triglycerides (TG), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) among the four groups of mice (fig. S5).
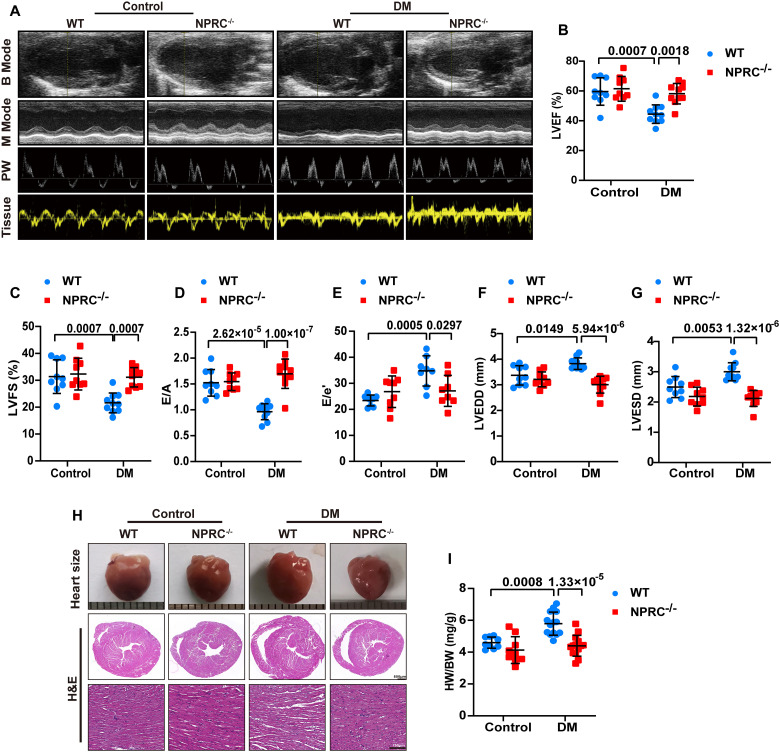
To explore the effect of NPRC deletion on cardiac function and remodeling, echocardiography was performed before the mice were euthanized. Compared with the mice in the WT + control group, mice in the WT + DM group developed cardiac systolic and diastolic dysfunction, as indicated by a significant decrease in left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), and the ratio of early to late diastolic mitral flow velocities (E/A) and an increase in the ratio of early diastolic mitral flow to mitral annular velocities (E/e′). NPRC deletion improved cardiac function induced by diabetes in the NPRC−/− + DM group, as demonstrated by restored LVEF, LVFS, E/A, and E/e′ compared with those in the WT + DM group. Furthermore, DM-induced enlargement in left ventricular end-diastolic diameter (LVEDD) and left ventricular end-systolic diameter (LVESD) in the WT + DM group was also restored by NPRC deletion in the NPRC−/− + DM group, indicating an improvement in cardiac remodeling (Fig. 2).
Fig. 2. NPRC deficiency improved left ventricular function and remodeling in diabetic mice.
(A) Representative echocardiographic images for four groups of mice. B Mode represents a two-dimensional echocardiogram showing left ventricular long-axis view; M Mode represents M-mode echocardiogram showing left ventricular dimensions; PW represents pulse-wave Doppler echocardiogram depicting diastolic mitral flow; Tissue represents tissue Doppler echocardiogram displaying mitral annular velocities. (B) Quantification of left ventricular ejection fraction (LVEF) in four groups of mice. n = 9 to 10 per group. (C) Quantification of left ventricular fractional shortening (LVFS) in four groups of mice. n = 9 to 10 per group. (D) Quantification of the ratio of early to late diastolic mitral flow velocities (E/A) in four groups of mice. n = 9 to 10 per group. (E) Quantification of the ratio of early diastolic mitral flow to early diastolic mitral annular velocities (E/e′) in four groups of mice. n = 8 to 9 per group. (F) Quantification of left ventricular end-diastolic diameter (LVEDD) in four groups of mice. n = 9 to 10 per group. (G) Quantification of left ventricular end-systolic diameter (LVESD) in four groups of mice. n = 9 to 10 per group. (H) Representative photographs of the hearts from four groups of mice (the first row), cross-sectional images of hematoxylin and eosin (H&E) staining at the papillary muscle level of the hearts (the second row, scale bar, 500 μm), and H&E-stained sections of hearts from four groups of mice (the third row, scale bar, 100 μm). (I) Quantification of the ratio of heart weight (HW) to body weight (BW) in four groups of mice. n = 9 to 14 per group. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Turkey’s post hoc test was used in (B) to (G) and (I).
In general, WT mice in the WT + DM group had a larger heart size than those in the WT + control group, which was attenuated by NPRC deletion in the NPRC−/− + DM group. This phenotype was also demonstrated by the ratio of heart weight to body weight (HW/BW). Morphological analysis showed that WT mice in the WT + DM group exhibited significant myocardial disarray under H&E staining, which was attenuated in the NPRC−/− + DM group (Fig. 2).
To examine the specific effect of NPRC knockdown on diabetic myocardium and eliminate the undesirable systemic effects of NPRC deletion, type 9 adeno-associated virus (AAV9) was applied, which delivers NPRC short hairpin RNA (shRNA) (shNPRC) or scramble shRNA (Scr) more specifically to the heart than other virus (30, 31), in the third part of the in vivo experiments. Western blot showed that cardiac NPRC expression was down-regulated in the AAV9-shNPRC + control and AAV9-shNPRC + DM groups, compared with those in the AAV9-Scr + control and AAV9-Scr + DM groups, respectively, which demonstrated the validity of AAV9-shNPRC transfection in our mouse model. Different from the results in NPRC−/− mice, AAV9-shNPRC delivery did not induce skeletal abnormalities including hunched backs, elongated tails, and increased body length in the AAV9-shNPRC + DM group compared with the AAV9-Scr + DM group. In addition, SBP was similar between the two groups (fig. S6). By contrast, AAV9-shNPRC delivery improved cardiac function and remodeling induced by diabetes in the AAV9-shNPRC + DM group, as demonstrated by restored LVEF, LVFS, E/A, E/e′, LVEDD, and LVESD in NPRC−/− mice, compared with those in the AAV9-Scr + DM group. Moreover, HW/BW and hematoxylin and eosin (H&E) staining showed that heart size and myocardial disarray of mice were alleviated in the AAV9-shNPRC + DM group relative to those in the AAV9-Scr + DM group (fig. S6).
NPRC deficiency alleviated cardiac fibrosis induced by diabetes in vivo
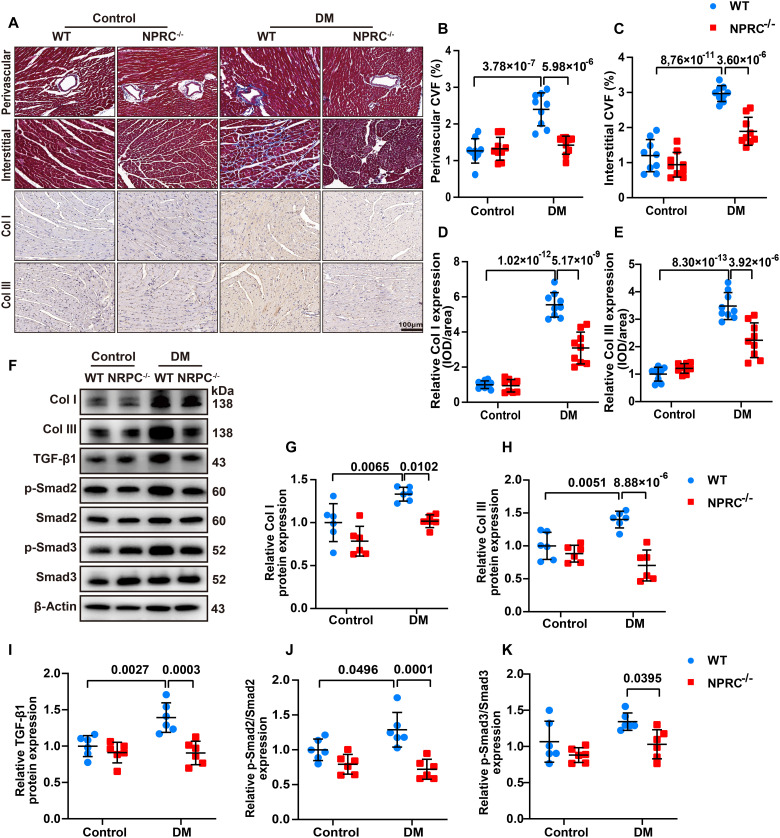
As cardiac fibrosis is a salient feature of DCM, we detected the extent of myocardial fibrosis in the second part of the in vivo experiments (WT + control, NPRC−/− + control, WT + DM, and NPRC−/− + DM). Masson’s trichrome staining showed that mice in the WT + control and NPRC−/− + control groups rarely displayed myocardial fibrosis. Diabetes increased collagen accumulation in the cardiac perivascular and interstitial spaces in WT + DM mice, compared with the NPRC−/− + DM mice. Immunohistochemistry (IHC) for collagen I and collagen III revealed higher collagen expression in the WT + DM group than in the NPRC−/− + DM group. Western blot revealed similar changes in collagen expression in the four groups of mice (Fig. 3). These findings suggested that NPRC deficiency alleviated cardiac fibrosis in diabetic hearts in vivo. We also examined the extent of left atrial fibrosis in the four groups of mice. Masson’s trichrome staining and IHC for collagen I and collagen III revealed that mice in the WT + control and NPRC−/− + control groups displayed similar extent of left atrial fibrosis, while mice in the NPRC−/− + DM group showed decreased left atrial collagen accumulation relative to that in the WT + DM group, which was similar to the changes in the left ventricle (fig. S7). In addition, the extent of the myocardial fibrosis in the left ventricle was also examined in the third part of the in vivo experiments. Masson’s trichrome staining and IHC for collagen I and collagen III displayed sparse collagen accumulation in the AAV9-Scr + control and AAV9-shNPRC + control groups, whereas mice in the AAV9-shNPRC + DM group showed decreased collagen accumulation relative to that in the AAV9-Scr + DM group. Western blot showed similar results (fig. S8).
Fig. 3. NPRC deficiency alleviated cardiac fibrosis induced by diabetes in vivo.
(A) Representative images of Masson’s trichrome staining of the myocardium (first and second rows) and immunohistochemistry (IHC) of collagen I (Col I) and collagen III (Col III) in the myocardium (third and fourth rows) in four groups of mice. Scale bar, 100 μm. (B) Quantification of perivascular collagen volume fraction (CVF) in four groups of mice. n = 9 per group. (C) Quantification of interstitial CVF in four groups of mice. n = 9 per group. (D) Quantification of IHC of collagen I in four groups of mice. n = 9 per group. (E) Quantification of IHC of collagen III in four groups of mice. n = 9 per group. (F) Representative Western blot images of collagen I, collagen III, TGF-β1, p-Smad2, and p-Smad3 in the hearts of the four groups of mice. (G to K) Quantification of the protein expression of collagen I, collagen III, TGF-β1, p-Smad2, and p-Smad3 in (F). n = 6 per group. IOD, integrated optical density. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Turkey’s post hoc test was used in (B) to (E) and (H) to (J). Two-way ANOVA with Bonferroni post hoc test was used in (G) and (K).
TGF-β1 is a key profibrotic cytokine implicated in the progression of fibrogenesis, which is partially related to the canonical Smad signaling pathway (32). Thus, we examined the expression levels of relevant mediators in the TGF-β1/Smad signaling pathway by Western blot. The results showed that compared with the WT + DM group, the expression levels of TGF-β1, p-Smad2, and p-Smad3 decreased in the NPRC−/− + DM group, indicating that NPRC deletion inhibited the activation of the TGF-β1/Smad signaling pathway (Fig. 3). Western blot analysis of the hearts in the AAV9-delivered mice showed similar results that the levels of TGF-β1, p-Smad2, and p-Smad3 decreased in the AAV9-shNPRC + DM group compared with that in the AAV9-Scr + DM group (fig. S8).
