Abstract
Endomembrane remodeling to form a viral replication complex (VRC) is crucial for a virus to establish infection in a host. Although the composition and function of VRCs have been intensively studied, host factors involved in the assembly of VRCs for plant RNA viruses have not been fully explored. TurboID-based proximity labeling (PL) has emerged as a robust tool for probing molecular interactions in planta. However, few studies have employed the TurboID-based PL technique for investigating plant virus replication. Here, we used Beet black scorch virus (BBSV), an endoplasmic reticulum (ER)–replicating virus, as a model and systematically investigated the composition of BBSV VRCs in Nicotiana benthamiana by fusing the TurboID enzyme to viral replication protein p23. Among the 185 identified p23-proximal proteins, the reticulon family of proteins showed high reproducibility in the mass spectrometry data sets. We focused on RETICULON-LIKE PROTEIN B2 (RTNLB2) and demonstrated its proviral functions in BBSV replication. We showed that RTNLB2 binds to p23, induces ER membrane curvature, and constricts ER tubules to facilitate the assembly of BBSV VRCs. Our comprehensive proximal interactome analysis of BBSV VRCs provides a resource for understanding plant viral replication and offers additional insights into the formation of membrane scaffolds for viral RNA synthesis.
TurboID-based proximity labeling reveals the viral replication complex structure of a plant virus, unveiling a proviral function of RETICULON-LIKE PROTEIN B2 in viral replication complex formation.
IN A NUTSHELL.
Background: Upon entry into host cells, positive-strand RNA viruses hijack host factors to remodel specific organelle membranes to form viral replication complexes (VRCs). VRCs provide an optimal microenvironment for protecting and generating progeny viral RNAs. Identifying host components of VRCs is crucial for understanding viral replication mechanisms and developing virus-resistant plants. However, VRCs are membrane associated and dynamic, making it technically challenging to study VRC components using traditional methods for examining protein–protein interactions in plant cells. The recently emerged TurboID-based proximity labeling (PL) approach has become a powerful tool for examining weak, transient, and dynamic molecular interactions in planta. However, TurboID-based PL has not been employed to investigate VRCs in plants.
Question: Can TurboID-based PL be used to identify key components and interaction networks of VRCs in plants?
Findings: Using Beet black scorch virus (BBSV) as a model, we systematically investigated the constituents of VRCs by fusing the TurboID enzyme to the BBSV replication protein p23. We identified 185 p23-proximal proteins, including several proteins that are known to interact with p23 or to be critical for BBSV replication. The reticulon family proteins were repeatedly identified. We further demonstrated that reticulon-like protein B2 (RTNLB2) interacts with p23 and plays a proviral role in BBSV replication by facilitating the establishment of VRCs. Moreover, RTNLB2 is crucial for replication of several other plant viruses that replicate on endoplasmic reticulum membranes.
Next steps: Whether RTNLB2 is involved in other aspects of virus infection, such as movement, remains to be investigated. Are other p23-proximal proteins involved in replications of BBSV and/or other plant viruses? If so, what are the underlying mechanisms?
Introduction
Plant viral diseases are serious threats to crop production and food security worldwide (Pennazio et al. 1996; Jones and Naidu 2019). A large number of these plant viruses belong to the class of positive-stranded RNA viruses (Scholthof et al. 2011). Usually, infection of host cells by a positive-strand RNA virus can be divided into several stages, including replication, virion assembly, and intracellular and intercellular movement (Saxena and Lomonossoff 2014). Among these steps, viral replication is one of the most crucial steps for the establishment of viral infection in host plants (Nagy and Pogany 2011).
Replication of positive-strand RNA viruses often occurs in organelle membranes (Verchot 2011), such as the endoplasmic reticulum (ER) (Cao et al. 2015), chloroplast (Jin et al. 2018b), mitochondrion (Gomez-Aix et al. 2015), peroxisome (Fernández de Castro et al. 2017), or vacuole (Wang et al. 2021). During replication, the virus remodels the host's endomembrane system, forming a gated and safe compartment that protects the viral genomes from host antiviral mechanisms (Laliberté and Sanfaçon 2010). Rearrangement of host membrane structures to form viral replication complexes (VRCs) is a well-organized and intricate process and requires the participation of numerous host factors (Nagy and Pogany 2011; Verchot 2011; Nagy 2016). Therefore, identifying and mining the host factors involved in VRCs formation is of great importance for in-depth understanding of viral replication and will aid in the development of mechanism-based antiviral strategies (Wang 2015).
During the last several decades, intensive studies have been performed to identify the host factors involved in the replication of plant viruses. For example, based on yeast (Saccharomyces cerevisiae) mutant libraries, many host factors have been found to participate in the replication of tombusviruses and bromoviruses (Diaz and Wang 2014; Wang 2015; Nagy 2016, 2017, 2022; Nagy and Feng 2021). Additionally, a combination of yeast 2-hybrid (Y2H) screening, co-immunoprecipitation (Co-IP), and/or mass spectrometry (MS) was often used to probe the interaction partners of the viral protein of interest. However, as mentioned above, replication proteins of positive-strand RNA viruses usually localize to organelle membranes, which makes the enrichment of VRC-associated proteins by Co-IP less efficient (Ngounou Wetie et al. 2014). Moreover, Co-IP is primarily employed to identify strongly interacting proteins and is not effective in identifying weak or transient protein–protein interactions (PPIs) (Lampugnani et al. 2018). Therefore, Co-IP is unable to capture dynamic PPIs during VRC formation. Y2H screening of viral replicase-interacting proteins requires the construction of a corresponding membrane protein-based yeast library (Iyer et al. 2005), which is laborious and time-consuming. However, the recently developed TurboID-based proximity labeling (PL) approach overcomes some of these drawbacks.
Compared with conventional Y2H and Co-IP methods, TurboID-based PL allows for rapid, nontoxic PL of target proteins in living cells at room temperature (RT) and shows a distinct advantage in identifying transient, weak, or hydrophobic PPIs (Branon et al. 2018; Mair et al. 2019; Zhang et al. 2019; Arora et al. 2020; Yang et al. 2021; Xu et al. 2023). Although TurboID-based PL has been used to dissect the VRCs of several animal viruses (Kumar et al. 2021; Fang et al. 2022a, b; Liu et al. 2022; Wei et al. 2022; Zhang et al. 2022), few studies have employed this technique to analyze VRCs in planta.
Beet black scorch virus (BBSV) is a member of the genus Betanecrovirus in the family Tombusviridae and is a positive-sense, single-stranded RNA virus consisting of 3,644 nucleotides (nt) (Cao et al. 2002). The BBSV genome contains 6 open reading frames (ORFs) that encode 2 replication-associated proteins (p23 and p82), 3 movement-related proteins (p7a, p7b, and p5′), and a coat protein (CP) (Fig. 1A) (Yuan et al. 2006). Among them, p23 is an auxiliary replication protein that cooperates with the viral RNA-dependent RNA polymerase (RdRp) p82 (Yuan et al. 2006). Our previous studies demonstrated that p23 is an ER-localized membrane protein and plays a key role in ER reorganization, a process essential for the formation of ER-invaginated VRCs during BBSV infection (Cao et al. 2015). Moreover, heat shock cognate 70 kDa protein (Hsc70-2) directly participates in BBSV replication by interacting with p23 (Wang et al. 2018). However, except for Hsc70-2, it is unclear what other host factors are involved in the formation of BBSV VRCs.
Figure 1.
Design of PL experiments to define the proximal interactome of the BBSV p23 replication protein and validation of biotinylation efficiency. A) Genome organization of BBSV. BBSV encapsidates a positive-sense, single-stranded genomic RNA (gRNA) of 3,644 nt. The boxes show predicted ORFs in the BBSV genome. P23 and its translational readthrough (RT) product, P82, are key components required for viral replication. Three ORFs (P7a, P7b, and P5′) are essential for viral movement. CP, coat protein. B) Schematic representation of the constructs used for identification of p23-proximal proteins. C) Subcellular localization of TurboID fusions used in this study. Confocal analysis was performed at 2.5 dpi. Scale bars, 10 μm. Arrowhead indicates the ER aggregate. D) Diagram of the experimental design and labeling conditions. Agrobacterium tumefaciens harboring the p23-Citrine-TurboID (p23) or the control 2C1-Citrine-TurboID (2C1) construct was co-infiltrated with Agrobacterium containing the pCB301-BBSV (+BBSV) plasmids or EV (−BBSV) into N. benthamiana leaves. At 60 hpi, 200 µm biotin was infiltrated into the same leaves. Leaf samples were harvested 8 h later. The biotinylated proteins were enriched with streptavidin beads followed by on-bead trypsin digestion and TMT labeling. LC–MS/MS analysis was then performed to identify the biotinylated proteins. Each treatment was conducted with 3 independent biological replicates (n = 3 plants for each replicate). The numbers 126 to 131 above the leaves indicate 6-plex TMT reporter ion masses at m/z 126 to 131. E) Immunoblot analysis of protein expression and biotinylation in the infiltrated leaves as shown in D). Input, total leaf lysate. F) Immunoblot analysis of streptavidin-based enrichment of biotin-tagged proteins. Strep pull-down of biotinylated proteins enriched by streptavidin beads. For E) and F), antibodies used for immunoblot analysis are indicated at the bottom right corner. Coomassie brilliant blue (CBB)–stained Rubisco large subunit (RbcL) is shown as a loading control.
Previously, we established the TurboID-based PL system for probing molecular interactions in planta (Zhang et al. 2019, 2020). Here, we aimed to identify the components constituting BBSV VRCs using TurboID-based PL. We used p23 as the bait protein and mapped the proximal interactome of p23. A number of p23-interacting proteins were identified from Nicotiana benthamiana. We focused on one of the candidates, RETICULON-LIKE PROTEIN B2 (NbRTNLB2), which showed high reproducibility in the MS data sets and whose function in plant virus replication remains uncharacterized. We demonstrated that NbRTNLB2 is recruited to BBSV VRCs by interacting with p23 and determined the key domain that is responsible for the proviral role of NbRTNLB2 in BBSV replication. Our study demonstrates that TurboID-based PL can be used to decipher the VRC composition of a plant virus and sheds light on the biogenesis of virus replication compartments.
Results
Establishment of PL procedures for identifying proteins proximal to BBSV p23
BBSV auxiliary replication protein p23 plays a key role in the formation of BBSV VRCs by inducing ER remodeling (Cao et al. 2015). To gain insight into the proteins that interact with BBSV p23, an improved yellow fluorescent protein (YFP) mutant (Citrine), together with the biotin ligase, TurboID, were fused to the C-terminus of BBSV p23. As a control, we utilized a 27-amino acid fragment derived from the native ER membrane protein cytochrome P450 2C1 (Ahn et al. 1993) to anchor the Citrine-TurboID fusion to the cytosolic face of the ER membrane (Fig. 1B). We then performed confocal microscopy analysis and found that both the p23-Citrine-TurboID and the 2C1-Citrine-TurboID showed a web-like structure and merged well with the mCherry-tagged ER marker HDEL in N. benthamiana leaf tissues when expressed alone (Yao et al. 2011) (Figs. 1C, panels −BBSV, and S1). We must note, however, that p23-Citrine-TurboID also forms a small number of aggregates (Fig. 1C, arrowhead). In leaf tissues infected with BBSV, launched by infiltration of an Agrobacterium tumefaciens culture harboring pCB301-BBSV (Gao et al. 2022), the number of ER aggregates increased in addition to the alterations of the ER membranes (Fig. 1C, panels +BBSV), consistent with what we previously reported (Cao et al. 2015).
To map the proximal interactome of p23 in the context of BBSV replication, Agrobacterium mixtures containing the infectious cDNA clone, pCB301-BBSV (Gao et al. 2022), and p23-Citrine-TurboID or 2C1-Citrine-TurboID were co-infiltrated into N. benthamiana leaves (Fig. 1D, Group I, +BBSV). Leaf tissues were infiltrated with Agrobacterium mixtures carrying either p23-Citrine-TurboID plus empty vector (EV) pCB301 or 2C1-Citrine-TurboID plus pCB301 as the control group (Fig. 1D, Group II, −BBSV). At 60 h post-infiltration (hpi), plant leaves were re-infiltrated with 200 µm biotin. After 8 h of incubation at RT, leaf samples were harvested followed by protein extraction, desalting, and streptavidin-based enrichment (Fig. 1D).
Immunoblot analysis with a homemade anti-TurboID antibody (Supplemental Fig. S2) and horseradish peroxidase-conjugated streptavidin (streptavidin-HRP) confirmed the expression and biotinylation of both p23-Citrine-TurboID and 2C1-Citrine-TurboID in various combinations (Fig. 1E, Input). The biotinylated proteins were efficiently enriched by affinity purification with streptavidin beads (Fig. 1F, Strep pull-down). The enriched proteins were then detached from the streptavidin beads using on-bead digestion, and the resulting peptides were chemically labeled with isotopically distinct tandem mass tag (TMT) followed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis (Fig. 1D).
Proximity proteomics defines the proximal interactome of BBSV p23
Bioinformatics analysis was performed on the raw MS data to map the p23-proximal interactome. Data analysis indicated that a total of 4,045 and 4,177 proteins without decoys and common contaminants were identified for p23-Citrine-TurboID in the presence and absence of BBSV infection, respectively, using Scaffold software for MS (Supplemental Data Sets 1 and 2). In addition, all proteins were visualized by the volcano plot (Fig. 2, A and B). Notably, among the significantly enriched proteins with a threshold of P < 0.05 and log2 (fold change, p23/2C1) ≥ 1.3, heat shock protein 70 (Hsp70), which had been verified to interact with BBSV p23 (Wang et al. 2018), was present in the MS data sets of both Group I (+BBSV) and Group II (−BBSV) (Fig. 2, A and B, blue dot, and Supplemental Data Sets 3 and 4), suggesting the reliability of this p23-proximal interactome generated by TurboID-based PL. Besides Hsp70, it is noteworthy that reticulon (RTN) family proteins were frequently enriched in the p23-Citrine-TurboID purification, either in the presence or absence of BBSV infection (Fig. 2, A and B, red dots, and Supplemental Data Sets 3 and 4).
Figure 2.
