SUMMARY
The role of genome organization in the control of gene expression persists as a central problem of regulatory biology. Most efforts have focused on the role of CTCF enriched boundary elements and TADs, which enable long-range DNA-DNA associations via loop extrusion processes. However, there is increasing evidence for long-range chromatin loops between promoters and distal enhancers formed through specific DNA sequences, including tethering elements, which bind GAGA-Associated Factor (GAF). Previous studies showed that GAF possesses amyloid properties in vitro, bridging separate DNA molecules. In this study we investigated whether GAF functions as a looping factor in Drosophila development. We employed Micro-C assays to examine the impact of defined GAF mutants on genome topology. These studies suggest that the N-terminal POZ/BTB oligomerization domain is important for long-range associations of distant GAGA-rich tethering elements, particularly those responsible for promoter-promoter interactions that coordinate the activities of distant paralogous genes.
eTOC Blurb
How genome organization controls gene expression remains unsolved. Li et al. present evidence that the pioneer factor GAF mediates long-range promoter-promoter interactions in the Drosophila genome. The POZ domain of GAF was found to be particularly important for these loops, and inactivation of this domain alters gene expression.
Graphical Abstract
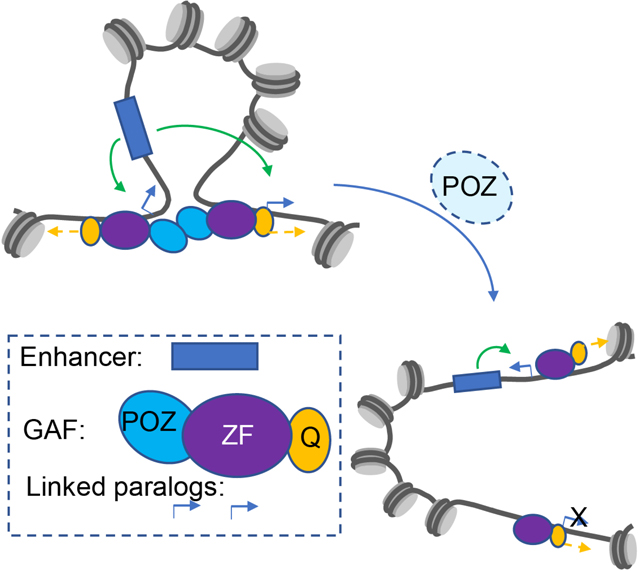
INTRODUCTION
How enhancers control gene expression over distances of tens to hundreds of kilobases persists as a major challenge in regulatory biology 1–5. A variety of studies have identified potential looping factors that might participate in this process, including YY1, Lbd1, and CTCF 6. Here, we explore the role of GAF (GAGA-Associated Factor) encoded by Trithorax-like (Trl) in the long-range associations of “tethering elements” that foster enhancer-promoter and promoter-promoter interactions during Drosophila development 7,8. Genome databases suggest that GAF binds to most or all of the nearly 400 tethering elements scattered across the Drosophila genome 7,8.
Past in vitro assays suggested that GAF possesses amyloid properties by virtue of an N-terminal POZ/BTB oligomerization domain 9–11 and a C-terminal low complexity moiety (Q-rich) 12,13. GAF aggregates were shown to mediate a potential looping activity by bridging separate DNA molecules 9,14. In addition, GAF can stimulate gene expression by distal regulatory sequences in yeast, consistent with its role as a looping factor 15. Here, we explore the possibility that GAF serves as an endogenous looping factor in Drosophila embryos and imaginal discs.
Using a combination of multiple quantitative measurements, including Micro-C assays, CUT&Tag, RNA-seq and HCR in situ detection methods, we present evidence that GAF-GAF associations are important for long-range looping of a subset of tethering elements, particularly those located in promoter-proximal regions that connect distant paralogous genes (Levo et al. 2022). We also show that mutant forms of GAF lacking a functional BTB/POZ oligomerization domain cause a disproportionate reduction in long-range looping as compared with mutants lacking the Q-rich moiety. We discuss the significance of these findings for long-range gene regulation.
RESULTS
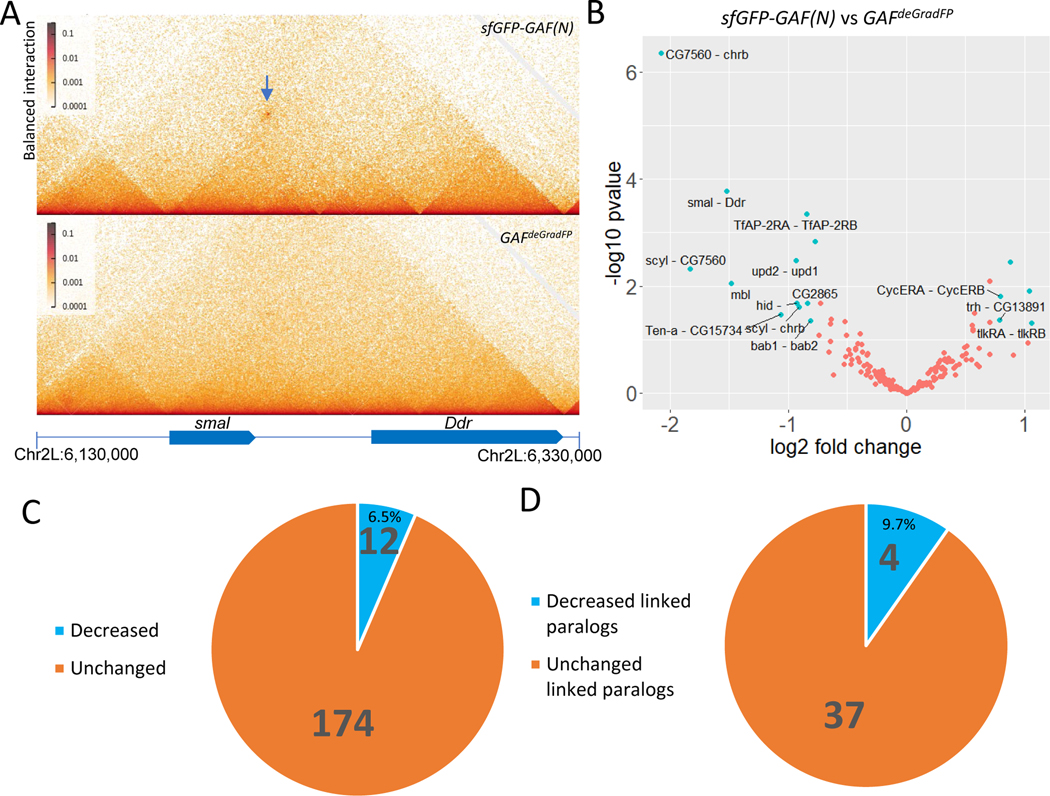
We used high resolution Micro-C assays to examine the consequences of depleting GAF in 2–2.5 hr Drosophila embryos (Fig. 1A, 1B). Depletion was achieved by expressing an F-box protein fused to a nanobody recognizing GFP to ubiquitinate GFP-tagged GAF, which was highly expressed in 0–2 hr embryos using the nanos (nos) promoter 16. The ubiquitinated GFP-GAF is degraded by the proteasome complex 17, and depletion was as efficient as previously reported (Fig. S1A, S1B, S1C). Whole-genome analysis identified losses of 12 of 186 long-range focal contacts in early embryos 8(Fig. 1B, 1C). Many of these occur at linked paralogous genes (also known as “topological operons” 8) (Fig. 1D). For example, the promoter regions of the smal (smoke alarm) and Ddr (Discoidin domain receptor) paralogs are associated by tethering elements (Fig. 1A; arrow, top panel). This contact is lost upon depletion of GAF (Fig. 1A, bottom panel).