NPRC deficiency inhibited collagen synthesis and proliferation of CFs
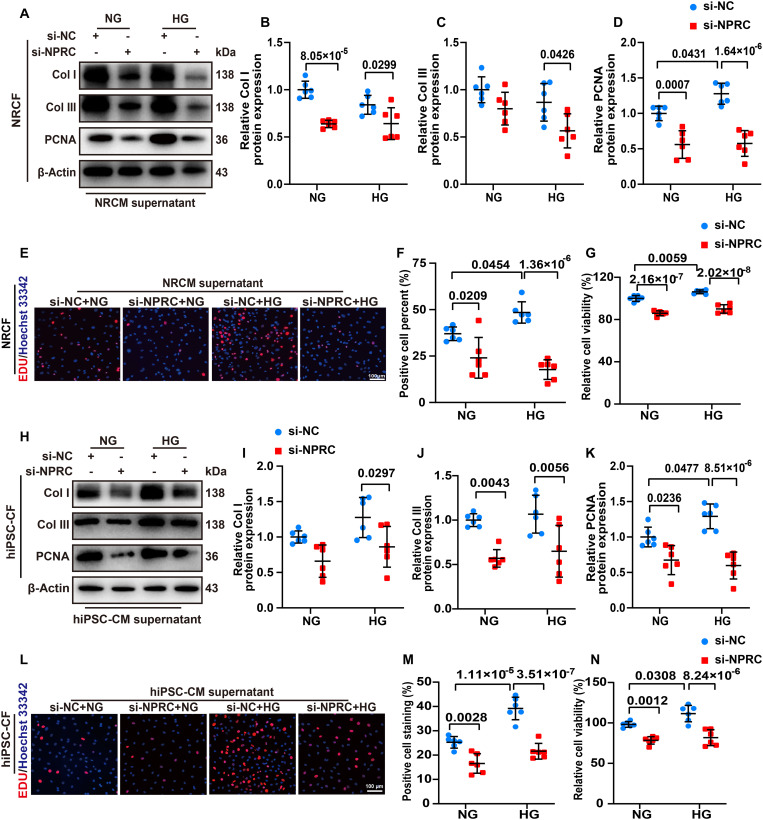
As NPRC is involved in the inhibition of the cAMP/PKA signaling pathway through its intracellular domain and the clearance of NPs from circulation (33), we assumed that NPRC deficiency may affect the intracellular cAMP/PKA signaling and extracellular NP clearance. To verify this hypothesis, we performed in vitro experiments in two parts. In the first part, the NPRC protein expression was knocked down by transfecting si-NPRC into NRCFs and hiPSC-CFs, and the cells were then treated with NG (5.5 mM) or HG (33.3 mM) for 72 hours. In the second part, considering that NPs are expressed and secreted not only by CFs but also by CMs, and the expression of NPRC in CMs was increased under HG stimulation (34), we first knocked down the protein expression of NPRC by si-NPRC transfection in NRCMs and hiPSC-CMs, followed by NG or HG stimulation for 48 hours. Subsequently, the NRCFs were treated with the NRCM supernatant, while the hiPSC-CFs were treated with the hiPSC-CM supernatant for 72 hours (fig. S2).
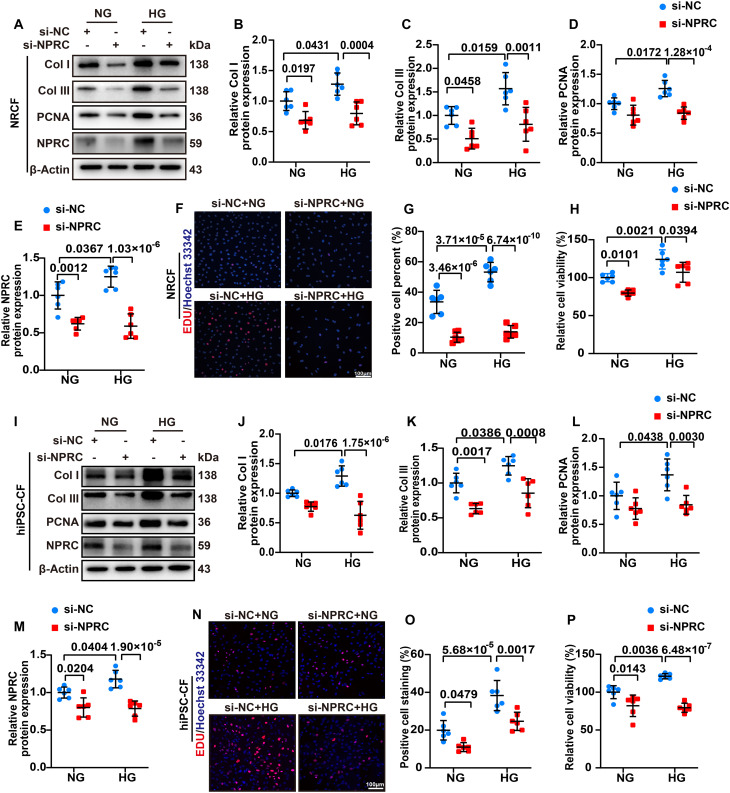
In the first part, the NPRC expression of NRCFs and hiPSC-CFs was significantly reduced in si-NPRC groups than in si-NC groups. NPRC knockdown significantly decreased the protein expression of collagen I and collagen III, which were initially increased by HG treatment. In addition, NRCFs and hiPSC-CFs were subjected to 5-ethynyl-2'-deoxyuridine (EDU) and cell counting kit-8 (CCK-8) assays to detect cell proliferation, which showed that HG treatment promoted the proliferation of CFs in the si-NC + HG group, which was decreased by NPRC knockdown in the si-NPRC + HG group. The protein expression of proliferating cell nuclear antigen (PCNA) showed similar changes, as determined by Western blot (Fig. 4). To further explore the effects of NPRC deletion on CF function in the DCM mice, we isolated adult mouse CFs (MCFs) from WT + DM mice and NPRC−/− + DM mice. Western blot showed that the expression of collagen I and collagen III decreased in MCFs from NPRC−/− + DM mice, compared with that from WT + DM mice (fig. S9).
Fig. 4. NPRC deficiency inhibited collagen synthesis and proliferation of CFs in vitro.
(A) Representative Western blot images of collagen I, collagen III, PCNA, and NPRC of NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. (B to E) Quantification of the protein expression of collagen I, collagen III, PCNA, and NPRC in (A). n = 6 per group. (F) Representative images of EDU (red) assays of NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. The nuclei were stained with Hoechst 33342. Scale bar, 100 μm. (G) Quantification of EDU-positive cell percent in (F). n = 6 per group. (H) Quantification of the relative cell viability detected by CCK-8 assay in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (I) Representative Western blot images of collagen I, collagen III, PCNA, and NPRC in hiPSC-CFs transfected with si-NC or si-NPRC and treated with NG or HG. (J to M) Quantification of the protein expression of collagen I, collagen III, PCNA, and NPRC in (I). n = 6 per group. (N) Representative images of EDU (red) assays of hiPSC-CFs transfected with si-NC or si-NPRC and treated with NG or HG. The nuclei were stained with Hoechst 33342. Scale bar, 100 μm. (O) Quantification of EDU-positive cell percent in (N). n = 6 per group. (P) Quantification of the relative cell viability detected by CCK-8 assay in hiPSC-CFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Turkey’s post hoc test was used in (E), (G), (J), (M), and (P). Two-way ANOVA with Bonferroni post hoc test was used in (B) to (D), (H), (K), (L), and (O).
As earlier described, the TGF-β1/Smad signaling pathway plays a crucial role in the regulation of collagen synthesis and proliferation in CFs (32). Therefore, we assessed changes in the TGF-β1/Smad signaling pathway of NRCFs. Western blot showed that the protein expression of TGF-β1 and phosphorylation of Smad2 and Smad3 (p-Smad2 and p-Smad3) were increased by HG treatment in the si-NC + HG group, which were reversed by NPRC knockdown in the si-NPRC + HG group. In contrast, the protein expression of TGF-β receptor 2 (TGF-βR2) did not exhibit significant changes. These results suggested that NPRC knockdown in NRCFs decreased the expression of TGF-β1 and phosphorylation of Smad2/3, leading to inhibited collagen synthesis and proliferation. Similar results were observed in hiPSC-CFs by Western blot (fig. S10).
In the second part, treatment of NRCFs with the NRCM supernatant in the si-NPRC + HG group significantly reduced the protein expression of collagen I and collagen III compared with that in the si-NC + HG group. Similar results were observed in hiPSC-CFs treated with hiPSC-CM supernatant. The EDU and CCK-8 assays also demonstrated that the proliferation of NRCFs and hiPSC-CFs was markedly inhibited by treatment with the NRCM and hiPSC-CM supernatant from the si-NPRC + HG group than from the si-NC + HG group. The protein expression of PCNA showed similar changes as determined by Western blot (Fig. 5). The changes in the TGF-β1/Smad signaling pathway were also detected by Western blot, which showed that TGF-β1 expression was not changed by treatment with NPRC-deficient NRCM supernatant. However, the expression of p-Smad2 and p-Smad3 was still reduced in NRCFs treated with the NRCM supernatant from the si-NPRC + HG group than from the si-NC + HG group. Similar results were observed in hiPSC-CFs treated with hiPSC-CM supernatant by Western blot (fig. S11). These results suggested that mechanisms regulating the TGF-β1/Smad signaling pathway may differ in NPRC-deficient CFs and CFs treated with NPRC-deficient CM supernatant.
Fig. 5. NPRC deficiency in CMs inhibited collagen synthesis and proliferation of CFs in vitro.
(A) Representative Western blot images of collagen I, collagen III, and PCNA in NRCFs treated with the supernatant of NRCMs. (B to D) Quantification of the protein expression of collagen I, collagen III, and PCNA in (A). n = 6 per group. (E) Representative images of EDU (red) assays of NRCFs treated with the supernatant of NRCMs. The nuclei were stained with Hoechst 33342. Scale bar, 100 μm. (F) Quantification of EDU-positive cell percent in (E). n = 6 per group. (G) Quantification of the relative cell viability in NRCFs treated with the supernatant of NRCMs using CCK-8 assay. (H) Representative Western blot images of collagen I, collagen III, and PCNA in hiPSC-CFs treated with the supernatant of hiPSC-CMs. (I to K) Quantification of the protein expression of collagen I, collagen III, and PCNA in (H). n = 6 per group. (L) Representative images of EDU (red) assays of hiPSC-CFs treated with the supernatant of hiPSC-CMs. The nuclei were stained with Hoechst 33342. Scale bar, 100 μm. (M) Quantification of EDU-positive cell percent in (L). n = 6 per group. (N) Quantification of the relative cell viability in hiPSC-CFs treated with the supernatant of hiPSC-CMs using CCK-8 assay. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Turkey’s post hoc test was used in (F), (K), and (M). Two-way ANOVA with Bonferroni post hoc test was used in (B) to (D), (G), (I), (J), and (N).
NPRC regulated collagen synthesis and proliferation of CFs by targeting TGIF1
To further explore the mechanism underlying the effect of NPRC on the regulation of collagen synthesis and proliferation, we performed transcriptome RNA sequencing using NRCFs in the first part of the in vitro experiments. The gene expression profiles were highly reproducible in the samples within each group. Gene ontology (GO) enrichment revealed that among the top 10 biological processes that exhibited significant differences between the si-NC + NG and si-NC + HG groups of NRCFs, chromosome segregation and cell cycles were mostly involved. This finding was consistent with the aforementioned results that the proliferation of NRCFs was increased in the si-NC + HG group versus that in the si-NC + NG group. Moreover, we found that response to organic substance, regulation of signal transduction, and cell proliferation were among the most affected biological processes in comparison with the si-NC + HG and si-NPRC + HG groups. Then, we selected 24 up-regulated and 16 down-regulated genes that were most frequently repeated in these processes. Among these genes, we focused on 7 up-regulated and 2 down-regulated genes related to fibrosis, and by analyzing their expression levels and fold changes in NRCFs, we finally chose Tgif1 for further investigation (fig. S12).
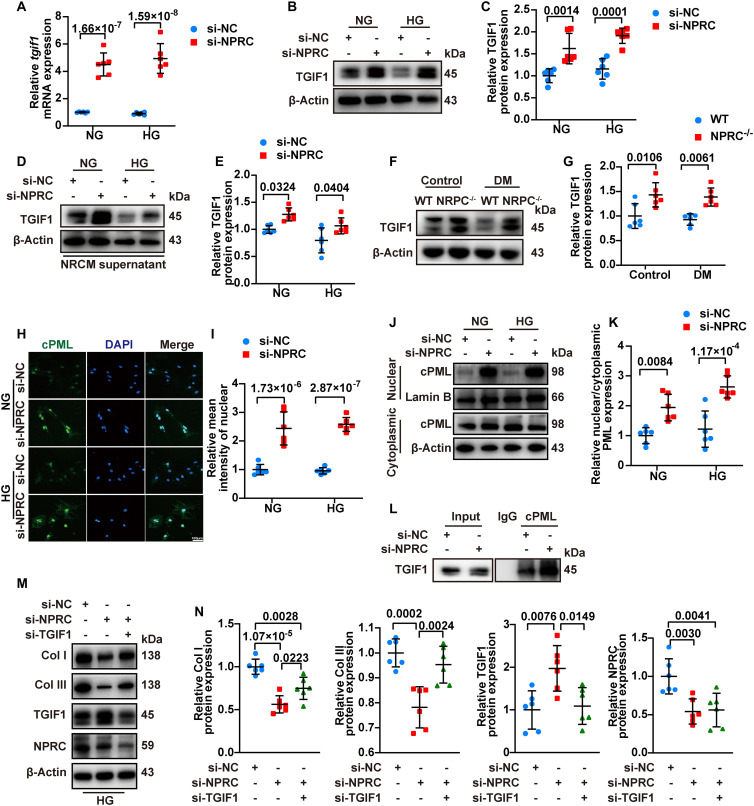
Tgif1 is responsible for encoding protein TGIF1, which is known as a nuclear co-repressor of TGF-β signaling through multiple mechanisms including recruitment of transcriptional repressor complex to repress the transcription of TGF-β signaling, promotion of ubiquitin-dependent proteasomal degradation of Smad2, and sequestering of cytoplasmic promyelocytic leukemia (cPML) in the nucleus to prevent its shuttling to the cytoplasm, where it promotes Smad2/3 phosphorylation (35). We found that NPRC knockdown increased the mRNA level of tgif1 in NRCFs in both NG and HG treatments in the first part of the in vitro experiments (Fig. 6, si-NC + NG versus si-NPRC + NG, and si-NC + HG versus si-NPRC + HG, all P < 0.001). Similarly, NPRC knockdown increased protein expression of TGIF1 in NRCFs (Fig. 6) and hiPSC-CFs (fig. S13). In the second part of the in vitro experiments, treatment with NRCM and hiPSC-CM supernatant from both the si-NPRC + NG or si-NPRC + HG groups increased the protein expression of TGIF1 in NRCFs (Fig. 6) and hiPSC-CFs (fig. S13). TGIF1 protein expression increased in the mice of the NPRC−/− + control and NPRC−/− + DM groups relative to the WT + control and WT + DM groups, respectively (Fig. 6). Similar results were observed in AAV9-shNPRC–delivered mice (fig. S13).