Proteomic analysis of biotinylated proteins enriched in p23-TurboID purification. A) and B) Volcano plot showing proteins identified by TurboID-based PL in 3 biologically independent experiments (n = 3 plants for each experiment). Mean log2 fold change of 3 replicates between p23-Citrine-TurboID and the 2C1-Citrine-TurboID control group in the presence of BBSV (+BBSV) or absence of BBSV (−BBSV) infection was plotted against the −log10 padj. The previously reported p23-interacting protein, Hsp70, and the NbRTNLB family of proteins were indicated in the panels. Values are provided in Supplemental Data Sets 1 and 2. C) Heatmap based on significantly upregulated data from individual samples of p23-Citrine-TurboID (n = 3) compared with 2C1-Citrine-TurboID controls (n = 3) in the presence or absence of BBSV infection. The protein clustering tree as indicated on the left was generated by using Gene Cluster 3.0 (de Hoon et al. 2004); unsupervised hierarchic clustering was carried out with average linkage algorithms and uncensored correlation as the similarity metrics. Heat maps were generated in Java Treeview. Relative protein expression values from the highest (red) to the lowest (blue) are shown, as indicated by the lower middle of the color scale bar (−0.8 to +0.8). D) Venn diagram showing overlaps among p23-proximal proteins in the presence or absence of BBSV infection. The RTNL protein, marked in red, represents the RTN protein that was further investigated, while the RTNL proteins in blue indicate other RTN proteins identified by the TurboID-based PL. E) A proximity-based interaction network for some of the proteins enriched in p23-Citrine-TurboID purification. The clusters are color-noted as indicated. Nodes represent individual proteins. Edges between the nodes represent interactions based on STRING with a confidence score equal to or above 0.4. Clusters are labeled on the basis of their common GO biological process assigned in UniProt (https://www.uniprot.org) or annotation in TAIR (https://www.arabidopsis.org). Red nodes represent the proteins derived from the intersection of 2 MS data sets (+BBSV and −BBSV). Blue nodes present the proteins derived from MS data set without BBSV infection (−BBSV). Green nodes represent the proteins derived from MS data set in the context of BBSV infection (+BBSV). Values are provided in Supplemental Data Set 6.
To further screen the p23-proximal proteome, we averaged 3 biological replicates and calculated the P-value to identify proteins that are in close proximity to p23. We then normalized the ratio of abundance of each protein in the p23-Citrine-TurboID samples to that in the 2C1-Citrine-TurboID control group, and obtained value-of-fold changes (Supplemental Data Sets 3 and 4). By comparing the protein level log2-normalized fold change of the p23-Citrine group with the 2C1-Citrine group, as shown in the heatmap (Fig. 2C), 104 and 119 p23-proximal proteins were significantly enriched for both Group I (+BBSV) and Group II (−BBSV), respectively, with a threshold of adjusted P < 0.05 and log2 (fold change) ≥ 1.3. We found 38 proteins that were present in both groups (Fig. 2D and Supplemental Data Set 5).
To gain insight into the functional associations among the proteins identified in close proximity to p23, we integrated data from the 2 sets of p23-proximal proteins obtained in Fig. 2D, resulting in a pool of 185 proteins. Subsequently, we performed Gene Ontology (GO) analysis on this set of proteins. Given the well-established connection between BBSV replication and ER membranes, 36 proteins, which belong to the GO terms of ER system, protein transport [including vesicle-mediated transport and the endosomal sorting complex required for transport (ESCRT)], and protein folding, were selected to construct a protein interaction network using the online STRING tool (https://cn.string-db.org) and Cytoscape software. This network revealed 51 interaction relationships (Supplemental Data Set 6) among the 36 selected proteins, as shown in Fig. 2E. These results suggest involvement of a variety of ER-associated host factors in BBSV replication.
BBSV p23 interacts with NbRTNLB2 on the ER
We noted that members of the RTNLB family of proteins, such as RTNLB2 [log2-normalized fold change (p23/2C1) ≥ 1.39] and RTNLB4 [log2-normalized fold change (p23/2C1) ≥ 1.37] (Fig. 2, A, B, and D, and Supplemental Data Sets 3 and 4), were frequently enriched in the p23-proximal interactome. The RTNLB protein family contains 14 representative putative paralogs proteins in N. benthamiana (Supplemental Fig. S3A), and each paralog has a different number of isoforms (Supplemental Fig. S4 and Data Set 7).
To test whether RTNLB2 and RTNLB4 were involved in BBSV infection, we cloned the full-length coding sequences of RTNLB2.1 and RTNLB4.1 from N. benthamiana and analyzed their effects on BBSV infection by transient overexpression. Compared with RTNLB2.1, which significantly increased BBSV infection upon overexpression, RTNLB4.1 had only a marginal effect on BBSV infection (Supplemental Fig. S5). Therefore, we focused on RTNLB2 for further analysis. Sequence alignment indicated that RTNLB2.1 shared 95.3% amino acid (96.2% nt) sequence identity with RTNLB2.3, and phylogenetic analysis indicated that both of them (Niben101Scf28925 22656-26422 and Niben101Scf01375 252209-256009) showed the highest similarity to Arabidopsis (Arabidopsis thaliana) RTNLB2 (Yang and Strittmatter 2007) (Supplemental Fig. S6); hence, the protein is designated as NbRTNLB2 hereafter (for convenience, NbRTNLB2 is abbreviated as RTNLB2 in the figures).
To investigate the relationship between BBSV p23 and NbRTNLB2, we fused red fluorescent protein (RFP) and cyan fluorescent protein (CFP) protein to the C-terminus of p23 and NbRTNLB2, respectively. Confocal microscopy analysis showed that p23-RFP co-localized well with the ER marker (GFP-HDEL) and induced the formation of aggregates in ER membranes. NbRTNLB2-CFP also localized to ER membranes and induced constriction of ER tubules. When NbRTNLB2-CFP and p23-RFP were co-expressed in leaf tissues, they co-localized well with the ER-derived aggregates (Fig. 3A). Similar results were obtained when we used another combination of fluorescent reporters [blue fluorescent protein (BFP)/green fluorescent protein (GFP)/mCherry red fluorescent protein] to analyze their subcellular localization (Supplemental Fig. S7). Moreover, due to the sequence and structural similarity between Rtn1p of S. cerevisiae and NbRTNLB2 (Supplemental Fig. S3, B and C), we also co-expressed p23 or mCherry-fused p23 and Rtn1p-GFP in yeast cells. Confocal analysis showed an apparent redistribution of Rtn1p-GFP upon p23 expression, and the p23-mCherry co-localized well with the Rtn1p-GFP (Supplemental Fig. S8).
Figure 3.
NbRTNLB2 interacts with BBSV p23 on the ER. A) Subcellular localization of p23-RFP and NbRTNLB2-CFP. Agrobacterium mixtures containing different combinations of constructs as indicated in the panel above were infiltrated into the leaves of GFP-transgenic N. benthamiana plants (Line 16C). Confocal analysis was performed at 2.5 dpi. Figures on the right panel indicate the normalized fluorescence intensities of the CFP (blue), RFP (magenta), and GFP (green) channels along the dashed white lines in the merged images. The relative intensity was calculated by using the Zen software on a Zeiss LSM 880 confocal microscope. Scale bars, 10 μm. B) BiFC assay to test the interaction between NbRTNLB2 and p23 in N. benthamiana cells. Agrobacterium mixtures containing different combinations of constructs as indicated on the top left were infiltrated into N. benthamiana leaves. Fluorescence signals were visualized by confocal microscopy at 2.5 dpi. BioID-YFPn or BioID-YFPc was used as negative controls. mCherry-HDEL served as the ER marker. +BBSV means leaves were agoinoculated with BBSV. Representative results of at least 3 independent experiments (n = 2 plants for each experiment) are shown. Scale bars, 50 μm. C) Y2H assay showing the interaction between p23 and NbRTNLB2. Various Y2H combinations are indicated below. Yeast cells harboring the plasmids pBT3N-p23 + pPR3N-p23, pTSU2-APP + pNubG-Fe65, and pBT3N-p23 + pOst-NubI served as positive controls (iii, iv, and v), and combinations of pBT3-N (EV) + pPR3N-RTNLB2 and pBT3N-p23 + pPR3-N (EV) constructs served as negative controls (ii and vi). SD media lacking corresponding nutrients were indicated beneath the plates. D) Co-IP assays to determine the interaction between NbRTNLB2 and p23. Nicotiana benthamiana leaves expressing NbRTNLB2-GFP and p23-3xFLAG were harvested at 3 dpi. Total proteins were immunoprecipitated with anti-FLAG beads and detected by immunoblotting with anti-GFP or anti-FLAG antibodies.
To test whether p23 interacts with NbRTNLB2 in plant cells, we performed a bimolecular fluorescence complementation (BiFC) assay. Agrobacterium mixtures containing plasmids expressing p23-YFPn and NbRTNLB2-YFPc or p23-YFPc and NbRTNLB2-YFPn were infiltrated into N. benthamiana leaves, either in the presence of BBSV or the presence of the replication-defective mutant of BBSV, BBSVGDDm. At 2.5 d post-infiltration (dpi), confocal microscopy analyses revealed the reconstitution of YFP signals on the ER for both of these combinations. Aggregation of the YFP signals observed in the BiFC assays may be attributed to the overexpression of p23, driven by the 35S promoter, which is consistent with our previous observation of p23's interaction with other proteins (Cao et al. 2015; Wang et al. 2018). As a control, neither p23-YFPn/BioID-YFPc nor p23-YFPc/BioID-YFPn generated observable YFP signals (Figs. 3B and S9A). Expression of all BiFC halves was verified by immunoblot analysis in the infiltrated leaves (Supplemental Fig. S9B).
Furthermore, we employed the split-ubiquitin membrane Y2H system to test the p23-NbRTNLB2 interaction. By using p23 as the bait and NbRTNLB2 as the prey, we observed the growth of yeast colonies containing pBT3N-p23 and pPR3N-NbRTNLB2 on the synthetic defined (SD) media lacking leucine, tryptophan, histidine, and adenine (SD/-Trp-Leu-His-Ade, Fig. 3C, i). As controls, neither the viral p23 protein nor the host NbRTNLB2 protein exhibited auto-activation in the Y2H assay (Fig. 3C, ii and vi). Yeast cells that were transformed with various combinations of plasmids including pTSU2-APP + pNubG-Fe65, pBT3N-p23 + pOst-NubI, and pBT3N-p23 + pPR3N-p23, all of which grew well on the selection plate and served as positive controls (Fig. 3C, iii, iv, and v). Moreover, the Co-IP assay showed that p23-3xFLAG was co-precipitated with RTNLB2-GFP but not with the GFP control (Fig. 3D). Altogether, these results demonstrate that p23 interacts with NbRTNLB2 in yeast cells and ER membranes in plant cells.
NbRTNLB2 acts as a proviral factor for BBSV replication
To characterize the functions of NbRTNLB2 in BBSV infection, we observed NbRTNLB2 localization in healthy and BBSV-infected N. benthamiana leaves. NbRTNLB2 presented a net-like structure and induced constrictions in ER tubules in healthy leaf tissue cells. In contrast, BBSV infection notably altered the subcellular distribution of NbRTNLB2, leading to the formation of multiple aggregates (Fig. 4A). This is consistent with the results from co-expression of RTNLB2 and p23 in plant cells (Fig. 3A) or Rtn1p and p23 in yeast cells (Supplemental Fig. S8). These results indicate that BBSV infection not only alters the structure of the ER membranes but also influences the morphology and distribution of NbRTNLB2.
Figure 4.
NbRTNLB2 is positively involved in BBSV replication. A) Subcellular localization of NbRTNLB2-GFP in healthy and BBSV-infected N. benthamiana leaves at 2.5 dpi. Scale bars, 10 μm. B) Levels of NbRTNLB2 mRNA in healthy and BBSV-infected N. benthamiana leaves as detected by RNA gel blot analysis. Leaves were harvested at different time points as indicated in the panel above. Sys., systemic leaves. Probes used for NbRTNLB2 and BBSV genomic RNA (gRNA) analyses are indicated on the right of each panel. Methylene blue–stained rRNAs served as RNA loading controls. C) The accumulated NbRTNLB2 protein in mock- or BBSV-inoculated N. benthamiana plants as detected by immunoblot analysis. Leaves were harvested at different time points as indicated in the panel above. Coomassie brilliant blue (CBB)–stained RbcL served as a loading control. D) The accumulated BBSV CP in N. benthamiana leaves transiently expressing NbRTNLB2 proteins. Nicotiana benthamiana leaves were infiltrated with Agrobacterium-carrying EV or NbRTNLB2-3xFLAG constructs (OD600 = 0.2 for each construct). One d later, the same leaves were inoculated with 400 ng BBSV virion and samples were collected at 3 dpi for immunoblotting with anti-CP or anti-FLAG antibodies. E) The accumulated BBSV CP in TRV-RTNLB2-inoculated N. benthamiana plants using anti-CP antibody. BBSV-inoculated systemic leaves from TRV-GFP-infected plants served as the control. F) Overexpression of NbRTNLB2 enhances BBSV replication. Nicotiana benthamiana leaves were infiltrated with Agrobacterium-carrying EV or Pro35S:NbRTNLB2-3xFLAG construct (OD600 = 0.2 for each construct). One d later, the movement-deficient BBSV mutant, BBSVm7a, was agroinoculated into the infiltrated leaves. Leaf samples from the inoculated leaves were collected at 3 dpi and detected by immunoblotting with anti-CP antibody. RbcL was used to monitor the protein loading. For D) to F), The band intensities were measured with ImageJ and normalized to RbcL levels (Supplemental Data Set 16). The relative ratios (%) were shown beneath each panel. The experiments were repeated 3 times (n = 3 plants for each biological replicate). ± means Sd. G) Silencing of NbRTNLB2 expression in N. benthamiana plants inhibits BBSV replication. Nicotiana benthamiana leaves were infiltrated with Agrobacterium carrying-EV or intron-hairpin NbRTNLB2 construct (OD600 = 0.2 for each construct). One d later, the NbRTNLB2-silenced leaves were agroinoculated with the BBSVm7a mutant. Leaf samples from the inoculated leaves were collected at 3 dpi and detected by immunoblotting with anti-CP antibody. The replication-deficient mutant, BBSVGDDm, served as the negative control. H) and I) The accumulated negative-strand (−) BBSV genomic RNA in the BBSVm7a-infected plants (as shown in F) and G)) as measured by RT-qPCR. For H) and I), error bars represent mean ± Sd (n = 3 plants for each biological replicate). J) Levels of the accumulated BBSV CP in the infiltrated leaves of NT or NbRTNLB2 KD N. benthamiana plants (NbRTNLB2-KD) plants using an anti-CP antibody in immunoblot analysis. Ino, inoculated leaves. For F) to I), the experiments were repeated 3 times with similar results. RbcL served as the loading control. K) The symptoms of NT and NbRTNLB2-KD infected with BBSV. Nicotiana benthamiana leaves were inoculated with 100 ng BBSV virions. Photographs were taken at 8 dpi and a representative result is shown. Systemically infected leaves (in white boxes) were enlarged and are shown below. L) Statistical analysis of symptom appearance in different N. benthamiana plants after inoculation with BBSV. Experiments were independently repeated 4 times with 6 plants in each experiment. Error bars represent mean ± Sd (n = 4 plants for each biological replicate). M) Levels of accumulated BBSV CP in the plants as shown in L). The upper uninoculated leaves (shown in L)) were collected at 8 dpi. Actin served as the loading control. These experiments were repeated 2 times with similar results. Sys, systemic leaves. For J) and M), the band intensities were measured with ImageJ and normalized to RbcL J) or Actin M) levels. The relative ratios of different bands are shown below the images, and the values of the first J) or second M) lane were set to 1.00. Mock in B) to E), G), and M) indicated N. benthamiana leaves inoculated with buffer without BBSV virion.