Fig. 1.
Contact changes in Trl knockout embryos. (A)The contact between paralog smal and Ddr is demolished (Arrow pointing to the contact); (B) Significantly changed contacts in Trl knockout embryos (labeled and in cyan); (C) Fraction of changed or unchanged contacts in Trl knockout embryos; (D) Fraction of changed or unchanged linked paralog contacts in Trl knockout embryos.
We observed similar losses or reduction of long-range focal contacts at a number of additional linked paralogs, including scyl (scylla)/chrb (charybde), bab1/2 (bric a brac 1/2), and upd1/2 (unpaired 1/2) (Fig. 1B). Moreover, several loops connecting distant intergenic regions with specific target promoters are also affected, as seen for the hid (head involution defective) locus (Fig. S1D). These observations raise the possibility that GAF functions as a looping factor in vivo. We further explored this possibility by examining mutations in the endogenous Trl locus (Fig. 2).
Fig. 2.
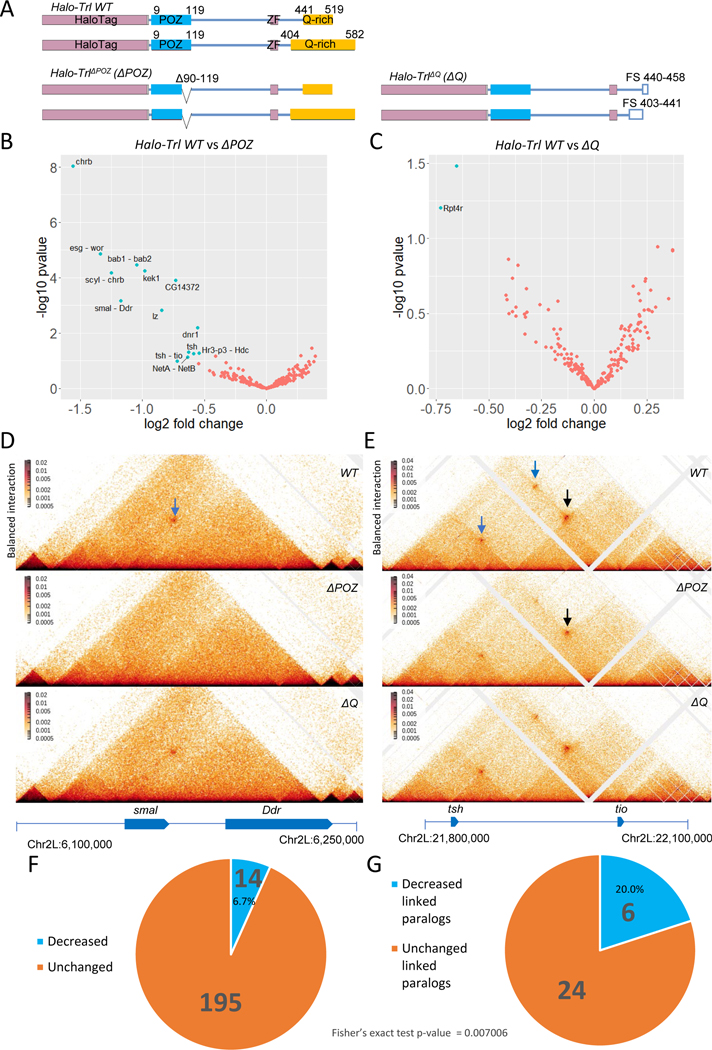
ΔPOZ but not ΔQ deletion causes decreased level of some contacts. (A) Illustration of Halo-Trl WT, ΔPOZ and ΔQ (ZF: Zinc Finger, FS: Frame Shift) ; Significantly changed (cyan) contacts in ΔPOZ (B) and ΔQ (C); Example Micro-C maps of decreased contacts between smal and Ddr (D) and tsh and tio (E) in wing imaginal discs (Arrow pointing to the contacts and the reduced ones are in blue); (F) Fraction of changed or unchanged contacts in ΔPOZ; (G) Fraction of changed or unchanged linked paralog contacts in ΔPOZ.
Previous studies demonstrated that high concentrations of bacterially expressed GAF can form amyloid fibrils in vitro and multimers link separate DNA molecules, as visualized by EM studies 9,13. These behaviors were shown to depend on two domains in GAF, the N-terminal BTB/POZ oligomerization domain, and the C-terminal LCR/IDR (Low Complexity Region / Intrinsically Disordered Region) containing a series of glutamine repeats (Q-rich) 9–13,18. To determine whether either domain is important for the formation of chromosomal loops in vivo, we used previously manipulated endogenous alleles of Trl (summarized in Fig. 2A) 19. A segment of DNA encoding the Halo fluorophore moiety was attached to the 5’ end of the GAF coding sequence and a series of deletions were introduced, including removal of 30 codons in the POZ coding sequence (ΔPOZ), as well as mutations causing frame shifts and truncations of the C-terminal Q-rich moiety (ΔQ). Homozygotes of the Halo-tagged wild-type and ΔQ alleles are viable, whereas ΔPOZ mutants are lethal during pupation. They nonetheless survive to sufficiently late stages to permit analysis of wing imaginal discs derived from climbing third instar larvae. We examined discs rather than embryos due to the high maternal stores of GAF that persist during early development.
High-resolution Micro-C contact maps were determined for both wild-type and mutant wing discs. These maps reveal a number of losses or reductions of focal contacts in ΔPOZ mutants (Fig. 2B, 2F), but most of these are unchanged in ΔQ (Fig. 2C). As seen in early embryos, there is a disproportionate loss of loops linking the promoter regions of paralogous genes (Fig. 2B, 2G). For example, there is a loss of the focal contacts linking smal and Ddr (Fig. 2D), as seen upon GAF depletion in embryos (Fig. 1A). We also observed a significant reduction in promoter-promoter focal contacts connecting tsh and tio (Fig. 2E, blue arrow on top). Interestingly, there are reduced associations of the tsh/tio intergenic region with the tsh promoter (Fig. 2E, blue arrow lower left), whereas focal contacts linking this region to tio is not significantly altered (Fig. 2E, black arrows). Altogether, 14 of 209 focal contacts are significantly reduced (6.7%), and 6 of these 14 altered focal contacts (>40%) link paralogous genes (Fig. 2F, G), a statistically significant bias (Fig. 2G; Fisher’s exact test p < 0.01). The contact maps also suggest that ΔPOZ mutants exhibit reduced insulation activities of anchor sequences at altered paralogs (Fig. S2A), but not unaffected loop anchors (Fig. S2A) or non-paralog loop anchors (Fig. S2B). These results highlight the asymmetric impact of POZ mutants on promoter-promoter associations as compared with all other focal contacts.