Fig. 6. NPRC regulated collagen synthesis and proliferation of CFs by targeting TGIF1.
(A) Relative mRNA levels of tgif1 in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (B and C) Representative Western blot images and quantification of the protein expression of TGIF1 in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (D and E) Representative Western blot images and quantification of the protein expression of TGIF1 in NRCFs treated with the supernatant of NRCMs. n = 6 per group. (F and G) Representative Western blot images and quantification of the protein expression of TGIF1 in the hearts of four groups of mice. n = 6 per group. (H) Representative immunofluorescence staining of cPML (green) in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. Scale bar, 100 μm. (I) Quantification of relative mean fluorescence intensity of nuclear cPML in (H). n = 6 per group. (J and K) Representative Western blot images of nuclear and cytoplasmic cPML expression and quantification of the ratio of nuclear to cytoplasmic cPML protein expression in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (L) Representative Western blot images of TGIF1 expression assayed by immunoprecipitation with cPML antibody in NRCFs transfected with si-NC or si-NPRC. (M and N) Representative Western blot images and quantification of collagen I, collagen III, TGIF1, and NPRC expression in NRCFs transfected with si-NC, si-NPRC, or si-NPRC + si-TGIF1. n = 6 per group. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Bonferroni post hoc test was used in (A), (C), (E), (G), (I), and (K). One-way ANOVA was applied in (N).
To further investigate the mechanisms through which elevated TGIF1 inhibited the TGF-β1/Smad signaling pathway in CFs, we transfected NRCFs with small interfering RNA (siRNA) (si-TGIF1) to knock down the expression of TGIF1 in NRCFs. Western blot showed that the protein expression of collagen I and collagen III increased significantly after TGIF1 knockdown. There was no significant change in the protein expression of Smad2 and Smad3, but the levels of p-Smad2 and p-Smad3 markedly increased after TGIF1 knockdown (fig. S14). These results indicated that TGIF1 inhibited TGF-β1/Smad signaling in NRCFs by attenuating Smad2 and Smad3 phosphorylation rather than degrading Smad2 and Smad3 per se.
Previous studies have shown that cPML is a crucial protein that shuttles between the nucleus and cytoplasm to facilitate phosphorylation of Smad2/3 (35). To explore whether NPRC knockdown in NRCFs was related to changes in cPML localization, immunofluorescent staining was performed in the first part of the in vitro experiments, which showed that nuclear localization of cPML was markedly enhanced in the si-NPRC + NG and si-NPRC + HG groups, relative to the si-NC + NG and si-NC + HG groups, respectively. In addition, we separated nuclear and cytoplasmic proteins, and found that the ratio of nuclear to cytoplasmic cPML expression levels was significantly higher in the si-NPRC + NG and si-NPRC + HG groups than in the si-NC + NG and si-NC + HG groups, respectively. Furthermore, the coimmunoprecipitation assay showed that NPRC knockdown in NRCFs resulted in an increased binding of cPML with TGIF1 (Fig. 6).
To further investigate whether TGIF1 mediates the effects of NPRC on collagen synthesis, we knocked down both NPRC and TGIF1 through transfection of siRNA, as described in the third part of the in vitro experiments. Compared with NRCFs transfected with si-NC, si-NPRC transfection decreased the protein expression of collagen I and collagen III and increased the protein expression of TGIF1 in the si-NPRC group. However, when the elevated levels of TGIF1 were knocked down by si-TGIF1, the protein expression of collagen I and collagen III was partially reversed in the si-NPRC + si-TGIF group (Fig. 6). Similar results were observed in hiPSC-CFs (fig. S15). These results suggested that TGIF1 played a crucial role in the mechanism through which NPRC deficiency regulated TGF-β1/Smad signaling as well as collagen synthesis and proliferation in CFs.
NPRC deficiency activated cAMP/PKA signaling in vivo and in vitro
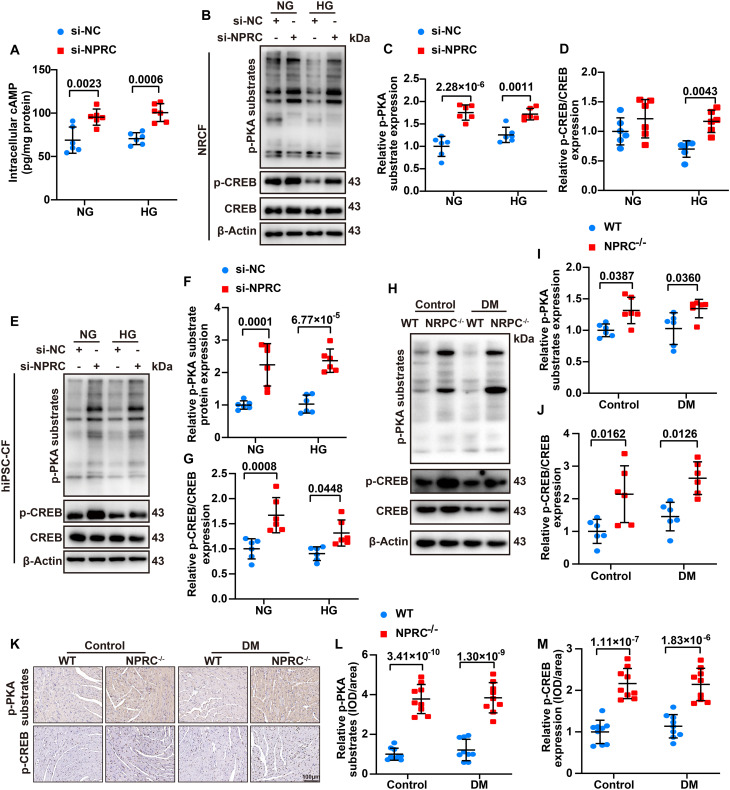
The aforementioned results confirmed that NPRC was involved in the regulation of TGF-β1/Smad signaling through TGIF1, but the mechanism through which NPRC deficiency increased TGIF1 expression remained to be elucidated. As previously reported, NPRC not only acts as a clearance receptor for ANP, BNP, and CNP but also has a binding domain of Gi in its intracellular portion, which can inhibit AC activity and reduce intracellular cAMP levels (34). To explore the effect of NPRC deficiency on cAMP levels, we first detected the intracellular cAMP levels of NRCFs in the first part of the in vitro experiments and found that the intracellular cAMP levels were substantially higher in the si-NPRC + HG group than in the si-NC + HG group. Furthermore, we investigated the intracellular PKA activity by measuring the phosphorylation level of PKA substrates (RRXS*/T*) and cAMP response element–binding protein (CREB). Consistent with the change in cAMP levels, the protein expression of p-PKA substrates and p-CREB increased in the si-NPRC + NG and si-NPRC + HG groups, in comparison with the si-NC + NG and si-NC + HG groups, respectively. We also detected the expression of p-PKA substrates and p-CREB in hiPSC-CFs and found that the level of p-PKA substrates and p-CREB increased in the si-NPRC–transfected groups, compared with the si-NC–transfected groups. In particular, mice in the NPRC−/− + control and NPRC−/− + DM groups exhibited increased protein expression of p-PKA substrates and p-CREB, relative to the WT + control and WT + DM groups, respectively, as detected by Western blot and IHC (Fig. 7). Similarly, the protein expression of p-PKA substrates and p-CREB increased in the AAV9-shNPRC–delivered groups, relative to the AAV9-Scr–delivered groups, as detected by Western blot and IHC (fig. S16).
Fig. 7. NPRC deficiency activated cAMP/PKA signaling in vivo and in vitro.
(A) Quantification of intracellular cAMP in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (B) Representative Western blot images of the protein expression of p-PKA substrates and p-CREB in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. (C and D) Quantification of the protein expression of p-PKA substrates and p-CREB in (B). n = 6 per group. (E) Representative Western blot images of the protein expression of p-PKA substrates and p-CREB in hiPSC-CFs transfected with si-NC or si-NPRC and treated with NG or HG. (F and G) Quantification of the protein expression of p-PKA substrates and p-CREB in (E). n = 6 per group. (H). Representative Western blot images of p-PKA substrates and p-CREB expression in the hearts of four groups of mice. (I and J) Quantification of the protein expression of p-PKA substrates and p-CREB in (H). n = 6 per group. (K) Representative IHC images of p-PKA substrates and p-CREB expression in the hearts of four groups of mice. Scale bar, 100 μm. (L and M) Quantification of the protein expression of p-PKA substrates and p-CREB in (K). n = 9 per group. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Bonferroni post hoc test was used in (A), (C), (D), (F), (G), (I), (J), (L), and (M).
NPRC deficiency activated cGMP/PKG signaling in vivo and in vitro
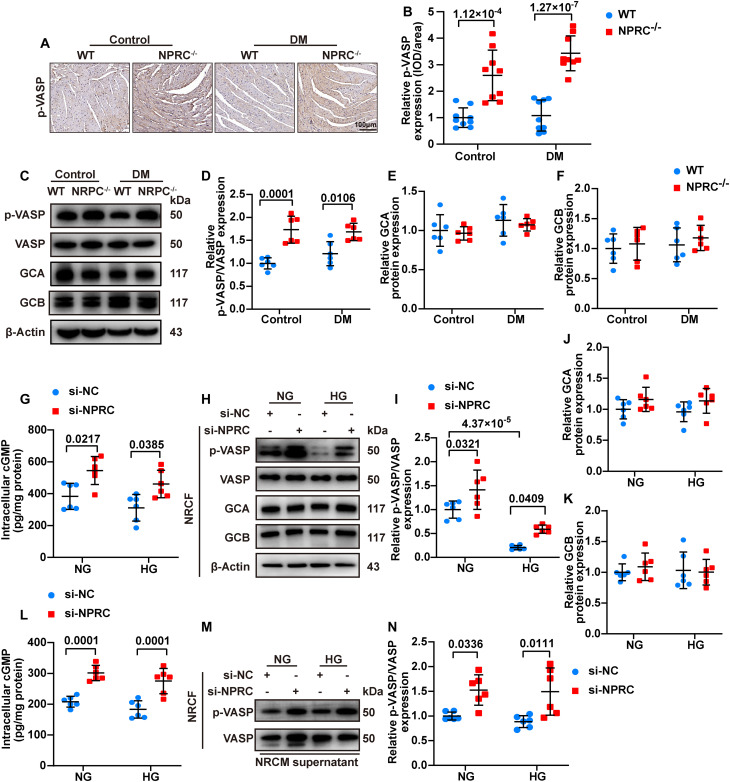
IHC and Western blot showed that the phosphorylation level of vasodilator-stimulated phosphoprotein (VASP), an indicator of cGMP/PKG activation, increased in the NPRC−/− + control and NPRC−/− + DM groups, in comparison with the WT + control and WT + DM groups, respectively, suggesting that the cGMP/PKG signaling pathway was activated in NPRC−/− mice (Fig. 8). Similarly, the protein expression of p-VASP increased in the AAV9-shNPRC–delivered groups, relative to the AAV9-Scr–delivered groups, as detected by Western blot and IHC (fig. S17).
Fig. 8. NPRC deficiency activated cGMP/PKG signaling in vivo and in vitro.
(A) Representative IHC images of p-VASP in the hearts of four groups of mice. (B) Quantification of IHC staining of p-VASP in (A). n = 9 per group. (C) Representative Western blot images of p-VASP, GCA, and GCB expression in the hearts of four groups of mice. (D to F) Quantification of the protein expression of p-VASP, GCA, and GCB in (C). n = 6 per group. (G) Quantification of intracellular cGMP in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. n = 6 per group. (H) Representative Western blot images of p-VASP, GCA, and GCB expression in NRCFs transfected with si-NC or si-NPRC and treated with NG or HG. (I to K) Quantification of the protein expression of p-VASP, GCA, and GCB in (H). n = 6 per group. (L) Quantification of intracellular cGMP of NRCFs treated with the supernatant of NRCMs. n = 6 per group. (M and N) Representative Western blot images and quantification of the protein expression of p-VASP in NRCFs treated with the supernatant of NRCMs. n = 6 per group. Normal distributions were tested by Shapiro-Wilk method. Two-way ANOVA with Bonferroni post hoc test was used in (B), (D) to (G), (I) to (L), and (N).
As NPRC is responsible for the clearance of ANP, BNP, and CNP in vivo, we assumed that the activated cGMP/PKG signaling in NPRC−/− mice may be due to the weakened ability of NPRC to degrade NPs. To test this hypothesis, we measured the protein expression levels of ANP, BNP, and CNP in the hearts of WT and NPRC−/− mice with or without DM by IHC. The results showed that mice in the NPRC−/− + control and NPRC−/− + DM groups had elevated levels of ANP, BNP, and CNP in the cardiac tissues, compared with the WT + control and WT + DM groups, respectively (fig. S18). Furthermore, serum levels of ANP and CNP were increased in the NPRC−/− + control and NPRC−/− + DM groups relative to the WT + control and WT + DM groups, respectively. On the other hand, although the serum level of BNP was higher in the WT + DM group than in the WT + control group, NPRC deletion did not significantly affect the serum level of BNP (fig. S18), likely due to the fact that the physiological serum level of BNP is considerably low, which did not increase significantly after NPRC deletion in the WT + control group. In contrast, the serum level of BNP increased dramatically in the WT + DM group as a consequence of cardiac dysfunction, which may have covered up the effect of NPRC deletion in this group of mice. As expected, there was no significant difference in the protein expression levels of GCA and GCB among the four groups of mice (Fig. 8). These results suggested that NPRC deletion in vivo resulted in elevated serum levels of NPs and local NP expression in the heart, which activated intracellular cGMP/PKG signaling through the unaffected GCA and GCB.