To test whether BBSV infection affects the expression of NbRTNLB2, we performed an RNA gel blot and RT-qPCR to analyze the changes in NbRTNLB2 mRNA levels during BBSV infection. We consistently found that the transcriptional level of NbRTNLB2 was upregulated with BBSV infection (Figs. 4B and S10). Moreover, we prepared NbRTNLB2-specific antiserum by immunizing a rabbit with synthetic peptides (Supplemental Fig. S11). NbRTNLB2 protein levels increased in N. benthamiana leaves upon BBSV infection, as determined by immunoblotting with anti-NbRTNLB2 antiserum (Fig. 4C).
To further determine the functional role of NbRTNLB2 in BBSV infection, we transiently expressed NbRTNLB2 in N. benthamiana leaves followed by BBSV inoculation. NbRTNLB2 overexpression significantly increased the accumulation of BBSV compared with the EV control (Fig. 4D). In contrast, when NbRTNLB2 expression was silenced by a Tobacco rattle virus (TRV)–based silencing system, BBSV accumulation significantly decreased compared with the nonsilenced control (TRV-GFP) plants (Fig. 4E). To further investigate whether NbRTNLB2 affected BBSV replication, a movement-deficient BBSV mutant, BBSVm7a (Gao et al. 2022), was tested in N. benthamiana leaves that overexpressed NbRTNLB2. The accumulated BBSV CP significantly increased compared with the EV control (Fig. 4F). In contrast, when the expression of NbRTNLB2 was silenced by infiltration of Agrobacteria carrying the intron-hairpin NbRTNLB2 (RTNLB2-RNAi) in N. benthamiana leaves, a substantial reduction of CP accumulation was observed in the BBSVm7a-infected leaves compared with the EV control (Fig. 4G).
Furthermore, RT-qPCR analysis of the total RNA extracted from the BBSVm7a-infected plants consistently revealed a significant augmentation of negative-sense BBSV genomic RNA in N. benthamiana leaves overexpressing NbRTNLB2 (Fig. 4H), while a decrease was observed in N. benthamiana leaves when the NbRTNLB2 expression was downregulated (Fig. 4I). Downregulation of NbRTNLB2 in TRV-NbRTNLB2 inoculated plants or leaves treated with NbRTNLB2-RNAi was confirmed by RT-qPCR (Supplemental Fig. S12, A and B). To confirm the specificity of RTNLB2 silencing, we demonstrated that the transcription levels of NbRTNLB4 and NbRTNLB5, which are evolutionarily closely related to NbRTNLB2, did not exhibit significant changes in the leaves silenced for RTNLB2 (Supplemental Fig. S12, C to E). These results indicate that RTNLB2 is required for BBSV replication.
We also generated NbRTNLB2 knockdown (KD) (RTNLB2-KD) transgenic N. benthamiana plants (Supplemental Fig. S13). BBSV was rub-inoculated onto the leaves of RTNLB2-KD plants. At 3 dpi, immunoblot analysis revealed a substantial decrease in accumulated BBSV in the inoculated leaves of NbRTNLB2-KD plants compared with the nontransgenic (NT) control plants (Fig. 4J). At 8 dpi, BBSV induced milder symptoms in the upper uninoculated leaves of NbRTNLB2-KD compared with the NT control plants (Fig. 4K). We also statistically analyzed the incidence of symptom appearance in the systemic leaves of different plants and found that the onset of symptoms in NbRTNLB2-KD plants was substantially delayed (Fig. 4L). Immunoblot analysis revealed substantially reduced BBSV accumulation in the systemic leaves of RTNLB2-KD plants compared with NT control plants at 8 dpi (Fig. 4M). Altogether, these results indicate that RTNLB2 plays a proviral role in BBSV replication.
The C-terminal amphipathic helix of RTNLB2 is crucial for promoting BBSV infection
To test whether overexpression of NbRTNLB2 remodels the ER, NbRTNLB2-mCherry and CFP-HDEL were co-expressed in N. benthamiana leaf epidermal cells. With gradually increased concentrations of NbRTNLB2-mCherry, an alteration of the morphology of ER membranes was seen, characterized by the aggregation of luminal CFP-HDEL (Fig. 5A). In addition, co-localization between CFP-HDEL and RTNLB2-mCherry was impaired under high concentrations of NbRTNLB2-mCherry as evidenced by the decline of Pearson's correlation coefficient (PCC) (Fig. 5B).
Figure 5.
An APH in the C-terminal NbRTNLB2 is crucial for promoting BBSV infection. A) Increasing expression of NbRTNLB2-mCherry alters the morphology of the ER. Nicotiana benthamiana plant leaves were agroinfiltrated with increasing concentrations of NbRTNLB2-mCherry as indicated in the panel above (OD600). CFP-HDEL was used as the luminal ER marker. Free mCherry served as the negative control. Scale bars, 10 μm. B) Statistical analyses of mCherry and CFP co-localization in A). Overlapping localization of NbRTNLB2-mCherry and CFP-HDEL with PCCs was measured with the ImageJ Pearson correlation coefficients (PSC) co-localization tool. The experiment was performed 3 times (n = 8 for each replicate), and representative results were shown. Error bars represent mean ± Sd. C) Multiple sequence alignment and WebLogo analysis of NbRTNLBs. Fourteen representative NbRTNLB putative paralogs were indicated on the left side. Sequence in the boxed region indicates the APH domain. The conserved Y and V residues across different NbRTNLBs are highlighted with arrowheads and stars. D) Helical wheel plot of the predicted hydrophobic moment of an APH-containing region within NbRTNLB2 and its variants. The directions of the hydrophobic moments of the helices are depicted in black arrows, and the corresponding magnitudes (μ) are shown below. E) Effects of overexpressed NbRTNLB2 and its derivatives on BBSV accumulation. Nicotiana benthamiana leaves were infiltrated with Agrobacterium containing different constructs or the empty pGD vector (EV) as indicated in the panel above. At 1 dpi, the same leaves were agroinoculated with BBSV. Leaf samples were harvested for immunoblot analysis after 3 d of BBSV inoculation. RbcL served as the loading control. The band intensities were measured with ImageJ and normalized to RbcL levels. The relative ratios of different bands are shown below the panel, and the value of the second lane was set to 1.00. F) Disruption of the hydrophobic face of the APH impairs NbRTNLB2 function. Nicotiana benthamiana epidermal cells were infiltrated with Agrobacterium mixtures containing the soluble ER marker, CFP-HDEL, and NbRTNLB2 or NbRTNLB2YV2K plasmids. Scale bars, 10 μm. G) Overlapping localization of NbRTNLB2-mCherry or NbRTNLB2YV2K-mCherry with CFP-HDEL as displayed by box and whisker plots. The solid lines within the boxes indicate median PCC values, and the upper and lower hinges indicate 75 and 25 percentiles, respectively. The whiskers show the largest/smallest PCC values that fall within a distance of 1.5 times interquartile range from the upper and lower hinges. Data are from values of PCC measured from 8 regions of interest (n = 8). H) The interaction between wild-type or mutant NbRTNLB2 and p23 as detected by BiFC. These proteins, fused to the N- or C-terminus of YFP, were transiently co-expressed in N. benthamiana leaves and analyzed by confocal microscopy at 3 dpi. Figures on the right indicate the normalized fluorescence intensities of YFP (green) or mCherry (magenta) channels along the dashed white lines shown in the merged images. Representative results of at least 3 independent experiments are displayed. The relative intensity was calculated by using the Zen software on a Zeiss LSM 880 confocal microscope. Scale bars, 20 µm. I) The interaction between wild-type or mutant NbRTNLB2 and p23 as tested using co-IP. Nicotiana benthamiana leave tissues transiently expressing combinations of different proteins were collected at 3 dpi. Total proteins were immunoprecipitated with anti-FLAG beads and detected by immunoblotting with anti-GFP or anti-FLAG antibody.
We subsequently examined whether NbRTNLB2 could affect the flow of luminal content using the fluorescence recovery after photobleaching (FRAP) assay. We found that NbRTNLB2 overexpression significantly slowed the recovery of ER retention signal (HDEL)-fused mCherry fluorescence after the mCherry signal was bleached compared with the EV control (Supplemental Fig. S14). These results suggest that overexpression of NbRTNLB2 induces remodeling of the ER membrane, resulting in constriction of ER tubules and reduced mobility of ER luminal content.
Previous studies identified an amphipathic helix (APH) at the C-terminal region of the Yeast homolog of the polyposis locus protein 1 (Yop1p) and Arabidopsis RETICULON 13 (AtRTN13) (Brady et al. 2015; Breeze et al. 2016). To investigate whether the APH domain exists in the NbRTNLB protein family of N. benthamiana, we analyzed amino acid sequences of 14 representative NbRTNLB proteins and found a region with high helical hydrophobic moment (Fig. 5C). To test whether the APH domain was critical for NbRTNLB2 to regulate BBSV infection, putative key sites were predicted with the HeliQuest online server (https://heliquest.ipmc.cnrs.fr/). Based on sequence similarity, tyrosine (Y) at the 213 site and valine (V) at the 217 site were conserved among the NbRTNLB family proteins (Fig. 5C, and Supplemental File 1) and were subsequently replaced with lysine (K) to generate Y213K, V217K, and YV2K mutants, in which both sites Y213 and V217 were replaced with K. These NbRTNLB2 variants were predicted to have reduced magnitude of the hydrophobic moment and amphipathicity in the helix (Fig. 5D).
We then transiently expressed NbRTNLB2 and its variants in N. benthamiana leaves followed by BBSV inoculation. Immunoblot analysis revealed that both NbRTNLB2V217K and NbRTNLB2YV2K failed to promote BBSV infection compared with wild-type NbRTNLB2 and NbRTNLB2Y213K (Fig. 5E). Interestingly, NbRTNLBYV2K not only failed to enhance but also severely inhibited BBSV infection compared with wild-type NbRTNLB2 (Fig. 5E). Subcellular localization analysis showed that although both wild-type NbRTNLB2 and NbRTNLB2YV2K have ER localization (Fig. 5F), the degree of co-localization of NbRTNLB2YV2K with CFP-HDEL increased significantly compared with wild-type NbRTNLB2 (Fig. 5G). To test whether the YV2K mutation affected the interaction between NbRTNLB2 and p23, BiFC and Co-IP assays were conducted. RTNLB2YV2K retained the ability to interact with p23 (Figs. 5, H and I, and S15), suggesting that the dysfunction of NbRTNLB2YV2K in BBSV infection was not due to disruption of the RTNLB2-p23 interaction. Altogether, these results indicate that the C-terminal APH domain is crucial for NbRTNLB2 to shape the ER membrane and promote BBSV infection.
NbRTNLB2 is required for the assembly of BBSV VRCs
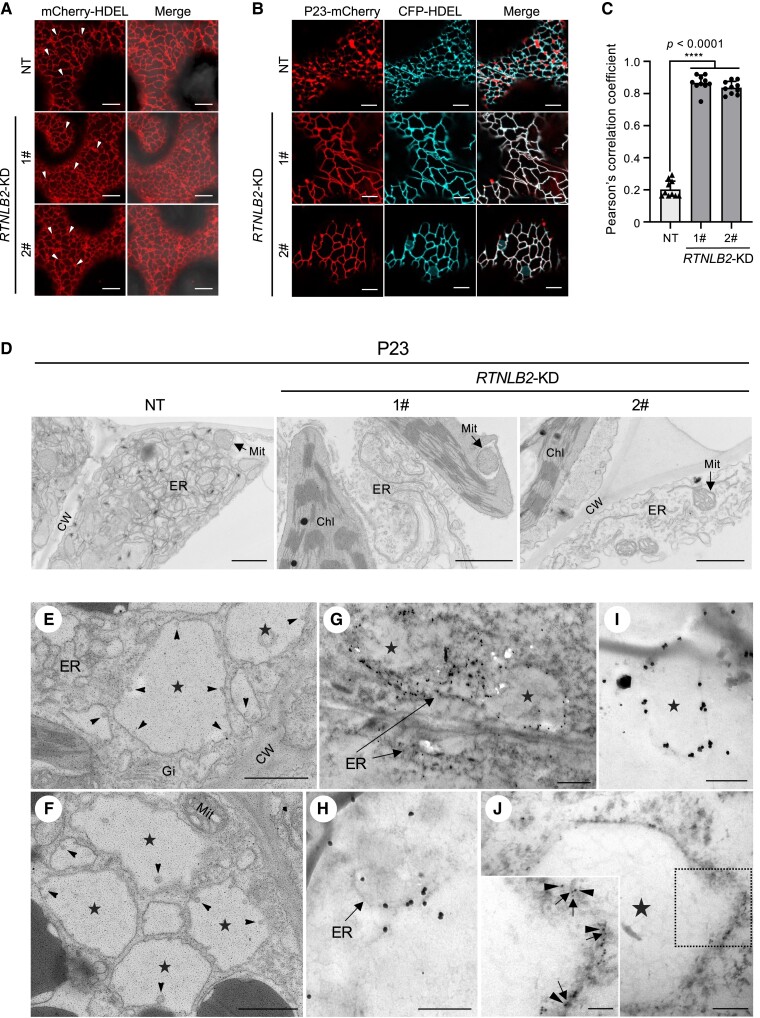
Given that BBSV forms VRCs by invaginating ER membranes and that p23 interacts with NbRTNLB2, we investigated whether NbRTNLB2 is directly involved in the assembly of BBSV VRCs. First, we expressed mCherry-HDEL in NT and NbRTNLB2-KD transgenic N. benthamiana plants. Confocal microscopy analysis showed obvious reduction of 3-way junctions in NbRTNLB2-KD plants compared with NT control plants (Figs. 6A and S16). To test the effect of NbRTNLB2 on p23-induced rearrangement of the ER membrane, we co-expressed p23-mCherry and CFP-HDEL in NT or NbRTNLB2-KD N. benthamiana leaves. ER-associated aggregates were severely decreased in RTNLB2-KD plants compared with NT plants (Fig. 6B). In contrast, co-localization of p23-mCherry with the CFP-HDEL marker was significantly enhanced in RTNLB2-KD plants as evidenced by the increased value of PCC (Fig. 6C). Furthermore, we used transmission electron microscopy (TEM) to analyze the ER membrane ultrastructure of NT and RTNLB2-KD leaf tissues transiently expressing p23. As shown in Fig. 6D, ER membranes were proliferated and became convoluted in NT leaf tissues upon p23 expression. In contrast, the extent of p23-induced ER deformation was substantially attenuated in RTNLB2-KD leaf tissues. These results indicate that NbRTNLB2 is required for p23 to remodel the ER membrane.