ChIP-seq datasets from previous publications 20–22 identified a number of transcription factors at tethering elements, including GAF and a few components of the Polycomb PRC1 complex such as Polyhomeotic (Ph) and Polycomb (Pc). These proteins are preferentially enriched at promoter-promoter focal contacts as compared with intergenic-promoter contacts (Fig. S2C). There is a particular enrichment of GAF and Ph at linked paralogs. They are more enriched at altered linked paralogs, as compared with unaltered focal contacts (Fig. S2D). By contrast, Pleiohomeotic (Pho) has a higher enrichment at unchanged loop anchors as compared with those that are reduced in ΔPOZ mutants (Fig. S2D). It would therefore appear that the most severely affected tethering elements mediating promoter-promoter contacts contain the highest levels of GAF.
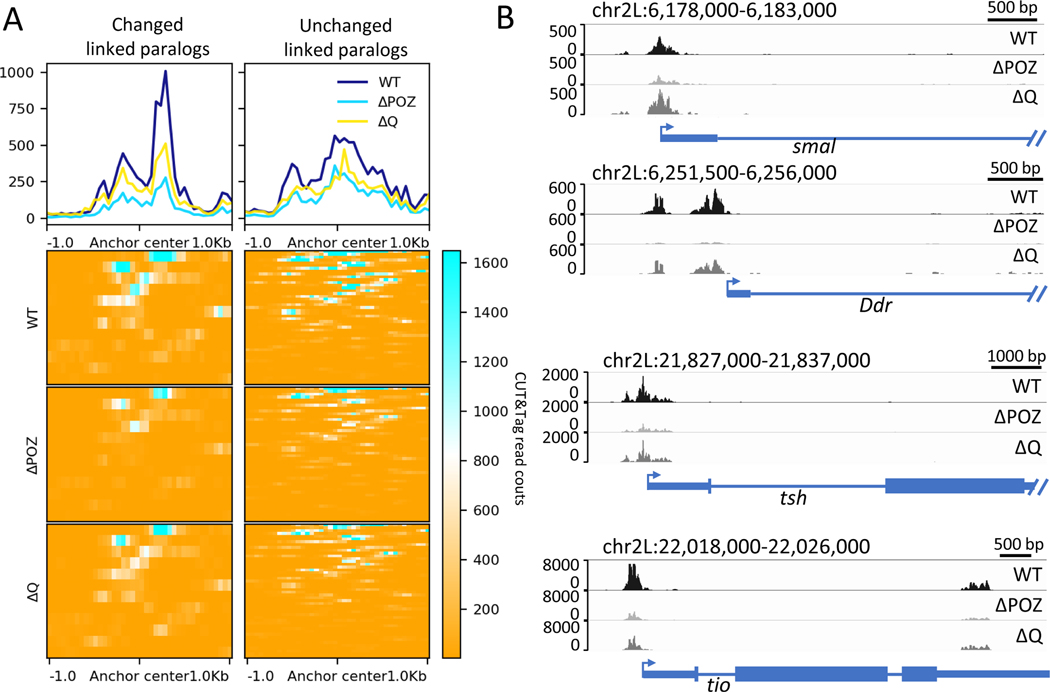
CUT&Tag assays were used to obtain a more quantitative assessment of the relationship between GAF binding and tether-tether loops. These experiments were performed with wing imaginal discs from Halo-tagged lines using anti-Halo antibodies. Over 85% of the peaks seen for all three forms of the GAF protein that were analyzed—wild-type (WT), ΔPOZ and ΔQ--contain at least one GAGAG motif (Fig. S3A), indicating there is no change in binding specificity for mutant proteins containing an intact DNA binding domain. Nonetheless, enrichment of ΔPOZ and ΔQ is significantly reduced when compared with WT peaks (Fig. S3B, S3D). The ΔPOZ mutant displays disproportionately lower median enrichments across non-overlapping GAGAG elements (Fig. S3C), consistent with previous findings that the POZ multimerization domain promotes cooperative binding at clustered GAGAG motifs 9.
There are higher levels of GAF at those loops that are lost or diminished in POZ mutants, as compared with loop anchors that are unaltered in these mutants (Fig. 3 and Fig. S2D). ΔPOZ proteins are greatly reduced at affected tethering elements (Fig. 3 and Fig. S3E, S3F), closely correlating with the loss of the corresponding loops in mutant wing discs. ΔQ mutants displayed milder reductions in binding at affected tethering elements, but did not cause significant changes in the resulting Micro-C contact maps (Fig. 3 and Fig. S3E, S3F). Moreover, there is a statistically significant correlation between reductions in the levels of ΔPOZ proteins at loop anchors and losses in corresponding focal contacts (Fig. S4A). By contrast, there is no such correlation for the ΔQ mutant (Fig. S4B), although ΔQ also affects GAF binding to DNA (Fig. 3 and Fig. S3B, S3D). These observations are consistent with the possibility that GAF-GAF interactions mediated by the POZ oligomerization domain are essential for both cooperative binding to tethering elements and long-range associations of these elements at linked paralogs (see Discussion).
Fig. 3.
POZ domain deletion reduced GAF binding to contact anchors dramatically. (A) Enrichment of CUT&Tag reads for different form of GAF proteins around the contact anchors. (B) Example of IGV views of genomic loci smal, Ddr, tsh, and tio bound by Halo-Trl WT and mutants.