Furthermore, we detected the intracellular cGMP/PKG activity in the first part of the in vitro experiments and found that the intracellular cGMP levels of NRCFs were significantly higher in the si-NPRC + NG and si-NPRC + HG groups than in the si-NC + NG and si-NC + HG groups, respectively. Consistently, the expression of p-VASP increased in the si-NPRC + NG and si-NPRC + HG groups, in comparison with the si-NC + NG and si-NC + HG groups, respectively. No significant difference was observed in the expression of GCA and GCB (Fig. 8). As CFs are capable of secreting NPs (7, 8, 11), we examined the NP levels in the supernatant of NRCFs and found that the three NPs were all increased in the si-NPRC + NG and si-NPRC + HG groups, compared with the si-NC + NG and si-NC + HG groups, respectively (fig. S18). Similar results were observed in hiPSC-CFs that the levels of p-VASP increased in the si-NPRC + NG and si-NPRC + HG groups, in comparison with the si-NC + NG and si-NC + HG groups, respectively (fig. S19).
Thereafter, we examined the intracellular levels of cGMP and p-VASP in NRCFs in the second part of the in vitro experiment. Treatment with the NRCM supernatant from the si-NPRC + NG or si-NPRC + HG groups increased the intracellular level of cGMP and protein level of p-VASP in NRCFs (Fig. 8), which might be attributable to the elevated levels of NPs in the NRCM supernatant (fig. S18). Similar results were observed in hiPSC-CFs that treatment with hiPSC-CM supernatant from the si-NPRC–transfected groups increased the expression of p-VASP (fig. S19). These results indicated that cGMP/PKG signaling was activated in NPRC−/− mice because of the elevated NP levels. In addition, the cGMP/PKG signaling was also activated in NPRC-deficient CFs by elevated NPs in an autocrine manner. Moreover, in CFs treated with the CM supernatant, NPRC deficiency in CMs also activated the cGMP/PKG signaling in CFs in a paracrine manner, suggesting the existence of CM-CF cross-talk. Western blot of MCFs from WT + DM and NPRC−/− + DM mice revealed that the levels of p-PKA substrates, p-CREB, and p-VASP increased in NPRC-deficient CFs derived from diabetic mice (fig. S20), indicating that NPRC deletion activated cAMP/PKA and cGMP/PKG signaling in CFs under diabetic condition.
To compare the protective effect of ANP, BNP, and CNP infusion versus NPRC knockout on diabetic cardiac fibrosis, we performed the fourth part of the in vivo experiment in which diabetic WT mice received ANP, BNP, or CNP infusion and the effects of ANP, BNP, and CNP were compared with those in diabetic NPRC−/− mice. Enzyme-linked immunosorbent assay (ELISA) showed that the serum levels of ANP, BNP, and CNP were substantially increased in the DM + ANP, DM + BNP, and DM + CNP infusion groups, respectively, in comparison with the DM + saline group, demonstrating the efficacy of NP infusion. By comparison, the serum levels of ANP, BNP, and CNP in the DM + NPRC−/− group were also elevated relative to the DM + saline group and similar to those in the DM + ANP, DM + BNP, or DM + CNP infusion group (fig. S21). Although ANP, BNP, and CNP infusion and NPRC deletion all decreased myocardial collagen accumulation, the NPRC−/− mice showed the most profound improvement among the four treatment groups (fig. S21). The results demonstrated that NPRC deletion exhibited a more potent cardioprotective effect than exogenous NP infusion. This interesting effect is probably due to the fact that NPRC deletion activates both cAMP/PKA and cGMP/PKG pathways, whereas NPs activate only the cGMP/PKG pathway, resulting in a better salutary effect by NPRC deletion.
Activation of cAMP/PKA or cGMP/PKG signaling increased TGIF1 expression
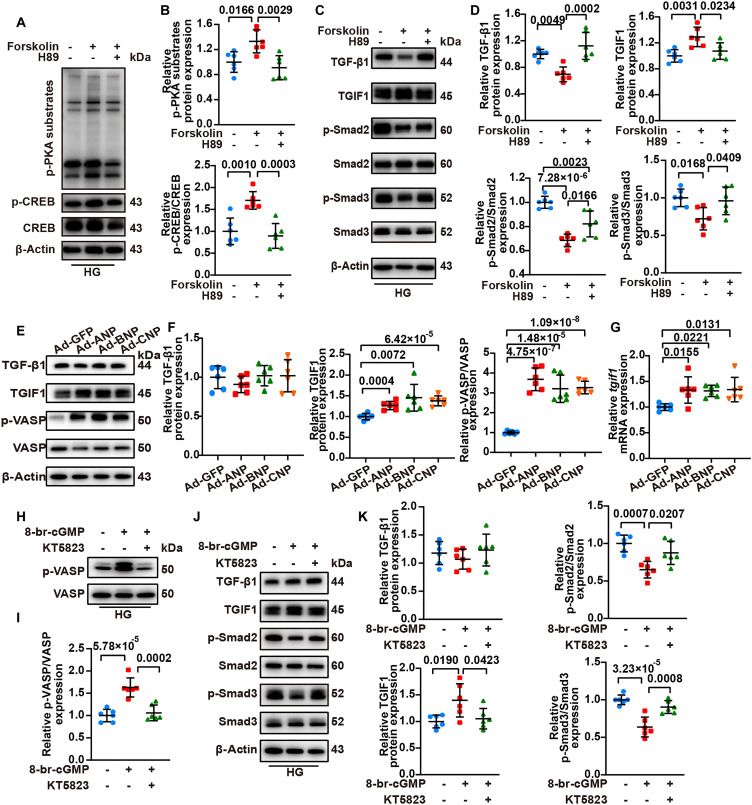
As both cAMP/PKA and cGMP/PKG signaling were activated by NPRC deficiency through different mechanisms, we hypothesized that both activated cAMP/PKA and cGMP/PKG signaling may up-regulate the expression of TGIF1. To test this hypothesis, we conducted the fourth part of the in vitro experiments (fig. S2) by first treating NRCFs or hiPSC-CFs with forskolin, a cAMP agonist, and H89, a PKA antagonist. The results revealed that forskolin substantially activated PKA signaling, as indicated by the increased levels of p-PKA substrates and p-CREB, and that H89 reversed the activation of PKA signaling induced by forskolin. In addition, forskolin increased the protein expression of TGIF1 but decreased that of p-Smad2/3, both of which were counteracted by treatment with H89. Moreover, forskolin decreased the expression level of TGF-β1, which was abrogated by H89 (Fig. 9 and fig. S22). These results suggested that the activation of PKA inhibited TGF-β1/Smad signaling by inhibiting TGF-β1 and enhancing TGIF1 expression.
Fig. 9. Activation of cAMP/PKA or cGMP/PKG signaling increased TGIF1 expression, but cAMP/PKA activation inhibited TGF-β1 expression.
(A) Representative Western blot images of p-PKA substrates and p-CREB expression in NRCFs, which were treated with either forskolin or forskolin combined with H89 followed by HG treatment. (B) Quantification of the protein expression of p-PKA substrates and p-CREB in (A). n = 6 per group. (C) Representative Western blot images of TGF-β1, TGIF1, p-Smad2, and p-Smad3 expression in NRCFs, which were treated with either forskolin or forskolin combined with H89 followed by HG treatment. (D) Quantification of the protein expression of TGF-β1, TGIF1, p-Smad2, and p-Smad3 in (C). n = 6 per group. (E) Representative Western blot images of TGF-β1, TGIF1, and p-VASP expression in NRCFs transfected with Ad-GFP, Ad-ANP, Ad-BNP, or Ad-CNP. (F) Quantification of the protein expression of TGF-β1, TGIF1, and p-VASP in (E). n = 6 per group. (G) Relative mRNA level of tgif1 in NRCFs transfected with Ad-GFP, Ad-ANP, Ad-BNP, or Ad-CNP. n = 6 per group. (H) Representative Western blot images of p-VASP in NRCFs treated with either 8-br-cGMP or 8-br-cGMP combined with KT5823 followed by HG treatment. (I) Quantification of the protein expression of p-VASP in (H). n = 6 per group. (J) Representative Western blot images of TGF-β1, TGIF1, p-Smad2, and p-Smad3 expression in NRCFs treated with either 8-br-cGMP or 8-br-cGMP combined with KT5823 followed by HG treatment. (K) Quantification of the protein expression of TGF-β1, TGIF1, p-Smad2, and p-Smad3 in (J). n = 6 per group. Normal distributions were tested by Shapiro-Wilk method. One-way ANOVA was used in (B), (D), (I), and (K). One-way ANOVA with Dunnett’s post hoc test was used in (F) and (G).
To further investigate the relationship between PKG signaling activation and changes in TGF-β1/Smad signaling, we transfected NRCFs by Ad-GFP (green fluorescent protein), Ad-ANP, Ad-BNP, and Ad-CNP, respectively, and found that overexpression of NPs increased p-VASP level, indicating an activation of PKG signaling in NRCFs. In contrast, there was no significant difference in TGF-β1 expression among the four groups of NRCFs. However, the protein and mRNA expression of TGIF1 increased in NP-overexpressing NRCFs (Fig. 9). In addition, we treated NRCFs or hiPSC-CFs with 8-br-cGMP, a cGMP agonist, to activate PKG signaling, followed by KT5823, a PKG antagonist, to reverse the activation of PKG induced by 8-br-cGMP, and these effects were confirmed by the changes in p-VASP expression (Fig. 9 and fig. S22). Treatment with 8-br-cGMP increased TGIF1 level and decreased p-Smad2/3 levels, and these effects were reversed by treatment with KT5823. Although no significant change was observed in TGF-β1 expression, p-Smad2/3 expression was significantly inhibited by activated PKG signaling through an increased level of TGIF1 (Fig. 9 and fig. S22). Together, these results suggested that both activation of PKA and PKG signaling increased the expression level of TGIF1, but PKA activation effectively inhibited TGF-β1 expression and TGF-β1/Smad signaling synergistically.
DISCUSSION
There were several important findings in the present study. First, NPRC expression was up-regulated in the hearts of patients with diabetes and HF and diabetic mice, and in HG-treated CFs. Second, NPRC deletion improved left ventricular function and remodeling by alleviating cardiac fibrosis in vivo. Third, NPRC deficiency decreased collagen synthesis and proliferation in CFs in vitro through the inhibition of TGF-β1/Smad signaling by targeting TGIF1 and TGF-β1. Finally, the effect of NPRC deletion on cardiac fibrosis and CFs was induced by activation of cAMP/PKA and cGMP/PKG signaling. To the best of our knowledge, our study is the first in the literature to report the beneficial effects of NPRC deletion on cardiac fibrosis in DM and the molecular mechanisms involved.
Among all NPRs, NPRC was the least studied receptor for NPs. In our previously reported multicenter study, six single-nucleotide polymorphisms (SNPs) of NPRC were found to be strongly associated with CAD in a large Chinese Han population (28), indicating a possible link between NPRC and cardiovascular diseases. Another study from our group demonstrated that NPRC deletion alleviated inflammation in the adipose tissue and promoted browning in the white adipose tissue through cAMP/PKA signaling in ApoE−/− mice (36), suggesting an underlying relationship between NPRC and metabolic diseases. Here, we demonstrated that NPRC expression was up-regulated in the hearts of patients with diabetes and HF and diabetic mice. It has been reported that NPRC expression in the left ventricle of patients with end-stage HF mostly induced by dilated cardiomyopathy was higher than that in healthy donors (37). In addition, NPRC expression was increased in experimental rats with isoproterenol-induced cardiac hypertrophy (38). Here, NPRC expression was up-regulated in the hearts of patients with diabetes and HF, and diabetic mice. In addition, NPRC expression was higher in CFs than in CMs, and high-glucose treatment enhanced NPRC expression in NRCFs and hiPSC-CFs, suggesting that NPRC was involved in the pathogenesis of DCM.