Figure 6.
NbRTNLB2 is recruited to BBSV replication sites to promote VRC assembly. A) Confocal microscopy analysis of the ER morphology in NT and NbRTNLB2 KD N. benthamiana plants (NbRTNLB2-KD). mCherry-HDEL serves as an ER marker. An enlarged version of this image is provided in Supplemental Fig. S16. Arrowheads indicate ER junctions. B) Subcellular localization of p23-mCherry and CFP-HDEL in NT and NbRTNLB2-KD N. benthamiana plants at 2.5 d post-infiltration (dpi). C) Overlapping localization of p23-mCherry and CFP-HDEL as shown in B) by chart of average PCCs. Asterisks indicate the significant difference (****P < 0.0001) based on a 1-sided unpaired Student's t test. Error bars represent mean ± Sd of PCC values from 10 regions of interest (n = 10). D) Effects of BBSV p23 on ER morphology and membrane remodeling. Agrobacterium-harboring plasmids expressing p23 were infiltrated into NT or NbRTNLB2-KD N. benthamiana leaves. TEM analysis was performed at 3 dpi. E) and F) ER-bound VRCs in BBSV-infected leaf tissues. Nicotiana benthamiana leaves displaying typical BBSV symptoms were sampled and observed under TEM. Arrowheads indicate the ER membrane–associated spherules, and pentagrams represent the vesicle packets. G) Immunogold labeling of the BBSV p23 in BBSV-infected N. benthamiana cells. H) and I) Immunogold labeling of the NbRTNLB2 in BBSV-infected N. benthamiana cells. J) Double immunogold labeling of the p23 (5 nm) and NbRTNLB2 (10 nm) in BBSV-infected N. benthamiana cells. Box inset is a zoom of the region inside the dashed square. An enlarged version of this image is provided in Supplemental Fig. S18. Arrowheads indicate the 10-nm gold particles, while arrows point to 5-nm gold particles. For A) and B), scale bars, 10 µm. For D) to F), scale bars, 1.0 µm. For G) to J), scale bars, 500 nm. Scale bars for the inset of J) represent 200 nm. CW, cell wall; Chl, chloroplast; Mit, mitochondria; Gi, Golgi.
To further investigate the association of NbRTNLB2 with the formation of BBSV VRCs, we used immunogold electron microscopy to observe NbRTNLB2 localization. Unlike healthy leaf tissues (Supplemental Fig. S17A), vesicle packets were readily observed in BBSV-infected leaf tissues in proliferated ER membranes (Fig. 6E) and vesicle-containing large bubbles (Fig. 6, E and F, pentagram), which is consistent with our previous studies (Cao et al. 2015). Immunogold labeling of p23 revealed that large numbers of gold particles were present along the ER and around the vesicle packets (Figs. 6G and S17B). Likewise, gold particles specifically bound to NbRTNLB2 can also be observed along the ER (Figs. 6H and S17C) and at the periphery of vesicle packets (Fig. 6I).
To further define the association of NbRTNLB2 with BBSV VRCs, we performed double immunogold labeling in BBSV-infected leaves transiently expressing NbRTNLB2-3xFLAG with anti-p23 and anti-FLAG antibodies and detected co-localization of NbRTNLB2 and p23 at both the ER membrane (Supplemental Fig. S17D) and the vesicle packet structure (Figs. 6J and S18). For the mock-inoculated control, a small number of gold particles near the ER could be observed when using anti-RTNLB2 in the immunogold labeling assay (Supplemental Fig. S17E); only a few gold particles were randomly distributed over the tissue sections used for immunogold labeling with anti-p23 and/or anti-FLAG antibodies (Supplemental Fig. S17, F and G). Collectively, these results indicate that BBSV co-opts NbRTNLB2 to induce ER membrane rearrangements and form VRCs.
NbRTNLB2 may play a role in other ER-replicating viruses
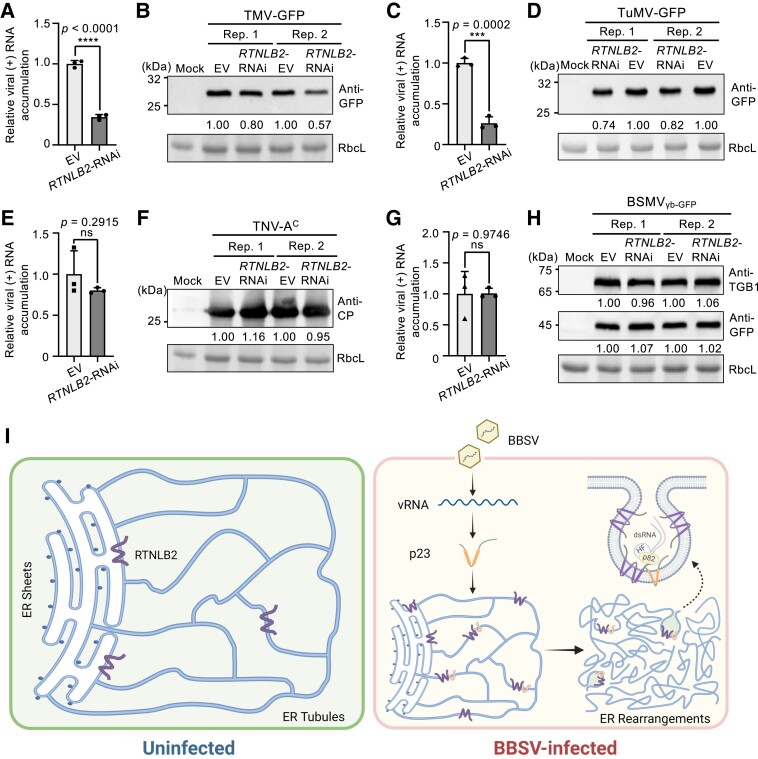
To investigate whether NbRTNLB2 functions in infections of other positive-strand RNA viruses, we selected Tobacco mosaic virus (TMV) and Turnip mosaic virus (TuMV), whose replication occurs on or associates with ER membranes (Más and Beachy 1999). Silencing the expression of NbRTNLB2 resulted in a significant decrease in the accumulation of TMV positive-strand (+) RNA compared with the EV control as determined by RT-qPCR (Fig. 7A). Consistently, a substantial reduction of accumulated GFP was observed in the TMV-GFP–infected NbRTNLB2-RNAi plants compared with the EV control (Fig. 7B). Similar results were obtained when NbRTNLB2-silenced and EV-inoculated leaves were inoculated with TuMV-GFP (Fig. 7, C and D). Furthermore, based on BiFC assays, the TMV P50 helicase fragment, TuMV NIb replicase, and TuMV 6K2 protein, which play important roles in TuMV VRC formation, interacted with NbRTNLB2 (Supplemental Fig. S19). Downregulation of NbRTNLB2 was confirmed by RT-qPCR (Supplemental Fig. S12, F to I). These results suggest that NbRTNLB2 plays a role in the infection of other ER-replicating plant viruses.
Figure 7.
NbRTNLB2 plays a proviral role in infections of several plant viruses that replicate in association with ER membranes. A) and B) Silencing of NbRTNLB2 impairs TMV infection. A total of 0.66 mg of purified GFP-tagged TMV (TMV-GFP) were rub-inoculated onto NbRTNLB2-silenced N. benthamiana leaves. At 3 dpi, RT-qPCR A) and immunoblot with anti-GFP antibody B) were conducted to analyze viral positive-strand (+) RNA and the accumulated GFP in TMV-GFP–infected leaves. C) to H) RT-qPCR and immunoblot analyses were used to measure the accumulated (+) RNAs and proteins of GFP-tagged turnip mosaic virus (TuMV-GFP) C) and D), tobacco necrosis virus-A Chinese isolate (TNV-AC) E) and F), and GFP-tagged BSMV (BSMVγb-GFP) G) and H) in the NbRTNLB2-silenced N. benthamiana plants. Leaf samples were harvested at 3 dpi, and antibodies used for immunoblot analysis are indicated on the right. For A) to H), experiments were independently repeated at least 2 times with similar results. The band intensities were measured with ImageJ and normalized to RbcL levels. The relative ratios of different bands are shown below the images, and the value of the EV lane in each replicate (Rep) was set to 1.00. For A), C), E), and G), each treatment was conducted with 3 independent biological replicates (n = 2 plants for each replicate). Error bars represent mean ± Sd (n = 3). For A) to H), leaves infiltrated with Agrobacterium carrying the empty pMDC32 vector (EV) served as the control. I) A proposed model illustrating the role of NbRTNLB2 in BBSV replication. Without virus infection, NbRTNLB2 induces the formation of tubular ER from the cisternal structure. Upon BBSV infection, the expression of NbRTNLB2 is induced and hijacked by BBSV p23 replication protein to promote ER remodeling and formation of vesicular structures to support BBSV replication. The model was partially created with BioRender (https://biorender.com). HF, host factor; dsRNA, double-stranded RNA; p82, BBSV RNA-dependent RNA polymerase.
We also included Tobacco necrosis virus-A Chinese isolate (TNV-AC) and Barley stripe mosaic virus (BSMV), whose replication occurs on vacuole and chloroplast membranes, respectively (Wan et al. 2015; Jin et al. 2018b; Wang et al. 2021). No significant changes were observed in the accumulation of either positive-strand RNA virus (Fig. 7, E and G), as determined by RT-qPCR, or the accumulated viral proteins of TNV-AC or BSMVγb-GFP when NbRTNLB2 was silenced (Fig. 7, F and H), indicating that NbRTNLB2 silencing had little effect on TNV-AC and BSMV infections.
Discussion
TurboID-based PL is an effective approach to study VRCs in planta
Both plant and animal positive-strand RNA viruses usually hijack diverse host factors to establish VRCs on specific organelle membranes (Verchot 2011; Mine and Okuno 2012; Nagy 2016; Huang et al. 2018). However, the formation of VRCs is a highly dynamic process and most VRC components localize to the viral replication protein-remodeled membranes, which often makes it difficult to identify these host factors using traditional protein purification methods. Although there are few studies on the comprehensive analysis of plant VRC components in living cells, to date, the only example of the application of TurboID-based PL to examine the plant virus infection process is the dissection of Bamboo mosaic virus movement complexes (Huang et al. 2023).
Here, we used BBSV, whose replication occurs in ER-derived membrane vesicles, as a model to systematically identify host factors that may constitute or participate in the formation of VRCs using the recently developed TurboID-based PL technique. We identified 185 candidate proteins that were in proximity to p23 that may play a role in the biogenesis of the BBSV replication compartment. We further characterized NbRTNLB2, which displayed high reproducibility in the MS data sets and whose role in plant virus replication remains uncharacterized. We validated the interaction between BBSV p23 and NbRTNLB2 and demonstrated that NbRTNLB2 played a proviral role in BBSV replication by directly facilitating the assembly of BBSV VRCs. Our study identifies a host factor that is directly involved in BBSV replication, providing an example of utilizing TurboID-based PL to investigate plant VRCs.
Some considerations about using TurboID-based PL to map the p23-proximal interactome
Like many methods for studying PPIs, TurboID-based PL has its own strengths and limitations (Yang et al. 2021). The TurboID method is more complicated to run compared with other methods for detecting PPIs and requires specialized equipment, such as mass spectrometry, as well as expertise. Also, the number of proteins obtained through the use of TurboID technology is often large; therefore, strict controls need to be set up to filter out background noise and prevent false positives.
Besides the cytosolic Citrine-TurboID, we also used the ER-localized 2C1-Citrine-TurboID as a control (Supplemental Fig. S20). We identified 610 proximity interactions with 320 host proteins (Supplemental Figs. S21 and S22 and Data Sets 8 to 13), which was more than the number of proteins obtained when using 2C1-Citrine-TurboID (Fig. 2 and Supplemental Data Sets 3 and 4). When Citrine-TurboID was used as a control, 49% of the enriched proteins originated from the endomembrane system (Supplemental Fig. S21C), while 15% of the enriched proteins were predicted to be localized to the nucleus. Conversely, when 2C1-Citrine-TurboID was used as a control, the proportion of endomembrane system–sourced proteins dropped to 21%, while the proportion of nuclear-localized proteins increased to 48% (Supplemental Fig. S23). These results indicate that the choice of control can affect the number and types of proteins obtained in the final MS data set. Nevertheless, analysis of the data from both rounds of MS showed that the RTN family of proteins were frequently identified in PL assays using different controls (Supplemental Fig. S24 and Data Set 14). These results pinpoint the involvement of NbRTNLB2 in BBSV infection and further demonstrate the rationality of our selection of NbRTNLB2 for further research.