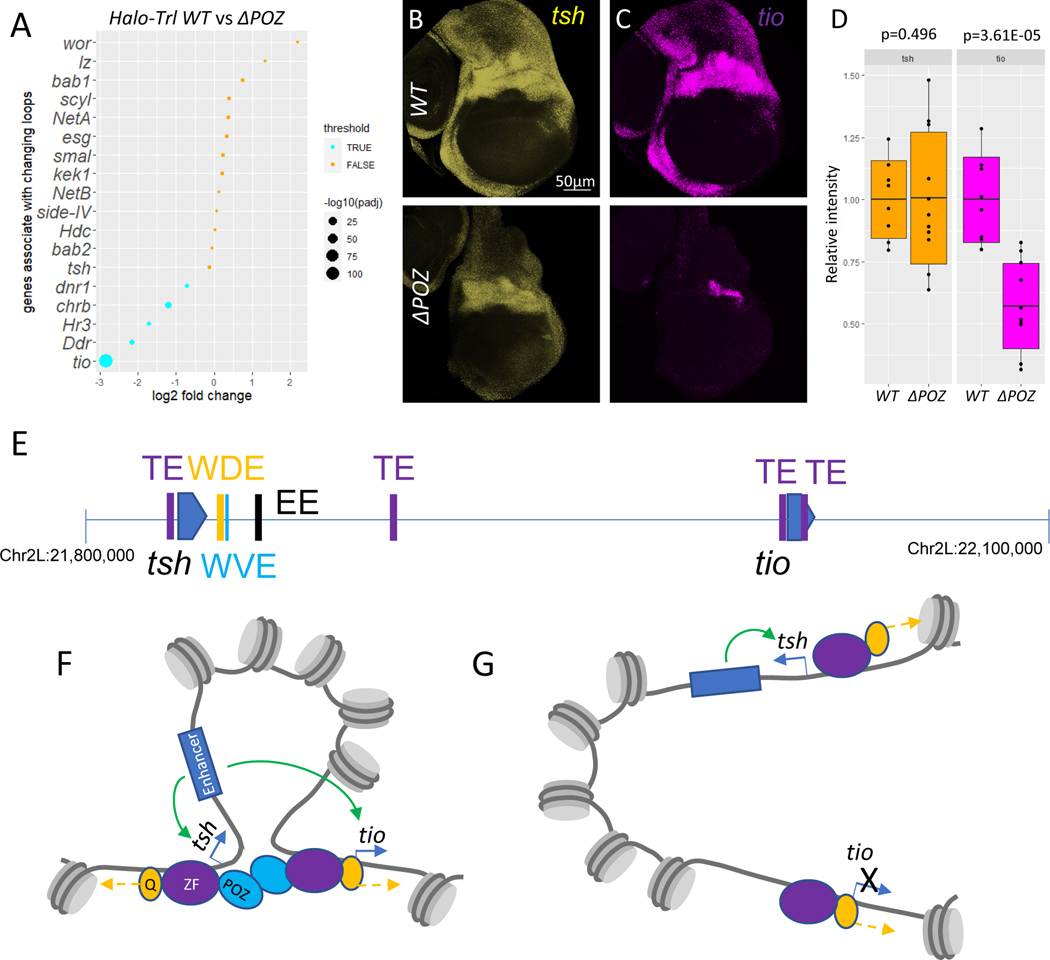
We next asked whether the loss of long-range loops in POZ mutants has a functional consequence on gene expression. RNA-Seq assays were performed on WT and POZ mutant wing imaginal discs. These assays identified substantial reductions in the expression of a number of linked paralogs (Fig. 4A). POZ mutants also exhibit changes in the expression of another ~1700 genes (Fig. S4C), and GAF binding is also reduced near the transcription start sites of these genes (Fig. S4D). The most affected paralog in ΔPOZ mutants is tiptop (tio), which is linked to teashirt (tsh) (Fig. 4A). There is a significant reduction in the promoter-promoter contacts of the two genes, which are separated by over 200 kb (Fig. 2E). Past studies have identified a series of shared wing margin enhancers that map just ~10–20 kb downstream of the tsh transcription start site, but are located over 150 kb away from tio (Fig. 4E) 23. Whole mount Hybridization Chain Reaction in situ fluorescent hybridization (HCR-FISH) assays reveal a significant loss of tio (Fig. 4C, 4D) expression in POZ mutant discs as compared with tsh (Fig. 4B, 4D). These results suggest that the long-range loops mediated by GAF are functionally significant.
Fig. 4.
Some of the contact changes correspond to the differential transcription in ΔPOZ. (A) Dotplot on transcription changes in ΔPOZ wing imaginal discs; HCR shows different expression level of tsh (yellow) (B) and tio (magenta) (C) and quantification (D) in wing imaginal discs of Halo-Trl WT (n=8) and ΔPOZ (n=11); The scale bar in B is 50μm; Boxplot in D are represented as mean ± SD; (E) Genome elements within tsh-tio loci Purple (TE): tethering elements; Orange (WDE): Wing disc dorsal hinge enhancer; Cyan (WVE): Wing disc ventral hinge enhancer; Black (EE): Embryonic enhancer; (F) Chromatin loop brings tio to distant enhancer through Trl/GAF POZ domain; (G) Loss of POZ domain disrupt the loop and reduced tio transcription. Blue arrow: active transcription; Green arrow: enhancer activates targe gene; Yellow arrow: chromatin remodeling.
DISCUSSION
Previous molecular and biochemical assays provided initial evidence that GAF can function as a looping factor in vitro 9,14, although it is primarily known as a pioneer factor that facilitates chromatin accessibility in close collaboration with chromatin remodelers 17,19,24–26. Using high-resolution Micro-C assays, we have provided evidence that it also participates in the formation of a subset of loops seen in Drosophila embryos and wing imaginal discs. The evolutionarily conserved POZ/BTB multimerization domain, but not the intrinsically disordered Q-rich moiety, was shown to be responsible for these long-range contacts, particularly long-range associations of the promoter regions of paralogous genes.
Overall, 20% of the best characterized linked paralogs are disrupted in Trl mutants (Fig. 2G), as compared with only 4.5% of all other loops (Fig. S4E). The enrichment of GAF at promoter-proximal tethering elements might reflect its dual role as a pioneer factor and looping factor (Fig. 4F, 4G). It fosters the formation of loops and also recruits chromatin remodelers to create accessible space for transcription initiation. Deletion of the POZ domain and the accompanying reduction of GAF multimers results in disruption of focal contacts and reduced levels of transcription by distant enhancers (e.g., Fig. 2E, 4C, 4D, 4G). It is possible that the ΔPOZ mutant retains at least some pioneering activity at target promoters. Although the total number of GAGAG repeats within all anchors is similar (Fig. S4F), affected anchors appear to contain a higher density of motifs than unaffected tethers (Fig. S4G), consistent with the cooperative binding activities of GAF 9,27.
The detailed mechanisms by which the POZ domain supports long-range tether-tether associations remains uncertain. There are two nonexclusive possibilities. POZ might permit GAF to form oligomers on individual tethering elements that subsequently interact to form tether-tether loops. Alternatively, it might help recruit additional factors required for looping. We favor the view that GAF functions as a looping factor since there is no correlation between diminished binding of ΔQ mutants and looping (Fig. S4B). These results suggest that GAF influences the formation of loops over and above its DNA binding activities.