To explore the regulatory role of NPRC in DCM, we generated NPRC−/− mice and established a diabetic phenotype by STZ injection in NPRC−/− and littermate WT mice. NPRC−/− mice exhibited improved cardiac remodeling and function, compared with WT diabetic mice. As no significant change was observed in blood glucose levels, NPRC deletion protected the mice from DCM likely through direct targeting effects rather than through glycemic control. Among the pathological mechanisms associated with DCM, cardiac fibrosis leads to cardiac dysfunction and ventricular arrhythmias, resulting in a poor prognosis (39). As CFs are known to play a key role in cardiac fibrosis, we focused on the relation between CFs and DCM in this study. We found that CFs expressed more NPRC than CMs and NPRC deletion inhibited cardiac fibrosis in mice with DCM. In addition, NPRC knockdown in human and rat CFs decreased collagen synthesis and proliferation of these cells, while NPRC knockdown in human and rat CMs also decreased collagen synthesis and proliferation in CFs through cell-cell cross-talk. Consistently, MCFs isolated from NPRC−/− mice showed decreased collagen synthesis relative to MCFs derived from WT mice. These results verified that NPRC deletion alleviated diabetes-induced cardiac fibrosis, but were contradictory to some previous studies. Egom et al. (40) demonstrated that NPRC-deleted mice exhibited impaired sinoatrial node function and increased susceptibility to atrial fibrillation owing to increased fibrosis in the atria. In addition, NPRC deletion promoted angiotensin II–induced sinoatrial node disease and atrial fibrillation by increasing atrial fibrosis (24, 41). In contrast, another study by Rahmutula et al. (25) revealed that NPRC deletion in mice attenuated atrial fibrosis and fibrillation under the condition of TGF-β1 overexpression or transverse aortic constriction–induced pressure overload, and the mechanism identified in primary atrial fibroblasts was related with the enhanced NP/GCA signaling induced by NPRC deletion. There were fundamental differences between our study and previous ones, which may explain these discrepant findings. First, previous studies focused on atrial fibrosis but our study focused on left ventricular fibrosis. Similar to our findings in the NPRC−/− + control group, Egom et al. (40) did not find any effect of NPRC deletion on left ventricular fibrosis. Here, we also examined the left atrial cross sections and found that NPRC deletion did not increase left atrial fibrosis in the NPRC−/− + control and NPRC−/− + DM mice versus the WT + control and WT + DM mice, respectively. Second, most previous studies used angiotensin II infusion, which may itself induce extensive atrial fibrosis. Third, no in vitro experiments in CFs were performed in previous studies. In contrast, our findings were not only based on the results from in vivo experiments but also supported by a series of in vitro molecular studies that demonstrated the associated mechanisms.
A major drawback of systemic NPRC deletion in mice is skeletal abnormalities and reduced SBP. Thus, here, we used AAV9 carrying shNPRC to specifically knock down the expression of NPRC in myocardium, as AAV9 has been reportedly the most effective virus for cardiac gene intervention (30, 31). Similar to the results observed in NPRC−/− mice, AAV9-shNPRC alleviated cardiac fibrosis and improved cardiac remodeling and function in diabetic mice while avoiding the undesirable effect on skeleton and blood pressure. These results demonstrated that the protective effects of NPRC deletion on myocardium in diabetic mice were independent of the effect of NPRC deletion on other tissues.
To study the specific mechanisms underlying the effect of NPRC deletion on DCM development, we focused on TGF-β1/Smad signaling, as it plays an essential role in cardiac fibrosis (42). We observed that NPRC deletion decreased TGF-β1 expression in the hearts of diabetic mice, accompanied by a decrease in p-Smad2 and p-Smad3 levels, which indicated that TGF-β1/Smad signaling was inhibited by NPRC deletion. We also observed a decrease in TGF-β1 expression and p-Smad2 and p-Smad3 levels in NPRC-deficient CFs in vitro. Our results also showed that NPRC-deficient CM supernatant decreased p-Smad2 and p-Smad3 levels but did not attenuate TGF-β1 expression in corresponding CFs, suggesting that different mechanisms may have been involved in the latter situation.
Here, RNA sequencing identified tgif1 as a target gene of NPRC deletion in CFs. Tgif1 encodes the protein TGIF1, which is related to TGF-β1/Smad signaling. TGF-β1 initiates the canonical Smad pathway by interacting with TGF-βRs. Activated TGF-βR transmits signals by phosphorylating Smad2 and Smad3, followed by binding to Smad4 to form a heteromeric Smad complex, which then translocates to the nucleus and regulates the transcription of target genes (32). The phosphorylation of Smad2 and Smad3 depends on their interaction with cPML, which bridges Smad2/3 to TGF-βR for phosphorylation. TGIF1 acts as a suppressor of TGF-β1 signaling through multiple mechanisms including the recruitment of the transcriptional repressor complexes on the Smad target promoter to repress the transcription of TGF-β signaling (43), promotion of the degradation of Smad2 by interacting with the E3 ubiquitin ligase (44), and sequestering of cPML in the nucleus to prevent its shuttling to the cytoplasm where cPML contributes to the phosphorylation of Smad2/3 (45). As previous studies on TGIF1 were conducted in the context of tumor progression, we first verified the aforementioned mechanisms by knocking down TGIF1 in CFs. We observed that TGIF1 knockdown did not alter the expression of Smad2 and Smad3, but increased the levels of p-Smad2 and p-Smad3, suggesting that TGIF1 inhibited TGF-β1/Smad signaling mainly by attenuating phosphorylation of Smad2/3, rather than promoting their degradation in CFs. Furthermore, we demonstrated that NPRC deficiency increased the binding of TGIF1 to cPML, resulting in the sequestration of cPML in the nucleus of CFs. Moreover, after TGIF1 knockdown, the effect of NPRC deficiency on CFs was counteracted. Together, these results revealed the following mechanism by which NPRC deficiency inhibited TGF-β1/Smad signaling: NPRC deficiency induced the up-regulation of TGIF1, which sequestered cPML in the nucleus, prevented its shuttling to the cytoplasm, and thus decreased the phosphorylation of Smad2 and Smad3.
Here, we explored the detailed mechanism through which NPRC deletion induced TGIF1 expression. As previously reported, NPRC comprises an intracellular domain that activates Gi proteins to inhibit AC activity and cAMP/PKA signaling when combined with NPs (15). Increases in cAMP levels induced an anti-fibrotic effect manifested as decreased cell proliferation and collagen synthesis (46). As one of the most crucial effectors of cAMP, PKA also exhibited an anti-fibrotic effect. Previous studies revealed that activation of PKA by different receptors suppressed DNA synthesis, collagen deposition, and myofibroblast differentiation, and activation of PKA in CFs inhibited the activity of ras homolog family member A (RhoA) (47), which regulated cytoskeleton dynamics and migration of CFs. Here, we demonstrated that cAMP levels were elevated and PKA was activated by NPRC deletion in vivo and by NPRC knockdown in CFs. In addition, activation of PKA by the cAMP agonist forskolin increased TGIF1 expression, whereas this effect was reversed by the PKA antagonist H89, indicating that NPRC deficiency up-regulated TGIF1 expression through cAMP/PKA signaling. These findings were in line with a study by Saito et al. (48) showing that parathyroid hormone (PTH) activated cAMP/PKA signaling, increased activator protein 1 (AP1) expression, and promoted TGIF1 transcription. We also observed that NPRC deficiency decreased TGF-β1 expression in CFs. Treatment with forskolin also decreased TGF-β1 expression, which was restored by treatment with H89, indicating that apart from TGIF1, NPRC deficiency also reduced TGF-β1 expression through activation of cAMP/PKA signaling.
An interesting finding in this study was that TGIF1 expression was up-regulated not only in NPRC-deficient CFs but also in CFs treated with the supernatant of NPRC-deficient CMs, indicating the involvement of intercellular cross-talk. As NPRC is responsible for the clearance of NPs, we believe that the intercellular cross-talk is probably undertaken by NPs, which was supported by the increased NP levels in the supernatant of CMs and increased intracellular cGMP levels and PKG activity in CFs. Furthermore, increased NPs in the supernatant of CFs also increased intracellular cGMP levels and PKG activity in CFs through an autocrine manner. As an upstream molecule that triggers the cGMP-PKG pathway, NPs exerted anti-fibrotic effects, such as inhibiting TGF-β1–induced myofibroblast transformation and suppressing stimulated DNA and collagen synthesis in CFs (49). Moreover, pharmacological therapies that enhance NPs and cGMP/PKG signaling, such as sacubitril/valsartan and nesiritide, have been recommended by international guidelines to treat patients with HF (22, 23). Here, we demonstrated that activation of cGMP by 8-br-cGMP increased the TGIF1 expression and decreased the levels of p-Smad2 and p-Smad3, which was counteracted by the PKG antagonist KT5823. However, TGF-β1 expression did not change during these treatments, suggesting that cGMP/PKG inhibited TGF-β1/Smad signaling mainly by increasing TGIF1 expression, independent of TGF-β1. Consistent with the in vitro results, intracellular PKG activity was also increased in vivo, which might be attributed to increased NP expression in the heart. Furthermore, the serum levels of ANP and CNP also increased significantly in NPRC−/− mice, inducing declined SBP and elongated bones consistent with previous reports (16, 50).
In the in vitro experiments of the present study, NRCFs isolated from rats and MCFs isolated from mice were used to explore the effect of NPRC deficiency on CFs. To further verify the experimental results obtained from animal cells, CFs and CMs differentiated from hiPSCs were also used, which have been increasingly applied for cardiac disease modeling and drug screening (51, 52). Here, NPRC deficiency in hiPSC-CFs activated cAMP/PKA and cGMP/PKG signaling, regulated TGIF1 and TGF-β1/Smad signaling, and decreased collagen synthesis under HG stimulation. Furthermore, NPRC deficiency in hiPSC-CMs activated cGMP/PKG signaling, regulated TGF-β1/Smad signaling, and decreased collagen synthesis under HG stimulation via CM-CF communications. These findings provide insights into NPRC-related signaling in human CFs and CMs under high-glucose stimulation and reveal a promising therapeutic target for cardiac fibrosis in DCM.
The present study has several limitations. First, we used systemic NPRC knockout mice and cardiac-specific AAV in the present study. As fibroblast is primarily involved in the pathogenesis of DCM, the generation of CF-specific NPRC knockout mice is necessary to further elucidate the mechanism underlying the effect of NPRC deletion on cardiac fibrosis. Unfortunately, the commercially available fibroblast-specific Cre mice induce gene deletion in the fibroblasts of multiple organs including kidney and lung, causing systemic side effects. Thus, CF-specific Cre mice are highly warranted. Second, although we demonstrated that a cross-talk between CM and CF was involved in the mechanism underlying the beneficial effect of NPRC deficiency, the specific molecular target of these cross-talks needs further elucidation.
In conclusion, NPRC expression was increased in DCM and NPRC deficiency attenuated cardiac fibrosis and improved cardiac remodeling and function. Loss of NPRC decreased collagen synthesis and proliferation in CFs by increasing TGIF1 expression and inhibiting TGF-β1/Smad signaling through activation of cAMP/PKA and cGMP/PKG pathways (Fig. 10). Thus, inhibition of NPRC by cardiac-specific gene or drug therapy provides a promising approach to the treatment of diabetes-associated cardiac fibrosis.
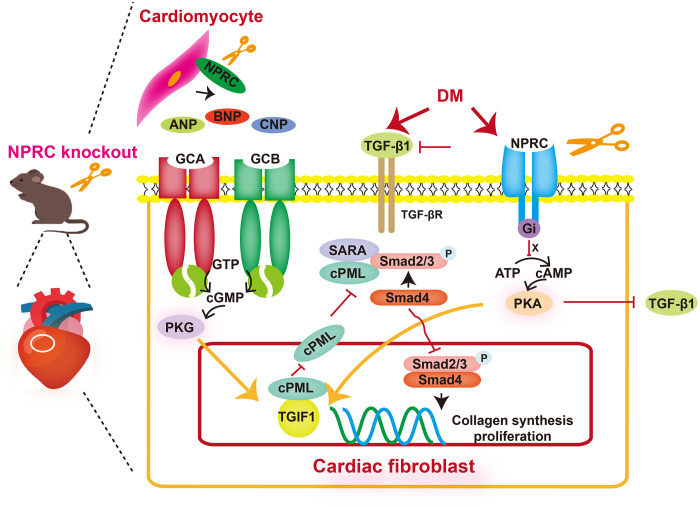
Fig. 10. Potential mechanisms of the effect of NPRC deficiency on cardiac fibrosis in DCM.
In the setting of DCM, NPRC is up-regulated, which leads to inhibited cAMP/PKA signaling. The deficiency of NPRC restores cAMP/PKA activity, which increases TGIF1 expression and inhibits TGF-β1 expression. In addition, NPRC deficiency decreases the clearance of NPs, which in turn enhances intracellular cGMP/PKG signaling through GCA and GCB. Activation of cGMP/PKG also increases TGIF1 expression but does not affect the expression of TGF-β1. Increased TGIF1 sequesters cPML in the nucleus to prevent its shuttling to the cytoplasm where it contributes to the phosphorylation of Smad2/3. Decreased TGF-β1 also inhibited the phosphorylation of Smad2/3. Altogether, these factors decrease collagen synthesis and proliferation in CFs, inhibit left ventricular fibrosis, and protect left ventricular function in DCM.
MATERIALS AND METHODS
Generation of NPRC knockout mice
NPRC knockout mice (NPRC−/−) were constructed from C57BL/6J background mice using CRISPR technology by Beijing Viewsolid Biotechnology Co. Ltd. (Beijing, China). On the basis of a previous report (16), we constructed a frameshift mutation at exon 1 (amino acids 1 to 252) of the npr3 genomic sequence by replacing GCTGGCGGCGCGAGCAGCGGCGCCG with CC before the GGGACACCCG sequence, resulting in an inactivation of protein function (fig. S1A). Genomic DNA was isolated from the mouse tails, and the fragments in the CRISPR-targeting region were amplified with the following primers for the genotyping of NPRC−/− mice: 5′-TTGGCGAGTTACTGAAGG-3′ (forward) and 5′-CGGTCCACAAGACTGAAG-3′ (reverse) by the PCR.