In addition, other potential drawbacks of TurboID-based PL include the potential effects of the TurboID tag on the function of target proteins. Typically, the target protein should retain its native function after being fused with TurboID; in other words, the TurboID-tagged version can substitute most of the functions of the unfused target protein (Kim et al. 2023). In this study, we expressed p23-Citrine-TurboID in BBSV-infected leaves, primarily because of the distinct advantages of PL technology, which relies on spatial proximity to identify proteins that may interact with a target protein. We first confirmed that p23-Citrine-TurboID localized to ER membranes and formed aggregates (Figs. 1C, 3A, and S7), which was consistent with the remodeling of ER membranes during BBSV infection. Our TEM analysis revealed that p23 alone can induce a disordered ER structure (Fig. 6D), similar to the ER morphology observed in BBSV-infected cells (Cao et al. 2015). Furthermore, we found that p23 has a strong self-interaction (Wang et al. 2018). Given these findings, it is reasonable to propose that p23-Citrine-TurboID can directly localize or be recruited to BBSV VRCs during BBSV infection. Most importantly, we successfully identified not only known p23 interacting proteins like Hsp70 (Wang et al. 2018) but also many other proteins such as NbRTNLB2 that are proven to interact with p23 (Figs. 3 and S25). These findings demonstrate the effectiveness of our system in examining the p23 interactome and its possible use in examining host proteins involved in viral infections of other plant viruses.
It should be pointed out that not all interactions are probed by TurboID-mediated PL. For example, the BBSV p82 replicase, which has been verified to interact with p23 (Cao et al. 2015), was not included in the MS data set. Since the p82 protein is generated through the read-through of the p23 protein (Fig. 1A), the amino acid sequence of the N-terminus of p23 and p82 is identical. Therefore, technically, it is difficult to determine whether the enriched peptides corresponding to p23 actually originated from p23 or from the p82 replicase. In addition, since the p82 protein is produced by read-through of p23, its expression level is very low, and there is often a bias against low abundance proteins during PL analysis (Kreis et al. 2022). All these factors may contribute to the low fold change value of p82 in the MS data set even though it interacts with p23. In addition, peptides derived from the BBSV CP was not present in the final MS data set, which is consistent with the fact that BBSV p23 does not interact with the CP (Wang et al. 2018).
Moreover, the enzymatic activity of TurboID, which covalently links biotin to adjacent proteins in living cells, can be influenced by intracellular factors such as an oxidative or acidic microenvironments or changes in temperature. TurboID-based PL is unsuitable for analyzing proteins involved in biotin synthesis and metabolism pathways. Finally, like other tools used for studying PPIs, the data obtained from TurboID-based spatial proteomics analysis primarily shows proximity between the identified proteins and a bait protein. Further experiments are necessary to validate their interactions. These limitations should be kept in mind when PL is used for the mapping of compartment-specific proteomes.
Although the Citrine-TurboID (cytosolic localization) is less suitable as a control than 2C1-Citrine-TurboID (ER membrane localization), the subcellular distribution of the Citrine-TurboID control at least partially overlaps with that of the p23-Citrine-TurboID, given that it is exposed toward the cytosol. In fact, except for NbRTNLB2, we also tested the interactions between p23 and randomly selected candidate proteins with a relatively high fold enrichment from the MS data set, which used the Citrine-TurboID as the control. The BiFC assay revealed that 8 out of 10 proteins were able to interact with p23 (Supplemental Fig. S25 and Data Set 9). Therefore, the data using Citrine-TurboID as the control can still provide important basic information and clues for our future in-depth investigation of the composition and function of BBSV VRCs.
Our MS data set also provided valuable information about the topology of ER membrane-localized proteins. For example, it cannot be determined whether the N-terminus of the MEMBRANE STEROID BINDING PROTEIN 1 (MSBP1) faces the cytosolic side or the ER lumen using only different online prediction tools. Intriguingly, the C-terminal peptides of MSBP1 were present in our MS data set, which clearly supported the notion that the N-terminus of MSBP1 is exposed to the ER lumen (Supplemental Fig. S26).
Agrobacterium-mediated transient expression can be used in TurboID-based PL analysis in planta
We also noted that several Arabidopsis RTN family proteins were reported to participate in Agrobacterium-mediated plant transformation (Hwang and Gelvin 2004; Huang et al. 2018, 2021). Nevertheless, this does not mean that the multiple RTN proteins obtained in our experiment were due to the agroinfiltration system, because (i) p23, but not VirB2, was fused with TurboID, and only those proteins adjacent to p23 could be biotinylated. (ii) No matter the p23-Citrine-TurboID group, or the Citrine-TurboID or 2C1-Citrine-TurboID control groups, the expression of these fusion proteins is all mediated by Agrobacterium. Theoretically, the potential influence of Agrobacterium on the PL proteome can be largely offset by quantitative proteomics analysis of the sample and the control groups. (iii) In our previous work, we also employed the Agrobacterium-mediated transient expression to perform TurboID-based PL analysis of the N protein as well as the N TIR domain. When we checked those MS data sets, we found that none of the RTN family proteins were significantly enriched in either the N or TIR proximal interactome (Zhang et al. 2019). Additionally, Agrobacterium-mediated transient expression has also been successfully applied to the TurboID-based PL analysis of PPIs in other studies (Mair et al. 2019; Arora et al. 2020). Taken together, these results suggest that the RTN family of proteins identified here were mainly due to their proximity to BBSV p23 rather than the agroinfiltration system used in our work.
A general role for RTNLB2 in the infection of ER-replicating viruses
RTNs are a group of integral membrane proteins that are commonly present in all eukaryotes and play an crucial role in the formation of ER tubules by inducing membrane curvature (Tolley et al. 2010). RTNs contain a reticulon homology domain (RHD) with 2 major hydrophobic segments that form a pair of V-shaped transmembrane wedges joined by a cytosolic loop, with both the N- and C-termini facing the cytoplasm (Supplemental Figs. S3A and S11B).
A growing body of evidence showed that RTNs are indispensable for viruses to establish infection in mammals and yeast (Tang et al. 2007; Diaz et al. 2010; Aktepe et al. 2017). In particular, yeast RTNs have been found to be recruited to build brome mosaic virus (BMV) VRCs. Deleting 1 or 2 RTNs affected the size and frequency of BMV replication vesicles; however, only all 3 RTN genes were deleted, and viral replication vesicles were not formed or formed in a very low rate (Diaz et al. 2010). Here, knocking down the NbRTNLB2 expression inhibited BBSV replication (Fig. 4, G and I) and mitigated the p23-induced proliferation and aggregation of ER membranes (Fig. 6, B to D). Whether the NbRTNLB2 KD affects the formation of BBSV replication vesicles like that of BMV in yeast cells needs further investigation. Recent studies showed that plant virus-encoded movement proteins can mimic or interact with RTNs to facilitate viral movement (Lazareva et al. 2021; Tilsner and Kriechbaumer 2022). However, whether and how RTNs participate in viral replication in planta remains unexplored. Here, using TurboID-based PL, several RTN-like proteins were identified to be proximal to BBSV p23. We focused on NbRTNLB2 due to its high reproducibility in the MS data sets (Fig. 2). Our data demonstrated a proviral role of NbRTNLB2 in BBSV replication (Fig. 4), and, more specifically, NbRTNLB2 was directly involved in the assembly of BBSV VRCs by inducing ER membrane rearrangements (Fig. 6), a process requiring the presence of a functional APH domain (Fig. 5). These results provide insight into the formation of membrane scaffolds for viral RNA replication.
Although different viruses may choose specific organelle membranes to establish replication sites, the morphology of VRCs shows great convergence, such as the common membrane of invaginated vesicles or spherules (Jin et al. 2018a). Therefore, our results have broad implications for understanding the mechanisms underlying the formation of VRCs, not only BBSV, which was studied here, but also for many other positive-strand RNA viruses. We have demonstrated the roles of NbRTNLB2 in its interaction with replication proteins from both TMV and TuMV (Supplemental Fig. S19) and its positive role in viral replication, suggesting a broad role for NbRTNLB2 in ER-replicating virus infection. Considering that there are 14 putative paralogs of the RTN family of proteins in N. benthamiana and each paralog has a varied number of isoforms and the role of NbRTNLB2 in TMV and TuMV infection appears to be less pronounced than that of BBSV infection, we cannot rule out the possibility that this difference may be due to the redundant role of other RTN paralogs in TMV and TuMV infection. Additionally, whether RTNLB2 or other RTN paralogs function in BBSV cell-to-cell movement is also an interesting question that warrants further investigation.
Based on our findings described above, we propose a model to illustrate the functional role of NbRTNLB2 in BBSV infection. Under normal conditions, NbRTNLB2 localizes to ER membranes to induce tubule formation in ER sheets and maintain normal architecture of the ER (Fig. 7I, left panel). Upon BBSV infection, NbRTNLB2 expression is upregulated significantly and co-opted by BBSV p23, thereby promoting remodeling of ER membranes and forming the membrane-invaginated spherules to support viral replication (Fig. 7I, right panel). Our results expand our knowledge of the function of RTNs in virus replication in plants, and the TurboID proteomics data for BBSV VRCs presented here provide a valuable resource for in-depth analysis of critical virus–host plant interactions that will help to discover promising targets for developing virus-resistant plants.
Materials and methods
Plant materials
Nicotiana benthamiana plants were grown in a growth-controlled chamber at 23 to 24°C under fluorescent lights at ∼75 μmol photons m−2 s−1 in a 15 h photoperiod (Philips Master TL5 HE 28W/865 lamps). Four-wk-old N. benthamiana plants were infiltrated with A. tumefaciens and used in the protein expression or gene silencing assays, as described previously (Zhang et al. 2023).
Plasmid construction
The pCB301-BBSV plasmids used for agroinoculation of BBSV into N. benthamiana plants were described in our previous work (Gao et al. 2021). For the construction of Pro35S:p23-Citrine-TurboID, the Citrine-fused TurboID fragment was amplified by using the ubiquitin-10 (UBQ10)::Citrine-TurboID as the template (Zhang et al. 2019), and the resulting fragments were cloned into the pMDC32-3xFLAG plasmid (Gao et al. 2021) at the KpnI and SpeI sites to generate Pro35S:Citrine-TurboID. The PCR-amplified p23 fragment was cloned into Pro35S:Citrine-TurboID at the KpnI and SpeI sites to yield Pro35S:p23-Citrine-TurboID. To generate Pro35S:2C1-Citrine-TurboID, the 2C1-fused Citrine fragment was cloned into Pro35S:TurboID at the KpnI and SpeI sites (Fig. 1B).
For transient expression in plants, the PCR-amplified NbRTNLB2 fragment was ligated into the KpnI- and SpeI-digested pMDC32-3xFLAG plasmid (Gao et al. 2021). To generate pGD-RTNLB2, pGD-RTNLB2Y213K, pGD-RTNLB2V217K, and pGD-RTNLB2YV2K, NbRTNLB2 was first cloned into the pMD19-T vector (Takara). The resulting plasmid was used for site-directed mutagenesis with a QuikChange Site-Directed Mutagenesis Kit, and then, the DNA fragment encoding wild-type or mutant NbRTNLB2 was amplified and inserted into pGD vectors (Goodin et al. 2002).
For BiFC assays, NbRTNLB2 and NbRTNLB2YV2K were inserted into the pSPYNE-35S or pSPYCE-35S vectors (Walter et al. 2004). The pSPYNE-p23 (p23-YFPn), pSPYCE-p23 (p23-YFPc), and P50-YFPc plasmids were described previously (Burch-Smith et al. 2007; Cao et al. 2015).
For subcellular localization and Co-IP assays, the wild-type or mutant NbRTNLB2 was cloned into pGDB, pGDGm, or pGD3G-mCherry vector (Goodin et al. 2002; Fan et al. 2014). To generate RTNLB4-GFP, RTNLB4 was cloned into the pCam35S-GFP vector at XhoI sites using the Seamless assembly cloning kit (Clone Smarter). To generate the p23-BFP vector, the PCR-amplified p23 fragment was ligated into BamHI- and PstI-digested pGD-CBFP plasmid (Xu and Nagy 2016). p23-RFP and pMDC32-p23-3xFLAG were described previously (Cao et al. 2015).
For Y2H assays, the coding sequence of p23 and NbRTNLB2 was cloned into the pBT3-N or pPR3-N vectors at the SfiI site to generate pBT3N-p23, pPR3N-p23, and pPR3N-RTNLB2, respectively.
For TRV-mediated gene silencing, the cDNA fragments corresponding to the putative NbRTNLB2 were amplified from N. benthamiana cDNA and cloned into the pYL156 vector (Liu et al. 2002).
To generate the NbRTNLB2-RNAi vector, 200 nt from NbRTNLB2 cDNA were predicted using the online SGN VIGS Tool (https://vigs.solgenomics.net) and amplified by PCR. The resulting fragments were then inserted into the pSK-In vector (Gao et al. 2022) at the KpnI and SpeI sites. DNA fragments that produce intron-hairpin RNAs were then amplified and introduced into the pMDC32 plasmid (Curtis and Grossniklaus 2003) at the KpnI and SpeI sites to generate the NbRTNLB2-RNAi vector.
All primers used for plasmid constructions are listed in Supplemental Data Set 15, and DNA sequencing was performed to confirm the identity of each construct.
Phylogenetic analysis
RTN protein family members from the N. benthamiana, Nicotiana tabacum, A. thaliana, and S. cerevisiae were identified through BLAST searches of SOL N. benthamiana database (https://solgenomics.net/organism/Nicotiana_benthamiana/genome), the NbC database at Oxford Research Archives (https://ora.ox.ac.uk/objects/uuid:f09e1d98-f0f1-4560-aed4-a5147bc7739d), the National Center for Biotechnology Information (NCBI), or The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org). Protein sequences were aligned using the Clustal W algorithm (Larkin et al. 2007) implemented in MEGA11 software (Kumar et al. 2018). Phylogenetic trees were constructed by MEGA11 with default parameters and 1,000 bootstraps. Alignment and tree files are provided in Supplemental Files 1 to 4.
Agroinfiltration
Agroinfiltration was performed according to procedures previously described (Jiang et al. 2018). Briefly, A. tumefaciens EHA105 was transformed with different constructs using a freeze–thaw method (Höfgen and Willmitzer 1988) and then cultured in Luria–Bertani (LB) medium containing appropriate antibiotics at 28°C for 10 to 16 h. Agrobacterium cells were collected and then resuspended in infiltration buffer [10 mm MgCl2, 150 μm acetosyringone, and 10 mm MES (pH 5.6)] followed by incubation at 28°C for 2 to 4 h before infiltration.
Immunoblot analysis
Proteins were separated on 10% or 12.5% SDS–PAGE gels based on the molecular weight of the tested proteins. Immunoblot analysis of the protein extracts was performed according to the procedures described previously (Gao et al. 2022). Briefly, plant tissue expressing the protein(s) of interest was harvested and ground in liquid nitrogen.