It seems likely that additional factors contribute to tether-tether interactions since most loops are unaffected in Trl mutants. Indeed, we observe a number of additional proteins associated with tethering elements, including Polycomb group proteins (Fig. S2C). Interestingly, Pho is preferentially enriched at anchors that are not disrupted by the ΔPOZ mutation (Fig. S2D), raising the possibility that it might serve as a looping factor at these anchors. Divergent protein binding profiles at different anchors raise the possibility of diverse subgroups of loops. We have focused on the subgroup that is formed by GAF, while others might depend primarily on Pho. Consistent with this possibility is the previous observation that Polycomb proteins can form repressive loops 28. Polycomb has also been implicated as a looping factor in mammalian stem cells 29.
There are additional candidate looping factors, including Clamp, Bab1/2 and Pipsqueak (Psq), which are known as oligomerizing proteins that bind to GA-repeats 30–37. Interestingly, Bab1/2 and Psq also contain POZ domains and can physically interact with GAF 30,38. These observations raise the possibility that mammalian transcription factors containing POZ domains, such as PLZF (promyelocytic leukemia zinc finger), could be involved in the formation of loops 39. It is possible that vertebrates also employ POZ-mediated long-range focal contacts as a mechanism for enhancer-promoter and promoter-promoter interactions.
LIMITATIONS OF THE STUDY
Our statistical analysis suggests that disruption of the POZ domain, but not the Q-rich moiety, affected the loops, although both mutants reduced GAF binding to the chromatin. These results are consistent with earlier in vitro studies implicating the POZ domain as an important mediator of loop formation. However, definitive evidence depends on the direct uncoupling of chromatin binding and loop formation.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources or reagents should be directed and filled by Michael Levine (msl2@princeton.edu).
Materials availability
All the materials generated in this study are accessible upon request.
Data and code availability
All sequencing data generated by this work has been deposited to GEO with accession code: GSE218168. All the images used for this work has been deposited to Mendeley: DOI: 10.17632/g4w8kxv2xh.2
All the data was analyzed with published pipelines and packages as described in the detailed methods section.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The detailed fly strain and origin of the lines are described in KEY RESOURCE TABLE. All the flies were raised at 25 °C with 12h-12h day-night cycle on standard food. The GAF depletion embryos is obtained by first crossing sfGFP-Trl(N) with nos-deGradFP; sfGFP-Trl(N)/TM6C, then crossing nos-deGradFP/+; sfGFP-Trl(N) female with sfGFP-Trl(N) male.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal Halotag antibody | Promega | G9281 |
|
| ||
| Rabbit anti-mouse IgG H&L | Abcam | ab46540 |
|
| ||
| Ginea pig α-rabbit antibody | Antibodie s online | ABIN101961 |
|
| ||
| Chemicals peptides and recombinant proteins | ||
|
| ||
| DSG (disuccinimidyl glutarate) | Thermo Fisher Scientific | 20593 |
|
| ||
| EGS (ethylene glycol bis(succinimidyl succinate)) | Thermo Fisher Scientific | 21565 |
|
| ||
| E. coli Spike-in DNA | EpiCypher | 18–1401 |
|
| ||
| pAG-Tn5 | EpiCyphe r | 15–1117 |
|
| ||
| Ampure XP beads | Beckman Coulter | A63880 |
|
| ||
| Hoechst 33342 | Sigma-Aldrich | 23491–52-3 |
|
| ||
| UltraPure™ SSC, 20X | Thermo Fisher Scientific |
15557044 |
|
| ||
| ProLong Gold Antifade Mountant | Thermo Fisher Scientific |
P36934 |
|
| ||
| TRIzol Reagents | Thermo Fisher Scientific |
15596026 |
|
| ||
| Critical commercial assays | ||
|
| ||
| HCR buffer set | Molecular Instruments |
www.molecularinstruments.com |
|
| ||
| HCR B1 488 amplifier | Molecular Instruments |
www.molecularinstruments.com |
|
| ||
| HCR B2 546 amplifier | Molecular Instruments | www.molecularinstruments.com |
|
| ||
| Deposited data | ||
|
| ||
| Micro-C for GAF depletion in embryo and Halo-Trl mutants | This study | GSE218168 |
|
| ||
| CUT&Tag for Halo-Trl mutants | This study | GSE218168 |
|
| ||
| RNA-seq for Halo-Trl mutants | This study | GSE218168 |
|
| ||
| Ph, Pc, Pho ChIP-seq | Brown et al., 2018 | GSE102339 |
|
| ||
| GAF ChIP-seq | Oh et al., 2013 | GSE38594 |
|
| ||
| CP190 ChIP-seq | Kyrchano va et al.,2021 | GSE175402 |
|
| ||
| Images for HCR in situ and GAF depletion effeciency | This study | DOI: 10.17632/g4w8kxv2xh.2 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Drosophila melanogaster/ Halo-Trl | Tang et al., 2022 | N/A |
|
| ||
| Drosophila melanogaster/ Halo-Trl ΔPOZ 90−119 /TM6B | Tang et al., 2022 | N/A |
|
| ||
| Drosophila melanogaster/ Halo-Trl ΔQ | Tang et al., 2022 | N/A |
|
| ||
| Drosophila melanogaster/ sfGFP-Trl(N) | Gaskill et al., 2021a | N/A |
|
| ||
| Drosophila melanogaster/ nos-deGradFP; sfGFP-Trl(N)/TM6C | Gaskill et al., 2021a | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| tsh probe | Molecular Instruments | www.molecularinstruments.com |
|
| ||
| tio probe | Molecular Instruments | www.molecularinstruments.com |
|
| ||
| Software and algorithms | ||
|
| ||
| BWA | Li and Durbin, 2009 | https://bio-bwa.sourceforge.net/ |
|
| ||
| pairtools | Goloboro dko et al., 2019 | https://github.com/open2c/pairtools |
|
| ||
| Cooler | Abdennur and Mirny, 2020 | https://github.com/open2c/cooler |
|
| ||
| HiGlass | Kerpedjiev et al., 2018 | https://higlass.io/ |
|
| ||
| SIP | Rowley et al., 2020 | https://github.com/PouletAxel/SIP |
|
| ||
| BEDtools | Quinlan and Hall,2010 | https://bedtools.readthedocs.io/en/latest/ |
|
| ||
| DESeq2 | Love et al., 2017 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
|
| ||
| ggplot2 | Villanueva and Chen, 2019 | https://ggplot2.tidyverse.org/ |
|
| ||
| cooltools | Sergey, 2022 | https://github.com/open2c/cooltools |
|
| ||
| deepTools | Ramírez et al.,2014 | https://deeptools.readthedocs.io/en/develop/ |
|
| ||
| nf-core/cutandrun v2.0 | Cheshire and West | https://nf-co.re/cutandrun |
|
| ||
| SEACR | Meers et al., 2019 | https://github.com/FredHutch/SEACR |
|
| ||
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
|
| ||
| featureCounts | Liao et al., 2014 | https://subread.sourceforge.net/ |
|
| ||
| Bowtie2 | Langmead and Salzberg, 2012 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
|
| ||
| SAMtools | Li et al., 2009 | http://www.htslib.org/ |
|
| ||
| MACS3 | Liu, 2015 | https://github.com/macs3-project/MACS |
|
| ||
| UCSC-tools | Kuhn et al., 2013 | https://genome.ucsc.edu/util.html |
|
| ||
| RNASTAR | Widmann et al., 2012 | https://github.com/alexdobin/STAR |
|
| ||
| ImageJ | Collins, 2007 | https://imagej.nih.gov/ij/ |
|
| ||
| t-test | Microsoft Office Excel | N/A |
|
| ||
| Linear regression | R | https://www.r-project.org/ |
METHOD DETAILS
Micro-C library preparation and data processing
GAF deGradFP depleted embryos and controls were collected on apple juice agar plate and harvested 2–2.5 hr after fertilization. Embryos were dechorionated for 2 min in 50% bleach and rinsed with water. Initial fixation was performed in 3.5 ml PBST (0.1% Triton-X in PBS), 6.5 ml N-heptane and 1 ml fresh 16% formaldehyde in a horizontal shaker for 15 min at 250 rpm. 3.7 ml of 2 M Tris-HCl pH 7.5 was added to terminate fixation. For wing imaginal discs, homozygous wandering 3rd instar larvae carrying Halo tagged GAF were collected and dissected in PBS. 90 discs were collected for each biological replicate and two replicates for each genotype were prepared. Discs were incubated in 500 μL 1% formaldehyde/PBST for 15 min, passively rotating at room temperature. The fixation was terminated by addition of 370 μL 2 M Tris-HCl pH 7.5. Secondary fixations with 3 mM DSG (Thermo Fisher Scientific) and EGS (Thermo Fisher Scientific) were performed as previously described 7,8.