Construction and delivery of AAV9
A type 9 AAV system (AAV9) carrying scramble shRNA (Scr) and NPRC shRNA (shNPRC) was constructed by Genechem Technologies (Shanghai, China) and applied to knock down the cardiac expression of NPRC. AAV9 has been reportedly the most effective virus for cardiac gene intervention (30, 31). Each mouse was injected with 4 × 1011 vector genomes (vg) AAV9-Scr or AAV9-shNPRC via the tail vein.
Animal protocols
Animal experiments were divided into four parts. In the first part of the animal experiment (fig. S1B), to examine the effect of diabetes on NPRC expression and cardiac fibrosis, 20 eight-week-old male C57BL/6J mice purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) were used. The mice were fed a chow diet and randomly divided into two groups (n = 10 per group): control and DM groups. Diabetes was induced in the DM group through intraperitoneal injection of STZ (Sigma-Aldrich, USA; 55 mg/kg per day) dissolved in citrate buffer (0.1 mM, pH 4.5) at a final concentration of 10 mg/ml for 5 consecutive days. In the control group, the mice were intraperitoneally injected with vehicle (citrate buffer) for 5 days. Two weeks after STZ injection, only mice with fasting blood glucose (FBG) ≥ 16.7 mM were considered diabetic who were continuously fed a chow diet for another 16 weeks before euthanasia (53). In the second part of the animal experiment (fig. S1C), to observe the effect of NPRC knockout on cardiac fibrosis and function in diabetes, NPRC−/− mice and their NPRC+/+ littermates serving as WT controls were selected, and these mice were randomly divided into four groups (n = 15 per group): WT nondiabetic group (WT + control), WT diabetic group (WT + DM), NPRC−/− nondiabetic group (NPRC−/− + control), and NPRC−/− diabetic group (NPRC−/− + DM). Diabetes was induced as earlier described. Sixteen weeks after diabetes was successfully induced, echocardiography and hemodynamic measurement were performed before the mouse hearts were harvested for pathological and molecular analysis. In the third part of the animal experiment (fig. S1D), to explore the specific effect of NPRC knockdown on diabetic myocardium, 24 eight-week-old male C57BL/6J mice were intraperitoneally injected with STZ or vehicle before receiving intravenous injection of AAV9 and divided into four groups (n = 6 per group): AAV9-scr (scramble shRNA) in control mice, AAV9-shNPRC in control mice, AAV9-scr in DM mice, and AAV9-shNPRC in DM mice. The mice were fed with a chow diet as earlier described for another 16 weeks before euthanasia. In the fourth part of the animal experiment, to compare the protective effect of ANP, BNP, and CNP infusion versus NPRC knockout on diabetic cardiac fibrosis, NPRC−/− mice and their NPRC+/+ littermates (WT) were selected and divided into six groups (n = 6 per group; fig. S1E): control group, DM + saline group, DM + ANP group, DM + BNP group, DM + CNP group, and DM + NPRC−/− group. After diabetes was successfully induced, mice were fed a chow diet for 12 weeks. At week 14, all mice were implanted with osmotic pumps (Alzet model 2004, Alza Corp., USA). A continuous subcutaneous infusion of saline was conducted in the DM + saline and DM + NPRC−/− groups, respectively, and infusion of ANP (ANP1-28, MCE, USA; 0.5 μg/kg per minute), BNP (BNP1-32, MCE, USA; 0.03 μg/kg per minute), and CNP (CNP1-22, TargetMol, USA; 0.1 μg/kg per minute) was performed in the DM + ANP, DM + BNP, and DM + CNP groups, respectively. The dose of ANP, BNP, and CNP was chosen by referring to previous studies (27, 54–58). After 28 days of infusion, these mice were euthanized for morphological and pathological detection. All the mice were housed under specific pathogen–free conditions on a 12-hour light/12-hour dark cycle with free access to food and water. The mice were randomly assigned to different groups, and the investigators were blinded to the grouping of animals when assessing the outcomes. Animal care and experimental procedures were approved by the Ethics Committee and the Scientific Investigation Board of Shandong University Qilu Hospital and performed according to the Animal Management Rules of the Chinese Ministry of Health.
Human heart samples
Human heart tissues were obtained from three healthy donors and two patients with diabetes and severe HF [New York Heart Association Class IV (NYHA IV)] without hypertension and coronary artery stenosis as revealed by coronary angiography. Informed consent was obtained from all participants. Human study was approved by the Ethical Committee of Qilu Hospital of Shandong University and conforms to the ethical guidelines of the Declaration of Helsinki. Protein was extracted from the cardiac samples and subjected to Western blot.
Echocardiography
Transthoracic echocardiography was performed at the end of the experiment using the Vevo 2100 Imaging System (VisualSonics Inc., Canada) to evaluate the cardiac function in mice. The mice were anesthetized with 2% inhaled isoflurane and placed on a heating platform at 37°C. LVEDD and LVESD were measured by M-mode echocardiography in the parasternal long-axis view, and LVFS and LVEF were calculated automatically using the imaging system. The peak mitral flow velocities in early diastole (E) and late diastole (A) were measured by pulsed-wave Doppler in the apical four-chamber view. The mitral annular velocities in early diastole (e′) were measured using tissue Doppler imaging in the apical four-chamber view. The ratios of E/A and E/e′ were calculated.
Body weight and biochemical assay
The body weight of the mice was measured by an electronic balance (Shimadzu Corp., Japan). FBG was measured in tail venous blood by an Accu-Chek glucose meter with matched blood glucose strips (Roche, Germany). Body weight and FBG levels were monitored every 4 weeks. Serum lipid levels, including TC, TG, LDL-C, and HDL-C, were measured by an automatic biochemical analyzer (Chemray 240, Shenzhen, China).
Blood pressure and heart rate measurement
At the end of the experiment, blood pressure and heart rate of the mice were measured by a mouse tail-cuff blood pressure analysis system (Softron BP-98A, Japan). All mice were first trained to get accustomed to the device for accurate and reproducible measurements. Measurements were conducted between 9 a.m. to noon in a warm and quiet environment by the same investigator. The mice were warmed inside of a hyperthermia cylinder for 5 min, and the cuff sensor was placed at the base of the tail to record the measurements. At least three consecutive measurements were recorded to get the average values from each mouse.
Histology and IHC
The hearts of mice were isolated and fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm cross-sections. H&E, Sirius red, and Masson’s trichrome staining were performed according to the manufacturer’s instructions using staining kits (Solarbio, Beijing, China).
For IHC, the sections were dewaxed and subjected to antigen retrieval with citrate buffer (pH 6.0), followed by treatment with 3% H2O2. The sections were then blocked with 5% goat serum for 30 min at 37°C and incubated with primary antibodies at 4°C overnight. The next day, the sections were incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies (ZSGB-Bio, Beijing, China) for 30 min at room temperature, and detection was performed using a 3′-diaminobenzidine (DAB) kit (ZSGB-Bio, Beijing, China). Hematoxylin was used for nuclear staining. All histological images were examined and photographed under a microscope (Ti-S, Nikon) and analyzed with the Image-Pro Plus 6.0 software (Media Cybernetics Inc., USA). The primary antibodies used are listed in table S1. Sections reacting with nonimmune immunoglobulin G (IgG) as well as secondary antibodies were used as negative controls.
Immunofluorescent staining
For immunofluorescent staining of tissue sections, the sections were dewaxed, blocked with 5% bovine serum albumin (BSA) for 30 min, and incubated with primary antibodies at 4°C overnight. For immunofluorescent staining of cells, the cells were first seeded on glass coverslips in the corresponding plates. Next, the cells were fixed in 4% paraformaldehyde for 15 min, permeabilized in 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min, blocked with 5% BSA, and incubated with primary antibodies at 4°C overnight. On the next day, the sections or cell coverslips were incubated with Alexa Fluor 488/594–conjugated secondary antibodies (Abcam, UK). The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Abcam, UK). Staining was observed and photographed using a fluorescent microscope (Ti-S, Nikon). The primary antibodies used are listed in table S1. Sections reacting with nonimmune IgG as well as secondary antibodies were used as negative controls.
Measurement of ANP, BNP, CNP, cGMP, and cAMP levels
ANP, BNP, and CNP levels in the serum of mice and in cell supernatants were measured using ANP EIA kits (EIAM-ANP-1, Raybiotech, USA), BNP EIA kits (EIAM-BNP-1, Raybiotech, USA), and CNP EIA kits (EIAM-CNP-1, Raybiotech, USA). The levels of intracellular cGMP and cAMP were measured using the corresponding kits (4339, 4360; Cell Signaling Technology, USA) according to the manufacturer’s instructions.
Cell culture
The following cells were used in the in vitro experiments: primary CMs (NRCMs) and CFs (NRCFs) from neonatal rat, and human CMs and CFs differentiated from hiPSCs (hiPSC-CM and hiPSC-CF). Primary NRCMs and NRCFs were isolated from the hearts of 1- to 3-day-old Sprague-Dawley rats as previously reported (59). Briefly, the hearts were removed and cut into small pieces in precooled D-Hank’s solution. The pieces were then transferred to a conical flask containing digestive solution (0.75 mg/ml collagenase type II in D-Hank’s solution) and spun on a magnetic stirrer at 37°C for 1 hour. Thereafter, the supernatant was collected and the pieces were digested. The supernatants from the whole digestion process were seeded into flasks, and NRCMs were isolated from NRCFs using a differential attachment technique. After culture at 37°C in 5% CO2 for 2 hours, most of the NRCFs were adherent to the flasks and the supernatant containing NRCMs was reseeded onto culture plates precoated with gelatin. Differentiated hiPSC-CM and hiPSC-CF were provided by NanJing Cosmos Biotechnology Co. Ltd. (Nanjing, China). NRCFs and hiPSC-CFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) containing 10% fetal bovine serum (Sigma-Aldrich, USA) and 1% penicillin/streptomycin (Gibco, USA). HiPSC-CMs were cultured in CM maintenance medium (Cosmos, Nanjing, China) and refreshed every 2 days. NRCMs were cultured in DMEM containing 8% fetal equine serum (Solarbio, Beijing, China), 5% newborn calf serum (Solarbio, Beijing, China), and 1% penicillin/streptomycin, and 5-bromo-2′-deoxyuridine (BrdU; 0.1 mM, Sigma-Aldrich, USA) was used to inhibit the proliferation of residual CFs at 37°C in 5% CO2.
In the first part of the in vitro experiments (fig. S2A), to investigate the effect of NPRC deficiency on CFs, siRNA targeting NPRC (si-NPRC) was transfected into NRCFs to knock down the expression of NPRC. Briefly, the primary NRCFs (F0 generation) were passaged to the corresponding plates. NRCFs were transfected with si-NPRC or negative control siRNA (si-NC) with Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s instructions. After 24 hours of transfection, the supernatant was replaced with fresh medium and the cells were treated with normal glucose (NG; 5.5 mM) or high glucose (HG; 33.3 mM) for 72 hours before cell collection. Similar experiments were performed for hiPSC-CFs.
In the second part of the in vitro experiments (fig. S2B), to investigate the effect of NPRC deficiency on the communication between CM and CF, the primary NRCMs were transfected with si-NPRC or si-NC as described earlier. After 24 hours of transfection, the supernatant was replaced with fresh medium and the cells were treated with NG (5.5 mM) or HG (33.3 mM) for 48 hours. The cell supernatant was collected and added to primary NRCFs for another 72 hours before cell collection. Similar experiments were conducted for hiPSC-CMs and hiPSC-CFs.
In the third part of the in vitro experiments (fig. S2C), to investigate the role of TGF-β–induced factor homeobox 1 (TGIF1) in NPRC deficiency–induced effects, the primary NRCFs were transfected with si-NC, si-NPRC, or combinational si-NPRC and si-TGIF1. After 24 hours of transfection, the supernatant was replaced with fresh medium and the cells were treated with HG (33.3 mM) for 72 hours before cell harvest. The sequences of the above siRNAs were listed in table S2. Similar experiments were carried out for hiPSC-CFs.
In the fourth part of the in vitro experiments (fig. S2D), to investigate the role of PKA and PKG activation in NPRC deficiency–induced effects, the primary NRCFs were treated with the corresponding agonist and antagonist. To study the role of PKA activation, the primary NRCFs were first treated with forskolin (20 μM), a cAMP agonist, for 15 min, followed by treatment with H89 (10 μM), a PKA antagonist. After another incubation for 15 min, the NRCFs were treated with HG (33.3 mM) for 24 hours before cell harvest. To study the role of PKG activation, the primary NRCFs were first treated with 8-br-cGMP (1 μM), a cGMP agonist, for 15 min, followed by treatment with KT5823 (1 μM), a PKG antagonist. After another incubation for 15 min, the NRCFs were treated with HG (33.3 mM) for 24 hours before cell harvest. Similar experiments were performed for hiPSC-CFs.
Isolation of adult MCFs
Adult CFs were isolated from diabetic WT and NPRC−/− mice as reported (60). Briefly, the hearts were removed and cut into small pieces, digested with collagenase type II (0.125 mg/ml in D-Hank’s solution) at 4°C overnight. On the next day, the supernatant was discarded and the pieces were digested with pancreatin (0.05% in PBS) for 2 min at 37°C. The supernatant was collected and the digestion was repeated for six to seven times until no visible tissue existed. Then, the supernatant from the whole digestion was seeded in plates and allowed cell attachment for 2 hours. After 48 hours of culture, the protein was extracted from the MCFs and subjected to Western blot.