Total protein extracts were prepared followed by SDS–PAGE, and proteins were then transferred to 0.45 μm nitrocellulose filters (GVS Life Sciences, USA, Cat. No. 1215471) using the Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Cat. No. 1703930). Membranes were blocked and incubated with the appropriate primary antibodies or streptavidin-HRP (Abcam, Cat. No. ab7403; 1:20,000 dilution), followed by incubation with corresponding secondary antibodies. Information about rabbit anti-TurboID and anti-RTNLB2 antibodies was available in Supplemental Figs. S2 and S11. Rabbit anti-P23 antibody was described previously (Cao et al. 2015). Bands were visualized by using NcmECL Ultra Luminol/Enhancer Reagent (New Cell & Molecullar Biotech, Cat. No. P10300A and P10300B) according to the manufacturer's instructions. Chemiluminescent signals were acquired by using a Sapphire RGBNIR Biomolecular Imager (Azure Biosystems, Dublin, CA, USA). Quantification of the band signal intensity was performed with ImageJ software.
Preparation of leaf samples for LC–MS/MS analysis
Leaf samples for LC–MS/MS analysis were prepared according to previously described procedures (Zhang et al. 2020). Briefly, leaves of 4- to 6-wk-old N. benthamiana were infiltrated with Agrobacterium mixtures harboring 2C1-Citrine-TurboID or p23-Citrine-TurboID and the pCB301-BBSV constructs. Three biological replicates (n = 3 plants for each replicate) for each sample were performed. At 60 hpi, 200 µM biotin in a 10 mm MgCl2 solution was infiltrated into these preinfiltrated leaves. After 8 h incubation at RT, infiltrated leaves from each biological replicate were flash-frozen in liquid nitrogen and stored at −80°C for subsequent use.
Immunoblot analysis was performed to confirm the expression and biotinylation of the TurboID fusions in these harvested leaves. These leaves were then ground into fine powders followed by protein extraction with RIPA lysis buffer [50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 1 mm EDTA, 1% IGEPAL630 (v/v), 0.1% SDS (w/v), 0.5% sodium deoxycholate (w/v), 1 mm DTT, and 2 tablets of cOmplete Protease Inhibitor Cocktail (Sigma, Cat. No. 11697489001)]. After vortex-mixing, the leaf homogenate was centrifuged at 12,000 × g for 10 min. The upper layer was desalted using a Zeba Spin Desalting Column (Thermo Fisher Scientific, Cat. No. 89893) to remove free biotin. Protein concentration in the desalted extracts was measured by the Bradford assay. The extracts containing equal amounts of proteins were then incubated with equilibrated streptavidin beads (Dynabeads MyOne Streptavidin C1, Invitrogen, Cat. No. 65001) overnight on a rotator at 4°C. The beads were then sequentially washed with buffer I (2% SDS, w/v), buffer II [50 mm HEPES (pH 7.5), 500 mm NaCl, 1 mm EDTA, 0.1% deoxycholic acid (w/v), and 1% Triton X-100], and buffer III [10 mm Tris-HCl (pH 7.5), 250 mm LiCl, 1 mm EDTA, 0.1% deoxycholic acid (w/v), and 1% IGEPAL-630 (v/v)]. To remove any detergent, the beads were thoroughly washed with 50 mm Tris-HCl (pH 7.5) and 50 mm ammonium bicarbonate before being sent for LC–MS/MS analysis.
On-bead trypsin digestion
Biotinylated proteins enriched with streptavidin beads were enzymatically digested into polypeptides and analyzed by LC–MS/MS. After absorbing the thawed beads with a magnetic frame, 100 µL of 5 mm dithiothreitol (DTT) in 50 mm triethylammonium bicarbonate (TEAB) (Sigma, Cat. No. T7408) was added, and the mixture was treated at 56°C for 30 min with shaking on a rotator. The beads were then washed with 200 µL 50 mm TEAB for 1 min and incubated at RT with 100 µL of 20 mm iodoacetamide (IAM) in 50 mm TEAB for 30 min under dark conditions. The beads were then washed once with 50 mm TEAB buffer. Finally, 1 µg of trypsin (Thermo Fisher Scientific, Cat. No. 90058) dissolved in 50 mm TEAB buffer (pH 8.0 to 8.5) was added to the beads followed by digestion overnight at 37°C. Magnetic beads were then collected on a magnetic separation rack, and the supernatant was added onto prewetted 10,000 MWCOHY ultrafiltration products of VIVACON 500 (Sartorius Stedim Biotech GmbH, Cat. No. VN01H02) followed by centrifugation at 16,160 × g for more than 20 min until all the peptides were collected into the tubes. Peptides were then quantified using the Pierce Quantitative Colorimetric Assay (Thermo Fisher Scientific, Cat. No. 23275). Samples were analyzed by Thermo Q-Exactive high-resolution MS (Thermo Scientific, Waltham, MA, USA) to examine the quality of the peptides.
TMT labeling
A total of 20 µg of each sample was taken out, and 50 mm TEAB solution was added to fill them up to a 70 µL volume. The powdered TMTsixplex Label Reagent (Thermo Fisher Scientific, Cat. No. 90066) was placed at RT for 30 min. The powders were then centrifuged at 16,160 × g and shaken until completely dissolved after adding 120 µL of acetonitrile. Then, 40 µL of TMT6 labels were taken out and added into the corresponding samples followed by incubation at RT for 2 h. A total of 20 µL of formic acid (FA) was then added to each tube followed by incubation at RT for 15 min to terminate the labeling reaction. After quenching, 5 µL from each TMT6 labeling reaction was pooled for mixing and dried at 45°C. The powder of digested peptides was re-dissolved with 20 µL of 0.1% FA (v/v). The ratio of the labeled components was calculated by MASCOT and Scaffold analysis (Koenig et al. 2008; Searle 2010). Equal amounts of peptides from each sample were combined according to the calculated ratio of the labeled components followed by air drying at 45°C. The labeled peptides were fractionated by a C18 stationary phase (2.1 mm 100 mm, 2.6 m, Kinetex, Phenomenex, Cat. No. 00D-4462-AN) using a gradient elution program of 20 mm ammonium acetate in water (pH 10.0) and 20 mm ammonium acetate in acetonitrile (pH 10.0). The fractionation process was carried out on a High-Performance Liquid Chromatography (HPLC) system (H-Class Bio, Waters, Milford, MA, USA). The resulting fractions were combined into 10 samples and re-dissolved with 0.1% FA for subsequent LC–MS analysis.
LC–MS/MS analysis
Protein identification was performed on a Q-Exactive high-resolution mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a Nano-Acquity HPLC (Waters, Milford, MA, USA). The peptides were loaded onto a trap column (Acclaim PepMap, 75 µm × 2 cm, 3 µm, C18, 100Å, Thermo Fisher Scientific, Waltham, MA, USA) and an analytical column (Aqua, 100 µm × 15 cm, 3 µm, C18, 125Å, Phenomenex, Los Angeles, CA, USA) at a flow rate of 400 nL/min. The method used to elute the peptides was a 125 min gradient with mobile phase A of 0.1% FA in water and mobile phase B of 0.1% FA in acetonitrile. The MS survey scan in the range of 300 to 1800 m/z was obtained with a resolution of 70,000. The 10 most intensive peptide signals of full scan resolution were 17,500 for higher-energy collisional dissociation. The dynamic exclusion time was 20 s.
MS data were processed with MASCOT (version 2.4; Matrix Science, London, UK) and validated with Scaffold software (version 3.6.5, Proteome Software, Portland, USA). MASCOT identifies proteins that include common contaminants, as well as proteins that are identified only by peptides that match to a decoy database of reversed peptide sequences, which were removed from our data. In addition, proteins that have been identified by fewer than 2 unique peptides or with a P-value < 0.01 of the BH test were also removed from the final results. The remaining proteins are thought to be “detected.” The value of a protein was averaged across 3 biological replicates and normalized against the distribution of biotinylated proteins from the control group. We then used the log2 (normalized ratios) for subsequent analysis. Proteins with fold changes above or equal to 1.3 (p23 vs 2C1-Citrine) or 1.5 (p23 vs Citrine) were considered to be proximal to the BBSV p23. Usually, a cutoff value of 1.5-fold enrichment compared with the control group during TMT labeling-based quantitative proteomics was used to select candidate interaction proteins for subsequent validation (Yang et al. 2021). In this study, we used the 1.3-fold enrichment as the cutoff value for p23/2C1-Citrine groups due to the fact that Hsp70, a known interactor of p23, showed a 1.3-fold change compared with the 2C1-Citrine-TurboID control (Supplemental Data Sets 3 and 4).
Data analysis for TurboID proteomics data
Gene Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm) and Java Treeview (Saldanha 2004) were employed to create heat maps representing proteins that were significantly enriched in p23-Citrine-TurboID purification. Subcellular localization of significant proteins was predicted using the online BUSCA web server (http://busca.biocomp.unibo.it).
The volcano plot, which represents all the enriched proteins, and dotplots or barplots were drawn using the R programming language and indicate the pathways in which the significantly enriched proteins were grouped. Since the NbC database does not have its own GO term annotations (Kourelis et al. 2019), we performed a 1-to-1 putative orthologs prediction between N. benthamiana and Arabidopsis (A. thaliana) proteins, thereby obtaining the GO entry database of N. benthamiana proteins. GO analyses were then performed using the online agriGO v2.0 (http://10.2.42.17/agriGO/).
Interaction network analysis
High-confidence [P-value < 0.01, log2-normalized fold change (p23/2C1) ≥ 1.3] N. benthamiana host proteins that were proximal to p23 were screened and subjected to PPI network construction. STRING interaction networks of A. thaliana orthologs corresponding to a subset of N. benthamiana host proteins were visualized in the Cytoscape program (version 3.9.1). The set of minimally required interaction scores is 0.4 for medium confidence on STRING. These host proteins are grouped into functional modules based on GO analysis and TAIR's functional annotations.
Confocal microscopy analysis
BiFC assays were performed as previously reported (Li et al. 2022). At 2.5 dpi, the infiltrated leaf samples were observed with a Zeiss LSM880 confocal microscope. YFP signals were excited at 488 nm and collected in the range of 500 to 530 nn. BFP, GFP, CFP, and mCherry were excited with a 380, 488, 405, and 561 nm laser line, respectively, and the collection of wavelengths used was 370 to 430 nm for BFP, 465 to 485 nm for CFP, 500 to 530 nm for GFP, and 600 to 630 nm for RFP and mCherry. The digital gain value was adjusted to a level that would eliminate fluorescence imaging of intrinsic autofluorescence and nonspecific fluorescence signals. Overlapping curves of fluorescence spectra were produced by line scan analysis using the pixel-based method with the ImageJ plot profile tool. To test the correlation of co-localization between NbRTNLB2-mCherry and CFP-HDEL, we used a PCC with the ImageJ PSC co-localization tool. Values of PCC close to 1 indicate that the co-location of the 2 images is highly correlated (Aaron et al. 2018).
Y2H assays
A DUAL-membrane system (Dualsystems Biotech AG, Zurich, Switzerland) was used to test the interaction between p23 and NbRTNLB2. According to manufacturer's instructions, the pBT3N-p23 plasmid was used as the bait and the pPR3N-RTNLB2 plasmid was used as the prey. Different bait and prey were co-transformed into NMY51 yeast (S. cerevisiae) strains. Co-transformations with pBT3N-p23/pPR3-N and pBT3-N/pPR3N-RTNLB2 served as negative controls, while pTSU2-APP/pNubG-Fe65 and pBT3N-p23/pPR3N-p23 served as positive controls. Transformed yeast cells were plated onto SD/-Trp-Leu drop-out media and cultured at 30°C for 4 d. Single colonies were picked and then cultured in SD/-Trp-Leu and SD/-Trp-Leu-His-Ade drop-out medium for 5 d at 30°C.
Co-IP assays
Co-IP assays were performed as previously described with slight modifications (Gao et al. 2022). Nicotiana benthamiana leaves expressing different combinations of proteins were harvested at 3 dpi. Leaves were then ground in liquid nitrogen and transferred to extraction buffer [10% glycerol, 25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 0.5% (v/v) NP40, 2% (w/v) PVP40, 10 mm DTT, 0.1% Triton X-100 (v/v), and 1 × Protease Inhibitor Cocktail (Sigma, Cat. No. 11697489001)]. After incubation in an ice bath for 30 min, the crude extract was centrifuged at 8,000 × g for 30 min. The supernatant was incubated with anti-FLAG M2 Affinity Gel (GNI, Cat. No. GNI4510-FG) on a rotator at 4°C for 4 h. The beads were pelleted and washed 3 times with IP buffer [10% glycerol, 25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, and 0.1% (v/v) NP40] at 4°C. Immunoblot analysis of the immunoprecipitated products was performed using anti-FLAG (Sigma-Aldrich, Cat. No. A2220, 1:5,000 dilution) and anti-GFP antibodies (Bioeasytech, Cat. No. BE2002, 1:5,000 dilution).
Viral inoculation
Agrobacterium tumefaciens EHA105 harboring pCB301-BBSV were infiltrated into the leaves of 3- to 4-wk-old N. benthamiana plants as described previously (Gao et al. 2022). Total proteins were extracted from infiltrated leaves at 2.5 or 3 dpi for immunoblot assays.
For mechanical inoculation of the virus, 400 ng of BBSV virions were mixed with FES inoculation buffer [0.1 m glycine, 0.06 m dipotassium phosphate, 1% sodium pyrophosphate decahydrate, 1% bentonite, and 1% celite (pH 8.5)] in a 1:1 ratio followed by rub-inoculation onto 5 to 6 leaf stage N. benthamiana plants.
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol Reagent (Invitrogen) based on manufacturer's instructions. RT-qPCR was performed as previously described with minor modifications (Liu et al. 2012). Briefly, 2.5 μg total RNA was treated with Recombinant DNase I (Takara) followed by reverse transcription with M-MLV reverse transcriptase (Promega, USA). Gene fragments were amplified with 2× SsoFast EvaGreen Supermix (Bio-Rad). The Protein Phosphatase 2A (PP2A) gene was used as the internal control, and data were analyzed using CFX Manager (Bio-Rad). Primers used for RT-qPCR analysis are listed in Supplemental Data Set 15.
Generation of transgenic N. benthamiana plants
The intron-hairpin construct pMDC32-RTNLB2-RNAi was introduced into Agrobacterium strain EHA105 and used to generate transgenic N. benthamiana plants by leaf disk transformation as previously described (Horsch et al. 1989). The leaf explants were cultured and regenerated. Genomic DNA was extracted using the hexadecyl cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987), and PCR analysis was conducted to screen the transgenic plants.