The libraries were sequenced by the Princeton University Genomics Core Facility with NovaSeq S1 100nt Flowcell v1.5. The raw data were mapped onto the Dm6 reference genome as described by bwa 40. Validated paired reads were filtered and finalized using pairtools 41, and further processed into multi-resolution matrix by Cooler 42. HiGlass 43 was used to visualize the map and generate the figures for example contacts. The detailed processing information is described in 7,8.
CUT&Tag library preparation and data processing
CUT&Tag libraries were prepared following the protocol described 44. 12 wing discs from homozygous wandering 3rd instar larvae of Halo-GAF lines were collected for each biological replicate and two replicates for each genotype were prepared. The discs were incubated with 1:100 rabbit polyclonal Halotag antibody (Promega G9281) for Halo-GAF lines, and 12 wildtype discs were incubated with 1:100 rabbit anti-mouse IgG H&L (Abcam ab46540) as an IgG control. Ginea pig α-rabbit antibody (Antibodies online ABIN101961) was used at 1:100 dilution as secondary antibody. 0.1ng of E. coli Spike-in DNA (EpiCypher 18–1401) was added to each sample at the same step where pAG-Tn5 (EpiCypher 15–1117) was added. DNA was purified by phenol-chloroform extraction and ethanol precipitation, then universal i5 primers and uniquely barcoded i7 primers were added to the DNA with 14 cycles of PCR. Individual libraries were purified with Ampure XP beads (Beckman Coulter A63880) at 1.3:1 ratio and eluted with 22 μL 10mM Tris-HCL, pH8. Mixed barcoded libraries were purified again with Ampure XP beads at 1.8:1 ratio and eluted with 20 μL μL 10mM Tris-HCL, pH8, the libraries were concentrated and the residual primers were eliminated after this step.
The libraries were sequenced by Johns Hopkins Genomics Core Facility with Illumina MiSeq. CUT&Tag data was processed using an integrated analysis pipeline for CUT&RUN and CUT&Tag, nf-core/cutandrun v2.0 45, with relax peak calling using SEACR 46. Non-overlapping GAGAG elements throughout the Drosophila genome were identified with HOMER seq2profile.pl and scanMotifGenomeWide.pl 47; Occurrences of GAGAG elements within peak or loop anchor regions were quantified with bedtools intersect 48. Read counts for WT, ΔPOZ, and ΔQ in WT peaks were generated by featureCounts 49, normalized by multiplying with the spike-in scaling factor calculated by nf-core/cutandrun 45. The profile of CUT&Tag flanking TSS was generated by deepTools computeMatrix and plotProfile 50. Violin and bar plots were generated by ggplot2 51.
ChIP-seq data processing
ChIP-seq from previous publications 20–22,52 was mapped to Dm6 reference genome with bowtie2 53 and sorted with SAMtools 54. The signals were then normalized to input by MACS3 55 and converted to Bigwig format by bedgraphToBigwig from UCSC-tools 56. The profile and heatmap was generated by deepTools 50.
RNA-sequencing
30 wing imaginal discs were dissected from wandering 3rd instar larvae of Halo tagged lines in PBS and transferred to TRIzol (Thermo Fisher Scientific) for each biological triplicate. The total RNA was extracted based on standard protocols. The library preparation and sequencing were performed by the Princeton University Genomics Core Facility.
In situ HCR
The split probes were designed and generated by Molecular Instruments. The HCR buffers and HCR amplifiers were also purchased from Molecular Instruments. The tsh probe was associated with B1 amplifier sequences and tio probe was associated with B2 amplifier sequences.
Wandering 3rd instar larvae were dissected in PBST and fixed in 4% formaldehyde/PBST for 15min at room temperature. The sample was briefly rinsed by PBST twice and resuspended in 100% methanol at room temperature. The sample was passively rotated for 5min and treated with 75% methanol/PBST, 50% methanol/PBST, 25% methanol/PBST at room temperature for 5min each. The samples were rinsed in PBST 3 times at room temperature for 5min. Then the samples were processed as described in https://files.molecularinstruments.com/MI-Protocol-RNAFISH-GenericSolution-Rev7.pdf with small modifications. Volumes for all the steps were reduced to 100 μL and concentrations for the probes were adjusted to 16 pmol. The hybridization incubation time was reduced to 90min. Hoechst 33342 was diluted in 5xSSCT at 1 μg/ml final concentration to label nuclear and applied at the first 30min final washing step. The sample was mounted in ProLong Gold Antifade Mountant.
Imaging
The discs were imaged by a Zeiss LSM 880 confocal microscope (Zen software 2.3 SP1) with a Plan-Apochromat ×40/1.3 N.A. oil-immersion objective. For sfGFP-GAF depletion related images, laser at 488nm was used. The z-stack was taken 1.71 μm apart for 18–26 steps and all steps were projected for the final figures and quantification. For the HCR in situ, three laser lines at 405 nm for Hoechst, 488 nm for B1–488 amplifier and 561 nm for B2–546 amplifier were used. The z-stack was taken 2 μm apart for 18–22 steps and 10 steps in the middle sections were projected for the final figures, 25 steps in the middle section were projected for quantification. The pictures were adjusted in ImageJ 57.