Adenovirus-mediated overexpression
To achieve overexpression of ANP, BNP, and CNP in NRCFs, adenovirus carrying plasmids expressing ANP, BNP, CNP, and GFP were added to the cell culture medium (multiplicity of infection = 300), followed by incubation for 12 hours. After 12 hours of transfection, the supernatant was replaced with fresh medium for another 48 hours before cell harvest.
EDU labeling and CCK-8 assay
The in vitro cell proliferation of NRCFs and hiPSC-CFs was detected using an EDU kit (RiboBio, Guangzhou, China). In the first and second parts of the in vitro experiments, the EDU labeling reagent was added to the culture medium (1:2500), followed by culture for 16 hours before cell collection. Next, the cells were fixed with 4% paraformaldehyde, incubated with Apollo staining solution, and counterstained with Hoechst 33342 for cell nuclei. Finally, fluorescence images were captured by a microscope (Ti-S, Nikon).
The cell viability of NRCFs and hiPSC-CFs was assessed by the CCK-8 (Proteintech, Wuhan, China) according to the manufacturer’s instructions. In the first and second parts of the in vitro experiments, the CCK8 reagent was added to a 96-well plate of NRCFs or hiPSC-CFs, and after incubation for 4 hours, the absorbance at 450 nm was measured by a microplate reader (BioTek, USA).
Extraction of cytoplasmic and nuclear proteins
Cytoplasmic and nuclear proteins were separated using Minute Cytoplasmic & Nuclear Extraction Kits (SC-003, Invent, USA) according to the manufacturer’s protocol. Briefly, cytoplasmic extraction buffer was added to the cell plates, and the plates were incubated on ice for 5 min, followed by a vigorous vortex for 15 s and centrifugation for 5 min at top speed. The supernatant (cytoplasmic fraction) was transferred to a new tube, and nuclear extraction buffer was added to the pellets. The pellets were vortexed vigorously for 15 s and then incubated on ice for 1 min, and this process was repeated for four times. The nuclear extract was transferred to a filter cartridge with a collection tube and centrifuged at top speed for 30 s. The cytoplasmic and nuclear proteins were further subjected to Western blot analysis.
Coimmunoprecipitation assay
NRCFs were cultured in a 100-mm cell dish and transfected with si-NC or si-NPRC. After 48 hours of culture, the cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (CWbio, China) supplemented with a protease inhibitor cocktail (CWbio, China) on ice for 30 min. After centrifugation, the supernatants were transferred to a new tube and incubated with 1 μg of primary antibodies or IgG (Proteintech, Wuhan, China) for 1 hour at 4°C. Protein A/G-agarose beads (Santa Cruz Biotechnology, USA) were then incubated with the mixture at 4°C overnight. On the second day, the beads were washed for five times through centrifugation with lysis buffer, and immunoprecipitated proteins were used for Western blot analysis after incubation with loading buffer at 99°C for 10 min.
Transcriptome RNA sequencing
Total RNA from NRCFs in the first part of the in vitro experiments was isolated using the RNeasy Mini Kit (Qiagen, Germany). After assessment of the integrity of total RNA using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., USA), the samples with RNA integrity number > 7.0 were used for sequencing. RNA-sequencing strand-specific libraries were constructed using the VAHTS Total RNA-seq (H/M/R) Library Prep Kit (Vazyme, Nanjing, China). The library was then diluted to 10 pM, after which clusters were generated and sequenced on the Illumina NovaSeq 6000 platform (Illumina, USA). Differential expression analysis of mRNA was performed using the R package edgeR.
Real-time PCR assay
Total RNA was extracted from cultured cells using RNA fast200 kits (220010, Fastagen, Shanghai, China). Total RNA was reverse-transcribed to cDNA using the HiScript RT Mix kit with a gDNA wiper (R323-01, Vazyme, Nanjing, China) following the manufacturer’s instructions. Reverse transcription products were amplified on a LightCycler 480 instrument (Roche, Switzerland) using SYBR Green qPCR Mix (Q711, Vazyme, Nanjing, China). All samples were measured in triplicates. β-Actin was used for normalization in each experiment, and relative gene expression was determined using the 2−ΔΔCt method. The primer sequences are listed in table S2.
Western blot analysis
Total protein was extracted from the left ventricular apex of mice using the Total Protein Extraction Kit (SD-001, Invent, USA). Protein lysates were extracted from cultured cells using RIPA lysis buffer (R0010, Solarbio, China) supplemented with a protease inhibitor cocktail (CWbio, China). Equal amounts of protein samples were separated on 10 to 12% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked with 5% skim milk for 1 hour at room temperature and incubated with primary antibodies at 4°C overnight. On the next day, the membranes were incubated with peroxidase-conjugated secondary antibodies (1:5000, Jackson ImmunoResearch Laboratories, USA) for 1 hour at room temperature, followed by detection using a chemiluminescent substrate (Millipore, USA) and chemiluminescent instrument (GE, Amersham Imager 680RGB, USA). Images were analyzed with ImageJ software (Media Cybernetics Inc., USA), and β-actin was used as a control to verify equal protein loading. The primary antibodies used were listed in table S1.
Statistical analysis
All data were presented as mean ± SEM of at least three independent experiments. All analyses were performed with GraphPad Prism 8 software (GraphPad, USA). The normality assumption of the data distribution was assessed using the Shapiro-Wilk test. Unpaired two-tailed Student’s t tests were used to determine the statistical difference between two groups. One-way analysis of variance (ANOVA) followed by Turkey’s post hoc test was performed to determine the statistical difference between multiple groups with one variable. Two-way ANOVA was used to compare multiple groups with more than one variable. For two-way ANOVA with significant interaction (P < 0.05), Turkey post hoc test was used. For two-way ANOVA without significant interaction, Bonferroni post hoc test was used. For all statistical comparisons, P < 0.05 was considered statistically significant.
Acknowledgments
We thank M. Zhang, W. Zhang, Q. Zhang, and S. Wang for their suggestions on experimental design. We thank N. Dong (Wuhan Union Hospital) for providing human heart samples.
Funding: This work was supported by grants from the National Natural Science Foundation of China (nos. 82241203, 81920108003, 82030051, 82000411, 82270487, and 81970373), State Key R&D Program of China (2021YFF0501403), Key R&D Program of Shandong Province (2021SFGC0503, 2021ZLGX02, 2021ZDSYS05, and 2020ZLYS05), Shandong Provincial Natural Science Foundation (ZR2020QH023 and ZR2020YQ53), and Taishan Scholars Program of Shandong Province (C.Z. and M.Z.).
Author contributions: L.M., Y.L., and X.W. performed the experiments, analyzed the data, and wrote the manuscript. C.C., F.X., L.X., and Yaoyuan Z. contributed to performing experiments and statistical analyses. W.S. and M.Z. contributed to revising the manuscript. Yun Z. and C.Z. designed the study and corrected the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Table S1 and S2
REFERENCES AND NOTES
- 1.R. H. Ritchie, E. D. Abel, Basic mechanisms of diabetic heart disease. Circ. Res. 126, 1501–1525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R. E. Gilbert, H. Krum, Heart failure in diabetes: Effects of anti-hyperglycaemic drug therapy. Lancet 385, 2107–2117 (2015). [DOI] [PubMed] [Google Scholar]
- 3.S. M. Shanbhag, A. M. Greve, T. Aspelund, E. B. Schelbert, J. J. Cao, R. Danielsen, G. Thornorgeirsson, S. Sigurethsson, G. Eiriksdottir, T. B. Harris, L. J. Launer, V. Guethnason, A. E. Arai, Prevalence and prognosis of ischaemic and non-ischaemic myocardial fibrosis in older adults. Eur. Heart J. 40, 529–538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Y. Kwong, H. Sattar, H. Wu, G. Vorobiof, V. Gandla, K. Steel, S. Siu, K. A. Brown, Incidence and prognostic implication of unrecognized myocardial scar characterized by cardiac magnetic resonance in diabetic patients without clinical evidence of myocardial infarction. Circulation 118, 1011–1020 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L. Dannenberg, S. Weske, M. Kelm, B. Levkau, A. Polzin, Cellular mechanisms and recommended drug-based therapeutic options in diabetic cardiomyopathy. Pharmacol. Ther. 228, 107920 (2021). [DOI] [PubMed] [Google Scholar]
- 6.T. Nishikimi, N. Maeda, H. Matsuoka, The role of natriuretic peptides in cardioprotection. Cardiovasc. Res. 69, 318–328 (2006). [DOI] [PubMed] [Google Scholar]
- 7.V. A. Cameron, M. T. Rademaker, L. J. Ellmers, E. A. Espiner, M. G. Nicholls, A. M. Richards, Atrial (ANP) and brain natriuretic peptide (BNP) expression after myocardial infarction in sheep: ANP is synthesized by fibroblasts infiltrating the infarct. Endocrinology 141, 4690–4697 (2000). [DOI] [PubMed] [Google Scholar]
- 8.T. Tsuruda, G. Boerrigter, B. K. Huntley, J. A. Noser, A. Cataliotti, L. C. Costello-Boerrigter, H. H. Chen, J. C. Burnett Jr., Brain natriuretic peptide is produced in cardiac fibroblasts and induces matrix metalloproteinases. Circ. Res. 91, 1127–1134 (2002). [DOI] [PubMed] [Google Scholar]
- 9.S. Suga, K. Nakao, H. Itoh, Y. Komatsu, Y. Ogawa, N. Hama, H. Imura, Endothelial production of C-type natriuretic peptide and its marked augmentation by transforming growth factor-beta. Possible existence of “vascular natriuretic peptide system”. J. Clin. Invest. 90, 1145–1149 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. del Ry, M. Cabiati, V. Lionetti, M. Emdin, F. A. Recchia, D. Giannessi, Expression of C-type natriuretic peptide and of its receptor NPR-B in normal and failing heart. Peptides 29, 2208–2215 (2008). [DOI] [PubMed] [Google Scholar]
- 11.T. Horio, T. Tokudome, T. Maki, F. Yoshihara, S. Suga, T. Nishikimi, M. Kojima, Y. Kawano, K. Kangawa, Gene expression, secretion, and autocrine action of C-type natriuretic peptide in cultured adult rat cardiac fibroblasts. Endocrinology 144, 2279–2284 (2003). [DOI] [PubMed] [Google Scholar]
- 12.X. Lin, J. Hänze, F. Heese, R. Sodmann, R. E. Lang, Gene expression of natriuretic peptide receptors in myocardial cells. Circ. Res. 77, 750–758 (1995). [DOI] [PubMed] [Google Scholar]
- 13.M. Moghtadaei, I. Polina, R. A. Rose, Electrophysiological effects of natriuretic peptides in the heart are mediated by multiple receptor subtypes. Prog. Biophys. Mol. Biol. 120, 37–49 (2016). [DOI] [PubMed] [Google Scholar]
- 14.L. R. Potter, S. Abbey-Hosch, D. M. Dickey, Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr. Rev. 27, 47–72 (2006). [DOI] [PubMed] [Google Scholar]
- 15.M. Pagano, M. B. Anand-Srivastava, Cytoplasmic domain of natriuretic peptide receptor C constitutes Gi activator sequences that inhibit adenylyl cyclase activity. J. Biol. Chem. 276, 22064–22070 (2001). [DOI] [PubMed] [Google Scholar]
- 16.N. Matsukawa, W. J. Grzesik, N. Takahashi, K. N. Pandey, S. Pang, M. Yamauchi, O. Smithies, The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc. Natl. Acad. Sci. U.S.A. 96, 7403–7408 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.K. Sabrane, M. N. Kruse, L. Fabritz, B. Zetsche, D. Mitko, B. V. Skryabin, M. Zwiener, H. A. Baba, M. Yanagisawa, M. Kuhn, Vascular endothelium is critically involved in the hypotensive and hypovolemic actions of atrial natriuretic peptide. J. Clin. Invest. 115, 1666–1674 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.F. Macheret, D. Heublein, L. C. Costello-Boerrigter, G. Boerrigter, P. McKie, D. Bellavia, S. Mangiafico, Y. Ikeda, K. Bailey, C. G. Scott, S. Sandberg, H. H. Chen, L. Malatino, M. M. Redfield, R. Rodeheffer, J. Burnett Jr., A. Cataliotti, Human hypertension is characterized by a lack of activation of the antihypertensive cardiac hormones ANP and BNP. J. Am. Coll. Cardiol. 60, 1558–1565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.B. K. Huntley, S. M. Sandberg, J. A. Noser, A. Cataliotti, M. M. Redfield, Y. Matsuda, J. C. Burnett Jr., BNP-induced activation of cGMP in human cardiac fibroblasts: Interactions with fibronectin and natriuretic peptide receptors. J. Cell. Physiol. 209, 943–949 (2006). [DOI] [PubMed] [Google Scholar]