TEM
TEM was performed as previously described (Cao et al. 2015). In brief, leaf tissue fragments (about 1 to 2 mm2) from healthy or BBMV-infected N. benthamiana were fixed overnight in fixation buffer [2.5% (v/v) glutaraldehyde, 0.05 m phosphate-buffered saline (PBS), pH 7.2)] at 4°C. After washing 3 times, samples were postfixed in 2% osmium tetroxide (OsO4) for 2 h at 4°C. The leaf pieces were embedded in Spurr's resin followed by dehydration in a graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%). Ultrathin sections were cut into 70 nm using a Leica EM UC7 ultramicrotome and sequentially stained with uranyl acetate for 30 min in the dark and Reynolds’ lead citrate for 5 min at RT. The sections were then viewed with a Hitachi 7650 transmission electron microscope (Hitachi, Tokyo, Japan) at 80 kV.
Immunogold labeling was performed as described previously with minor modifications (Jin et al. 2018b). Briefly, samples from healthy or BBMV-infected N. benthamiana were vacuum-infiltrated in fixative buffer [4% paraformaldehyde, 0.1% glutaraldehyde, 4% sucrose, and 0.1 m PBS (pH 7.2)] and fixed for 2 to 3 h at 4°C. After washing with 0.1 m PBS containing 4% sucrose, the samples were dehydrated using a gradient ethanol series (30%, 50%, 70%, 80%, 90%, 95%, and 100%) followed by infiltration in London Resin (LR) white resin. Samples were polymerized for 48 h at 50°C. Ultrathin sections were prepared from blocks and collected on Formvar-coated 150-mesh nickel grids. The grids were washed by placing drops of 0.01 m PBS (pH 7.2) containing 0.02 m glycine (PBG) onto the grids for 5 min to quench residual aldehyde groups, after which a drop of blocking solution [0.01 m PBS (pH 7.2), 0.01% Triton X-100, 0.01% Tween 20, and 1% BSA] was added for 5 min at RT to reduce nonspecific binding of antibodies. After diluting homemade rabbit anti-P23 (Cao et al. 2015) and anti-RTNLB2 (Supplemental Fig. S11) antibodies by 1:500 and 1:800 in blocking solution, the grids were incubated with the primary antibody solutions overnight at 4°C. The grids were then incubated with goat anti-rabbit secondary antibodies conjugated with 5 nm or 10 nm gold particles (Sigma-Aldrich, Cat. No. G7277, G7652, 1:20 dilution) for 1.5 h at RT followed by rinsing with blocking solution and 0.02 m PBG. Grids were incubated in silver sensitizing solution for 15 min in the dark to enhance the signal and then washed with ddH2O. After staining for 15 min with uranyl acetate, the sections were examined with a Hitachi 7650 transmission electron microscope (Hitachi, Tokyo, Japan) at 80 kV.
Statistical analysis
For simple pair-wise data analysis, two-tailed Student's t test was performed using Microsoft Excel. For multiple comparison statistical analysis, 1-way ANOVA was performed using Duncan’s Multiple Range Test module embedded in GraphPad Prism (version 8.0). All ANOVA and Student's t test results are provided in Supplemental Data Set 16.
Accession numbers
Sequence data from this article can be found in the SOL N. benthamiana database (https://solgenomics.net/organism/Nicotiana_benthamiana/genome) or the NbC database at Oxford Research Archives (https://ora.ox.ac.uk/objects/uuid:f09e1d98-f0f1-4560-aed4-a5147bc7739d). The gene accession numbers are available in Supplemental Figs. S4 andS6, and Supplemental Files 3 and 4. All NbRTNLB protein sequence information in this study is also provided in Supplemental Data Set 7.
Supplementary Material
Acknowledgments
We would like to thank Dr. Kai Xu at Nanjing Normal University for kindly providing the pGD-CBFP plasmid, Dr. Fangfang Li at the Chinese Academy of Agricultural Sciences for providing the 6K2-YFPc and NIb-YFPc plasmids, and Dr. Zejian Guo at China Agricultural University for the pCam35S-GFP plasmid. We thank Drs. Xian-Bing Wang, Jialin Yu, Chenggui Han, Ying Wang, and Meng Yang at China Agricultural University for their helpful comments. We would like to express our gratitude to Dr. Wenhao Zhao from Virginia Tech for providing technical assistance and to Dr. Janet Webster from Virginia Tech for editing the language of our manuscript.
Contributor Information
Qianshen Zhang, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zhiyan Wen, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xin Zhang, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jiajie She, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiaoling Wang, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zongyu Gao, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Ruiqi Wang, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiaofei Zhao, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zhen Su, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zhen Li, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Dawei Li, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiaofeng Wang, School of Plant and Environmental Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
Yongliang Zhang, State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Author contributions
Y.Z., Xiaof.W., Q.Z., and D.L. designed the research. Q.Z., Z.W., X.Zhang, Xiaol.W., Z.G., R.W., J.S., and X.Zhao carried out the experiments. Y.Z., Z.W., Q.Z., X.Zhang, Z.S., and Z.L. analyzed the data. Y.Z., Q.Z., Z.W., and X.Zhang wrote the manuscript. Q.Z., Z.W., and X.Zhang contributed equally to this work.
Supplemental data
Supplemental Figure S1. Subcellular localization of 2C1-Citrine-TurboID and Citrine-TurboID.
Supplemental Figure S2. Preparation and validation of TurboID-specific antibody.
Supplemental Figure S3. Characterization of different NbRTNLBs.
Supplemental Figure S4. Phylogenetic tree of RTN family proteins from N. benthamiana.
Supplemental Figure S5. Analysis of the involvement of RTNLB putative paralogs in BBSV infection.
Supplemental Figure S6. Phylogenetic tree of RTN protein family from multiple plant species.
Supplemental Figure S7. Subcellular localization of p23-BFP and RTNLB2-mCherry.
Supplemental Figure S8. Confocal analysis of Rtn1p-GFP localization in yeast cells.
Supplemental Figure S9. Analysis of the interaction between NbRTNLB2 and p23 in the absence of BBSV infection.
Supplemental Figure S10. RT-qPCR analysis of NbRTNLB2 in mock- or BBSV-inoculated N. benthamiana plants shown in Fig. 4B.
Supplemental Figure S11. Preparation and validation NbRTNLB2-specific antibody.
Supplemental Figure S12. RT-qPCR to confirm the silencing of RTNLB2.
Supplemental Figure S13. Characterization of NbRTNLB2-KD transgenic N. benthamiana plants.
Supplemental Figure S14. ER luminal mobility is reduced by NbRTNLB2 overexpression.
Supplemental Figure S15. Immunoblot analysis to confirm the protein expression in the BiFC assay shown in Fig. 5H.
Supplemental Figure S16. An enlarged version of the image shown in Fig. 6A.
Supplemental Figure S17. TEM analysis of mock or BBSV-infected N. benthamiana leaf tissues.
Supplemental Figure S18. An enlarged version of the image shown in Fig. 6J.
Supplemental Figure S19. NbRTNLB2 interacts with replication proteins of TuMV and TMV on the ER.
Supplemental Figure S20. Characterization of the TurboID fusion proteins and schematic representation of the PL experiment.
Supplemental Figure S21. Proteomic analysis of biotinylated proteins enriched in p23-TurboID purification.
Supplemental Figure S22. A proximity-based interaction network for the proteins enriched in p23-TurboID purification.
Supplemental Figure S23. Pie chart showing the proportion of identified p23-proximal proteins with different subcellular locations.
Supplemental Figure S24. Venn diagram shows overlaps among the identified p23-proximal proteins using different controls in the presence or absence of BBSV infection.
Supplemental Figure S25. BiFC assay to test the interaction between the candidate proteins identified in the p23-proximal interactome and BBSV p23.
Supplemental Figure S26. Topology of ER-localized MSBP1 and Sec62.
Supplemental Data Set 1. Full list of enriched identifications with p23-Citrine-TurboID using 2C1-Citrine-TurboID as the control without decoys and common contaminants in the presence of BBSV infection.
Supplemental Data Set 2. Full list of enriched identifications with p23-Citrine-TurboID using 2C1-Citrine-TurboID as the control without decoys and common contaminants in the absence of BBSV infection.
Supplemental Data Set 3. List of significantly enriched p23-proximal proteins in the presence of BBSV infection.
Supplemental Data Set 4. List of significantly enriched p23-proximal proteins in the absence of BBSV infection.
Supplemental Data Set 5. Thirty-eight commonly enriched identifications of p23-proximal proteins in Group I (+BBSV) and Group II (−BBSV).
Supplemental Data Set 6. List of selected p23-proximal proteins used for interactome analysis using the online STRING website.
Supplemental Data Set 7. Sequences of 14 representative NbRTNLB putative paralogs and their isoforms that were used in this study.
Supplemental Data Set 8. Full list of enriched identifications with p23-Citrine-TurboID using Citrine-TurboID as the control without decoys and common contaminants in the presence of BBSV infection.
Supplemental Data Set 9. List of significantly enriched identifications with p23-Citrine-TurboID versus Citrine-TurboID in the presence of BBSV infection with a threshold of P < 0.05 and log2 (fold change, p23/Citrine) ≥ 1.5).
Supplemental Data Set 10. The terms of GO enrichment analysis of the biological process (P ≤ 0.01, FDR ≤ 0.01) in p23-proximal proteins using Citrine-TurboID as the control.
Supplemental Data Set 11. The terms of GO enrichment analysis of a cellular component (P ≤ 0.01, FDR ≤ 0.01) in p23-proximal proteins using Citrine-TurboID as the control.
Supplemental Data Set 12. The terms of GO enrichment analysis of molecular function (P ≤ 0.01, FDR ≤ 0.01) in p23-proximal proteins using Citrine-TurboID as the control.
Supplemental Data Set 13. List of p23-proximal proteins (Citrine-TurboID as the control) used for interactome analysis using the online STRING website.
Supplemental Data Set 14. Eighteen commonly enriched identifications of p23-proximal proteins among Group I (+BBSV vs 2C1), Group II (−BBSV vs 2C1), and Group II (+BBSV vs Citrine).
Supplemental Data Set 15. List of primers used in this study.
Supplemental Data Set 16. Statistical analyses in this study.
Supplemental File 1. Alignment for phylogenetic analysis in Fig. 5C.
Supplemental File 2. Alignment for phylogenetic analysis in Supplemental Fig. S3B.
Supplemental File 3. Alignment for phylogenetic analysis in Supplemental Fig. S4.
Supplemental File 4. Alignment for phylogenetic analysis in Supplemental Fig. S6.
Funding
This work was supported by the National Natural Science Foundation of China (32122070 and 31872637).
Data availability
The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the iProX (Ma et al. 2019; Chen et al. 2022) partner repository (https://www.iprox.cn/page/SSV024.html;url=1676210060741VRY0, project ID: IPX0005908001).