QUANTIFICATION AND STATISTICAL ANALYSIS
Systematic loop calling and differential looping analysis
We used previous lists of loops in the early embryo for our analysis 8. Focal contacts in wing imaginal discs expressing Halo-GAF were detected by SIP 58 using newly created Micro-C maps. The parameters were set as -g 4.0 -min 2.0 -max 2.0 -mat 5000 -d 15 -res 400 –sat 0.01 -t 2800 -nbZero 4 -factor 2 -fdr 0.1 -del true -cpu 16 -isDroso false. The called loops were further filtered with a “value” > 4.25 and with enough micro-C data coverage (filtered out anchors with marginals < 500). The loops were further filtered by removing loops associated with high CP190 (>10000) 52 by intersecting with BEDtools 48. The loops were first mapped to transcription start sites within 2500 bp of the loop anchor center using BEDtools 48. They were subsequently manually inspected to determine whether linked promoters correspond to paralogs.
The standard loop lists were used to obtain paired reads from individual samples. The dumped matrix with 100bp bin size from the cooler files were compared with the standard loop list with anchors normalized to 1.6kb for embryos and 3.2kb for discs using the pairToPair function from BEDtools 48. The reads were summed for each standard loop and summarized into a count table. The count tables were imported for differential looping analysis with DESeq2 59. P-value < 0.05 and log2FoldChange > 0.77 or < −0.77 was used for significant looping changes in embryo samples and p-value < 0.12and log2FoldChange > 0.5 or < −0.5 was used for changes in wing discs. The differential looping results were volcano plotted by ggplot2 51.
The insulation score was obtained with 3.2kb window from 400bp resolution Micro-C maps by cooltools 60 and plotted with deepTools 50.
Correlation between protein binding and looping level analysis
Correlation between log2FoldChange of Micro-C contact counts and log2FoldChange of CUT&Tag read counts were tested with lm (linear regression) function and plotted in R 61.
RNA-sequencing analysis
The sequence was aligned to Dm6 with RNASTAR 62 and count table was generated by featureCounts 49. The differential gene analysis was performed with DESeq2 59. Significantly differentially expressed genes were defined by adjusted p-value < 0.05. The results were visualized by ggplot2 51.
Imaging quantification
The total intensity of each channel was measured in ImageJ 57. The statistic was performed in excel with t.test, one-tailed, unpaired and plotted by ggplot2 51. The number of measured embryo and wing imaginal discs are included in the figure legend of Fig. S1 and Fig. 4. The individual measurements were included as single dots in Fig. S1C and Fig. 4D, and a boxplot was used in each panel to present mean ± SD as described in responding figure legends.
Supplementary Material
Highlights.
GAF mediates long-range promoter-promoter interactions in the Drosophila genome
Deletion of the POZ domain results in the loss of promoter-promoter interactions
Loss of long-range loops correlates with reductions in gene expression
ACKNOWLEDGEMENTS
We thank Melissa Harrison and Marissa Gaskill for sharing degradFP-GAF reagents for the depletion of maternal GAF. We also thank Michal Levo for invaluable advice and support. We are grateful to Kami Ahmad, David Mohr, and Yiming Andy Mao for advice and assistance on CUT&Tag library preparation and sequencing. This work was funded by grants from the NIH, U01 DA047730 and R35 GM118147 to ML and U01 DK127432 to CW. This study represents a collaboration that was fostered by the 4D Nucleome project.
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Long HK, Prescott SL, and Wysocka J. (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, and Corces VG (2017). Evolutionarily conserved principles predict 3D chromatin organization. Molecular cell 67, 837–852. e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robson MI, Ringel AR, and Mundlos S. (2019). Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Molecular cell 74, 1110–1122. [DOI] [PubMed] [Google Scholar]
- 4.Hou C, Li L, Qin ZS, and Corces VG (2012). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Molecular cell 48, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, and Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. [DOI] [PubMed] [Google Scholar]
- 6.Furlong EE, and Levine M. (2018). Developmental enhancers and chromosome topology. Science 361, 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batut PJ., Bing XY, Sisco Z, Raimundo J, Levo M, and Levine MS (2022). Genome organization controls transcriptional dynamics during development. Science 375, 566–570. 10.1126/science.abi7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levo M, Raimundo J, Bing XY, Sisco Z, Batut PJ, Ryabichko S, Gregor T, and Levine MS (2022). Transcriptional coupling of distant regulatory genes in living embryos. Nature 605, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsani KR, Hajibagheri MA, and Verrijzer CP (1999). Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. The EMBO Journal 18, 698–708. 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell VJ, and Treisman R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes & Development 8, 1664–1677. 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 11.Espinas ML, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, and Azorin F. (1999). The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. Journal of Biological Chemistry 274, 16461–16469. 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins RC, and Lis JT (1999). DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. Journal of molecular biology 285, 515–525. [DOI] [PubMed] [Google Scholar]
- 13.Agianian B, Leonard K, Bonte E, Van der Zandt H, Becker PB, and Tucker PA (1999). The glutamine-rich domain of the Drosophila GAGA factor is necessary for amyloid fibre formation in vitro, but not for chromatin remodelling. Journal of Molecular Biology 285, 527–544. 10.1006/jmbi.1998.2355. [DOI] [PubMed] [Google Scholar]
- 14.Mahmoudi T, Katsani KR, and Verrijzer CP (2002). GAGA can mediate enhancer function in trans by linking two separate DNA molecules. The EMBO Journal 21, 1775–1781. 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, and Barberis A. (2005). DNA looping induced by a transcriptional enhancer in vivo. Nucleic acids research 33, 3743–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, and Lehmann R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637–647. [DOI] [PubMed] [Google Scholar]
- 17.Gaskill MM, Gibson TJ, Larson ED, and Harrison MM (2021). GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. eLife 10, e66668. 10.7554/eLife.66668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonchuk A, Denisov S, Georgiev P, and Maksimenko O. (2011). Drosophila BTB/POZ Domains of “ttk Group” Can Form Multimers and Selectively Interact with Each Other. Journal of Molecular Biology 412, 423–436. 10.1016/j.jmb.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 19.Tang X, Li T, Liu S, Wisniewski J, Zheng Q, Rong Y, Lavis LD, and Wu C. (2022). Kinetic principles underlying pioneer function of GAGA transcription factor in live cells. Nature Structural & Molecular Biology, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, and Irvine KD (2013). Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell reports 3, 309–318. 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JL., Sun M. a., and Kassis JA (2018). Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho. Proceedings of the National Academy of Sciences 115, E1839–E1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevil M, Bondra ER, Schulz KN, Kaplan T, and Harrison MM (2017). Stable binding of the conserved transcription factor grainy head to its target genes throughout Drosophila melanogaster development. Genetics 205, 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soanes KH, and Bell JB (2001). The drosophila aeroplane mutant is caused by an I-element insertion into a tissue-specific teashirt enhancer motif. Genome 44, 919–928. [DOI] [PubMed] [Google Scholar]