- 20.P. M. Oliver, J. E. Fox, R. Kim, H. A. Rockman, H. S. Kim, R. L. Reddick, K. N. Pandey, S. L. Milgram, O. Smithies, N. Maeda, Hypertension, cardiac hypertrophy, and sudden death in mice lacking natriuretic peptide receptor A. Proc. Natl. Acad. Sci. U.S.A. 94, 14730–14735 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Nakao, K. Kuwahara, T. Nishikimi, Y. Nakagawa, H. Kinoshita, T. Minami, Y. Kuwabara, C. Yamada, Y. Yamada, T. Tokudome, C. Nagai-Okatani, N. Minamino, Y. M. Nakao, S. Yasuno, K. Ueshima, M. Sone, T. Kimura, K. Kangawa, K. Nakao, Endothelium-derived C-Type natriuretic peptide contributes to blood pressure regulation by maintaining endothelial integrity. Hypertension 69, 286–296 (2017). [DOI] [PubMed] [Google Scholar]
- 22.T. A. McDonagh, M. Metra, M. Adamo, R. S. Gardner, A. Baumbach, M. Bohm, H. Burri, J. Butler, J. Celutkiene, O. Chioncel, J. G. F. Cleland, A. J. S. Coats, M. G. Crespo-Leiro, D. Farmakis, M. Gilard, S. Heymans, A. W. Hoes, T. Jaarsma, E. A. Jankowska, M. Lainscak, C. S. P. Lam, A. R. Lyon, J. J. V. McMurray, A. Mebazaa, R. Mindham, C. Muneretto, M. Francesco Piepoli, S. Price, G. M. C. Rosano, F. Ruschitzka, A. Kathrine Skibelund; E. S. C. S. D. Group , 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 42, 3599–3726 (2021). [DOI] [PubMed] [Google Scholar]
- 23.P. A. Heidenreich, B. Bozkurt, D. Aguilar, L. A. Allen, J. J. Byun, M. M. Colvin, A. Deswal, M. H. Drazner, S. M. Dunlay, L. R. Evers, J. C. Fang, S. E. Fedson, G. C. Fonarow, S. S. Hayek, A. F. Hernandez, P. Khazanie, M. M. Kittleson, C. S. Lee, M. S. Link, C. A. Milano, L. C. Nnacheta, A. T. Sandhu, L. W. Stevenson, O. Vardeny, A. R. Vest, C. W. Yancy, 2022 AHA/ACC/HFSA guideline for the management of heart failure: Executive summary: A report of the American College of Cardiology/American Heart Association Joint Committee on clinical practice guidelines. Circulation 145, e876–e894 (2022). [DOI] [PubMed] [Google Scholar]
- 24.H. J. Jansen, M. Mackasey, M. Moghtadaei, Y. Liu, J. Kaur, E. E. Egom, J. M. Tuomi, S. A. Rafferty, A. W. Kirkby, R. A. Rose, NPR-C (Natriuretic peptide receptor-C) modulates the progression of angiotensin II-mediated atrial fibrillation and atrial remodeling in mice. Circ. Arrhythm. Electrophysiol. 12, e006863 (2019). [DOI] [PubMed] [Google Scholar]
- 25.D. Rahmutula, H. Zhang, E. E. Wilson, J. E. Olgin, Absence of natriuretic peptide clearance receptor attenuates TGF-β1-induced selective atrial fibrosis and atrial fibrillation. Cardiovasc. Res. 115, 357–372 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Z. Kovacova, W. G. Tharp, D. Liu, W. Wei, H. Xie, S. Collins, R. E. Pratley, Adipose tissue natriuretic peptide receptor expression is related to insulin sensitivity in obesity and diabetes. Obesity (Silver Spring) 24, 820–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Coue, P. M. Badin, I. K. Vila, C. Laurens, K. Louche, M. A. Marques, V. Bourlier, E. Mouisel, G. Tavernier, A. C. Rustan, J. E. Galgani, D. R. Joanisse, S. R. Smith, D. Langin, C. Moro, Defective natriuretic peptide receptor signaling in skeletal muscle links obesity to type 2 diabetes. Diabetes 64, 4033–4045 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Q. Hu, Q. Liu, S. Wang, X. Zhen, Z. Zhang, R. Lv, G. Jiang, Z. Ma, H. He, D. Li, X. Liu, F. Gao, J. Li, L. Li, M. Zhang, X. Ji, Y. Chen, D. Wang, D. Huang, A. Ma, W. Huang, Y. Zhao, Y. Gong, C. Zhang, Y. Zhang, NPR-C gene polymorphism is associated with increased susceptibility to coronary artery disease in Chinese Han population: A multicenter study. Oncotarget 7, 33662–33674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Camelliti, T. K. Borg, P. Kohl, Structural and functional characterisation of cardiac fibroblasts. Cardiovasc. Res. 65, 40–51 (2005). [DOI] [PubMed] [Google Scholar]
- 30.C. A. Pacak, C. S. Mah, B. D. Thattaliyath, T. J. Conlon, M. A. Lewis, D. E. Cloutier, I. Zolotukhin, A. F. Tarantal, B. J. Byrne, Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 99, e3–e9 (2006). [DOI] [PubMed] [Google Scholar]
- 31.J. M. Kieserman, V. D. Myers, P. Dubey, J. Y. Cheung, A. M. Feldman, Current landscape of heart failure gene therapy. J. Am. Heart Assoc. 8, e012239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Y. Yue, K. Meng, Y. Pu, X. Zhang, Transforming growth factor beta (TGF-β) mediates cardiac fibrosis and induces diabetic cardiomyopathy. Diabetes Res. Clin. Pract. 133, 124–130 (2017). [DOI] [PubMed] [Google Scholar]
- 33.R. A. Rose, W. R. Giles, Natriuretic peptide C receptor signalling in the heart and vasculature. J. Physiol. 586, 353–366 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. Bie, Natriuretic peptides and normal body fluid regulation. Compr. Physiol. 8, 1211–1249 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Y. D. Lang, Y. S. Jou, PSPC1 is a new contextual determinant of aberrant subcellular translocation of oncogenes in tumor progression. J. Biomed. Sci. 28, 57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. Cheng, F. Xue, W. Sui, L. Meng, L. Xie, C. Zhang, J. Yang, Y. Zhang, Deletion of natriuretic peptide receptor C alleviates adipose tissue inflammation in hypercholesterolemic Apolipoprotein E knockout mice. J. Cell. Mol. Med. 25, 9837–9850 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.T. Ichiki, J. A. Schirger, B. K. Huntley, F. V. Brozovich, J. J. Maleszewski, S. M. Sandberg, S. J. Sangaralingham, S. J. Park, J. C. Burnett Jr., Cardiac fibrosis in end-stage human heart failure and the cardiac natriuretic peptide guanylyl cyclase system: Regulation and therapeutic implications. J. Mol. Cell. Cardiol. 75, 199–205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.B. Venkatesan, A. Tumala, V. Subramanian, E. Vellaichamy, Transient silencing of Npr3 gene expression improved the circulatory levels of atrial natriuretic peptides and attenuated β-adrenoceptor activation-induced cardiac hypertrophic growth in experimental rats. Eur. J. Pharmacol. 782, 44–58 (2016). [DOI] [PubMed] [Google Scholar]
- 39.I. Tuleta, N. G. Frangogiannis, Fibrosis of the diabetic heart: Clinical significance, molecular mechanisms, and therapeutic opportunities. Adv. Drug Deliv. Rev. 176, 113904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. E. Egom, K. Vella, R. Hua, H. J. Jansen, M. Moghtadaei, I. Polina, O. Bogachev, R. Hurnik, M. Mackasey, S. Rafferty, G. Ray, R. A. Rose, Impaired sinoatrial node function and increased susceptibility to atrial fibrillation in mice lacking natriuretic peptide receptor C. J. Physiol. 593, 1127–1146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Mackasey, E. E. Egom, H. J. Jansen, R. Hua, M. Moghtadaei, Y. Liu, J. Kaur, M. D. McRae, O. Bogachev, S. A. Rafferty, G. Ray, A. W. Kirkby, R. A. Rose, Natriuretic peptide receptor-C protects against angiotensin II-mediated sinoatrial node disease in mice. JACC Basic Transl. Sci. 3, 824–843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.I. Tuleta, N. G. Frangogiannis, Diabetic fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166044 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Wotton, R. S. Lo, S. Lee, J. Massagué, A Smad transcriptional corepressor. Cell 97, 29–39 (1999). [DOI] [PubMed] [Google Scholar]
- 44.S. R. Seo, F. Lallemand, N. Ferrand, M. Pessah, S. L'Hoste, J. Camonis, A. Atfi, The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 23, 3780–3792 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. R. Seo, N. Ferrand, N. Faresse, C. Prunier, L. Abécassis, M. Pessah, M. F. Bourgeade, A. Atfi, Nuclear retention of the tumor suppressor cPML by the homeodomain protein TGIF restricts TGF-β signaling. Mol. Cell 23, 547–559 (2006). [DOI] [PubMed] [Google Scholar]
- 46.P. A. Insel, F. Murray, U. Yokoyama, S. Romano, H. Yun, L. Brown, A. Snead, D. Lu, N. Aroonsakool, cAMP and Epac in the regulation of tissue fibrosis. Br. J. Pharmacol. 166, 447–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. Oishi, N. Makita, J. Sato, T. Iiri, Regulation of RhoA signaling by the cAMP-dependent phosphorylation of RhoGDIα. J. Biol. Chem. 287, 38705–38715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H. Saito, A. Gasser, S. Bolamperti, M. Maeda, L. Matthies, K. Jahn, C. L. Long, H. Schluter, M. Kwiatkowski, V. Saini, P. D. Pajevic, T. Bellido, A. J. van Wijnen, K. S. Mohammad, T. A. Guise, H. Taipaleenmaki, E. Hesse, TG-interacting factor 1 (Tgif1)-deficiency attenuates bone remodeling and blunts the anabolic response to parathyroid hormone. Nat. Commun. 10, 1354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Park, P. Sandner, T. Krieg, cGMP at the centre of attention: Emerging strategies for activating the cardioprotective PKG pathway. Basic Res. Cardiol. 113, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Y. Kanai, A. Yasoda, K. P. Mori, H. Watanabe-Takano, C. Nagai-Okatani, Y. Yamashita, K. Hirota, Y. Ueda, I. Yamauchi, E. Kondo, S. Yamanaka, Y. Sakane, K. Nakao, T. Fujii, H. Yokoi, N. Minamino, M. Mukoyama, N. Mochizuki, N. Inagaki, Circulating osteocrin stimulates bone growth by limiting C-type natriuretic peptide clearance. J. Clin. Invest. 127, 4136–4147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O. K. Choong, C. Y. Chen, J. Zhang, J. H. Lin, P. J. Lin, S. C. Ruan, T. J. Kamp, P. C. H. Hsieh, Hypoxia-induced H19/YB-1 cascade modulates cardiac remodeling after infarction. Theranostics 9, 6550–6567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.F. Seibertz, H. Sutanto, R. Dulk, J. R. D. Pronto, R. Springer, M. Rapedius, A. Liutkute, M. Ritter, P. Jung, L. Stelzer, L. M. Husgen, M. Klopp, T. Rubio, F. E. Fakuade, F. E. Mason, N. Hartmann, S. Pabel, K. Streckfuss-Bomeke, L. Cyganek, S. Sossalla, J. Heijman, N. Voigt, Electrophysiological and calcium-handling development during long-term culture of human-induced pluripotent stem cell-derived cardiomyocytes. Basic Res. Cardiol. 118, 14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. J. King, The use of animal models in diabetes research. Br. J. Pharmacol. 166, 877–894 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Y. Oi, T. Nagoshi, H. Kimura, Y. Tanaka, A. Yoshii, R. Yasutake, H. Takahashi, Y. Kashiwagi, T. D. Tanaka, T. Tachibana, M. Yoshimura, Exogenous ANP treatment ameliorates myocardial insulin resistance and protects against ischemia-reperfusion injury in diet-induced obesity. Int. J. Mol. Sci. 23, 8373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.H. Kimura, T. Nagoshi, Y. Oi, A. Yoshii, Y. Tanaka, H. Takahashi, Y. Kashiwagi, T. D. Tanaka, M. Yoshimura, Treatment with atrial natriuretic peptide induces adipose tissue browning and exerts thermogenic actions in vivo. Sci. Rep. 11, 17466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Tauscher, H. Nakagawa, K. Volker, F. Werner, L. Krebes, T. Potapenko, S. Doose, A. L. Birkenfeld, H. A. Baba, M. Kuhn, β cell-specific deletion of guanylyl cyclase A, the receptor for atrial natriuretic peptide, accelerates obesity-induced glucose intolerance in mice. Cardiovasc. Diabetol. 17, 103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.K. Michel, M. Herwig, F. Werner, K. Spiranec Spes, M. Abesser, K. Schuh, S. Dabral, A. Mugge, H. A. Baba, B. V. Skryabin, N. Hamdani, M. Kuhn, C-type natriuretic peptide moderates titin-based cardiomyocyte stiffness. Insight 5, e139910 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. J. Bubb, A. A. Aubdool, A. J. Moyes, S. Lewis, J. P. Drayton, O. Tang, V. Mehta, I. C. Zachary, D. J. Abraham, J. Tsui, A. J. Hobbs, Endothelial C-type natriuretic peptide is a critical regulator of angiogenesis and vascular remodeling. Circulation 139, 1612–1628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.F. Xue, J. Cheng, Y. Liu, C. Cheng, M. Zhang, W. Sui, W. Chen, P. Hao, Y. Zhang, C. Zhang, Cardiomyocyte-specific knockout of ADAM17 ameliorates left ventricular remodeling and function in diabetic cardiomyopathy of mice. Signal Transduct. Target. Ther. 7, 259 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.X. Xu, H. Jiang, Y. Lu, M. Zhang, C. Cheng, F. Xue, M. Zhang, C. Zhang, M. Ni, Y. Zhang, Deficiency of NONO is associated with impaired cardiac function and fibrosis in mice. J. Mol. Cell. Cardiol. 137, 46–58 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Table S1 and S2