References
- Aaron JS, Taylor AB, Chew T-L. Image co-localization—co-occurrence versus correlation. J Cell Sci. 2018:131(3):jcs211847. 10.1242/jcs.211847 [DOI] [PubMed] [Google Scholar]
- Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem. 1993:268(25):18726–18733. 10.1016/S0021-9258(17)46690-7 [DOI] [PubMed] [Google Scholar]
- Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep. 2017:21(6):1639–1654. 10.1016/j.celrep.2017.10.055 [DOI] [PubMed] [Google Scholar]
- Arora D, Abel NB, Liu C, Van Damme P, Yperman K, Eeckhout D, Vu LD, Wang J, Tornkvist A, Impens F, et al. Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 2020:32(11):3388–3407. 10.1105/tpc.20.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JP, Claridge JK, Smith PG, Schnell JR. A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proc Natl Acad Sci U S A. 2015:112(7):E639–E648. 10.1073/pnas.1415882112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018:36(9):880–887. 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Dzimitrowicz N, Kriechbaumer V, Brooks R, Botchway SW, Brady JP, Hawes C, Dixon AM, Schnell JR, Fricker MD, et al. A C-terminal amphipathic helix is necessary for the in vivo tubule-shaping function of a plant reticulon. Proc Natl Acad Sci U S A. 2016:113(39):10902–10907. 10.1073/pnas.1605434113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007:5(3):e68. 10.1371/journal.pbio.0050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Cai Z, Ding Q, Li D, Han C, Yu J, Liu Y. The complete nucleotide sequence of Beet black scorch virus (BBSV), a new member of the genus Necrovirus. Arch Virol. 2002:147(12):2431–2435. 10.1007/s00705-002-0896-1 [DOI] [PubMed] [Google Scholar]
- Cao X, Jin X, Zhang X, Li Y, Wang C, Wang X, Hong J, Wang X, Li D, Zhang Y. Morphogenesis of endoplasmic reticulum membrane-invaginated vesicles during Beet black scorch virus infection: role of auxiliary replication protein and new implications of three-dimensional architecture. J Virol. 2015:89(12):6184–6195. 10.1128/JVI.00401-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ma J, Liu Y, Chen Z, Xiao N, Lu Y, Fu Y, Yang C, Li M, Wu S, et al. Iprox in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 2022:50(D1):D1522–D1527. 10.1093/nar/gkab1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003:133(2):462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 2004:20(9):1453–1454. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- Diaz A, Wang X. Bromovirus-induced remodeling of host membranes during viral RNA replication. Curr Opin Virol. 2014:9:104–110. 10.1016/j.coviro.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Diaz A, Wang X, Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci U S A. 2010:107(37):16291–16296. 10.1073/pnas.1011105107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987:19(1):11–15. [Google Scholar]
- Fan H, Sun H, Wang Y, Zhang Y, Wang X, Li D, Yu J, Han C. Deep sequencing-based transcriptome profiling reveals comprehensive insights into the responses of Nicotiana benthamiana to Beet necrotic yellow vein virus infections containing or lacking RNA4. PLoS One 2014:9(1):e85284. 10.1371/journal.pone.0085284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Pietzsch C, Tsaprailis G, Crynen G, Cho KF, Ting AY, Bukreyev A, de la Torre JC, Saphire EO. Functional interactomes of the Ebola virus polymerase identified by proximity proteomics in the context of viral replication. Cell Rep. 2022a:38(12):110544. 10.1016/j.celrep.2022.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Pietzsch C, Witwit H, Tsaprailis G, Crynen G, Cho KF, Ting AY, Bukreyev A, Saphire EO, de la Torre JC. Proximity interactome analysis of Lassa polymerase reveals eRF3a/GSPT1 as a druggable target for host-directed antivirals. Proc Natl Acad Sci U S A. 2022b:119(30):e2201208119. 10.1073/pnas.2201208119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Castro I, Fernández JJ, Barajas D, Nagy PD, Risco C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J Cell Sci. 2017:130(1):260–268. 10.1242/jcs.181586 [DOI] [PubMed] [Google Scholar]
- Gao Z, Pu H, Liu J, Wang X, Zhong C, Yue N, Zhang Z, Wang X-B, Han C, Yu J, et al. Tobacco necrosis virus-AC single coat protein amino acid substitutions determine host-specific systemic infections of Nicotiana benthamiana and soybean. Mol Plant Microbe Interact. 2021:34(1):49–61. 10.1094/MPMI-07-20-0184-R [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang D, Wang X, Zhang X, Wen Z, Zhang Q, Li D, Dinesh-Kumar SP, Zhang Y. Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKα-mediated antiviral immunity in plants. Nat Commun. 2022:13(1):716. 10.1038/s41467-022-28395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Aix C, Garcia-Garcia M, Aranda MA, Sanchez-Pina MA. Melon necrotic spot virus replication occurs in association with altered mitochondria. Mol Plant Microbe Interact. 2015:28(4):387–397. 10.1094/MPMI-09-14-0274-R [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002:31(3):375–383. 10.1046/j.1365-313X.2002.01360.x [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988:16(20):9877. 10.1093/nar/16.20.9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT. Leaf disc transformation. In: Gelvin SB, Schilperoort RA, Verma DPS, editors. Plant molecular biology manual. Dordrecht: Springer Netherlands; 1989. p. 63–71 [Google Scholar]
- Huang F-C, Chi S-F, Chien P-R, Liu Y-T, Chang H-N, Lin C-S, Hwang H-HJP, Physiology C. Arabidopsis RAB8A, RAB8B and RAB8D proteins interact with several RTNLB proteins and are involved in the Agrobacterium tumefaciens infection process. Plant Cell Physiol. 2021:62(10):1572–1588. 10.1093/pcp/pcab112 [DOI] [PubMed] [Google Scholar]
- Huang F-C, Fu B-J, Liu Y-T, Chang Y-R, Chi S-F, Chien P-R, Huang S-C, Hwang H-H. Arabidopsis reticulon-like3 (RTNLB3) and RTNLB8 participate in Agrobacterium-mediated plant transformation. Int J Mol Sci. 2018:19(2):638. 10.3390/ijms19020638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-W, Sun C-I, Hu C-C, Tsai C-H, Meng M, Lin N-S, Dinesh-Kumar SP, Hsu Y-H. A viral movement protein co-opts endoplasmic reticulum luminal-binding protein and calreticulin to promote intracellular movement. Plant Physiol. 2023:191(2):904–924. 10.1093/plphys/kiac547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-H, Gelvin SBJTPC. Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 2004:16(11):3148–3167. 10.1105/tpc.104.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer K, Bürkle L, Auerbach D, Thaminy S, Dinkel M, Engels K, Stagljar I. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein–protein interactions of integral membrane proteins. Sci STKE. 2005:2005(275):pl3. 10.1126/stke.2752005pl3 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Li Z, Yue N, Zhang K, Li D, Zhang Y. Construction of infectious clones of Lychnis ringspot virus and evaluation of its relationship with Barley stripe mosaic virus by reassortment of genomic RNA segments. Virus Res. 2018:243:106–109. 10.1016/j.virusres.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Jin X, Cao X, Wang X, Jiang J, Wan J, Laliberté JF, Zhang Y. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front Plant Sci. 2018a:9:57. 10.3389/fpls.2018.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Jiang Z, Zhang K, Wang P, Cao X, Yue N, Wang X, Zhang X, Li Y, Li D, et al. Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiol. 2018b:176(1):282–294. 10.1104/pp.17.00871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RAC, Naidu RA. Global dimensions of plant virus diseases: current status and future perspectives. Annu Rev Virol. 2019:6(1):387–409. 10.1146/annurev-virology-092818-015606 [DOI] [PubMed] [Google Scholar]
- Kim TW, Park CH, Hsu CC, Kim YW, Ko YW, Zhang Z, Zhu JY, Hsiao YC, Branon T, Kaasik K, et al. Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics. Plant Cell 2023:35(3):975–993. 10.1093/plcell/koad013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig T, Menze BH, Kirchner M, Monigatti F, Parker KC, Patterson T, Steen JJ, Hamprecht FA, Steen H. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J Proteome Res. 2008:7(9):3708–3717. 10.1021/pr700859x [DOI] [PubMed] [Google Scholar]
- Kourelis J, Kaschani F, Grosse-Holz FM, Homma F, Kaiser M, van der Hoorn RAL. A homology-guided, genome-based proteome for improved proteomics in the alloploid Nicotiana benthamiana. BMC Genomics. 2019:20(1):722. 10.1186/s12864-019-6058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis E, König K, Sommer F, Schroda M. TurboID reveals the proxiomes of CGE1, VIPP1, and VIPP2 in Chlamydomonas reinhardtii. bioRxiv 518767. 10.1101/2022.12.01.518767, 5 December 2022, preprint: not peer reviewed. [DOI]
- Kumar A, Salemi M, Bhullar R, Guevara-Plunkett S, Lyu Y, Wang K-H, Izumiya C, Campbell M, Nakajima K-I, Izumiya Y. Proximity biotin labeling reveals Kaposi's sarcoma-associated herpesvirus interferon regulatory factor networks. J Virol. 2021:95(9):e02049-02020. 10.1128/JVI.02049-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018:35(6):1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté JF, Sanfaçon H. Cellular remodeling during plant virus infection. Annu Rev Phytopathol. 2010:48(1):69–91. 10.1146/annurev-phyto-073009-114239 [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Wink RH, Persson S, Somssich M. The toolbox to study protein−protein interactions in plants. Crit Rev Plant Sci. 2018:37(4):308–334. 10.1080/07352689.2018.1500136 [DOI] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007:23(21):2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lazareva EA, Lezzhov AA, Chergintsev DA, Golyshev SA, Dolja VV, Morozov SY, Heinlein M, Solovyev AG. Reticulon-like properties of a plant virus-encoded movement protein. New Phytol. 2021:229(2):1052–1066. 10.1111/nph.16905 [DOI] [PubMed] [Google Scholar]
- Li Z, Yang X, Li W, Wen Z, Duan J, Jiang Z, Zhang D, Xie X, Wang X, Li F, et al. SAMDC3 enhances resistance to Barley stripe mosaic virus by promoting the ubiquitination and proteasomal degradation of viral γb protein. New Phytol. 2022:234(2):618–633. 10.1111/nph.17993 [DOI] [PubMed] [Google Scholar]
- Liu Y, Katoh H, Sekizuka T, Bae C, Wakata A, Kato F, Sakata M, Yamaji T, Wang Z, Takeda M. SNARE protein USE1 is involved in the glycosylation and the expression of mumps virus fusion protein and important for viral propagation. PLoS Pathog. 2022:18(12):e1010949. 10.1371/journal.ppat.1010949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002:31(6):777–786. 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- Liu D, Shi L, Han C, Yu J, Li D, Zhang Y. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 2012:7(9):e46451. 10.1371/journal.pone.0046451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, Li K, Zhang G, Jin Z, He F, et al. Iprox: an integrated proteome resource. Nucleic Acids Res. 2019:47(D1):D1211–d1217. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair A, Xu S-L, Branon TC, Ting AY, Bergmann DC. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife. 2019:8:e47864. 10.7554/eLife.47864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Beachy RN. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol. 1999:147(5):945–958. 10.1083/jcb.147.5.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A, Okuno T. Composition of plant virus RNA replicase complexes. Curr Opin Virol. 2012:2(6):669–675. 10.1016/j.coviro.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Nagy PD. Tombusvirus-host interactions: co-opted evolutionarily conserved host factors take center court. Annu Rev Virol. 2016:3(1):491–515. 10.1146/annurev-virology-110615-042312 [DOI] [PubMed] [Google Scholar]
- Nagy PD. Exploitation of a surrogate host, Saccharomyces cerevisiae, to identify cellular targets and develop novel antiviral approaches. Curr Opin Virol. 2017:26:132–140. 10.1016/j.coviro.2017.07.031 [DOI] [PubMed] [Google Scholar]
- Nagy PD. Co-opted membranes, lipids, and host proteins: what have we learned from tombusviruses? Curr Opin Virol. 2022:56:101258. 10.1016/j.coviro.2022.101258 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Feng Z. Tombusviruses orchestrate the host endomembrane system to create elaborate membranous replication organelles. Curr Opin Virol. 2021:48:30–41. 10.1016/j.coviro.2021.03.007 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2011:10(2):137–149. 10.1038/nrmicro2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngounou Wetie AG, Sokolowska I, Woods AG, Roy U, Deinhardt K, Darie CC. Protein−protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches. Cell Mol Life Sci. 2014:71(2):205–228. 10.1007/s00018-013-1333-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennazio S, Roggero P, Conti M. Yield losses in virus-infected crops. Arch Phytopathol Plant Protect. 1996:30(4):283–296. 10.1080/03235409609383178 [DOI] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics 2004:20(17):3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Saxena P, Lomonossoff GP. Virus infection cycle events coupled to RNA replication. Annu Rev Phytopathol. 2014:52(1):197–212. 10.1146/annurev-phyto-102313-050205 [DOI] [PubMed] [Google Scholar]
- Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011:12(9):938–954. 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010:10(6):1265–1269. 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- Tang W-F, Yang S-Y, Wu B-W, Jheng J-R, Chen Y-L, Shih C-H, Lin K-H, Lai H-C, Tang P, Horng J-T. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J Biol Chem. 2007:282(8):5888–5898. 10.1074/jbc.M611145200 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Kriechbaumer V. Reticulons 3 and 6 interact with viral movement proteins. Mol Plant Pathol. 2022:23(12):1807–1814. 10.1111/mpp.13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, Frigerio L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010:64(3):411–418. 10.1111/j.1365-313X.2010.04337.x [DOI] [PubMed] [Google Scholar]
- Verchot J. Wrapping membranes around plant virus infection. Curr Opin Virol. 2011:1(5):388–395. 10.1016/j.coviro.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004:40(3):428–438. 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- Wan J, Basu K, Mui J, Vali H, Zheng H, Laliberté JF. Ultrastructural characterization of Turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J Virol. 2015:89(24):12441–12456. 10.1128/JVI.02138-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol. 2015:53(1):45–66. 10.1146/annurev-phyto-080614-120001 [DOI] [PubMed] [Google Scholar]
- Wang X, Cao X, Liu M, Zhang R, Zhang X, Gao Z, Zhao X, Xu K, Li D, Zhang Y. Hsc70-2 is required for Beet black scorch virus infection through interaction with replication and capsid proteins. Sci Rep. 2018:8(1):4526. 10.1038/s41598-018-22778-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ma J, Jin X, Yue N, Gao P, Mai KKK, Wang X-B, Li D, Kang B-H, Zhang Y. Three-dimensional reconstruction and comparison of vacuolar membranes in response to viral infection. J Integr Plant Biol. 2021:63(2):353–364. 10.1111/jipb.13027 [DOI] [PubMed] [Google Scholar]
- Wei X-F, Fan S-Y, Wang Y-W, Li S, Long S-Y, Gan C-Y, Li J, Sun Y-X, Guo L, Wang P-Y, et al. Identification of STAU1 as a regulator of HBV replication by TurboID-based proximity labeling. iScience 2022:25(6):104416. 10.1016/j.isci.2022.104416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Nagy PD. Enrichment of phosphatidylethanolamine in viral replication compartments via co-opting the endosomal Rab5 small GTPase by a positive-strand RNA virus. PLoS Biol. 2016:14(10):e2000128. 10.1371/journal.pbio.2000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S-L, Shrestha R, Karunadasa SS, Xie P-Q. Proximity labeling in plants. Annu Rev Plant Biol. 2023:74(1):1. 10.1146/annurev-arplant-070522-052132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007:8(12):234. 10.1186/gb-2007-8-12-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wen Z, Zhang D, Li Z, Li D, Nagalakshmi U, Dinesh-Kumar SP, Zhang Y. Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Commun. 2021:2(2):100137. 10.1016/j.xplc.2020.100137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Zhang T, Tian Z, Wang Y, Tao X. Construction of Agrobacterium-mediated Cucumber mosaic virus infectious cDNA clones and 2b deletion viral vector. Scientia Agricultura Sinica. 2011:44(14):3060–3068. [Google Scholar]
- Yuan X, Cao Y, Xi D, Guo L, Han C, Li D, Zhai Y, Yu J. Analysis of the subgenomic RNAs and the small open reading frames of Beet black scorch virus. J Gen Virol. 2006:87(Pt 10):3077–3086. 10.1099/vir.0.81928-0 [DOI] [PubMed] [Google Scholar]
- Zhang D, Gao Z, Zhang H, Yang Y, Yang X, Zhao X, Guo H, Nagalakshmi U, Li D, Dinesh-Kumar SP, et al. The MAPK-Alfin-like 7 module negatively regulates ROS scavenging genes to promote NLR-mediated immunity. Proc Natl Acad Sci U S A. 2023:120(3):e2214750120. 10.1073/pnas.2214750120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Y, Yang X, Wen Z, Nagalakshmi U, Dinesh-Kumar SP. TurboID-based proximity labeling for in planta identification of protein–protein interaction networks. J Vis Exp. 2020:159:e60728. 10.3791/60728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shang L, Zhang J, Liu Y, Jin C, Zhao Y, Lei X, Wang W, Xiao X, Zhang X, et al. An antibody-based proximity labeling map reveals mechanisms of SARS-CoV-2 inhibition of antiviral immunity. Cell Chem Biol. 2022:29(1):5–18.e6. 10.1016/j.chembiol.2021.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Song G, Lal NK, Nagalakshmi U, Li Y, Zheng W, Huang P-J, Branon TC, Ting AY, Walley JW, et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun. 2019:10(1):3252. 10.1038/s41467-019-11202-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the iProX (Ma et al. 2019; Chen et al. 2022) partner repository (https://www.iprox.cn/page/SSV024.html;url=1676210060741VRY0, project ID: IPX0005908001).