- 24.Chetverina D, Erokhin M, and Schedl P. (2021). GAGA factor: a multifunctional pioneering chromatin protein. Cellular and molecular life sciences. CMLS 78, 4125–4141. 10.1007/s00018-021-03776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judd J, Duarte FM, and Lis JT (2021). Pioneer-like factor GAF cooperates with PBAP (SWI/SNF) and NURF (ISWI) to regulate transcription. Genes & Development 35, 147–156. 10.1101/gad.341768.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moshe A, and Kaplan T. (2017). Genome-wide search for Zelda-like chromatin signatures identifies GAF as a pioneer factor in early fly development. Epigenetics & chromatin 10, 33. 10.1186/s13072-017-0141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Steensel B, Delrow J, and Bussemaker HJ (2003). Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proceedings of the National Academy of Sciences of the United States of America 100, 2580–2585. 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, and Cavalli G. (2018). Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Molecular Cell 71, 73–88.e75. 10.1016/j.molcel.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 29.Crispatzu G, Rehimi R, Pachano T, Bleckwehl T, Cruz-Molina S, Xiao C, Mahabir E, Bazzi H, and Rada-Iglesias A. (2021). The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nature communications 12, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwendemann A, and Lehmann M. (2002). Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proceedings of the National Academy of Sciences of the United States of America 99, 12883–12888. 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SY, Chang YL, Swamy KB, Chiang RL, and Huang DH (2016). GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics & chromatin 9, 32. 10.1186/s13072-016-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan J, Rieder L, Colonnetta MM, Huang A, McKenney M, Watters S, Deshpande G, Jordan W, Fawzi N, and Larschan E. (2021). CLAMP and Zelda function together to promote Drosophila zygotic genome activation. eLife 10, e69937. 10.7554/eLife.69937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tikhonova E, Mogila V, Georgiev P, and Maksimenko O. (2021). The Role of CLAMP Binding Sites in Maintaining of Distant Interactions in Drosophila Transgenic Lines. Russian Journal of Genetics 57, 1229–1232. [Google Scholar]
- 34.Urban JA., Urban JM, Kuzu G, and Larschan EN (2017). The Drosophila CLAMP protein associates with diverse proteins on chromatin. PLoS ONE 12, e0189772. 10.1371/journal.pone.0189772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaye EG, Booker M, Kurland JV, Conicella AE, Fawzi NL, Bulyk ML, Tolstorukov MY, and Larschan E. (2018). Differential Occupancy of Two GA-Binding Proteins Promotes Targeting of the Drosophila Dosage Compensation Complex to the Male X Chromosome. Cell reports 22, 3227–3239. 10.1016/j.celrep.2018.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodgson JW, Argiropoulos B, and Brock HW (2001). Site-specific recognition of a 70-base-pair element containing d(GA)[[n]] repeats mediates bithoraxoid polycomb group response element-dependent silencing. Molecular and Cellular Biology 21, 4528–4543. 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez-Perez I, Rowley MJ, Lyu X, Valadez-Graham V, Vallejo DM, Ballesta-Illan E, Lopez-Atalaya JP, Kremsky I, Caparros E, Corces VG, and Dominguez M. (2019). Ecdysone-Induced 3D Chromatin Reorganization Involves Active Enhancers Bound by Pipsqueak and Polycomb. Cell reports 28, 2715–2727.e2715. 10.1016/j.celrep.2019.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shokri L, Inukai S, Hafner A, Weinand K, Hens K, Vedenko A, Gisselbrecht SS, Dainese R, Bischof J, Furger E, et al. (2019). A Comprehensive Drosophila melanogaster Transcription Factor Interactome. Cell reports 27, 955–970.e957. 10.1016/j.celrep.2019.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaharbakhshi E, and Jemc JC (2016). Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: Critical regulators of development. genesis 54, 505–518. 10.1002/dvg.22964. [DOI] [PubMed] [Google Scholar]
- 40.Li H, and Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goloborodko A, Abdennur N, and Venev S. (2019). hbbrandao & gfudenberg. mirnylab/pairtools: v0. 2.2. Zenodo. [Google Scholar]
- 42.Abdennur N, and Mirny LA (2020). Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, and Kumar N. (2018). HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome biology 19, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad K, and Henikoff S. (2020). CUT&Tag with Drosophila tissues. [Google Scholar]
- 45.Cheshire C, and West C. Analysis pipeline for CUT&RUN and CUT&TAG experiments that includes QC, support for spike-ins, IgG controls, peak calling and downstream analysis. [Google Scholar]
- 46.Meers MP, Tenenbaum D, and Henikoff S. (2019). Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics & chromatin 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- 50.Ramírez F, Dündar F, Diehl S, Grüning BA, and Manke T. (2014). deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research 42, W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villanueva RAM, and Chen ZJ (2019). ggplot2: elegant graphics for data analysis. Taylor & Francis. [Google Scholar]
- 52.Kyrchanova O, Klimenko N, Postika N, Bonchuk A, Zolotarev N, Maksimenko O, and Georgiev P. (2021). Drosophila architectural protein CTCF is not essential for fly survival and is able to function independently of CP190. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1864, 194733. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T. (2015). MACS–Model-based Analysis of ChIP-Seq. Github. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhn RM, Haussler D, and Kent WJ (2013). The UCSC genome browser and associated tools. Briefings in bioinformatics 14, 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins TJ (2007). ImageJ for microscopy. Biotechniques 43, S25–S30. [DOI] [PubMed] [Google Scholar]
- 58.Rowley MJ, Poulet A, Nichols MH, Bixler BJ, Sanborn AL, Brouhard EA, Hermetz K, Linsenbaum H, Csankovszki G, and Aiden EL (2020). Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome research 30, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Anders S, and Huber W. (2017). Analyzing RNA-seq data with DESeq2. Bioconductor 2, 1–63. [Google Scholar]
- 60.Sergey V, NezarGoloborodko, AntonFlyamer, IlyaFudenberg, GeoffreyNuebler, JohannesGalitsyna, AleksandraAkgol, BetulAbraham, SameerKerpedjiev, Peter Imakaev, Maksim (2022). open2c/cooltools: v0.5.1. 10.5281/zenodo.6324229. [DOI] [Google Scholar]
- 61.Team RC (2013). R: A language and environment for statistical computing. [Google Scholar]
- 62.Widmann J, Stombaugh J, McDonald D, Chocholousova J, Gardner P, Iyer MK, Liu Z, Lozupone CA, Quinn J, and Smit S. (2012). RNASTAR: an RNA STructural Alignment Repository that provides insight into the evolution of natural and artificial RNAs. Rna 18, 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated by this work has been deposited to GEO with accession code: GSE218168. All the images used for this work has been deposited to Mendeley: DOI: 10.17632/g4w8kxv2xh.2
All the data was analyzed with published pipelines and packages as described in the detailed methods section.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.